Background: The N-terminal transactivation domain (N-TAD) is requisite for GATA1 function in vivo.
Results: The C-terminal region of GATA1 works as TAD, and fetal hematopoiesis is perturbed in the GATA1-deficient mice rescued with GATA1 lacking C-terminal TAD activity.
Conclusion: GATA1 has two TADs, which differentially work.
Significance: The complex nature of molecular mechanisms is underlying how GATA1 works for the maintenance of hematopoietic homeostasis.
Keywords: Erythropoeisis, Hematopoiesis, Transcription Factors, Transcription Target Genes, Transgenic Mice, Megakaryopoiesis
Abstract
Transcription factor GATA1 regulates the expression of a cluster of genes important for hematopoietic cell differentiation toward erythroid and megakaryocytic lineages. Three functional domains have been identified in GATA1, a transactivation domain located in the N terminus (N-TAD) and two zinc finger domains located in the middle of the molecule. Although N-TAD is known as a solitary transactivation domain for GATA1, clinical observations in Down syndrome leukemia suggest that there may be additional transactivation domains. In this study, we found in reporter co-transfection assays that transactivation activity of GATA1 was markedly reduced by deletion of the C-terminal 95 amino acids without significant attenuation of the DNA binding activity or self-association potential. We therefore generated transgenic mouse lines that expressed GATA1 lacking the C-terminal region (GATA1-ΔCT). When we crossed these transgenic mouse lines to the Gata1-deficient mouse, we found that the GATA1-ΔCT transgene rescued Gata1-deficient mice from embryonic lethality. The embryos rescued with an almost similar level of GATA1-ΔCT to endogenous GATA1 developed beyond embryonic 13.5 days, showing severe anemia with accumulation of immature erythroid cells, as was the case for the embryos rescued by endogenous levels of GATA1 lacking N-TAD (GATA1-ΔNT). Distinct sets of target genes were affected in the embryos rescued by GATA1-ΔCT and GATA1-ΔNT. We also found attenuated GATA1 function in cell cycle control of immature megakaryocytes in both lines of rescued embryos. These results thus demonstrate that GATA1 has two independent transactivation domains, N-TAD and C-TAD. Both N-TAD and C-TAD retain redundant as well as specific activities for proper hematopoiesis in vivo.
Introduction
The GATA family of transcription factors plays critical roles in cell proliferation, differentiation, and survival. Six GATA factors have been identified in vertebrates. Of the six GATA factors, GATA1, GATA2, and GATA3 are abundantly expressed in hematopoietic tissues and are referred to as hematopoietic GATA factors. Each GATA factor is nonredundantly required in the specific cell lineages (1), such that GATA3 is expressed in Th2 lymphoid cells (2) and GATA2 is expressed in immature multipotent progenitors and hematopoietic stem cells (3). GATA1 is expressed in erythroid lineage cells, megakaryocytic lineage cells, eosinophils, and mast cells. Unique expression profiles of individual GATA factor genes are defined by elaborate regulation to these GATA factor genes (1).
The expression profiles of GATA1 and GATA2 are clearly distinct but somewhat overlapping. GATA1 is mainly required for erythropoiesis and megakaryopoiesis (4–6). The complete lack of Gata1 expression leads mice to embryonic lethality by embryonic 11.5 days (E11.5)3 due to insufficient primitive erythropoiesis (7). Strict regulations of Gata1 and Gata2 genes by a transcription factor network, including GATA factors, are important for the maintenance of hematopoietic homeostasis (8).
We established Gata1 knockdown mice, in which expression of the Gata1 gene deteriorated to 5% of the endogenous level by disruption of the upstream regulatory region of the Gata1 gene (9). We refer this Gata1 allele as Gata1.05. Because the Gata1 gene is localized in the X-chromosome, male mice harboring the Gata1.05 allele die by E11.5 (9). This phenotype is similar to that of Gata1 gene knock-out embryos (10), indicating that 5% expression of GATA1 is insufficient to support the GATA1 function in vivo. By replacing the Gata1 gene upstream enhancer region with the neomycin gene cassette, another GATA1 knockdown mouse line was generated that showed ∼20% of the gene expression level compared with the endogenous Gata1 level. These mice showed potentially a prolonged life span (11), indicating that the GATA1 expression level is critical for the embryonic erythropoiesis.
GATA1 contains three functional domains as follows: an N-terminal transcription activation domain (N-TAD) rich in acidic amino acid residues; an N-terminal zinc finger domain (N-finger), which is important for interaction with co-factor FOG1 and association of DNA, and a C-terminal zinc finger domain (C-finger), which is essential for binding to the GATA box. We have examined the activities of these domains by using the transgenic complementation rescue approach and found that these domains independently and cooperatively act during embryogenesis (12).
Of particular interest is that although transactivation activity of GATA1 was significantly reduced by the N-TAD deletion (13), a family harboring an inherited GATA1 mutation leading to the production of GATA1 without N-TAD (GATA1-ΔNT) has been reported (14). We found in a mouse system that transgenic expression of GATA1-ΔNT at an endogenous level has the potential to rescue the Gata1.05 male mice from embryonic lethality (12). Similarly, GATA1-ΔNT expression under the endogenous Gata1 gene regulation rescued Gata1-null mice that were born alive (15). Thus, GATA1-ΔNT can activate GATA1 target genes sufficiently to sustain erythropoiesis in GATA1-deficient mice.
Considering these situations, there emerges a question how GATA1-ΔNT activates target gene expression. The most plausible explanation for this question is that there may be an additional transactivation domain that supports the GATA1-ΔNT activity. Therefore, to identify new transactivation domains within GATA1, in this study we examined the C-terminal end of GATA1, as the proline-rich C-terminal region is a prime candidate of the transactivation domain. We found that the GATA1 C-terminal region acts as a new transactivation domain. Transgenic complementation rescue analyses clearly demonstrated that lack of the C-terminal region indeed perturbed normal mouse hematopoiesis in vivo. Results of this study support the notion that two independent transactivation domains not only share redundant activities but that the domains also retain specific activities. We propose that GATA1 participates in the regulation of diverse sets of target genes utilizing these elaborate transactivation mechanisms.
EXPERIMENTAL PROCEDURES
Reporter Assays
Gata1 mutant cDNAs were constructed by PCR-mediated mutagenesis and cloned into pEF-1 (16) or pcDNA-pGBT9 (17) expression vectors. QT6 (a quail fibroblast-derived) cells were cultured in low glucose DMEM medium (WAKO) supplemented with 10% FBS (Invitrogen) and 100 IU/ml penicillin/streptomycin (Invitrogen). pEF-1-based or pcDNA-pGBT9-based expression vectors were transfected into QT6 cells concomitant with firefly luciferase reporter gene driven by either the triplicate mouse GATA boxes from mouse α-globin promoter (18) or the upstream activating sequence (17). Transfection experiments were performed utilizing Lipofectamine 2000 (Invitrogen) following the manufacturer's procedure. pRL-TK was co-transfected to normalize the transfection efficiency. The luciferase activities were measured using Dual-Luciferase reporter assay system (Promega) with luminometer (Berthold). pEGFP-N2 (Clontech) was occasionally co-transfected to measure the level of protein expression. Immunoblot analysis was performed using anti-GAL4 (Santa Cruz Biotechnology) and anti-GFP (Medical and Biological Laboratories Co. Ltd.) antibodies.
Recombinant Proteins, DNA Binding Assays, and Self-association Assays
Maltose-binding protein (MBP)-fused GATA1 and GST-fused GATA1 were produced in Escherichia coli BL21(DE3)pLysS-competent cells (Novagen) as described (19) and purified using glutathione-Sepharose beads (GE Healthcare) and amylose resin (New England Biolabs). Protein concentration was determined by using the Bradford assay (Bio-Rad).
The binding reaction for EMSA was performed using MBP fusion GATA1 proteins in the binding buffer containing 10 mm Tris-HCl (pH 7.5), 1 mm EDTA, 4% Ficoll, 1 mm DTT, and 75 mm KCl (20). Oligonucleotide (5′-AAGATCTCCGGCAACTGATAAGGATTCCCTG-3′) from mouse α-globin promoter (18) was end-labeled with T4 polynucleotide kinase. Poly[d(I-C)] (Sigma) was used as a nonspecific competitor. Radioactive signals were detected by autoradiography.
For the GST-pulldown assay, a graded amount of MBP fusion GATA1 (0, 1, 2, 5 and 10 μg) was mixed with GST fusion GATA1 fragments corresponding to amino acids 200–305, 200–319, and 200–322. These peptides were bound to glutathione-Sepharose beads (19). MBP (10 μg) was used as a negative control. Immunoblot analysis was performed using anti-MBP and anti-GST antibodies (Santa Cruz Biotechnology).
Animals
All experimental procedures have been approved by the Institutional Animal Experiment Committee, and experiments have been performed in accordance with the Regulation for Animal Experiments of Tohoku University. Gata1ΔNeoΔHS/Y mice lacking GATA1 expression in megakaryocytes (21) were supplied from The Jackson Laboratory and maintained in our animal facility. For microinjection, GATA1 mutant cDNAs were subcloned into the Gata1 gene hematopoietic regulatory domain (G1HRD) expression vector (22), and these constructs were microinjected into fertilized BDF1 eggs by utilizing standard procedures (23). Founders and offspring were screened by PCR. Sense (5′-AAGCGGCCGCATCGTCATTTGTG-3′) and antisense (5′-TAGGCCTCAGCTTCTCTGTA-3′) primer set were used to identify the transgene. The Gata1 gene knockdown Gata1.05 allele and the Y chromosome were detected by primer sets described previously (9).
Flow Cytometry
Cells were incubated with antibodies for 1 h on ice and washed with cold phosphate-buffered saline (PBS) supplemented with 2% fetal bovine serum. The samples were analyzed with CellQuest program using FACS CaliburTM (BD Biosciences). Dead cells were excluded with BD Via-Probe or propidium iodide (Sigma). Fluorescein isothiocyanate-conjugated anti-CD71 and-CD41 antibodies, and phycoerythrin-conjugated anti-Ter119 and -CD61 antibodies were purchased from eBiosciences. Allophycocyanin-conjugated anti-c-Kit antibody was from BD Biosciences.
Quantitative RT-PCR
Total RNA was prepared by using ISOGEN (Nippon Gene). cDNA was synthesized using Superscript III (Invitrogen), and quantitative RT-PCR was carried out using Power SYBR® Green PCR Master Mix (Invitrogen) with PRISM 7300 (Applied Biosystems). Expression levels were normalized to Gapdh. Primer sequences are described in supplemental Table 1.
Chromatin Immunoprecipitation (ChIP) Assay
Chromatin fixation and purification procedures were performed as described previously (24) with the exception that mouse erythroleukemia (MEL) cells were used. MEL cells were cultured in a same manner as the QT6 cells. Rat anti-GATA1 N6 antibody (Santa Cruz Biotechnology) (25) was used for immunoprecipitation. Normal rat IgG was utilized as a negative control. DNAs were amplified using primers described in supplemental Table 2.
Retroviral Infection
cDNAs encoding wild type and GATA1 mutants were inserted into the retroviral vector pMX-puro (kindly provided by Dr. Toshio Kitamura (26)). After excluding lineage-positive cells using BioMag-streptavidin (BD Biosciences) and biotin-conjugated anti-CD4, -CD8, -B220, -Mac1, -Gr1, and -Ter119 antibodies (Pharmingen), cells from E13.5 fetal livers of Gata1ΔNeoΔHS/Y embryos were expanded with a combination of thrombopoietin (10 ng/ml), stem cell factor (50 ng/ml), and interleukin-3 (IL-3; 10 ng/ml) for 1 day. Subsequently 1-day transduction with retroviral vector was performed using RetroNectin (Takara-Bio)-coated plates. After a 24-h puromycin treatment (6–10 μg/ml), viable cells were cultured with thrombopoietin (50 ng/ml) and stem cell factor (50 ng/ml) for the days indicated. The last day of the puromycin treatment was set to day 0. The number of living cells was counted with a Countess automated cell counter (Invitrogen). The total number of living c-Kit+CD41+ cells was calculated by multiplying the frequency of c-Kit+CD41+ cells by the number of living cells.
Statistical Analysis
Statistical analyses were performed with a Student's t test. Probability values of p < 0.05 were considered statistically significant.
RESULTS
C-terminal Region Is Vital for Transactivation Activity of GATA1
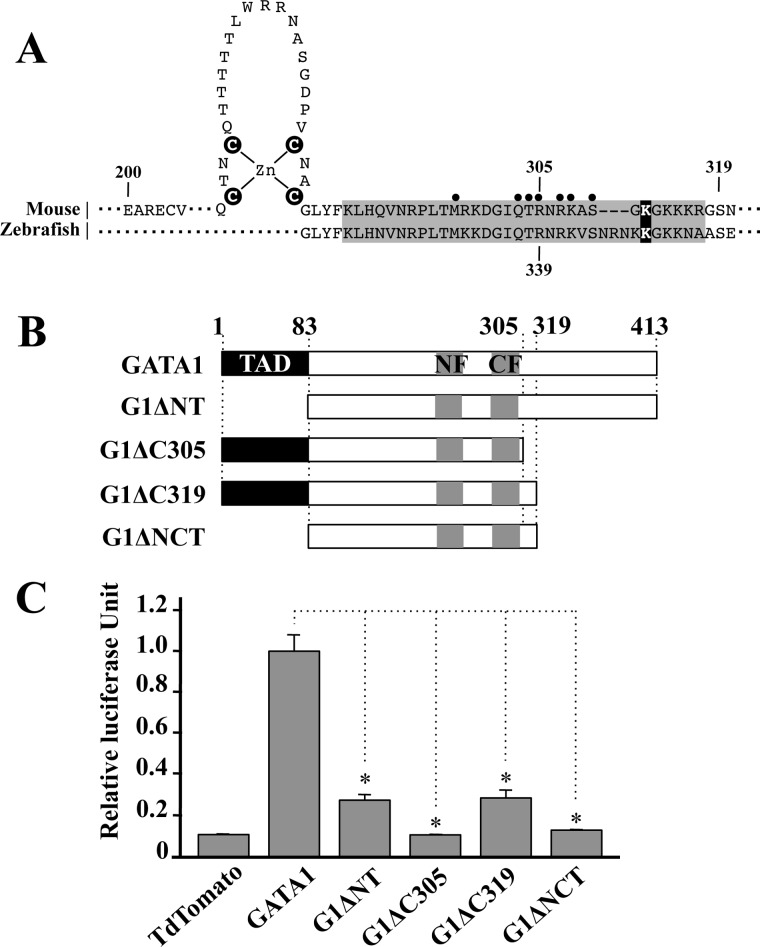
To search for a new transactivation domain, we focused on the C-terminal region of GATA1, as the structure of the region fulfills the criteria of the transactivation domain. In addition, this region is phylogenetically conserved in the human and mouse. On the contrary, it has been reported that the truncated type of GATA1 lacking the C-terminal region could not sustain erythropoiesis in zebrafish mutant Vlad tepes (27). A nonsense mutation at codon 339 in zebrafish GATA1 gene gave rise to massive deletion of GATA1 from the C-finger tail to the C terminus (Fig. 1A). This deletion resulted in weakened interaction of mutant GATA1 with the GATA box and caused the bloodless phenotype (27, 28). Therefore, to evaluate the function of the C-terminal region, we generated two C-terminal truncated types of mouse GATA1 (Fig. 1B). One was the GATA1 mutant similar to the zebrafish vlad tepes mutant (truncated at amino acid 305), and the other was GATA1 lacking 95 amino acids of the C-terminal region by introducing nonsense mutation at codon 319 (Fig. 1A). We referred to these GATA1 mutants to as G1ΔC305 and G1ΔC319, respectively (Fig. 1B). In addition, we constructed G1ΔNT and G1ΔNCT mutants by removing 83 amino acids of the N-terminal region from wild-type GATA1 and G1ΔC319, respectively.
FIGURE 1.
Deletion of C-terminal region attenuates the transactivation activity of GATA1. A, sequence alignment of mouse and zebrafish GATA1 at the C-finger and basic tail regions. Basic region is highlighted with a gray background, and the closed circles indicate the amino acid residues that make direct interaction with the minor groove of DNA (28). Cysteine residues in the C-finger domain and lysine residue responsible for self-association potential are marked with a black background. B, GATA1 deletion mutants are schematically illustrated. Numbers indicate position of amino acid residues. C, transactivation activity of GATA1 mutant proteins. Note that transactivation activity is significantly reduced by the deletion of C-terminal region. Data are presented as mean ± S.D. (*, p < 0.05).
To examine the contribution of the C-terminal region for transcriptional regulation, we executed luciferase reporter assays with these GATA1 deletion mutants. Transactivation activity of G1ΔNT was decreased to the ∼30% level of the wild-type GATA1 activity (Fig. 1C). This result is consistent with our previous study (12). Of note, transactivation activity of G1ΔC319 was also markedly decreased to ∼30% level of the wild-type GATA1 activity (Fig. 1C). Although there still remained certain transactivation activity in these two mutants, such activity had disappeared from the G1ΔC305 mutant because of the lack of DNA binding. Importantly, concomitant deletion of the 83 amino acids of N-terminal region from the G1ΔC319 completely abolished its transcriptional activity. These results indicate that the C-terminal region (319–413) of GATA1 retains transactivation activity. We therefore referred this region to as C-terminal transactivation domain (C-TAD).
DNA Binding and Self-association Activities of GATA1 Are Not Influenced by the C-terminal Deletion
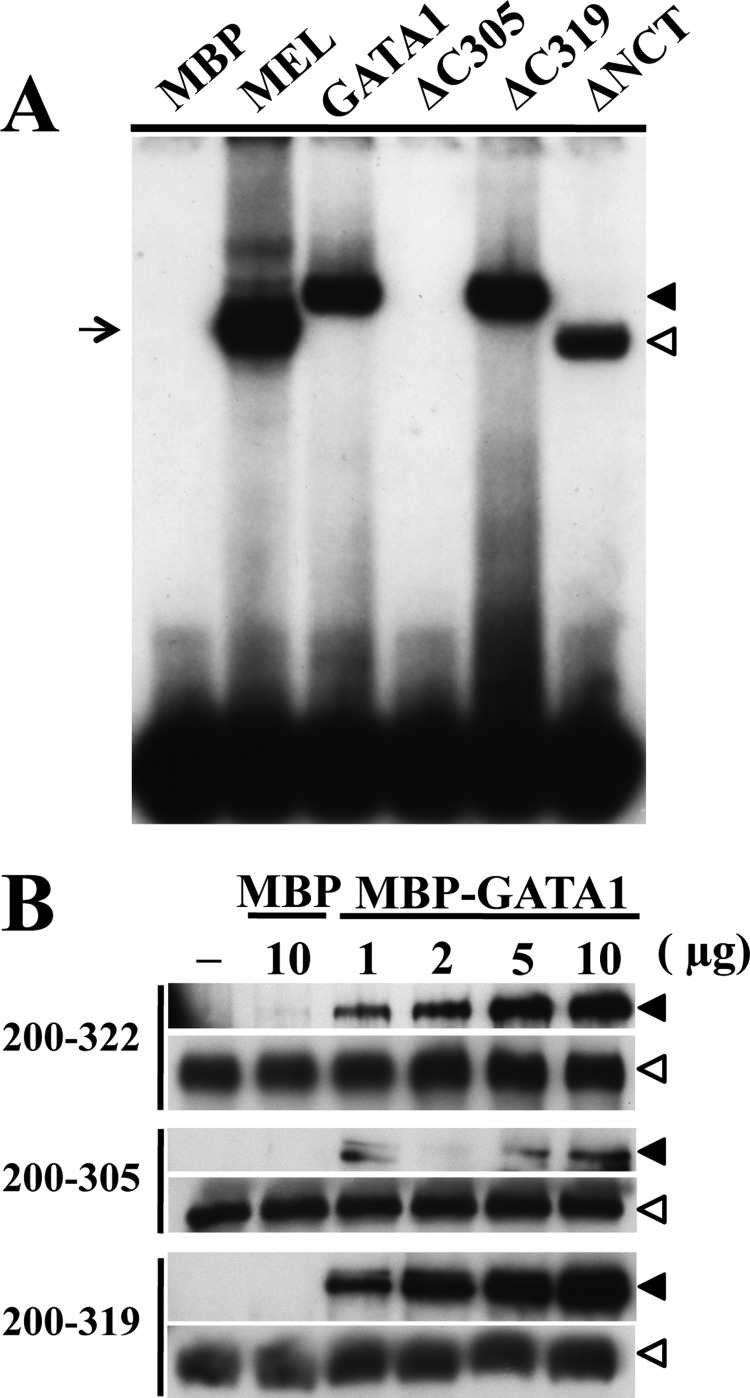
Because G1ΔC319 possesses the entire basic tail region of GATA1 C-finger, the DNA binding ability of G1ΔC319 should be equivalent to that of the wild-type GATA1. To verify this point, we examined the DNA binding ability of G1ΔC305 and G1ΔC319. To this end, MBP was fused to the N termini of these mutants, and MBP fusion proteins were purified. We then conducted EMSA using a single GATA site of the mouse α-globin gene promoter (18) and the purified proteins. Band corresponding to the G1ΔC305-probe complex was not detectable in this EMSA analysis (Fig. 2A). This is in very good agreement with the previous report (27). In contrast, bands corresponding to the complexes containing G1ΔC319 or G1ΔNCT mutant as well as wild-type GATA1 were clearly detectable (Fig. 2A).
FIGURE 2.
DNA binding and self-association capacity are intact in GATA1 lacking C-terminal 95 amino acids. A, EMSA of mutant GATA1 binding to a single GATA box. Closed and open arrowheads indicate DNA and MBP fusion GATA1 complexes of wild type and mutant, respectively. Arrows indicate DNA complexes with endogenous GATA1 extracted from mouse erythroleukemia (MEL) cells. B, GST-pulldown assays performed using GST-fused GATA1 fragments consisting of the indicated amino acids. These peptides were incubated with gradated amounts of MBP fusion GATA1. The amount of captured proteins was estimated by immunoblotting to detect MBP fusion protein utilizing anti-MBP antibody (closed arrowhead). Equal volumes of the GST-fused GATA1 fragments-Sepharose bead fractions were used for the pulldown assay, and protein amounts were assessed by immunoblotting against GST (open arrowhead).
Because the N-finger domain of GATA1 contributes to DNA binding with palindromic GATA sequences in collaboration with the C-finger domain (29), there is a possibility that deletion of the C-terminal region might affect the DNA binding ability of GATA1 to a certain type of GATA boxes. To assess this possibility, we performed EMSA using a palindromic GATA motif in the chicken Gata1 gene promoter, whose expression was attenuated by N-finger dysfunction (30). We also examined a double GATA motif artificially synthesized from the palindromic GATA motif in chicken Gata1 gene promoter. The result demonstrated that G1ΔC319 and G1ΔNCT bound to the palindromic and double GATA motifs as similarly as the wild-type GATA1 did. In contrast, we could not detect the band of the G1ΔC305 complex (supplemental Fig. S1). These results are consistent with those obtained by using the single GATA motif. We also confirmed that deletion of the C-terminal region (ΔC319) did not affect the off-rates of GATA-DNA binding onto any type of GATA motifs by means of a competitive dissociation assay (data not shown).
GATA1 self-association (or dimerization) is known to be important for the GATA1 function (31). The domain for GATA1 dimerization overlaps with basic tail region of the C-finger domain, and the Lys-312 residue is important for the self-association ability (19). Therefore, we examined whether the lack of the C-terminal region affected the self-association ability of GATA1. For this purpose, we carried out GST-pulldown assays using MBP fusion GATA1 and GST fusion GATA1 fragments corresponding to amino acids 200–305, 200–319, and 200–322. These peptides were bound to glutathione-Sepharose beads and pulled down MBP fusion GATA1, and immunoblot analyses were performed with anti-MBP antibody. As shown in Fig. 2B, a comparable amount of GATA1 protein was captured by 200–319 peptide to that by 200–322 peptide. In contrast, a lesser amount of MBP-GATA1 was precipitated by the 200–305 peptide, indicating that self-association potential was preserved in G1ΔC319, but the ability was disturbed in G1ΔC305. Thus, these results demonstrate that the marked decrease of transactivation activity of G1ΔC319 was irrelevant to the impairment of the DNA binding ability and self-association potential.
N-TAD and C-TAD Are Two Independent Transactivation Domains with Degron Activity
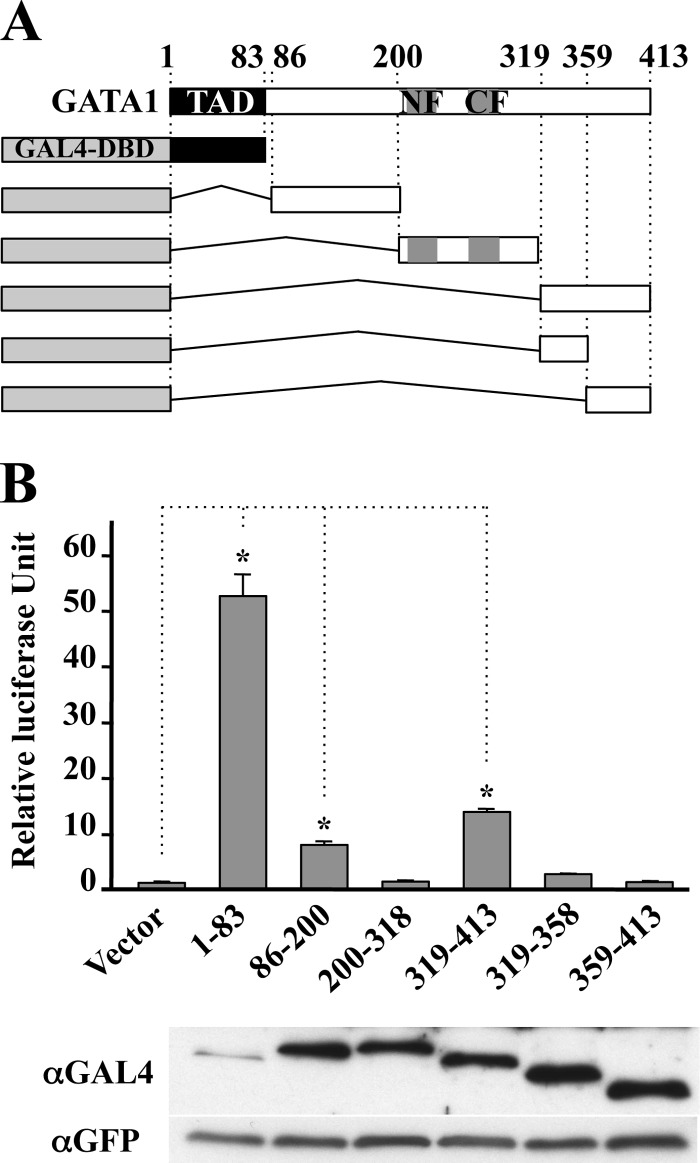
Results thus far support the notion that the C-terminal region of GATA1 acts as the C-TAD. To further verify this notion, we examined the nature of the transactivation activity of C-TAD. We fused the C-TAD of GATA1 to GAL4 DNA-binding domain (GAL4-DBD) within the pcDNA-pGBT9 expression vector (Fig. 3A) and transfected it into QT6 cells with reporter plasmid directed by the upstream activating sequence (17).
FIGURE 3.
C-terminal region (319–413) confers transactivation activity to GAL4-DBD. A, schematic illustration of GAL4-DBD fusion constructs used in transactivation assay. B, QT6 cells were transfected with reporter plasmid and the indicated effectors. Expression of GAL4-DBD fusion constructs in QT6 cells was detected by anti-GAL4 antibody. Transfection efficiency was estimated by expression of green fluorescence protein derived from co-transfected pEGFP-N2. Data are presented as mean ± S.D. (*, p < 0.05).
In this experiment, we noticed that proteins containing either the N- or C-terminal region of GATA1 were easily degraded (supplemental Fig. S2). Although no lysine residue for ubiquitination was located in the N- and C-terminal regions, a proteasome inhibitor MG132 stabilized the GAL4-DBD-GATA1 fusion proteins containing the C-terminal region (supplemental Fig. S2). Consistent with this finding, we previously found that the transactivation domain located in the C terminus of GATA2 was easily degraded through the proteasome pathway upon fusing with GAL4-DBD, although no lysine residue was found in the region (17). We speculate that the C-terminal region of GATA1 acts as a degron functional in the proteasome pathway. Less effect of MG132 in the expression of protein containing N-TAD suggests an important possibility that this region may transduce the signal to an alternative intracellular protein degradation pathway, which concomitantly controls the level of this protein.
We conducted luciferase reporter assay in the presence of 10-μm MG132 during the last 5 h. The expression levels of proteins were equivalent except for that of GAL4-DBD fused with N-terminal region (1–83). To our expectation, the N-terminal 83-amino acid (i.e. N-TAD) possesses strong transactivation activity as reported previously (13). In addition, we found that the internal region (86–200) and the C-terminal 95 amino acids conferred a 9- and 15-fold increase in the reporter expression, respectively (Fig. 3B), indicating that these two regions also retain transactivation activity. In contrast, the finger region of GATA1 (200–318) did not lead to the transactivation. Interestingly, the transactivation activity in the C-terminal 95 amino acids (i.e. C-TAD) completely vanished when this segment was fragmented into two (319–358 and 359–413), suggesting that entire C-terminal region is required for enhancing the heterologous promoter activity. Considering the results of GAL4-DBD assays and markedly weaker transactivation activity in G1ΔNCT than in G1ΔNT (Fig. 1C), we concluded that the transactivation activity of GATA1 is supported by at least two transactivation domains, N-TAD and C-TAD.
C-TAD Is Essential for Embryonic Hematopoiesis
Targeted disruption of mouse Gata1 gene causes embryonic lethality due to the perturbation of primitive erythropoiesis (7, 9, 10). To examine the importance of the transactivation activity conferred by C-TAD in vivo, we exploited transgenic complementation rescue analysis utilizing GATA1 knockdown male (Gata11.05/Y) mice. As this Gata1.05 knockdown allele supports GATA1 expression only 5% of the endogenous level, male mutant embryos (Gata11.05/Y) die by E11.5. On the contrary, heterozygous female (Gata11.05/X) mice are viable and fertile, bearing various degrees of anemia and thrombocytopenia due to random inactivation of X chromosome (9). Transgenic expression of GATA1 using G1HRD rescues the Gata11.05/Y mice from hematopoietic abnormalities and embryonic lethality (32).
We established multiple transgenic mouse lines that expressed G1ΔC319 under the G1HRD regulation. These mouse lines were categorized into three groups depending on the expression level of transgene mRNA (12). We selected three mouse lines, in which the transgene was expressed at a higher, an almost equivalent, and a lower than the endogenous GATA1 in the spleen (supplemental Fig. S3A). We referred these mouse lines to as G1ΔC319-H, G1ΔC319-M, and G1ΔC319-L. The expression levels of the G1ΔC319 protein were comparable with the levels of the transcript (supplemental Fig. S3B).
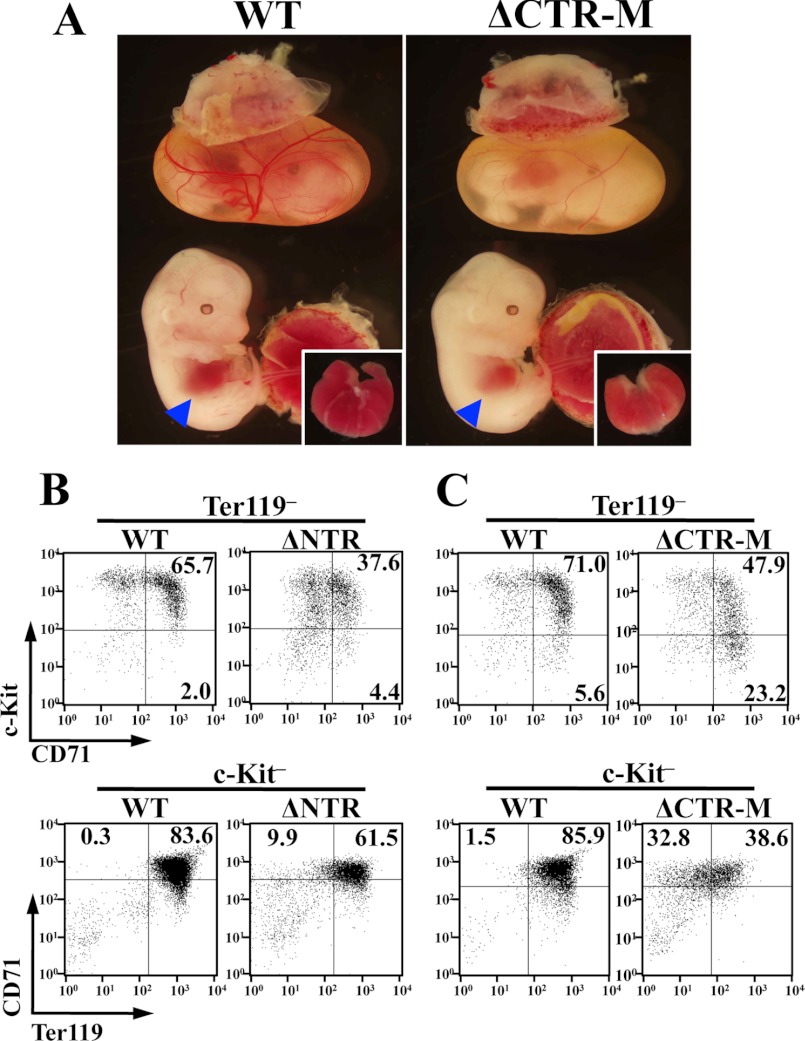
The progeny of crossing the G1ΔC319-H male mice with heterozygous Gata1 knockdown female (Gata11.05/X) mice showed complete conformity with Mendelian expectations. We obtained 11 rescued pups (ΔCTR-H) out of 84 pups. Hematopoietic indices in 3-day-old pups showed slightly anemic parameters in rescued pups, which were out of statistical significance (supplemental Fig. S4A). On the contrary, ΔCTR-M and ΔCTR-L embryos were readily distinguishable from their wild-type littermates by their anemic appearance (Fig. 4A). We obtained 7 living ΔCTR-M embryos at E13.5 out of 57 embryos. The appearance of ΔCTR-M embryos varied embryo-to-embryo (data not shown). Circulating erythrocytes were decreased, and fetal liver was small and pale compared with those in the wild-type littermates (Fig. 4A). Histological examination revealed that the number of hemoglobinized erythrocytes was decreased in the ΔCTR-M embryos (supplemental Fig. S4B). Importantly, severity of anemia was in inverse correlation with the expression level of the transgenic ΔCTR. ΔCTR-L embryos hardly survived beyond E11.5 due to the severe defect in both primitive (yolk sac) and definitive (fetal liver) erythropoiesis (supplemental Fig. S4C). Thus, these results indicate that C-TAD is responsible in both primitive and definitive erythropoiesis, and the defect in GATA1 function caused by the lack of C-TAD can be compensated for depending on the amount of G1ΔC319 expressed.
FIGURE 4.
Impaired erythroid differentiation in ΔCTR embryos. A, ΔCTR-M embryo (right panel) is distinguished from its wild-type (WT) littermate (left panel) by the anemic appearance. Small and pale livers of ΔCTR-M embryos are shown in the inset. B and C, flow cytometry evaluation of erythroid differentiation. Fetal liver monoclonal cells of E13.5 embryos of ΔNTR (B) and ΔCTR-M (C) were analyzed with the expressions of c-Kit and CD71 (upper panels) or CD71 and Ter119 (lower panels) in a Ter119-negative or c-Kit-negative fraction, respectively. Wild-type littermate was used as control.
In the erythroid differentiation process, c-Kit-positive immature progenitors gradually express transferrin receptor (CD71). Subsequently, the c-Kit expression is down-regulated concomitant with the expression of glycophorin A, which is one of the major membrane proteins of mature erythrocyte. Ter119 antibody recognizes a molecule associated with glycophorin A, and the intensity of Ter119 corresponds to the expression level of glycophorin A on the surface of erythroid cells (33). Therefore, we assessed erythroid differentiation in E13.5 fetal livers of ΔCTR-M and ΔNTR, which was Gata11.05/Y mice rescued with transgenic expression of G1ΔNT at a comparable level to endogenous GATA1, by measuring the expression of c-Kit, CD71, and Ter119 (12). As shown in Fig. 4, CD71 and Ter119 expression was weak in both ΔCTR-M and ΔNTR embryos compared with that in their wild-type littermates.
C-TAD Is Required to Regulate a Set of GATA1 Target Genes
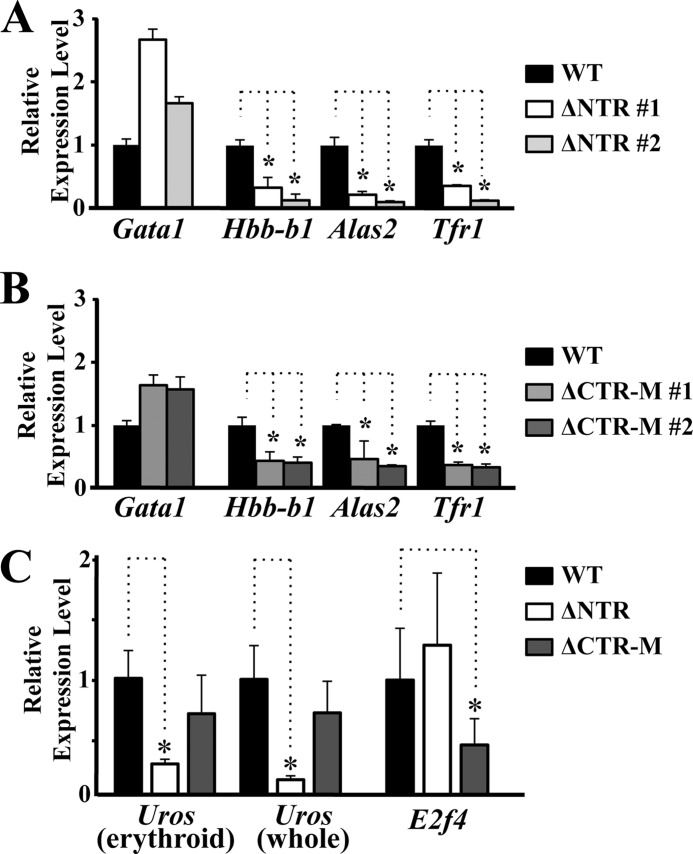
We next examined roles C-TAD plays during the erythroid cell differentiation. As reduced expression of GATA1 target genes is predicted, we examined the expressions of hemoglobin-β1 (Hbb-b1), erythroid 5-aminolevulinate synthase (Alas2), and transferrin receptor 1 (Tfr1) in the liver of E13.5 ΔCTR-M and ΔNTR embryos by means of quantitative RT-PCR analyses. As expected, expressions of Hbb-b1, Alas2, and Tfr1 were severely impaired in both embryos, although the transgene-derived Gata1 gene expression was maintained (Fig. 5, A and B).
FIGURE 5.
Overlapping and unique expression profiles of GATA1 target genes in the ΔCTR and ΔNTR embryos. A and B, quantitative RT-PCR analyses of Hbb-b1, Alas2, and Tfr1 genes using E13.5 fetal livers of ΔNTR (A) and ΔCTR-M (B). Data are shown relative to the expression of wild-type embryo. C, quantitative RT-PCR analyses of Uros and E2f4 genes using E13.5 fetal livers of four wild-type, three ΔNTR, and five ΔCTR-M embryos. Two primer sets corresponding to erythroid-specific 5′-untranslated region in exon 2 and common coding region in exon 9–10 of Uros gene were utilized for amplifying the erythroid-specific (erythroid) and total (whole) Uros transcripts, respectively. Data are shown relative to the average expression of wild-type embryos. (*, p < 0.05).
We searched for GATA1 target genes whose expression was differentially regulated by the N-TAD and C-TAD. Importantly, we found that the expression of uroporphyrinogen III synthase (Uros) was specifically reduced in the ΔNTR embryos, although the expression was maintained in the ΔCTR-M embryos (Fig. 5C). In stark contrast, the expression of E2F transcription factor 4 (E2f4) gene was reduced only in ΔCTR-M embryos (Fig. 5C). Uros is a gene responsible for congenital erythropoietic porphyria (CEP). Two alternative promoters were identified in the UROS/Uros gene in humans and mice, by which housekeeping and erythroid-specific transcripts were produced (34, 35). Various mutations in close proximity of the GATA-binding sequences in the erythroid-specific promoter were reported in the families with CEP (36). In addition, an inherited mutation in the GATA1 gene leading to the production of GATA1 with weak DNA binding activity was identified to cause CEP (37). These broad observations suggest that the GATA1 mutation leading to the decrease of the N-TAD activity may give rise to CEP.
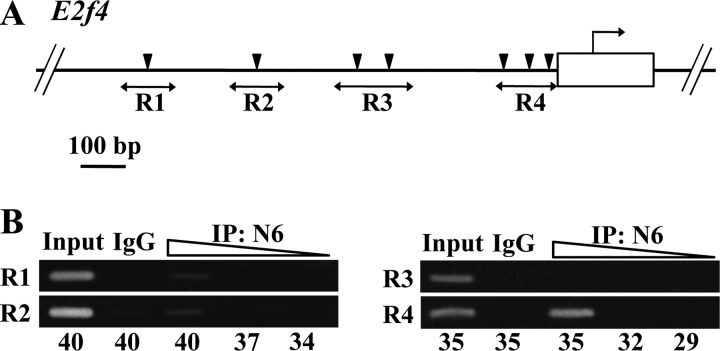
Because E2F4 contributes to the cell cycle regulation as a repressor for the E2F-responsive gene expression (38), proliferation of erythroid cells is specifically affected by the lack of E2F4 in mice without attenuation of differentiation and cell survival process (39). Although seven GATA sites are scattered within 1 kbp upstream to the mouse E2f4 gene (Fig. 6A), we found strong occupancy of GATA1 at a site proximal to the E2f4 gene (R4 in Fig. 6B). We did not identify significant GATA1 occupancy in other regions of the E2f4 gene. This observation shows very good agreement with the data in University of California Santa Cruz genome browser. Thus, E2f4 is one of the GATA1 target genes whose expression depends on the C-TAD activity. Taken together, these results support the contention that C-TAD and N-TAD contribute to the erythroid differentiation program through regulation of overlapping as well as unique target genes.
FIGURE 6.
GATA1 regulates expression of E2f4 gene. A, schematic illustration of the upstream region of mouse E2f4 gene. Arrowheads indicate putative GATA-binding sites. Regions (R1 to R4) amplified in the ChIP analysis are shown with bidirectional arrows. B, ChIP analysis for detection of GATA1 occupancy in the upstream region of E2f4 gene. DNA fragments immunoprecipitated (IP) with GATA1-N6 antibody rendered for indicated cycles of amplification. A 10 times diluted input DNA was used as a positive control for PCR. Samples of control immunoprecipitation reaction with control IgG were also included. Note that the DNA fragments immunoprecipitated with antibody to GATA1 were amplified in the R4 region. Representative data from three independent experiments are shown.
C-TAD Is Essential to Control Megakaryocytic Proliferation
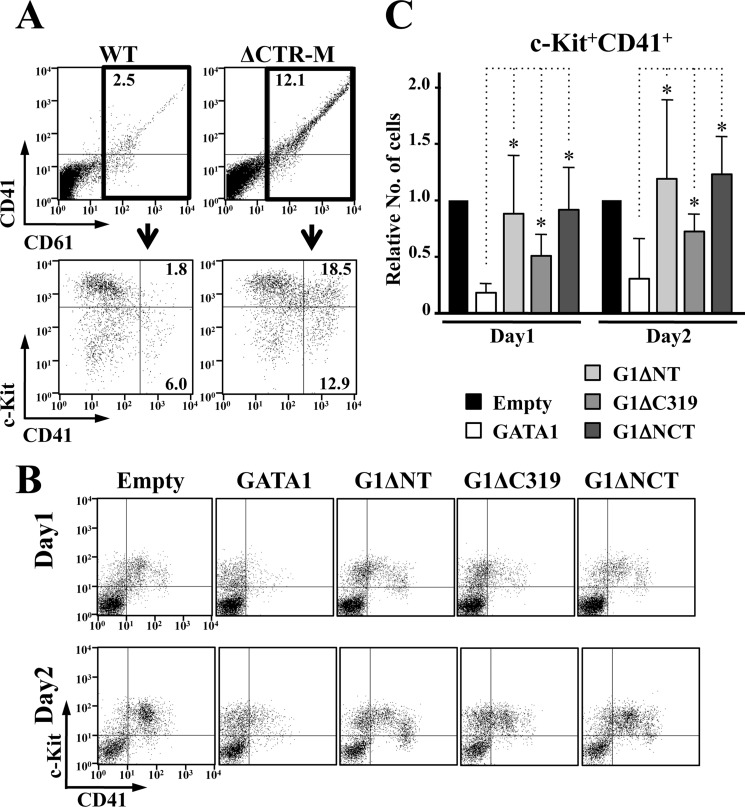
The expression profiles of the platelet glycoprotein IIb/IIIa complex were examined using anti-CD41 and -CD61 antibodies. The number of CD41+CD61+ cells was significantly increased in the ΔCTR-M embryos compared with the wild-type littermates (Fig. 7A, upper panels). Most CD41+CD61+ cells in the ΔCTR-M embryos were c-Kit positive, although CD41+CD61+ cells in wild-type embryos were not (Fig. 7A, lower panels), suggesting that the number of immature megakaryocytes was increased.
FIGURE 7.
Both N-TAD and C-TAD are involved in the GATA1-mediated megakaryocyte growth control. A, flow cytometry analysis of E13.5 fetal livers. CD41+CD61+ cells are abundant in ΔCTR-M embryo (upper panels). Note that c-Kit+CD41+CD61+ cells are dominantly increased in number. B and C, anti-proliferative activity of GATA1 for GATA1-deficient megakaryocytes is attenuated by C-TAD deletion, as is the case for N-TAD. Representative flow cytometry analysis (B) and statistical examination of the number of c-Kit+CD41+ cells from four independent experiments at day 1 and day 2 (C) are shown. The value in the cells introduced empty vector is set to one in every experiment in C. Data are presented as mean ± S.D. (*, p < 0.05).
This hyperproliferative phenotype of megakaryocyte is surprising, as similar phenotype occurs in humans and mice because of the lack of N-TAD (40, 41). To assess the relevance of these observations, we executed in vitro megakaryocyte differentiation assays. We isolated hematopoietic cells from E13.5 fetal livers of Gata1ΔNeoΔHS/Y mice, which lack GATA1 expression specifically in megakaryocytes (21), and retrovirally transduced a series of GATA1 mutants. After a 1- or 2-day incubation in the presence of 50 ng/ml thrombopoietin and 50 ng/ml stem cell factor, we conducted flow cytometry analyses. Consistent with a previous report (42), the number of c-Kit+CD41+ immature megakaryocytes was decreased by the expression of wild-type GATA1, but G1ΔNT did not influence the proliferation of c-Kit+CD41+ cells, similar to the case for empty vector (Fig. 7, B and C). Although the expression of G1ΔC319 partially reduced the number of c-Kit+CD41+ cells, it could not recapitulate the entire function of wild-type GATA1. These findings thus indicate that both N-TAD and C-TAD contribute to the regulation of early megakaryocyte proliferation.
DISCUSSION
Domain function of GATA1 has been studied extensively, as the characterization of the molecular basis for the GATA1 activity is vital for our better understanding of the pathogenesis of human diseases. In this study, we identified for the first time the importance of the C-terminal region of GATA1 as a transactivation domain, and we named this region C-TAD. As is the case for N-TAD, C-TAD is indispensable for the GATA1 activity in both erythroid and megakaryocytic lineage development in embryos. Thus, in this study we demonstrated that GATA1 has two transactivation domains, N-TAD and C-TAD, each of which contributes to the erythroid and megakaryocytic cell differentiation through regulation of overlapping as well as unique target genes.
We have revealed by exploiting genetically manipulated mouse lines that three functional domains of GATA1, i.e. N-TAD, N-finger, and C-finger domains, are indispensable for operating the full activity of GATA1 (12, 41, 43). Dysfunction of these domains leads to characteristic features. For instance, somatic mutations in the GATA1 gene, which result in the production of GATA1-ΔNT (or GATA1s), are a prerequisite for the onset of transient myeloproliferative disorder (TMD) and subsequent acute megakaryoblastic leukemia (AMkL) in Down syndrome patients (44). Similarly, GATA1 mutations in the N-finger domain were found in multiple cases with familial anemia and thrombocytopenia (45–48). In the latter cases, the association potential of GATA1 with either FOG1 or GATA-box DNA was attenuated, indicating that the N-finger domain is important for the GATA1 activity. In contrast, the importance of the C-terminal region was first assessed in the Vlad tepes mutant that leads to the bloodless phenotype in zebrafish (27). However, this phenotype is because of the complete loss of DNA binding ability. Therefore, the genuine function of C-terminal regions of GATA1 remained to be clarified.
Primary structures of transactivation domains are categorized into three types, i.e. acidic, glutamine-rich, or proline-rich (49). The GATA1 N-TAD is categorized into the acidic transactivation domain. The acidic domain can form amphipathic α-helical structure and interacts with TFIID (50). It has been proposed that interaction of the acidic domain with components of the transcription initiation complex stabilizes the general transcription apparatus and promotes transcription initiation (51). Primary structures of N-TAD and C-TAD are relatively diversified in vertebrate homologues compared with other hematopoietic GATA factors, GATA2 and GATA3 (supplemental Fig. S5). The latter GATA factors also possess an acidic transactivation domain in the N-terminal region (13, 17, 52). In contrast, the C-terminal regions of the hematopoietic GATA factors are rich in proline residues.
In this study we demonstrated that the C-terminal region of GATA1 indeed functions as a transactivation domain. This is consistent with our previous observation suggesting that the C-terminal proline-rich region of GATA2 works as a transactivation domain (17). Thus, hematopoietic GATA factors appear to harbor two independent transactivation domains, the acidic domain in the N-terminal region and the proline-rich domain in the C-terminal region. We propose that these two transactivation domains orchestrate the regulation of the GATA1 target genes cooperatively and/or independently in vivo, and this machinery is important for the erythroid and megakaryocytic lineage development (supplemental Fig. S6).
We found that GAL4-DBD fused with N-TAD or C-TAD is unstable, suggesting that these domains act as degrons in QT6 cells. It is interesting to note that deletion of either N- or C-terminal region rather made GATA1 unstable in hematopoietic cells (53). One simple explanation for this discrepancy is that truncations of these domains render GATA1 a less folded protein to be easily recognized by intracellular protein quality control systems. The other and more intriguing possibility is that the transactivation complex consisting of N- or C-TAD in hematopoietic cells is different from that in nonhematopoietic cells. Transactivation domains generally lack stable structure, but they can fold into stable complex with the components of the basal transcription machinery (54). Therefore, the transcription initiation complex on the endogenous GATA1 target genes may fold tightly in hematopoietic cells, but not so on the promoter region of the artificial reporter gene in the nonhematopoietic cells. We also found that the proteasome inhibitor MG132 effectively stabilized GAL4-DBD fused with C-TAD. Because the unstructured region in the ubiquitinated substrate is required for the initiation of degradation in the proteasome system (55), GATA1-C-TAD may act as an initiation mark of degradation in nonhematopoietic cells.
Although the phylogenic tree analysis revealed that N-TAD and C-TAD are diversified regions (supplemental Fig. S5), we previously found that the GATA1 knockdown (Gata11.05/Y) mice were effectively rescued by the transgenic expression of GATA2 or GATA3 if expressed under the regulatory control of G1HRD (32). These results suggest that functional similarity of N-TAD and C-TAD is preserved and acts to overcome the structural diversity of these regions. As a consequence, expression of GATA2 or GATA3 successfully compensates for the GATA1 deficiency during embryogenesis, although the incidence of anemia with aging was increased in these paralog rescued mice (32).
On the contrary, transgenic expression of GATA4 cannot rescue Gata11.05/Y mice from embryonic lethality. GATA4-rescued mice survive only few days longer than Gata11.05/Y (56). GATA4 belongs to the endodermal GATA factor family, in which two independent proline-rich transactivation domains are localized in the N-terminal region, but no transactivation domain is identified in C-terminal region (57). Genome-wide sequence analyses revealed that the hematopoietic and endodermal GATA factors share high similarity in the GATA binding core sequences (4, 5, 58), suggesting that GATA4 expressed in hematopoietic lineages effectively binds to the GATA boxes of target genes.
We surmise one of the major reasons why GATA4 cannot rescue effectively the Gata11.05/Y mice is that transactivation mechanisms of endodermal GATA factors do not operate well under the hematopoietic environment. Interestingly, transgenic expression of the chimeric GATA factor, in which N-terminal half (i.e. N-TAD and N-finger) of GATA4 was fused with C-terminal half (C-finger and C-TAD) of GATA1, gave rise to a longer life span of Gata11.05/Y rather than simple GATA4 did (56). It seems that the transactivation activity of GATA1 C-TAD made certain contributions to the regulation of GATA1 target genes.
Most of the mutations found in the Down syndrome-related TMD and AMkL cases are localized in or neighboring exon 2 of the GATA1 gene. These mutations result in the forced use of the second methionine in the exon 3 as a translation initiation codon, leading to the production of short form GATA1 (GATA1s) lacking the N-terminal 83 amino acids fragment or N-TAD. Therefore, it has been considered that the impaired transactivation activity of GATA1 is one of the most important causes for the pathogenesis of TMD and AMkL. On the contrary, in this study we have revealed that GATA1-ΔNT (or GATA1s) still retains transactivation activity supported by C-TAD. This finding provides evidence that a skewed transactivation activity caused by C-TAD is involved in the onset of Down syndrome TMD and AMkL.
Acknowledgments
We thank Drs. Kinuko Ohneda and Mikiko Suzuki for the kind advice. We also thank Hiromi Yamazaki-Nakayama, Atsushi Hasegawa, Yukie Kawatani, Eriko Naganuma, and Aya Gotoh for their comments and support. We also thank Biomedical Research Core of Tohoku University Graduate School of Medicine for technical support.
This work was supported in part by grants-in-aid for scientific research on priority areas (to M. Y.), for scientific research (to R. S. and M. Y.), and for exploratory research (to M. Y.) from the Ministry of Education, Science, Sports and Culture, the Naito Foundation (to M. Y.), the Takeda Foundation (to R. S. and M. Y.),the Mitsubishi Foundation (to R. S. and M. Y.), the Asahi Glass Foundation (to R. S.) and the Network Medicine Global-COE program to conquest diseases.

This article contains supplemental Figs. 1–6 and Tables 1 and 2.
- E
- embryonic day
- N-TAD
- N-terminal transactivation domain
- C-TAD
- C-terminal transactivation domain
- MBP
- maltose-binding protein
- G1HRD
- Gata1 hematopoietic regulatory domain
- GAL4-DBD
- GAL4-DNA binding domain
- UROS
- uroporphyrinogen III synthase
- CEP
- congenital erythropoietic porphyria
- TMD
- transient myeloproliferative disorder
- AMkL
- acute megakaryoblastic leukemia
- CEP
- congenital erythropoietic porphyria.
REFERENCES
- 1. Ohneda K., Yamamoto M. (2002) Roles of hematopoietic transcription factors GATA-1 and GATA-2 in the development of red blood cell lineage. Acta Haematol. 108, 237–245 [DOI] [PubMed] [Google Scholar]
- 2. Zheng W., Flavell R. A. (1997) The transcription factor GATA-3 is necessary and sufficient for Th2 cytokine gene expression in CD4 T cells. Cell 89, 587–596 [DOI] [PubMed] [Google Scholar]
- 3. Suzuki N., Ohneda O., Minegishi N., Nishikawa M., Ohta T., Takahashi S., Engel J. D., Yamamoto M. (2006) Combinatorial Gata2 and Sca1 expression defines hematopoietic stem cells in the bone marrow niche. Proc. Natl. Acad. Sci. U.S.A. 103, 2202–2207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fujiwara T., O'Geen H., Keles S., Blahnik K., Linnemann A. K., Kang Y. A., Choi K., Farnham P. J., Bresnick E. H. (2009) Discovering hematopoietic mechanisms through genome-wide analysis of GATA factor chromatin occupancy. Mol. Cell 36, 667–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yu M., Riva L., Xie H., Schindler Y., Moran T. B., Cheng Y., Yu D., Hardison R., Weiss M. J., Orkin S. H., Bernstein B. E., Fraenkel E., Cantor A. B. (2009) Insights into GATA-1-mediated gene activation versus repression via genome-wide chromatin occupancy analysis. Mol. Cell 36, 682–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Takayama M., Fujita R., Suzuki M., Okuyama R., Aiba S., Motohashi H., Yamamoto M. (2010) Genetic analysis of hierarchical regulation for Gata1 and NF-E2 p45 gene expression in megakaryopoiesis. Mol. Cell. Biol. 30, 2668–2680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fujiwara Y., Browne C. P., Cunniff K., Goff S. C., Orkin S. H. (1996) Arrested development of embryonic red cell precursors in mouse embryos lacking transcription factor GATA-1. Proc. Natl. Acad. Sci. U.S.A. 93, 12355–12358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kaneko H., Shimizu R., Yamamoto M. (2010) GATA factor switching during erythroid differentiation. Curr. Opin. Hematol. 17, 163–168 [DOI] [PubMed] [Google Scholar]
- 9. Takahashi S., Onodera K., Motohashi H., Suwabe N., Hayashi N., Yanai N., Nabesima Y., Yamamoto M. (1997) Arrest in primitive erythroid cell development caused by promoter-specific disruption of the GATA-1 gene. J. Biol. Chem. 272, 12611–12615 [DOI] [PubMed] [Google Scholar]
- 10. Ferreira R., Wai A., Shimizu R., Gillemans N., Rottier R., von Lindern M., Ohneda K., Grosveld F., Yamamoto M., Philipsen S. (2007) Dynamic regulation of Gata factor levels is more important than their identity. Blood 109, 5481–5490 [DOI] [PubMed] [Google Scholar]
- 11. McDevitt M. A., Shivdasani R. A., Fujiwara Y., Yang H., Orkin S. H. (1997) A “knockdown” mutation created by cis-element gene targeting reveals the dependence of erythroid cell maturation on the level of transcription factor GATA-1. Proc. Natl. Acad. Sci. U.S.A. 94, 6781–6785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shimizu R., Takahashi S., Ohneda K., Engel J. D., Yamamoto M. (2001) In vivo requirements for GATA-1 functional domains during primitive and definitive erythropoiesis. EMBO J. 20, 5250–5260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Martin D. I., Orkin S. H. (1990) Transcriptional activation and DNA binding by the erythroid factor GF-1/NF-E1/Eryf 1. Genes Dev. 4, 1886–1898 [DOI] [PubMed] [Google Scholar]
- 14. Hollanda L. M., Lima C. S., Cunha A. F., Albuquerque D. M., Vassallo J., Ozelo M. C., Joazeiro P. P., Saad S. T., Costa F. F. (2006) An inherited mutation leading to production of only the short isoform of GATA-1 is associated with impaired erythropoiesis. Nat. Genet. 38, 807–812 [DOI] [PubMed] [Google Scholar]
- 15. Li Z., Godinho F. J., Klusmann J. H., Garriga-Canut M., Yu C., Orkin S. H. (2005) Developmental stage-selective effect of somatically mutated leukemogenic transcription factor GATA1. Nat. Genet. 37, 613–619 [DOI] [PubMed] [Google Scholar]
- 16. Mizushima S., Nagata S. (1990) pEF-BOS, a powerful mammalian expression vector. Nucleic Acids Res. 18, 5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Minegishi N., Suzuki N., Kawatani Y., Shimizu R., Yamamoto M. (2005) Rapid turnover of GATA-2 via ubiquitin-proteasome protein degradation pathway. Genes Cells 10, 693–704 [DOI] [PubMed] [Google Scholar]
- 18. Igarashi K., Kataoka K., Itoh K., Hayashi N., Nishizawa M., Yamamoto M. (1994) Regulation of transcription by dimerization of erythroid factor NF-E2 p45 with small Maf proteins. Nature 367, 568–572 [DOI] [PubMed] [Google Scholar]
- 19. Shimizu R., Trainor C. D., Nishikawa K., Kobayashi M., Ohneda K., Yamamoto M. (2007) GATA-1 self-association controls erythroid development in vivo. J. Biol. Chem. 282, 15862–15871 [DOI] [PubMed] [Google Scholar]
- 20. Yamamoto M., Ko L. J., Leonard M. W., Beug H., Orkin S. H., Engel J. D. (1990) Activity and tissue-specific expression of the transcription factor NF-E1 multigene family. Genes Dev. 4, 1650–1662 [DOI] [PubMed] [Google Scholar]
- 21. Shivdasani R. A., Fujiwara Y., McDevitt M. A., Orkin S. H. (1997) A lineage-selective knockout establishes the critical role of transcription factor GATA-1 in megakaryocyte growth and platelet development. EMBO J. 16, 3965–3973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Onodera K., Takahashi S., Nishimura S., Ohta J., Motohashi H., Yomogida K., Hayashi N., Engel J. D., Yamamoto M. (1997) GATA-1 transcription is controlled by distinct regulatory mechanisms during primitive and definitive erythropoiesis. Proc. Natl. Acad. Sci. U.S.A. 94, 4487–4492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wassarman P. M., DePamphilis M. L. (eds) (1993) Methods in Enzymology, Guide to Techniques in Mouse Development, Vol. 225, pp. 747–799, Academic Press, New York [Google Scholar]
- 24. Hasegawa A., Shimizu R., Mohandas N., Yamamoto M. (2012) Mature erythrocyte membrane homeostasis is compromised by loss of the GATA1-FOG1 interaction. Blood 119, 2615–2623 [DOI] [PubMed] [Google Scholar]
- 25. Ito E., Toki T., Ishihara H., Ohtani H., Gu L., Yokoyama M., Engel J. D., Yamamoto M. (1993) Erythroid transcription factor GATA-1 is abundantly transcribed in mouse testis. Nature 362, 466–468 [DOI] [PubMed] [Google Scholar]
- 26. Kitamura T., Koshino Y., Shibata F., Oki T., Nakajima H., Nosaka T., Kumagai H. (2003) Retrovirus-mediated gene transfer and expression cloning. Powerful tools in functional genomics. Exp. Hematol. 31, 1007–1014 [PubMed] [Google Scholar]
- 27. Lyons S. E., Lawson N. D., Lei L., Bennett P. E., Weinstein B. M., Liu P. P. (2002) A nonsense mutation in zebrafish gata1 causes the bloodless phenotype in Vlad tepes. Proc. Natl. Acad. Sci. U.S.A. 99, 5454–5459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Omichinski J. G., Clore G. M., Schaad O., Felsenfeld G., Trainor C., Appella E., Stahl S. J., Gronenborn A. M. (1993) NMR structure of a specific DNA complex of Zn-containing DNA binding domain of GATA-1. Science 261, 438–446 [DOI] [PubMed] [Google Scholar]
- 29. Trainor C. D., Omichinski J. G., Vandergon T. L., Gronenborn A. M., Clore G. M., Felsenfeld G. (1996) A palindromic regulatory site within vertebrate GATA-1 promoters requires both zinc fingers of the GATA-1 DNA-binding domain for high affinity interaction. Mol. Cell. Biol. 16, 2238–2247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yu C., Niakan K. K., Matsushita M., Stamatoyannopoulos G., Orkin S. H., Raskind W. H. (2002) X-linked thrombocytopenia with thalassemia from a mutation in the amino finger of GATA-1 affecting DNA binding rather than FOG-1 interaction. Blood 100, 2040–2045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Crossley M., Merika M., Orkin S. H. (1995) Self-association of the erythroid transcription factor GATA-1 mediated by its zinc finger domains. Mol. Cell. Biol. 15, 2448–2456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Takahashi S., Shimizu R., Suwabe N., Kuroha T., Yoh K., Ohta J., Nishimura S., Lim K. C., Engel J. D., Yamamoto M. (2000) GATA factor transgenes under GATA-1 locus control rescue germline GATA-1 mutant deficiencies. Blood 96, 910–916 [PubMed] [Google Scholar]
- 33. Kina T., Ikuta K., Takayama E., Wada K., Majumdar A. S., Weissman I. L., Katsura Y. (2000) The monoclonal antibody TER-119 recognizes a molecule associated with glycophorin A and specifically marks the late stages of murine erythroid lineage. Br. J. Haematol. 109, 280–287 [DOI] [PubMed] [Google Scholar]
- 34. Aizencang G., Solis C., Bishop D. F., Warner C., Desnick R. J. (2000) Human uroporphyrinogen-III synthase. Genomic organization, alternative promoters, and erythroid-specific expression. Genomics 70, 223–231 [DOI] [PubMed] [Google Scholar]
- 35. Aizencang G. I., Bishop D. F., Forrest D., Astrin K. H., Desnick R. J. (2000) Uroporphyrinogen III synthase. An alternative promoter controls erythroid-specific expression in the murine gene. J. Biol. Chem. 275, 2295–2304 [DOI] [PubMed] [Google Scholar]
- 36. Solis C., Aizencang G. I., Astrin K. H., Bishop D. F., Desnick R. J. (2001) Uroporphyrinogen III synthase erythroid promoter mutations in adjacent GATA1 and CP2 elements cause congenital erythropoietic porphyria. J. Clin. Invest. 107, 753–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Phillips J. D., Steensma D. P., Pulsipher M. A., Spangrude G. J., Kushner J. P. (2007) Congenital erythropoietic porphyria due to a mutation in GATA1. The first trans-acting mutation causative for a human porphyria. Blood 109, 2618–2621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chen H. Z., Tsai S. Y., Leone G. (2009) Emerging roles of E2Fs in cancer. An exit from cell cycle control. Nat. Rev. Cancer 9, 785–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kinross K. M., Clark A. J., Iazzolino R. M., Humbert P. O. (2006) E2f4 regulates fetal erythropoiesis through the promotion of cellular proliferation. Blood 108, 886–895 [DOI] [PubMed] [Google Scholar]
- 40. Roy A., Roberts I., Norton A., Vyas P. (2009) Acute megakaryoblastic leukemia (AMKL) and transient myeloproliferative disorder (TMD) in Down syndrome. A multistep model of myeloid leukemogenesis. Br. J. Haematol. 147, 3–12 [DOI] [PubMed] [Google Scholar]
- 41. Shimizu R., Kobayashi E., Engel J. D., Yamamoto M. (2009) Induction of hyperproliferative fetal megakaryopoiesis by an N-terminally truncated GATA1 mutant. Genes Cells 14, 1119–1131 [DOI] [PubMed] [Google Scholar]
- 42. Kuhl C., Atzberger A., Iborra F., Nieswandt B., Porcher C., Vyas P. (2005) GATA1-mediated megakaryocyte differentiation and growth control can be uncoupled and mapped to different domains in GATA1. Mol. Cell. Biol. 25, 8592–8606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nakano M., Ohneda K., Yamamoto-Mukai H., Shimizu R., Ohneda O., Ohmura S., Suzuki M., Tsukamoto S., Yanagawa T., Yoshida H., Takakuwa Y., Yamamoto M. (2005) Transgenic overexpression of GATA-1 mutant lacking N-finger domain causes hemolytic syndrome in mouse erythroid cells. Genes Cells 10, 47–62 [DOI] [PubMed] [Google Scholar]
- 44. Wechsler J., Greene M., McDevitt M. A., Anastasi J., Karp J. E., Le Beau M. M., Crispino J. D. (2002) Acquired mutations in GATA1 in the megakaryoblastic leukemia of Down syndrome. Nat. Genet. 32, 148–152 [DOI] [PubMed] [Google Scholar]
- 45. Nichols K. E., Crispino J. D., Poncz M., White J. G., Orkin S. H., Maris J. M., Weiss M. J. (2000) Familial dyserythropoietic anaemia and thrombocytopenia due to an inherited mutation in GATA1. Nat. Genet. 24, 266–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mehaffey M. G., Newton A. L., Gandhi M. J., Crossley M., Drachman J. G. (2001) X-linked thrombocytopenia caused by a novel mutation of GATA-1. Blood 98, 2681–2688 [DOI] [PubMed] [Google Scholar]
- 47. Freson K., Devriendt K., Matthijs G., Van Hoof A., De Vos R., Thys C., Minner K., Hoylaerts M. F., Vermylen J., Van Geet C. (2001) Platelet characteristics in patients with X-linked macrothrombocytopenia because of a novel GATA1 mutation. Blood 98, 85–92 [DOI] [PubMed] [Google Scholar]
- 48. Freson K., Matthijs G., Thys C., Mariën P., Hoylaerts M. F., Vermylen J., Van Geet C. (2002) Different substitutions at residue Asp-218 of the X-linked transcription factor GATA1 lead to altered clinical severity of macrothrombocytopenia and anemia and are associated with variable skewed X inactivation. Hum. Mol. Genet. 11, 147–152 [DOI] [PubMed] [Google Scholar]
- 49. Mitchell P. J., Tjian R. (1989) Transcriptional regulation in mammalian cells by sequence-specific DNA binding proteins. Science 245, 371–378 [DOI] [PubMed] [Google Scholar]
- 50. Horikoshi M., Carey M. F., Kakidani H., Roeder R. G. (1988) Mechanism of action of a yeast activator: direct effect of GAL4 derivatives on mammalian TFIID-promoter interactions. Cell 54, 665–669 [DOI] [PubMed] [Google Scholar]
- 51. Ma J., Ptashne M. (1987) Deletion analysis of GAL4 defines two transcriptional activating segments. Cell 48, 847–853 [DOI] [PubMed] [Google Scholar]
- 52. Yang Z., Gu L., Romeo P. H., Bories D., Motohashi H., Yamamoto M., Engel J. D. (1994) Human GATA-3 trans-activation, DNA binding, and nuclear localization activities are organized into distinct structural domains. Mol. Cell. Biol. 14, 2201–2212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lurie L. J., Boyer M. E., Grass J. A., Bresnick E. H. (2008) Differential GATA factor stabilities. Implications for chromatin occupancy by structurally similar transcription factors. Biochemistry 47, 859–869 [DOI] [PubMed] [Google Scholar]
- 54. Lavery D. N., McEwan I. J. (2005) Structure and function of steroid receptor AF1 transactivation domains. Induction of active conformations. Biochem. J. 391, 449–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Prakash S., Tian L., Ratliff K. S., Lehotzky R. E., Matouschek A. (2004) An unstructured initiation site is required for efficient proteasome-mediated degradation. Nat. Struct. Mol. Biol. 11, 830–837 [DOI] [PubMed] [Google Scholar]
- 56. Hosoya-Ohmura S., Mochizuki N., Suzuki M., Ohneda O., Ohneda K., Yamamoto M. (2006) GATA-4 incompletely substitutes for GATA-1 in promoting both primitive and definitive erythropoiesis in vivo. J. Biol. Chem. 281, 32820–32830 [DOI] [PubMed] [Google Scholar]
- 57. Morrisey E. E., Ip H. S., Tang Z., Parmacek M. S. (1997) GATA-4 activates transcription via two novel domains that are conserved within the GATA-4/5/6 subfamily. J. Biol. Chem. 272, 8515–8524 [DOI] [PubMed] [Google Scholar]
- 58. He A., Kong S. W., Ma Q., Pu W. T. (2011) Co-occupancy by multiple cardiac transcription factors identifies transcriptional enhancers active in heart. Proc. Natl. Acad. Sci. U.S.A. 108, 5632–5637 [DOI] [PMC free article] [PubMed] [Google Scholar]