Background: An oxidized guanine, 8-oxo-7,8-dihydroguanine, induces base mispairing, thereby altering genetic information.
Results: Human MTH3 degrades 8-oxoguanine-containing nucleoside diphosphates to prevent misincorporation of 8-oxoguanine into DNA and RNA.
Conclusion: MTH3 is closely related to MTH1 and MTH2 but differs in substrate specificity.
Significance: MTH3 may be involved in maintaining the high fidelity of DNA replication as well as transcription under oxidative stress.
Keywords: Hydrolases, Mutagenesis, Nucleoside/Nucleotide Metabolism, Nucleotide, Oxidative Stress, MutT, Nucleic Acid Precursor, Nudix
Abstract
Most of the proteins carrying the 23-residue MutT-related sequence are capable of hydrolyzing compounds with a general structure of nucleoside diphosphate linked to another moiety X and are called the Nudix hydrolases. Among the 22 human Nudix proteins (identified by the sequence signature), some remain uncharacterized as enzymes without a defined substrate. Here, we reveal that the NUDT18 protein, whose substrate was unknown, can degrade 8-oxo-7,8-dihydroguanine (8-oxo-Gua)-containing nucleoside diphosphates to the monophosphates. Because this enzyme is closely related to MTH1 (NUDT1) and MTH2 (NUDT15), we propose that it should be named MTH3. Although these three human proteins resemble each other in their sequences, their substrate specificities differ considerably. MTH1 cleaves 8-oxo-dGTP but not 8-oxo-dGDP, whereas MTH2 can degrade both 8-oxo-dGTP and 8-oxo-dGDP, although the intrinsic enzyme activity of MTH2 is considerably lower than that of MTH1. On the other hand, MTH3 is specifically active against 8-oxo-dGDP and hardly cleaves 8-oxo-dGTP. Other types of oxidized nucleoside diphosphates, 2-hydroxy-dADP and 8-hydroxy-dADP, were also hydrolyzed by MTH3. Another notable feature of the MTH3 enzyme is its action toward the ribonucleotide counterpart. MTH3 can degrade 8-oxo-GDP as efficiently as 8-oxo-dGDP, which is in contrast to the finding that MTH1 and MTH2 show a limited activity against the ribonucleotide counterpart, 8-oxo-GTP. These three enzymes may function together to help maintain the high fidelity of DNA replication and transcription under oxidative stress.
Introduction
Reactive oxygen species, such as superoxide and hydroxyl radicals, are generated in normally growing cells under physiological conditions. Although most of these radicals are eliminated by the actions of antioxidant systems, some remain in the cell and damage its constituents, including proteins, lipids, and nucleic acids (1–3). Among the various types of oxidized purine and pyrimidine bases thus produced, 8-oxo-7,8-dihydroguanine (8-oxo-Gua)2 is the most abundant and seems to play a critical role that affects the maintenance and transfer of genetic information (4–8). Unlike other types of oxidized bases, 8-oxo-Gua does not block nucleic acid synthesis but rather induces base mispairing. When such mispairing occurs during DNA replication, AT-to-CG and GC-to-TA transversions are induced. Likewise, 8-oxo-Gua mispairing that occurs in RNA can cause translational errors, resulting in the formation of abnormal proteins in the cell.
Organisms are equipped with elaborate mechanisms for counteracting such deleterious effects of 8-oxo-Gua. In Escherichia coli, MutT plays a major role in preventing mutations caused by misincorporation of 8-oxo-Gua into DNA. The MutT protein hydrolyzes the 8-oxo-Gua-containing deoxyribonucleoside tri- and diphosphates, 8-oxo-7,8-dihydro-2′-deoxyguanosine triphosphate (8-oxo-dGTP) and 8-oxo-dGDP, to the monophosphate, thereby preventing the incorporation of 8-oxo-Gua-containing nucleotides into DNA (9, 10). In addition to this antimutagenic activity, MutT possesses the ability to degrade the 8-oxo-Gua-containing RNA precursors, 8-oxo-GTP and 8-oxo-GDP, to prevent the formation of 8-oxo-Gua-containing RNA (11, 12). Therefore, MutT is responsible for ensuring accurate DNA replication as well as preventing erroneous protein synthesis under oxidative stress.
In our search for an enzyme with activity similar to that of MutT, we found that certain human tumor-derived cell lines contain a potent activity to degrade 8-oxo-dGTP (13). The enzyme that was identified, which was named MTH1 for MutT homologue 1, was purified to physical homogeneity, and based on its amino acid sequence, its cDNA was cloned (14, 15). Comparison of the sequences of human MTH1 and E. coli MutT revealed that the two proteins are similar in size and carry a conserved 23-amino acid sequence, named the MutT box. Subsequently, this sequence (GX5EX7REUXEEXGU, where U is a bulky hydrophobic residue and X is any residue) was found in many proteins in various species. Because most of these proteins possess catalytic activities to hydrolyze a group of compounds characterized as nucleoside diphosphates linked to another moiety X, it was proposed that these should be called Nudix hydrolases (16–18). In human, 22 Nudix proteins are presently known, the substrates of which range from sugar phosphates to mRNA cap nucleotides. Referring to its substrate specificity as well as its structural feature, MTH1 clearly belongs to the Nudix hydrolase family and was assigned the name NUDT1. We found subsequently that mammalian cells possess another enzyme, named MTH2 (NUDT15), which also hydrolyzes 8-oxo-dGTP (19). Although the intrinsic enzyme activity of MTH2 is considerably lower than that of MTH1, these two enzymes are closely related because their sizes and primary structures resemble each other. In addition to these enzymes, which act primarily on 8-oxo-dGTP, it was found that NUDT5, which was originally characterized as an enzyme that cleaves sugar phosphates (20–22), is capable of degrading 8-oxo-dGDP to the monophosphate form (23–25). However, the physiological role of NUDT5-associated 8-oxo-dGDPase activity may be limited because the optimum pH for the 8-oxo-dGDP cleavage reaction is 10.5, which is in contrast to the finding that the cleavage of ADP-ribose efficiently occurs around pH 7.5 (24).
The error rates of RNA transcription are usually high compared with those of DNA replication because RNA polymerase enzymes lack editing nuclease activities. Under aerobic conditions, where the oxidation of substrate guanine nucleotides occurs, the fidelity of transcription would be expected to be worse. It has been shown that E. coli RNA polymerase can utilize 8-oxo-Gua-containing ribonucleotides as substrates (11), and mammalian RNA polymerase II also possesses this ability (26). The existence of 8-oxo-Gua in mRNA may cause translational errors, eventually resulting in the accumulation of a large amount of abnormal proteins in the cell. The MutT protein of E. coli is capable of degrading 8-oxo-Gua-containing ribonucleoside di- and triphosphates as efficiently as the deoxyribonucleotide counterparts, thus preventing the formation of erroneous proteins in cells growing under an aerobic state (11, 12). In mammalian cells with longer life spans, the elimination of 8-oxo-Gua-containing RNA precursors may be more important, and in this regard, it is worth noting that the MTH1 protein possesses a low but significant level of activity to degrade 8-oxo-GTP (25). Reflecting the fact that most mammalian cells contain larger amounts of RNA compared with DNA and also that RNA is metabolically more active, the pool size of RNA precursors is larger than that of DNA. This raises the question as to whether mammalian cells possess an enzyme acting on 8-oxo-Gua-containing ribonucleotides, besides MTH1.
Among the 22 human Nudix proteins (identified on the basis of the existence of certain sequence motifs), some remain uncharacterized as enzymes (17). Considering the substrate specificities of the other Nudix enzymes, some of these might act on oxidized nucleotides. Because the sequence of NUDT18 is closely related to those of MTH1 and MTH2, we have focused on this protein and explored its enzyme activity. We present the results of these studies.
EXPERIMENTAL PROCEDURES
Purification of MTH3 and Related Proteins
The cDNA encoding human MTH3 was isolated from a SuperScript premade cDNA library constructed from HeLa cells (Invitrogen 11254018) by PCR amplification and inserted into the SphI/HindIII sites of E. coli expression vector pQE80L (Qiagen). The resulting plasmid was named pQE80L(Hs_MTH3) and was used to produce the human MTH3 protein with six repeats of the His tag sequence at its N terminus. E. coli strain BL21 carrying pQE80L(Hs_MTH3) was grown at 37 °C for 3 h in the presence of 1 mm isopropyl β-d-thiogalactopyranoside. The cells (5 g, wet weight) were collected by centrifugation, and the lysate was prepared by sonication in 100 ml of 20 mm sodium phosphate buffer (pH 7.4) containing 500 mm NaCl, 10% glycerol, 20 mm imidazole, and 1 mm DTT. After clarification by low speed centrifugation, 100 ml of crude cell extract was loaded on a HisTrap HP column (1-ml bed volume). The column was washed with 5 ml of buffer containing 20 mm sodium phosphate (pH 7.4), 500 mm NaCl, 10% glycerol, 1 mm DTT, and 20 mm imidazole and then developed by a linear gradient (20–250 mm) of imidazole in 20 ml of the same buffer. Each fraction was monitored by Western blot analysis using a primary antibody against the His tag (Abcam ab5000), and the fractions containing the protein were combined. The sample was dialyzed against 20 mm sodium phosphate buffer (pH 7.4) containing 10% glycerol, 1 mm DTT, and 50 mm NaCl and applied to a HiTrap heparin HP column (1 ml). Proteins were eluted with 10 ml of 400 mm NaCl, and fractions containing MTH3 (NUDT18) were collected. The sample was dialyzed against equilibration buffer (20 mm sodium phosphate (pH 7.4), 10% glycerol, 1 mm DTT, and 25 mm NaCl) and applied to a Mono S 5/50 GL column. After washing the column with 5 volumes of equilibration buffer, proteins were eluted with a linear gradient of NaCl (25–800 mm) in 20 column volumes of equilibration buffer. Finally, the second cycle of HisTrap HP column chromatography was applied.
To produce N-terminally His-tagged human MTH1, cDNA for human MTH1, which was cloned previously (15), was inserted into the NdeI/XbaI sites of E. coli expression vector pCold1 (TaKaRa). The pCold1(Hs_MTH1) plasmid was introduced into E. coli strain Rosetta 2 (Novagen), which carries extra tRNAs for the codons rarely used in E. coli. The cDNA encoding human MTH2 (GenBankTM accession number BC107875.1) was amplified by PCR from the I.M.A.G.E. clone (I.M.A.G.E. ID 6451308; purchased from DNAFORM) and inserted into the NdeI/XbaI sites of pCold1. The plasmid was named pCold1(Hs_MTH2) and introduced into E. coli BL21. Cells carrying each of the plasmids were cultured at 15 °C for 24 h according to the manual for the pCold DNA cold shock expression system (TaKaRa). His6-tagged MTH1 and MTH2 were affinity-purified using TALON metal affinity resin (Clontech) according to the manufacturer's instructions. All of the purified proteins were concentrated using a Microcon YM-3 unit (Amicon) in 10 mm Tris-HCl buffer (pH 8.0) containing 0.1 mm EDTA, 5 mm DTT, and 10% glycerol.
Preparation of Oxidized Nucleotides
Oxidized forms of nucleotides were prepared as described (24) with slight modifications. The reaction was performed in a mixture containing 100 mm sodium phosphate (pH 6.8), 6 mm nucleotide (dGTP, dGDP, dGMP, GTP, GDP, or GMP), 30 mm ascorbic acid, and 100 mm H2O2 at 37 °C for 5 h in the dark. The mixture was then loaded onto an ion-exchange column (Mono Q HR 5/5, 5 × 50 mm, GE Healthcare) equilibrated with 5% triethylammonium hydrogen carbonate and developed with a 5–80% gradient of triethylammonium hydrogen carbonate for 40 min. To obtain purified materials, the sample was monitored with UV absorption for two cycles of the separation procedure. Fractions containing oxidized nucleotides were lyophilized, dissolved in deionized distilled water, and stored at −20 °C. Oxidized dADP derivatives, 2-hydroxy-2′-deoxyadenosine diphosphate (2-OH-dADP) and 8-OH-dADP, were prepared as described by Fujikawa et al. (27).
Enzyme Reactions
The reaction mixture contained an enzyme, one of the substrates, 80 μg/ml BSA, 8 mm MgCl2, 40 mm NaCl, 5 mm DTT, 2% glycerol, and 20 mm Tris-HCl (pH 8.0). The reactions were carried out at 30 °C for various times and terminated by the addition of SDS (final concentration, 1%). After the hydrolytic reaction, nucleotides were separated by HPLC using a TSKgel DEAE-2SW column (4.6 × 250 mm, Tosoh Corp.) in an isocratic solution of 0.12 m sodium phosphate (pH 6.0) containing 40% acetonitrile at a flow rate of 0.7 ml/min. Nucleotides were quantified by measuring the area of UV absorbance using the Empower 2 software program (Waters). The Km and Vmax values for hydrolysis of nucleotides were determined from the intercepts of both axes, x (1/S) and y (1/v), of Lineweaver-Burk plots, respectively.
Phylogenetic Analysis
The primary structures of human Nudix proteins were analyzed using the ClustalW software program, which is equipped with a sequence analysis application, MacVector (MacVector, Inc.).
RESULTS
Purification of Enzymes Acting on Oxidized Guanine-containing Nucleotides
The N-terminally His-tagged forms of human MTH1, MTH2, and MTH3 proteins were produced in E. coli cells harboring plasmids with the respective cDNAs. The proteins were purified to apparent physical homogeneity by a series of affinity and ion-exchange column chromatographic separations. On SDS-polyacrylamide gel, each of the proteins exhibited a single band at positions corresponding to the molecular masses of the fusion proteins (Fig. 1).
FIGURE 1.
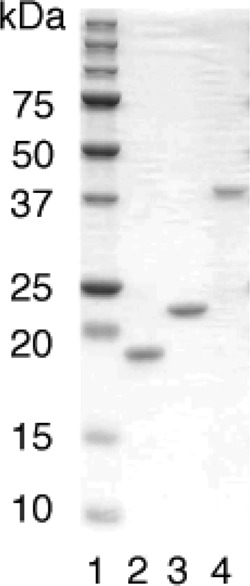
SDS-PAGE of purified preparations of N-terminally His-tagged human enzymes. Purified proteins (800 ng each) were subjected to 15% SDS-PAGE. Lane 1, molecular mass markers; lane 2, MTH1; lane 3, MTH2; lane 4, MTH3.
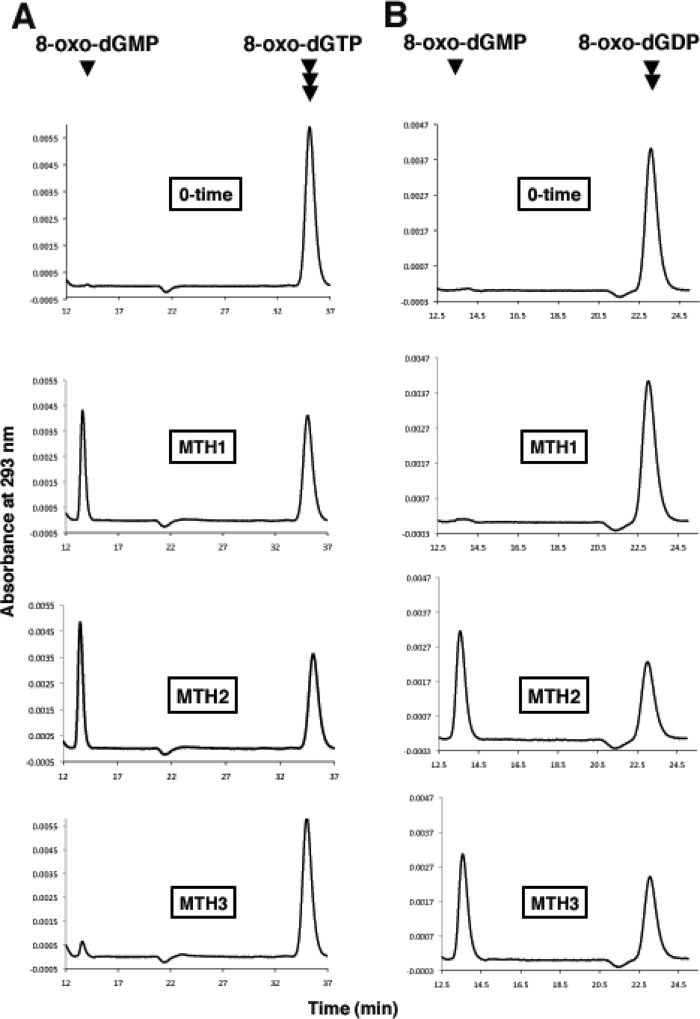
Using these preparations, we examined their enzyme activities against 8-oxo-Gua-containing deoxyribonucleotides. The reactions were carried out at pH 8.0 in the presence of Mg2+, and the products were analyzed by HPLC. Fig. 2 shows the elution profiles of nucleotides before and after the enzyme treatment. As we reported previously (13, 19), 8-oxo-dGTP was degraded to the monophosphate form by treatment with MTH1 as well as with MTH2. Under these conditions, 8-oxo-dGDP was hardly degraded by MTH1, but a considerable degree of cleavage of 8-oxo-dGDP was observed with MTH2 treatment. MTH3 cleaved 8-oxo-dGDP but hardly degraded 8-oxo-dGTP. Therefore, MTH1 and MTH3 exhibit substrate specificities that are complementary to each other.
FIGURE 2.
Actions of MTH1, MTH2, and MTH3 on 8-oxo-dGTP and 8-oxo-dGDP. 8-Oxo-dGTP (A) and 8-oxo-dGDP (B), both at a concentration of 10 μm, were incubated with 3 ng of MTH1, 400 ng of MTH2, or 120 ng of MTH3 in a reaction mixture (40 μl) containing 20 mm Tris-HCl (pH 8.0), 80 μg/ml BSA, 8 mm MgCl2, 40 mm NaCl, 5 mm DTT, and 2% glycerol. The reactions were carried out at 30 °C for 15, 6, and 9 min for MTH1, MTH2, and MTH3, respectively.
Comparison of the Three Types of Enzymes
We first determined the optimum pH for the 8-oxo-dGDP cleavage reaction by MTH3 and the 8-oxo-dGTP cleavage by MTH1 and MTH2. As shown in Fig. 3, all of the enzymes exhibited similar pH dependence curves, with the highest values at pH 8.5. We also measured the pH dependence of the ribonucleotide cleavage reactions with these enzymes, and the results are also shown in Fig. 3. Essentially similar curves were obtained for deoxyribo- and ribonucleotides when each of the maximum values was set at 100.
FIGURE 3.
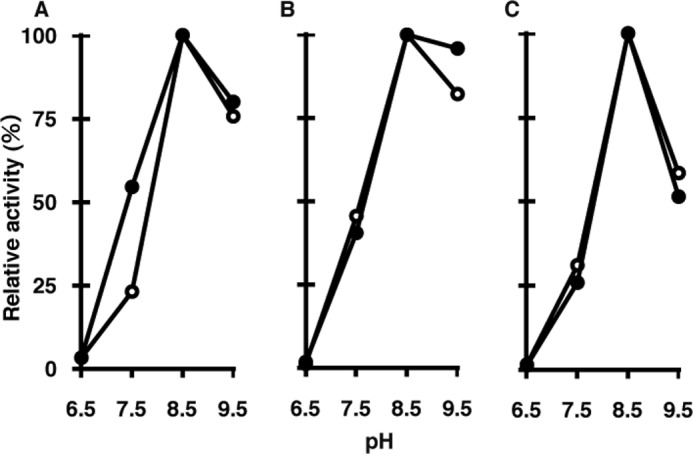
Effects of pH on hydrolysis of 8-oxo-Gua-containing nucleotides by MTH3, MTH1, and MTH2. The reaction mixture (10 μl) contained 20 mm Tris-HCl (pH 6.5–9.5), 80 μg/ml BSA, 8 mm MgCl2, 40 mm NaCl, 5 mm DTT, 2% glycerol, 10 μm nucleotides, and appropriate amounts of enzymes. The reaction was performed at 30 °C and terminated by the addition of SDS (final concentration, 1%). A, hydrolysis of 8-oxo-dGDP and 8-oxo-GDP by MTH3. For both 8-oxo-dGDP (●) and 8-oxo-GDP (○), 10 ng of MTH3 was used for 15- and 21-min reactions, respectively. B, hydrolysis of 8-oxo-dGTP and 8-oxo-GTP by MTH1. For the cleavage of 8-oxo-dGTP (●) and 8-oxo-GTP (○), 0.5 and 1.5 ng of MTH1 were used for 10- and 20-min reactions, respectively. C, hydrolysis of 8-oxo-dGTP and 8-oxo-GTP by MTH2. 8-Oxo-dGTP (●) and 8-oxo-GTP (○) were treated with 20 and 60 ng of MTH2 protein in 10- and 20-min reactions, respectively.
Fig. 4 shows the time course of cleavage of 8-oxo-Gua-containing deoxyribo- and ribonucleotides by the three types of enzymes. It was evident that MTH3 cleaved 8-oxo-dGDP and 8-oxo-GDP with almost the same efficiencies. On the other hand, MTH1 and MTH2 were less active on 8-oxo-Gua-containing ribonucleotides compared with the deoxyribonucleotide counterparts. The rate of cleavage of 8-oxo-GTP by these two enzymes was <5% of that of 8-oxo-dGTP.
FIGURE 4.
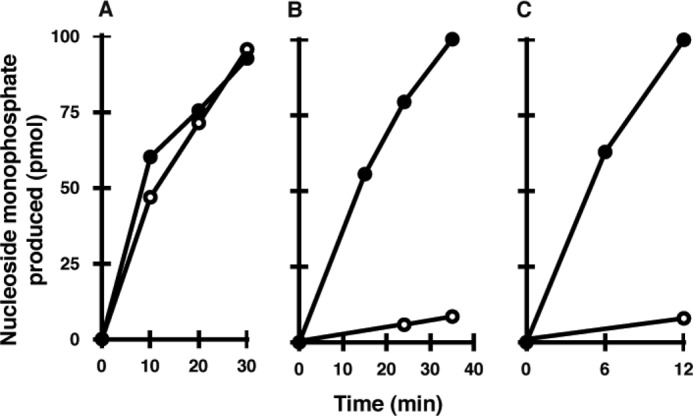
Actions of the three enzymes on 8-oxo-Gua-containing deoxy- and ribonucleotides. The reaction mixture contained 20 mm Tris-HCl (pH 8.0), 80 μg/ml BSA, 8 mm MgCl2, 40 mm NaCl, 5 mm DTT, 2% glycerol, 10 μm substrate nucleotide, and appropriate amounts of enzymes, and the reaction was performed at 30 °C. A, hydrolysis of 8-oxo-dGDP (●) and 8-oxo-GDP (○) by MTH3. B, hydrolysis of 8-oxo-dGTP (●) and 8-oxo-GTP (○) by MTH1. C, hydrolysis of 8-oxo-dGTP (●) and 8-oxo-GTP (○) by MTH2.
Substrate Specificity of MTH3
A purified preparation of MTH3 was applied to eight different kinds of nucleotides, and the time courses of conversion to the monophosphate forms were determined. As shown in Fig. 5, 8-oxo-dGDP and 8-oxo-GDP were rapidly converted to the corresponding monophosphates, whereas no significant cleavage of 8-oxo-dGTP and 8-oxo-GTP was observed. Under these conditions, MTH3 exhibited no or minimal activity toward normal nucleotides.
FIGURE 5.
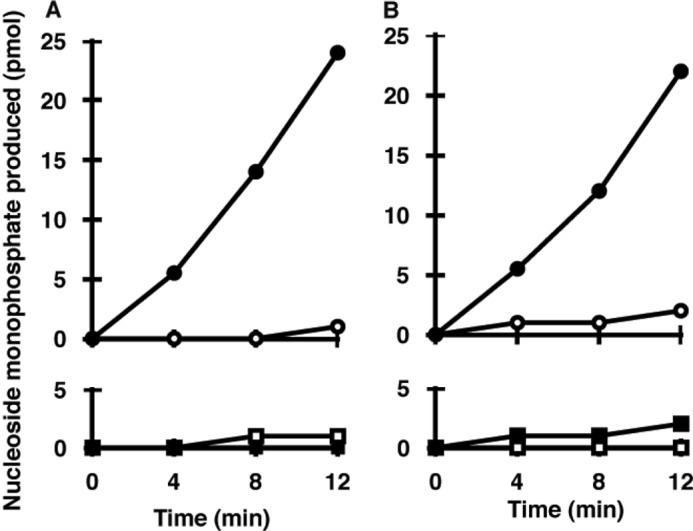
Substrate specificity of MTH3. The reactions were carried out with 10 ng of MTH3 and 10 μm substrate nucleotide in 20 mm Tris-HCl (pH 8.0) containing 80 μg/ml BSA, 8 mm MgCl2, 40 mm NaCl, 5 mm DTT, and 2% glycerol. A, hydrolysis of 8-oxo-dGDP (●) and dGDP (○) (upper panel) and hydrolysis of 8-oxo-dGTP (■) and dGTP (□) (lower panel). B, hydrolysis of 8-oxo-GDP (●) and GDP (○) (upper panel) and hydrolysis of 8-oxo-GTP (■) and GTP (□) (lower panel).
The kinetic parameters of the MTH3 enzyme for the hydrolysis of 8-oxo-dGDP and 8-oxo-GDP were determined, and the results are summarized in Table 1. The apparent Km and Vmax values for the deoxyribo- and ribonucleotides were essentially the same. It seems that MTH3 removes oxidized guanine nucleotides from both DNA and RNA precursor pools.
TABLE 1.
Kinetic parameters of MTH3 on 8-oxo-Gua-containing nucleotides
The reactions were performed at different concentrations of substrates, 2.2–20 μm 8-oxo-dGDP and 8-oxo-GDP, using 10 ng of MTH3 protein in Tris-HCl buffer (pH 8.0) as described under “Experimental Procedures.” The assays were done in triplicate, and the means ± S.D. are shown.
| Substrate | Km | Vmax | Vmax/Km |
|---|---|---|---|
| μm | pmol/min/μg | ||
| 8-Oxo-dGDP | 10.7 ± 3.1 | 212 ± 30 | 19.8 ± 9.7 |
| 8-Oxo-GDP | 11.7 ± 2.9 | 246 ± 49 | 22.4 ± 16.9 |
Actions of MTH3 on Other Types of Oxidized Nucleotides
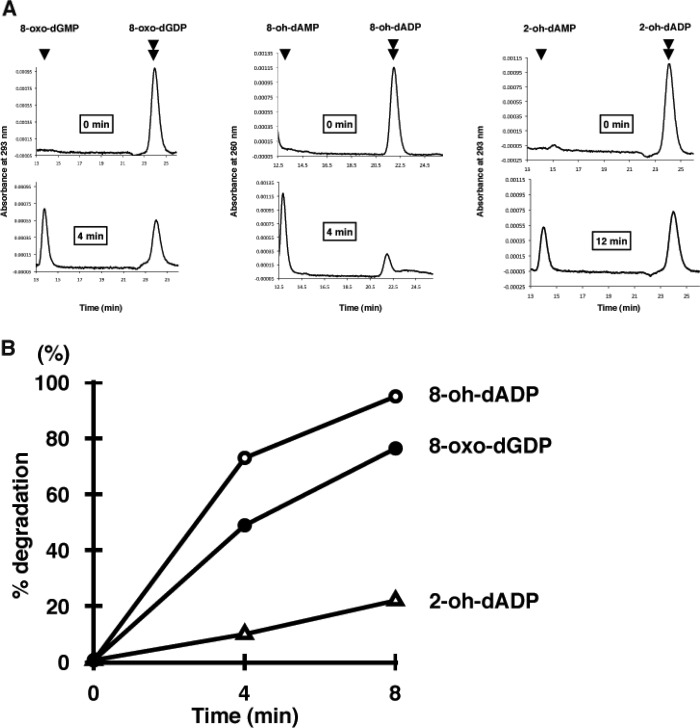
It has been shown that the human MTH1 and NUDT5 proteins possess the ability to hydrolyze other types of oxidized nucleotides besides those containing 8-oxo-Gua (27, 28). To determine whether MTH3 has such an activity, three types of oxidized nucleoside diphosphates were treated with MTH3, and the reaction products were analyzed by HPLC. As shown in Fig. 6A, 8-OH-dADP and 2-OH-dADP were converted to the monophosphates, as was 8-oxo-dGDP. Fig. 6B shows the time courses of the reactions when the three types of oxidized nucleotides were treated with the same amount of MTH3. The cleavage rate of 8-OH-dADP was slightly higher than that of 8-oxo-dGDP, whereas 2-OH-dADP was cleaved more slowly by MTH3. In this respect, MTH3 resembles NUDT5, although the optimum pH levels for the two enzymes differed considerably.
FIGURE 6.
Hydrolysis of oxidized forms of deoxyribonucleoside diphosphates by MTH3. The reaction mixture (10 μl) contained 20 mm Tris-HCl (pH 8.0), 80 μg/ml BSA, 8 mm MgCl2, 40 mm NaCl, 5 mm DTT, 2% glycerol, 30 ng of MTH3 protein, and 10 μm nucleoside diphosphate (8-oxo-dGDP, 2-OH-dGDP, or 8-OH-dADP). The mixture was incubated at 30 °C, and the reaction was terminated by the addition of SDS (final concentration, 1%). A, HPLC elution profiles of the nucleotides before and after treatment for 4 or 12 min. B, time-dependent increases of the degradation products.
DISCUSSION
The MutT protein of E. coli is capable of hydrolyzing a wide range of 8-oxo-Gua-containing nucleotides (8-oxo-dGTP, 8-oxo-GTP, 8-oxo-dGDP, and 8-oxo-GDP) to forms that are unusable for nucleic acid syntheses (12). Because cells lacking this protein exhibit an extremely high frequency of spontaneous mutations, it seems that MutT is solely responsible for eliminating the oxidized guanine nucleotides from the precursor pool (9, 10). On the other hand, in mammalian cells, multiple species of enzymes appear to sanitize the nucleotide pool. Several mammalian counterparts of MutT, including MTH1 (NUDT1), MTH2 (NUDT15), and NUDT5, have been identified, all of which carry the 23-residue MutT-related sequence (Nudix box) (15, 19–21). Despite their structural similarities, these mammalian enzymes exhibit different preferences for substrates. MTH1 and MTH2 preferentially degrade 8-oxo-dGTP, but their potencies are considerably different; MTH1 is ∼40 times more active than MTH2, as calculated from the data shown in Fig. 2. It was further noted that MTH2 possesses a low but significant activity for hydrolysis of 8-oxo-dGDP, whereas MTH1 does not appear to hydrolyze this substrate. In addition to MTH1 and MTH2, NUDT5 was found to cleave 8-oxo-Gua-containing nucleotides. NUDT5 was originally characterized as an enzyme that hydrolyzes ADP-ribose to AMP and ribose 5′-phosphate (20–22). Because high concentrations of free ADP-ribose could result in non-enzymatic ADP-ribosylation of proteins, which is hazardous to cells, the role of NUDT5 was assumed to be its control of the intracellular level of ADP-ribose. Subsequently, it was found that NUDT5 has the ability to degrade 8-oxo-dGDP to the monophosphate form (23, 24). The cleavage of 8-oxo-dGDP was competitively inhibited by ADP-ribose and vice versa, in accord with the idea that the substrates share the same binding site for catalysis. The crystal structures of the NUDT5 complex with 8-oxo-dGDP and ADP-ribose revealed that this is indeed the case (30–32). Nevertheless, their modes of binding and subsequent cleavage reactions are considerably different. Moreover, it was shown that the optimum pH for the cleavage of 8-oxo-dGDP is ∼10, whereas that for ADP-ribose is 7.5 (24). This raised the question as to whether NUDT5 plays a physiological role in the sanitization of the nucleotide pool. In this regard, it is important to note that MTH3 has the ability to cleave 8-oxo-dGDP in the neutral pH range. In this study, we have shown that the pH dependence curve of MTH3 is essentially the same as those of MTH1 and MTH2. Our previous studies on gene-targeted mice revealed that MTH1 is involved in suppression of spontaneous tumorigenesis in certain organs (29).
Another notable feature of MutT-related proteins is their actions on 8-oxo-Gua-containing ribonucleotides. Although E. coli MutT is capable of degrading 8-oxo-GTP and 8-oxo-GDP as efficiently as their deoxyribonucleotide counterparts (12), the actions of human enzymes toward oxidized deoxyribo- and ribonucleotides are somewhat different. As shown in this study, MTH1 and MTH2 degrade 8-oxo-GTP at rates <5% of those for 8-oxo-dGTP. On the other hand, MTH3 cleaves 8-oxo-GDP and 8-oxo-dGDP with almost the same efficiency. In this regard, it is important to note that ribonucleotide reductase, which catalyzes the conversion of ribonucleotides to deoxyribonucleotides, is totally inactive for 8-oxoguanine-containing ribonucleotides (26). It is therefore expected that 8-oxo-GDP and 8-oxo-dGDP are generated independently in the RNA and DNA precursor pools. This may be related to the fact that MTH3 cleaves 8-oxo-GDP and 8-oxo-dGDP with almost the same high efficiency.
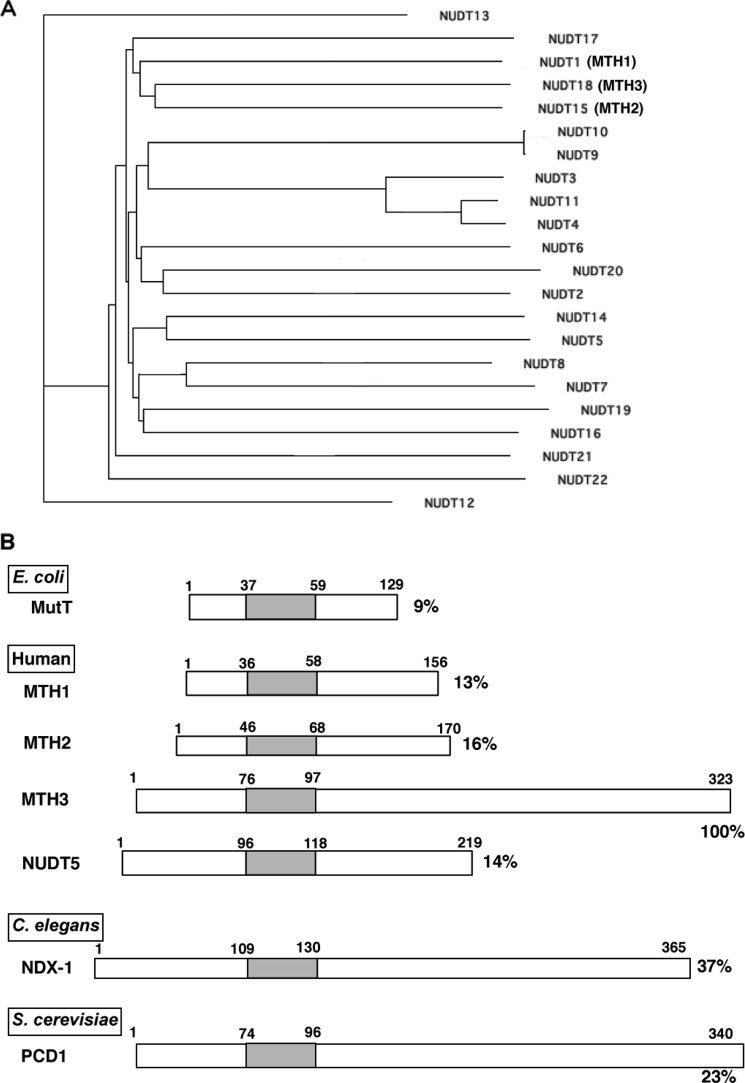
The sequence motif called the MutT or Nudix box is present in many proteins in >250 species, including viruses, bacteria, archaea, and eukaryotes (16–18). Based on human genome sequence analyses, 22 proteins with this motif have been identified and are collectively called Nudix proteins because most of these proteins possess Mg2+-requiring enzyme activities catalyzing the hydrolysis of nucleoside diphosphates linked to another moiety X. The phylogenic tree of these proteins is shown in Fig. 7A. Although the substrate specificities of most of these proteins have been defined, some of them, including NUDT17, NUDT18, NUDT21, and NUDT22, were uncharacterized. In this study, we revealed that NUDT18 has a unique activity, wherein it degrades 8-oxo-Gua-containing deoxyribo- and ribonucleoside diphosphates to the corresponding monophosphates. Because this enzyme activity is closely related to those of MTH1 and MTH2, we named it MTH3. Inspection of the phylogenic tree reveals that these three proteins are closely related. NUDT5, which exhibits a potent activity against ADP-sugars and also is capable of cleaving 8-oxo-dGDP under alkaline conditions (21, 24), is also present in this region, although it is somewhat remote from the MTH3-related proteins.
FIGURE 7.
Sequence characteristics of MTH3-related proteins. A, phylogenic tree of human Nudix family proteins. Structural alignment and the construction of a phylogenic tree were performed using the ClustalW program. NUDT1, NUDT15, and NUDT18 (characterized by their structural features) correspond to MTH1, MTH2, and MTH3, respectively, the names of which were assigned on the basis of their similar biochemical activities. B, comparison of the structures of the MutT family proteins. The numbers correspond to the positions of the amino acid residues from the N termini. The positions of the conserved MutT box are shown by shaded boxes. The relative numbers of amino acid residues identical to those of MTH3 are shown to the right of each sequence.
Fig. 7B shows the sequence alignments of MutT-related proteins, in which the conserved MutT sequences were placed in the same positions. In the overall structures, MTH3 is about twice as large as the other human proteins (MTH1, MTH2, and NUDT5), which are slightly larger than E. coli MutT. MTH3 is similar in size to Pcd1 (ORFYLR151c) of Saccharomyces cerevisiae and NDX-4 of Caenorhabditis elegans, which have capacities to cleave 8-oxo-dGTP and 8-oxo-dGDP, respectively (30, 31). When the amino acid sequence similarity was determined based on the whole sequence of MTH3, the highest values were obtained with the NDX-1 and Pcd1 proteins (37 and 23%, respectively). Because these three proteins possess long C-terminal regions, this might contribute to the relatively high scores for the sequence similarity of these proteins. It is noteworthy that MTH3 and NDX-1 exhibit the same substrate specificity, mainly acting on 8-oxo-Gua-containing nucleoside diphosphates, whereas Pcd1 acts primarily on the nucleoside triphosphates.
We also examined the proteins for a possible relationship between their primary structures and their ranges of substrate specificity. The three human proteins MTH1, MTH3, and NUDT5, as well as the S. cerevisiae protein Pcd1, can cleave various types of oxidized purine nucleotides, whereas the E. coli MutT and C. elegans NDX-1 proteins act strictly on 8-oxo-Gua-containing nucleotides (27, 28, 30), yet specific sequence motifs for distinguishing these two groups of proteins have not been identified. The substrate recognition and binding may be achieved by a few amino acid residues located near or within the active center, and thus, elucidation of the three-dimensional structures is essential for solving this problem. Analyses of the crystal structures of MutT, MTH1, and NUDT5, all of which are complexed with 8-oxo-Gua-containing nucleotides, have been performed (32–37). In the cases of MutT and MTH1, the reaction product 8-oxo-dGMP binds to the same substrate-binding pocket composed of two β-sheets and one α-helix. Nevertheless, these two proteins exhibit significantly different 8-oxo-dGMP-binding modes: MutT binds to 8-oxo-dGMP with large ligand-induced conformational changes, which would contribute to its high substrate specificity for 8-oxo-Gua-containing nucleotides, whereas the structure of MTH1 does not change after ligand binding. Furthermore, the 8-oxo-Gua base-binding mode and the glycosidic conformation are quite different in MutT and MTH1, although the sugar and phosphate positions of the two 8-oxo-dGMPs are similar. In contrast to the finding that MutT and MTH1 exist as monomers (32, 34, 36), NUDT5 forms a homodimer with substantial domain swapping (33, 35). In NUDT5, the nucleoside moiety of 8-oxo-dGDP binds to a completely different region near the dimer interface without ligand-induced conformational changes. The only common feature among these proteins is the α-phosphate positions in the Nudix motif. Elucidation of the structures of MTH3 in complex with 8-oxo-Gua-containing nucleotides is necessary to determine whether it also shares this feature.
Acknowledgments
We thank Hiroshi Hayakawa and Masumi Hidaka for critical reading of the manuscript and Aya Fujikane for technical assistance.
This work was supported by grants from the Ministry of Education, Culture, Sports, Science and Technology of Japan, including the MEXT-supported Program for the Strategic Research Foundation at Private Universities (2008–2012).
- 8-oxo-Gua
- 8-oxo-7,8-dihydroguanine.
REFERENCES
- 1. Bjelland S., Seeberg E. (2003) Mutagenicity, toxicity, and repair of DNA base damage induced by oxidation. Mutat. Res. 531, 37–80 [DOI] [PubMed] [Google Scholar]
- 2. Evans M. D., Cooke M. S. (2004) Factors contributing to the outcome of oxidative damage to nucleic acids. BioEssays 26, 533–542 [DOI] [PubMed] [Google Scholar]
- 3. Imlay J. A. (2003) Pathways of oxidative damage. Annu. Rev. Microbiol. 57, 395–418 [DOI] [PubMed] [Google Scholar]
- 4. Grollman A. P., Moriya M. (1993) Mutagenesis by 8-oxoguanine: an enemy within. Trends Genet. 9, 246–249 [DOI] [PubMed] [Google Scholar]
- 5. Kuchino Y., Mori F., Kasai H., Inoue H., Iwai S., Miura K., Ohtsuka E., Nishimura S. (1987) Misreading of DNA templates containing 8-hydroxydeoxyguanosine at the modified base and at adjacent residues. Nature 327, 77–79 [DOI] [PubMed] [Google Scholar]
- 6. Sekiguchi M. (1996) MutT-related error avoidance mechanism for DNA synthesis. Genes Cells 1, 139–145 [DOI] [PubMed] [Google Scholar]
- 7. Sekiguchi M., Tsuzuki T. (2002) Oxidative nucleotide damage: consequences and prevention. Oncogene 21, 8895–8904 [DOI] [PubMed] [Google Scholar]
- 8. Tsuzuki T., Nakatsu Y., Nakabeppu Y. (2007) Significance of error-avoiding mechanisms for oxidative DNA damage in carcinogenesis. Cancer Sci. 98, 465–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Maki H., Sekiguchi M. (1992) MutT protein specifically hydrolyzes a potent mutagenic substrate for DNA synthesis. Nature 355, 273–275 [DOI] [PubMed] [Google Scholar]
- 10. Tajiri T., Maki H., Sekiguchi M. (1995) Functional cooperation of MutT, MutM, and MutY proteins in preventing mutations caused by spontaneous oxidation of guanine nucleotide in Escherichia coli. Mutat. Res. 336, 257–267 [DOI] [PubMed] [Google Scholar]
- 11. Taddei F., Hayakawa H., Bouton M., Cirinesi A., Matic I., Sekiguchi M., Radman M. (1997) Counteraction by MutT protein of transcriptional errors caused by oxidative damage. Science 278, 128–130 [DOI] [PubMed] [Google Scholar]
- 12. Ito R., Hayakawa H., Sekiguchi M., Ishibashi T. (2005) Multiple enzyme activities of Escherichia coli MutT protein for sanitization of DNA and RNA precursor pools. Biochemistry 44, 6670–6674 [DOI] [PubMed] [Google Scholar]
- 13. Mo J. Y., Maki H., Sekiguchi M. (1992) Hydrolytic elimination of a mutagenic nucleotide, 8-oxo-dGTP, by human 18-kilodalton protein: sanitization of nucleotide pool. Proc. Natl. Acad. Sci. U.S.A. 89, 11021–11025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Furuichi M., Yoshida M. C., Oda H., Tajiri T., Nakabeppu Y., Tsuzuki T., Sekiguchi M. (1994) Genomic structure and chromosome location of the human mutT homologue gene MTH1 encoding 8-oxo-dGTPase for prevention of A:T to C:G transversion. Genomics 24, 485–490 [DOI] [PubMed] [Google Scholar]
- 15. Sakumi K., Furuichi M., Tsuzuki T., Kakuma T., Kawabata S., Maki H., Sekiguchi M. (1993) Cloning and expression of cDNA for a human enzyme that hydrolyzes 8-oxo-dGTP, a mutagenic substrate for DNA synthesis. J. Biol. Chem. 268, 23524–23530 [PubMed] [Google Scholar]
- 16. Bessman M. J., Frick D. N., O'Handley S. F. (1996) The MutT proteins or “Nudix” hydrolases, a family of versatile, widely distributed, “housecleaning” enzymes. J. Biol. Chem. 271, 25059–25062 [DOI] [PubMed] [Google Scholar]
- 17. McLennan A. G. (2006) The Nudix hydrolase superfamily. Cell. Mol. Life Sci. 63, 123–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mildvan A. S., Xia Z., Azurmendi H. F., Saraswat V., Legler P. M., Massiah M. A., Gabelli S. B., Bianchet M. A., Kang L. W., Amzel L. M. (2005) Structures and mechanisms of Nudix hydrolases. Arch. Biochem. Biophys. 433, 129–143 [DOI] [PubMed] [Google Scholar]
- 19. Cai J. P., Ishibashi T., Takagi Y., Hayakawa H., Sekiguchi M. (2003) Mouse MTH2 protein, which prevents mutations caused by 8-oxoguanine nucleotides. Biochem. Biophys. Res. Commun. 305, 1073–1077 [DOI] [PubMed] [Google Scholar]
- 20. Gasmi L., Cartwright J. L., McLennan A. G. (1999) Cloning, expression, and characterization of YSA1H, a human adenosine 5′-diphosphosugar pyrophosphatase possessing a MutT motif. Biochem. J. 344, 331–337 [PMC free article] [PubMed] [Google Scholar]
- 21. Yang H., Slupska M. M., Wei Y. F., Tai J. H., Luther W. M., Xia Y. R., Shih D. M., Chiang J. H., Baikalov C., Fitz-Gibbon S., Phan I. T., Conrad A., Miller J. H. (2000) Cloning and characterization of a new member of the Nudix hydrolases from human and mouse. J. Biol. Chem. 275, 8844–8853 [DOI] [PubMed] [Google Scholar]
- 22. Rafty L. A., Schmidt M. T., Perraud A. L., Scharenberg A. M., Denu J. M. (2002) Analysis of O-acetyl-ADP-ribose as a target for Nudix ADP-ribose hydrolases. J. Biol. Chem. 277, 47114–47122 [DOI] [PubMed] [Google Scholar]
- 23. Ishibashi T., Hayakawa H., Sekiguchi M. (2003) A novel mechanism for preventing mutations caused by oxidation of guanine nucleotides. EMBO Rep. 4, 479–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ito R., Sekiguchi M., Setoyama D., Nakatsu Y., Yamagata Y., Hayakawa H. (2011) Cleavage of oxidized guanine nucleotide and ADP sugar by human NUDT5 protein. J. Biochem. 149, 731–738 [DOI] [PubMed] [Google Scholar]
- 25. Ishibashi T., Hayakawa H., Ito R., Miyazawa M., Yamagata Y., Sekiguchi M. (2005) Mammalian enzymes for preventing transcriptional errors caused by oxidative damage. Nucleic Acids Res. 33, 3779–3784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hayakawa H., Hofer A., Thelander L., Kitajima S., Cai Y., Oshiro S., Yakushiji H., Nakabeppu Y., Kuwano M., Sekiguchi M. (1999) Metabolic fate of oxidized guanine ribonucleotides in mammalian cells. Biochemistry 38, 3610–3614 [DOI] [PubMed] [Google Scholar]
- 27. Fujikawa K., Kamiya H., Yakushiji H., Fujii Y., Nakabeppu Y., Kasai H. (1999) The oxidized forms of dATP are substrates for the human MutT homologue, the hMTH1 protein. J. Biol. Chem. 274, 18201–18205 [DOI] [PubMed] [Google Scholar]
- 28. Kamiya H., Hori M., Arimori T., Sekiguchi M., Yamagata Y., Harashima H. (2009) NUDT5 hydrolyzes oxidized deoxyribonucleoside diphosphates with broad substrate specificity. DNA Repair 8, 1250–1254 [DOI] [PubMed] [Google Scholar]
- 29. Tsuzuki T., Egashira A., Igarashi H., Iwakuma T., Nakatsuru Y., Tominaga Y., Kawate H., Nakao K., Nakamura K., Ide F., Kura S., Nakabeppu Y., Katsuki M., Ishikawa T., Sekiguchi M. (2001) Spontaneous tumorigenesis in mice defective in the Mth1 gene encoding 8-oxo-dGTPase. Proc. Natl. Acad. Sci. U.S.A. 98, 11456–11461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sanada U., Yonekura S., Kikuchi M., Hashiguchi K., Nakamura N., Yonei S., Zhang-Akiyama Q. M. (2011) NDX-1 protein hydrolyzes 8-oxo-7,8-dihydrodeoxyguanosine 5′-diphosphate to sanitize oxidized nucleotides and prevent oxidative stress in Caenorhabditis elegans. J. Biochem. 150, 649–657 [DOI] [PubMed] [Google Scholar]
- 31. Nunoshiba T., Ishida R., Sasaki M., Iwai S., Nakabeppu Y., Yamamoto K. (2004) A novel Nudix hydrolase for oxidized purine nucleoside triphosphates encoded by ORFYLR151c (PCD1 gene) in Saccharomyces cerevisiae. Nucleic Acids Res. 32, 5339–5348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nakamura T., Kitaguchi Y., Miyazawa M., Kamiya H., Toma S., Ikemizu S., Shirakawa M., Nakabeppu Y., Yamagata Y. (2006) Crystallization and preliminary x-ray analysis of human MTH1 complexed with two oxidized nucleotides, 8-oxo-dGMP and 2-oxo-dATP. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 62, 1283–1285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zha M., Guo Q., Zhang Y., Yu B., Ou Y., Zhong C., Ding J. (2008) Molecular mechanism of ADP-ribose hydrolysis by human NUDT5 from structural and kinetic studies. J. Mol. Biol. 379, 568–578 [DOI] [PubMed] [Google Scholar]
- 34. Nakamura T., Meshitsuka S., Kitagawa S., Abe N., Yamada J., Ishino T., Nakano H., Tsuzuki T., Doi T., Kobayashi Y., Fujii S., Sekiguchi M., Yamagata Y. (2010) Structural and dynamic features of the MutT protein in the recognition of nucleotides with the mutagenic 8-oxoguanine base. J. Biol. Chem. 285, 444–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zha M., Zhong C., Peng Y., Hu H., Ding J. (2006) Crystal structures of human NUDT5 reveal insights into the structural basis of the substrate specificity. J. Mol. Biol. 364, 1021–1033 [DOI] [PubMed] [Google Scholar]
- 36. Svensson L.M., Jemth A.S., Desroses M., Loseva O., Helleday T., Högbom M., Stenmark P. (2011) Crystal structure of human MTH1 and the 8-oxo-dGMP product complex. FEBS Lett. 585, 2617–2621 [DOI] [PubMed] [Google Scholar]
- 37. Arimori T., Tamaoki H., Nakamura T., Kamiya H., Ikemizu S., Takagi Y., Ishibashi T., Harashima H., Sekiguchi M., Yamagata Y. (2011) Diverse substrate recognition and hydrolysis mechanisms of human NUDT5. Nucleic Acids Res. 39, 8972–8983 [DOI] [PMC free article] [PubMed] [Google Scholar]