Abstract
Systematic transrectal ultrasound (TRUS)-guided biopsy is the standard method for a definitive diagnosis of prostate cancer. However, this biopsy approach uses two-dimensional (2D) ultrasound images to guide biopsy and can miss up to 30% of prostate cancers. We are developing a molecular image-directed, three-dimensional (3D) ultrasound image-guided biopsy system for improved detection of prostate cancer. The system consists of a 3D mechanical localization system and software workstation for image segmentation, registration, and biopsy planning. In order to plan biopsy in a 3D prostate, we developed an automatic segmentation method based wavelet transform. In order to incorporate PET/CT images into ultrasound-guided biopsy, we developed image registration methods to fuse TRUS and PET/CT images. The segmentation method was tested in ten patients with a DICE overlap ratio of 92.4% ± 1.1 %. The registration method has been tested in phantoms. The biopsy system was tested in prostate phantoms and 3D ultrasound images were acquired from two human patients. We are integrating the system for PET/CT directed, 3D ultrasound-guided, targeted biopsy in human patients.
Keywords: Molecular imaging, PET/CT, image segmentation, nonrigid registration, 3D ultrasound, prostate biopsy
1. INTRODUCTION
Prostate cancer is the second leading cause of cancer mortality in American men [1]. Systematic transrectal ultrasound (TRUS)-guided prostate biopsy is considered as the standard method for prostate cancer detection. The current biopsy technique has a significant sampling error and can miss up to 30% of cancers [2]. As a result, a patient may be informed of a negative biopsy result but may in fact be harboring an occult early-stage cancer. It is a difficult challenge for physicians to manage patients with false negative biopsies who, in fact, harbor curable prostate cancer as indicated by biochemical measurements such as rising prostate specific antigen (PSA), as well as patients diagnosed with early-stage disease. Although ultrasound imaging is a preferred method for image-guided biopsy because it is performed in real time and because it is portable and cost effective, current ultrasound imaging technology has difficulty to differentiate carcinoma from benign prostate tissue. Various PET imaging agents have been developed for prostate cancer detection and staging, these include 11C-choline [3, 4], 18F-fluorocholine [5], 11C-acetate [6], 11C-methionine [7], and other PET agents. Of particularly, PET imaging with new molecular imaging tracers such as anti-1-amino-3-18F-fluorocyclobutane-1-carboxylic acid (18F-FACBC) has shown promising results for detecting and localizing prostate cancer in humans [8, 9]. PET imaging with 18F-FACBC show focal uptake at the tumor and thus can provide location information to direct targeted biopsy of the prostate. By combining PET/CT with 3D ultrasound images, multimodality image-guided targeted biopsy may be able to improve the detection of prostate cancer.
In order to use 3D models to guide the biopsy, segmentation of the prostate is a key component of the 3D ultrasound image-guided biopsy system. However, segmentation of the prostate in ultrasound images can be difficult because of the shadows from the bladder and because of a low contrast between the prostate and non-prostate tissue. Many methods were proposed to automatically segment the prostate in ultrasound images [10–28]. Particularly, various shape model based methods are used to guide the segmentation. Gong et al. modeled the prostate shape using superellipses with simple parametric deformations [15]. Ding et al. described a slice-based 3D prostate segmentation method based on a continuity constraint, implemented as an autoregressive model [13]. Hu et al. used a model-based initialization and mesh refinement for prostate segmentation [29]. Hodge et al. described 2D active shape models for semi-automatic segmentation of the prostate and extended the algorithm to 3D segmentation using rotational-based slicing [30]. Tutar et al. proposed an optimization framework where the segmentation process is to fit the best surface to the underlying images under shape constraints [21]. Zhan et al. proposed a deformable model for automatic segmentation of the prostates from 3D ultrasound images using statistical matching of both shape and texture and Gabor support vector machines [27]. Ghanei et al. proposed a 3D deformable surface model for automatic segmentation of the prostate [14]. Pathak et al. used anisotropic diffusion filter and prior knowledge of the prostate for the segmentation [20]. Others proposed wavelet-based methods for the segmentation of the prostate. Knoll et al. used snakes with shape restrictions based on the wavelet transform for outlining the prostate [31]. Chiu et al. introduced a semi-automatic segmentation algorithm based on the dyadic wavelet transform and the discrete dynamic contour [12]. Zhang et al. improved the prostate boundary detection system by tree-structured nonlinear filter, directional wavelet transforms and tree-structured wavelet transform [28]. In this study, we developed a new segmentation method that based on the wavelet support vector machine (W-SVM) and statistical shape model in order to improve the robustness of the segmentation.
In order to incorporate other images such as PET/CT into the 3D ultrasound-guided biopsy, image registration plays a key role in combining multimodality images. Various registration methods have been proposed for the prostate [32–40]. However, deformable registration of ultrasound and PET/CT images is difficult for the following reasons: (1) Neither PET nor ultrasound has enough structure information about the prostate for direct intensity-based image registration; (2) Ultrasound provides only a small field of view that covers just the prostate and surrounding tissue which is merely a small portion of a PET image that includes the entire pelvic region; (3) The registration should be fast enough to be practical for biopsy guidance; and (4) The significant prostate deformation caused by the transrectal probe disqualifies registration algorithms that assume small deformation. In our preliminary study, we used CT images as the bridge to register PET with TRUS because both PET and CT images are acquired from a combined PET/CT system and they will be aligned before the biopsy procedure [33]. Although CT and ultrasound image registration has been used for the prostate using rigid registration methods with the aid from additional tracking device [41], they could not be applied to this application because of significant deformation at the prostate and rectum during ultrasound imaging. Using an articulator arm or electromagnetic positioning device, ultrasound-CT registration has been used in radiotherapy [42] and in radiofrequency ablation [43]. Although deformable CT-ultrasound registration methods have been used in other applications such as cardiac procedures [44] and computer-aided orthopedic surgery [45], when applying them to the prostate and the pelvic region, several challenges raise because the abdomen has irregular boundaries, unlike the bone or head to which registration has been most often applied and because a normal prostate is small. In addition, there are potential factors such as different patient positions, and rectal and bladder filling that can stress registration. Furthermore, it is more difficult to evaluate pelvic and/or prostate registration because no external markers are available. In this study, we developed and evaluate a nonrigid registration method for this particular application.
2. MATERIALS AND METHODS
2.1 PET-directed, 3D ultrasound-guided biopsy
The protocol for our PET/CT directed, 3D ultrasound-guided biopsy of the prostate is described as follows. (1) Before undergoing prostate biopsy, the patient undergoes a PET/CT scan with 18F-FACBC as part of the examinations [8, 9]. The anatomic CT images are combined with the PET images for improved localization of the prostate and suspicious tumors. (2) The patient has a 3D ultrasound scan before the actual biopsy appointment. The ultrasound image is called “pre-biopsy” image. This scan can be one week before the biopsy or on the same day of the PET/CT scan. (3) The PET/CT and pre-biopsy ultrasound images are registered offline before biopsy. (4) Immediately before biopsy, another 3D ultrasound image volume is acquired before the biopsy planning. This “intra-biopsy” ultrasound images are registered with the pre-biopsy ultrasound images. As the pre-biopsy ultrasound image has been registered with the PET/CT data, in turn, the PET/CT image is also registered with the intra-biopsy ultrasound image for tumor targeting. Three-dimensional visualization tools are then used to guide the biopsy needle to a suspicious lesion. (5) At the end of each core biopsy, the position of the needle tip is recorded on real-time ultrasound images during the procedure. The location information of biopsy cores is saved and can be restored in a re-biopsy procedure when the patient is followed up for prostate cancer examination. This allows the physician to re-biopsy the same area and check the possible progression or regression of a lesion. The location information of the biopsy cores can also be used to guide another additional biopsy to different locations if the original biopsy was negative.
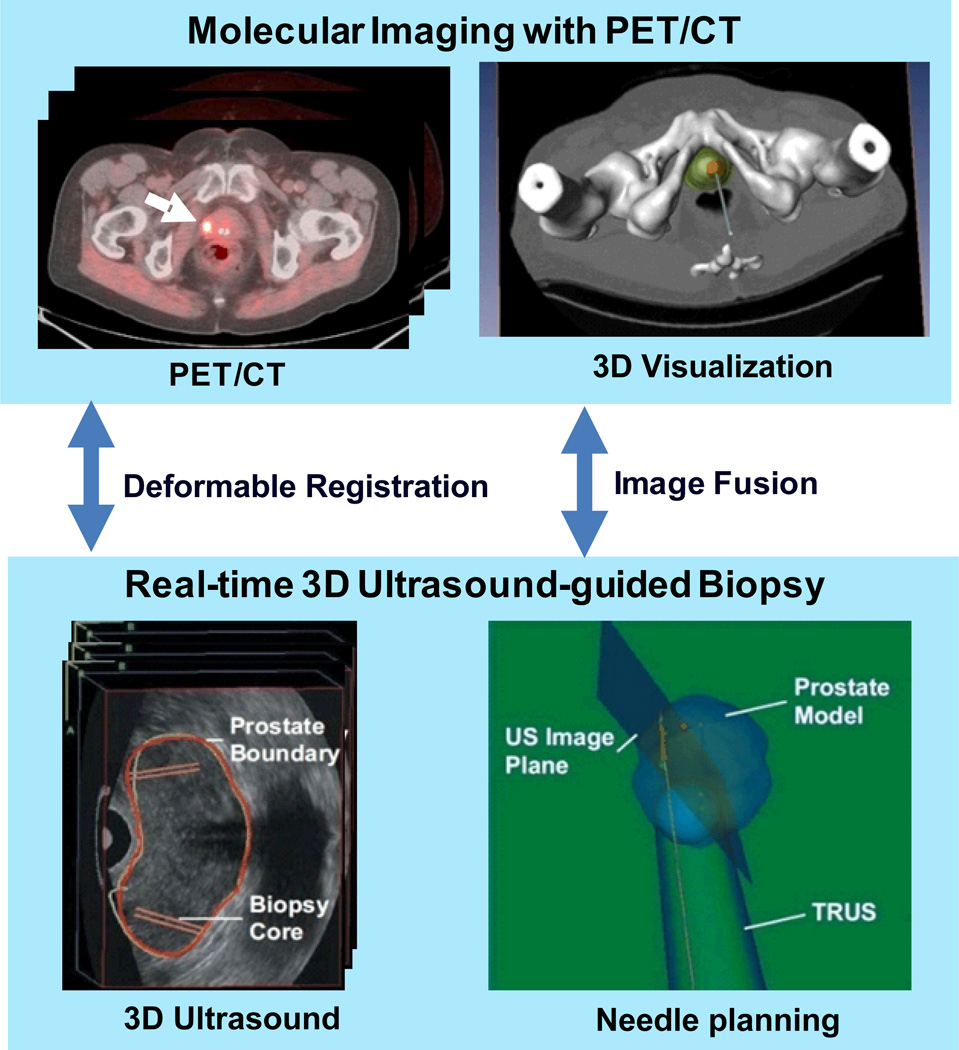
Figure 1 shows the diagram of our molecular image-directed, 3D ultrasound-guided biopsy system. The system uses: (1) Passive mechanical components for guiding, tracking, and stabilizing the position of a commercially available, end-firing, transrectal ultrasound probe. We use the BK Flex Focus 400 (BK Medical, Peabody, MA) and the Artemis system (Eigen, Grass Valley, CA). (2) Software components for acquiring, storing, and reconstructing real- time, a series of 2D TRUS images into a 3D image; and (3) Software that segment the prostate in 3D TRUS images and displays a model of the 3D scene to guide a biopsy needle in three dimensions. The system allows real-time tracking and recording of the 3D position and orientation of the biopsy needle as a physician manipulates the ultrasound transducer. (4) An offline workstation system that is used to register and fuse PET/CT and ultrasound images.
Figure 1.
Molecular image-directed, 3D ultrasound-guided biopsy. Top: The PET/CT images with 18F-FACBC were acquired from a prostate patient at our institution. PET/CT images show a focal lesion within the prostate (white arrow). The 3D visualization of the pelvis and the prostate can be used to aid the insertion of the biopsy needle into a suspicious tumor target. Bottom: During biopsy, a mechanically assisted navigation device was used to acquire 3D TRUS images from patients. The prostate boundaries on TRUS images were segmented from each slice and were used to generate a 3D model of the prostate. The 3D prostate model and real-time TRUS images are used to guide the biopsy in human patients.
2.2. 3D segmentation of the prostate in TRUS images
In order to build a 3D model for biopsy guidance, we developed a 3D segmentation method to segment the prostate in 3D TRUS images. This method utilizes Wavelet-based texture extraction technique followed by support vector machines (SVMs) to adaptively collect texture priors of prostates and non-prostate tissues and classify tissues in different sub-regions around the prostate boundary by statistically analyzing their textures using wavelet features.
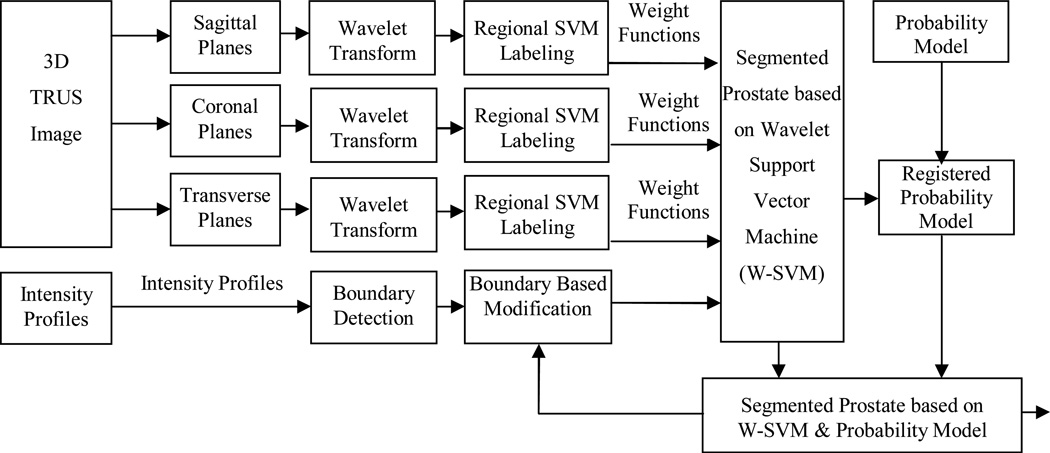
Figure 2 shows the flowchart of the segmentation method. The segmentation method has three main components. (1) Each Wavelet support vector machine (W-SVM) consists of two elements, i.e. Wavelet filter collection and SVM. The Wavelet filter collection is utilized to extract and characterize texture features in TRUS images. The Wavelet filter collection has Wavelet filters at multi-scales and multi-orientations. Therefore, it has the ability to characterize textures with different dominant sizes and orientations from noisy TRUS images. The wavelet transforms are applied to 3D transrectal ultrasound images in three planes. The wavelet is used in 2D sagittal, coronal, and transverse images. (2) The SVMs have been trained by a set of 3D TRUS image samples in the coronal, sagittal, and transverse planes and at each sub-region to label the voxels based on the captured Wavelet texture features. Each voxel is labeled by three sub-regional SVMs in three planes separately. Each voxel in each plane is labeled by a real value between 0 and 1 that represents the likelihood of a voxel belonging to the prostate tissue. (3) A statistical prostate shape model (Figure 3) is incorporated in the segmentation process. Therefore, each voxel has a label of the statistical shape model and three labels for the SVM in three planes. After applying the weight functions that were defined in the SVM training step, the segmented prostate is used for registering the probability model. After defining special weight for each label at each region, each voxel tentatively labeled as prostate or non-prostate voxel. Consequently, the surfaces of the prostate are modified based on the intensity profiles of the model. To improve the robustness of the method a manual intervention was employed. This step was performed through defining a bounding box for the prostate in one middle slice or two orthogonal slices. The probability model was scaled to the size of the box. The segmented prostate is updated and compared to the shape model. The modification and update steps are repeated until the algorithm converges.
Figure 2.
Flowchart of the wavelet support vector machine (W-SVM) based segmentation method.
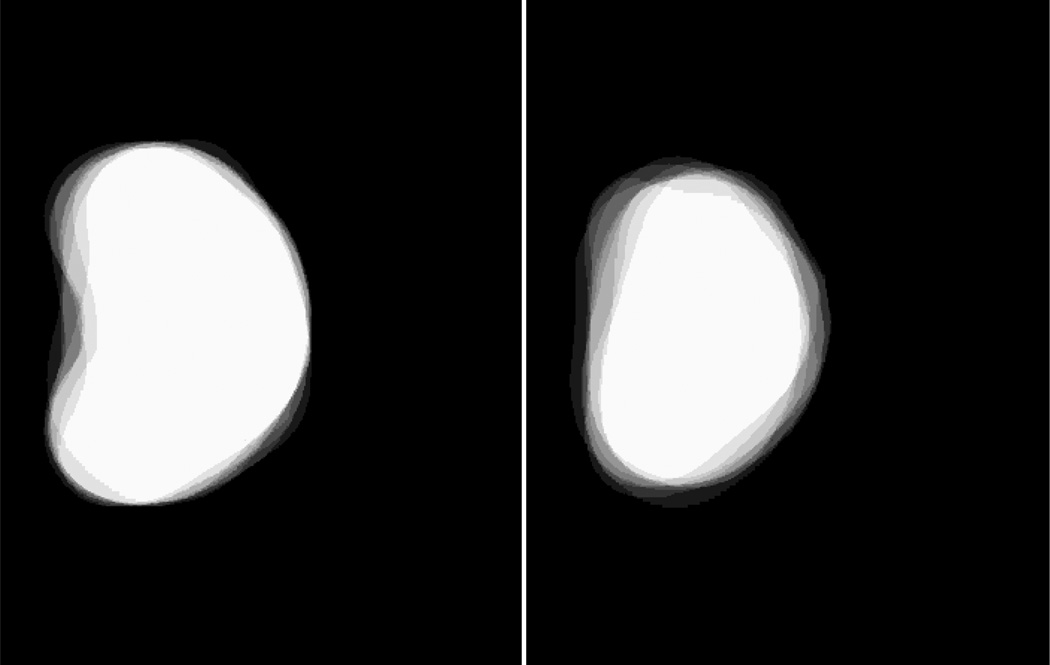
Figure 3.
The 3D probability model in two orthogonal orientations. A white voxel has a probability of 1 as being a prostate voxel while a black voxel has the probability of 0 as a prostate voxel.
Quantitative performance assessment of the method was done by comparing the results with the corresponding gold standard from manual segmentation. The Dice similarity was employed as a performance assessment metric for the prostate segmentation algorithm [46, 47]. Sensitivity represents the proportion of the segmented prostate volume, S, that is correctly overlapped with the gold standard (G), i.e. Sensitivity = TP/G × 100%, where TP is the true positive volume and represents the overlapped volume of the segmented prostate and the gold standard.
2.3 Image registration of TRUS and PET/CT images
Our nonrigid registration method was previously described [33]. Briefly, the registration method is a hybrid approach that simultaneously optimizes the similarities from point-based registration and volume matching methods. The 3D registration is obtained by minimizing the distances of corresponding points at the surface and within the prostate and by maximizing the overlap ratio of the bladder neck on both images. The hybrid approach not only capture deformation at the prostate surface and internal landmarks but also the deformation at the bladder neck regions. The registration uses a soft assignment and deterministic annealing process. The correspondences are iteratively established in a fuzzy-to-deterministic approach. B-splines are used to generate a smooth non-rigid spatial transformation. The similarity function includes three terms: (1) surface landmark matching, (2) internal landmark matching, (3) volume overlap matching. The transformation between CT and TRUS images are represented by a general function, which can be modeled by various function basis. In our study, we choose B-splines as the transformation basis.
3. RESULTS
We developed a PET-directed, 3D ultrasound-guided biopsy system for the prostate. The biopsy system has been tested in prostate phantoms. The proposed 3D segmentation method was applied to 10 TRUS image volumes from 10 human patients. The size of the images was 448×448×350 voxels. The prostate was manually segmented on individual image slices. The experimental data for segmentation differed from the data set for the SVM training. Table 1 shows the quantitative evaluation results from 10 image volumes. The DICE overlap ratios were 92.4% ± 1.1 % and the sensitivity was 93.2% ± 2.6 %.
Table 1.
Quantitative evaluation of four segmentation method.
| Patient# | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | Mean ± STD |
|---|---|---|---|---|---|---|---|---|---|---|---|
| DICE | 92.3% | 91.5% | 93.2% | 91.8% | 92.0% | 93.9% | 90.5% | 92.7% | 92.6% | 94.0% | 92.4% ± 1.1% |
| Sensitivity | 90.4% | 91.2% | 96.6% | 90.8% | 92.9% | 96.9% | 91.4% | 95.2% | 91.2% | 95.5% | 93.2% ± 2.6% |
Our nonrigid registration method was tested with pre- and post-biopsy TRUS data of five patients. We used pre-biopsy images as the reference images and registered the post-biopsy images of the same patient. For five sets of patient data, the target registration error (TRE) was 0.88±0.16 mm and the maximum TRE is 1.08 ± 0.21 mm.
4. DISCUSSION AND CONCLUSION
We developed a molecular image-directed, 3D ultrasound-guided biopsy system for the prostate. In order to include other imaging modality such as PET/CT into 3D ultrasound-guided biopsy, we developed a 3D non-rigid registration method that combines point-based registration and volume overlap matching methods. The registration method was evaluated for TRUS and MR images. The registration method was also used to register 3D TRUS images acquired at different time points and thus can be used for potential use in TRUS-guided prostate re-biopsy. Our next step is to apply this registration method to CT and TRUS images and then incorporate PET/CT images into ultrasound image-guided targeted biopsy of the prostate in human patients. In order to build a 3D model of the prostate, a set of Wavelet-based support vector machines and a shape model are developed and evaluated for automatic segmentation of the prostate TRUS images. Wavelet transform was employed for prostate texture extraction. A probability prostate model was incorporated into the approach to improve the robustness of the segmentation. With the model, even if the prostate has diverse appearance in different parts and weak boundary near bladder or rectum, the method is able to produce a relatively accurate segmentation in 3-D TRUS images.
ACKNOWLEDGEMENT
This research is supported in part by NIH grant R01CA156775 (PI: Fei), Georgia Cancer Coalition Distinguished Clinicians and Scientists Award (PI: Fei), SPORE in Head and Neck Cancer (NIH P50CA128613), and the Emory Molecular and Translational Imaging Center (NIH P50CA128301).
REFERENCES
- 1.Siegel R, Ward E, Brawley O, et al. Cancer statistics, 2011: the impact of eliminating socioeconomic and racial disparities on premature cancer deaths. CA Cancer J Clin. 2011;61(4):212–236. doi: 10.3322/caac.20121. [DOI] [PubMed] [Google Scholar]
- 2.Roehl KA, Antenor JA, Catalona WJ. Serial biopsy results in prostate cancer screening study. J Urol. 2002;167(6):2435–2439. [PubMed] [Google Scholar]
- 3.Schilling D, Schlemmer HP, Wagner PH, et al. Histological verification of 11C-choline-positron emission/computed tomography-positive lymph nodes in patients with biochemical failure after treatment for localized prostate cancer. BJU.Int. 2008;102(4):446–451. doi: 10.1111/j.1464-410X.2008.07592.x. [DOI] [PubMed] [Google Scholar]
- 4.Fei B, Wang H, Wu C, et al. Choline PET for monitoring early tumor response to photodynamic therapy. Journal of Nuclear Medicine. 2010;51(1):130. doi: 10.2967/jnumed.109.067579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DeGrado TR, Coleman RE, Wang S, et al. Synthesis and evaluation of 18F-labeled choline as an oncologic tracer for positron emission tomography: initial findings in prostate cancer. Cancer Res. 2001;61(1):110–117. [PubMed] [Google Scholar]
- 6.Oyama N, Akino H, Kanamaru H, et al. 11C-acetate PET imaging of prostate cancer. J.Nucl.Med. 2002;43(2):181–186. [PubMed] [Google Scholar]
- 7.Nunez R, Macapinlac HA, Yeung HW, et al. Combined 18F-FDG and 11C-methionine PET scans in patients with newly progressive metastatic prostate cancer. J.Nucl.Med. 2002;43(1):46–55. [PubMed] [Google Scholar]
- 8.Schuster DM, Votaw JR, Nieh PT, et al. Initial experience with the radiotracer anti-1-amino-3-18F-fluorocyclobutane-1-carboxylic acid with PET/CT in prostate carcinoma. J.Nucl.Med. 2007;48(1):56–63. [PubMed] [Google Scholar]
- 9.Schuster D, Fei B, Fox T, et al. Histopathologic Correlation of Prostatic Adenocarcinoma on Radical Prostatectomy with Pre-Operative Anti-18F Fluorocyclobutyl-Carboxylic Acid Positron Emission Tomography/Computed Tomography. Laboratory Investigation. 2011;91:222A–223A. [Google Scholar]
- 10.Akbari H, Yang X, Halig L, et al. 3D Segmentation of Prostate Ultrasound Images using Wavelet Transform. Proceedings of SPIE. 2011;7962:79622K. doi: 10.1117/12.878072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Betrouni N, Vermandel M, Pasquier D, et al. Segmentation of abdominal ultrasound images of the prostate using a priori information and an adapted noise filter. Comput.Med.Imaging Graph. 2005;29(1):43–51. doi: 10.1016/j.compmedimag.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 12.Chiu B, Freeman GH, Salama MMA, et al. Prostate segmentation algorithm using dyadic wavelet transform and discrete dynamic contour. Physics in Medicine and Biology. 2004;49(21):4943–4960. doi: 10.1088/0031-9155/49/21/007. [DOI] [PubMed] [Google Scholar]
- 13.Ding MY, Chiu B, Gyacskov I, et al. Fast prostate segmentation in 3D TRUS images based on continuity constraint using an autoregressive model. Medical Physics. 2007;34(11):4109–4125. doi: 10.1118/1.2777005. [DOI] [PubMed] [Google Scholar]
- 14.Ghanei A, Soltanian-Zadeh H, Ratkewicz A, et al. A three-dimensional deformable model for segmentation of human prostate from ultrasound images. Med.Phys. 2001;28(10):2147–2153. doi: 10.1118/1.1388221. [DOI] [PubMed] [Google Scholar]
- 15.Gong LX, Pathak SD, Haynor DR, et al. Parametric shape modeling using deformable superellipses for prostate segmentation. Ieee Transactions on Medical Imaging. 2004;23(3):340–349. doi: 10.1109/TMI.2004.824237. [DOI] [PubMed] [Google Scholar]
- 16.Kachouie NN, Fieguth P. A medical texture local binary pattern for TRUS prostate segmentation. Conf Proc IEEE Eng Med Biol Soc. 2007;2007:5605–5608. doi: 10.1109/IEMBS.2007.4353617. [DOI] [PubMed] [Google Scholar]
- 17.Liu X, Langer DL, Haider MA, et al. Prostate cancer segmentation with simultaneous estimation of Markov random field parameters and class. IEEE Trans Med Imaging. 2009;28(6):906–915. doi: 10.1109/TMI.2009.2012888. [DOI] [PubMed] [Google Scholar]
- 18.Lixin G, Pathak SD, Haynor DR, et al. Parametric shape modeling using deformable superellipses for prostate segmentation. IEEE Transactions on Medical Imaging. 2004;23:340–349. doi: 10.1109/TMI.2004.824237. [DOI] [PubMed] [Google Scholar]
- 19.Nanayakkara ND, Samarabandu J, Fenster A. Prostate segmentation by feature enhancement using domain knowledge and adaptive region based operations. Phys.Med.Biol. 2006;51(7):1831–1848. doi: 10.1088/0031-9155/51/7/014. [DOI] [PubMed] [Google Scholar]
- 20.Pathak SD, Haynor DR, Kim Y. Edge-guided boundary delineation in prostate ultrasound images. IEEE Transactions on Medical Imaging. 2000;19(12):1211–1219. doi: 10.1109/42.897813. [DOI] [PubMed] [Google Scholar]
- 21.Tutar IB, Pathak SD, Gong LX, et al. Semiautomatic 3-D prostate segmentation from TRUS images using spherical harmonics. IEEE Transactions on Medical Imaging. 2006;25(12):1645–1654. doi: 10.1109/tmi.2006.884630. [DOI] [PubMed] [Google Scholar]
- 22.Xu RS, Michailovich O, Salama M. Information tracking approach to segmentation of ultrasound imagery of the prostate. IEEE Trans Ultrason Ferroelectr Freq Control. 2010;57(8):1748–1761. doi: 10.1109/TUFFC.2010.1613. [DOI] [PubMed] [Google Scholar]
- 23.Yan P, Xu S, Turkbey B, et al. Adaptively Learning Local Shape Statistics for Prostate Segmentation in Ultrasound. IEEE Trans Biomed Eng. 2010;58:633–641. doi: 10.1109/TBME.2010.2094195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang F, Suri J, Fenster A. Segmentation of prostate from 3-D ultrasound volumes using shape and intensity priors in level set framework. Conf.Proc.IEEE Eng Med Biol.Soc.2006. 2006;1:2341–2344. doi: 10.1109/IEMBS.2006.260000. [DOI] [PubMed] [Google Scholar]
- 25.Yang XF, Schuster D, Master V, et al. Automatic 3D Segmentation of Ultrasound Images Using Atlas Registration and Statistical Texture Prior. Proceedings of SPIE. 2011;7964:796432. doi: 10.1117/12.877888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu Y, Molloy JA, Acton ST. Segmentation of the prostate from suprapubic ultrasound images. Med.Phys. 2004;31(12):3474–3484. doi: 10.1118/1.1809791. [DOI] [PubMed] [Google Scholar]
- 27.Zhan Y, Shen D. Deformable segmentation of 3-D ultrasound prostate images using statistical texture matching method. IEEE Transactions on Medical Imaging. 2006;25(3):256–272. doi: 10.1109/TMI.2005.862744. [DOI] [PubMed] [Google Scholar]
- 28.Zhang Y, Sankar R, Qian W. Boundary delineation in transrectal ultrasound image for prostate cancer. Comput.Biol.Med. 2007;37(11):1591–1599. doi: 10.1016/j.compbiomed.2007.02.008. [DOI] [PubMed] [Google Scholar]
- 29.Hu N, Downey DB, Fenster A, et al. Prostate boundary segmentation from 3D ultrasound images. Med.Phys. 2003;30(7):1648–1659. doi: 10.1118/1.1586267. [DOI] [PubMed] [Google Scholar]
- 30.Hodge AC, Fenster A, Downey DB, et al. Prostate boundary segmentation from ultrasound images using 2D active shape models: optimisation and extension to 3D. Comput. Methods. Programs. Biomed. 2006;84:99–113. doi: 10.1016/j.cmpb.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 31.Knoll C, Alcaniz M, Grau V, et al. Outlining of the prostate using snakes with shape restrictions based on the wavelet transform (Doctoral Thesis: Dissertation) Pattern Recognition. 1999;32(10):1767–1781. [Google Scholar]
- 32.Fei B, Wang H, Muzic RF, et al. Deformable and rigid registration of MRI and microPET images for photodynamic therapy of cancer in mice. Med Phys. 2006;33(3):753–760. doi: 10.1118/1.2163831. [DOI] [PubMed] [Google Scholar]
- 33.Yang XF, Akbari H, Halig L, et al. 3D Non-rigid Registration Using Surface and Local Salient Features for Transrectal Ultrasound Image-guided Prostate Biopsy. Proceedings of SPIE. 2011;7964:79642V. doi: 10.1117/12.878153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fei B, Duerk JL, Sodee DB, et al. Semiautomatic Nonrigid Registration for the Prostate and Pelvic MR Volumes1. Academic Radiology. 2005;12(7):815–824. doi: 10.1016/j.acra.2005.03.063. [DOI] [PubMed] [Google Scholar]
- 35.Guo Y, Werahera PN, Narayanan R, et al. Image registration accuracy of a 3-dimensional transrectal ultrasound-guided prostate biopsy system. J.Ultrasound Med. 2009;28(11):1561–1568. doi: 10.7863/jum.2009.28.11.1561. [DOI] [PubMed] [Google Scholar]
- 36.Fei B, Lee Z, Duerk JL, et al. Registration and Fusion of SPECT, High Resolution MRI, and interventional MRI for Thermal Ablation of the Prostate Cancer. IEEE Transactions on Nuclear Science. 2004;51(1):177–183. [Google Scholar]
- 37.Karnik VV, Fenster A, Bax J, et al. Evaluation of intersession 3D-TRUS to 3D-TRUS image registration for repeat prostate biopsies. Medical Physics. 2011;38(4):1832–1843. doi: 10.1118/1.3560883. [DOI] [PubMed] [Google Scholar]
- 38.Fei B, Lee S, Boll D, et al. Image Registration and Fusion for Interventional MRI Guided Thermal Ablation of the Prostate Cancer. Medical Image Computing And Computer-Assisted Intervention (MICCAI 2003) - Lecture Notes in Computer Science. 2011;2879:364–372. [Google Scholar]
- 39.Baumann M, Mozer P, Daanen V, et al. Prostate Biopsy Assistance System with Gland Deformation Estimation for Enhanced Precision. Medical Image Computing and Computer-Assisted Intervention (MICCAI 2009) - Lecture Notes in Computer Science. 2009;5761:67–74. doi: 10.1007/978-3-642-04268-3_9. [DOI] [PubMed] [Google Scholar]
- 40.Fei B, Duerk JL, Wilson DL. Automatic 3D registration for interventional MRI-guided treatment of prostate cancer. Computer Aided Surgery. 2002;7(5):257–267. doi: 10.1002/igs.10052. [DOI] [PubMed] [Google Scholar]
- 41.Hummel J, Figl M, Bax M, et al. 2D/3D registration of endoscopic ultrasound to CT volume data. Phys Med Biol. 2008;53(16):4303–4316. doi: 10.1088/0031-9155/53/16/006. [DOI] [PubMed] [Google Scholar]
- 42.Molloy JA, Oldham SA. Benchmarking a novel ultrasound-CT fusion system for respiratory motion management in radiotherapy: Assessment of spatio-temporal characteristics and comparison to 4DCT. Medical Physics. 2008;35(1):291–300. doi: 10.1118/1.2818732. [DOI] [PubMed] [Google Scholar]
- 43.Crocetti L, Lencioni R, Debeni S, et al. Targeting liver lesions for radipfrequency ablation - An experimental feasibility study using a CT-US fusion imaging system. Investigative Radiology. 2008;43(1):33–39. doi: 10.1097/RLI.0b013e31815597dc. [DOI] [PubMed] [Google Scholar]
- 44.Huang X, Moore J, Guiraudon G, et al. Dynamic 2D Ultrasound and 3D CT Image Registration of the Beating Heart. IEEE Trans Med Imaging. 2009;28:1179–1189. doi: 10.1109/TMI.2008.2011557. [DOI] [PubMed] [Google Scholar]
- 45.Barratt DC, Chan CS, Edwards PJ, et al. Instantiation and registration of statistical shape models of the femur and pelvis using 3D ultrasound imaging. Med.Image Anal. 2008;12(3):358–374. doi: 10.1016/j.media.2007.12.006. [DOI] [PubMed] [Google Scholar]
- 46.Wang HS, Fei BW. A modified fuzzy C-means classification method using a multiscale diffusion filtering scheme. Medical Image Analysis. 2009;13(2):193–202. doi: 10.1016/j.media.2008.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang XF, Fei BW. A multiscale and multiblock fuzzy C-means classification method for brain MR images. Medical Physics. 2011;38(6):2879–2891. doi: 10.1118/1.3584199. [DOI] [PMC free article] [PubMed] [Google Scholar]