Abstract
Background
Chronic rhinosinusitis (CRS) is a disease characterized by inflammation of the nasal mucosa and paranasal sinuses. This inflammation may result in part from decreased epithelial barrier and innate immune responses, leading to frequent bacterial and fungal colonization. The objectives of this study were to investigate the expression of innate immune proteins of the Palate Lung and Nasal epithelium Clone (PLUNC) family in patients with CRS.
Methods
Nasal tissue samples were collected from control subjects and CRS patients with and without nasal polyps. Expression of the members of the PLUNC family was analyzed by real-time PCR. Expression of SPLUNC1 and LPLUNC2 proteins was analyzed by ELISA, immunoblot and immunohistochemical analysis.
Results
Levels of mRNA for most of the members of the PLUNC family were profoundly reduced in nasal polyps (NPs) compared to uncinate tissue from control subjects or CRS patients. LPLUNC2 and SPLUNC1 proteins were decreased in NPs of CRS patients compared to uncinate tissue from control subjects. Immunohistochemical data revealed that within submucosal glands of sinonasal tissues, SPLUNC1 and LPLUNC2 were differentially expressed, in serous and mucous cells, respectively. The decrease in expression of these molecules is probably explained by a decrease in the number of glands in NPs as revealed by correlations with levels of the glandular marker lactoferrin.
Conclusions
Decreased SPLUNC1 and LPLUNC2 in NPs reflects a profound decrease in the number of submucosal glands. Decreased glands may lead to a localized defect in the production and release of glandular innate defense molecules.
Keywords: Chronic rhinosinusitis, innate immunity, LPLUNC2, nasal polyps, SPLUNC1
INTRODUCTION
Chronic rhinosinusitis (CRS) is one of the most highly prevalent chronic diseases. CRS afflicts up to 14 percent of Americans, resulting in an expenditure of over 4.3 billion dollars and 200,000 sinus surgeries annually (1–3). In Europe, a multicentre study involving 12 countries (GA2LEN), found the overall prevalence of CRS (by EP3OS criteria) to be 10.9% (4). CRS is characterized by chronic mucosal inflammation of the nose and paranasal sinuses confirmed by nasal endoscopy or sinus CT scan (5). This condition can be further subdivided into 2 entities: CRS with nasal polyps (CRSwNP) and CRS without nasal polyps (CRSsNP).
The pathogenesis of CRS is thought to reflect a complex interplay between host factors, consisting of the innate and adaptive immune responses, and environmental factors, including fungal or bacterial colonization, biofilms, presence of superantigens, osteitis, and allergen exposure (6). Epithelium plays a role beyond providing a physical barrier as a source of constitutive and induced antimicrobial factors. We have proposed that dysregulation of defense pathways results in a defective response to environmental triggers, thus contributing to the pathogenesis of CRS (7, 8).
Short, palate, lung, and nasal epithelial clone 1(SPLUNC1) is a member of the PLUNC family of proteins, which are structural homologues of the LPS binding innate defense proteins lipopolysacccharide binding protein (LBP) and Bactericidal/permeability-increasing protein (BPI) (9, 10). SPLUNC1 is expressed in the epithelium and submucosa of the nasopharynx and is secreted into the nasal cavity, as evidenced by the high concentrations detected in nasal lavage fluid (9, 11–13). Functional studies have revealed that SPLUNC1 can bind to the lipid A portion of LPS and inhibit the growth of Pseudomonas aeruginosa and Mycoplasma pneumonia (Mp) and presumably other gram negative organisms (14–18). Moreover, SPLUNC1 has been shown to suppress inflammation and conversely, inflammatory cytokines also reduce SPLUNC1 expression and Mp clearance (16, 19). Recent evidence has elucidated a role of SPLUNC1 as an extracellular inhibitor of epithelial Na Channel (ENaC) activity, thus altering airway hydration and increasing mucous clearance (20). The hydrophobicity of SPLUNC1 allows it to act as a surfactant, capable of dispersing matrix encased-biofilms of P. aeruginosa in vitro (21). Thus, SPLUNC1 has immunoregulatory, antimicrobial and surfactant properties that make it an important molecule in the lining fluid of the nasal cavity.
In the current study, we tested the PLUNC family of proteins for impairment in CRS. Despite the obvious importance of this family of molecules in the nasal mucosa, this represents the first comprehensive evaluation of the family in the nose and sinuses. After an initial screen of mRNA levels of PLUNC family proteins in disease and normal tissue, we focused on the two most highly expressed, SPLUNC1 and LPLUNC2, to elucidate their role in CRS.
METHODS
Patients and Specimens
CRS patients were recruited from the clinics at Northwestern University using protocols that were approved by the Institutional Review Board of Northwestern University and all subjects gave informed consent. Patients were diagnosed with CRS using task force guidelines (6, 22). Nasal tissues were obtained from defined anatomical site (uncinate and nasal polyps) by functional endoscopic sinus surgery from CRS patients who failed conservative medical therapy (saline irrigations, decongestants, prolonged treatments with antibiotic and/or steroids). Some patients had been on steroids within 2 weeks of surgery. Normal control nasal tissues were similarly obtained from patients who underwent skull based tumor excision. Control patients did not have any history of upper airway inflammatory diseases. Subjects with fungal sinusitis, established immunodeficiency, Churg-Strauss syndrome or cystic fibrosis were excluded from the study. Characteristics of the study population are shown in Table I.
Table I.
Subjects’ characteristics
| Control | CRSsNP | CRSwNP | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Total no. of subjects | 48 (25M) | 59(27M) | 81 (59M) | ||||||
| Age (y), median (range) | 43 (16–77) | 38(18–64) | 41(22–74) | ||||||
| Y | N | U | Y | N | U | Y | N | U | |
| Atopy | 3 | 44 | 1 | 30 | 23 | 6 | 45 | 26 | 10 |
| Asthma | 0 | 47 | 1 | 11 | 44 | 4 | 38 | 43 | 0 |
| PCR | Uncinate | Uncinate | Uncinate | Polyp | |||||
| No. of subjects | 10 (3M) | 11 (6M) | 11 (9M) | 10 (7M) | |||||
| Age (y), median (range) | 42 (16–62) | 34 (23–64) | 39 (27–61) | 48.5 (28–74) | |||||
| SPLUNC1 (PLUNC) | |||||||||
| ELISA-Lavage | |||||||||
| No. of subjects | 18 (9M) | 18 (6M) | 15 (11M) | ||||||
| Age (y), median (range) | 33(17–63) | 38 (28–60) | 44 (30–70) | ||||||
| ELISA-Extracts | |||||||||
| No. of subjects | 10 (4M) | 9 (1M) | 10 (7M) | 10 (7M) | |||||
| Age (y), median (range) | 52.5 (16–72) | 36 (28–62) | 40 (22–74) | 41 (26–74) | |||||
| IHC | |||||||||
| No. of subjects | 10 (3M) | 9 (5M) | 9 (8M) | 10 (7M) | |||||
| Age (y), median (range) | 54 (25–77) | 27 (21–50) | 35 (29–53) | 35.5 (22–63) | |||||
| LPLUNC2 (BPIL-1) | |||||||||
| IB-Lavage | |||||||||
| No. of subjects | 10 (6M) | 10 (4M) | 6 (5M) | ||||||
| Age (y), median (range) | 33 (21–44) | 38.5 (28–60) | 52.5 (33–68) | ||||||
| IB-Extracts | |||||||||
| No. of subjects | 6 (1M) | 7 (3M) | 5 (3M) | 7 (7M) | |||||
| Age (y), median (range) | 48.5 (16–63) | 49 (18–63) | 41 (34–52) | 42 (32–74) | |||||
| IHC | |||||||||
| No. of subjects | 9 (6M) | 10 (6M) | 8 (6M) | 7 (6M) | |||||
| Age (y), median (range) | 58 (19–64) | 34 (21–54) | 54 (34–72) | 50 (27–72) | |||||
| Lactoferrin | |||||||||
| ELISA-Extracts | |||||||||
| No. of subjects | 7 (5M) | 12 (5M) | 10 (6M) | 11 (8M) | |||||
| Age (y), median (range) | 45 (19–62) | 36 (21–62) | 40 (30–68) | 40 (26–64) | |||||
M:male, Y:yes, U: unknown, IHC: immunohistochemistry, IB: immunoblot
Microarray and Real-time PCR
A comprehensive microarray analysis was performed as described previously and gene expression was measured with GeneChip Human U133 Plus 2.0 probe arrays (Affymetrix) (23). Detailed protocols for microarray and real-time PCR are provided in the supporting information. All microarray data has been deposited to gene expression omnibus : GSE36830
ELISA and Immunoblots
A SPLUNC1 sandwich ELISA was developed in our laboratory with anti-SPLUNC1 antibodies (R&D Systems, MN). A lactoferrin ELISA was purchased from Oxis International Inc (CA). As there was no commercially available ELISA for LPLUNC2, we used immunoblot analysis to detect LPLUNC2 (Proteintech, IL). Detailed procedures are provided in the supporting information.
Collection and extraction of proteins from sinus tissue and nasal lavage fluids (NLF)
NLF and sinonasal tissue proteins were collected and extracted as described previously (24). Detail procedures are provided in the supporting information.
Immunohistochemistry
The basic protocol for Immunohistochemistry has been described previously (24). Detailed procedures are in the supporting information.
Statistical analysis
All data are presented as mean±SEM. Comparisons were made using a Mann-Whitney U test. Correlations were assessed by Spearman Rank correlation. All statistical analyses were performed using GraphPad prism 5.0 software. A P value of less that 0.05 was considered statistically significant.
RESULTS
Screen of the PLUNC family in sinonasal tissues; decreased levels in nasal polyps
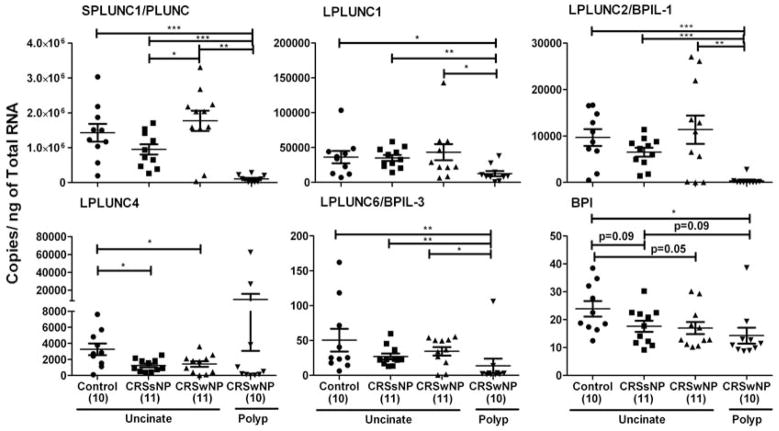
We performed a microarray analysis to compare global gene expression in uncinate tissue from control, CRSsNP, CRSwNP patients and polyp tissue from CRSwNP patients. We observed that mRNA levels of the PLUNC family were differentially expressed in various regions of sinonasal mucosa. We also found that the expression of some of these proteins was decreased in polyps of patients with CRSwNP (Fig E1). We confirmed decreased expression of these proteins using real-time PCR (Fig 1). Expression of mRNA for SPLUNC1, LPLUNC1, LPLUNC2, LPLUNC6 and BPI was significantly reduced in the polyps of patients with CRSwNP compared to uncinates of either control subjects or patients with CRS (P<0.05, n=10–11). Surprisingly, we found no differences in the expression of most of these molecules when comparing only uncinates of control subjects and patients with CRS, with the exception of BPI (P=0.05) and LPLUNC4 (P<0.05), indicating a possible global decrease of these two molecules in patients with CRS. As SPLUNC1 and LPLUNC2 were most highly expressed in uncinate tissues and their expression was decreased in polyps of patients with CRSwNP, we decided to confirm the decrease in their expression at the protein level.
Figure 1. Evaluation of mRNA expression of the BPI-PLUNC family of proteins in sinonasal tissues.
Total RNA was extracted from uncinates of control subjects and patients with CRSsNP or CRSwNP, and polyps of patients with CRSwNP. Expression of mRNA was analyzed by real-time PCR. Expression was normalized to the median value of β-glucuronidase and expressed as copies/ng of total RNA. *P<0.05, **P<0.01, ***P<0.001
Decreased protein expression of SPLUNC1 and LPLUNC2 in polyps of patients with CRSwNP
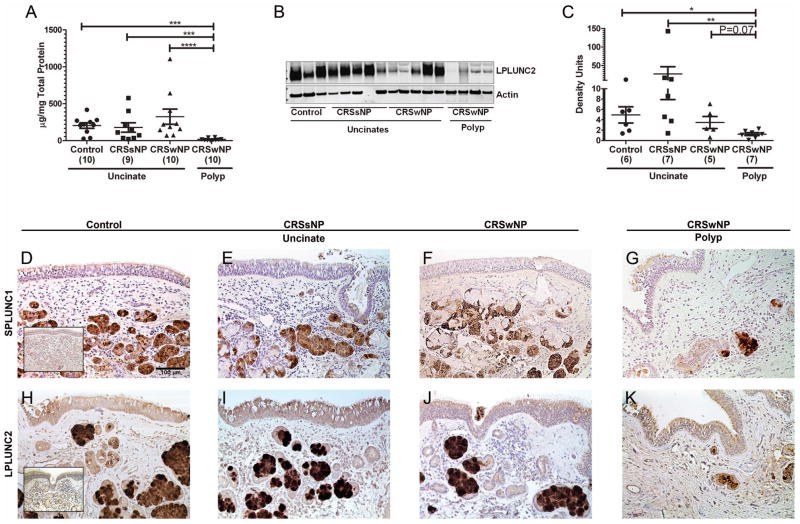
To confirm the mRNA findings, we analyzed the expression of SPLUNC1 protein by ELISA in extracts of 39 sinonasal tissues from control, CRSsNP and CRSwNP patients. We detected substantial expression of SPLUNC1 but there was no difference in the expression of SPLUNC1 protein in uncinates of normal controls and patients with CRS, supporting our mRNA data. Importantly, we found a 90% reduction in the expression of SPLUNC1 protein in nasal polyps of patients with CRSwNP compared to normal control uncinate tissue (Fig 2, A, P<0.05, n= 9–10). We used immunoblot analysis to analyze the expression of LPLUNC2 in 25 sinonasal tissues from control, CRSsNP and CRSwNP patients. As observed for SPLUNC1, we did not find any difference in the expression of LPLUNC2 among the various uncinate tissues. However, we found polyps to have a significant decrease in the level of expression of LPLUNC2 protein compared to control uncinate tissue, supporting our mRNA data (Fig 2, B and C, P<0.05, n≥5). Due to semi-quantitative nature of western blots, errors due to differential transfer and quantitation may affect the linearity of the assay; future studies required involve developing ELISA assays for quantifying LPLUNC2. These data collectively indicate that polyps of patients with CRSwNP have profoundly decreased expression of SPLUNC1 and LPLUNC2 compared to control uncinate tissue.
Figure 2. Evaluation of protein expression and localization of the two highly expressed PLUNC family members by ELISA (SPLUNC1), immunoblot (LPLUNC2) and immunohistochemistry.
Tissue extracts of uncinates from control subjects and patients with CRSsNP or CRSwNP, and polyps from patients with CRSwNP were used to analyze the concentration of SPLUNC1 protein by ELISA (A) and LPLUNC2 protein by immunoblot analysis (B and C). A representative immunoblot for LPLUNC2 (49 kDa) and Actin (42 kDa) (B). Densitometry analysis of LPLUNC2 immunoblots (C). Representative SPLUNC1 (D–G) and LPLUNC2 (H–K) immunostaining in uncinate of a control subject (D and H), a CRSsNP patient (E and I), a CRSwNP patient (F and J) and a polyp from a CRSwNP patient (G and K). Inset in D and H shows control IgG staining for SPLUNC1 and LPLUNC2 respectively. Representative of 7–10 donors per group shown as 200X. *P<0.05, **P<0.01, ***P<0.001
Immunohistochemical analysis indicates that SPLUNC1 and LPLUNC2 are differentially expressed in submucosal gland cell types and epithelium of sinonasal tissues
We performed immunohistochemistry to determine the localization of SPLUNC1 and LPLUNC2 in sinonasal tissues. We compared the expression of SPLUNC1 and LPLUNC2 in uncinates of controls, CRSsNP, CRSwNP patients and in polyps of CRSwNP patients. Our staining was specific, as we did not observe any staining with an IgG control (Fig 2, D and H inset). Both SPLUNC1 and LPLUNC2 were highly expressed in the submucosal glands and moderately expressed in the respiratory epithelial cells of the normal uncinate tissues. Upon scoring for staining intensity, we found no differences in glandular and epithelial intensity of SPLUNC1 staining in polyps of patients with CRSwNP compared to uncinate tissues of control subjects or CRS patients (Fig 2, D–G and Fig E2, Top). In contrast, glandular staining intensity of LPLUNC2 was slightly reduced in polyps of patients with CRSwNP compared to control uncinates (P=0.06, Fig 2, H–K and Fig E2, Bottom). These observations indicate that submucosal glands may be the major source of both SPLUNC1 and LPLUNC2 in sinonasal tissues.
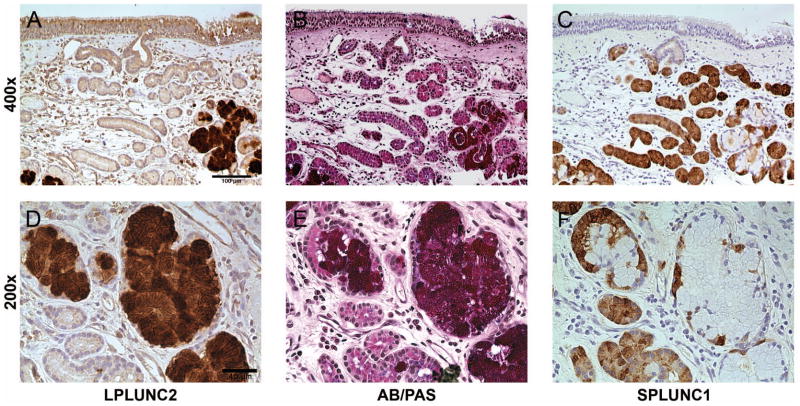
To explore the cellular localization of SPLUNC1 and LPLUNC2 within the submucosal glands, we used both immunohistochemical analysis and AB/PAS staining in serial sections of control uncinate tissues. AB/PAS staining, which stains neutral and acidic mucins, indicated that both mucous and serous cells were found within the glands of the sinonasal tissues (Fig 3, B and E). SPLUNC1 staining was observed in regions with minimal or no AB/PAS staining, suggesting that SPLUNC1 is mainly produced by the serous cells of the submucosal glands (Fig 3, C and F). In contrast to SPLUNC1 staining, LPLUNC2 staining was restricted to regions that stained with AB/PAS, indicating that LPLUNC2 is contained mainly within the mucous cells of the submucosal glands (Fig 3, A and D). This indicates that SPLUNC1 and LPLUNC2 are produced by different cells lining the submucosal glands. Of note, we analyzed expression of SPLUNC1 and LPLUNC2 in NLF of control subjects and patients with CRSsNP and CRSwNP by ELISA and immunoblotting, respectively. However, we did not observe any differences in the levels of SPLUNC1 or LPLUNC2 in NLF of patients with CRS compared to control (Fig E3). Thus from these observations, we conclude that the suppression of levels of SPLUNC1 and LPLUNC2 in patients with CRS is restricted to the nasal polyp itself and not global within the sinonasal cavity.
Figure 3. Evaluation of sequential staining for AB/PAS, SPLUNC1 and LPLUNC2 in sinonasal tissue.
Serial sections (3 μm) of uncinate tissues from normal patients were used to stain with LPLUNC2 antibody (A and D), AB/PAS (B and E) or SPLUNC1 antibody (C and F). Shown is a representative subject out of 4 control subjects, represented as 200X (A–C) and 400X (D–F).
Decrease in the number of glands explains reduced SPLUNC1 and LPLUNC2 expression in polyps
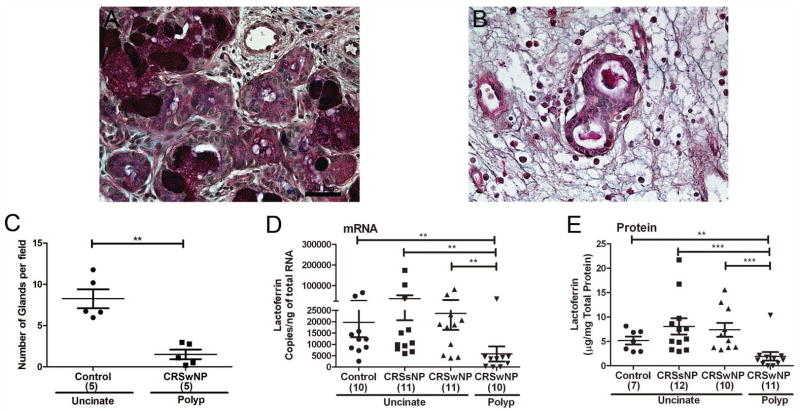
We tested whether reduced expression of SPLUNC1 and LPLUNC2 may be due to a decrease in the number of glands in polyps. We stained tissues with AB/PAS and determined the number and morphology of glands. Nasal polyps from patients with CRSwNP had an 81% decrease in the number of glands compared to control uncinates (Fig 4, A–C). We also observed that polyps had glands that appeared to be stretched and elongated.
Figure 4. Evaluation of the number of glands and expression of lactoferrin in sinonasal tissue.
Representative AB/PAS staining in uncinate from a control subject (A) and a polyp from a patient with CRSwNP (B). Quantitation of the number of glands in tissues stained with AB/PAS in uncinate from control subjects and polyps from patients with CRSwNP (C) (n=5, 10 fields per slide). Lactoferrin expression in sinonasal tissue was assessed by real-time PCR (D) and ELISA (E). **P<0.01, ***P<0.001
Expression of lactoferrin, a serous cell marker, is decreased in polyps of patients with CRS
To confirm the decreased number of glands in polyps, we analyzed the expression of a glandular marker lactoferrin, which is mainly produced by the serous cells of the submucosal glands (25). In support of our findings with SPLUNC1, we found reduced levels of mRNA for lactoferrin (Fig 4, D) in nasal polyps of patients with CRSwNP compared to uncinate tissues from control or CRSsNP or CRSwNP patients. We confirmed this finding at the protein level for lactoferrin (Fig 4, E). Levels of mRNA for SPLUNC1 significantly correlated with lactoferrin (r=0.664, P<0.0001, n=42) supporting the conclusion that lactoferrin and SPLUNC1 are contained within the serous cells of the submucosal glands (Fig E4, A). Although neutrophils are often present in sinonasal tissues and have been shown to produce lactoferrin, we observed a negative correlation between the expression of a neutrophil marker (CXCR1) and lactoferrin (r=−0.403, P=0.0081, n=42), suggesting that neutrophils are not a major source of lactoferrin in sinonasal tissue (Fig E4, B). These data collectively suggest that the decreased expression of SPLUNC1 in nasal polyp tissues of patients with CRSwNP is due to a decrease in the number of glands in nasal polyps.
DISCUSSION
Recent studies have suggested that dysfunction of the innate immune system may play a permissive role in CRS pathogenesis, creating an environment that fosters increased microbial colonization (7, 8). The PLUNC family of proteins came into interest initially due to reports of high concentrations of SPLUNC1 in NLF and to their structural similarities to the antimicrobial protein BPI, suggesting potential antimicrobial and endotoxin neutralization functions (9). Subsequent studies demonstrated a profound antimicrobial effect of SPLUNC1 (14, 18). The present study demonstrates decreased mRNA expression for most, members of the PLUNC family in nasal polyps when compared with uncinate tissue of either control or CRS patients. This decrease in PLUNC family expression was generally restricted to the polyp itself. The PLUNC family members LPLUNC4 and BPI were exceptions, with lower expression in uncinate tissue from both CRSsNP and CRSwNP patients. Further study will be required to assess whether there are global defects in production of these two molecules in CRS.
In the present study, we focused our efforts on the two most highly expressed PLUNC family molecules in sinonasal tissue, SPLUNC1 and LPLUNC2. Using SPLUNC1 ELISA and LPLUNC2 immunoblot analysis, we were able to confirm reduced levels in nasal polyp tissue. In a previous report using proteomics technology, Min-man et al. concluded that the intensity of SPLUNC1 staining was decreased in nasal polyp tissue (26). The current study confirms this finding and extends it to demonstrate that the decreased expression in polyp tissue was due to a decreased number of glands. It has been previously reported that some nasal polyps have a lower density of glandular structures than the surrounding ethmoid tissue (27). SPLUNC1 and LPLUNC2 were strongly expressed in submucosal glands and moderately expressed in surface epithelium. Remarkably, we observed that SPLUNC1 was localized in the serous cells and LPLUNC2 was selectively localized in the mucous cells within the submucosal glands. This suggests differential release of these host defense proteins during basal and stimulated conditions.
Based on our findings of decreased expression of glandular proteins in nasal polyp tissue, we propose that glandular hyperplasia associated with CRS, if it occurs, may be restricted to certain forms of polyps or to tissues other than the polyp itself (27). Our findings strongly suggest that the decreased SPLUNC1 in polyps is an effect of formation of the polyp without an accompanying expansion of submucosal glands. We detected substantial amounts of SPLUNC1 and LPLUNC2 in NLF and despite the decreased production of these molecules in nasal polyps, we failed to observe any differences in expression of these molecules in NLF of patients with CRS compared to control subjects. Since NLF is collected as a wash of the entire sinonasal cavity, the presumed reduction of SPLUNC1 and LPLUNC2 on the surface of the polyps may not influence the total collection. We speculate that other methods of collection of nasal lining fluid, such as using filter paper discs or sponges will detect reduced SPLUNC1 on the polyp surface (28, 29).
Some current theories of CRS pathogenesis have focused on defects in production of innate host defense proteins, barrier defects, mucociliary dysfunction, and a role for superantigens. More recently our laboratory published reduction in innate immune molecules of the S100 family in CRS (30). In the present study we show that polyps of patients with CRS have a defect in the local production of proteins of the PLUNC family and lactoferrin. Despite SPLUNC1’s homology to BPI and its ability to bind LPS, it has been tested only with a few organisms (14, 16–18, 31). Recent findings, however suggest that SPLUNC1 may also be involved in suppressing Pseudomonas biofilm formation (21). Biofilms are a leading cause of decreased efficacy of antibiotics, which in turn may lead to increased colonization by bacteria in CRS (32, 33). Our findings thus suggest that reduced PLUNC proteins on the surface of a nasal polyp may increase susceptibility to colonization by microorganisms that form biofilms or are otherwise sensitive to PLUNCs. Based on its LPS binding ability, we speculate that SPLUNC1 may impact additional organisms other than Mycoplasma and Pseudomonas.
There are other mechanisms by which reduced PLUNC might alter sinonasal physiology or inflammation. Published studies suggest SPLUNC1 is an anti-inflammatory protein, suppressing the ova-alum model of allergic inflammation (19). It could be speculated that the increased inflammation seen in polyps may thus relate in part to decreased SPLUNC1 expression. Also of interest is recent research showing that SPLUNC1 has a role in suppressing the activation of epithelial sodium channels (ENaC), which has implications in mucociliary clearance as well as ion and fluid influx into mucosal tissue (20, 34). A decrease of SPLUNC1 in polyp tissue may result in the local over-activation of ENaC, leading to transepithelial water transport and contributing to polyp formation. Of note, cultured nasal polyp epithelial cells show a greater rate of transepithelial ion transport, suggesting that the movement of water into the cell and into the interstitial tissue could explain the edema commonly seen in polyps (35). The dysregulation of ENaC and surface liquid levels also impairs mucociliary clearance, another mechanism by which pathogens are able to thrive and cause chronic inflammation. Recent studies have implicated SPLUNC1 in mucociliary clearance in vivo (31). Based on the multifaceted functional abilities of SPLUNC1, we hypothesize that reduction in SPLUNC1 may contribute to CRS pathogenesis via loss of its physicochemical effects as much as via loss of its antimicrobial or LPS neutralizing effects.
The functions of other proteins of the PLUNC family are not yet elucidated. Based on protein sequence homology with BPI, and their abundance at mucosal surfaces and NLF, it is thought that this family may be involved in mucosal innate host defense (7, 8). Further functional analysis needs to be undertaken to elucidate the specific roles of each of these molecules in host defense and in CRS. Our results and their implications for localization of host defense molecules are summarized in Figure 5. To the extent that they are important in immunity, the decrease in the expression of PLUNCs, and other glandular proteins such as lactoferrin and lysozyme, in nasal polyps may play a contributory role in the increased bacterial colonization of the nasal mucosa in CRSwNP patients. In conclusion, this study demonstrates markedly suppressed expression of PLUNC family members in nasal mucosa of CRS patients, particularly in nasal polyp tissue. Decreased SPLUNC1, theoretically associated with decreased ability to clear pathogens and dysregulation of ionic balance, may contribute to the chronic inflammatory response and subsequent pathogenesis of nasal polyps in CRSwNP patients.
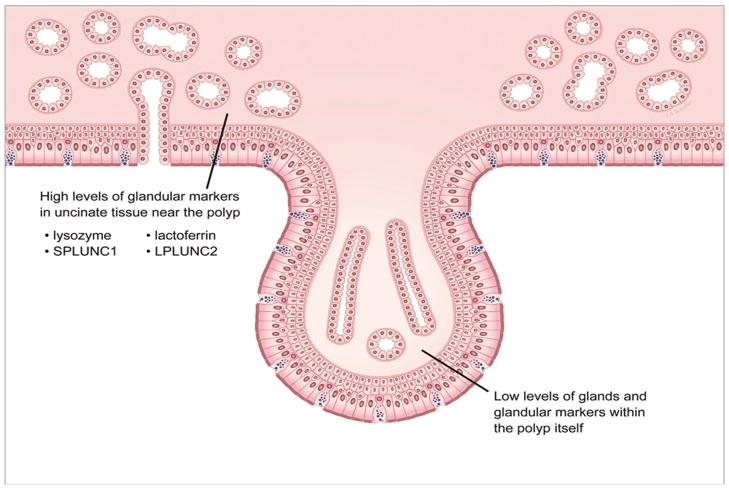
Figure 5.
Decrease in the number of glands in nasal polyps of patients with CRSwNP may lead to a localized defect in the production of antimicrobial proteins such as lactoferrin, lysozyme and proteins of the PLUNC family.
Supplementary Material
Acknowledgments
This research was supported in part by NIH grants R01 HL068546, R01 HL078860, and R01 AI072570 and by the Ernest S. Bazley Trust.
Abbreviations
- CRS
Chronic rhinosinusitis
- CRSwNP
CRS with nasal polyps
- CRSsNP
CRS without nasal polyps
- UT
Uncinate tissue
- PLUNC
Palate LUng Nasal epithelial Clone
- SPLUNC1
Short PLUNC1
- LPLUNC2
Long PLUNC2
- BPI
Bactericidal/Permeability-Increasing Protein
- NLF
Nasal lavage fluid
Footnotes
Conflict of Interest
All authors declare no conflict of interest.
Author Contribution
SS and RPS designed the study. SS, DCL, MR and RGC performed the experiments; SS analyzed the data with the help of DCL and MR. AK performed the microarray analysis. JEN, LS, KEH, RKC, ATP, BKT, DBC, LCG, RCK helped in sample collection and evaluation. HWC provided SPLUNC1 standard for ELISA. MR, RKC, LCG, RCK and BKT critically revised the manuscript. All authors have read and approved the final form of the manuscript. SS, DCL and RPS wrote the manuscript.
References
- 1.Adams PF, Hendershot GE, Marano MA. Current estimates from the National Health Interview Survey, 1996. Vital Health Stat. 1999;10(200):1–203. [PubMed] [Google Scholar]
- 2.Murphy MP, Fishman P, Short SO, Sullivan SD, Yueh B, Weymuller EA., Jr Health care utilization and cost among adults with chronic rhinosinusitis enrolled in a health maintenance organization. Otolaryngol Head Neck Surg. 2002;127(5):367–376. doi: 10.1067/mhn.2002.129815. [DOI] [PubMed] [Google Scholar]
- 3.Hemmerdinger SA, Jacobs JB, Lebowitz RA. Accuracy and cost analysis of image-guided sinus surgery. Otolaryngol Clin North Am. 2005;38(3):453–460. doi: 10.1016/j.otc.2004.10.024. [DOI] [PubMed] [Google Scholar]
- 4.Hastan D, Fokkens WJ, Bachert C, Newson RB, Bislimovska J, Bockelbrink A, et al. Chronic rhinosinusitis in Europe--an underestimated disease. A GA(2)LEN study. Allergy. 2011;66(9):1216–1223. doi: 10.1111/j.1398-9995.2011.02646.x. [DOI] [PubMed] [Google Scholar]
- 5.Rosenfeld RM, Andes D, Bhattacharyya N, Cheung D, Eisenberg S, Ganiats TG, et al. Clinical practice guideline: adult sinusitis. Otolaryngol Head Neck Surg. 2007;137(3 Suppl):S1–31. doi: 10.1016/j.otohns.2007.06.726. [DOI] [PubMed] [Google Scholar]
- 6.Meltzer EO, Hamilos DL, Hadley JA, Lanza DC, Marple BF, Nicklas RA, et al. Rhinosinusitis: establishing definitions for clinical research and patient care. J Allergy Clin Immunol. 2004;114(6 Suppl):155–212. doi: 10.1016/j.jaci.2004.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kern RC, Conley DB, Walsh W, Chandra R, Kato A, Tripathi-Peters A, et al. Perspectives on the etiology of chronic rhinosinusitis: an immune barrier hypothesis. Am J Rhinol. 2008;22(6):549–559. doi: 10.2500/ajr.2008.22.3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tieu DD, Kern RC, Schleimer RP. Alterations in epithelial barrier function and host defense responses in chronic rhinosinusitis. J Allergy Clin Immunol. 2009;124(1):37–42. doi: 10.1016/j.jaci.2009.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bingle CD, Craven CJ. PLUNC: a novel family of candidate host defence proteins expressed in the upper airways and nasopharynx. Hum Mol Genet. 2002;11(8):937–943. doi: 10.1093/hmg/11.8.937. [DOI] [PubMed] [Google Scholar]
- 10.Canny G, Levy O. Bactericidal/permeability-increasing protein (BPI) and BPI homologs at mucosal sites. Trends Immunol. 2008;29(11):541–547. doi: 10.1016/j.it.2008.07.012. [DOI] [PubMed] [Google Scholar]
- 11.Ghafouri B, Kihlstrom E, Stahlbom B, Tagesson C, Lindahl M. PLUNC (palate, lung and nasal epithelial clone) proteins in human nasal lavage fluid. Biochem Soc Trans. 2003;31(Pt 4):810–814. doi: 10.1042/bst0310810. [DOI] [PubMed] [Google Scholar]
- 12.Casado B, Pannell LK, Iadarola P, Baraniuk JN. Identification of human nasal mucous proteins using proteomics. Proteomics. 2005;5(11):2949–2959. doi: 10.1002/pmic.200401172. [DOI] [PubMed] [Google Scholar]
- 13.Campos MA, Abreu AR, Nlend MC, Cobas MA, Conner GE, Whitney PL. Purification and characterization of PLUNC from human tracheobronchial secretions. Am J Respir Cell Mol Biol. 2004;30(2):184–192. doi: 10.1165/rcmb.2003-0142OC. [DOI] [PubMed] [Google Scholar]
- 14.Gally F, Di YP, Smith SK, Minor MN, Liu Y, Bratton DL, et al. SPLUNC1 promotes lung innate defense against Mycoplasma pneumoniae infection in mice. Am J Pathol. 2011;178(5):2159–2167. doi: 10.1016/j.ajpath.2011.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ghafouri B, Kihlstrom E, Tagesson C, Lindahl M. PLUNC in human nasal lavage fluid: multiple isoforms that bind to lipopolysaccharide. Biochim Biophys Acta. 2004;1699(1–2):57–63. doi: 10.1016/j.bbapap.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 16.Chu HW, Thaikoottathil J, Rino JG, Zhang G, Wu Q, Moss T, et al. Function and regulation of SPLUNC1 protein in Mycoplasma infection and allergic inflammation. J Immunol. 2007;179(6):3995–4002. doi: 10.4049/jimmunol.179.6.3995. [DOI] [PubMed] [Google Scholar]
- 17.Zhou HD, Li XL, Li GY, Zhou M, Liu HY, Yang YX, et al. Effect of SPLUNC1 protein on the Pseudomonas aeruginosa and Epstein-Barr virus. Mol Cell Biochem. 2008;309(1–2):191–197. doi: 10.1007/s11010-007-9659-3. [DOI] [PubMed] [Google Scholar]
- 18.Lukinskiene L, Liu Y, Reynolds SD, Steele C, Stripp BR, Leikauf GD, et al. Antimicrobial activity of PLUNC protects against Pseudomonas aeruginosa infection. J Immunol. 2011;187(1):382–390. doi: 10.4049/jimmunol.1001769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wright PL, Yu J, Di YP, Homer RJ, Chupp G, Elias JA, et al. Epithelial reticulon 4B (Nogo-B) is an endogenous regulator of Th2-driven lung inflammation. J Exp Med. 2010;207(12):2595–2607. doi: 10.1084/jem.20100786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garcia-Caballero A, Rasmussen JE, Gaillard E, Watson MJ, Olsen JC, Donaldson SH, et al. SPLUNC1 regulates airway surface liquid volume by protecting ENaC from proteolytic cleavage. Proc Natl Acad Sci U S A. 2009;106(27):11412–11417. doi: 10.1073/pnas.0903609106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gakhar L, Bartlett JA, Penterman J, Mizrachi D, Singh PK, Mallampalli RK, et al. PLUNC is a novel airway surfactant protein with anti-biofilm activity. PLoS One. 2010;5(2):e9098. doi: 10.1371/journal.pone.0009098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pearlman AN, Conley DB. Review of current guidelines related to the diagnosis and treatment of rhinosinusitis. Curr Opin Otolaryngol Head Neck Surg. 2008;16(3):226–230. doi: 10.1097/MOO.0b013e3282fdcc9a. [DOI] [PubMed] [Google Scholar]
- 23.Kato A, Chustz RT, Ogasawara T, Kulka M, Saito H, Schleimer RP, et al. Dexamethasone and FK506 inhibit expression of distinct subsets of chemokines in human mast cells. J Immunol. 2009;182(11):7233–7243. doi: 10.4049/jimmunol.0801375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peters AT, Kato A, Zhang N, Conley DB, Suh L, Tancowny B, et al. Evidence for altered activity of the IL-6 pathway in chronic rhinosinusitis with nasal polyps. J Allergy Clin Immunol. 2010;125(2):397–403. e310. doi: 10.1016/j.jaci.2009.10.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Basbaum CB, Jany B, Finkbeiner WE. The serous cell. Annu Rev Physiol. 1990;52:97–113. doi: 10.1146/annurev.ph.52.030190.000525. [DOI] [PubMed] [Google Scholar]
- 26.Min-man W, Hong S, Zhi-qiang X, Xue-ping F, Chang-qi L, Dan L. Differential proteomic analysis of nasal polyps, chronic sinusitis, and normal nasal mucosa tissues. Otolaryngol Head Neck Surg. 2009;141(3):364–368. doi: 10.1016/j.otohns.2009.04.022. [DOI] [PubMed] [Google Scholar]
- 27.Berger G, Kattan A, Bernheim J, Ophir D. Polypoid mucosa with eosinophilia and glandular hyperplasia in chronic sinusitis: a histopathological and immunohistochemical study. Laryngoscope. 2002;112(4):738–745. doi: 10.1097/00005537-200204000-00026. [DOI] [PubMed] [Google Scholar]
- 28.Lu FX, Esch RE. Novel nasal secretion collection method for the analysis of allergen specific antibodies and inflammatory biomarkers. J Immunol Methods. 2010;356(1–2):6–17. doi: 10.1016/j.jim.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 29.Chawes BL, Edwards MJ, Shamji B, Walker C, Nicholson GC, Tan AJ, et al. A novel method for assessing unchallenged levels of mediators in nasal epithelial lining fluid. J Allergy Clin Immunol. 2010;125(6):1387–1389. e1383. doi: 10.1016/j.jaci.2010.01.039. [DOI] [PubMed] [Google Scholar]
- 30.Tieu DD, Peters AT, Carter RG, Suh L, Conley DB, Chandra R, et al. Evidence for diminished levels of epithelial psoriasin and calprotectin in chronic rhinosinusitis. J Allergy Clin Immunol. 2010;125(3):667–675. doi: 10.1016/j.jaci.2009.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McGillivary G, Bakaletz LO. The multifunctional host defense peptide SPLUNC1 is critical for homeostasis of the mammalian upper airway. PLoS One. 2010;5(10):e13224. doi: 10.1371/journal.pone.0013224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suh JD, Cohen NA, Palmer JN. Biofilms in chronic rhinosinusitis. Curr Opin Otolaryngol Head Neck Surg. 2010;18(1):27–31. doi: 10.1097/MOO.0b013e328334f670. [DOI] [PubMed] [Google Scholar]
- 33.Al-Mutairi D, Kilty SJ. Bacterial biofilms and the pathophysiology of chronic rhinosinusitis. Curr Opin Allergy Clin Immunol. 2011;11(1):18–23. doi: 10.1097/ACI.0b013e3283423376. [DOI] [PubMed] [Google Scholar]
- 34.Rollins BM, Garcia-Caballero A, Stutts MJ, Tarran R. SPLUNC1 expression reduces surface levels of the epithelial sodium channel (ENaC) in Xenopus laevis oocytes. Channels (Austin) 2010;4(4):255–259. doi: 10.4161/chan.4.4.12255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bernstein JM, Yankaskas JR. Increased ion transport in cultured nasal polyp epithelial cells. Arch Otolaryngol Head Neck Surg. 1994;120(9):993–996. doi: 10.1001/archotol.1994.01880330071013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.