SUMMARY
Structural studies of human G protein-coupled receptors (GPCRs) have recently been accelerated through the use of the T4 lysozyme fusion partner that was inserted into the third intracellular loop. Using chimeras of the human β2-adrenergic and human A2A adenosine receptors, we present the methodology and data for the selection of five new fusion partners for crystallizing GPCRs. In particular, the use of the thermostabilized apocytochrome b562RIL as a fusion partner displays certain advantages over the previously utilized T4 lysozyme, resulting in a significant improvement in stability and structure in GPCR-fusion constructs.
Keywords: GPCR, stabilization, chimera, adenosine receptor, fusion domain, protein engineering
INTRODUCTION
G protein-coupled receptors (GPCRs) encompass the largest family of cell surface receptors, and are responsible for the cell’s ability to recognize a variety of external stimuli, including hormones, neurotransmitters, lipids, peptides and light (Bjarnadottir et al., 2006). Despite the enormous efforts devoted to studying the biology of these receptors, progress on their structural characterization remains comparatively slow. Major hurdles associated with crystallizing GPCRs manifest at all stages in the process, including protein expression, purification, and crystallization. Receptors of immediate interest may not always be easily expressed to sufficient quantities required for crystallization studies, and even if that hurdle is overcome, the stability of GPCRs may be compromised at the purification stage, during which the protein must be removed from its native bilayer environment (Chae et al., 2010; Serrano-Vega et al., 2008). Additionally, GPCRs are inherently dynamic proteins, sampling a range of conformations between active and inactive signaling states. This flexibility, along with the scarcity of hydrophilic regions available to form potential crystal contacts, are major impediments to obtaining diffraction quality GPCR crystals.
The level of difficulty associated with GPCR crystallization is evidenced by the fact that despite significant effort, rhodopsin is the only wild-type (WT) receptor to have been successfully crystallized (Palczewski et al., 2000). For all other GPCRs, crystallization has required protein engineering using a combination of the following approaches: 1) truncation of flexible N- and/or C-terminal domains, 2) implementation of point mutations in order to stabilize the receptor in a specific conformation (Lebon et al., 2011; Miller and Tate, 2011; Serrano-Vega et al., 2008), and 3) insertion of T4 Lysozyme (T4L) into the third intracellular loop (ICL3) (Engel et al., 2002; Rosenbaum et al., 2007), or more recently at the receptor N-terminus (Rasmussen et al., 2011b). While the truncation of termini has been employed for most GPCR constructs, successful crystallization has also required further protein engineering or the addition of stabilizing antibodies as in case of the β2-adrenergic receptor (β2AR) (Cherezov et al., 2007; Day et al., 2007), the first non-rhodopsin GPCR structure to be determined. More recently, the antibody method has also successfully enabled the crystallization of the A2A adenosine receptor (A2AAR) (Hino et al., 2012). Point mutations found via exhaustive alanine-scanning technology helped to stabilize and crystallize the β1-adrenergic receptor (β1AR) (Warne et al., 2008), as well as A2AAR (Dore et al., 2011; Lebon et al., 2011). To date, the most successful methodology has been the T4L engineering method, which enabled the crystallization of the β2AR (Alexandrov et al., 2008; Cherezov et al., 2007; Rasmussen et al., 2011a; Wacker et al., 2010), A2AAR (Jaakola et al., 2008; Xu et al., 2011b), chemokine CXCR4 receptor (Wu et al., 2010), dopamine D3 receptor (Chien et al., 2010), histamine H1 receptor (Shimamura et al., 2011), sphingosine phosphate S1P1 receptor (Hanson et al., 2012), muscarinic acetylcholine M2 (Haga et al., 2012) and M3 (Kruse et al., 2012) receptors, μ-opioid receptor (Manglik et al., 2012), and κ-opioid receptor (Wu et al., 2012).
While the replacement of ICL3 with T4L has already proven its utility in GPCR crystallization, we found that a number of GPCRs were not amenable to this approach due to deleterious effects on the expression or stability of the chimeric protein. With approximately 800 GPCRs in humans (Bjarnadottir et al., 2006) and a growing demand for higher resolution structures, the determination of GPCR crystal structures remains in its infancy. The continuous development of new approaches in order to expand the repertoire of GPCR crystallization tools will be paramount in moving the field forward. Here we report the identification and development of novel GPCR fusion proteins to facilitate GPCR crystallization efforts. Starting with a search of the Protein Data Bank (PDB) and past literature of non GPCR fusion proteins (Engel et al., 2002; Prive et al., 1994), a number of fusion candidates were evaluated that possessed characteristics deemed suitable for enhancing crystallizability of membrane proteins. From this effort five new fusion proteins were selected and examined. Of the five considered in this study, thermostabilized apocytochrome b562RIL showed great utility in the crystallization of multiple GPCRs with superior characteristics relative to T4L that has been used previously.
RESULTS
Selection of chimeric partners and design of constructs
ICL3 has highly variable length and pronounced structural flexibility in many GPCRs as shown by its high proteolytic susceptibility and high hydrogen-deuterium exchange rate (West et al., 2011). When present in a construct sequence, it is often found disordered in crystallized GPCRs (Palczewski et al., 2000; Warne et al., 2008). Based on past successes, and for the reasons mentioned above, this receptor region was therefore chosen as the site for new fusion partner insertion. Optimal insertion into ICL3 requires the N- and C- termini of a fusion partner to be separated by a distance similar to the distance between the intracellular ends of helices V and VI (6-14 Å) as found in existing GPCR structures. This allows the new domain to be inserted without significantly disrupting the fold of the fusion domain or the core structure of the membrane spanning helices in the GPCR of interest.
We used the following search criteria to define fusion partners from suitable single domain proteins in the PDB: 1) crystal structure at higher than 2.0 Å resolution and mean B-factor lower than 40, factors that usually correlate with an acceptable stability of the domain and its propensity to form crystal contacts; 2) a molecular weight between 5-20 kD, to exclude both small unstable peptides and large/multidomain proteins with complex folding kinetics; 3) a difference between the number of basic and acidic residues N(Arg,Lys) - N(Asp,Glu) in the range of −20 to 10, to exclude highly charged protein domains; and 4) a distance between the N- and C-terminal Cα atoms of less than 15 Å for optimal insertion. The search was also limited to bacterial proteins to avoid issues with posttranslational modification, and to entries with a single protein chain to eliminate oligomerization interfaces. A search of the PDB with the above criteria yielded 193 non-redundant protein domains (Table S1). Further selection of the domains was performed manually by first excluding domain fragments (except for the C-terminal fragment of T4 lysozyme) and uncharacterized protein hits to narrow the list to proteins that have been previously characterized biochemically and validated to be members of their predicted family. Greater weight was placed on hits that had high representation within the PDB and the availability of apo X-ray or NMR structural data. Structures were manually reviewed to ensure that the N- and C-termini were not buried or part of a highly structured region of the fold. Additionally, disulfide containing proteins were eliminated due to the cytoplasmic location of the ICL3 fusions. From that reduced set, emphasis was placed on maximizing diversity of both fold and molecular weight.
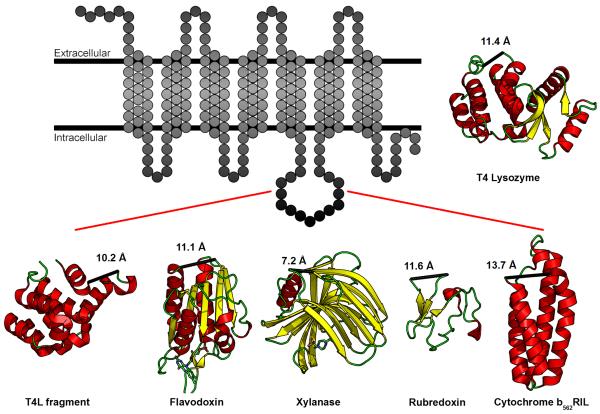
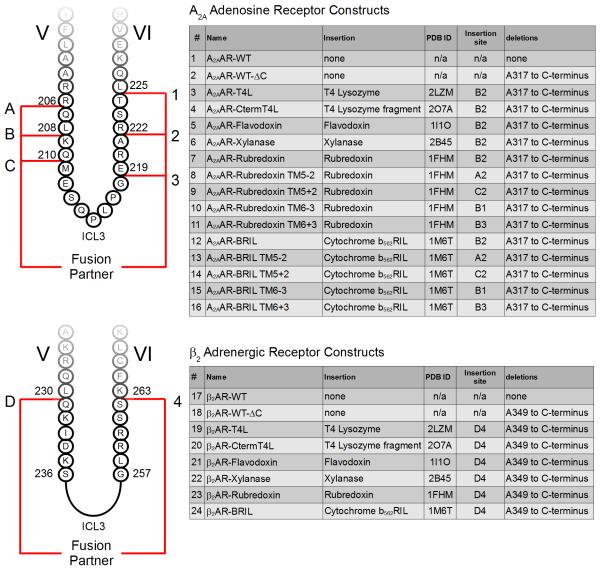
To test the selection criteria, five initial fusion partners were considered: a C-terminal fragment of T4 lysozyme (with mutation C39A, PDB ID 2O7A, henceforth referred to as CtermT4L), flavodoxin (PDB ID 1I1O), xylanase (PDB ID 2B45), rubredoxin (PDB ID 1FHM), and thermostabilized apocytochrome b562 (with mutations M7W, H102I, R106L, PDB ID 1M6T, henceforth referred to as BRIL), (Figure 1). This set of domains exhibits a variety of secondary structure elements ranging from being almost completely composed of loops (rubredoxin), β-sheets (xylanase), α-helices (BRIL and CtermT4L), and a combination of all these elements (flavodoxin). The ICL3 of A2AAR and β2AR were replaced with each of these domains to assess their utility in expression, purification, stability, and crystallization. For a complete list of constructs and details regarding insertion sites into the respective receptors, please refer to Figure 2. Construct numbers from Figure 2 will also be referenced in the remainder of the text to preclude ambiguity.
Figure 1. Five fusion domains selected for fusion into the third intracellular loop of A2AAR and β2AR.
Figure illustrating the insertion of five new domains into the ICL3 of a prototypical GPCR, represented as a transmembrane snakeplot. The five domains are a C-terminal fragment of T4L (PDB ID 2O7A, MW 15.9 kD), flavodoxin (PDB ID 1I1O, MW 14.9 kD), xylanase (PDB ID 2B45, MW 19.1 kD), rubredoxin (PDB ID 1FHM, MW 5.5 kD), and cytochrome b562RIL (PDB ID 1M6T, MW 10.9). These domains exhibit a variety of secondary structures consisting of either α-helices, β-sheets, or a combination of both. Numbers indicate distance (Å) between the N- and C-termini of each domain. T4 Lysozyme (PDB ID 3G3V, MW 18.6 kD) is shown for reference. See also Table S1.
Figure 2. Diagram of fusion partner insertion sites for both A2AAR and β2AR constructs.
The construct number in the first column as well as the alphanumeric insertion site are used as a reference for all the constructs referred to in the main text. For example, insertion site B2 refers to the initial insertion site of T4L into A2AAR-T4L, between residues L208 and R222.
Assessment of A2A adenosine receptor chimeras
Expression and purification
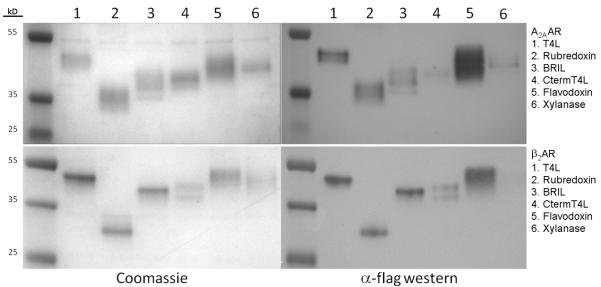
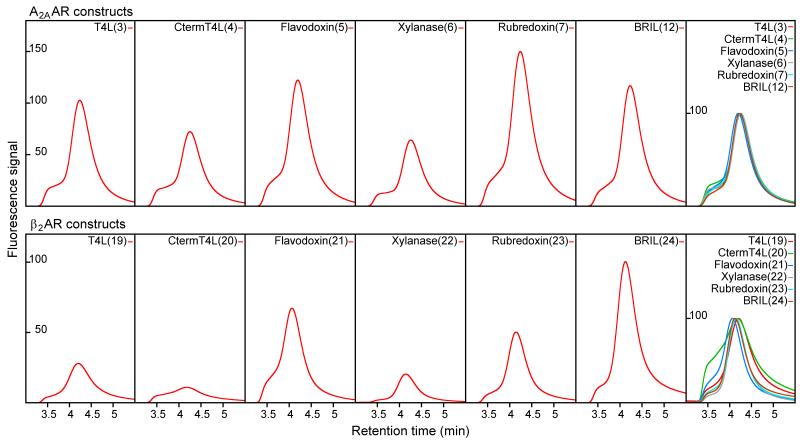
Spodoptera frugiperda (Sf9) cells expressing the A2AAR chimeras were disrupted by repeated dounce homogenization, followed by solubilization with 0.5% (w/v) n-Dodecyl-β-D-Maltoside (DDM, Affymetrix) and 0.1% (w/v) cholesteryl hemisuccinate (CHS, Sigma). Solubilized chimeras were purified by immobilized metal affinity column (IMAC) purification against a C-terminal 10x His tag. Among the five new A2AAR chimeric constructs, A2AAR-rubredoxin, A2AAR-BRIL, and A2AAR-flavodoxin had greater recovery of purified protein compared to that of A2AAR-CtermT4L and A2AAR-xylanase as evidenced by immunoblotting (Figure 3). Analytical size exclusion chromatography (aSEC) of the five A2AAR chimeras showed that each eluted mainly as a single peak, preceded by a minor component, which we attributed to protein aggregation (Figure 4). While the elution profiles of each construct were very similar in terms of shape, the relative peak sizes corroborated the better purification yield of A2AAR-flavodoxin, A2AAR-rubredoxin, and A2AAR-BRIL. Due to the inferior purification recovery of A2AAR-CtermT4L and A2AAR-xylanase, these two constructs were removed from consideration in subsequent assays where purified protein was required.
Figure 3. Purified chimeras analyzed by SDS-PAGE and visualized by coomassie staining and α-flag Western Immunoblots.
Coomassie stained gels are on the left panels, and the equivalent Western Immunoblots on the right. Multiple receptor bands are in part due to differential glycosylation states of the receptor, and can be consolidated after deglycosylation with PNGaseF and reduction by reducing agent (Figure S1).
Figure 4. Analytical size exclusion chromatography analysis of each chimera.
Signals represent fluorescence emission at 350 nm. The rightmost box of each row shows the normalized and overlaid profiles for comparative purposes. Construct numbers are in parenthesis after construct name.
Thermostability
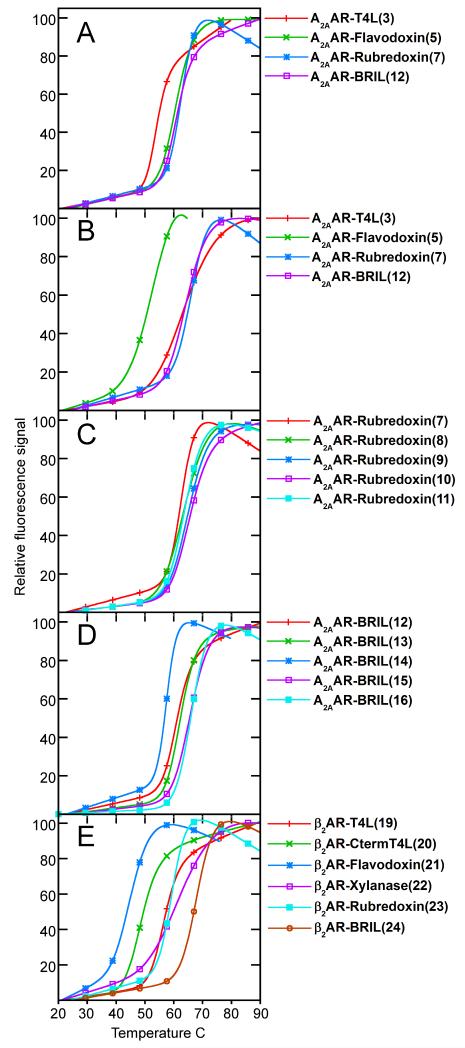
Due to their inherent flexibility, high thermostability is an important metric in the crystallization of GPCRs. To test the effect of the fusion domains on the protein, a fluorescence based thermostability assay (Alexandrov et al., 2008) was carried out. A2AAR constructs were incubated with either an antagonist, ZM241385, or an agonist, UK432,097, to stabilize the receptor prior to heating. We found that when bound with ZM241385, A2AAR-flavodoxin, A2AAR-rubredoxin (construct 7), and A2AAR-BRIL (construct 12) all had almost identical thermal transition temperatures, about 6 °C higher than that of the previously studied A2AAR-T4L (Jaakola et al., 2008) (Figure 5A). When bound with UK432,097, A2AAR-BRIL and A2AAR-rubredoxin showed similar thermal transition temperatures, while the transition temperature of A2AAR-flavodoxin was reduced by over 10 °C when compared with A2AAR-T4L (Figure 5B). The improved thermostability of A2AAR-BRIL (construct 12) and A2AAR-rubredoxin (construct 7) in either antagonist or agonist bound conformations makes them attractive candidates for crystallization trials, as the availability of multiple highly stabilizing ligands increases the likelihood of finding a successful crystallizing condition. A2AAR-BRIL and A2AAR-rubredoxin were thus selected for further crystallization studies.
Figure 5. Normalized fluorescence based thermostability profiles of receptor-fusion chimeras.
Thermostability profiles for (A) initial A2AAR chimera screen (constructs 3, 5, 7, 12) with ZM241385, (B) initial A2AAR chimera screen (constructs 3, 5, 7, 12) with UK432,097, (C) junction optimization for A2AAR-Rubredoxin with ZM241385 (constructs 7-11), (D) junction optimization for A2AAR-BRIL with ZM241285 (constructs 12-16), and (E) initial β2AR chimera (constructs 19-24) screen with timolol. Construct numbers in parenthesis after name.
Junction optimization
Initial crystallization trials with either A2AAR-rubredoxin (construct 7) or A2AAR-BRIL (construct 12) in lipidic cubic phase (LCP) did not produce any crystals using crystallization screens described previously (Xu et al., 2011a). While the reason for the absence of crystal growth cannot be known for certain, we further optimized these two A2AAR chimeras by modifying the junction between the receptor and the insertion domain. The interface on the A2AAR side of the junction was originally optimized for the insertion of T4L, which has a different distance between the N- and C- termini (10.1 Å) than that of either rubredoxin (11.6 Å) or BRIL (13.7 Å). The secondary structure at the interface is also different between all three domains: the T4L interface consists of two α-helices perpendicular to each other, BRIL consists of two adjacent α-helices anti-parallel to each other, and rubredoxin is composed of loops. These considerations led us to test if adding or removing native residues on the A2AAR side of the junction could improve the thermostability of the protein. Multiple alternative junction sites that improved the stability of both A2AAR-rubredoxin and A2AAR-BRIL (Figure 5 C,D) were identified. Of these, the most stabilizing junctions for A2AAR-rubredoxin were the addition of 2 residues on helix V (construct 9) or the removal of 3 residues on helix VI (construct 10), both of which raised the stability of the protein by approximately 4 °C. For A2AAR-BRIL, the most stabilizing junctions were either the removal (construct 15) or addition (construct 16) of 3 residues on the helix VI side of the junction, which increased the stability of the protein by approximately 4 °C.
Protein diffusion and crystallization in LCP
The thermostabilized A2AAR-BRIL (construct 16) and A2AAR-rubredoxin (construct 7) constructs in complex with ZM241385 were further assessed by the high-throughput LCP-FRAP assay (Xu et al., 2011a). Proteins were preferentially labeled at the N-terminus with Cy3-mono NHS ester at pH 7.5 to minimize interferences with the protein core. All protein samples were evaluated for purity, monodispersity, and labeling efficiency by aSEC prior to LCP-FRAP sample preparation. Labeled protein was reconstituted in LCP by mixing protein solution with molten lipid in a final ratio of 40% (w/w) protein solution, 54% (w/w) monoolein, 6% (w/w) cholesterol, and incubated with homemade screens as introduced previously (Xu et al., 2011a). The pH for the screens were adjusted to 4.0, 5.0 and 6.0 based on previous A2AAR crystallization conditions (Jaakola et al., 2008; Xu et al., 2011b).
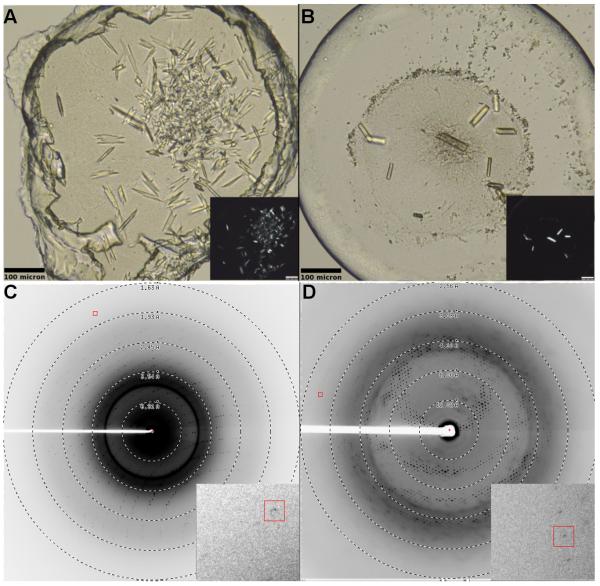
A2AAR-BRIL showed optimal mobile fractions at pH 5.0, which reached 70% diffusion recovery with certain precipitants (Figure S2). We found that the average mobile fraction of A2AAR-BRIL throughout the 96-well plate was consistently higher than that of A2AAR-rubredoxin (Figure S2 C), and proceeded with A2AAR-BRIL for further crystallization trials with a focus on conditions that produced a mobile fraction recovery higher than 70%, including citrate, tartrate, nitrate, and thiocyanate. Crystallization trials were performed in 96-well glass sandwich plates (Marienfeld) by an NT8-LCP crystallization robot (Formulatrix) using 40-50 nL protein-laden LCP overlaid with 0.8 μL precipitant solution in each well, and sealed with a glass coverslip (Caffrey and Cherezov, 2009; Cherezov et al., 2004). The best diffraction quality crystals were obtained within 7 days in 25-28% (v/v) PEG 400, 0.04 to 0.06 M sodium thiocyanate, 2% (v/v) 2,5-hexanediol, 100 mM sodium citrate pH 5.0. Crystals grew to an average size of 60 × 10 × 3 μm (Figure 6) and diffracted to 1.7 Å resolution. The high-resolution crystal structure of A2AAR-BRIL was solved at 1.8 Å resolution, the best resolution of all GPCRs to date, allowing us to appreciate new details that were previously unobservable at lower resolution (Liu et al., 2012).
Figure 6. Representative crystals and diffraction of A2AAR-BRIL and β2AR-BRIL in LCP.
(A) A2AAR-BRIL/ZM241385 (construct 16) and (B) β2AR-BRIL/timolol (construct 24). Crystals grew to approximately 60 × 10 × 3 μm for A2AAR-BRIL and 80 × 15 × 5 μm for β2AR-BRIL. Inset shows the same image under cross polarized light. Diffraction patterns for (C) A2AAR-BRIL and (D) β2AR-BRIL. Inset shows magnified view around diffraction spot enclosed by the red box.
Assessment of β2AR chimeras
Expression and purification
Each of the five potential fusion partners was cloned into the T4L insertion site of β2AR and expressed in Sf9 cells. The integrity of the fusion protein was assessed in a similar manner as with the A2AAR constructs after purification. β2AR constructs were incubated with 1 mM timolol prior to solubilization with 0.5% (w/v) DDM and 0.1% (w/v) CHS, followed by IMAC purification against a C-terminal 10x His tag. β2AR-rubredoxin, β2AR-BRIL, and β2AR-flavodoxin showed purification recovery levels similar to that of β2AR-T4L, while the β2AR-CtermT4L signal was weak, and β2AR-xylanase was almost undetectable (Figure 3). aSEC analysis showed that the elution profiles of most of the chimeras were very similar to β2AR-T4L in terms of peak shape, with the exception of β2AR-CtermT4L, which had a more prominent aggregate component relative to the primary peak (Figure 4).
Thermostability
The thermostability of the different β2AR constructs was then evaluated. Assays were carried out in the presence of 1 mM timolol to maintain receptor stability during heating (Figure 5E). Relative to β2AR-T4L, β2AR-flavodoxin and β2AR-CtermT4L were both less stable by approximately 10 °C, while β2AR-xylanase and β2AR-rubredoxin had similar stability compared to β2AR-T4L. β2AR-BRIL was more stable than β2AR-T4L by approximately 13 °C. Thus, β2AR-BRIL was selected for further crystallization studies.
Protein diffusion and crystallization in LCP
We were able to grow crystals of β2AR-BRIL (construct 24) in LCP without the need for junction site optimization, as was necessary with A2AAR-BRIL. The LCP-FRAP assay was carried out in similar fashion to A2AAR-BRIL in order to guide crystallization trials. Proteins were labeled with Cy3-mono NHS ester, in pH 7.5 buffer, and likewise evaluated for purity, monodispersity, and labeling efficiency by aSEC prior to LCP-FRAP sample preparation. Labeled protein was reconstituted into LCP by mixing protein solution with molten lipid in a final ratio of 40% (w/w) protein solution, 54% (w/w) monoolein, 6% (w/w) cholesterol, and incubated with homemade screens as introduced previously. The pH for the screens were adjusted to 6.0, 7.0, and 8.0 based on previous β2AR crystallization conditions (Alexandrov et al., 2008; Cherezov et al., 2007; Wacker et al., 2010). Among all three pHs, β2AR-BRIL displayed optimal mobile fractions at pH 7.0, which reached up to 50% diffusion recovery with certain precipitants (Figure S2). Crystals were grown in 0.1 M Bis-Tris propane pH 7.5, 25-30% (v/v) PEG 400, 0.1 to 0.3 M ammonium formate, and 25-30% (w/v) trimethylamine N-oxide. Optimized crystals grew to approximately 80 × 15 × 5 μm (Figure 6) and diffracted anisotropically to a maximum resolution of 2.8 Å along the best axis and ~3.7 Å along the worst axis.
DISCUSSION
The methodology for identification of new fusion partners was validated by the successful crystallization and X-ray diffraction of A2AAR-BRIL and β2AR-BRIL fusions in the ICL3 to 1.8 and 2.8 Å resolution, respectively. More recently, we have successfully determined the structure of the opioid Nociceptin/Orphanin FQ peptide receptor at 3.0 Å resolution by placing BRIL on the N-terminus of the receptor demonstrating the utility of fusion partners at receptor termini (Thompson et al., 2012).
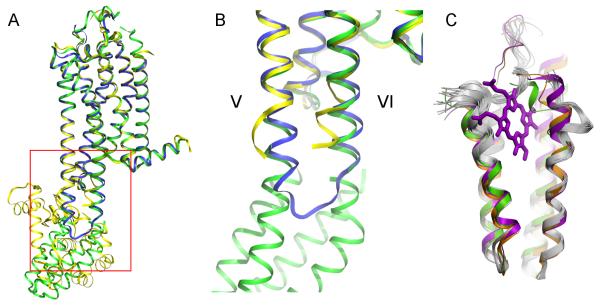
Although the biological impact of these new structures are beyond the scope of this methods study, a number of structural details regarding the fusion regions are important for understanding how the fusion proteins are engineered into the receptor. For example, in the 1.8 Å A2AAR-BRIL structure, the interface between BRIL and A2AAR consists of receptor helices V and VI that extend seamlessly into the α-helices of BRIL (Figure 7 A,B). This is in contrast to the variable loop interfaces connecting the receptor with the T4L domain observed in previous GPCR-T4L structures. From a structural perspective, this has the advantage that the residues on the receptor side are able to maintain their helical secondary structure, as opposed to being artificially forced into a different conformation by the fusion domain. The availability of inactive A2AAR structures solved by different methods (T4L fusion: PDB ID 3EML; thermostabilized, with intact ICL3: PDB ID 3PWH) provides a unique opportunity to examine this. Compared to the structure of A2AAR with an intact ICL3, the cytoplasmic end of helix V on A2AAR-T4L breaks into a loop, while the end of helix VI becomes shifted towards helix V. These distortions are presumably necessary to accommodate the transition from the receptor to T4L. Conversely, in the A2AAR-BRIL structure, the protein backbone conformation at the cytoplasmic ends of helices V and VI closely follows the unmodified ICL3 structure (3PWH), with the helices continuing into BRIL. The BRIL chimera thus exhibits minimal distortion of the intracellular portions of helices V and VI, matching more closely what is seen in the non-chimeric receptor structure than in the T4L chimera. From a crystallographic perspective, the junction flexibility in previous GPCR-T4L chimeras has allowed a wide range of T4L domain orientations, permitting their crystallization in different possible crystal packing contacts. While this has been beneficial in the past, the structure in the immediate vicinity of the junction is affected unpredictably by the crystal packing lattice. A well-defined α-helical junction on the other hand is expected to be more resistant to such crystallographic disturbances, providing a more rigid protein overall, but potentially at the expense of being able to readily crystallize in different forms. It will be instructive to see how the α-helical junction impacts the stability and final resolution of other GPCR-BRIL crystal structures. Insights from such structures and comparisons with GPCR-T4L structures may allow us to rationally design improved GPCR-fusion domain junctions for stabilization and crystallization.
Figure 7. Structural comparison of A2AAR solved by different engineering methods.
(A) High-resolution crystal structure of A2AAR-BRIL (green) superimposed with A2AAR-T4L (PDB ID 3EML, yellow) and thermostabilized A2AAR (PDB ID 3PWH, blue). (B) Close up view of the junction site enclosed by red box in panel A. The cytoplasmic ends of helix V and helix VI of A2AAR-BRIL and thermostabilized A2AAR superimpose very well, while the helices of A2AAR-T4L must diverge to accommodate the insertion of T4L (T4L domain not shown). (C) Superimposition of the fused BRIL (green) with standalone BRIL (PDB ID 1M6T, orange), cytochrome b562 with heme (PDB ID 256B, magenta) and NMR models of apocytochrome b562 (PDB ID 1YYX, grey) suggest high rigidity in the protein core (bottom half) and high flexibility in the termini and loop2 (top half), especially in the apo structures.
Despite the well-defined helical structure and stability of the BRIL hydrophobic core, the GPCR-BRIL junction site also affords some level of plasticity due to inherent flexibility of the heme-binding region in apo-BRIL that may be beneficial in terms of accommodating its insertion into adjacent GPCR α-helices. Thus, though the connection between receptor helix V and BRIL helix I are almost perfectly aligned, receptor helix VI connects with BRIL helix IV at an angle of ~40°. This bending distortion is accommodated by the last two turns of helix IV in BRIL and is highly flexible in apocytochrome b562, as it exhibits large conformational variations between NMR models (Feng et al., 2004) as well as between crystal structures of the apo- and heme-bound cytochrome b562 (Lederer et al., 1981) (Figure 7 C). Additionally, the long loop connecting helices II and III in the heme-binding region of BRIL has 16 disordered residues, which also reflects its highly flexible behavior described previously in apo-BRIL NMR studies. These observations reflect a dual nature of BRIL, which is highly flexible in the heme-binding region, but has a very stable and rigid core in the distal part that ensures fast folding kinetics and stabilizes the fold (Kimura et al., 2009). This duality makes BRIL an attractive partner for fusions in the loop regions between α-helices of membrane proteins, as it can easily accommodate minor lateral misalignments between the α-helices of the two proteins, and at the same time help with folding kinetics and stability.
With this new toolchest of fusion partners for membrane protein structural biology and the knowledge gained, it is of interest to see if the initial PDB search criteria could be refined to incorporate what we have learned thus far to enable the rational selection of improved fusion domains. Such improvements could include fluorescent or colored proteins, or ones with other advantageous signaling capabilities for biophysical investigations. Furthermore, we have evaluated the use of fusion partners in the termini of GPCRs to aid in the expression, stabilization, and crystal packing, including the successful structure determination of the Nociceptin/Orphanin FQ peptide receptor (Thompson et al., 2012). Notwithstanding the different considerations involved in selecting termini fusion domains, the scope of crystallizable membrane proteins could be vastly expanded by examining fusions in all extracellular and intracellular loops and termini. The successful matching of fusion partners with membrane proteins will not only improve forthcoming GPCR crystal structure determination, but also serve as a platform to push forward the crystallization of other membrane proteins and even larger protein assemblies that to date have been recalcitrant to structure characterization.
EXPERIMENTAL PROCEDURES
Construct design and expression
Constructs of all chimeras were synthesized by GenScript and then cloned into a modified pFastBac1 vector (Invitrogen) containing an HA signal sequence and a FLAG tag at the N-terminus, and a 10x His tag at the C-terminus. For A2AAR constructs, fusion proteins in the ICL3 were initially inserted between residues L208 and R222. The C-terminus was truncated to residue A317 to minimize flexibility. For β2AR, fusion proteins in the ICL3 were inserted between residues L230 and K263. The C-terminus was truncated to residue K348. The N-terminus was left intact in both receptors. Recombinant baculoviruses were generated with the Bac-to-Bac system (Invitrogen) and used to infect Sf9 cells. 50 μL of P0 baculovirus was used to infect 5 mL of Sf9 cells at a density of 2-3 × 106 cells/mL. Cells were grown at 27 °C for 48 h prior to being harvested.
Purification
For small scale analysis of chimeric receptors, 5 mL of frozen Sf9 cells were thawed in buffer containing 20 mM HEPES pH 7.5, 10 mM NaCl, 10 mM MgCl2, 20 mM KCl, and Protease Inhibitor cocktail tablets (1 tablet per 50 mL buffer, Roche). Dounce homogenization and centrifugation at 14,000 × g was carried out to disrupt intact cells and to remove unwanted soluble and membrane associated proteins, and repeated twice with the addition of 1 M NaCl in the buffer. Remaining membranes containing receptors were resuspended in buffer containing 50 mM HEPES pH 7.5, 150 mM NaCl, 2 mg/mL iodoacetamide, and an appropriate ligand (1 mM timolol for β2AR constructs and 4 mM theophylline for A2AAR constructs). Membranes were allowed to rock for 30 min at 4 °C, followed by solubilization with 0.5% (w/v) n-Dodecyl-β-D-Maltoside (DDM, Affymetrix) and 0.1% (w/v) cholesteryl hemisuccinate (CHS, Sigma) for 2-4 h at 4 °C. Solubilized membranes were spun down at 14,000 × g for 45 min, and the resulting supernatant was incubated with 50-100 μL TALON IMAC resin (Clontech) overnight in the presence of 20 mM imidazole. TALON resin was spun down at 500 × g, resuspended in buffer containing 50 mM HEPES pH 7.5, 150 mM NaCl, 0.05% (w/v) DDM, 0.01% (w/v) CHS, 25 mM imidazole, and applied to a gravity column (Poly-Prep, Bio-Rad). Protein was washed with an additional 20 column volumes (CV) of the same resuspension buffer, and then eluted with buffer containing 50 mM HEPES pH 7.5, 150 mM NaCl, 0.05% (w/v) DDM, 0.01% (w/v) CHS, and 250 mM imidazole. Fractions containing protein as determined by colorimetric Bradford assay were pooled together. For A2AAR constructs, 800 mM NaCl was used in place of 150 mM NaCl. All purification buffers were pre-chilled on ice and purifications carried out at 4 °C.
For purifications intended for crystallization trials, the above procedure was used with the following modifications: 1) 1 L of frozen Sf9 cells was used, and TALON resin bed volume was increased to 0.5 mL, typically providing approximately 1-2 mg of purified receptor, 2) for β2AR constructs, 1 mM timolol was present in all buffers during and after solubilization, up until reconstitution in LCP. 20 μL PNGaseF (New England Biolabs) was added after the initial 20 CV wash and allowed to incubate overnight before elution to deglycosylate the receptor. 3) for A2AAR constructs, 4 mM theophylline was present during solubilization, and exchanged for 100 μM ZM241385 on TALON resin and maintained in all subsequent steps up until reconstitution in LCP.
For purifications intended for FRAP studies, the following modifications were made: Packed TALON resin was resuspended in 5 mL wash buffer, followed by the addition of 5-10 μL of 5 mg/mL Cy3-mono NHS ester (GE Healthcare) dissolved in dimethylformamide. Protein was allowed to incubate with the dye at 4 °C for 2-3 h in darkness. Free dye was then removed by flowing additional wash buffer through resin until the pink color could no longer be visually observed in the flowthrough, followed by elution as usual. All steps after dye labeling were carried out in darkness to prevent photobleaching.
SDS-PAGE and immunoblotting
Protein samples were mixed with LDS sample buffer (Novex), separated on 10% Bis-Tris gels (Novex) at 130 volts, and stained with Coomassie SimplyBlue SafeStain (Novex). For Western blots, proteins were transferred from gels to nitrocellulose membranes using the iBlot Dry Blotting System and probed with monoclonal anti-FLAG M2 alkaline phosphatase antibody (Sigma) for at least 1 h. Membranes were then washed in PBS, and antibodies were visualized with SIGMAFAST BCIP/NBT (Sigma).
Analytical size exclusion chromatography
Purified receptor constructs were applied to a Sepax Nanofilm SEC-250 column (4.6 × 250 mm) using an Agilent model 1200 HPLC system at a flowrate of 0.5 mL/min and signal detection set to 350 nm. Prior to sample injection the column was equilibrated in 50 mM HEPES pH 7.5, 500 mM NaCl, 2% (v/v) glycerol, 0.05% (w/v) DDM (Affymetrix) and 0.01% (w/v) CHS (Sigma). Sample reservoirs and column were maintained at 4 °C throughout analysis.
Thermostability assay
N-[4-(7-diethylamino-4-methyl-3-coumarinyl)phenyl]maleimide (CPM) dye (Invitrogen) was dissolved in DMSO (Sigma) at 4 mg/mL as a stock solution for future use. The stock solution was kept at -80°C and diluted 1:40 in dye dilution solution (10 mM buffer, 500 mM NaCl, 10% (v/v) glycerol, 0.025% (w/v) DDM and 0.005% (w/v) CHS) before use. The thermal denaturation assay was performed with total volume of 200 μL sample in a quartz fluorometer cuvette (Starna Cells, Inc., Atascadero, CA). Receptor (4 μg) was diluted in the appropriate buffer solution to a final volume of 200 μL. 5 μL of the diluted dye solution was then added to the protein solution and incubated for 30 min at 4 °C in darkness. The mixed solution was transferred to a cuvette and the data were collected by a Cary Eclipse spectrofluorometer (Varian, USA) with a temperature ramping rate of 2 °C/min. The excitation wavelength was 387 nm and the emission wavelength was 463 nm. All assays were performed over a temperature range starting from 20 °C to 90 °C. The stability data were processed with GraphPad Prism (GraphPad Software, San Diego, CA, USA).
Crystallization
In meso crystallization methods for membrane proteins have been described in detail (Caffrey and Cherezov, 2009). Protein solution was mixed with a molten lipid mixture of 10:1 (w/w) monoolein:cholesterol, at a ratio of 40% (w/w) protein with 60% (w/w) lipid in a custom syringe mixer. A syringe containing reconstituted LCP was loaded onto an automated crystallization robot (NT8-LCP, Formulatrix) and 35-50 nL of LCP dispensed onto 96-well glass sandwich plates (Marienfeld), and then overlaid with 800 nL of precipitant solution. Drops were manually sealed with a coverslip and incubated and imaged at 20 °C in an automatic incubator/imager, RockImager 1000 (Formulatrix). Precipitants were all from homemade screens made and aliquoted into 96-well plates. Crystals of both β2AR-BRIL and A2AAR -BRIL grew within 7 d.
LCP-FRAP
Cy3-labeled protein for LCP-FRAP analysis was reconstituted into LCP and dispensed onto glass sandwich plates as described in the crystallization section. The setup for automated high-throughput LCP-FRAP analysis has been described (Xu et al., 2011a). Briefly, LCP sandwich plates were loaded onto a custom built LCP-FRAP station consisting of a Zeiss AxioImager A1 fluorescent microscope, a Micropoint dye cell laser (Photonic Instruments), a cooled CCD FireWire camera CoolSnap HQ2 (Photometrics), and an automated XYZ microscope stage MS-2000 (Applied Scientific Instrumentation). Each LCP drop was then bleached by firing 15-20 laser pulses at a 25 Hz pulse rate. Fluorescence images were taken immediately before and after bleaching. After a recovery period of 30 minutes, images were taken again to determine fluorescence recovery, and analyzed by ImagePro Advanced Microscopy Suite (Media Cybernetics).
X-ray data collection
Crystallographic data were collected on the 23ID-B/D beamline (GM/CA CAT) of the Advanced Photon Source at the Argonne National Laboratory using a 10 μm collimated minibeam at a wavelength of 1.0330 Å and a MarMosaic 300 detector. To reduce radiation damage, crystals were translated to a fresh position, if possible, or replaced after collecting 5-10 frames at 3 s exposure and 1° oscillation with unattenuated beam. For details on the A2AAR-BRIL structure determination, please refer to Liu et al. (2012).
Supplementary Material
Highlights.
A method was developed for the selection of fusion domains for GPCR crystallization.
Apocytochrome b562RIL has advantages over previously utilized T4 lysozyme.
Diffraction quality crystals of two new engineered GPCRs were successfully grown.
The method led to the crystal structure of the A2A adenosine receptor at 1.8Å.
Acknowledgements
This work was supported by NIH Common Fund in Structural Biology grant P50 GM073197 and by the NCRR and NIGMS grant R21 GM103972 for technology development. The authors thank J. Velasquez for help on molecular biology; T. Trinh, K. Allin and M. Chu for help on baculovirus expression; A. Walker for assistance with manuscript preparation; L. Heitman and A. IJzerman for critical review of the manuscript.
References
- Alexandrov AI, Mileni M, Chien EY, Hanson MA, Stevens RC. Microscale fluorescent thermal stability assay for membrane proteins. Structure. 2008;16:351–359. doi: 10.1016/j.str.2008.02.004. [DOI] [PubMed] [Google Scholar]
- Bjarnadottir TK, Gloriam DE, Hellstrand SH, Kristiansson H, Fredriksson R, Schioth HB. Comprehensive repertoire and phylogenetic analysis of the G protein-coupled receptors in human and mouse. Genomics. 2006;88:263–273. doi: 10.1016/j.ygeno.2006.04.001. [DOI] [PubMed] [Google Scholar]
- Caffrey M, Cherezov V. Crystallizing membrane proteins using lipidic mesophases. Nat Protoc. 2009;4:706–731. doi: 10.1038/nprot.2009.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae PS, Rasmussen SG, Rana RR, Gotfryd K, Chandra R, Goren MA, Kruse AC, Nurva S, Loland CJ, Pierre Y, et al. Maltose-neopentyl glycol (MNG) amphiphiles for solubilization, stabilization and crystallization of membrane proteins. Nat Methods. 2010;7:1003–1008. doi: 10.1038/nmeth.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherezov V, Peddi A, Muthusubramaniam L, Zheng YF, Caffrey M. A robotic system for crystallizing membrane and soluble proteins in lipidic mesophases. Acta Crystallogr D Biol Crystallogr. 2004;60:1795–1807. doi: 10.1107/S0907444904019109. [DOI] [PubMed] [Google Scholar]
- Cherezov V, Rosenbaum DM, Hanson MA, Rasmussen SG, Thian FS, Kobilka TS, Choi HJ, Kuhn P, Weis WI, Kobilka BK, Stevens RC. High-resolution crystal structure of an engineered human beta2-adrenergic G protein-coupled receptor. Science. 2007;318:1258–1265. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien EY, Liu W, Zhao Q, Katritch V, Han GW, Hanson MA, Shi L, Newman AH, Javitch JA, Cherezov V, Stevens RC. Structure of the human dopamine D3 receptor in complex with a D2/D3 selective antagonist. Science. 2010;330:1091–1095. doi: 10.1126/science.1197410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day PW, Rasmussen SG, Parnot C, Fung JJ, Masood A, Kobilka TS, Yao XJ, Choi HJ, Weis WI, Rohrer DK, Kobilka BK. A monoclonal antibody for G protein-coupled receptor crystallography. Nat Methods. 2007;4:927–929. doi: 10.1038/nmeth1112. [DOI] [PubMed] [Google Scholar]
- Dore AS, Robertson N, Errey JC, Ng I, Hollenstein K, Tehan B, Hurrell E, Bennett K, Congreve M, Magnani F, et al. Structure of the adenosine A(2A) receptor in complex with ZM241385 and the xanthines XAC and caffeine. Structure. 2011;19:1283–1293. doi: 10.1016/j.str.2011.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel CK, Chen L, Prive GG. Insertion of carrier proteins into hydrophilic loops of the Escherichia coli lactose permease. Biochim Biophys Acta. 2002;1564:38–46. doi: 10.1016/s0005-2736(02)00398-x. [DOI] [PubMed] [Google Scholar]
- Feng H, Vu ND, Bai Y. Detection and structure determination of an equilibrium unfolding intermediate of Rd-apocytochrome b562: native fold with non-native hydrophobic interactions. J Mol Biol. 2004;343:1477–1485. doi: 10.1016/j.jmb.2004.08.099. [DOI] [PubMed] [Google Scholar]
- Haga K, Kruse AC, Asada H, Yurugi-Kobayashi T, Shiroishi M, Zhang C, Weis WI, Okada T, Kobilka BK, Haga T, Kobayashi T. Structure of the human M2 muscarinic acetylcholine receptor bound to an antagonist. Nature. 2012 doi: 10.1038/nature10753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson MA, Roth CB, Jo E, Griffith MT, Scott FL, Reinhart G, Desale H, Clemons B, Cahalan SM, Schuerer SC, et al. Crystal structure of a lipid G protein-coupled receptor. Science. 2012;335:851–855. doi: 10.1126/science.1215904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hino T, Arakawa T, Iwanari H, Yurugi-Kobayashi T, Ikeda-Suno C, Nakada-Nakura Y, Kusano-Arai O, Weyand S, Shimamura T, Nomura N, et al. G-protein-coupled receptor inactivation by an allosteric inverse-agonist antibody. Nature. 2012;482:237–240. doi: 10.1038/nature10750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaakola VP, Griffith MT, Hanson MA, Cherezov V, Chien EY, Lane JR, Ijzerman AP, Stevens RC. The 2.6 angstrom crystal structure of a human A2A adenosine receptor bound to an antagonist. Science. 2008;322:1211–1217. doi: 10.1126/science.1164772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura T, Lee JC, Gray HB, Winkler JR. Folding energy landscape of cytochrome cb562. Proc Natl Acad Sci U S A. 2009;106:7834–7839. doi: 10.1073/pnas.0902562106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse AC, Hu J, Pan AC, Arlow DH, Rosenbaum DM, Rosemond E, Green HF, Liu T, Chae PS, Dror RO, et al. Structure and dynamics of the M3 muscarinic acetylcholine receptor. Nature. 2012;482:552–556. doi: 10.1038/nature10867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebon G, Warne T, Edwards PC, Bennett K, Langmead CJ, Leslie AG, Tate CG. Agonist-bound adenosine A2A receptor structures reveal common features of GPCR activation. Nature. 2011;474:521–525. doi: 10.1038/nature10136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lederer F, Glatigny A, Bethge PH, Bellamy HD, Matthew FS. Improvement of the 2.5 A resolution model of cytochrome b562 by redetermining the primary structure and using molecular graphics. J Mol Biol. 1981;148:427–448. doi: 10.1016/0022-2836(81)90185-6. [DOI] [PubMed] [Google Scholar]
- Liu W, Chun E, Thompson AA, Chubukov P, Xu F, Katritch V, Han GW, Roth CB, Heitman LH, IJzerman AP, et al. Structural basis for allosteric regulation in GPCRs. Science Submitted. 2012 doi: 10.1126/science.1219218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manglik A, Kruse AC, Kobilka TS, Thian FS, Mathiesen JM, Sunahara RK, Pardo L, Weis WI, Kobilka BK, Granier S. Crystal structure of the micro-opioid receptor bound to a morphinan antagonist. Nature. 2012 doi: 10.1038/nature10954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JL, Tate CG. Engineering an ultra-thermostable beta(1)-adrenoceptor. J Mol Biol. 2011;413:628–638. doi: 10.1016/j.jmb.2011.08.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palczewski K, Kumasaka T, Hori T, Behnke CA, Motoshima H, Fox BA, Le Trong I, Teller DC, Okada T, Stenkamp RE, et al. Crystal structure of rhodopsin: A G protein-coupled receptor. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- Prive GG, Verner GE, Weitzman C, Zen KH, Eisenberg D, Kaback HR. Fusion proteins as tools for crystallization: the lactose permease from Escherichia coli. Acta Crystallogr D Biol Crystallogr. 1994;50:375–379. doi: 10.1107/S0907444993014301. [DOI] [PubMed] [Google Scholar]
- Rasmussen SG, Choi HJ, Fung JJ, Pardon E, Casarosa P, Chae PS, Devree BT, Rosenbaum DM, Thian FS, Kobilka TS, et al. Structure of a nanobody-stabilized active state of the beta(2) adrenoceptor. Nature. 2011a;469:175–180. doi: 10.1038/nature09648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen SG, DeVree BT, Zou Y, Kruse AC, Chung KY, Kobilka TS, Thian FS, Chae PS, Pardon E, Calinski D, et al. Crystal structure of the beta2 adrenergic receptor-Gs protein complex. Nature. 2011b;477:549–555. doi: 10.1038/nature10361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum DM, Cherezov V, Hanson MA, Rasmussen SG, Thian FS, Kobilka TS, Choi HJ, Yao XJ, Weis WI, Stevens RC, Kobilka BK. GPCR engineering yields high-resolution structural insights into beta2-adrenergic receptor function. Science. 2007;318:1266–1273. doi: 10.1126/science.1150609. [DOI] [PubMed] [Google Scholar]
- Serrano-Vega MJ, Magnani F, Shibata Y, Tate CG. Conformational thermostabilization of the beta1-adrenergic receptor in a detergent-resistant form. Proc Natl Acad Sci U S A. 2008;105:877–882. doi: 10.1073/pnas.0711253105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimamura T, Shiroishi M, Weyand S, Tsujimoto H, Winter G, Katritch V, Abagyan R, Cherezov V, Liu W, Han GW, et al. Structure of the human histamine H1 receptor complex with doxepin. Nature. 2011;475:65–70. doi: 10.1038/nature10236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson AA, Liu W, Chun E, Katritch V, Wu H, Vardy E, Huang XP, Trapella C, Guerrini R, Calo G, et al. Structure of the nociceptin/orphanin FQ receptor in complex with a peptide mimetic. Nature. 2012 doi: 10.1038/nature11085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wacker D, Fenalti G, Brown MA, Katritch V, Abagyan R, Cherezov V, Stevens RC. Conserved binding mode of human beta2 adrenergic receptor inverse agonists and antagonist revealed by X-ray crystallography. J Am Chem Soc. 2010;132:11443–11445. doi: 10.1021/ja105108q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warne T, Serrano-Vega MJ, Baker JG, Moukhametzianov R, Edwards PC, Henderson R, Leslie AG, Tate CG, Schertler GF. Structure of a beta1-adrenergic G-protein-coupled receptor. Nature. 2008;454:486–491. doi: 10.1038/nature07101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West GM, Chien EY, Katritch V, Gatchalian J, Chalmers MJ, Stevens RC, Griffin PR. Ligand-dependent perturbation of the conformational ensemble for the GPCR beta2 adrenergic receptor revealed by HDX. Structure. 2011;19:1424–1432. doi: 10.1016/j.str.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B, Chien EY, Mol CD, Fenalti G, Liu W, Katritch V, Abagyan R, Brooun A, Wells P, Bi FC, et al. Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists. Science. 2010;330:1066–1071. doi: 10.1126/science.1194396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Wacker D, Mileni M, Katritch V, Han GW, Vardy E, Liu W, Thompson AA, Huang XP, Carroll FI, et al. Structure of the human kappa-opioid receptor in complex with JDTic. Nature. 2012 doi: 10.1038/nature10939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F, Liu W, Hanson MA, Stevens RC, Cherezov V. Development of an Automated High Throughput LCP-FRAP Assay to Guide Membrane Protein Crystallization in Lipid Mesophases. Cryst Growth Des. 2011a;11:1193–1201. doi: 10.1021/cg101385e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F, Wu H, Katritch V, Han GW, Jacobson KA, Gao ZG, Cherezov V, Stevens RC. Structure of an agonist-bound human A2A adenosine receptor. Science. 2011b;332:322–327. doi: 10.1126/science.1202793. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.