Background: The Hsp70 chaperone cycle mediates stress-denatured protein refolding.
Results: We present the structure of a DnaK-GrpE complex containing the DnaK interdomain linker and substrate-binding domain.
Conclusion: Interaction between the DnaK linker/lid regions and the GrpE N-terminal α-helix and disordered region are essential for function.
Significance: The structure provides a framework for studies concerning interaction of full-length DnaK and GrpE.
Keywords: Crystal Structure, Crystallography, Heat Shock Protein, Protein Folding, Protein-Protein Interactions, Hsp70, Chaperone Cycle
Abstract
The conserved, ATP-dependent bacterial DnaK chaperones process client substrates with the aid of the co-chaperones DnaJ and GrpE. However, in the absence of structural information, how these proteins communicate with each other cannot be fully delineated. For the study reported here, we solved the crystal structure of a full-length Geobacillus kaustophilus HTA426 GrpE homodimer in complex with a nearly full-length G. kaustophilus HTA426 DnaK that contains the interdomain linker (acting as a pseudo-substrate), and the N-terminal nucleotide-binding and C-terminal substrate-binding domains at 4.1-Å resolution. Each complex contains two DnaKs and two GrpEs, which is a stoichiometry that has not been found before. The long N-terminal GrpE α-helices stabilize the linker of DnaK in the complex. Furthermore, interactions between the DnaK substrate-binding domain and the N-terminal disordered region of GrpE may accelerate substrate release from DnaK. These findings provide molecular mechanisms for substrate binding, processing, and release during the Hsp70 chaperone cycle.
Introduction
The evolutionarily conserved 70-kDa heat shock proteins (Hsp70)3 are molecular chaperones (1, 2) involved in the assembly of protein complexes, refolding of stress-denatured proteins, and transport of newly synthesized peptides across membranes; they act by binding and releasing protein substrates in an ATP-dependent manner (3–6). The importance of Hsp70 in numerous cancers and neurological disorders including Alzheimer, Parkinson, and Huntington diseases, has been evaluated (7–9). In Escherichia coli, DnaK, DnaJ, and GrpE, corresponding to Hsp70, the J-domain ATPase-activating protein (Hsp40 family), and the nucleotide exchange factor, respectively, participate in a Hsp70 chaperone cycle. The affinity of the DnaK C-terminal substrate-binding domain (SBD) toward substrates is governed by the nucleotide status of its N-terminal nucleotide-binding domain (NBD) (10, 11). A conserved hydrophobic linker connects NBD and SBD. When ADP is bound to NBD, the substrate affinity of SBD is high, whereas substrate affinity is lower when ATP is bound to NBD (12, 13). However, how the two DnaK domains, DnaJ and GrpE, communicate during the chaperone cycle is unclear.
GrpE accelerates exchange of ADP for ATP in DnaK 5000-fold (14, 15). Previous biochemical and thermodynamic studies have emphasized the importance of full-length DnaK and GrpE for formation of a ternary complex and for substrate processing (12, 13, 16, 17). The GrpE N-terminal disordered region may accelerate the release of substrate bound to DnaK (12, 13). So far, the structural information is available only for a truncated form of the E. coli (Eco) GrpE dimer in complex with the E. coli NBD of DnaK (EcoDnaK_NBD) (18). The complex structure contains neither the SBD nor the DnaK interdomain linker, which are necessary for substrate association (12). Moreover, an EcoGrpE that contains a point mutation (G122D) has a decreased affinity for EcoDnaK (16). To understand how GrpE mediates the release of substrate and nucleotide from DnaK, the mechanism(s) underlying its intermolecular communication with DnaK must be elucidated. Toward this goal, the three-dimensional structure of a full-length DnaK and GrpE complex must be solved.
For the study reported here, we solved the crystal structure of a nearly full-length DnaK (GkDnaK, residues 1–509) complexed with the full-length GrpE (GkGrpE) from the eubacteria Geobacillus kaustophilus HTA426. GkDnaK and GkGrpE are, respectively, the structural and functional homologs of the well characterized EcoDnaK and EcoGrpE. The GkDnaK construct used in this study contains the NBD, the SBD, and the interdomain linker, but not the 10-kDa C-terminal lid that causes GkDnaK to aggregate (19). Strikingly, the structure of the complex suggests two possible modes of interaction between GkDnaK and GkGrpE during the nucleotide exchange step, and it offers insights into how the long GrpE α-helices and interaction between the DnaK lid domain and the N-terminal disordered region of GrpE affect the chaperone cycle.
EXPERIMENTAL PROCEDURES
Cloning, Protein Expression, and Purification
The isolation and purification of GkDnaK have been reported (20). The G. kaustophilus HTA426 gene encoding full-length GkGrpE (residues 1–213) was PCR-amplified using the forward primer 5′-GGAATTCCATATGGAGCAAGGAGAAAAACAAG-3′ and the reverse primer 5′-CCGCTCGAGTTATTGGCTTACTTTGACCATGG-3′. The product was cloned between the NdeI-XhoI restriction sites in a pET21b vector (Novagen) for GkGrpE expression. The gene encoding GkGrpE was expressed in E. coli BL21 (DE3) cultured in Luria-Bertani (LB) broth at 37 °C.
All purification procedures were performed at 4 °C. The cell pellets were pooled and then suspended in 10 ml of 20 mm Tris-HCl, pH 8.0, 20 mm NaCl (buffer A) per gram of cell paste, passed through an M-110L Microfluidizer apparatus (Microfluidics), and then centrifuged at 205,572 × g for 1 h. The supernatant was loaded onto a HiTrap Q HP column (GE Healthcare) equilibrated with buffer A. The column was washed with 30 ml of buffer A, and bound GkGrpE was eluted with a linear gradient of 20–500 mm NaCl in buffer A. Fractions containing GkGrpE were pooled, dialyzed against 20 mm Tris-HCl, pH 8.0, 4 m NaCl (buffer B), and applied to a HiTrap Phenyl FF column (GE Healthcare) equilibrated with buffer B. The column was washed with 20 ml of buffer B, and bound GkGrpE was eluted with a 4.0–0.0 m NaCl gradient in buffer B. The purity of GkGrpE was >90% as assessed by SDS-PAGE.
GkDnaK-GkGrpE Complex Preparation
The GkDnaK-GkGrpE complex was prepared by mixing GkDnaK with a 2-fold molar excess of GkGrpE in 20 mm Tris-HCl, pH 7.9, 500 mm NaCl, 5 mm imidazole (buffer C), incubating the mixture for 1 h at 4 °C, and then applying the mixture to a HisTrap Ni2+-chelating column (GE Healthcare) equilibrated with buffer C. The bound complex was eluted with a linear imidazole gradient of 5–300 mm imidazole in buffer C. Peak fractions were subjected to size exclusion chromatography through a HiLoad 16/600 Superdex 200-pg column (GE Healthcare) equilibrated with buffer C. Eluted fractions containing a protein of ∼320 kDa-, the expected molecular mass of the GkDnaK-GkGrpE complex, were pooled, subjected to buffer exchange (20 mm sodium citrate, pH 5.5; buffer D), and concentrated to 10 mg/ml for crystallization trials.
Crystallization of GkDnaK-GkGrpE Complex
Crystallization screening was performed at 16 °C using the hanging-drop vapor-diffusion method. A mixture of equal volumes of protein solution and reservoir buffer (0.1 m HEPES, pH 7.5, 2% (v/v) polyethylene glycol 400, 2.0 m ammonium sulfate) produced an urchin-shaped GkDnaK-GkGrpE crystal. Crystallization conditions were optimized using the hanging-drop vapor-diffusion method and 1:1 mixtures of protein and reservoir solutions (1 μl each) in the wells of 24-well plates. For data collection, a single octagonal crystal was used that had been grown for 5 days in a solution of the aforementioned composition plus 0.2 mm sodium thiocyanate, and crystal dehydration was performed by serially transfer complex crystal to reservoir drop containing increasing amounts of glycerol (increased in steps of 5% up to 15% (v/v)).
Data Collection, Structure Determination, and Refinement
The crystal was flash frozen in liquid nitrogen and kept under a stream of cold nitrogen (100 K) during data collection, which used the synchrotron radiation x-ray source (1.0000 Å) at the National Synchrotron Radiation Research Center (Taiwan) beamline BL13B1 and an ADSC Quantum-315 CCD detector. Data were integrated and scaled using the HKL-2000 suite of programs (21). The crystal belongs to the space group I4122, has unit-cell dimensions of a = b = 280.0 Å and c = 278.8 Å, and diffracted to 4.1-Å resolution. Multiwavelength anomalous diffraction data for an Au-derivative (potassium tetrachloroaurate (III) hydrate) were used to determine the initial phase. PHENIX (22) was used to locate the gold site and derive the experimental phase. Phase improvement was carried out with density modification by PHENIX. The protein backbones were traced using the truncated GkDnaK (Protein Data Bank, (PDB) code 2V7Y), Thermus thermophilus TthGrpE (PDB code 3A6M), and EcoGrpE (PDB code 1DKG) structures as reference models. There are two GkDnaK-GkGrpE complexes in an asymmetric unit, with each complex containing two GkGrpE and two GkDnaK molecules (assigned as chains A–D in complex 1 and chains E–H in complex 2). Structural refinement was carried out using PHENIX and a random set of 5% of the reflections, which was set aside for cross-validation and calculation of Rfree. Manual adjustments to the model were carried with XtalView (23) and the (2Fo − Fc) and (Fo − Fc) electron density maps. Probably because they were flexible, the N-terminal regions (residues 1–58) in the four GkGrpE molecules, and certain loops in chain E (Lys-127 to Glu-133, Ile-161 to Lys-166, and Gln-178 to Glu-185), chain F (Leu-126 to Glu-133, His-173 to Glu-185, and Val-211 to Gln-213), chain G (Ala-257 to Leu-263), and chain H (Ser-256 to Leu-263) in complex 2 were untraceable. After refinement, the R factor was 27.2% for all reflections between 26.2- and 4.1-Å resolution, and Rfree was 34.4% for the 5% randomly distributed reflections. The Ramachandran plot for the complex contains allowed torsion angles for all observable residues. PyMOL was used to generate the figures (24). Statistics for data collection and structure refinement are listed in Table 1. The atomic coordinates for the GkDnaK-GkGrpE complex have been deposited in the PDB under the accession code 4ANI.
TABLE 1.
Data collection and refinement statistics for GkDnaK-GkGrpE complex
| Parameters | Native | Derivative (gold) | |
|---|---|---|---|
| Data collection | |||
| Space group | I4122 | ||
| Cell dimensions a/b, c(Å) | 280.0, 278.8 | 280.0, 277.2 | |
| High remote | Inflection | ||
|---|---|---|---|
| Wavelength (Å) | 1.00000 | 1.02249 | 1.03964 |
| Resolution (Å) | 26.2-4.1 (4.35-4.1)a | 26.2-4.8 | (4.97-4.8) |
| Rmergeb (%) | 9.3 (49.4) | 9.6 (44.7) | 9.2 (42.9) |
| I/σ | 32.3 (2.8) | 25.2 (5.4) | 26.1 (5.4) |
| Completeness (%) | 94.7 (94.5) | 99.8 (99.9) | 99.8 (100.0) |
| Redundancy | 8.5 (8.7) | 9.4 (8.8) | 9.4 (8.7) |
| Refinement | |||
| Resolution (Å) | 26.2-4.1 | ||
| Number of reflections | 41,248 | ||
| Rwork/Rfree | 27.2/34.4 | ||
| Number of atoms | |||
| Protein | 18,773 | ||
| B-factor (Å2) | |||
| Protein | 136.9 | ||
| r.m.s.d.c | |||
| Bond length (Å) | 0.015 | ||
| Bond angles (º) | 1.960 | ||
a Number in parentheses refer to the highest resolution shell.
b Rmerge = ΣhklΣi|Ii(hkl) − <I(hkl)>|/ΣhklΣi Ii(hkl), where <I(hkl)> is the mean of the observations Ii(hkl) of reflection hkl.
c r.m.s.d., root mean square deviation.
Isothermal Titration Calorimetry (ITC)
The interaction between GkGrpE and GkDnaK or GkDnaK_NBD (residues 1–352) was monitored by ITC at 25 °C with an ITC200 calorimeter (Microcal, GE Healthcare). The protein samples contained 25 mm HEPES, pH 7.0, 50 mm KCl, 2 mm TCEP. A solution of GkDnaK_NBD or GkDnaK plus the C-terminal lid (GkDnaK_FL) (15 μm each in 25 mm HEPES, pH 7.0, 50 mm KCl, 2 mm TCEP) in the calorimeter cell was titrated with 20 consecutive injections of 200 μm GkGrpE. Injections of 200 μm GkGrpE into buffer were used to subtract the heat of dilutions from the corresponding experimental heats. The binding isotherms, ΔH versus molar ratio, were plotted using Microcal ORIGIN software and a single-site binding model.
In Vivo Complementation Assay
GkDnaK mutants containing point mutations in the interdomain linker were subjected to an in vitro viability test (25). Nine codons encoding the GkDnaK residues E354, V355, D357, V358, V359, L360, L361, D362, and V363 were individually mutated to an alanine codon using QuikChange Site-directed Mutagenesis kit reagents (Stratagene). pRSF-Duet plasmids (KanR; Novagen) carrying full-length GkDnaK (GkDnaK_FL), full-length EcoDnaK (EcoDnaK_FL), or a GkDnaK mutant were individually transformed into an E. coli dnak-deletion strain (JW0013) that was derived from the wild-type strain BW25113 (26, 27). Before transformation, the antibiotic marker in the plasmids was removed using the PCP20 vector-mediated method (Gene Bridges) (28). Transformants were selected at 30 °C on agar plates that contained LB and 50 μg/ml kanamycin. Fresh overnight cultures were grown from each single colony, and then the A600 of each culture was adjusted to 0.2 by addition of LB. Serial dilutions (10-fold) of these cultures were spotted onto agar plates that contained LB, 20 mm isopropyl-thio-β-d-galactopyranoside, 50 mm kanamycin, and then incubated overnight at 37 °C or 42 °C to evaluate growth behavior.
RESULTS
Overall Structure of GkDnaK-GkGrpE Complex
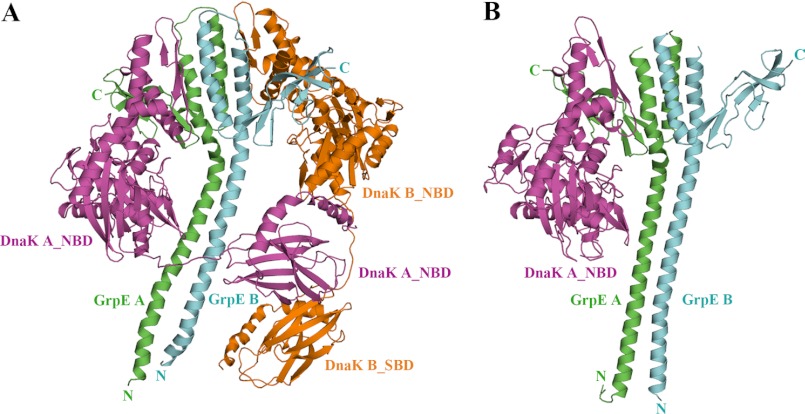
The GkDnaK-GkGrpE crystals belong to the I4122 space group. Because of the high solvent content (71%) and large unit cell dimensions of the crystal, the structure could only be determined to 4.1-Å resolution using multiwavelength anomalous dispersion obtained with the gold derivative (Table 1). Interestingly, there are two GkDnaK-GkGrpE complexes per asymmetric unit, and the interdomain linker in GkDnaK interacts with the substrate-binding site in a neighboring complex (supplemental Fig. 1). Both complexes have similar conformations with a root mean square deviation of 1.6 Å for the Cα coordinates. For simplicity, the structure of only one of the complexes in the asymmetric unit is displayed in Fig. 1A. Each complex contains two GkDnaK molecules (NBD and SBD) and a GkGrpE dimer (Fig. 1A). This is the first structure of the DnaK-GrpE complex to include a nearly full-length DnaK and a full-length GrpE dimer in the nucleotide-free and substrate-bound configuration. The stoichiometry of GkDnaK and GkGrpE in a complex is 2:2, i.e. two GkDnaK molecules bind to one GkGrpE dimer, which was unexpected because the stoichiometry of EcoGrpE to EcoDnaK had been reported as 2:1, implying that one EcoGrpE dimer interacts with only one EcoDnaK molecule (Fig. 1B) (18).
FIGURE 1.
Overall structure of the GkDnaK-GkGrpE complex and comparison with the EcoDnaK_NBD-EcoGrpE complex. A, ribbon diagram of the GkDnaK-GkGrpE complex showing the GkGrpE homodimer (green, GrpE A; cyan, GrpE B) bound by two GkDnaK molecules (magenta, DnaK A; orange, DnaK B). B, ribbon diagram of the EcoDnaK_NBD-EcoGrpE complex (PDB code 1DKG). The color coding is the same as in A.
We labeled the two GkGrpE monomers in the dimer as GkGrpE A and B and the two GkDnaK molecules that dock onto the opposite sides of the two GkGrpE C-terminal β-sheet domains as DnaK A and B. Because two GkDnaK molecules bind to one GkGrpE dimer, one might expect that both GkDnaK molecules would have the same conformation; however, that is not the case. NBD and SBD are oriented side by side in GkDnaK A, but almost linearly in GkDnaK B.
Stoichiometry of DnaK and GrpE
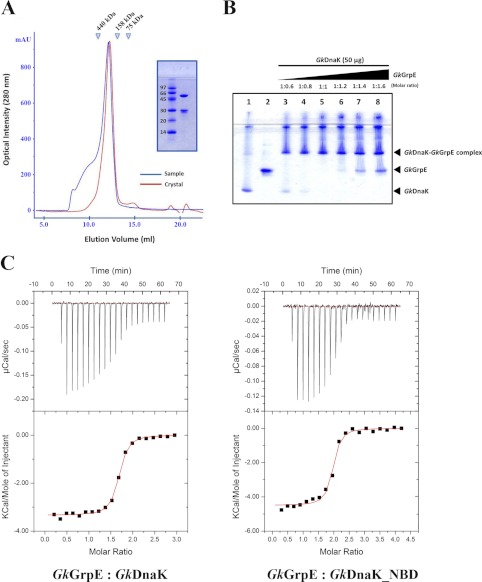
To determine whether GkDnaK and GkGrpE bind in a 2:2 stoichiometry in solution, we determined the size exclusion chromatographic profiles of the proteins that had been dissolved in the same buffer before and after crystallization. The proteins in the two chromatograms eluted at the same positions (Fig. 2A), which suggests that the stoichiometry of the solution complex is also 2:2. We also determined the binding ratio of GkDnaK and GkGrpE using native gel electrophoresis. At an equimolar protein ratio (GkDnaK:GkGrpE = 1:1), bands corresponding to GkDnaK and GkGrpE were not present (Fig. 2B), although a band corresponding to free GkGrpE was observed when excess GkGrpE was present in the starting sample, which indicates that higher order complexes had not formed.
FIGURE 2.
Stoichiometry of GkDnaK with GkGrpE homodimer. A, gel filtration analysis of GkDnaK-GkGrpE complex. The elution profiles colored in blue and in red represent the complex protein solution before crystallization and the complex solution dissolved from protein crystals, respectively. The x-axis and y-axis represent the elution volume and optical intensity by UV spectrometer at 280 nm, respectively. Inset, SDS-PAGE analysis of GkDnaK-GkGrpE complex dissolved from crystals. B, interprotein interactions GkDnaK and GkGrpE performed by increasing concentrations of GkGrpE protein at different molar ratios (lanes 3-8) as indicated with control proteins (protein sample do not contain partner protein; lanes 1 and 2). The individual bands corresponding to GkDnaK and GkGrpE were observed to disappear at equimolar ratio (lane 5). A further increase in GkGrpE concentration (lanes 6-8) over GkDnaK does not result in further binding indicating that the interaction follows 2:2 stoichiometry, not 2:1 as speculated earlier in Gk species. C, upper panels, heat release/s after addition of aliquots of the GkGrpE homodimer into a calorimetry cell containing GkDnaK or GkDnaK_NBD. Lower panels, integrated binding isotherms (black circles, derived from upper panel) and experimental fits (solid red lines) to a single-site model. The best-fit molar-binding stoichiometry values are 1.59 and 1.88 for GkGrpE with GkDnaK and GkDnaK_NBD, respectively.
The interacting interface conformations of the two GkDnaK_NBDs and the GkGrpE dimer are similar to that of the EcoGrpE-EcoDnaK_NBD complex. We calculated the surface areas for the interfaces between GrpE and DnaK_NBD from G. kaustophilus and E. coli. The total buried interface surfaces area for the GkGrpE dimer and GkDnaK_NBD of DnaK A and B are 2778.1 Å2 and 2126.5 Å2, respectively, whereas the buried interface surface area in the EcoDnaK_NBD-EcoGrpE complex (PDB code 1DKG) is 2034.7 Å2. Notably, there is a 23% difference in surface area between GkDnaK_NBD A-GkGrpE and GkDnaK_NBD B-GkGrpE, although these surface areas are both greater than that for the EcoDnaK_NBD-EcoGrpE complex.
Even though our chromatographic and electrophoretic studies suggested a 2:2 stoichiometry for the GkDnaK-GkGrpE complex, our ITC experiments indicated binding stoichiometries between 2:1 and 2:2 (Fig. 2C). Conversely, the ITC study of GkDnaK_NBD-GkGrpE complex formation indicated a binding stoichiometry of 2:1. These different stoichiometries suggest a possible asymmetric binding which resulted from the weak interactions of the second DnaK (DnaK B, note the smaller interacting surface area) with GkGrpE (GkGrpE B). In summary, GkDnaK and GkGrpE appear to form a 2:2 complex, which is not caused by a crystal-packing artifact. The 2:1 stoichiometry observed for the E. coli complex may be a result of the structural/functional differences between the two species because E. coli and G. kaustophilus are a Gram-negative mesophile and a Gram-positive thermophile, respectively.
Flexible DnaK Interdomain Linker Allows Dynamic Motion of DnaK_SBD
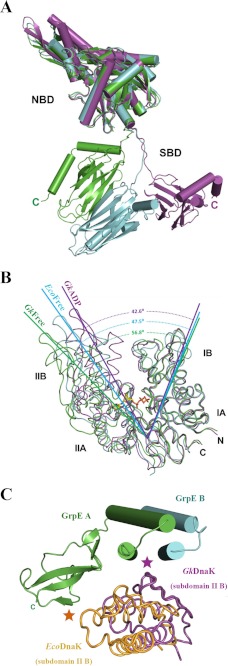
DnaKs exist in a domain-joined conformation (ATP-bound states in which the SBD and NBD interact) or a domain-disjoined conformation (ADP-bound and nucleotide-free states in which the two domains are disengaged (20, 29). Although the SBD and NBD in GkDnaK A and B in our structure do not interact (domain-disjoined conformation) and resemble the ADP-bound GkDnaK structure (PDB code 2V7Y) (20), significant differences in position and conformation are found for the SBDs discussed in this paper when the individual DnaK_NBDs are superimposed (Fig. 3A). The differences in the SBD orientations may be related to the flexibilities of the GkDnaK linkers, which would allow for large domain movements during the various nucleotide-binding events in the chaperone cycle.
FIGURE 3.
Structural comparison of DnaK molecules in different nucleotide-binding states from different species. A, NBDs of GkDnaK in the ADP/Mg2+/Pi state (magenta; PDB code 2V7Y) and the nucleotide-free state (present study; green, DnaK A; cyan, DnaK B) superimposed to show the large scale movement of the SBDs. B, Superpositioning of GkDnaK_NBD in the ADP/Mg2+/Pi state (magenta) and EcoDnaK_NBD (cyan) and GkDnaK_NBD (green), both in nucleotide-free state. The bound nucleotide (ADP) in GkDnaK_NBD is shown as a stick model. The dashed lines indicate the angle(s) (°) of the open nucleotide-binding pockets among the different structures. C, interaction of the DnaK_NBD subdomain IIB with the GrpE four-helix bundle or the C-terminal β-sheet domain. GkGrpE A and B are shown as green and cyan cylinders, respectively. Subdomains IIB of nucleotide-free GkDnaK_NBD and EcoDnaK_NBD are colored in magenta and orange. Magenta and orange asterisks indicate major interacting regions between GrpE and the GkDnaK and EcoDnaK subdomains IIB, respectively.
Nucleotide-binding Pocket Widens upon GrpE Binding
When bound to their respective GrpEs, the nucleotide-binding pocket of EcoDnaK_NBD in the nucleotide-free state (PDB code 1DKG) is wider than that for the ADP-bound state of GkDnaK (PBD code 2V7Y). These states have been denoted “open” and “closed,” respectively (20). To further explore the general pocket-opening phenomenon when DnaK binds dimeric GrpE, we superimposed the nucleotide-free GkDnaK structure from the complex reported in this study with the nucleotide-free EcoDnaK (PDB code 1DKG) and the ADP-bound GkDnaK (PDB code 2V7Y) structures. Interestingly, the opening of the nucleotide-binding pocket for our GkDnaK structure is 56.8°, which is 9.3° wider than that of the EcoDnaK (Fig. 3B) even though both pockets are open and free of nucleotides.
We also found differences between the DnaK/GrpE interfaces of GkDnaK-GkGrpE and the EcoDnaK-EcoGrpE complex. Subdomain IIB in EcoDnaK_NBD interacts with the β-sheet domain in EcoGrpE A (Fig. 3C). For GkDnaK_NBD, subdomain IB interacts with GkGrpE A in a manner similar to that found for EcoDnaK_NBD (data not shown in Fig. 3C), although the GkDnaK subdomain IIB interacts with the four-helix bundle formed by both molecules in the GkGrpE dimer. This interaction might pull subdomain IIB away from subdomain IB, resulting in a wider opening of the nucleotide-binding pocket compared with that of the EcoDnaK-EcoGrpE complex. The differences in interactions of DnaK and GrpE may be a consequence of the presence of the C-terminal SBD and the linker region that interact with the long N-terminal α-helices of the GrpE dimer (Fig. 1A). The observed structural differences suggest that the conformation observed in our structure is more realistic than that of the EcoDnaK-EcoGrpE complex. The interaction of the GkDnaK subdomain IIB with both chains in the GkGrpE dimer also provides another explanation as to why GrpE functions as a dimer aside from its thermosensing ability.
Similarities and Differences among GrpE Structures
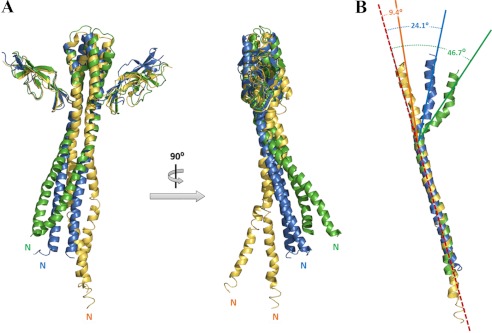
The overall structural features of GkGrpE are consistent with those of EcoGrpE (18) and TthGrpE (30) and include the long N-terminal α-helix, the central four-helix bundle, and the C-terminal β-sheet domain (Fig. 4A). In contrast to the rigid and parallel conformation of the long N-terminal α-helices seen in the EcoGrpE structure (colored blue in Fig. 4A), a coiled-coil structure is seen for this region in the GkGrpE dimer (colored green in Fig. 4A), which is similar to that observed for the TthGrpE structure (colored yellow in Fig. 4A). Furthermore, upon aligning the N-terminal α-helices of these molecules, the four-helix bundle in GkGrpE has a greater curvature (46.7°) than that found in EcoGrpE (24.1°) and TthGrpE (9.4°) (Fig. 4B). Perhaps, the curvature of the GkGrpE α-helices is a consequence of the interaction between the GkDnaK C-terminal region (linker and SBD) and the GkGrpE helices, which is not observed in EcoDnaK-EcoGrpE structure. The increased curvature may cause the DnaK nucleotide-binding pocket to open further and cause the altered interactions between the DnaK_NBD subdomains as described above.
FIGURE 4.
Structural comparison of GrpEs from different species. A, front and orthogonal views of GkGrpE (green) aligned with TthGrpE (orange; PDB code 3A6M) and EcoGrpE (blue; PDB code 1DKG). Large shifts in the orientations and positions occur mainly in the long N-terminal α-helices. B, superpositioning of the long GrpE N-terminal α-helices showing substantially more curvature for the four-helix bundle in GkGrpE (46.7°, green, chain A) compared with four-helix bundle in EcoGrpE (24.1°, blue) and TthGrpE (9.4°, yellow). Red dashed lines indicate central axes.
A previous report on the four-helix bundle in TthGrpE (30) proposed a topology different from that observed for the corresponding structures in EcoGrpE (18). The description of the topology for the EcoGrpE relied on discontinuous and weak electron density in the region connecting the α-helices of the four-helix bundle in the EcoGrpE-EcoDnaK complex (18). Although the resolution of the structure reported here is only 4.1 Å, continuous electron density is present in the 2 Fo − Fc electron density map, and the density contoured at 2.0 σ in this map is distinguishable and seemingly in accordance with the topology of the EcoDnaK-EcoGrpE structure (Fig. 5). Therefore, these two regions in the EcoGrpE and GkGrpE dimers have the same topology. However, the possibility of a different topology observed in TthGrpE cannot be excluded. The difference may be caused by structural and functional diversities across different species as the thermosensor mechanisms are known to be different between T. thermophilus and E. coli.
FIGURE 5.
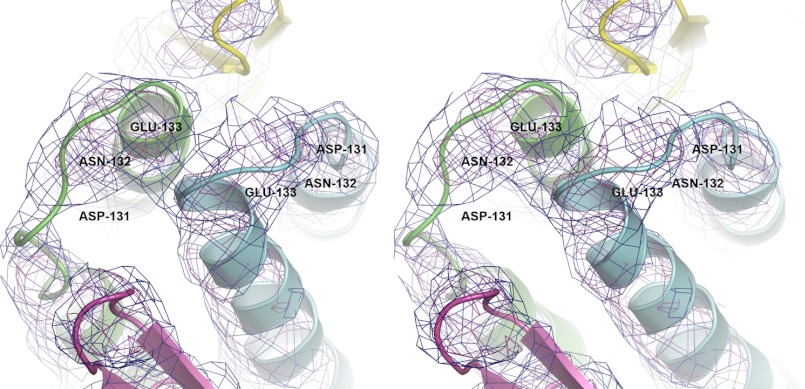
Topology of GkGrpE homodimer. Stereo view of GkGrpE initial experimental electron density map around the linker region connecting α-helices at the four-helix bundle domain. Chains A and B of GkGrpE are represented in green and blue, respectively. The observed electron density is shown in blue and purple mesh at 1.0σ and 2.0σ, respectively, and the residues involved in connecting the helices at four-helix bundle domain are represented with legends.
Long N-terminal α-Helices of GkGrpE Stabilize Ternary Complex
Residues 34–68 in EcoGrpE (residues 54–88 in GkGrpE) interact with the EcoDnaK interdomain linker (13), help stabilize the DnaK-substrate complex, and facilitate nucleotide exchange (12, 17, 31). The interaction between the long GrpE N-terminal α-helices and DnaK is therefore important to substrate processing.
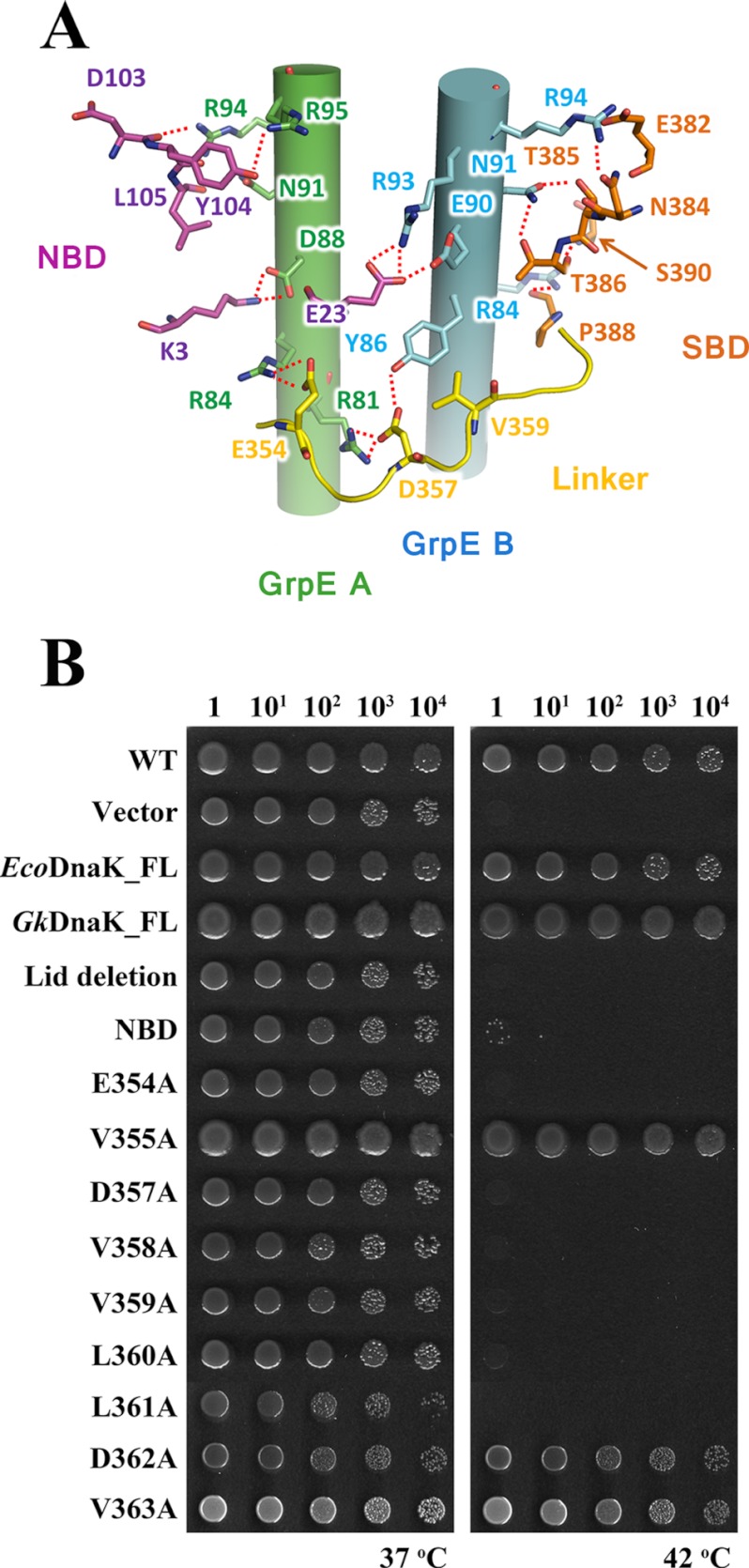
In the GkDnaK-GkGrpE structure, GkDnaK A passes over the GkGrpE dimer as a result of the extended GkDnaK linker, which suggests that the linker can interact with the long GkGrpE N-terminal α-helices (Figs. 1A and 6A). To test whether this interaction exists, we selected residues to mutate by carefully scrutinizing the structures of GkDnaK (PDB code 2V7Y), EcoGrpE (PDB code 1DKG), and TthGrpE (PDB code 3A6M) (see below). According to an examination of the structure, the interdomain linker seems to interact with the long N-terminal α-helices via hydrogen bonding and van der Waals interactions. Additionally, several nonlinker residues in NBD and SBD may interact with the GkGrpE long N-terminal α-helices also. The alignment of the GkGrpE, EcoGrpE, and TthGrpE sequences also identified several conserved residues involved in the aforementioned possible interactions (supplemental Fig. 2).
FIGURE 6.
Interactions between GkDnaK and the GkGrpE homodimer. A, possible side chain interactions between GkDnaK A and the GkGrpE homodimer. GkGrpE A (green) and B (blue) are shown as cylinders with side chains. GkDnaK residues in the NBD (magenta), interdomain linker (yellow), and SBD (orange) are shown in stick models. Possible hydrogen bonds are shown as dashed lines. B, complementation assay for DnaK. Serial dilutions of fresh E. coli cultures were spotted onto agar containing LB and incubated at 37 °C or 42 °C overnight. Empty vector and full-length DnaK (EcoDnaK_FL and GkDnaK_FL) vectors were used as negative and positive controls, respectively. Dilution factors for the E. coli cultures are labeled over the panels.
The linker region between NBD and SBD may be a pseudosubstrate for DnaK because of its hydrophobic nature (32–34). In agreement with this hypothesis, in the ADP-bound GkDnaK structure, the hydrophobic region of the linker is within the substrate-binding pocket of a neighboring GkDnaK molecule (20), which is also found for the current structure. The GkGrpE dimer and the chain A GkDnaK_SBD interact with the neighboring GkDnaK linker, thereby mimicking a ternary complex configuration (data not shown). This result agrees with previous findings that substrate recognition by DnaK increases when the GrpE dimer binds and subsequently stabilizes the linker (12, 17). The spatial relationship between GkDnaK A and the GkGrpE dimer suggests that the allosteric communication between DnaK SBD and NBD occurs via the linker which is co-regulated by the long GrpE N-terminal α-helices.
In Vivo Complementation Assays for DnaK
In the GkDnaK-GkGrpE structure, the linker surrounds and appears to interact with the GkGrpE long N-terminal α-helices via side chain interactions (Fig. 6A). To determine whether these interactions are crucial for the function of DnaK and GrpE, we prepared two mutants containing deletions and several containing point mutations in the linker sequence for use in in vivo complementary assays. DnaK is essential for cell viability at elevated temperatures or under heat shock conditions (42 °C) but is not essential at 37 °C. The GkDnaK mutants encoded on plasmids were transformed into the dnak-deletion E. coli strain JW0013 (26), and cell viability was assessed at 42 °C. Transformed cells cultured at 37 °C served as the controls. Full-length EcoDnaK or GkDnaK rescued the temperature-dependent viability defect (Fig. 6B). However, GkDnaK_NBD (residues 1–352) and GkDnaK (the lid domain-deletion mutant used in this study, residues 1–509) did not rescue cells cultured at 42 °C, which indicates the functional importance of the lid domain and the SBD. The importance of the lid domain for function, as identified by the complementation assay, is consistent with previous reports that suggested its interaction with the N-terminal disordered region of GrpE is necessary for function (13, 32, 35).
For the GkDnaK single-point mutants, only V355A, D362A, and V363A maintained cell viability after heat shock (Fig. 6B). In the GkDnaK-GkGrpE structure, Val-355, Asp-362, and Val-363 do not interact with GkGrpE, which is consistent with the abilities of their alanine mutants to maintain cell viability. In addition to the four well studied consecutive hydrophobic residues (Val-358, Val-359, Leu-360, and Leu-361) in the linker (the corresponding residues are VLLL in E. coli), we identified two additional residues (Asp-354 and Asp-357) that are necessary for cell viability after heat shock and are therefore important for DnaK chaperone activity (Fig. 6B). Using the same rationale, we attempted to develop an E. coli strain that is defective in GrpE activity for complementary studies. Unfortunately, we could not obtain such a mutant probably because GrpE is necessary for cell viability (26, 36, 37).
Given the relationship between the positions of the mutations in the GkDnaK-GkGrpE complex and their effects in the complementation assay, interactions between the DnaK linker and the long GrpE α-helices and/or those between the DnaK lid domain and the GrpE N-terminal disordered region are probably essential for DnaK function.
DISCUSSION
The association and dissociation of substrates are coupled to conformational changes in DnaK and are controlled by nucleotide exchange, which raises the issue of why DnaK requires GrpE homodimer for nucleotide exchange. Previous reports have suggested a 2:1 stoichiometry for EcoGrpE and EcoDnaK (18). In our present study, however, we found a 2:2 stoichiometry for the GkDnaK-GkGrpE complex in the nucleotide-free state, suggesting that the chaperone systems differ for Gram-positive thermophiles and the mesophilic E. coli. The 2:2 stoichiometry can be rationalized as follows. After association of DnaK A and GrpE A, there is room for DnaK B and GrpE B to bind. Additionally, the types of nucleotide exchange factors available are structurally more diverse than are the Hsp70- and Hsp40-type proteins. Finally, the extended GrpE N or C termini found for even different prokaryotes might still influence DnaK-GrpE binding (38, 39).
We cannot exclude the possibility that the 2:1 stoichiometry of GrpE and DnaK does exist in nature, although under heat stress or mechanical stress conditions the 2:2 stoichiometry would be advantageous because substrate could be processed more rapidly. Furthermore, uneven expression of GrpE and DnaK has been reported. When the Gram-positive Lactococcus spp. or an Archaea (40, 41) are initially subjected to heat shock, their DnaK expression levels are significantly higher than are those of GrpE. Conceivably, when excess DnaK is available, a 2:2 stoichiometry is preferred.
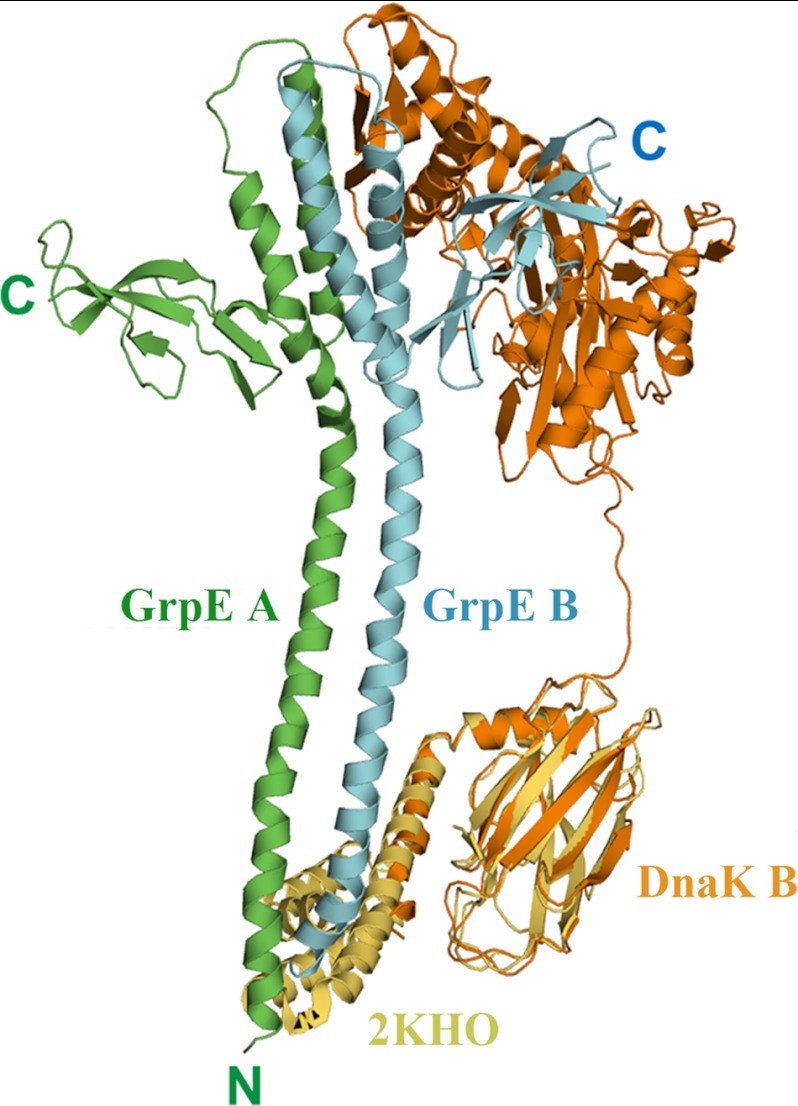
It has been proposed that, in addition to the nucleotide exchange mechanism, the N-terminal disordered region (residues 1–33) of GrpE can trigger substrate release from DnaK (12, 13). Possibly the interaction of the N-terminal disordered region of GrpE interacts with DnaK to activate chaperone activity (13, 32, 38). How this interaction occurs is unclear, however, because the conformation of the GrpE participant has not been delineated. Because the lid domain is not present in the GkDnaK construct used in our study and because electron density for the GkGrpE N-terminal disordered region was not found, we could not observe an interaction between these two regions directly. Nonetheless, GkDnaK B is parallel to the GkGrpE dimer, and its SBD faces the expected position for the GkGrpE N-terminal disordered region (Fig. 1A), suggesting that SBD and the GrpE N-terminal disordered region might interact. In addition, the lid domain is crucial for DnaK function as shown by our complementation assays. We therefore propose that the GkDnaK lid domain interacts with the GkGrpE N-terminal disordered region during a certain stage of the chaperone cycle. To strengthen our hypothesis, we replaced the GkDnaK_SBD structure with the full EcoDnaK_SBD structure (residues 389–607) (PDB code 1DKX or 2KHO) through molecular modeling approach (42, 43) (Fig. 7). The extended C-terminal lid domain of EcoDnaK is located in the vicinity of the GkGrpE N-terminal disordered region and the N-terminal region of the GkGrpE long α-helix domain in the modeling result, suggesting that the lid domain interacts with the N-terminal region of GrpE, which agrees with published reports suggesting a role for the GrpE N-terminal disordered region in accelerating the release of bound substrate from DnaK.
FIGURE 7.
Molecular modeling between GkDnaK and GkGrpE homodimer. Ribbon diagram describes the modeling result of GkDnaK (orange, DnaK B) aligned with the complete EcoDnaK_SBD structure (yellow; PDB code 2KHO). The mimic GkDnaK full-length model shows the possible interaction between the extended α-helical lid domain of GkDnaK and the N-terminal disordered and long helical region of GkGrpE dimer (green, GrpE A; cyan, GrpE B).
In summary, we determined the crystal structure of a two-domain construct of GkDnaK in complex with its nucleotide exchange factor GkGrpE from the Gram-positive G. kaustophilus HTA426. The structure suggests a novel 2:2 stoichiometric arrangement for chaperone proteins that would promote effective substrate processing and also provides insights into the intermolecular communication between DnaK and GrpE in the nucleotide-free and substrate-bound configuration.
Acknowledgments
We are grateful for the access to the synchrotron radiation beamline 13B1 at the National Synchrotron Radiation Research Center (NSRRC) in Taiwan.
This work was supported by Research Grant NSC98-2311-B-001-010-MY3 from Academia Sinica and the National Science Council Taiwan, Republic of China (to C.-D. H.).

This article contains supplemental Figs. 1 and 2.
The atomic coordinates and structure factors (code 4ANI) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- Hsp70
- 70-kDa heat shock protein
- Eco
- Escherichia coli
- FL
- full-length
- Gk
- Geobacillus kaustophilus HTA426
- ITC
- isothermal titration calorimetry
- NBD
- nucleotide-binding domain
- PDB
- Protein Data Bank
- SBD
- substrate-binding domain
- TCEP
- tris(2-carboxyethyl)phosphine
- Tth
- Thermus thermophilus.
REFERENCES
- 1. Mayer M. P., Bukau B. (2005) Hsp70 chaperones: cellular functions and molecular mechanism. Cell. Mol. life Sci. 62, 670–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Meimaridou E., Gooljar S. B., Chapple J. P. (2009) From hatching to dispatching: the multiple cellular roles of the Hsp70 molecular chaperone machinery. J. Mol. Endocrinol. 42, 1–9 [DOI] [PubMed] [Google Scholar]
- 3. Hartl F. U. (1996) Molecular chaperones in cellular protein folding. Nature 381, 571–579 [DOI] [PubMed] [Google Scholar]
- 4. Hartl F. U., Hayer-Hartl M. (2002) Molecular chaperones in the cytosol: from nascent chain to folded protein. Science 295, 1852–1858 [DOI] [PubMed] [Google Scholar]
- 5. Rohde M., Daugaard M., Jensen M. H., Helin K., Nylandsted J., Jäättelä M. (2005) Members of the heat-shock protein 70 family promote cancer cell growth by distinct mechanisms. Genes Dev. 19, 570–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Su P. H., Li H. M. (2010) Stromal Hsp70 is important for protein translocation into pea and Arabidopsis chloroplasts. Plant Cell 22, 1516–1531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gestwicki J. E., Crabtree G. R., Graef I. A. (2004) Harnessing chaperones to generate small-molecule inhibitors of amyloid β aggregation. Science 306, 865–869 [DOI] [PubMed] [Google Scholar]
- 8. Jinwal U. K., Miyata Y., Koren J., 3rd, Jones J. R., Trotter J. H., Chang L., O'Leary J., Morgan D., Lee D. C., Shults C. L., Rousaki A., Weeber E. J., Zuiderweg E. R., Gestwicki J. E., Dickey C. A. (2009) Chemical manipulation of Hsp70 ATPase activity regulates tau stability. J. Neurosci. 29, 12079–12088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Novoselova T. V., Margulis B. A., Novoselov S. S., Sapozhnikov A. M., van der Spuy J., Cheetham M. E., Guzhova I. V. (2005) Treatment with extracellular Hsp70/Hsc70 protein can reduce polyglutamine toxicity and aggregation. J. Neurochem. 94, 597–606 [DOI] [PubMed] [Google Scholar]
- 10. Raviol H., Bukau B., Mayer M. P. (2006) Human and yeast Hsp110 chaperones exhibit functional differences. FEBS Lett. 580, 168–174 [DOI] [PubMed] [Google Scholar]
- 11. Schmid D., Baici A., Gehring H., Christen P. (1994) Kinetics of molecular chaperone action. Science 263, 971–973 [DOI] [PubMed] [Google Scholar]
- 12. Brehmer D., Gässler C., Rist W., Mayer M. P., Bukau B. (2004) Influence of GrpE on DnaK-substrate interactions. J. Biol. Chem. 279, 27957–27964 [DOI] [PubMed] [Google Scholar]
- 13. Moro F., Taneva S. G., Velázquez-Campoy A., Muga A. (2007) GrpE N-terminal domain contributes to the interaction with DnaK and modulates the dynamics of the chaperone substrate binding domain. J. Mol. Biol. 374, 1054–1064 [DOI] [PubMed] [Google Scholar]
- 14. Mally A., Witt S. N. (2001) GrpE accelerates peptide binding and release from the high affinity state of DnaK. Nat. Struct. Biol. 8, 254–257 [DOI] [PubMed] [Google Scholar]
- 15. Packschies L., Theyssen H., Buchberger A., Bukau B., Goody R. S., Reinstein J. (1997) GrpE accelerates nucleotide exchange of the molecular chaperone DnaK with an associative displacement mechanism. Biochemistry 36, 3417–3422 [DOI] [PubMed] [Google Scholar]
- 16. Grimshaw J. P., Siegenthaler R. K., Züger S., Schönfeld H. J., Z'graggen B.R., Christen P. (2005) The heat-sensitive Escherichia coli grpE280 phenotype: impaired interaction of GrpE(G122D) with DnaK. J. Mol. Biol. 353, 888–896 [DOI] [PubMed] [Google Scholar]
- 17. Han W., Christen P. (2001) Mutations in the interdomain linker region of DnaK abolish the chaperone action of the DnaK/DnaJ/GrpE system. FEBS Lett. 497, 55–58 [DOI] [PubMed] [Google Scholar]
- 18. Harrison C. J., Hayer-Hartl M., Di Liberto M., Hartl F., Kuriyan J. (1997) Crystal structure of the nucleotide exchange factor GrpE bound to the ATPase domain of the molecular chaperone DnaK. Science 276, 431–435 [DOI] [PubMed] [Google Scholar]
- 19. Chou C. C., Forouhar F., Yeh Y. H., Shr H. L., Wang C., Hsiao C. D. (2003) Crystal structure of the C-terminal 10-kDa subdomain of Hsc70. J. Biol. Chem. 278, 30311–30316 [DOI] [PubMed] [Google Scholar]
- 20. Chang Y. W., Sun Y. J., Wang C., Hsiao C. D. (2008) Crystal structures of the 70-kDa heat shock proteins in domain disjoining conformation. J. Biol. Chem. 283, 15502–15511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Otwinowski Z., Minor W. (1997) Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 20, 307–326 [DOI] [PubMed] [Google Scholar]
- 22. Zwart P. H., Afonine P. V., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., McKee E., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., Storoni L. C., Terwilliger T. C., Adams P. D. (2008) Automated structure solution with the PHENIX suite. Methods Mol. Biol. 426, 419–435 [DOI] [PubMed] [Google Scholar]
- 23. McRee D. E. (1999) XtalView/Xfit: a versatile program for manipulating atomic coordinates and electron density. J. Struct. Biol. 125, 156–165 [DOI] [PubMed] [Google Scholar]
- 24. Delano W. L. (2010) The PyMOL Molecular Graphics System, version 1.3r1, Schrödinger, LLC, New York [Google Scholar]
- 25. Kumar D. P., Vorvis C., Sarbeng E. B., Cabra Ledesma V. C., Willis J. E., Liu Q. (2011) The four hydrophobic residues on the Hsp70 interdomain linker have two distinct roles. J. Mol. Biol. 411, 1099–1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Baba T., Ara T., Hasegawa M., Takai Y., Okumura Y., Baba M., Datsenko K. A., Tomita M., Wanner B. L., Mori H. (2006) Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Systems Biol. 2, 2006.0008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yamamoto N., Nakahigashi K., Nakamichi T., Yoshino M., Takai Y., Touda Y., Furubayashi A., Kinjyo S., Dose H., Hasegawa M., Datsenko K. A., Nakayashiki T., Tomita M., Wanner B. L., Mori H. (2009) Update on the Keio collection of Escherichia coli single-gene deletion mutants. Mol. Systems Biol. 5, 335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Datsenko K. A., Wanner B. L. (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 97, 6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wisniewska M., Karlberg T., Lehtiö L., Johansson I., Kotenyova T., Moche M., Schüler H. (2010) Crystal structures of the ATPase domains of four human Hsp70 isoforms: HSPA1L/Hsp70-hom, HSPA2/Hsp70–2, HSPA6/Hsp70B′, and HSPA5/BiP/GRP78. PLoS One 5, e8625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nakamura A., Takumi K., Miki K. (2010) Crystal structure of a thermophilic GrpE protein: insight into thermosensing function for the DnaK chaperone system. J. Mol. Biol. 396, 1000–1011 [DOI] [PubMed] [Google Scholar]
- 31. Bogdanov M., Sun J., Kaback H. R., Dowhan W. (1996) A phospholipid acts as a chaperone in assembly of a membrane transport protein. J. Biol. Chem. 271, 11615–11618 [DOI] [PubMed] [Google Scholar]
- 32. Chesnokova L. S., Slepenkov S. V., Protasevich I. I., Sehorn M. G., Brouillette C. G., Witt S. N. (2003) Deletion of DnaK's lid strengthens binding to the nucleotide exchange factor, GrpE: a kinetic and thermodynamic analysis. Biochemistry 42, 9028–9040 [DOI] [PubMed] [Google Scholar]
- 33. Haslbeck M., Walke S., Stromer T., Ehrnsperger M., White H. E., Chen S., Saibil H. R., Buchner J. (1999) Hsp26: a temperature-regulated chaperone. EMBO J. 18, 6744–6751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rüdiger S., Schneider-Mergener J., Bukau B. (2001) Its substrate specificity characterizes the DnaJ co-chaperone as a scanning factor for the DnaK chaperone. EMBO J. 20, 1042–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Schlecht R., Erbse A. H., Bukau B., Mayer M. P. (2011) Mechanics of Hsp70 chaperones enables differential interaction with client proteins. Nat. Struct. Mol. Biol. 18, 345–351 [DOI] [PubMed] [Google Scholar]
- 36. Ang D., Chandrasekhar G. N., Zylicz M., Georgopoulos C. (1986) Escherichia coli grpE gene codes for heat shock protein B25.3, essential for both λDNA replication at all temperatures and host growth at high temperature. J. Bacteriol. 167, 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ang D., Georgopoulos C. (1989) The heat shock-regulated grpE gene of Escherichia coli is required for bacterial growth at all temperatures but is dispensable in certain mutant backgrounds. J. Bacteriol. 171, 2748–2755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Harrison C. (2003) GrpE, a nucleotide exchange factor for DnaK. Cell Stress Chaperones 8, 218–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Liu Y., Gierasch L. M., Bahar I. (2010) Role of Hsp70 ATPase domain intrinsic dynamics and sequence evolution in enabling its functional interactions with NEFs. PLoS Computational Biol. 6, e1000931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Conway De Macario E., Clarens M., Macario A. J. (1995) Archaeal grpE: transcription in two different morphologic stages of Methanosarcina mazei and comparison with DnaK and DnaJ. J. Bacteriol. 177, 544–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Arnau J., Sørensen K. I., Appel K. F., Vogensen F. K., Hammer K. (1996) Analysis of heat shock gene expression in Lactococcus lactis MG1363. Microbiology 142, 1685–1691 [DOI] [PubMed] [Google Scholar]
- 42. Bertelsen E. B., Chang L., Gestwicki J. E., Zuiderweg E. R. (2009) Solution conformation of wild-type E. coli Hsp70 (DnaK) chaperone complexed with ADP and substrate. Proc. Natl. Acad. Sci. U.S.A. 106, 8471–8476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhu X., Zhao X., Burkholder W. F., Gragerov A., Ogata C. M., Gottesman M. E., Hendrickson W. A. (1996) Structural analysis of substrate binding by the molecular chaperone DnaK. Science 272, 1606–1614 [DOI] [PMC free article] [PubMed] [Google Scholar]