Abstract
We are developing a molecular image-directed, 3D ultrasound-guided, targeted biopsy system for improved detection of prostate cancer. In this paper, we propose an automatic 3D segmentation method for transrectal ultrasound (TRUS) images, which is based on multi-atlas registration and statistical texture prior. The atlas database includes registered TRUS images from previous patients and their segmented prostate surfaces. Three orthogonal Gabor filter banks are used to extract texture features from each image in the database. Patient-specific Gabor features from the atlas database are used to train kernel support vector machines (KSVMs) and then to segment the prostate image from a new patient. The segmentation method was tested in TRUS data from 5 patients. The average surface distance between our method and manual segmentation is 1.61 ± 0.35 mm, indicating that the atlas-based automatic segmentation method works well and could be used for 3D ultrasound-guided prostate biopsy.
Keywords: Automatic 3D segmentation, atlas registration, Gabor filter, support vector machine, ultrasound imaging, prostate cancer
1. INTRODUCTION
Prostate cancer affects one in six men [1]. Systematic transrectal ultrasound (TRUS)-guided biopsy is considered as the standard method for definitive diagnosis of prostate cancer. However, the current biopsy technique has a significant sampling error and can miss up to 30% of cancers. We are developing a molecular image-directed, 3D ultrasound-guided biopsy system with the aim of improving cancer detection rate. Accurate segmentation of the prostate plays a key role in biopsy needle placement [2], treatment planning [3], and motion monitoring [4]. As ultrasound images have a relatively low signal-to-noise ratio, automatic segmentation of the prostate is difficult. However, manual segmentation during biopsy or treatment can be time consuming. We are developing automated methods to address this technical challenge.
A number of segmentation methods have been reported for TRUS images [4–7]. A semiautomatic method by warping an ellipse to fit the prostate on TRUS images was presented [8,9]. A 2D semiautomatic discrete dynamic contour model was used to segment the prostate [10]. A level set based method [11,12] and shape model-based minimal path method [13] also were used to detect the prostate surface from 3D ultrasound images. Gabor support vector machine (G-SVM) and statistical shape model were used to extract the prostate boundary [2,14]. In this paper, we propose an automatic 3D segmentation method based on atlas registration and statistical texture priors. Our method does not require initialization of a shape model but use a subject-specific prostate atlas to train SVMs. The detailed steps of our method and its evaluation results are reported in the following sections.
2. METHODS
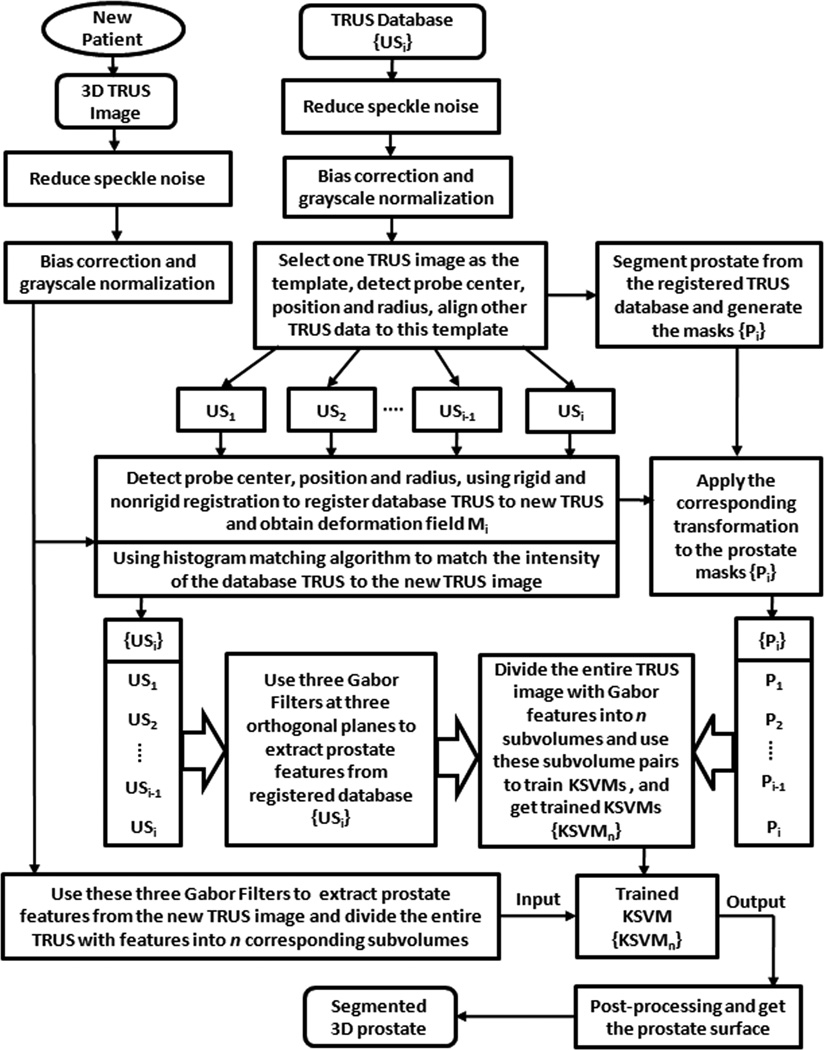
Our segmentation method consists of three major components: (1) Atlas-based registration, (2) Hierarchical representation of image features using Gabor filter banks, and (3) Training of kernel support vector machine (KSVM). Fig. 1 shows a schematic flow chart of our method. The steps are briefly described below.
Build a 3D TRUS image database. Every 3D TRUS image in the database will be preprocessed. In order to align all data in the database, one TRUS image is randomly selected as the template, and others are registered to the template. The prostate surface is manually segmented from all registered TRUS data, which will be used as the prostate mask in Step 3. The atlas database includes pairs of 3D TRUS and the segmented prostate surface from 15 patients.
Register the images in the atlas database to a newly acquired TRUS image. We previously developed several registration methods for the prostate [15–20]. We first align this template to the new data, and then apply this rigid-body transformation matrix to all other data in the atlas database. Deformable registration methods are then used to obtain the spatial deformation field between the new TRUS image and the images in the database. The same transformation is applied to the segmented prostate surface in the database. Use a histogram matching algorithm, the intensity of every TRUS image in the database will be matched to the intensity of the new TRUS image.
Apply three Gabor filters at three orthogonal planes to extract prostate features from the registered image in the database as well as from the newly acquired TRUS image. Every TRUS image is divided into n subvolumes overlapped with each other. Every subvolume has three corresponding subvolumes with Gabor features at different planes. The three subvolumes are combined as the input of a training pairs for KSVM [15]. The corresponding subvolume at the same position in the prostate mask is used as the output of the training pairs. KSVM is applied to each subvolume training pairs with Gabor texture features. The KSVMs are then applied to the corresponding subvolumes from the newly acquired TRUS image in order to segment the prostate of the new patient.
Figure. 1.
Schematic flow chart of the proposed algorithm for the 3D prostate segmentation.
We validate our segmentation methods using a variety of evaluation methods [15,21–23]. Particularly, our segmentation is compared with the manual results. In order to get a quantitative evaluation of this comparison, we used the Dice overlap ratio and volume overlap error as defined below:
| (1) |
| (2) |
where V1 and V2 are binary prostate segmented volumes.
3. RESULTS
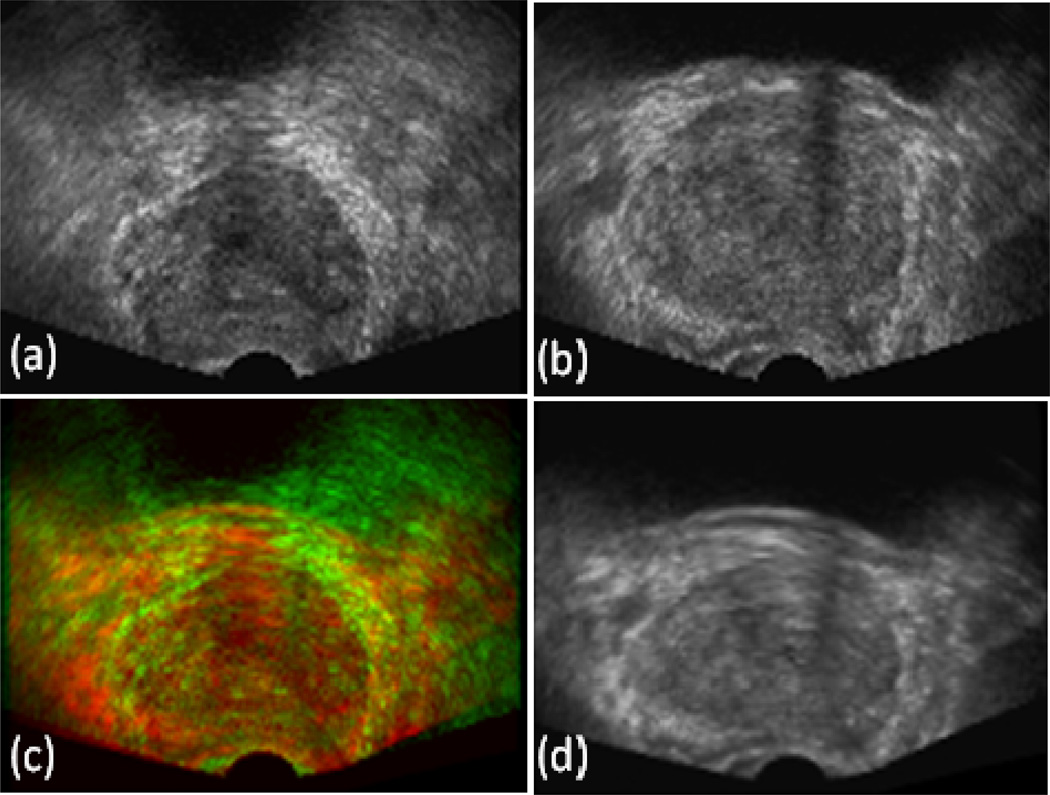
We apply the segmentation method to TRUS images of five patients (matrix: 448×448×350, 0.190×0.190×0.195 mm3/voxel). In our implementation, 36 KSVMs are attached to 36 subvolumes. The number of orientations and scales are 6 and 3 for the Gabor filter, respectively. In our experiment, five images were used for evaluation. Fig. 2 shows the non-rigid registration result between the newly acquired TRUS image and one image in the atlas database. As different patients have different sizes of the prostate and their TRUS images may be acquired at slightly different position and orientation, the non-rigid registration (translations, rotations, scaling, and deformation) is able to normalize the image with respect to the template and thus build the atlas database. As demonstrated in Fig. 3 and Fig. 4, the proposed automatic segmentation method works well for 3D TRUS images of the prostate and achieved similar results as compared to manual segmentation. Table 1 provides quantitative evaluation of the segmentation method for five patients.
Figure. 2.
3D TRUS registration. (a) Reference image, (b) Floating image, (d) Registered image, and (c) Fusion of the reference and registered images.
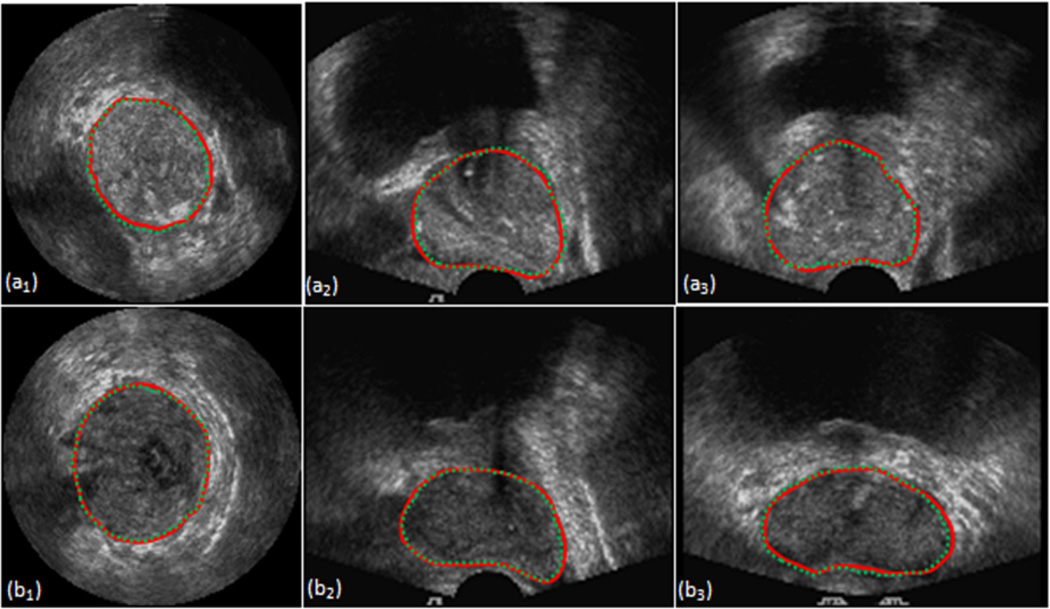
Figure. 3.
Comparison between proposed and manual segmentation. Images on the top and bottom are from two patients. Images from left to right are in three orientations of the same TRUS image volume. The line in red is the manual segmentation result. The dashed line in green is the segmentation result of the proposed method.
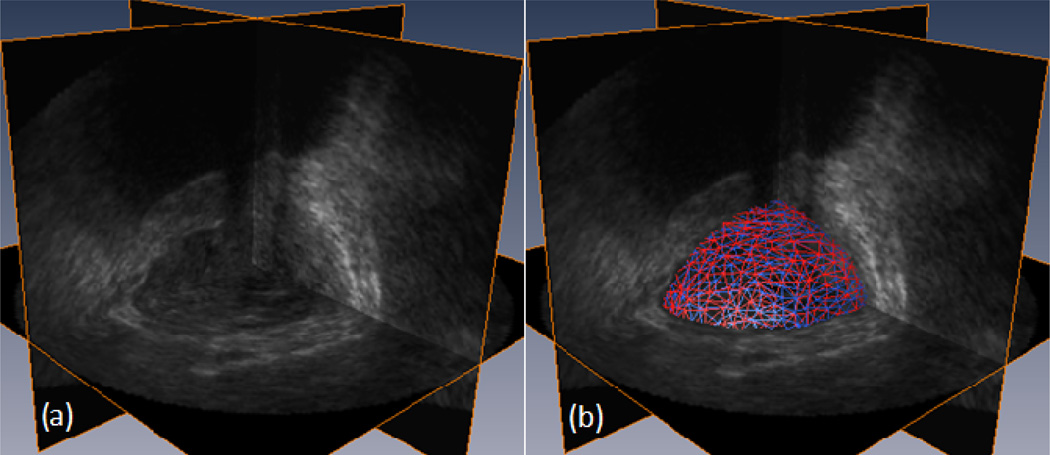
Figure. 4.
3D visualization of TRUS images and the segmented prostate, (a) Original 3D TRUS images. (b) Segmented prostate by manual (red) and by our method (blue).
Table 1.
Comparison between our and manual segmentation of images from 5 patients
| Patient | Surface Distance (mm) | Dice (%) | Error (%) | ||
|---|---|---|---|---|---|
| Average | RMS | Max | |||
| 01 | 1.19 | 1.14 | 3.57 | 92.06 | 13.87 |
| 02 | 1.86 | 1.76 | 5.31 | 90.25 | 17.65 |
| 03 | 1.36 | 1.56 | 3.91 | 91.38 | 15.64 |
| 04 | 1.57 | 1.73 | 6.40 | 91.26 | 16.16 |
| 05 | 2.05 | 2.45 | 6.01 | 89.08 | 18.90 |
| Mean±Std | 1.61±0.35 | 1.72±0.47 | 5.04±1.26 | 90.81±1.16 | 16.44±1.93 |
4. DISCUSSIONS AND CONCLUSIONS
We propose an automatic 3D segmentation method based on atlas registration and statistical texture priors. Patient-specific Gabor features from the atlas database are used to train kernel support vector machines (KSVMs) and then to segment the prostate image from a new patient. Our method is based on multi-atlas segmentation but is different from standard atlas-based segmentation. It does not require any initialization but uses a subject-specific prostate atlas database to train KSVMs, and uses machine-learning to combine registered atlas database. Validation on 5 patients’ images shows that our approach is able to accurately segment the prostate from TRUS images. As a patient-specific TRUS atlas database is used to train kernel support vector machines for automatic segmentation, the robustness and accuracy of the algorithm is improved. The segmentation reliability does not depend on any initialization step. We are exploring parallel processing for the registration tasks in order to integrate the proposed segmentation method into our 3D ultrasound-guided biopsy system.
ACKNOWLEDGEMENT
This research is supported in part by NIH grant R01CA156775 (PI: Fei), Coulter Translational Research Grant (Pis: Fei and Hu), Georgia Cancer Coalition Distinguished Clinicians and Scientists Award (PI: Fei), Emory Molecular and Translational Imaging Center (NIH P50CA128301), and Atlanta Clinical and Translational Science Institute (ACTSI) that is supported by the PHS Grant UL1 RR025008 from the Clinical and Translational Science Award program.
References
- 1.Prostate Cancer Foundation, http://www.prostatecancerfoundation.org/ 2008
- 2.Shen D, Zhan Y, Davatzikos C. Segmentation of prostate boundaries from ultrasound images using statistical shape model. IEEE Trans. Med. Imaging. 2003;22(4):539–551. doi: 10.1109/TMI.2003.809057. [DOI] [PubMed] [Google Scholar]
- 3.Hodge KK, McNeal JE, Terris MK, Stamey TA. Random systematic versus directed ultrasound guided transrectal core biopsies of the prostate. J. Urol. 1989;142:71–74. doi: 10.1016/s0022-5347(17)38664-0. [DOI] [PubMed] [Google Scholar]
- 4.Yan P, Xu S, Turkbey B, Kruecker J. Discrete Deformable Model Guided by Partial Active Shape Model for TRUS Image Segmentation. IEEE Transactions on Biomedical Engineering. 2010;57:1158–1166. doi: 10.1109/TBME.2009.2037491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Akbari H, Yang X, Halig L, Fei B. 3D segmentation of prostate ultrasound images using wavelet transform. SPIE Medical Imaging 2011: Image Processing. 2011;7962 doi: 10.1117/12.878072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Noble JA, Boukerroui D. Ultrasound image segmentation: a survey. IEEE Trans. Med. Imaging. 2006;25:987–1010. doi: 10.1109/tmi.2006.877092. [DOI] [PubMed] [Google Scholar]
- 7.Yang F, Suri J, Fenster A. Segmentation of prostate from 3-D ultrasound volumes using shape and intensity priors in level set framework. Conf. Proc. IEEE Eng Med Biol. Soc. 2006;1:2341–2344. doi: 10.1109/IEMBS.2006.260000. [DOI] [PubMed] [Google Scholar]
- 8.Badiei S, Salcudean SE, Varah J, Morris WJ. Prostate segmentation in 2D ultrasound images using image warping and ellipse fitting. MICCAI. 2006;4191:17–24. doi: 10.1007/11866763_3. [DOI] [PubMed] [Google Scholar]
- 9.Lixin G, Pathak SD, Haynor DR, Cho PS, Yongmin K. Parametric shape modeling using deformable superellipses for prostate segmentation, Medical Imaging. IEEE Transactions on. 2004;23(3):340–349. doi: 10.1109/TMI.2004.824237. [DOI] [PubMed] [Google Scholar]
- 10.Ladak HM, Mao F, Wang Y, Downey DB, Steinman DA, Fenster A. Prostate boundary segmentation from 2D ultrasound images. Med. Phys. 2000;27(8):1777–1788. doi: 10.1118/1.1286722. [DOI] [PubMed] [Google Scholar]
- 11.Shao F, ling KV, Ng WS. 3D Prostate Surface Detection from Ultrasound Images Based on Level Set Method. MICCAI. 2003;4191:389–396. [Google Scholar]
- 12.Zhang H, Bian Z, Guo Y, Fei B, Ye M. An efficient multiscale approach to level set evolution. Proceedings of the 25th Annual International Conference of the IEEE Engineering in Medicine and Biology Society. 2003;1:694–697. [Google Scholar]
- 13.Guo S, Fei B. A minimal path searching approach for active shape model (ASM)-based segmentation of the lung. Proc. SPIE 2009: Image Processing. 2009;7259(1):72594B. doi: 10.1117/12.812575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhan Y, Shen D. Deformable segmentation of 3-D ultrasound prostate images using statistical texture matching method. IEEE Trans. Med. Imaging. 2006;25(3):256–272. doi: 10.1109/TMI.2005.862744. [DOI] [PubMed] [Google Scholar]
- 15.Akbari H, Kosugi Y, Kojima K, Tanaka N. Detection and analysis of the intestinal ischemia using visible and invisible hyperspectral imaging. IEEE Trans Biomed. Eng. 2010;57(8):2011–2017. doi: 10.1109/TBME.2010.2049110. [DOI] [PubMed] [Google Scholar]
- 16.Fei B, Lee Z, Boll DT, Duerk JL, Lewin JS, ilson DL. Image Registration and Fusion for Interventional MRI Guided Thermal Ablation of the Prostate Cancer. The Sixth Annual International Conference on Medical Imaging Computing & Computer Assisted Intervention (MICCAI 2003) - published in Lecture Notes in Computer Science (LNCS) 2003;2879:364–372. [Google Scholar]
- 17.Fei B, Lee Z, Duerk JL, Wilson DL. Image Registration for Interventional MRI Guided Procedures: Similarity Measurements, Interpolation Methods, and Applications to the Prostate. The Second International Workshop on Biomedical Image Registration - Published in Lecture Notes in Computer Science (LNCS) 2003;2717:321–329. [Google Scholar]
- 18.Fei B, Lee Z, Duerk JL, Lewin J, Sodee D, Wilson DL. Registration and Fusion of SPECT, High Resolution MRI, and interventional MRI for Thermal Ablation of the Prostate Cancer. IEEE Transactions on Nuclear Science. 2004;51(1):177–183. [Google Scholar]
- 19.Fei B, Duerk JL, Sodee DB, Wilson DL. Semiautomatic nonrigid registration for the prostate and pelvic MR volumes. Acad. Radiol. 2005;12:815–824. doi: 10.1016/j.acra.2005.03.063. [DOI] [PubMed] [Google Scholar]
- 20.Yang X, Akbari H, Halig L, Fei B. 3D Non-rigid Registration Using Surface and Local Salient Features for Transrectal Ultrasound Image-guided Prostate Biopsy. SPIE Medical Imaging 2011: Visualization, Image-Guided Procedures, and Modeling. 2011;7964 doi: 10.1117/12.878153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang H, Feyes D, Mulvihill J, Oleinick N, MacLennan G, Fei B. Multiscale fuzzy C-means image classification for multiple weighted MR images for the assessment of photodynamic therapy in mice. Proceedings of the SPIE - Medical Imaging. 2007;6512 doi: 10.1117/12.710188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang H, Fei B. A modified fuzzy C-means classification method using a multiscale diffusion filtering scheme. Med. Image Anal. 2009;13(2):193–202. doi: 10.1016/j.media.2008.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang X, Fei B. A skull segmentation method for brain MR images based on multiscale bilateral filtering scheme. Proc. SPIE 2010: Image Processing. 2010;7623(1):76233K. [Google Scholar]