Abstract
RNA interference (RNAi) is a remarkable endogenous regulatory pathway that can bring about sequence-specific gene silencing. If harnessed effectively, RNAi could result in a potent targeted therapeutic modality with applications ranging from viral diseases to cancer. The major barrier to realizing the full medicinal potential of RNAi is the difficulty of delivering effector molecules, such as small interfering RNAs (siRNAs), in vivo. An effective delivery strategy for siRNAs must address limitations that include poor stability and non-targeted biodistribution, while protecting against the stimulation of an undesirable innate immune response. The design of such a system requires rigorous understanding of all mechanisms involved. This article reviews the mechanistic principles of RNA interference, its potential, the greatest challenges for use in biomedical applications, and some of the work that has been done toward engineering delivery systems that overcome some of the hurdles facing siRNA-based therapeutics.
Keywords: RNA interference, siRNA, therapeutics, delivery, chemical modification, liposome, nanoparticle, targeting
Mechanisms of RNAi
In 1998, Fire and Mello uncovered the world of RNA interference (RNAi) and revolutionized the contemporary understanding of gene regulation when they made the discovery that the silencing effectors in Caenorhabditis elegans were double stranded RNAs [1]. Shortly thereafter, small interfering RNAs (siRNAs) were discovered in plants [2] and similarly demonstrated to guide sequence-dependent endonucleolytic cleavage of the mRNAs that they regulate in mammalian cells [3,4]. By 2001, Elbashir et al. had successfully used synthetic siRNAs for silencing and determined the basic principles of siRNA structure and RNAi mechanics, providing the foundation for developing RNAi applications [5,6]. Since then, selectively silencing genes by hijacking the endogenous RNAi pathway with synthetic constructs has become a widely used technique for the study of gene function, and further, this approach has shown impressive therapeutic potential that is expected to be realized in the near future.
RNAi Machinery
RNAi can be effected when short (~22nt), double-stranded fragments of RNA — known as small interfering RNAs (siRNAs) — are loaded into the RNA-Induced Silencing Complex (RISC), where the strands are separated, and one strand guides cleavage by Argonaute of target mRNAs in a sequence homology-dependent manner [3].
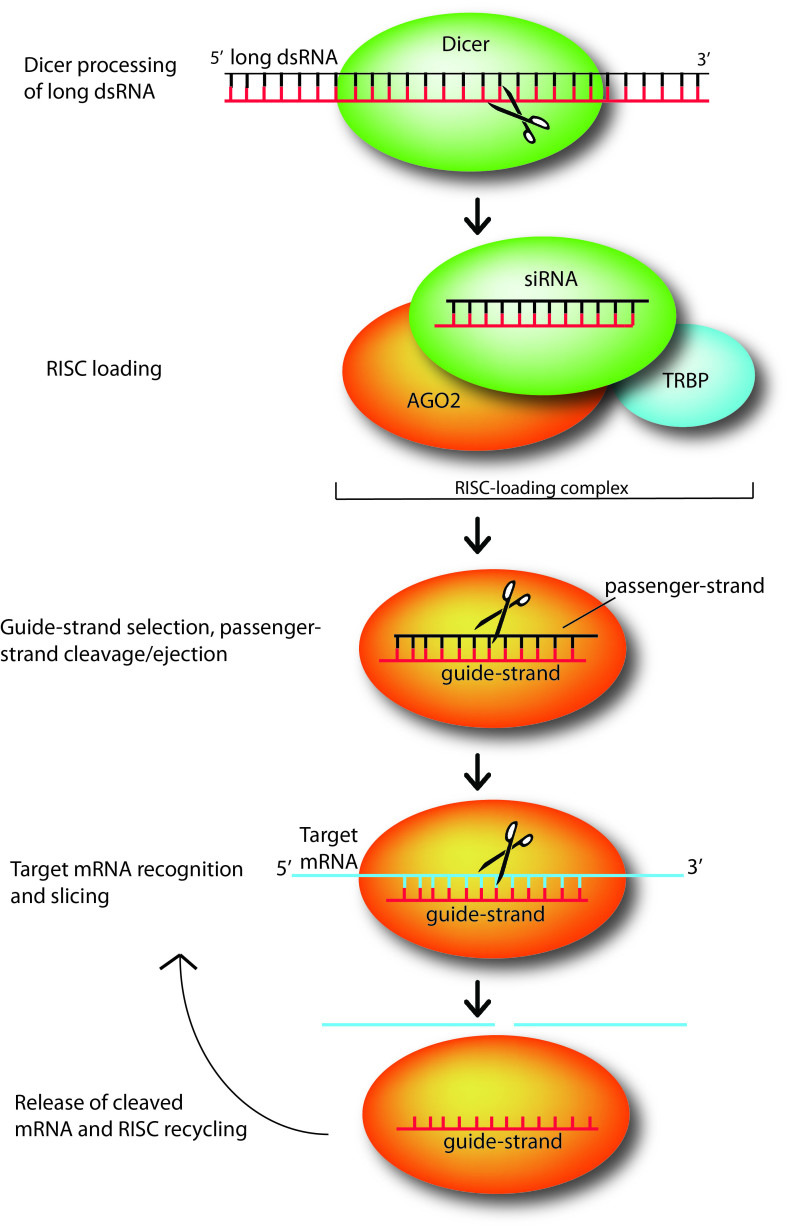
In mammalian cells, siRNAs are produced via endonucleolytic processing by the ribonuclease Dicer of exogenously introduced long, double-stranded RNA [12]. Dicer is an endonuclease of the RNase III family (Figure 1); it acts as a “molecular ruler” and precisely produces RNA duplexes ~21-25 nucleotides in length with characteristic termini. The 3’ end carries a dinucleotide overhang, while the 5’ end terminates in a monophosphate group [6,7,8,9,12]. The siRNA duplex length and distinctive ends are necessary features for efficient recognition by and integration into the RISC. Furthermore, recent biochemical studies show that Dicer processing itself is coupled with RISC loading through the tight association of Dicer with TRBP (the human immunodeficiency trans activating response RNA-binding protein) [10,11].
Figure 1.
Small interfering RNAs (siRNAs) mediate silencing of target genes by guiding sequence dependent slicing of their target mRNAs. These non-coding, silencing RNAs begin as long double-stranded RNA (dsRNA) molecules, which are processed by endonuclease Dicer into short, active ~21-25 nt constructs. Once generated, a siRNA duplex is loaded by Dicer, with the help of RNA-binding protein TRBP, onto Argonaute (AGO2), the heart of the RNA-induced silencing complex (which here is represented just by AGO2). Upon loading, AGO2 selects the siRNA guide strand, then cleaves and ejects the passenger strand. While tethered to AGO2, the guide strand subsequently pairs with its complementary target mRNAs long enough for AGO2 to slice the target. After slicing, the cleaved target mRNA is released and RISC is recycled, using the same loaded guide strand for another few rounds of slicing [12].
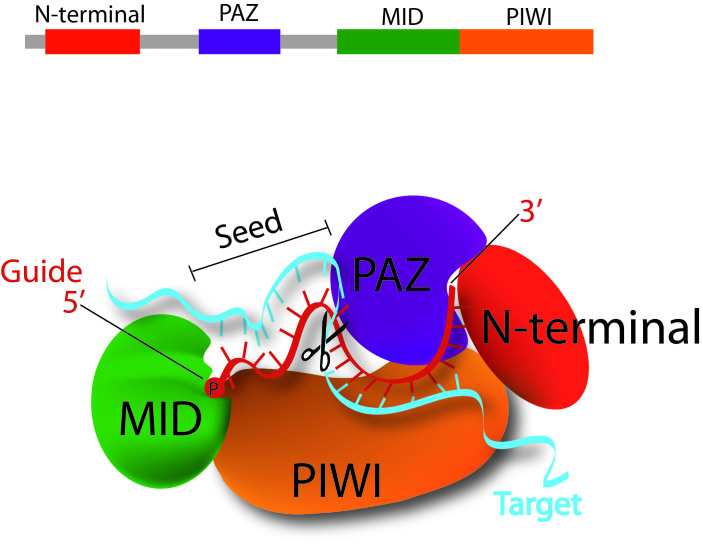
The heart of the RISC complex and principal executer of RNAi-mediated silencing is the Argonaute protein [13,14]. There are four Argonaute proteins in humans (AGO 1-4), and silencing by siRNAs is accomplished via AGO2 [13]. To bring about siRNA-mediated silencing, AGO2 must tether the guide siRNA strand, extrude the passenger strand, and then undergo several cycles of target mRNA recognition, cleavage, and release while the guide strand remains bound (Figure 1) [12]. Structural studies revealed some of the mechanisms underlying AGO2’s activity. AGO2 has three functional domains, PAZ, MID, and PIWI, of which PIWI adopts an RNase H fold and is the powerhouse behind RISC’s “slicer” activity [15]. For RISC loading, structural evidence suggests that the characteristic terminal moieties of siRNA serve anchoring functions: the 3’dinucleotide is specifically recognized by the PAZ domain of Argonaute. The overhang burrows deep into a hydrophobic pocket of the domain, where the base of the terminal nucleotide can stack with an aromatic ring of one of the numerous aromatic residues that line the pocket [16-18]. Meanwhile, the 5’ phosphate group inserts between the MID and PIWI domains, binding to a magnesium ion that itself is coordinated to the C-terminus of the protein (Figure 2) [19,21]. For guide-strand selection, thermodynamic data indicates that Argonaute selects the guide strand as the one with the less thermodynamically stable 5’ end and subsequently slices the passenger strand to encourage its ejection [20].
Figure 2.
A closer look at the model for siRNA guide-strand tethering by AGO2 and target-mRNA recognition and slicing. The terminal 5’ monophosphate group of the guide strand tucks in between the MID and PIWI domains of AGO2. Meanwhile, AGO2’s PAZ domain has a hydrophobic pocket that specifically recognizes the guide-strands 3’ dinucleotide overhang. This positioning opens up siRNA guide nucleotides 2-8, the “seed region,” for base pairing with complementary target mRNA, and next base pairing at nucleotides 10-11 correctly orients the scissile phosphate between these two for cleavage by AGO2’s PIWI domain, which houses the protein’s “slicer” activity [12].
Structural, biochemical, and computational studies of RISC in complex with the guide-target duplex provide rationale for the specificity of slicer activity. In this complex, guide strand bases 2-8 (from the 5’ end) are uncovered and free to participate in Watson-Crick base-pairing with the mRNA target. This “seed region,” as it is known, is essential for specific target recognition and places the target’s scissile phosphate group at the slicer active site [21]. Meanwhile, base-pairing between guide and target at nucleotides 10-11 has similarly been shown to be crucial for properly orienting the scissile phosphate group for slicer cleavage, explaining the fixed distance at which slicing takes place with respect to the guide’s 5’end (Figure 2) [22].
RNAi machinery also can be engaged by endogenously encoded short RNA molecules known as microRNAs (miRNAs) [23]. The initial precursors of miRNAs, pri-miRNAs are generated in the nucleus where they are processed by RNase III-family enzyme Drosha to yield pre-miRNAs with a hairpin structure (two base-paired arms linked by a loop), a 5’ phosphate group, and a 3’ two-nucleotide. Pre-miRNAs are then exported into the cytoplasm, where they are further cleaved by Dicer to remove the loop and produce a duplex with the same characteristic termini as siRNAs that is then loaded into RISC [24]. Unlike siRNAs, however, miRNAs are only partially complementary to their target mRNAs’ 3’UTR region. MiRNAs regulate their targets via all four Argonaute proteins, and while they can sometimes bring about mRNA cleavage and degradation like siRNAs, they primarily accomplish gene silencing through downstream translational repression and mRNA decay by deadenylation [25].
RNAi Potential
The power of RNAi lies in the key discovery that endogenous RNAi gene silencing machinery can be hijacked to artificially regulate genes of interest. RISC is triggered with the introduction of an active RNAi effector, and delivery of such an effector to a cell brings about potent and specific knockdown of its target. Theoretically, siRNAs can be designed for any gene of interest based on its mRNA sequence alone. Such unlimited potential has made RNAi a favorite gene knockdown strategy in mammalian cells [26]. And more importantly, RNAi-based silencing potentially can be applied to design a powerful line of therapeutics for the vast number of human diseases caused by one or a few genes, such as genetic defects, viral diseases, autoimmune disorders, and cancers [26]. Furthermore, while miRNAs are not targetable to genes of choice the way siRNAs are, many endogenous miRNAs function as oncogenes and thus themselves may serve as therapeutic targets [27].
Potency is a tremendous consideration in drug design, and another advantage of RNAi is that it is significantly more potent than other nucleic acid-based antisense technologies [28]. Due to the fact that a single siRNA guide strand can be recycled for several rounds of mRNA cleavage, the RNAi pathway can achieve surprising efficiency given the right trigger. The efficiency of RNAi mediated silencing for any particular gene can vary greatly, and a number of factors are involved in the efficacy of any given siRNA. RNAi makes use of a complicated, endogenous, biological process that is contingent on a number of specific interactions between effector molecules and natural machinery. Clear understanding of these interactions is indispensible to siRNA design, and while features such as thermodynamic end stability [29], target accessibility [30], position-related characteristics, and other structural features [41] are known to play a role [26,31-33], current knowledge is still limited. Better understanding of these features may someday lead to super-active siRNAs requiring less target-site accumulation of the siRNA, which can be a challenge [26].
Challenges of Therapeutic siRNA
The therapeutic application of siRNA is extremely promising due to efficient and specific gene silencing, as demonstrated in selected in vitro and in vivo studies. However, to be generally applicable, a number of intracellular and extracellular barriers still need to be overcome to harness the full potential of this technology.
siRNA Stability and Targeting
Extracellularly, siRNAs are highly susceptible to degradation by enzymes found in serum and tissues. The half-life of naked siRNAs in serum ranges from several minutes to an hour [34]. As a result, target-site accumulation to therapeutically appropriate levels is a major challenge [35]. To be effective in a disease-relevant setting, siRNAs must not only survive in the serum, but also reach their target cells in the specific tissues that express the aberrant gene (or genes) of interest. Then having reached their target cells, siRNAs still face a number of hurdles before they can exert their gene silencing activity. The large size and negative charge of naked siRNAs thwarts their diffusion across the plasma membrane and prevents intracellular accumulation. Meanwhile, siRNA delivery strategies that take advantage of endocytosis also must provide for endosomal escape. And even once in the cell cytoplasm, siRNAs remain vulnerable to degradation by intracellular RNAses and still need to be recognized by and incorporated into RISC with high efficiency.
Off-Target Silencing
Leaving aside the problems of delivery, the RNAi paradigm of specific silencing unfortunately breaks down somewhat in reality [26]. Microarray analysis has revealed that siRNA treatment can result in off-target gene silencing, i.e., suppression of genes other than the desired gene targets [36]. Off-target silencing is undesirable as it can lead to dangerous mutation of gene expression and unexpected cell transformation. Recent studies have demonstrated that most off-target silencing is a result of homology with six to seven nucleotides in the “seed region” of the siRNA sequence [37,38]. This makes sense in the context of what is known about miRNA gene silencing. Recall that these constructs exert their silencing activity having only partial sequence complementarity (~6-8 nucleotides) with the 3’UTR of their mRNA targets [38]. In addition, because mRNA degradation is only one of several ways in which miRNAs affect gene expression, some of which are at the level of translational repression, experimental screens for off-targets that focus exclusively on analyzing mRNA levels may miss genes suppressed during translation rather than by means of mRNA degradation [26]. Still further, some siRNA sequences may cause altered gene expression because of “seed region” complementarity with endogenous miRNAs, which themselves regulate a family of genes [26]. Poor selection of guide strand over passenger strand by RISC can further lead to an even higher probability of matching undesired targets for an siRNA duplex. Off-target silencing cannot be ignored in developing siRNA-based therapeutics, and all potential therapeutic siRNA candidate sequences must be heavily tested for perturbation of normal protein expression profiles. Also, as our knowledge of siRNA mechanism advances, predictive bioinformatic approaches implemented at the stage of siRNA design promise to significantly reduce and eventually eradicate off-target silencing.
Activation of Immune Response
Though largely well-tolerated, especially compared to long dsRNAs, siRNAs can, in some cases, trigger an immune response. A recent report by Dharmacon demonstrates that siRNA duplexes 23 nucleotides long can activate interferon responses and cause cell death in culture [39]. Another recent study shows that certain siRNAs can bind to and activate Toll-like receptor 7 (TLR7) if they contain the professed “danger motif” (5’-GUCCUUCAA-3’) or similar GU-rich sequences that also can be recognized by TLR7 [40]. Because immune reactions can vary among different cell types, it is difficult to anticipate all in vivo responses based on in vitro work. Understandably, immunogenicity and toxicity are grounds for concern that must be addressed in developing RNAi for therapeutic use.
siRNA Delivery
A potent gene-silencing agent has no utility if it cannot be delivered to its intended cell type, tissue, or organ. Delivery of genetic material in vivo is the biggest obstacle faced by siRNA therapies [26,34]. And virus-based delivery systems, while efficient, may be fatally flawed due to the safety concerns they raise as they induce mutations and trigger immunogenic and inflammatory responses [42]. As a result, extensive work has been done to develop efficacious non-viral delivery systems, including direct chemical modification of siRNA, liposome formulations, nanoparticles, and targeting moieties. These novel strategies provide ways to safely overcome obstacles facing siRNA.
Chemical Modification
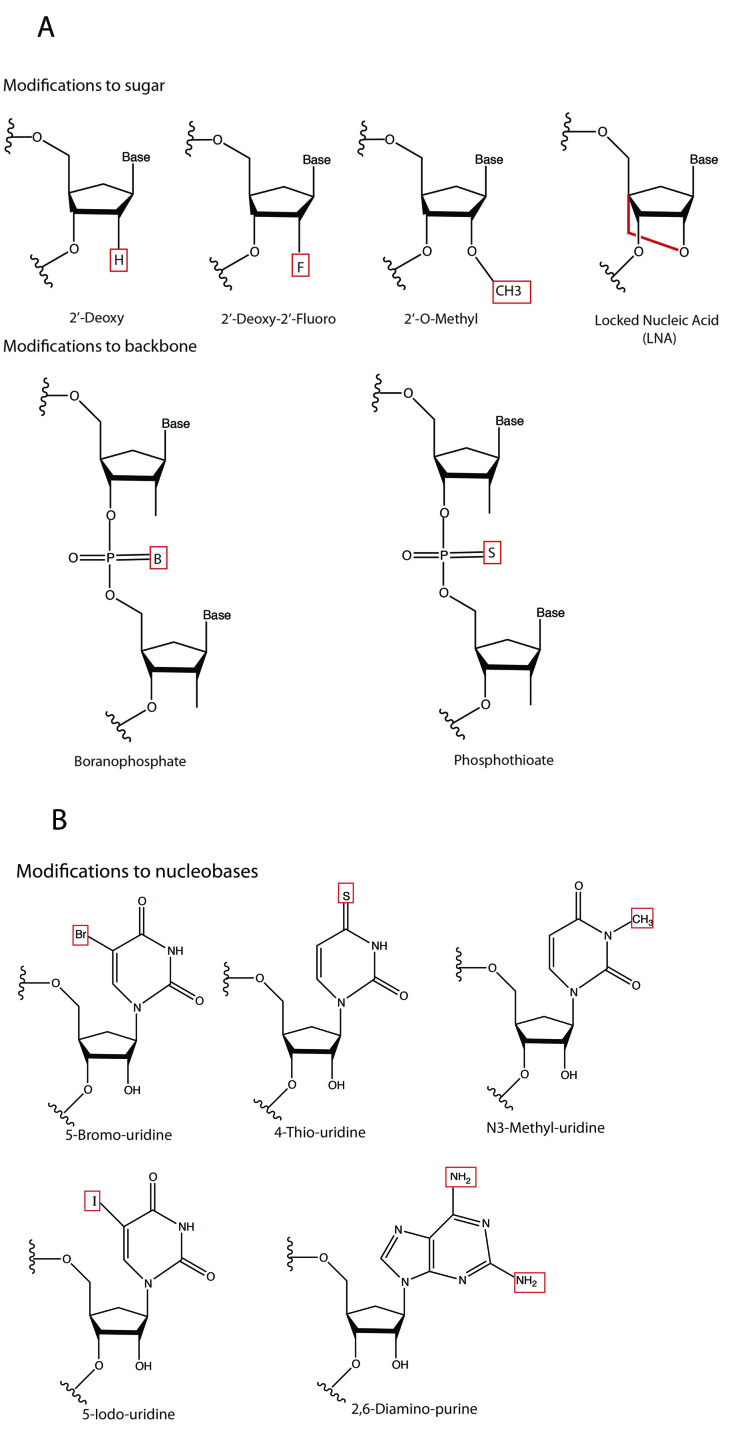
Chemical modifications can significantly enhance the stability and uptake of naked siRNAs [43]. Importantly, siRNAs can be directly modified without crippling their ability to silence their targets [43]. Chemical modifications have been rigorously investigated for virtually every part of siRNA molecules, from the termini and backbone to the sugars and bases, with the goal of engineering siRNA with prolonged half-life and increased cellular uptake. Most commonly, the sugar moiety is modified. For example, the incorporation of a 2’-fluoro (2’-F) [44], 2’-O-methyl [45], 2’-halogen, 2’-amine [46], or 2’-deoxy [47] can significantly increase the stability of siRNA in serum, as can the bridging of the sugar’s 2’- and 4’-positions with a –O-CH2 linker (producing what is called a “locked nucleic acid” or LNA) (Figure 3a) [48]. Among these, only the 2’-F can be introduced through endogenous transcription as opposed to chemical synthesis. Another caveat is that when the sugars of both strands of an siRNA duplex are replaced with 2’-O-methyl moieties, the duplex loses its silencing ability. However, 2’-O-methyl modification of only the sense strand leaves silencing activity intact as long as certain positions in the “seed” region of the sense strand are not modified [49]. Also, recent studies have shown that while heavy modification of siRNA duplexes with LNAs prolongs half-life in serum to as much as 90 hours, this is not without adverse affects on the gene-silencing activity, suggesting that the natural RNAi machinery can only accommodate moderate alterations of the chemical structure of siRNAs [50].
Figure 3.
A) Common chemical modifications to siRNA sugars and backbone. B) Chemical modifications to nucleobases.
Backbone modifications in siRNA duplexes can protect against nucleases in both the serum and cytoplasm. For example, modifying the internucleotide phosphate linkage in siRNA with phosphothioate (P = S) (Figure 3a) results in moderate stability improvement in a nuclease-containing milieu [43], while facilitating cellular uptake and preserving silencing function [51]; this is not without problems, as, in some cases, P = S substitution causes cytotoxicity [52]. Perhaps a better choice is modification of the backbone with boranophosphate (P = B) (Figure 3a), which enhances nuclease resistance by more than 10-fold as compared to unmodified constructs, without causing cytotoxicity or damage to siRNA silencing function [53].
A number of modifications to siRNA nucleobases have been explored with variable success. Replacement with 5-(3-aminoally)-uridine residues eradicates gene-silencing activity, whereas 4-thiouridine and 5-bromouridine modified duplexes remain functional [35,54]. Other common nucleobase modifications, such as 5-iodouridine, N-3-Me-uridine, and 2,6-diaminopurine residues (Figure 3b), can be tolerated if they are on the passenger strand or terminal area of the siRNA duplex, but not on the guide strand or seed region of the siRNA [55]. Interestingly, siRNA seed region nucleotides 2-8 (from the 5’ end of the guide) can be replaced with DNA nucleotides without adversely affecting silencing activity [55].
Modification of siRNA termini can be used for tuning pharmacokinetic properties, as well as for imparting new functionalities to siRNA duplexes. Tagging the ends of siRNAs with moieties such as cholesterol, folate, various peptides, and aptamers can aid in transport across cellular barriers or targeting to specific cells and organs (also see section Targeting) [56,57]. Likewise, fluorescent molecules can be attached to study siRNA biodistribution and uptake [35]. These modifications must preserve certain characteristics of the 5’ and 3’ ends of siRNAs. In particular, the 5’-phosphate group on the sense strand is necessary for gene silencing by RNAi. However, terminal modifications that leave the 5’-phosphodiester intact are able to retain their silencing ability [58]. Consequences of including modifications at the 3’-end are less consistent and depend on the particular modification [58]. For example, 3’-biotin has no adverse effects on silencing, whereas 3’-2-hydroxyethyl phosphate abolishes silencing activity [58]. SiRNAs containing 3’-ends with dinucleotide overhangs that mimic Dicer cleavage products are substantially more stable and efficient than those without; thus, most currently used synthetic siRNAs are made with 3’ overhangs [58].
Liposomes
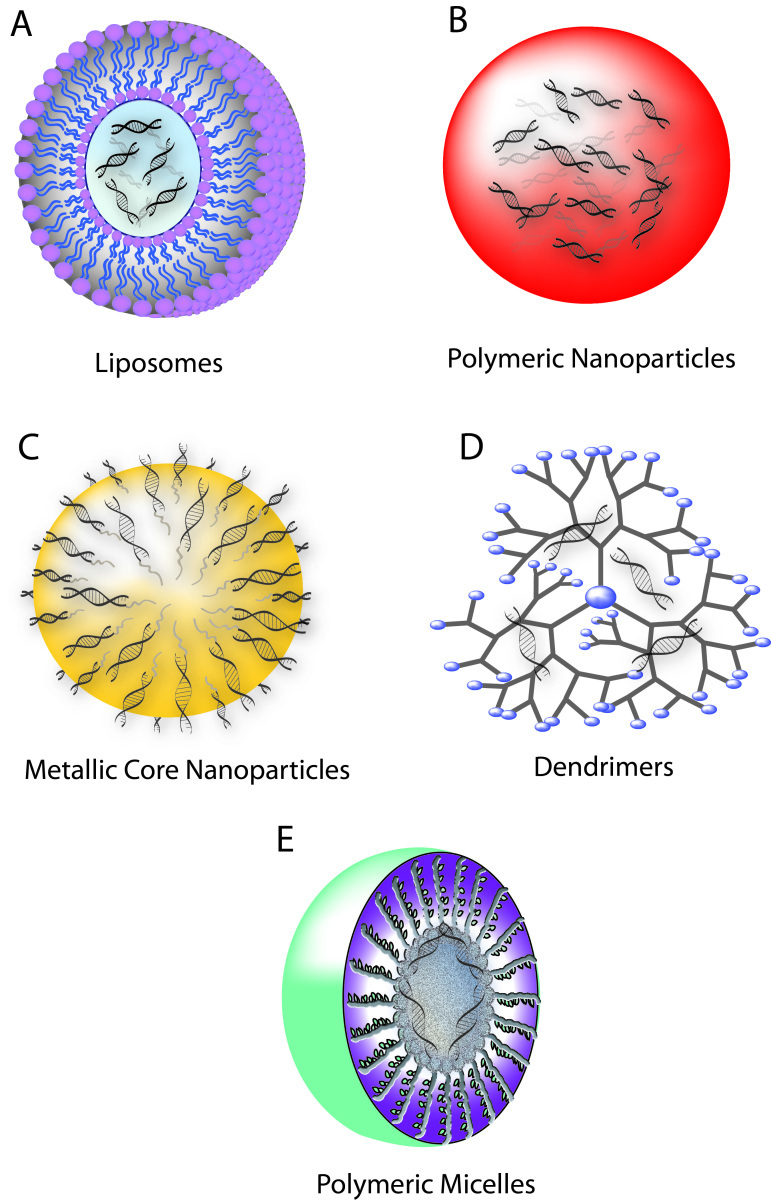
Loading siRNA cargo into liposomes — vesicles consisting of a phospholid bilayer that circumscribes an inner aqueous compartment — is a prominent strategy for delivery to target cells (Figure 4a). Developed early on in the pursuit of an efficient non-viral delivery approach, these vectors have since been rigorously explored and characterized [59]. Liposomes facilitate efficient internalization of their siRNA cargo via membrane fusion with the host cell [42]. Lipid encapsulation is an attractive delivery approach because of the biocompatibility of the constituents and facile assembly of the complexes, which requires only mixing and incubation of components [35]. In addition, these complexes can be engineered for specific delivery through conjugation of targeting moieties directly to the lipid molecules prior to liposome production. Neutral lipids are highly non-toxic and do not activate an immune response. 1,2-Oleoyl-sn-Glycero-3-phosphocholine (DOPC) and 1,2-Dioleoyl-sn-Glycero-3-phosphoethanolamine (DOPE) are among the most widely used neutral lipids. Simply mixing siRNA with DOPC results in more than 65 percent encapsulation, and these complexes have been shown to bring about siRNA-mediated silencing in cancer cells in vivo [60]. Generally, however, neutral liposomes yield relatively low transfection efficiency. Cationic lipids, such as 1-oleoyl-2-[6-[(7-nitro-2-1,3-benzoxadiazol-4-yl) amino]hexanoyl]-3-trimethylammonium propane (DOTAP), can complex electrostatically with siRNAs and be used to create a more effective liposome as the positively charged lipids provide enhanced cell entry and increased protection against serum enzymes [61]. But incorporation of positive charge to increase transfection efficiency must be carefully balanced against inflammatory effects that the polycations create in vivo, as well as unwanted interaction with negatively charged serum proteins, which can lead to opsonization and clearance of the lipocomplex [62].
Figure 4.
Schematic of siRNA nanocarriers. A) Liposomes. B) Polymeric nanoparticles. C) Metallic core nanoparticles. D) Dendrimers. E) Polymeric micelles.
Recently, a new wave of work in lipid-based delivery systems has demonstrated that some synthetic lipid-like materials (termed “lipidoids”) form complexes with siRNA or miRNA that facilitate intracellular delivery of the oligonucleotides [100]. In fact, lipidoids that are individually ineffective at delivering siRNA become surprisingly effective when formulated together as binary combinations into single delivery vehicles [63]. The rationale behind the synergy of certain combinations of materials is that while neither individual component material is capable of mediating every part of siRNA delivery (i.e., cellular entry, endosomal escape), each may facilitate one distinct step and together may be able to accomplish complete delivery [63]. The discovery of synergy among materials significantly expands the material space available for engineering a therapeutic delivery system and may produce important systems for siRNA delivery.
Nanoparticles
Polymeric nanoparticles are promising gene delivery systems because they offer stability and controlled release, have the capacity to encapsulate large amounts of genetic material, allow for co-delivery, and can readily be surface-modified to enhance stability, transport properties, targeting, or uptake. Polymers that are biodegradable, biocompatible, and non-toxic make attractive candidates for constructing in vivo delivery vehicles. Chitosan, cyclodextrin, polyethyleneimine (PEI), poly(lactic-co-glycolic) acid (PLGA), dendrimers, and metallic core nanoparticles have become popular for use in delivery systems, although none of these materials possess all of the desirable properties [35,42].
Chitosan is a natural, cationic polysaccharide harvestable from crustacean exoskeletons. It is an extensively studied biomaterial due to its biocompatibility, mucoadhesive properties, and nuclease resistance [35,64]. Optimal cationic charge for maximal siRNA encapsulation in chitosan can be attained by tuning the ratio of amines to phosphates (N:P). In two separate studies, optimized chitosan-siRNA nanoparticles have been successfully administered intranasally to silence GAPDH and EGFP in the lungs of mice [65,66].
Cyclodextrin-based polycations (CDPs) are another class of highly non-toxic polymer-based complexes used to deliver siRNAs, as well as other therapeutic compounds such as plasmids and small molecule drugs. These constructs consist of cationic polymer complexed with siRNA duplexes and interdigitated with funnel-like cyclodextrin molecules, which, in turn, can be linked to functionalized adamantane molecules [35]. Impressive in vivo results were recently reported with siRNA-loaded CDPs functionalized with adamantane-transferrin and adamantane-PEG. Functionalized CDPs loaded with siRNAs targeting fusion oncoprotein EWS-FLI1 were administered to non-human primates and demonstrated to bring about shrinkage of implanted tumors [67]. Significantly, early results also suggest that these materials can produce RNAi in humans [98].
Extensive branching and dense cationic charge gives synthetic polymer polyethyleneimine (PEI) the capacity to condense siRNAs, protect them from degradation by RNases, and facilitate their cellular uptake via endocytosis [68]. An added feature is the ability of PEI to act as a proton sponge, because its extensive amine groups buffer the acidic inner compartment of an endosome causing water to swell the endosome to the point of rupture, thereby facilitating endosomal escape of its encapsulated siRNA [35]. Some wariness surrounds PEI use in vivo, however, due to in vitro evidence of high cytotoxicity [69]. In an effort to reduce toxic effects of PEI, the polymer has been modified with polyethylene glycol (PEG) (previously demonstrated to slow clearance and reduce toxicity) and the PEI-PEG/siRNA complex shown to exhibit decreased toxicity, but drastically increased particle size [70].
The degradable polymer PLGA is another attractive choice for siRNA encapsulation toward in vivo delivery of siRNA. FDA-approved PLGA breaks down by hydrolysis of its ester bonds into lactic and glycolic acids — natural metabolic breakdown products of the body. In addition to the biocompatibility advantage, PLGA also can be easily assembled into a carrier system for large amounts of siRNA that offers controlled and sustained release [71]. PLGA nanoparticles (~100nm in diameter) are capable of achieving intracellular delivery of DNA plasmids [72], siRNAs [71], and chemotherapeutic agents [73]. Still further, PLGA nanoparticles are favorable siRNA delivery systems because they can readily be surface-modified to enhance targeting or uptake [97,99].
Dendrimers are heavily branched polymeric molecules that can be engineered to form modular, nano-sized, spherical strutures for siRNA delivery (Figure 4d). Packaging siRNAs in dendrimer structures can be accomplished by positively charging the core while abolishing surface charge [74]. Alternatively, siRNAs can be caged within dendrimer polyplexes via disulfide linkages, which incidentally also provide for controlled release in the reducing intracellular milieu. These structures can be additionally stabilized through the incorporation of PEG [75]. The modularity of dendrimers allows for dendrimer-siRNA polyplexes to be further improved for siRNA delivery by combining them with targeting ligands and technologies that provide for endosomal release [76].
Polymeric micelles share some characteristics with liposomes and polymeric nanoparticles, providing the stealth properties of liposomes with their hydrophilic shells while simultaneously offering protective stability within their hydrophobic cores (Figure 4e) [77,79]. These self-assembled nanostructures composed of amphiphilic block copolymers can be tuned for siRNA delivery by grafting them with amines that can complex siRNA. Alternatively, siRNA can be “reversibly” conjugated to the amphiphilic polymers through disulfide bonds, which are then reduced intracellularly to release the siRNA [80]. The ability of polymeric micelles to both remain stable through dilution in biological fluids and shelter siRNA from degradation makes them promising carriers for therapeutic development [77].
Another siRNA delivery strategy involves metallic core nanoparticles (Figure 4c) [83]. Metal cores of iron oxide, iron cobalt, iron gold, or iron nickel are coated with a layer of sugars or other polymers generating a core-shell structure to which siRNA can be externally conjugated through linking molecules such as thiols [78], dextran [79], cationic polymers [80,82], or biotin-streptavadin [35]. Contingent upon the metal used, the cores of these particles can impart properties that allow for study of biodistribution upon injection using magnetic resonance imaging or targeting to specific tissues by applying external magnets. These systems provide unique advantages other approaches lack, but in vivo toxicity may prove prohibitory. More recently, multifunctional platforms, such as iron oxide nanoparticles-dendrimer complexes [81], have been shown to effectively deliver siRNA in vivo representing headway of novel combinatorial strategies.
Targeting
Selectively targeting siRNAs to diseased cells or tissues increases accumulation of the therapeutic at the site of interest, increasing the silencing potency, thereby making a given treatment dose more effective. In addition, targeting to cells adds another layer of specificity: avoiding off-target effects by decreasing the probability of uptake by healthy cells. A common drug delivery strategy for targeting cells of interest is conjugation to ligands such as antibodies, aptamers, small molecules, and peptides that specifically interact with corresponding surface moieties of target cells [90,91].
Antibodies have been widely used for targeting in a number of drug delivery applications. Popular for their specificity, diversity, and ability to modulate biodistribution, antibodies against targets ranging from oncogenes such as HER2 [84] to HIV envelope proteins [29] have been appended to nanoparticles for in vivo target-site delivery. An unfortunate difficulty of this approach is the large size of antibody targeting moieties, which can make them difficult to conjugate to particle surfaces at high concentrations. And if translated to the clinics, long-term administration of treatment may be limited by immune responses to the targeting reagent. However, incorporation of humanized antibodies may provide the solution to this problem.
As an alternate to antibodies, aptamers have been explored for targeted siRNA delivery. Aptamers are able to bind their respective ligand molecules with an affinity and specificity on the same scale as antibody-antigen partners, but without recognition by native antibodies, which makes them more amenable to long-term treatment schedules [35]. However, the number of aptamers known to bind targets that are markers for disease is extremely limited. And moreover, aptamer binding to targets does not always lead to cargo internalization [35]. Aptamer-based targeting may become more widespread as more aptamers for disease-relevant are targets are discovered.
Cholesterol and its derivatives have been effectively employed as targeting ligands. The surfaces of hepatocytes are heavily populated with cholesterol receptors, which internalize cholesterol through endocytosis [85,86]. This property has been used for liver-targeting. In a notable study, siRNA against apolipoprotein B (apoB) was directly conjugated to cholesterol and administered intravenously in mice. The constructs were shown to accumulate in the liver and to reduce apoB levels by more than 57 percent [87]. In addition to targeting, cholesterol conjugation has been shown to impart generally desirable “drug-like” properties such as stability and bioavailability.
Folate receptors are highly overexpressed in a number of cancers but are virtually absent in all other normal tissues, with the exception of the kidneys [88,89]. Active folate receptor facilitates cellular uptake of folate compounds and folate conjugates [92]. Its abilities to mediate internalization and its narrow expression range make it an attractive surface target, and conjugating folate to the surface of particles has become a common strategy in cancer drug delivery [93]. Similarly to folate receptors, some cancer cells overexpress surface receptors responsive to larger protein ligands such as transferrin. SiRNA-loaded particles conjugated with transferrin moities have been shown to preferentially accumulate in certain metastasized tumors and bring about significant reduction of tumor growth in mice and have been successfully tested in non-human primates [94,95].
A general surface modification shown to be tremendously effective in a wide variety of systems is the attachment of a polyethylene glycol (PEG) moiety. PEG has been shown to generally reduce toxicity and slow clearance from the blood, thereby profoundly prolonging half-life and bioavailability and allowing for longer-term treatment administration and resulting in an overall increase in drug activity achieved [96]. Extension in half-life of materials in the circulation can improve passive targeting to tumors by the enhanced permeability and retention (EPR) effect [101,102].
Although small, nanoparticles are generally too large to easily transverse the plasma membrane. Cell membrane translocation can be facilitated by conjugating cell-penetrating peptides (CPPs) to the nanoparticle surface to aid transduction. Well-studied CPPs penetratin (ANTP) and TAT have been demonstrated to significantly enhance uptake in a number of applications [97]. Further, there is evidence for a synergistic effect between these CPPs and folate, which may also be found among other combinations of surface moieties [97].
Conclusion
The discovery of siRNAs — constructs that can be designed to specifically and efficiently silence genes of interest — has stirred considerable excitement. Most exciting is the potential therapeutic application of this technology. Though a number of challenges stand in the way of realizing this potential, the biggest bottleneck in siRNA delivery, over a decade of innovative engineering has resulted in solutions to a number of these challenges, laying down a foundation for continuing headway toward making widespread therapeutic siRNA a reality hopefully in the not too distant future.
Abbreviations
- RNAi
RNA interference
- siRNA
small interfering RNA
- AGO2
Argonaute
- RISC
RNA-induced silencing complex
- miRNA
micro RNA
- TLR7
toll-like receptor 7
- LNA
Locked Nucleic Acid
- DOPC
1,2-Oleoyl-sn-Glycero-3-phosphocholine
- DOPE
1,2-Dioleoyl-sn-Glycero-3-phosphoethanolamine
- DOTAP
1-oleoyl-2-[6-[(7-nitro-2-1,3-benzoxadiazol-4-yl) amino]hexanoyl]-3-trimethylammonium propane
- PEI
polyethyleneimine
- PLGA
poly(lactic-co-glycolic) acid
- CDPs
Cyclodextrin-based polycations
- PEG
polyethylene glycol
- EPR
enhanced permeability and retention
- CPPs
cell-penetrating peptides
References
- Fire A, Xu S, Montgomery MK, Kostas SK, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391(6669):806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Hamilton AJ, Baulcombe DC. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science. 1999;286(5441):950–952. doi: 10.1126/science.286.5441.950. [DOI] [PubMed] [Google Scholar]
- Zamore PD, Tuschl T, Sharp PA, Bartel DP. RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell. 2000;101(1):25–33. doi: 10.1016/S0092-8674(00)80620-0. [DOI] [PubMed] [Google Scholar]
- Hammond SM, Bernstein E, Beach D, Hannon GJ. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. 2000;404(6775):293–296. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411(6836):494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- Elbashir SM, Lendeckel W, Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 2001;15(2):188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409(6818):363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- Nykanen A, Haley B, Zamore PD. ATP requirements and small interfering RNA structure in the RNA interference pathway. Cell. 2001;107(3):309–321. doi: 10.1016/s0092-8674(01)00547-5. [DOI] [PubMed] [Google Scholar]
- Elbashir SM, Martinez J, Patkaniowska A, Lendeckel W, Tuschl T. Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J. 2001;20(23):6877–6888. doi: 10.1093/emboj/20.23.6877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory RI, Chendrima TP, Cooch N, Shiekhattar R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell. 2005;123(4):631–640. doi: 10.1016/j.cell.2005.10.022. [DOI] [PubMed] [Google Scholar]
- Macrae IJ, Ma E, Zhou M, Robinson CV, Doudna JA. In vitro reconstitution of the human RISC-loading complex. Proc Natl Acad Sci USA. 2008;105(2):512–517. doi: 10.1073/pnas.0710869105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Doudna JA. A three-dimensional view of the molecular machinery of RNA interference. Nature. 2009;457(7228):405–412. doi: 10.1038/nature07755. [DOI] [PubMed] [Google Scholar]
- Hutvagner G, Simar MJ. Argonaute proteins: key players in RNA silencing. Nat Rev Mol Cell Biol. 2008;9(1):22–32. doi: 10.1038/nrm2321. [DOI] [PubMed] [Google Scholar]
- Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ. et al. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305(5689):1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- Song JJ, Smith SK, Hannon GJ, Joshua-Tor L. Crystal structure of Argonaute and its implications for RISC slicer activity. Science. 2004;305(5689):1434–1437. doi: 10.1126/science.1102514. [DOI] [PubMed] [Google Scholar]
- Song JJ, Liu J, Tolia NH, Schneiderman J, Smith SK, Martienssen RA. et al. The crystal structure of the Argonaute2 PAZ domain reveals an RNA binding motif in RNAi effector complexes. Nat Struct Biol. 2003;10(12):1026–1032. doi: 10.1038/nsb1016. [DOI] [PubMed] [Google Scholar]
- Lingel A, Simon B, Izaurralde E, Sattler M. Structure and nucleic-acid binding of the Drosophila Argonaute 2 PAZ domain. Nature. 2003;426(6965):465–469. doi: 10.1038/nature02123. [DOI] [PubMed] [Google Scholar]
- Yan KS, Yan S, Farooq A, Han A, Zeng L, Zhou MM. Structure and conserved RNA binding of the PAZ domain. Nature. 2003;426(6965):468–474. doi: 10.1038/nature02129. [DOI] [PubMed] [Google Scholar]
- Ma JB, Yuan YR, Meister G, Pei Y, Tuschl T, Patel DJ. Structural basis for 5’-end-specific recognition of guide RNA by the A. fulgidus Piwi protein. Nature. 2005;434(7033):666–670. doi: 10.1038/nature03514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz DS, Hutvagner G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115(2):199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- Parker JS, Roe SM, Barford D. Structural insights into mRNA recognition from a PIWI domain-siRNA guide complex. Nature. 2005;434(7033):663–666. doi: 10.1038/nature03462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu YL, Rana TM. RNAi in human cells: basic structural and functional features of small interfering RNA. Mol Cell. 2009;10(3):549–561. doi: 10.1016/s1097-2765(02)00652-4. [DOI] [PubMed] [Google Scholar]
- Carthew RW, Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell. 2009;136(4):642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11(9):597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- Bagga S, Bracht J, Hunter S, Massirer K, Holtz J, Eachus R. et al. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005;122(4):553–563. doi: 10.1016/j.cell.2005.07.031. [DOI] [PubMed] [Google Scholar]
- Aagaard L, Rossi JJ. RNAi therapeutics: principles, prospects and challenges. Adv Drug Deliv Rev. 2007;59(2-3):75–86. doi: 10.1016/j.addr.2007.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond SM. MicroRNA therapeutics: a new niche for antisense nucleic acids. Trends Mol Med. 2006;12(3):99–101. doi: 10.1016/j.molmed.2006.01.004. [DOI] [PubMed] [Google Scholar]
- Bertrand JR, Pottier M, Vekris A, Opolon P, Maksimenko A, Malvy C. Comparison of antisense oligonucleotides and siRNAs in cell culture and in vivo. Biochem Biophys Res Commun. 2002;296(4):1000–1004. doi: 10.1016/s0006-291x(02)02013-2. [DOI] [PubMed] [Google Scholar]
- Song E, Zhu P, Lee SK, Chowdhury D, Kussman S, Dykxhoorn DM. et al. Antibody mediated in vivo delivery of small interfering RNAs via cell-surface receptors. Nat Biotechnol. 2005;23(6):709–717. doi: 10.1038/nbt1101. [DOI] [PubMed] [Google Scholar]
- Heale BS, Soifer HS, Bowers C, Rossi JJ. siRNA target site secondary structure predictions using local stable substructures. Nucleic Acid Res. 2005;33(3):e30. doi: 10.1093/nar/gni026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patzel V, Rutz S, Dietrich I, Koberle C, Scheffold A, Kaufmann SH. Design of siRNAs producing unstructured guide-RNAs results in improved RNA interference efficiency. Nat Biotechnol. 2005;23(11):1440–1444. doi: 10.1038/nbt1151. [DOI] [PubMed] [Google Scholar]
- Amarzguioui M, Prydz H. An algorithm for selection of functional siRNA sequences. Biochem Biophys Res Commun. 2004;316(4):1050–1058. doi: 10.1016/j.bbrc.2004.02.157. [DOI] [PubMed] [Google Scholar]
- Reynolds A, Leake D, Boese Q, Scaringe S, Marshall WS, Khvorova A. Rational siRNA design for RNA interference. Nat Biotechnol. 2004;22(3):326–330. doi: 10.1038/nbt936. [DOI] [PubMed] [Google Scholar]
- Behlke MA. Progress towards in vivo use of siRNAs. Mol Ther. 2006;13(4):644–670. doi: 10.1016/j.ymthe.2006.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo P, Coban O, Snead NM, Trebley J, Hoeprich S, Guo S. et al. Engineering RNA for targeted siRNA delivery and medical application. Adv Drug Deliv Rev. 2010;62(6):650–666. doi: 10.1016/j.addr.2010.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson AL, Bartz SR, Schelter J, Kobayashi SV, Burchard J, Mao M. et al. Expression profiling reveals off-target gene regulation by RNAi. Nat Biotechnol. 2003;21(6):635–637. doi: 10.1038/nbt831. [DOI] [PubMed] [Google Scholar]
- Birmingham A, Anderson EM, Reynolds A, Ilsley-Tyree D, Leake D, Fedorov Y. et al. 3' UTR seed matches, but not overall identity, are associated with RNAi off-targets. Nat Methods. 2006;3(3):199–204. doi: 10.1038/nmeth854. [DOI] [PubMed] [Google Scholar]
- Jackson AL, Burchard J, Schelter J, Chau BN, Cleary M, Lim L. et al. Widespread siRNA “off-target” transcript silencing mediated by seed region sequence complementarity. RNA. 2006;12(7):1179–1187. doi: 10.1261/rna.25706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds A, Anderson EM, Vermeulen A, Fedorov Y, Robinson K, Leake D. et al. Induction of the interferon response by siRNA is cell type- and duplex length- dependent. RNA. 2006;12(6):988–993. doi: 10.1261/rna.2340906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornung V, Guenthner-Biller M, Bourquin C, Ablasser A, Schlee M, Uematsu S. et al. Sequence-specific potent induction of IFN-alpha by short interfering RNA in plasmacytoid dendritic cells through TLR7. Nat Med. 2005;11(3):263–270. doi: 10.1038/nm1191. [DOI] [PubMed] [Google Scholar]
- Kim DH, Behlke MA, Rose SD, Chang MS, Choi S, Rossi JJ. et al. Synthetic dsRNA Dicer substrates enhance RNAi potency and efficacy. Nat Biotechnol. 2005;23(2):227–231. doi: 10.1038/nbt1051. [DOI] [PubMed] [Google Scholar]
- Gao Y, Liu XL, Li XR. Research progress on siRNA delivery with nonviral carriers. Int J Nanomedicine. 2011;6:1017–1025. doi: 10.2147/IJN.S17040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choung S, Kim YJ, Kim S, Park HO, Choi YC. Chemical modification of siRNAs to improve serum stability without loss of efficacy. Biochem Biophys Res Commun. 2006;342(3):919–927. doi: 10.1016/j.bbrc.2006.02.049. [DOI] [PubMed] [Google Scholar]
- Kawasaki AM, Casper MD, Freier SM, Lesnik EA, Zounes MC, Cummins LL. et al. Uniformly modified 2’-deoxy-2’-fluoro phosphorothioate oligonucleotides as nuclease-resistant antisense compounds with high affinity and specificity for RNA targets. J Med Chem. 1993;36(7):831–841. doi: 10.1021/jm00059a007. [DOI] [PubMed] [Google Scholar]
- Rusckowski M, Qu T, Roskey A, Agrawal S. Biodistribution and metabolism of a mixed backbone oligonucleotide (GEM 231) following single and multiple dose administration in mice. Antisense Nucleic Acid Drug Dev. 2000;10(5):333–345. doi: 10.1089/oli.1.2000.10.333. [DOI] [PubMed] [Google Scholar]
- Pieken WA, Olsen DB, Benseler F, Aurup H, Eckstein F. Kinetic characterization of ribonuclease-resistant 2’-modified hammerhead ribozymes. Science. 1991;253(5017):314–317. doi: 10.1126/science.1857967. [DOI] [PubMed] [Google Scholar]
- Parrish S, Fleenor J, Xu S, Mello C, Fire A. Functional anatomy of a dsRNA trigger: differential requirement for the two trigger strands in RNA interference. Mol Cell. 2000;6(5):1077–1087. doi: 10.1016/s1097-2765(00)00106-4. [DOI] [PubMed] [Google Scholar]
- Elmen J, Thonberg K, Ljungberg K, Frieden M, Westergaard M, Xu Y. et al. Locked nucleic acid (LNA) mediated improvements in siRNA stability and functionality. Nucleic Acids Res. 2005;33(1):439–447. doi: 10.1093/nar/gki193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen PY, Weinmann L, Gaidatzis D, Pei Y, Zavolan M, Tuschl T. et al. Strand-specific 5’-O-methylation of siRNA duplexes controls guide strand selection and targeting specificity. RNA. 2008;14(2):263–274. doi: 10.1261/rna.789808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mook OR, Baas F, de Wissel MB, Fluiter K. Evaluation of locked nucleic acid-modified small interfering RNA in vitro and in vivo. Mol Cancer Ther. 2007;6(3):833–843. doi: 10.1158/1535-7163.MCT-06-0195. [DOI] [PubMed] [Google Scholar]
- Overhoff M, Sczakiel G. Phosphorothioate-stimulated uptake of short interfering RNA by human cells. EMBO Rep. 2005;6(12):1176–1181. doi: 10.1038/sj.embor.7400535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stessl M, Machetti-Deschmann M, Winkler J, Lachmann B, Allmaier G, Noe CR. A proteomic study reveals unspecific apoptosis induction and reduction of glycolytic enzymes by the phosphorothioate antisense oligonucleotide oblimersen in human melanoma cells. J Proteomics. 2009;72(6):1019–1030. doi: 10.1016/j.jprot.2009.06.001. [DOI] [PubMed] [Google Scholar]
- Hall AH, Wan J, Shaughnessy EE, Ramsay Shaw B, Alexander KA. RNA interference using boranophosphate siRNAs: structure-activity relationships. Nucleic Acids Res. 2004;32(20):5991–6000. doi: 10.1093/nar/gkh936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrish JZ, Xue D. Functional genomic analysis of apoptotic DNA degradation in C. elegans. Mol Cell. 2003;11(4):987–996. doi: 10.1016/s1097-2765(03)00095-9. [DOI] [PubMed] [Google Scholar]
- Ui-Tei K, Naito Y, Zenno S, Nishi L, Yamato K, Takashi F. et al. Functional dissection of siRNA sequence by systematic DNA substitution: modified siRNA with a DNA seed arm is a powerful tool for mammalian gene silencing with significantly reduced off-target effect. Nucleic Acids Res. 2008;36(7):2136–2151. doi: 10.1093/nar/gkn042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz C, Hadwiger P, John M, Vornlocher HP, Unverzagt C. Steroid and lipid conjugates of siRNAs to enhance cellular uptake and gene silencing. Bioorg Med Chem Lett. 2004;14(19):4975–4977. doi: 10.1016/j.bmcl.2004.07.018. [DOI] [PubMed] [Google Scholar]
- Guo S, Huang F, Guo P. Construction of folate-conjugated pRNA of bacteriophage phi29 DNA packaging motor for delivery of chimeric siRNA to nasopharyngeal carcinoma cells. Gene Ther. 2006;13(10):814–820. doi: 10.1038/sj.gt.3302716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manoharan M. RNA interference and chemically modified small interfering RNAs. Curr Opin Chem Biol. 2004;8(6):570–579. doi: 10.1016/j.cbpa.2004.10.007. [DOI] [PubMed] [Google Scholar]
- Li W, Szoka FC Jr.. Lipid-based nanoparticles for nucleic acid delivery. Pharm Res. 2007;24(3):438–449. doi: 10.1007/s11095-006-9180-5. [DOI] [PubMed] [Google Scholar]
- Landed CN Jr., Chavez-Reyes A, Bucana C, Schmandt R, Deavers MT, Lopez-Berestein G. et al. Therapeutic EphA2 gene targeting in vivo using neutral liposomal small interfering RNA delivery. Cancer Res. 2005;65(15):6910–6918. doi: 10.1158/0008-5472.CAN-05-0530. [DOI] [PubMed] [Google Scholar]
- Zhang C, Tang N, Liu X, Liang W, Xu W, Torchilin VP. siRNA-containing liposomes modified with polyarginine effectively silence the targeted gene. J Control Release. 2006;112(2):229–239. doi: 10.1016/j.jconrel.2006.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris TJ, Green JJ, Fung PW, Langer R, Anderson DG, Bhatia SN. Tissue-specific gene delivery via nanoparticle coating. Biomaterials. 2010;31(5):998–1006. doi: 10.1016/j.biomaterials.2009.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead KA, Sahay G, Li GZ, Love KT, Alabi CA, Ma M. et al. Synergistic silencing: combinations of lipid-like materials for efficacious siRNA delivery. Mol Ther. 2011;19(9):1688–1694. doi: 10.1038/mt.2011.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard K. Unlocking the money-making potential of RNAi. Nat Biotechnol. 2003;21(12):1441–1446. doi: 10.1038/nbt1203-1441. [DOI] [PubMed] [Google Scholar]
- Ghosn B, Singh A, Li M, Vlassov AV, Burnett C, Puri N. et al. Efficient gene silencing in lungs and liver using imidazole-modified chitosan as a nanocarrier for small interfering RNA. Oligonucleotides. 2010;20(3):163–172. doi: 10.1089/oli.2010.0235. [DOI] [PubMed] [Google Scholar]
- Howard KA, Rahbek UL, Liu X, Damgaard CK, Glud SZ, Andersen MO. et al. RNA interference in vitro and in vivo using a novel chitosan/siRNA nanoparticle system. Mol Ther. 2006;14(4):476–484. doi: 10.1016/j.ymthe.2006.04.010. [DOI] [PubMed] [Google Scholar]
- Hu-Lieskovan S, Heidel JD, Bartlett DW, Davis ME, Triche TJ. Sequence-specific knockdown of EWS-FLI1 by targeted, nonviral delivery of small interfering RNA inhibits tumor growth in a murine model of metastatic Ewing’s sarcoma. Cancer Res. 2005;65(19):8984–8992. doi: 10.1158/0008-5472.CAN-05-0565. [DOI] [PubMed] [Google Scholar]
- Urban-Klein B, Werth S, Abuharbeid S, Czubayko F, Aigner A. RNAi-mediated gene-targeting through systemic application of polyethylenimine (PEI)-complexed siRNA in vivo. Gene Ther. 2004;12(5):461–466. doi: 10.1038/sj.gt.3302425. [DOI] [PubMed] [Google Scholar]
- Xie FY, Woodle MC, Lu PY. Harnessing in vivo siRNA delivery for drug discovery and therapeutic development. Drug Discov Today. 2006;11(1-2):67–73. doi: 10.1016/S1359-6446(05)03668-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiffelers RM, Mixson AJ, Ansari AM, Fens MH, Tang Q, Zhou Q. et al. Transporting silence: design of carriers for siRNA to angiogenic endothelium. J Control Release. 2005;109(1-3):5–14. doi: 10.1016/j.jconrel.2005.05.018. [DOI] [PubMed] [Google Scholar]
- Woodrow KA, Cu Y, Booth CJ, Saucier-Sawyer JK, Wood MJ, Saltzman WM. Intravaginal gene silencing using biodegradable polymer nanoparticles densely loaded with small-interfering RNA. Nat Mater. 2009;8(6):526–533. doi: 10.1038/nmat2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum JS, Saltzman WM. High loading efficiency and tunable release of plasmid DNA encapsulated in submicron particles fabricated from PLGA conjugated with poly-L-lysine. J Control Release. 2008;129(1):66–72. doi: 10.1016/j.jconrel.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Fong PM, Lu J, Russell KS, Booth CJ, Saltzman WM. et al. PEGylated PLGA nanoparticles for the improved delivery of doxorubicin. Nanomedicine. 2009;5(4):410–418. doi: 10.1016/j.nano.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil ML, Zhang M, Betigeri S, Taratula O, He H, Minko T. Surface-modified and internally cationic polyamidoamine dendrimers for efficient siRNA delivery. Bioconjugate Chem. 2008;19(7):1396–1403. doi: 10.1021/bc8000722. [DOI] [PubMed] [Google Scholar]
- Taratula O, Garbuzenko OB, Kirkpatrick P, Pandya I, Savla R, Pozharov VP. et al. Surface-engineered targeted PPI dendrimer for efficient intracellular and intratumoral siRNA delivery. J Control Release. 2009;140(3):284–293. doi: 10.1016/j.jconrel.2009.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal A, Min DH, Singh N, Zhu H, Birjiniuk A, von Maltzahn G. et al. Functional delivery of siRNA in mice using dendriworms. ACS Nano. 2009;3(9):2495–2504. doi: 10.1021/nn900201e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis MF, Cristea M, Winnik FM. Polymeric micelles for oral drug delivery: why and how. Pure Appl Chem. 2004;76(7-8):1321–1335. [Google Scholar]
- Kataoka K, Harada A, Nagasaki Y. Block copolymer micelles for drug delivery: design, characterization and biological significance. Adv Drug Deliv Rev. 2001;47(1):113–131. doi: 10.1016/s0169-409x(00)00124-1. [DOI] [PubMed] [Google Scholar]
- Moore A, Medarova Z, Potthast A, Dai G. In vivo targeting of underglycosylated MUC-1 tumor antigen using a multimodal imaging probe. Cancer Res. 2004;64(5):1821–1827. doi: 10.1158/0008-5472.can-03-3230. [DOI] [PubMed] [Google Scholar]
- Medarova Z, Pham W, Farrar C, Petkova V, Moore A. In vivo imaging of siRNA delivery and silencing in tumors. Nat Med. 2007;13(3):372–377. doi: 10.1038/nm1486. [DOI] [PubMed] [Google Scholar]
- Taratula O, Garbuzenko O, Savla R, Wang YA, He H, Minko T. Multifunctional nanomedicine platform for cancer specific delivery of siRNA by superparamagnetic iron oxide nanoparticles-dendrimer complexes. Curr Drug Deliv. 2011;8(1):59–69. doi: 10.2174/156720111793663642. [DOI] [PubMed] [Google Scholar]
- Liu G, Xie J, Zhang F, Wang Z, Luo K, Zhu L. et al. N-Alkyl-PEI-functionalized iron oxide nanoclusters for efficient siRNA delivery. Small. 2011;7(19):2742–2749. doi: 10.1002/smll.201100825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin S, Leach JC, Ye K. Nanoparticle-mediated gene delivery. Methods Mol Biol. 2009;544:547–557. doi: 10.1007/978-1-59745-483-4_34. [DOI] [PubMed] [Google Scholar]
- Tan WB, Jiang S, Zhang Y. Quantum-dot based nanoparticles for targeted silencing of HER2/neu gene via RNA interference. Biomaterials. 2007;28(8):1565–1571. doi: 10.1016/j.biomaterials.2006.11.018. [DOI] [PubMed] [Google Scholar]
- Lorenz C, Hadwiger P, John M, Vornlocher HP, Unverzagt C. Steroid and lipid conjugates of siRNAs to enhance cellular uptake and gene silencing in liver cells. Bioorg Med Chem Lett. 2004;14(19):4975–4977. doi: 10.1016/j.bmcl.2004.07.018. [DOI] [PubMed] [Google Scholar]
- McGookey DJ, Fagerberg K, Anderson RG. Filipin-cholesterol complexes form in uncoated vesicle membrane derived from coated vesicles during receptor-mediated endocytosis. J Cell Bio. 1983;96(5):1273–1278. doi: 10.1083/jcb.96.5.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soutschek J, Akinc A, Bramlage B, Charisse K, Constien R, Donoghue M. et al. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature. 2004;432(7014):173–178. doi: 10.1038/nature03121. [DOI] [PubMed] [Google Scholar]
- Mathias CJ, Wang S, Lee RJ, Waters DJ, Low PS, Green MA. Tumor-selective radiopharmaceutical targeting via receptor-mediated endocytosis of gallium-67-deferoxamine-folate. J Nucl Med. 1996;37(6):1003–1008. [PubMed] [Google Scholar]
- Lee RJ, Low PS. Delivery of liposomes into cultured KB cells via folate receptor-mediated endocytosis. J Biol Chem. 1994;269(5):3198–3204. [PubMed] [Google Scholar]
- Gu FX, Karnik R, Wang AZ, Alexis F, Levy-Nissenbaum E, Hong S. et al. Targeted nanoparticles for cancer therapy. Nano Today. 2007;2(3):14–21. [Google Scholar]
- Allen TM. Ligand-targeted therapeutics in anticancer therapy. Nat Rev Cancer. 2002;2(10):750. doi: 10.1038/nrc903. [DOI] [PubMed] [Google Scholar]
- Wang H, Zheng X, Behm FG, Ratnam M. Differentiation-independent retinoid induction of folate receptor type beta, a potential tumor target in myeloid leukemia. Blood. 2000;96(10):3529–3536. [PubMed] [Google Scholar]
- Yoshizawa T, Hattori Y, Hakoshima M, Koga K, Maitani Y. Folate-linked lipid-based nanoparticles for synthetic siRNA delivery in KB tumor xenografts. Eur J Pharm Biopharm. 2008;70(3):718–725. doi: 10.1016/j.ejpb.2008.06.026. [DOI] [PubMed] [Google Scholar]
- Hu-Lieskovan S, Heidel JD, Bartlett DW, Davis ME, Triche TJ. Sequence-specific knockdown of EWS-FLI1 by targeted, nonviral delivery of small interfering RNA inhibits tumor growth in a murine model of metastatic Ewing’s sarcoma. Cancer Res. 2005;65(19):8984–8992. doi: 10.1158/0008-5472.CAN-05-0565. [DOI] [PubMed] [Google Scholar]
- Heidel JD, Yu Z, Liu JY, Rele SM, Liang Y, Zeidan RK. et al. Administration in non-human primates of escalating intravenous doses of targeted nanoparticles containing ribonucleotide reductase subunit M2 siRNA. PNAS. 2007;104(14):5715–5721. doi: 10.1073/pnas.0701458104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dassie JP, Liu XY, Thomas GS, Whitaker RM, Thiel KW, Stockdale KR. et al. Systemic administration of optimized aptamer-siRNA chimeras promotes regression of PSMA-expressing tumors. Nat Biotechnol. 2009;27(9):839–849. doi: 10.1038/nbt.1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng CJ, Saltzman WM. Enhanced siRNA delivery into cells by exploiting the synergy between targeting ligands and cell-penetrating peptides. Biomaterials. 2011;32(26):6194–6203. doi: 10.1016/j.biomaterials.2011.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis ME, Zuckerman JE, Choi CH, Seligson D, Tolcher A, Alabi CA. et al. Evidence of RNAi in humans from systemically administered siRNA via targeted nanoparticles. Nature. 2010;464:1067–1070. doi: 10.1038/nature08956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Patel TR, Fu M, Bertram JP, Saltzman WM. Octa-functional PLGA nanoparticles for targeted and efficient siRNA delivery to tumors. Biomaterials. 2012;33(2):583–591. doi: 10.1016/j.biomaterials.2011.09.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akinc A, Zumbuehl A, Goldberg M, Leshciner ES, Busini V, Hossain N. et al. A combinatorial library of lipid-like materials for delivery of RNAi therapeutics. Nat Biotechnol. 2008;26(5):561–569. doi: 10.1038/nbt1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda H, Wu J, Sawa T, Matsumura Y, Hori K. Tumor vascular permeability and the EPR effect in macromolecular therapeutics: a review. J Control Release. 2000;65(1-2):271–284. doi: 10.1016/s0168-3659(99)00248-5. [DOI] [PubMed] [Google Scholar]
- Maeda H. The enhanced permeability and retention (EPR) effect in tumor vasculature: the key role of tumor-selective macromolecular drug targeting. Adv Enzyme Regul. 2001;41:189–207. doi: 10.1016/s0065-2571(00)00013-3. [DOI] [PubMed] [Google Scholar]