Abstract
African trypanosomes have plastic genomes with a great degree of variability at the chromosome ends. These telomeres are where the genes encoding the expressed major surface protein of the infective bloodstream form stages of Trypanosoma brucei are located. These telomeric expression site transcription units are turning out to be surprisingly polymorphic in structure and sequence.
Keywords: Antigenic variation, telomere, Trypanosoma brucei, gene conversion, genome plasticity
Introduction
The African trypanosome Trypanosoma brucei relies on population diversity in order to maintain the chronic infections characteristic of African Sleeping Sickness. The extracellular bloodstream form trypanosome, covered by a single Variant Surface Glycoprotein (VSG), is an easy target for the immune system. Once antibodies against a given VSG are raised, all trypanosomes wearing this VSG coat are effectively eradicated. However, trypanosomes continuously arise wearing new (temporarily) unrecognisable VSG coats, which allow a chronic infection to be maintained. Each VSG coat is encoded by a single gene, transcribed in a telomeric VSG expression site. Changing the active VSG gene can involve DNA rearrangements slotting a new VSG gene into an active expression site. Alternatively, as there are twenty bloodstream form VSG expression sites, VSGs can be changed by switching between expression sites. Antigenic variation in the bloodstream form trypanosome is reviewed in: [1-4]. The infective metacyclic trypanosome injected by the tsetse fly into the mammalian host also wears a VSG coat, but the genes encoding metacyclic VSGs are expressed in telomeric metacyclic VSG expression sites. These are structurally quite distinct from bloodstream form expression sites, and contain only a VSG gene (reviewed in [5-6]).
Research in recent years into T. brucei genome structure has shown that the genome is extraordinarily plastic, particularly at the chromosome ends. Both within a trypanosome and between trypanosome strains, chromosomal alleles can differ from each other drastically in size. This is caused to a great extent by a large degree of variability at the chromosome ends. In addition, within a given trypanosome there is a surprising degree of variability within the telomeric VSG expression sites. Both the bloodstream and metacyclic form VSG expression sites are extremely polymorphic in structure. The challenge comes in trying to understand why this is the case.
The plastic T. brucei genome
The karyotype of T. brucei varies enormously between strains, and identical karyotypes are seldom found in the field. The T. brucei genome is divided over a large number of chromosomes: eleven pairs of megabase chromosomes (1-6 Mb), a number of intermediate chromosomes (200-900 kb) and approximately one hundred minichromosomes (50-150 kb) [7]. The minichromosomes appear to consist primarily of repeats and a telomeric VSG gene [8], and the intermediate chromosomes have expression sites, but do not appear to have unique genes [7]. The megabase chromosomes contain the housekeeping genes. Although the gene synteny within the core of these larger chromosomes is generally conserved between stocks, the size of sister alleles can vary enormously both within parasites and between strains [7].
This variability in size appears to arise to a large extent from expansions and contractions within the repetitive regions of the subtelomeric and telomeric regions of chromosomes [9]. First of all, although the megabase chromosomes are present as diploid pairs, the telomeres of sister alleles are not necessarily similar with regards to the presence of VSG expression sites. As expression sites appear to be glued on to chromosomal cores in a variable fashion, the chromosome ends are therefore segmentally aneuploid [7]. It has been shown that expression sites can be easily lost during switching [10], presumably after breakages in the subtelomeric region. Upstream of the telomeric expression sites, the subtelomeric regions can contain large stretches of repetitive sequence that extend for hundreds of kilobases [9]. It is possible that this provides a barren region protecting the housekeeping genes in the chromosomal core from the damaging effects of DNA rearrangements and deletions occuring at the telomeric VSG expression sites during switching.
Polymorphic bloodstream form expression sites
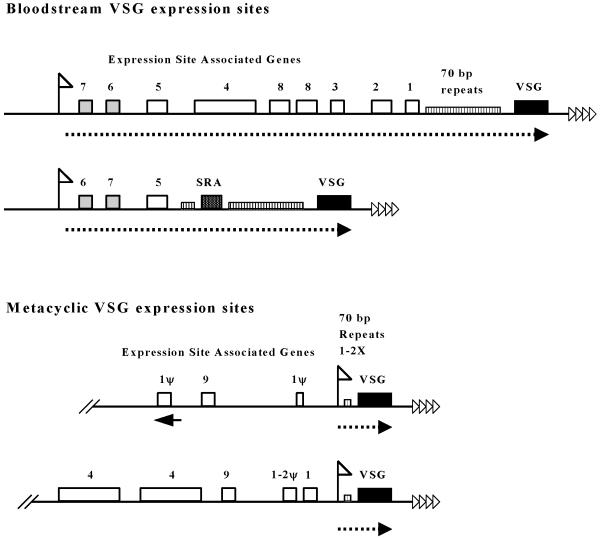
The telomeric bloodstream form expression sites are typically large (40-60 kb) transcription units, containing various Expression Site Associated Genes (ESAG)s in addition to the telomeric VSG gene (Figure 1). The function of most of these ESAGs is unknown. ESAG 6 and 7 encode the subunits of a transferrin receptor [11], and ESAG 4 an adenylate cyclase [12]. Although ESAG 6 and 7 appear to be present in all expression sites and don’t appear to have copies elsewhere, some ESAGs (like ESAG 1) have a large number of related copies elsewhere in the genome and can be disrupted in the VSG expression site without generating a phenotype [13]. Currently, eleven ESAGs have been described, but there are undoubtedly more.
Figure 1.
The two T. brucei bloodstream form VSG expression sites are drawn according to Xong et al., 1998, and the schematic of the two metacyclic VSG expression sites is redrawn approximately according to Barry et al, 1998. The expression site promoters are indicated with white flags, and transcription with a dotted line. Expression site associated genes (ESAGs) are indicated with boxes: ESAGs 6 and 7 (encoding the transferrin receptor subunits) are grey, The SRA (human serum resistance) gene is indicated with dark stipples, and the other ESAGs are white. The ψ symbol indicates a pseudogene, and the black arrowhead under the ESAG 1 pseudogene indicates that it is present in an inverted orientation. The telomeric VSG gene is indicated with a black box. Characteristic 70 bp repeat arrays are shown as vertically striped boxes. In the metacyclic VSG expression sites the 70 bp repeats are present as either one or two copies (1X or 2X). The telomere repeats are indicated with white triangles.
The architecture of bloodstream form VSG expression sites is turning out to be surprisingly variable. The full complement of eight ESAGs as found in the AnTat 1.3A expression site [14] does not appear to be essential for life in the bloodstream form. Bloodstream form trypanosomes transcribing a truncated VSG expression site containing only ESAGs 7, 6 and 5 and an ESAG conferring resistance to human serum (SRA), have recently been described [15]. This indicates that life in the bloodstream is possible without the transcription of ESAGs 1, 2, 3, 4 and 8 from the active VSG expression site. Other expression sites, rather than being truncated, have more ESAG genes than usual. The 221 VSG expression site has undergone massive gene amplifications, resulting in duplications of ESAG 4 and triplications of ESAGs 2 and 8 [16, and unpublished results].
Why is there this degree of variability in VSG expression site architecture? Possibly the variation found between different expression sites allows the trypanosome to switch between the expression of different sets of genes allowing adaptation to life in different hosts [17]. T. brucei infects a large range of mammals in Africa. Alternating between different expression sites, allows the trypanosome to vary between the expression of different polymorphic transferrin receptor subunits encoded in the different expression sites. This could allow the trypanosome to optimise uptake of the variable transferrin molecules present in the serum of different hosts [17]. It is possible that bloodstream form expression sites contain different permutations and combinations of a variable number of ESAG genes in order to vary the dosage of a range of ESAG products necessary for life in different hosts. Presumably specialised ESAGs (like the SRA gene), are only present in a subset of expression sites. It is clear that we need the sequence of a broader range of T. brucei expression sites in order to determine which ESAGs are ubiquitous, and the extent of ESAG sequence variability. This will probably necessitate a concerted effort to clone telomeric T. brucei sequences, as these tend to be underrepresented in standard genomic libraries.
The telomeric location of bloodstream form VSG expression sites provides a recombinogenic location allowing the telomeric VSG genes to be easily exchanged. In addition, frequent DNA rearrangements at the telomeres presumably allow genetic diversity to be easily generated in both the VSG genes necessary for antigenic variation, and the ESAG genes which appear to play a role in the phenotypic variation necessary for host range adaptation [18].
In addition, it is highly likely that the telomeric location of bloodsteam form VSG expression sites plays a critical role in their control. It has been shown that bloodstream form expression sites are turned on and off in a promoter sequence nonspecific manner [19, 20]. Bloodstream form VSG expression sites are silenced as domains, as the endogenous promoter far upstream from the chromosome end, can not be uncoupled from promoters inserted at the telomere [20]. However, the extent to which expression site downregulation operates at the level of transcription initiation is still unclear. There is a low amount of transcription around the promoter and the ESAG 6 and 7 genes of “silent” bloodstream form VSG expression sites [21 and 22]. However, the transcripts generated from these “inactive” VSG expression sites appear to be primarily nonpolyadenylated [22]. This could indicate that VSG expression site activation involves not only enhanced transcription, but also more efficient RNA processing of the transcripts generated from the activated site. How this could be mediated is unclear.
Polymorphic metacyclic expression sites
In contrast to the bloodstream form, the metacyclic trypanosome is the infective stage that establishes a foothold in a mammal after transmission by the bite of a tsetse fly. Of critical importance for this life-cycle stage, is the innoculation of the host with a heterogeneous population of trypanosomes. Metacyclic VSG coats are encoded by genes in metacyclic VSG expression sites, transcribed as monocistronic transcription units containing just the VSG gene (Figure 1) [23]. Both bloodstream and metacyclic VSG expression sites are always telomeric, and are transcribed in a mutually exclusive fashion ensuring that only one VSG expression site is maximally active in a single cell [24, 25].
It is striking that upstream of metacyclic VSG expression sites, one finds ESAG genes and pseudogenes characteristic of bloodstream form expression sites (ESAGs 1, 2, 4 and 9) [5, 6, 26]. These ESAGs are not part of the metacyclic transcription unit, and could indicate that metacyclic expression sites have evolved from bloodstream form ones [5]. Alternatively, recombination between telomere ends could have resulted in metacyclic VSG expression sites being glued onto the ends of bloodstream form VSG expression sites, which later became disfunctional.
Metacyclic expression sites have turned out to be surprisingly polymorphic with regards to the sequence of their promoter regions [26, 27]. It is unclear why this is the case. As metacyclic expression site promoters are so close (within ten kilobases) of the chromosome end, the critical factor controlling their expression could be a position effect operating at the telomeres. Control at the level of position effect could allow more leeway in the sequences that operate as functional metacyclic promoters.
Similar to the bloodstream form expression sites, the telomeric location of metacyclic VSG expression sites presumably plays a critical role in expression site control, though this has not been formally shown. In addition, the telomeric location of metacyclic VSG genes is presumably critical for maintenance of diversity in the metacyclic VSG gene repertoire. The metacyclic VSG gene repertoire of T. brucei strains has been shown to change through time [28], and this instability is presumably due to occasional VSG gene replacements by gene conversion.
Counting at telomeres
How does the trypanosome manage to activate only one of the estimated forty to fifty telomeres containing either bloodstream or metacyclic VSG expression sites? There appears to be a severe restriction on double expression of bloodstream form VSG expression sites. Using trypanosomes with drug resistance genes inserted into two different bloodstream form expression sites, it was shown that even under double drug selection, stable maximal transcription of two expression sites can not be achieved [25]. Instead, the double-resistant trypanosomes generated behaved as if they were rapidly switching between the two tagged sites. This would imply that double expressors are not stable intermediates in expression site switching, and that expression sites are not independently switched on and off. Additional evidence for this coupled mechanism of VSG expression site (in)activation comes from experiments where trypanosomes with expression sites genetically modified to be driven by (an inadequate) T7 promoter rapidly switched to transcription of another expression site [29]. Similarly, disruption of the active VSG expression site telomere resulted in an increase in the switching frequency [30]. Inactivation of an expression site could be the first obligatory step in switching [10, 31], allowing a “pre-active” expression site to take over [25]. The rapidity of the switch would indicate that silent bloodstream VSG expression sites are potentially active rather tightly repressed. This “pre-active” state is compatible with the low level of transcription from “silent” expression site promoters [21, 22].
It remains to be determined whether the subnuclear location of the active VSG expression site plays a critical role in its activation. It has been shown that T. brucei telomeres are present as clusters in the cell [32]. However, no evidence has yet been found for a distinct “activating region” in the cell nucleus. Fluorescent in situ hybridisation (FISH) experiments have shown that the active bloodstream form expression site is not colocalised with the nucleolus [33]. FISH experiments detecting the two marked VSG expression sites of trypanosomes selected for double expression showed two dots [25]. These dots, although located closer together than the sister alleles of housekeeping genes, were not obviously colocalised. However, as these trypanosomes might have been switching back and forth between two expression sites, these experiments don’t exclude the presence of a discrete subnuclear “activating region”.
Parasite telomeres
Transcription units with genes involved in antigenic (and phenotypic) variation are frequently at telomeres. This includes the vmp genes of Borrelia sp. [34], most of the VAR gene family of Plasmodium [reviewed in 35] and the Major Surface Glycoprotein (MSG) genes of Pneumocystis carinii [36]. DNA recombination at these telomeres can be involved in the switch of the active antigen gene [37, 38], and the generation of antigen gene repertoire diversity [39, 40]. In trypanosomes, DNA rearrangements at the telomeres are also involved in switching, and in the generation of antigen gene diversity [41]. The challenge will be in determining whether this telomeric location plays a role in expression site activation.
Acknowledgements
I would like to thank Piet Borst, Tomoko Isobe, Ramit Mehr and Daniëlle te Vruchte for comments on the manuscript. G.R. is a Wellcome Senior Fellow in the Basic Biomedical Sciences funded by the Wellcome Trust.
Abbreviations
- VSG
Variant Surface Glycoprotein
- ESAG
Expression site associated gene
- SRA
Serum Resistance Associated
- FISH
Fluorescent in situ hybridisation
References
- 1.Borst P, Bitter W, Blundell PA, Chaves I, Cross M, Gerrits H, van Leeuwen F, McCulloch R, Taylor M, Rudenko G. Control of VSG gene expression sites in Trypanosoma brucei. Mol. Biochem. Parasitol. 1998;91:67–76. doi: 10.1016/s0166-6851(97)00184-9. [DOI] [PubMed] [Google Scholar]
- 2.Cross GAM, Wirtz LE, Navarro M. Regulation of vsg expression site transcription and switching in Trypanosoma brucei. Mol. Biochem. Parasitol. 1998;91:77–91. doi: 10.1016/s0166-6851(97)00186-2. [DOI] [PubMed] [Google Scholar]
- 3.Pays E, Nolan DP. Expression and function of surface proteins in Trypanosoma brucei. Mol. Biochem. Parasitol. 1998;91:3–36. doi: 10.1016/s0166-6851(97)00183-7. [DOI] [PubMed] [Google Scholar]
- 4.Rudenko G, Cross M, Borst P. Changing the end: antigenic variation orchestrated at the telomeres of African trypanosomes. Trends Micro. 1998;6:113–117. doi: 10.1016/s0966-842x(97)01200-6. [DOI] [PubMed] [Google Scholar]
- 5.Barry JD, Graham SV, Fotheringham M, Graham VS, Kobryn K, Wymer B. VSG gene control and infectivity strategy of metacyclic stage Trypanosoma brucei. Mol. Biochem. Parasitol. 1998;91:93–105. doi: 10.1016/s0166-6851(97)00193-x. [DOI] [PubMed] [Google Scholar]
- 6.Donelson JE, Hill KL, El-Sayed NMA. Multiple mechanisms of immune evasion by African trypanosomes. Mol. Biochem. Parasitol. 1998;91:51–66. doi: 10.1016/s0166-6851(97)00209-0. [DOI] [PubMed] [Google Scholar]
- 7.Melville SE, Leech V, Gerrard CS, Tait A, Blackwell JM. The molecular karyotype of the megabase chromosomes of Trypanosoma brucei and the assignment of chromosome markers. Mol. Biochem. Parasitol. 1998;94:155–173. doi: 10.1016/s0166-6851(98)00054-1. [DOI] [PubMed] [Google Scholar]
- 8.Weiden M, Osheim YN, Beyer A, van der Ploeg LHT. Chromosome structure: DNA nucleotide sequence elelments of a subset of the minichromosomes of the protozoan Trypanosoma brucei. Mol. Cell Biol. 1991;11:3823–3834. doi: 10.1128/mcb.11.8.3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Melville SE, Gerrard CS, Blackwell JM. Multiple causes of size variation in the diploid megabase chromosomes of African trypanosomes. Chrom. Res. 1999;7:191–203. doi: 10.1023/a:1009247315947. [DOI] [PubMed] [Google Scholar]
- 10.Rudenko G, Chaves I, Dirks-Mulder A, Borst P. Selection for the activation of a new variant surface glycoprotein gene expression site in Trypanosoma brucei can result in deletion of the old one. Mol. Biochem. Parasitol. 1998;95:97–109. doi: 10.1016/s0166-6851(98)00099-1. [DOI] [PubMed] [Google Scholar]
- 11.Ligtenberg MJL, Bitter W, Kieft R, Steverding D, Janssen H, Calafat J, Borst P. Reconstitution of a surface transferrin binding complex in insect form Trypanosoma brucei. EMBO J. 1994;13:2565–2573. doi: 10.1002/j.1460-2075.1994.tb06546.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alexandre S, Paindavoine P, Hanocq-Quertier J, Paturiaux-Hanocq F, Tebabi P, Pays E. Families of adenylate cyclase genes in Trypanosoma brucei. Mol. Biochem. Parasitol. 1996;77:173–182. doi: 10.1016/0166-6851(96)02591-1. [DOI] [PubMed] [Google Scholar]
- 13.Carruthers VB, Navarro M, Cross GAM. Targeted disruption of expression site-associated gene-1 in bloodstream-form Trypanosoma brucei. Mol. Biochem. Parasitol. 1996;81:65–79. doi: 10.1016/0166-6851(96)02672-2. [DOI] [PubMed] [Google Scholar]
- 14.Lips S, Revelard P, Pays E. Identification of a new expression site-associated gene in the complete 30.5 kb sequence from the AnTat 1.3A variant sruface protein gene expression site of Trypanosoma brucei. Mol. Biochem. Parasitol. 1993;62:135–138. doi: 10.1016/0166-6851(93)90189-5. [DOI] [PubMed] [Google Scholar]
- 15.Xong HV, Vanhamme L, Chamekh M, Chimfwembe CE, van den Abbeele J, Pays A, van Meirvenne N, Hamers R, de Baetselier P, Pays E. A VSG expression-site associated gene confers resistance to human serum in Trypanosoma rhodesiense. Cell. 1998;95:839–846. doi: 10.1016/s0092-8674(00)81706-7. [DOI] [PubMed] [Google Scholar]
- 16.Kooter JM, van der Spek HJ, Wagter R, d’Oliveira CE, van der Hoeven F, Johnson PJ, Borst P. The anatomy and transcription of a telomeric expression site for variant-specific surface antigens in T. brucei. Cell. 1987;51:261–272. doi: 10.1016/0092-8674(87)90153-x. [DOI] [PubMed] [Google Scholar]
- 17.Bitter W, Gerrits H, Kieft R, Borst P. The role of transferrin-receptor variation in the host range of Trypanosoma brucei. Nature. 1998;391:499–502. doi: 10.1038/35166. [DOI] [PubMed] [Google Scholar]
- 18.Rudenko G. Genes involved in phenotypic and antigenic variation in African trypanosomes and malaria. Curr. Op. Micro. 1999;2:651–656. doi: 10.1016/s1369-5274(99)00039-9. [DOI] [PubMed] [Google Scholar]
- 19.Rudenko G, Blundell PA, Dirks-Mulder A, Kieft R, Borst P. A ribosomal DNA promoter replacing the promoter of a telomeric VSG gene expression site can be efficiently switched on and off in T. brucei. Cell. 1995;83:547–553. doi: 10.1016/0092-8674(95)90094-2. [DOI] [PubMed] [Google Scholar]
- 20.Horn D, Cross GAM. A developmentally regulated position effect at a telomeric location in Trypanosoma brucei. Cell. 1995;83:555–561. doi: 10.1016/0092-8674(95)90095-0. [DOI] [PubMed] [Google Scholar]
- 21.Ansorge I, Steverding D, Melville S, Hartmann C, Clayton C. Transcription of ‘inactive’ expression sites in African trypanosomes leads to expression of multiple transferrin receptor RNAs in bloodstream forms. Mol. Biochem. Parasitol. 1999;101:81–94. doi: 10.1016/s0166-6851(99)00060-2. [DOI] [PubMed] [Google Scholar]
- 22.Vanhamme L, Poelvoorde P, Pays A, Tebabi P, Xong HV, Pays E. Differential RNA elongation controls the variant surface glycoprotein gene expression sites of Trypanosoma brucei. Mol. Micro. 2000;36:328–340. doi: 10.1046/j.1365-2958.2000.01844.x. [DOI] [PubMed] [Google Scholar]
- 23.Alarcon CM, Son HJ, Hall T, Donelson JE. A monocistronic transcript for a trypanosome variant surface glycoprotein. Mol. Cell Biol. 1994;14:5579–5591. doi: 10.1128/mcb.14.8.5579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lenardo MJ, Esser KM, Moon AM, van der Ploeg LHT, Donelson JE. Metacyclic variant surface glycoprotein genes of Trypanosoma brucei subsp. rhodesiense are activated in situ, and their expression is transcriptionally regulated. Mol. Cell. Biol. 1986;6:1991–1997. doi: 10.1128/mcb.6.6.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chaves I, Rudenko G, Dirks-Mulder A, Cross M, Borst P. Control of variant surface glycoprotein gene-expression sites in Trypanosoma brucei. EMBO J. 1999;18:4846–4855. doi: 10.1093/emboj/18.17.4846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Graham SV, Terry S, Barry JD. A structural and transcription pattern for variant surface glycoprotein gene expression sites used in metacyclic stage Trypanosoma brucei. Mol. Biochem. Parasitol. 1999;103:141–154. doi: 10.1016/s0166-6851(99)00128-0. [DOI] [PubMed] [Google Scholar]
- 27.Nagoshi Y,L, Alarcon CM, Donelson JE. The putative promoter for a metacyclic VSG gene in African trypanosomes. Mol. Biochem. Parasitol. 1995;72:33–45. doi: 10.1016/0166-6851(95)00062-6. [DOI] [PubMed] [Google Scholar]
- 28.Barry JD, Crowe JS, Vickerman K. Instability of the Trypanosoma brucei rhodesiense metacyclic variable antigen repertoire. Nature. 1983;306:699–701. doi: 10.1038/306699a0. [DOI] [PubMed] [Google Scholar]
- 29.Navarro M, Cross GAM, Wirtz E. Trypanosoma brucei variant surface glycoprotein regulation involves coupled activation/inactivation and chromatin remodeling of expression sites. EMBO J. 1999;18:2265–2272. doi: 10.1093/emboj/18.8.2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Davies K, Carruthers VB, Cross GAM. Manipulation of the vsg co-transposed region increases expression-site switching in Trypanosoma brucei. Mol. Biochem. Parasitol. 1997;86:163–177. doi: 10.1016/s0166-6851(97)02853-3. [DOI] [PubMed] [Google Scholar]
- 31.Cross M, Taylor MC, Borst P. Frequent loss of the active site during Variant Surface Glycoprotein expression site switching in vitro in Trypanosoma brucei. Mol. Cell. Biol. 1998;18:198–205. doi: 10.1128/mcb.18.1.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chung HM, Shea C, Fields S, Taub RN, van der Ploeg LHT. Architectural organization in the interphase nucleus of the protozoan Trypanosoma brucei: location of telomeres and mini-chromosomes. EMBO J. 1990;9:2611–2619. doi: 10.1002/j.1460-2075.1990.tb07443.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chaves I, Zomerdijk J, Dirks-Mulder A, Dirks RW, Rapp AK, Borst P. Subnuclear localization of the active variant surface glycoprotein gene expression site Trypanosoma brucei. Proc. Natl. Acad. Sciences USA. 1998;95:12328–12333. doi: 10.1073/pnas.95.21.12328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kitten T, Barbour A. Juxtaposition of expressed variable antigen genes with a conserved telomere in the bacterium Borrelia hermsii. Proc. Natl. Acad. Sci. USA. 1990;87:6077–6081. doi: 10.1073/pnas.87.16.6077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Newbold CI. Antigenic variation in Plasmodium falciparum: mechanisms and consequences. Curr. Op. Micro. 1999;2:420–425. doi: 10.1016/S1369-5274(99)80074-5. [DOI] [PubMed] [Google Scholar]
- 36.Wada N, Nakamura Y. Unique telomeric expression site of major-surface-glycoprotein genes of Pneumocystis carinii. DNA Res. 1996;3:55–64. doi: 10.1093/dnares/3.2.55. [DOI] [PubMed] [Google Scholar]
- 37.Sunkin SM, Stringer JR. Translocation of surface antigen genes to a unique telomeric expression site in Pneumocystis carinii. Mol. Micro. 1996;19:283–295. doi: 10.1046/j.1365-2958.1996.375905.x. [DOI] [PubMed] [Google Scholar]
- 38.Plasterk RHA, Simon MI, Barbour AG. Transposition of structural genes to an expression sequence on a linear plasmid causes antigenic variation in the bacterium Borrelia hermsii. Nature. 1985;318:257–263. doi: 10.1038/318257a0. [DOI] [PubMed] [Google Scholar]
- 39.Zhang JR, Hardham JM, Barbour AG, Norris SJ. Antigenic variation in Lyme disease Borreliae by promiscuous recombination of VMP-like sequence cassettes. Cell. 1997;89:275–285. doi: 10.1016/s0092-8674(00)80206-8. [DOI] [PubMed] [Google Scholar]
- 40.Ward CP, Clottey GT, Dorris M, Ji D-D, Arnot DE. Analysis of Plasmodium falciparum PfEMP-1/ var genes suggests that recombination rearranges constrained sequences. Mol. Biochem. Parasitol. 1999;102:167–177. doi: 10.1016/s0166-6851(99)00106-1. [DOI] [PubMed] [Google Scholar]
- 41.Roth C, Bringaud F, Layden RE, Baltz T, Eisen H. Active late-appearing variable surface antigen genes in Trypanosoma equiperdum are constructed entirely from pseudogenes. Proc. Natl. Acad. Sci. USA. 1989;86:9375–9379. doi: 10.1073/pnas.86.23.9375. [DOI] [PMC free article] [PubMed] [Google Scholar]