Abstract
Mass spectrometry has already become an indispensable tool in the analytical armamentarium of the biopharmaceutical industry, although its current uses are limited to characterization of covalent structure of recombinant protein drugs. However, the scope of applications of mass spectrometry-based methods is beginning to expand to include characterization of the higher order structure and dynamics of biopharmaceutical products, a development which is catalyzed by the recent progress in mass spectrometry-based methods to study higher order protein structure. The two particularly promising methods that are likely to have the most significant and lasting impact in many areas of biopharmaceutical analysis, direct ESI MS and hydrogen/deuterium exchange, are focus of this article.
Introduction
Biopharmaceutical products, the majority of which are recombinant proteins or protein-polymer conjugates, remain the most dynamic sector of the pharmaceutical industry with ten new protein therapeutics approved in 2010 alone. Many protein-based drugs have already become standard treatments for a variety of diseases ranging from pathologies affecting large population groups, such as diabetes (e.g., insulin and glucagon-based products) and multiple sclerosis (e.g., interferon β), to rare genetic disorders, such as Gaucher's disease (glucocerebrosidase) [1]. At the same time, protein therapeutics emerging from the biotech pipeline provide new hope to doctors and patients even in the case of illnesses that were deemed incurable only a decade ago, e.g. targeted treatment of various types of cancer with therapies based on recombinant antibody technology.
However, the spectacular success of these innovative medicines also focuses the attention of all stakeholders on the unique challenges related to the analytical characterization of protein drugs, which is of paramount importance not onlyfor the quality control, but also for various stages of discovery and development. Indeed, the large size of protein therapeutics (from several kDa to nearly 1 MDa, well beyond the molecular weight range of classical medicines - small molecule drugs) leads to an important distinction between the small molecule drugs (where the covalent structure alone is the sole determinant of the three-dimensional structure and, ultimately, the therapeutic properties of the drug) and the protein pharmaceuticals (where the large physical size makes the multitude of non-covalent contacts not only inevitable, but in fact the defining element of their three-dimensional structure). Correct folding is vital not only for the ability of a protein to execute its biological function, but also for many other aspects of its behavior. Failure to fold or maintain the native conformation obviously has a negative impact on the efficacy of the protein drug, since the recognition of a range of physiological targets requires that the native conformation be maintained throughout the lifecycle of a protein molecule. Proteins that are not folded properly are prone to aggregation both in vitro and in vivo, and are targeted by proteases both inside and outside the cell, which obviously impacts bioavailability of the protein drug. Furthermore, misfolding and aggregation may trigger an immune response, thereby adversely affecting the safety profile of the protein drug. Critical dependence of the protein drug potency, stability and safety on conformation makes its characterization an essential element throughout the drug development process from design to manufacturing to post-approval monitoring.
Although protein conformation and dynamics can be probed at the greatest level of resolution by X-ray crystallography and high-resolution NMR, inherent limitations of these techniques often makes it very difficult to carry out the analyses of biopharmaceutical products under relevant production/storage conditions (e.g., protein drug substance, product or dosing solution) or physiological conditions (e.g., mimicking the environment encountered by the protein post-administration). It is not therefore surprising that routine analyses of conformation and stability of protein drugs still rely on classical biophysical methods, such as optical spectroscopy (circular dichroism, fluorescence, UV-absorption and FTIR spectroscopy), light scattering, calorimetry, as well as analytical centrifugation and size exclusion chromatography [2]. While these techniques are capable of probing conformational properties of protein drugs in relevant environments, they are typically focused on one particular aspect of higher order structure (e.g., cumulative content of secondary structure produced, exposure of aromatic residues to solvent, etc.) and fail to characterize protein conformation in sufficient detail.
Mass spectrometry (MS) allows biopolymer structures to be characterized at a variety of levels, including conformation, dynamics and interaction with physiological partners [3], although most applications of MS in the biopharmaceutical industry are still focused almost exclusively on characterizing the covalent structure [4, 5]. Since in many cases there is no clear correlation between the occurrence of non-enzymatic PTMs and protein misfolding, focusing characterization of a biopharmaceutical product exclusively on PTMs may generate both false-positive and false-negative outcomes vis-a-vis conformational integrity of therapeutic proteins. Furthermore, structural complexity and heterogeneity exhibited by the majority of protein drugs mak es precise mapping of all PTMs in those cases a truly gargantuan task. Therefore, introducing MS techniques that focus on detection and characterization of changes in the higher order structure and dynamics of protein drugs without the need for exhaustive cataloging of all PTMs will be a boon for analytical characterization of biopharmaceutical products.
To date, an impressive experimental arsenal of MS-based techniques targeting non-covalent structure has been amassed, many of which proved highly successful in dealing with a variety of problems in biophysics and structural biology (Table 1). While some techniques listed in Table 1 are not well suited for the specific needs of biopharmaceutical analysis for a variety of reasons (e.g., intrinsically low yields in chemical cross-linking, etc.), several other have been used successfully in the past several years to look at various properties of protein therapeutics. These are hydrogen/deuterium exchange (HDX) with MS detection [6] and a suite of methods involving direct ESI MS analysis of biopolymers and their complexes in solution [7-9]. The great potential of HDX MS in the field of biopharmaceutical analysis has been already recognized [10-12], and the utility of complementary methods relying on direct ESI MS measurements is also beginning to be appreciated [13]. This paper provides an overview of both techniques and discusses current trends as related to the analytical characterization of biopharmaceutical products.
Table 1. Characterization of higher order protein structure by MS.
| Specific features of protein higher order structure | Does the analysis require alterations in the protein environment? | |
|---|---|---|
| protein environment is significantly altered (transfer to a volatile solvent system is required) | protein environment should not be altered to ensure its integrity | |
| Secondary and ternary structure: overall physical dimensions/compactness | Direct ESI MS: analysis of protein ion charge state distributions [7] | |
| Secondary and ternary structure: protein backbone flexibility maps | HDX MS | HDX MS |
| Ternary structure: integrity of disulfide networks | Disulfide bond mapping [61] | |
| Ternary structure: side chain solvent exposure maps | Photo- [62], radiolytic [63], oxidative [64] and selective chemical [65] labeling | |
| Quaternary structure: composition and stoichiometry of non-covalent assemblies | Direct ESI MS [8] | Chemical cross-linking [66] |
| Quaternary structure: inter-subunit interfaces | HDX MS | HDX MS; chemical cross-linking; chemical labeling |
| Interaction with physiological partners | Direct ESI MS [32, 67] | Chemical cross-linking |
Direct ESI MS measurement of proteins in solution: charge state distributions
Multiple charging is a phenomenon that is inherent to the electrospray ionization (ESI) of all macromolecules, including biopolymers, and it was not long after the introduction of ESI as an ionization technique in biological MS that observations were made linking dramatic changes of charge state distributions of protein ions to protein denaturation [14, 15]. These observations brought about appreciation of the tremendous potential of ESI MS as a means of probing protein higher order structure and detecting large-scale conformational transitions in solution. Natively folded proteins undergo ESI to produce ions carrying a relatively small number of charges, because the compact shape of tightly folded polypeptide chains in solution does not allow the accommodation of a significant number of protons on their surface upon transition from solution to the gas phase [7]. For this reason, ion peaks in ESI mass spectra of proteins in aqueous solutions at neutral pH typically dominate the high m/z regions of the mass spectra and are almost always characterized by having narrow distribution of charge states (Figure 1A).
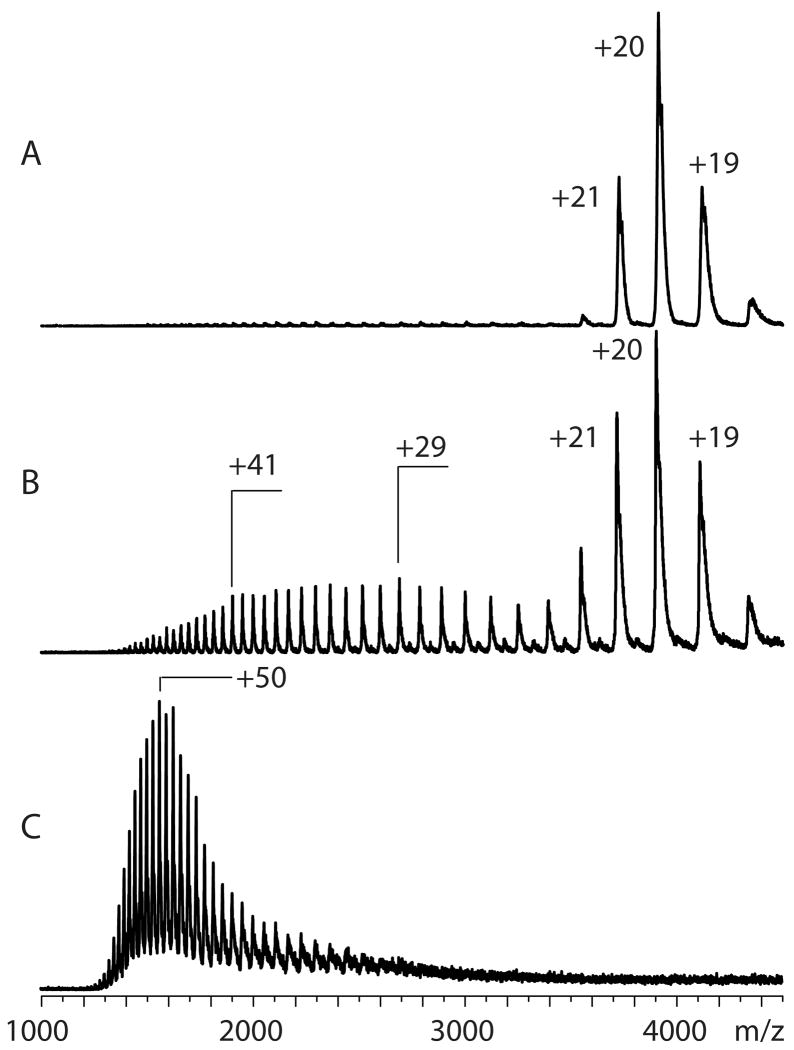
Figure 1.
ESI mass spectra of an 80 kDa protein bovine transferrin acquired under near-native (10mM ammonium acetate, pH 7.0, panel (A)), mildly denaturing (10 mM ammonium acetate, pH adjusted to 5.0, panel (B)) and strongly denaturing (water/methanol/acetic acid, 47:50:3, v:v:v, panel (C)) conditions. Emergence of non-native (partially unfolded) states is evident in (B) as the charge state distribution becomes bimodal. Further unfolding of the protein (population of significantly less compact states) is manifested in (C) by a dramatic increase of the abundance of highly charged protein ions. Reproduced with permission from [68].
Unlike folded proteins, conformers lacking native structure (i.e., those that are either partially or fully unfolded in solution as a result of denaturation) give rise to ions carrying a significantly larger number of charges and their charge-state distributions are significantly broader. This is due to the fact that once a protein molecule loses its compactness upon unfolding, a significantly larger number of charges can be accommodated on its surface. Native and non-native protein states often coexist at equilibrium under mildly denaturing conditions. In such situations protein ion charge state distributions become bimodal, reflecting the presence of both native and denatured states (Figure 1B), and more complete unfolding typically results in disappearance of the low-charge density part of the charge state distribution (Figure 1C). Therefore, dramatic changes of protein charge-state distributions can be used as gauges of large-scale conformational changes in processes ranging from protein folding [16, 17] to small ligand binding [18, 19] to multi-unit protein assembly [20, 21] and interaction with other biopolymers [22]. Incorporation of chemometric tools expand the capabilities of this technique further by allowing the individual conformers to be detected and monitored across a range of conditions [23, 24]. The extent of multiple charging can also be used to estimate the physical size of proteins in solution [25].
Although the simplicity of practical implementation of this technique and the ease of data analysis make it very appealing as a tool to monitor protein behavior in solution, its acceptance within the biopharmaceutical industry was somewhat limited until recently due to the need to carry out all measurements in the so-called “ESI-friendly” solvent systems. Indeed, the presence of non-volatile electrolytes (which are present in all biopharmaceutical formulations) has a detrimental effect on the quality of ESI MS data even at relatively low concentrations. As a result, sample preparation for direct ESI MS measurements almost always contains an obligatory solvent exchange step. During this step the protein is transferred to a volatile electrolyte solution (typically ammonium acetate or ammonium bicarbonate) at desired ionic strength, which often raises concerns that the observed protein behavior may be at least partially attributed to the influence of the “foreign” environment, rather than the intrinsic features of the biopharmaceutical product itself.
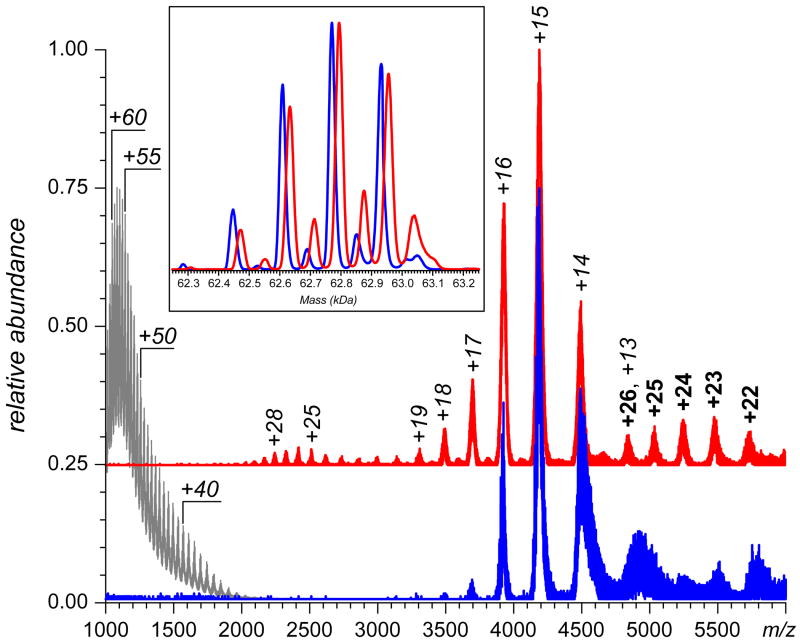
However, there is growing evidence, at least in the case of stressed protein therapeutics, that detection of unfolding events by direct ESI MS is not affected by the solvent exchange step [10, 26]. An example is shown in Figure 2, where oxidation of a recombinant form of acid-β-glucocerebrosidase (GCase, a recombinant glycoprotein used for treatment of Gaucher's disease by enzyme replacement therapy [27]) results in a noticeable change of the protein ion charge state distribution in ESI MS, revealing the presence of a less compact conformation in solution (represented by charge states +17 through +28).
Figure 2.
Nano-ESI mass spectra (inset contains the deconvoluted spectra) of 6 μM GCase (blue) and GCase-ox (red) solutions in 50mM ammonium acetate, pH 4.5. The gray trace shows a reference mass spectrum of GCase under denaturing conditions (50% acetonitrile, 5% acetic acid). Reproduced with permission from [26].
Direct ESI MS measurement of proteins in solution: interactions with physiological partners and therapeutic targets
Preservation of physiologically relevant non-covalent interactions is another feature of ESI MS, which was noted soon after acceptance of this ionization technique in the arsenal of biological mass spectrometry [28, 29]. In the two decades passed since the publication of these initial reports, direct ESI MS has been used successfully to study interactions of proteins with a variety of physiological partners, ranging from metal ions [30] and small organic molecules [31] to other biopolymers, including large macromolecular assemblies in the MDa range [8, 32]. The tremendous promise of direct ESI MS as a tool facilitating the drug discovery process was also recognized [33-35], although wider acceptance of this technique remained rather slow due to the fact that the drug-target binding must be studied in a volatile solvent system compatible with the ESI process (vide supra) and concerns over the possibility of non-specific binding [36]. Recently, the utility of direct ESI MS for characterizing interactions of protein drugs with their therapeutic targets was demonstrated using recombinant antibodies [37]. The ability of direct ESI MS to rank the affinities of various forms of recombinant proteins to their cognate receptors was also demonstrated using interferon β [13] and transferrin [38].
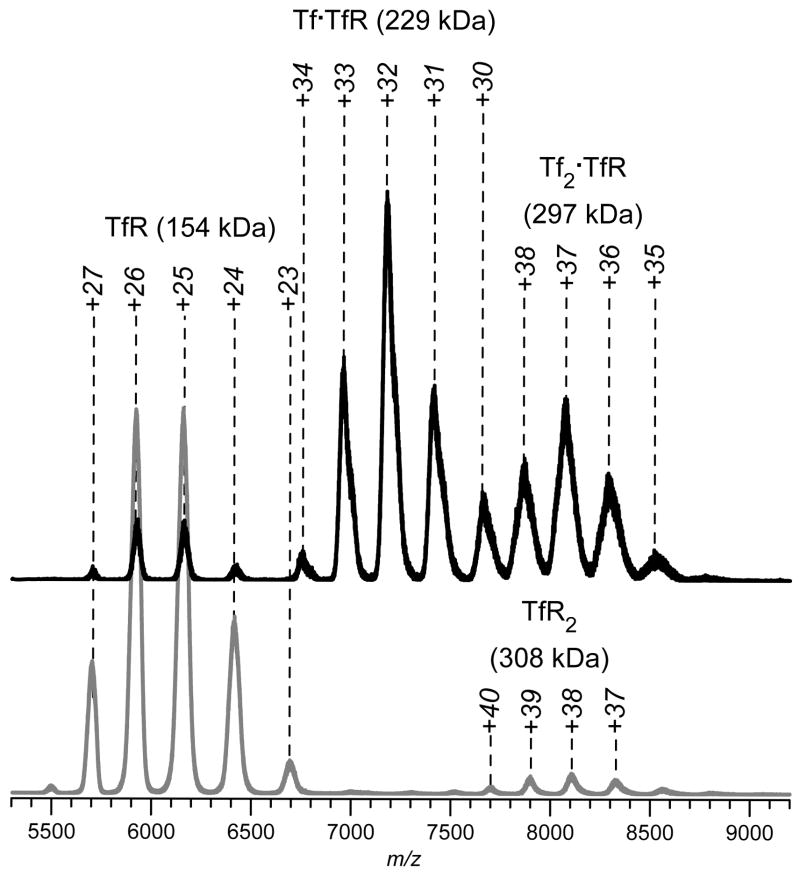
An example of using direct ESI MS for detection and characterization of interaction of a recombinant protein with its physiological partner is shown in Figure 3, where a recombinant form of human serum transferrin (Tf) is shown to bind specifically to transferrin receptor (TfR). Tf (an 80 kDa glycoprotein that transports iron) is one of the very few plasma proteins that can enter the cell in the process of receptor-mediated endocytosis. It is a component of several anti-cancer therapies in various stages of development, where it is used for precise routing of protein drugs to malignant cells by exploiting their dramatically elevated levels of iron consumption [39-41]. Tf/TfR binding is an obligatory step for delivery of Tf and Tf-based therapeutics to malignant cells, and detailed information on Tf/TfR interaction at the molecular level is extremely important for the drug development process. As can be seen in Figure 3, formation of Tf/TfR complexes is readily detected by ESI MS; furthermore, this approach can be used to obtain receptor affinity ranking for various forms of Tf [38].
Figure 3.
Nano-ESI MS of TfR (gray trace) and Tf/TfR mixture (black trace) in 130 mM ammonium acetate/10 mM ammonium bicarbonate solution at pH 7.4. Protein concentrations (where appropriate): TfR, 4 μM, and Tf, 0.5 μM. Each TfR molecule has two Tf binding sites, hence the presence of both 1:1 and 2:1 complexes in the mass spectra (signal corresponding to free Tf can be seen in the mass spectrum of Tf/TfR mixture, indicating that the binding goes to completion under these conditions).
HDX MS in the analysis of conformation and dynamics of recombinant proteins and related biopharmaceutical products
One of the shortcomings of analytical characterization of protein therapeutics using methods based on direct ESI MS discussed in the preceding sections is relatively low resolution. For example, while the protein conformational integrity can be monitored and the onset of structure loss can be confidently detected, little or no information is provided regarding the specific protein segments that become affected. Likewise, monitoring protein interaction with physiological partners and therapeutic targets using direct ESI MS rarely reveals the location of binding epitopes. These shortcomings (as well as the necessity to carry out all measurements in volatile salt systems compatible with ESI MS) are overcome by HDX MS, which already demonstrated its utility vis-à-vis conformational analysis of biopharmaceutical products [10-12]. This experimental technique is reliable, robust and sensitive, and is capable not only of detecting the presence of (partially) unfolded species in solution, but also localizing protein segments with anomalous protection levels. Furthermore, the ability of HDX MS to detect and characterize structurally compromised proteins on the background of the natively folded species in highly complex matrices makes it a very promising tool for conformational characterization of protein drugs and may open up new and exciting opportunities in discovery and formulation of biopharmaceutical products.
HDX provides information on conformational properties of proteins in solution by monitoring exchange of their labile hydrogen atoms with labile hydrogen atoms of solvent molecules. The labile hydrogen atoms in proteins comprise two distinct groups, namely those found in polar and ionic side chains and protein termini, and those attached to backbone amide nitrogen atoms. Absent any protection from solvent, the labile hydrogen atoms from each of the two groups will exchange with the solvent protons. The rate of this chemical reaction (termed intrinsic exchange rate, kint) is determined by the identity of the exchanging proton (and, to a lesser extent, by the neighboring amino acid residues), as well as solution temperature and pH [42, 43]:
| (1) |
The base catalysis term is usually a more significant contributor, so that the minima of the kint vs. pH plots always occur at pH < 7. However, the location of the minimum for any specific proton is determined by its identity: labile hydrogen atoms located in polar and ionic side chains and protein termini have the kint minima in the pH range 4-6, while the slowest intrinsic exchange for the backbone amide protons occurs in a more acidic range, pH 2.5-3 [44]. Adjusting the protein solution pH to this level and lowering its temperature result in a very slow exchange rate kint for the backbone amide hydrogen atoms (as low as 10−4 s−1 at 0 °C), conditions that are frequently referred to as “quenching.”
While the exchange of labile hydrogen atoms in short unstructured polypeptides is determined by the their intrinsic exchange rates [43], presence of higher order structure can alter the observed exchange rates in a very significant way. Protons that are involved in hydrogen bonding or sequestered from solvent in the protein interior will not be able to exchange with the solvent unless they become exposed to it through a local conformational fluctuation or a more global (complete or partial) protein unfolding event [42]. Although proton exchange with the solvent does not result in any changes that can be detected with mass spectrometry directly, substitution of H2O with D2O provides a straightforward means to monitor the progress of the exchange reactions. Indeed, each individual H/D exchange event will result in a mass increase of 1.01 Da, a difference that can be easily detected by MS even in the context of large proteins. Therefore, monitoring the mass change of an initially unlabeled protein placed in D2O-based solvent (or, conversely, a fully deuterated protein in H2O-based solvent system) allows the extent of hydrogen bonding network within the protein and its stability to be evaluated. This forms a basis of a popular experimental technique, hydrogen/deuterium exchange MS (HDX MS), which is increasingly used to evaluate both higher order structure and dynamics of proteins and protein complexes.
In principle, HDX MS can provide information on solvent protection of all labile hydrogen atoms. In reality, however, polar and charged side chains are frequently exposed to the solvent, and exchange very fast, although there are rare examples of polar and charged side chains buried in the protein interior and, therefore, protected from exchange with the solvent. The labile hydrogen atoms from the second group (backbone amides) are typically better reporters of higher order structure due to their intimate involvement in H-bonding networks that maintain the integrity of secondary structural elements. Although the distinction between the contributions of hydrogen atoms from the two groups to overall HDX pattern cannot be made in a straightforward measurement of a protein mass shift following its exposure to the exchange solvent, the contribution of the side chains can be eliminated prior to the mass measurement. Indeed, the exchange of the backbone amide hydrogen atoms can be quenched, by bringing the solution pH to the 2.5-3.0 interval and lowering the temperature to 0-4 °C (vide supra). These acidic conditions also result in facile unfolding of most of the proteins, thereby exposing all labile hydrogen atoms to the solvent. While such exposure is relatively harmless to the backbone amide hydrogen atoms (due to the very slow rates of intrinsic exchange), the exchange of the protons from the first group (side chains and termini) would still proceed at relatively high rates. This means that all information on the protection of the backbone amides prior to quenching the exchange will be retained, while the isotopic labels at the side chains will be quickly lost to the so-called back-exchange, allowing the exchange patterns of the protein backbone to be probed selectively, without any contribution from the side chains.
Protein properties influencing HDX: reflection of protein behavior in HDX MS
Interpretation of the results of HDX measurements is somewhat complicated by the fact that the exchange rate of each labile hydrogen atom involved in hydrogen bonding or sequestered from the solvent in the protein interior is not zero, but is determined by both conformation and dynamics. Indeed, even the most stable elements of protein higher order structure are not frozen entities. They still undergo conformational fluctuations, and may in fact unfold partially or completely in a transient fashion. Both frequencies and durations of such events vary, depending on the shape of the protein free energy landscape (energy differences between the ground and excited states and the potential energy barriers separating them). While transient loss of structure may expose one or several labile hydrogen atoms to the solvent, the extent of the exchange in each case (and, ultimately, the appearance and time evolution of the overall HDX MS pattern) will depend on how the time scale of the unfolding event (kcl) compares to the chemical exchange time scale in the absence of any protection (kint):
| (2) |
The detailed discussion of these issues lies outside of the scope of this chapter, but can be found elsewhere in the literature [45]. In brief, there are two extreme cases, i.e. the so-called EX1 (kcl (symbol) kint), and EX2 (kcl (symbol) kint) exchange regimes. The latter case is usually more relevant under physiological conditions (the protein is relatively stable and the refolding rate leading to the protected state is much higher than the intrinsic exchange rate). The short residence time at the local free energy minimum (1/kcl) corresponding to the “open” state is much shorter than the characteristic time of exchange of an unprotected labile hydrogen atom (1/kint), leading to a very low (but finite) probability of exchange for a given hydrogen atom during a single unfolding event.
The overall rate of exchange is defined in this case by both the frequency of unfolding events (kop) and the probability of exchange during a single opening event:
| (3) |
where KU is an effective equilibrium constant for the unfolding reaction and is determined by the free energy difference between the “protected” and “open” states of the protein. While in NMR measurements kHDX is simply a rate of depletion of a number of protein molecules labeled with 1H at a specific amide, HDX MS measurements would regard this rate constant as a cumulative rate of exchange. In other words, kHDX is an ensemble-averaged rate of loss of the entire 1H content (for all amides combined), when the isotopic distribution of a protein ion gradually shifts towards higher m/z [45, 46].
The EX1 exchange regime EX1 (kcl (symbol) kint) is frequently observed when the protein is destabilized either externally (e.g., due to the presence of chaotropes in the solvent) or internally (e.g., due to a destabilizing mutation, chemical modification or removal of structure-stabilizing ligands). Under these conditions the protein becomes trapped in the unfolded (exchange-competent) state long enough to allow all labile hydrogen atoms to be exchanged during a single unfolding event, and the exchange rate is determined only by the unfolding rate:
| (4) |
Although the EX1 exchange regime is frequently associated with the so-called correlated exchange patterns (bimodal isotopic distributions in HDX MS experiments), one must be very careful in equating the EX1 regime and correlated exchange patterns. While it is true that the bimodal isotopic distributions are observed when the exchange follows the EX1 regime, the opposite is not necessarily true. In fact, unfolding events exposing only one (or very few) labile hydrogen atoms to the solvent (e.g., local conformational fluctuations) may favor the EX1 exchange regime, particularly at elevated pH (when kint is very high). Nevertheless, no bimodal distribution would be observed in this case due to very modest deuterium incorporation resulting from each individual small-scale opening event.
Finally, in the intermediate exchange regime (when the values of kint and kcl are comparable) the residency time in the exchange-competent state is long enough to have one or more protons exchanged during each unfolding event, but too short to have the entire set of all labile hydrogen atoms exchanged at once. HDX MS measurements under these conditions produce very convoluted exchange patterns [45]. The isotopic distribution may exhibit a bimodal character, but the distance (or mass difference) between the two isotopic clusters will increase as the exchange progresses. In principle, values of both kop and kcl can be extracted from the HDX MS measurements carried out under such conditions, which are termed EXX (or intermediate) exchange regime [45]. In larger proteins, presence of multiple non-native (partially unfolded states) may further complicate the analysis of HDX MS data [46, 47].
Local measurements with HDX MS
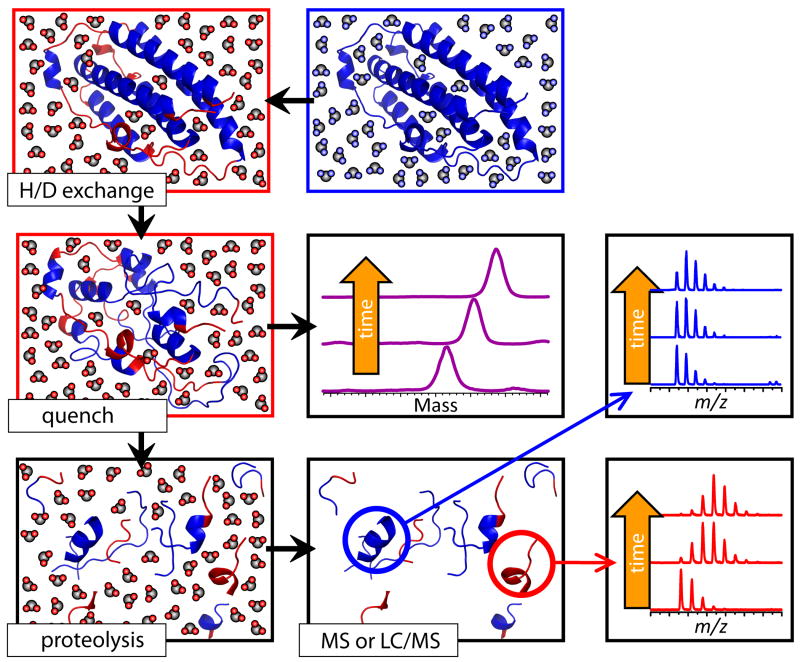
Local information on protein higher order structure and dynamics can be obtained by carrying out proteolysis under the slow exchange conditions prior to MS analysis. Pepsin, as well as few other acidic proteases, is active within the pH range 2.5-3.0, which makes it suitable for probing backbone amide protection following the quench of the exchange reactions [48, 49]. Since the amide hydrogen atoms continue to exchange under the slow exchange conditions, the local exchange details can be maintained only if the sample handling is relatively fast and is carried out at low solution temperature (typically < 4 °C). Following the quench of the exchange reactions, all sample handling steps (including proteolysis and LC/MS measurements) can be carried out on-line. A typical workflow of HDX MS measurements is shown in Figure 4.
Figure 4.
Schematic representation of HDX MS work flow to examine protein stability. The exchange is initiated by placing the unlabeled protein into a D2O-based solvent system (e.g., by a rapid dilution). Unstructured and highly dynamic protein segments undergo fast exchange (blue and red colors represent protons and deutrons, respectively). Following the quench step (rapid solution acidification and temperature drop), the protein loses its native conformation, but the spatial distribution of backbone amide protons and deuterons across the backbone is preserved (all labile hydrogen atoms at side chains undergo fast back-exchange at this step). Rapid clean-up followed by MS measurement of the protein mass reports the total number of backbone amide hydrogen atoms exchanged under native conditions (a global measure of the protein stability under native conditions), as long as the quench conditions are maintained during the sample work-up and measurement. Alternatively, the protein can by digested under the quench conditions using acid-stable protease(s), and LC/MS analysis of masses of individual proteolytic fragments will provide information on the backbone protection of corresponding protein segments under the native conditions.
The majority of recombinant therapeutic proteins structural have disulfide bonds, a structural feature that has a negative impact on the spatial resolution achievable in HDX MS measurements. Apart from limiting the efficiency of proteolysis, it also prevents physical separation of peptic fragments connected by the cysteine-cysteine bridges even if the enzymatic reaction is successful. Common disulfide-reducing agents (such as dithiothreitol) are inactivated at acidic pH, and, therefore, cannot be used under the slow exchange conditions. The task of reducing disulfides under such conditions can be successfully carried out in many cases by another agent, tris(2-carboxyethyl)phosphine, dubbed TCEP, which retains its disulfide-reducing capacity at pH as low as 1.5 [50].
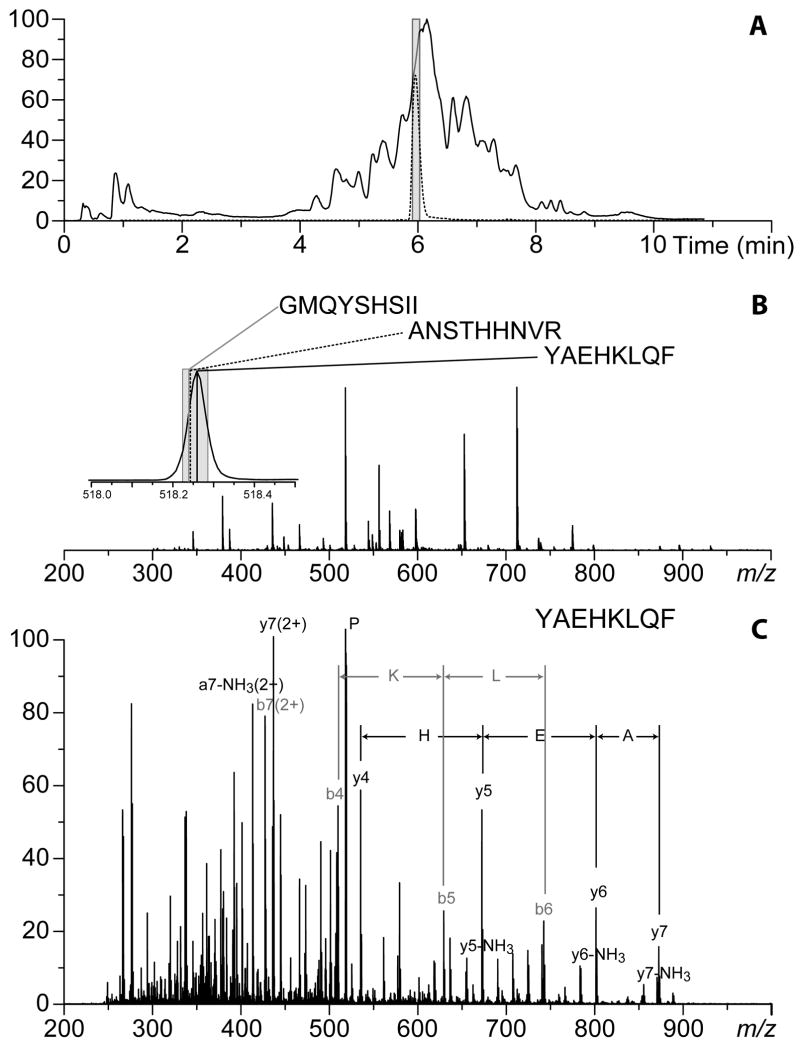
Since all proteases used in HDX MS work are non-specific, a priori prediction of the expected cleavage sites is impossible, and successful identification of proteolytic fragments becomes essential for meaningful interpretation of HDX MS data. Typically, this is not a big problem for smaller proteins, where most peptide fragments can be confidently identified by their unique masses. However, large protein size coupled with extensive glycosylation and/or the presence of multiple disulfide bonds that are not completely reduced prior to the digestion step may complicate the task of identifying proteolytic fragments. In fact, it is not uncommon to observe a proteolytic fragment whose mass would fit more than one putative isobaric peptides derived from the protein sequence. An example is shown in Figure 5, where a measured mass of one of the peptides produced upon digestion of GCase matches within 30 ppm three possible fragments derived from the sequence of this protein. In cases like this an unequivocal identification of the proteolytic fragment in question requires more advanced MS tools, such as ion fragmentation in the gas phase. The fragmentation pattern of the peptide ion (Figure 5C) exhibits series of abundant b- and y-ions, whose masses are consistent with the sequence of only one of the candidate peptides (incidentally, the best match in MS1 measurements). Definitive identification of peptide fragments can also be carried out using ultra-high accuracy mass measurements with a high-resolution MS (e.g., Fourier transform ion cyclotron resonance MS).
Figure 5.
Identification of a fragment peptide produced by peptic digestion of a 60 kDa glycoprotein GCase. A: total ion chromatogram of a peptic digest of unlabeled GCase run under the slow-exchange conditions. B: MS averaged across the time span indicated in panel A with a gray box. The inset in panel B shows a zoomed view of ionic signal at m/z 518; the shaded area shows the 30 ppm confidence interval, and the sequence-based masses of three isobaric peptides fitting in this interval are indicated with a solid black (220-227, YAEHKLQF), solid gray (265-274, GPTLADSTHH) and dotted black (360-368, GMQYSHSII) lines. Reconstructed ions chromatogram of the doubly charged ion at m/z 518.3 is shown in panel A with a gray line. C: a fragmentation mass spectrum (MS/MS) produced upon collision-induced dissociation of the doubly charged ion at m/z 518.
Localization of structural instability regions within stressed protein drugs by HDX MS: influence GCase oxidation on its conformation and dynamics
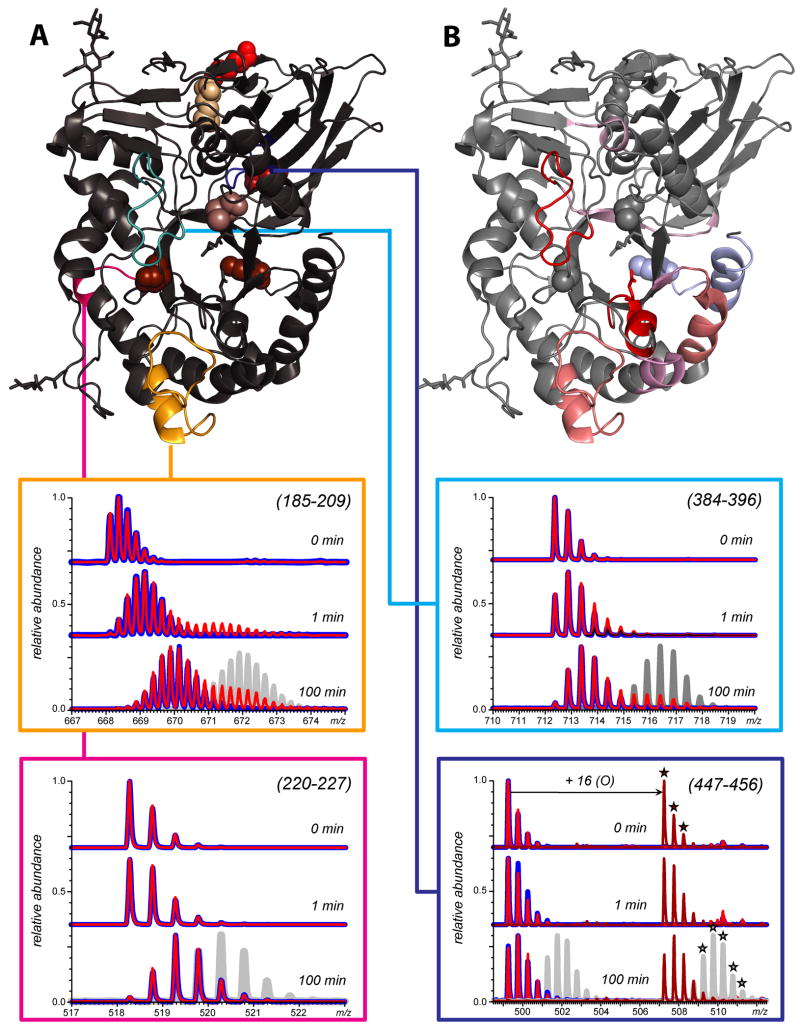
Several recent reports highlight the power of HDX MS as a tool providing detailed information on changes in conformation and dynamics of recombinant protein therapeutics caused by either non-enzymatic PTMs under stress conditions [10, 26, 51] or engineered PTMs [52]. An example is shown in Figure 6, where HDX MS is used to probe local conformational dynamics of intact and oxidized GCase [26]. The uneven distribution of flexibility across the backbone of intact GCase highlights the importance of the delicate balance between protein structure and dynamics. Oxidation clearly shifts this balance by adversely affecting stability of several protein segments. Surprisingly, the segments containing the oxidation sites (e.g., 447-456 in Figure 6) are not affected by the covalent modifications, while several segments that are remote from the modified residues (e.g., 185-209) exhibit noticeable decrease in backbone protection.
Figure 6.
Isotopic distributions of peptide ions representing peptic fragments (185-209, 220-2270, 384-396, and 447-456) of GCase (blue) and GCase-ox (red) following 1 and 100 min of HDX in solution. The gray traces represent isotopic distributions within the same peptide for acid denatured GCase, which allowed for complete exchange (e.g., the maximum level of amide exchange for the peptide) and was analyzed under the same conditions as native GCase. Location of these segments within the crystal structure of GCase is shown in structure A. Local backbone amide protection deduced from HDX MS measurements mapped to GCase structure are colored based on ΔHDX (difference between oxidized and intact forms of GCase, exchange time 100 min) is shown in structure B.
A detailed analysis of HDX MS data for the intact and oxidized forms of GCase [26] indicate that the segments whose protection is affected by oxidation tend to form a contiguous cluster on the protein surface (Figure 6B). This suggests that the greatly diminished activity of GCase caused by extensive oxidation results from the decrease of its conformational stability, while the catalytic function is not affected directly [26].
Protein interaction with physiological partners and therapeutic targets probed by HDX MS
The preceding section focused on applications of HDX MS that aim at understanding how conformation of recombinant therapeutic proteins is affected by non-enzymatic PTMs under stress conditions or purposefully engineered “designer” PTMs, but the utility of HDX MS in biopharmaceutical analysis extends far beyond the quality control and comparability issues. Nearly a decade ago, Woods et al. pointed out a great potential of this technique in design of small molecule pharmaceuticals, where it can be used for optimizing the drug candidate structure by providing information on structure and dynamics of the binding site at the therapeutic target [53]. The idea of incorporation of MS in the workflow of the discovery and development of protein therapeutics is also beginning to enjoy attention following recent reports that it may aid epitope mapping [54] and, therefore, facilitate rational design of biopharmaceutical products with enhanced binding to therapeutic targets.
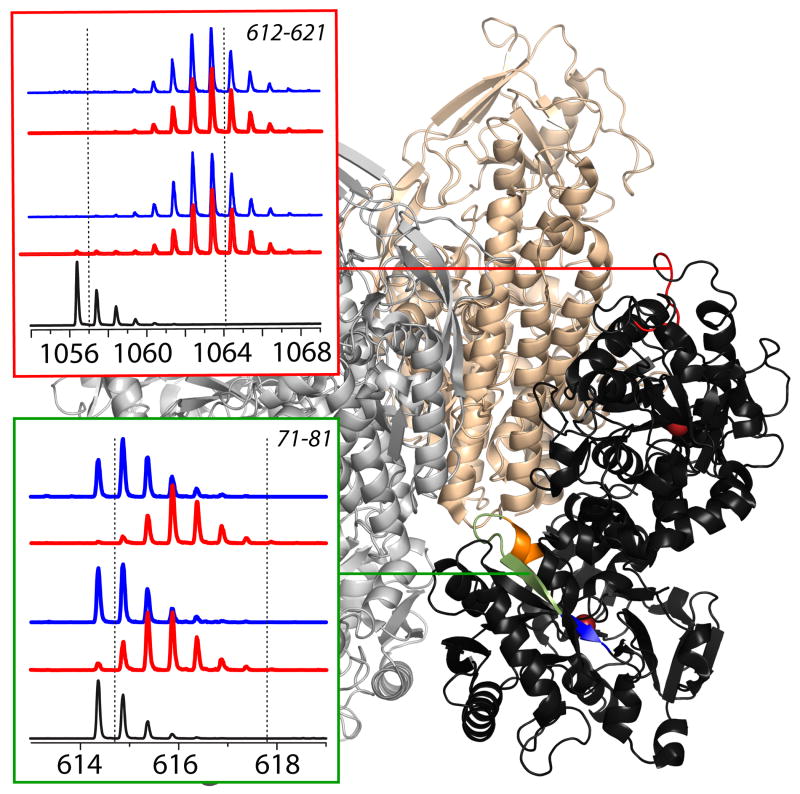
An example of using HDX MS to obtain detailed information on how biopharmaceuticals interact with their specific receptors is shown in Figure 7, where HDX is used to probe conformational dynamics of Tf alone and in the receptor-bound form. As mentioned earlier, the ability of Tf or any Tf-conjugate drug to enter the cells critically depends on its recognition by the receptor (TfR); therefore, detailed information on Tf/TfR interaction at the molecular level is extremely important for the drug development process. Unfortunately, the large size and the presence of paramagnetic ferric ion (Fe3+) places the Tf/TfR complex out of reach of NMR, while it's dynamic character and high carbohydrate content make acquisition of high-resolution X-ray diffraction data highly problematic, leaving HDX MS as the only technique capable of providing structural information on Tf/TfR interaction.
Figure 7.
Left: HDX MS within two segments of Tf in the presence (blue) and the absence (red) of the cognate receptor. The exchange was carried out by diluting the protein stock solution 1:10 in exchange solution (100 mM NH4HCO3 in D2O, pH adjusted to 7.4) and incubating for a certain period of time as indicated on each diagram followed by rapid quenching (lowering pH to 2.5 and temperature to near 0°C). The black trace at the bottom of each diagram shows unlabeled protein and the dotted lines represent the end-point of the exchange reaction. Location of these segments in Tf is shown within the context of the low-resolution structure of Tf/TfR [69].
Indeed, HDX MS readily provides information on involvement of various protein segments in Tf/TfR interaction. For example, two similarly sized peptic fragments of Tf (71-81 and 612-621) whose deuterium content evolution is shown in Figure 7 represent protein regions with increased backbone mobility in the absence of the receptor. The 612-621 fragment (which maps to a loop region in Tf C-lobe in the crystal structure of the protein) has very low backbone protection, and the other peptide (which maps to a helix-loop-strand segment in the N-lobe) exhibits intermediate level of protection. It is important to remember, however, that the data presented in Figure 7 show the protection levels averaged across the entire lengths of the peptides, and the “intermediate” level of deuterium uptake exhibited by (71-81) may be a result of smearing a very uneven protection pattern across its backbone, a suspicion that is reinforces by the fact that this segment incorporates three distinct structural elements (a helix, a loop and a strand). In cases like this, higher spatial resolution is needed in order to deduce individual contributions of various structural elements to the overall exchange kinetics within the entire segment, which can be achieved either using emerging top-down HDX MS technology [55] or in a more traditional way by analyzing deuterium uptake kinetics within the overlapping peptic fragments [56]. This latter approach allows the entire (71-81) segment to be broken into three parts (as colored in the protein structure in Figure 7), where the blue-colored part of the β-strand shows very high protection level, and the two other segments are more flexible. Further enhancement of special resolution identifies the loop region as the most flexible element within this peptide (Abzalimov, et al., manuscript in preparation).
The highly uneven distribution of the backbone protection exhibited by the (71-81) segment of Tf is completely eliminated upon binding to the receptor, with the entire peptide becoming highly protected (see blue traces in Figure 7). This suggests strong involvement of this flexible loop region in forming the protein/receptor interface. In contrast, the flexible loop in the C-lobe of the protein (segment 612-621 discussed earlier) is not affected at all by the Tf/TfR interaction. This type of analysis, particularly in the absence of a reliable crystal structure of a protein drug/therapeutic target complex will be invaluable in designing new and enhancing existing biopharmaceutical products (e.g., in optimizing conjugation of a payload to a carrier protein).
Conclusion: Current challenges and future directions in the field
While biopharmaceutical products cover a wide molecular weight range, a significantly larger fraction of recombinant therapeutic proteins exceed 50 kDa in mass [57], and even smaller protein drugs are frequently conjugated to polymers in order to increase their physical size and, therefore, enhance their pharmacokinetic profiles [58]. Larger protein systems are more challenging targets for analytical characterization for many reasons, not the least because size increase of a protein drug inevitably translates into higher frequency of non-enzymatic PTMs. Precise mapping of these changes within a large protein is a gargantuan task, and availability of robust, easy-to-use and reasonably high-throughput methods to probe conformational integrity of such large systems directly without the need for exhaustive characterization of their covalent structure would obviously be a boon to the biopharmaceutical industry.
Implementation of both direct ESI MS measurements and HDX MS for conformational analysis of large (>50 kDa) protein drugs faces several challenges. Larger mass and high degree of structural heterogeneity (e.g., due to extensive glycosylation or conjugation to a synthetic polymer) make even protein ion charge state assignment in ESI mass spectra a challenging task, although methods of ion chemistry may provide elegant solutions to this problem [59]. HDX MS measurements of large heterogeneous protein systems are also complicated due to the fact that non-specificity of pepsin cleavage combined with the large physical size of the protein typically results in a large number of fragment peptides, which must be identified in order to extract meaningful HDX information. This problem is magnified by the frequent occurrence of disulfide bonds in protein drugs, which are typically only partially reduced during protein handling prior to MS analysis, thereby multiplying the sheer number of candidate peptides that fit a given mass. Even complete identification of all observed peptic fragments does not necessarily result in complete sequence coverage of a large protein. Indeed, the necessity to minimize the sample handling time prior to MS analysis as a means to limit the extent of back-exchange typically translates to very short (and, therefore, crowded) LC runs. While MS detection itself (and particularly high-resolution MS) solves the problem of detection of multiple co-eluting peaks, signal suppression is likely to result in loss of some of the peptides, thereby leaving gaps in sequence coverage. Despite these challenges, significant progress was made recently in this field, as demonstrated by successful use of HDX MS to probe conformational properties of protein therapeutics exceeding 50 kDa, such as GCase [26, 60] and monoclonal antibodies [11, 12].
Ultimately, the success of both direct ESI MS and HDX MS as tools to probe conformation of recombinant therapeutic proteins will be determined by their adaptability to the specific needs of the biopharmaceutical industry. For example, development of high-throughput methods of analysis with complete automation of the entire procedure (from sample handling to data processing) seems to be a key to the general adoption of MS-based methods of conformational analysis. Reproducibility and robustness of these measurements is another issue that needs to be addressed in the near future. The issue of sensitivity as related to detecting small conformational changes and/or alterations of the higher order structure affecting only a small fraction of the protein molecules also awaits careful exploration and evaluation. Addressing these questions will require extensive efforts, but the end result is well worth it. Indeed, adoption of mass spectrometry by the biopharmaceutical industry in this new role will not only become a boon to analytical characterization and quality control, but also greatly catalyze development of new protein-based therapies.
Acknowledgments
Various aspects of the work presented in this manuscript were supported by the National Institutes of Health (grant R01 GM061666), National Science Foundation (grant CHE-0750389), and Shire Human Genetic Therapies (Cambridge, MA).
References
- 1.Walsh G. Biopharmaceutical benchmarks 2010. Nature Biotechnol. 2010;28:917–294. doi: 10.1038/nbt0910-917. [DOI] [PubMed] [Google Scholar]
- 2.Lundblad RL. Approaches to the Conformational Analysis of Biopharmaceuticals. In: Lundblad RL, editor. Approaches to the conformational analysis of biopharmaceuticals. Boca Raton: CRC Press/Taylor & Francis; 2010. [Google Scholar]
- 3.Kaltashov IA, Eyles SJ. Studies of biomolecular conformations and conformational dynamics by mass spectrometry. Mass Spectrom Rev. 2002;21:37–71. doi: 10.1002/mas.10017. [DOI] [PubMed] [Google Scholar]
- 4.Zhang Z, Pan H, Chen X. Mass spectrometry for structural characterization of therapeutic antibodies. Mass Spectrom Rev. 2009;28:147–176. doi: 10.1002/mas.20190. [DOI] [PubMed] [Google Scholar]
- 5.Srebalus-Barnes CA, Lim A. Applications of mass spectrometry for the structural characterization of recombinant protein pharmaceuticals. Mass Spectrom Rev. 2007;26:370–388. doi: 10.1002/mas.20129. [DOI] [PubMed] [Google Scholar]
- 6.Englander SW. Hydrogen exchange and mass spectrometry: A historical perspective. J Am Soc Mass Spectrom. 2006;17:1481–1489. doi: 10.1016/j.jasms.2006.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaltashov IA, Abzalimov RR. Do ionic charges in ESI MS provide useful information on macromolecular structure? J Am Soc Mass Spectrom. 2008;19:1239–1246. doi: 10.1016/j.jasms.2008.05.018. [DOI] [PubMed] [Google Scholar]
- 8.Hernandez H, Robinson CV. Determining the stoichiometry and interactions of macromolecular assemblies from mass spectrometry. Nat Protoc. 2007;2:715–726. doi: 10.1038/nprot.2007.73. [DOI] [PubMed] [Google Scholar]
- 9.Yin S, Loo JA. Mass spectrometry detection and characterization of noncovalent protein complexes. Methods Mol Biol. 2009;492:273–282. doi: 10.1007/978-1-59745-493-3_16. [DOI] [PubMed] [Google Scholar]
- 10.Bobst CE, et al. Detection and characterization of altered conformations of protein pharmaceuticals using complementary mass spectrometry-based approaches. Anal Chem. 2008;80:7473–7481. doi: 10.1021/ac801214x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Houde D, et al. Characterization of IgG1 conformation and conformational dynamics by hydrogen/deuterium exchange mass spectrometry. Anal Chem. 2009;81:2644–2651. doi: 10.1021/ac802575y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burkitt W, Domann P, O'Connor G. Conformational changes in oxidatively stressed monoclonal antibodies studied by hydrogen exchange mass spectrometry. Protein Science. 2010;19:826–835. doi: 10.1002/pro.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaltashov IA, et al. Conformation and dynamics of biopharmaceuticals: transition of mass spectrometry-based tools from academe to industry. J Am Soc Mass Spectrom. 2010;21:323–37. doi: 10.1016/j.jasms.2009.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chowdhury SK, Katta V, Chait BT. Probing Conformational Changes in Proteins by Mass Spectrometry. J Am Chem Soc. 1990;112:9012–9013. [Google Scholar]
- 15.Loo JA, et al. Solvent-Induced Conformational Changes of Polypeptides Probed by Electrospray-Ionization Mass Spectrometry. Rapid Commun Mass Spectrom. 1991;5:101–105. doi: 10.1002/rcm.1290050303. [DOI] [PubMed] [Google Scholar]
- 16.Konermann L, Douglas DJ. Equilibrium unfolding of proteins monitored by electrospray ionization mass spectrometry: distinguishing two-state from multi-state transitions. Rapid Commun Mass Spectrom. 1998;12:435–442. doi: 10.1002/(SICI)1097-0231(19980430)12:8<435::AID-RCM181>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 17.Grandori R. Detecting equilibrium cytochrome c folding intermediates by electrospray ionisation mass spectrometry: Two partially folded forms populate the molten-globule state. Protein Sci. 2002;11:453–458. doi: 10.1110/ps.45102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gumerov DR, Kaltashov IA. Dynamics of iron release from transferrin N-lobe studied by electrospray ionization mass spectrometry. Anal Chem. 2001;73:2565–2570. doi: 10.1021/ac0015164. [DOI] [PubMed] [Google Scholar]
- 19.van den Bremer ET, et al. Probing metal ion binding and conformational properties of the colicin E9 endonuclease by electrospray ionization time-of-flight mass spectrometry. Protein Sci. 2002;11:1738–1752. doi: 10.1110/ps.0200502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Griffith WP, Kaltashov IA. Highly asymmetric interactions between globin chains during hemoglobin assembly revealed by electrospray ionization mass spectrometry. Biochemistry. 2003;42:10024–33. doi: 10.1021/bi034035y. [DOI] [PubMed] [Google Scholar]
- 21.Simmons DH, et al. Subunit Disassembly and Unfolding Kinetics of Hemoglobin Studied by Time-Resolved Electrospray Mass Spectrometry. Biochemistry. 2004;43:14792–14801. doi: 10.1021/bi048501a. [DOI] [PubMed] [Google Scholar]
- 22.Griffith WP, Kaltashov IA. Mass spectrometry in the study of hemoglobin: from covalent structure to higher order assembly. Curr Org Chem. 2006;10:535–553. [Google Scholar]
- 23.Dobo A, Kaltashov IA. Detection of multiple protein conformational ensembles in solution via deconvolution of charge state distributions in ESI MS. Anal Chem. 2001;73:4763–4773. doi: 10.1021/ac010713f. [DOI] [PubMed] [Google Scholar]
- 24.Mohimen A, et al. A chemometric approach to detection and characterization of multiple protein conformers in solution using electrospray ionization mass spectrometry. Anal Chem. 2003;75:4139–4147. doi: 10.1021/ac034095+. [DOI] [PubMed] [Google Scholar]
- 25.Kaltashov IA, Mohimen A. Estimates of protein surface areas in solution by electrospray ionization mass spectrometry. Anal Chem. 2005;77:5370–5379. doi: 10.1021/ac050511+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bobst CE, et al. Impact of oxidation on protein therapeutics: Conformational dynamics of intact and oxidized acid-β-glucocerebrosidase at near-physiological pH. Protein Sci. 2010;19:2366–2378. doi: 10.1002/pro.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grabowski GA. Phenotype, diagnosis, and treatment of Gaucher's disease. Lancet. 2008;372:1263–1271. doi: 10.1016/S0140-6736(08)61522-6. [DOI] [PubMed] [Google Scholar]
- 28.Katta V, Chait BT. Observation of the Heme Globin Complex in Native Myoglobin by Electrospray-Ionization Mass-Spectrometry. J Am Chem Soc. 1991;113:8534–8535. [Google Scholar]
- 29.Smith RD, et al. Preservation of noncovalent associations in electrospray ionization mass spectrometry - Multiply charged polypeptide and protein dimers. Org Mass Spectrom. 1992;27:811–821. [Google Scholar]
- 30.Kaltashov IA, et al. Investigation of structure, dynamics and function of metalloproteins with electrospray ionization mass spectrometry. Anal Bioanal Chem. 2006;386:472–481. doi: 10.1007/s00216-006-0636-6. [DOI] [PubMed] [Google Scholar]
- 31.Loo JA. Studying noncovalent protein complexes by electrospray ionization mass spectrometry. Mass Spectrom Rev. 1997;16:1–23. doi: 10.1002/(SICI)1098-2787(1997)16:1<1::AID-MAS1>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 32.Heck AJR. Native mass spectrometry: a bridge between interactomics and structural biology. Nat Meth. 2008;5:927–933. doi: 10.1038/nmeth.1265. [DOI] [PubMed] [Google Scholar]
- 33.Bruce JE, et al. Bio-affinity characterization mass spectrometry. Rapid Commun Mass Spectrom. 1995;9:644–650. doi: 10.1002/rcm.1290090805. [DOI] [PubMed] [Google Scholar]
- 34.Hofstadler SA, Sannes-Lowery KA. Applications of ESI-MS in drug discovery: interrogation of noncovalent complexes. Nat Rev Drug Discov. 2006;5:585–595. doi: 10.1038/nrd2083. [DOI] [PubMed] [Google Scholar]
- 35.Hannah VV, et al. Native MS: an ‘ESI’ way to support structure- and fragment-based drug discovery. Fut Med Chem. 2010;2:35–50. doi: 10.4155/fmc.09.141. [DOI] [PubMed] [Google Scholar]
- 36.Aplin RT, et al. Does the observation of noncovalent complexes between biomolecules by electrospray-ionization mass-spectrometry necessarily reflect specific solution interactions. J Chem Soc Chem Comm. 1994;1994:2415–2417. [Google Scholar]
- 37.Atmanene C, et al. Extending mass spectrometry contribution to therapeutic monoclonal antibody lead optimization: Characterization of immune complexes using noncovalent ESI-MS. Anal Chem. 2009;81:6364–6373. doi: 10.1021/ac9007557. [DOI] [PubMed] [Google Scholar]
- 38.Leverence R, Mason AB, Kaltashov IA. Noncanonical interactions between serum transferrin and transferrin receptor evaluated with electrospray ionization mass spectrometry. Proc Natl Acad Sci USA. 2010;107:8123–8128. doi: 10.1073/pnas.0914898107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Singh M. Transferrin as a targeting ligand for liposomes and anticancer drugs. Curr Pharm Des. 1999;5:443–451. [PubMed] [Google Scholar]
- 40.Qian ZM, et al. Targeted drug delivery via the transferrin receptor-mediated endocytosis pathway. Pharmacol Rev. 2002;54:561–87. doi: 10.1124/pr.54.4.561. [DOI] [PubMed] [Google Scholar]
- 41.Daniels TR, et al. The transferrin receptor part II: Targeted delivery of therapeutic agents into cancer cells. Clin Immunol. 2006;121:159–176. doi: 10.1016/j.clim.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 42.Krishna MMG, et al. Hydrogen exchange methods to study protein folding. Methods. 2004;34:51–64. doi: 10.1016/j.ymeth.2004.03.005. [DOI] [PubMed] [Google Scholar]
- 43.Bai Y, et al. Primary structure effects on peptide group hydrogen exchange. Proteins. 1993;17:75–86. doi: 10.1002/prot.340170110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dempsey CE. Hydrogen exchange in peptides and proteins using NMR-spectroscopy. Progr Nucl Magn Res Spectrosc. 2001;39:135–170. [Google Scholar]
- 45.Xiao H, et al. Mapping protein energy landscapes with amide hydrogen exchange and mass spectrometry: I. A generalized model for a two-state protein and comparison with experiment. Protein Sci. 2005;14:543–557. doi: 10.1110/ps.041001705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kaltashov IA. Probing protein dynamics and function under native and mildly denaturing conditions with hydrogen exchange and mass spectrometry. Int J Mass Spectrom. 2005;240:249–259. [Google Scholar]
- 47.Hoerner JK, Xiao H, Kaltashov IA. Structural and dynamic characteristics of a partially folded state of ubiquitin revealed by hydrogen exchange mass spectrometry. Biochemistry. 2005;44:11286–94. doi: 10.1021/bi0509548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rosa JJ, Richards FM. An experimental procedure for increasing the structural resolution of chemical hydrogen-exchange measurements on proteins: application to ribonuclease S peptide. J Mol Biol. 1979;133:399–416. doi: 10.1016/0022-2836(79)90400-5. [DOI] [PubMed] [Google Scholar]
- 49.Englander JJ, Rogero JR, Englander SW. Protein hydrogen exchange studied by the fragment separation method. Anal Biochem. 1985;147:234–44. doi: 10.1016/0003-2697(85)90033-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Han JC, Han GY. A procedure for quantitative determination of Tris(2-carboxyethyl)phosphine, an odorless reducing agent more stable and effective than dithiothreitol. Anal Biochem. 1994;220:5–10. doi: 10.1006/abio.1994.1290. [DOI] [PubMed] [Google Scholar]
- 51.Burkitt W, Domann P, O'Connor G. Conformational changes in oxidatively stressed monoclonal antibodies studied by hydrogen exchange mass spectrometry. Protein Sci. 2010;19:826–35. doi: 10.1002/pro.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Houde D, et al. Post-translational modifications differentially affect IgG1 conformation and receptor binding. Mol Cel Proteom. 2010;9:1716–1728. doi: 10.1074/mcp.M900540-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Woods VL, Jr, Hamuro Y. High resolution, high-throughput amide deuterium exchange-mass spectrometry (DXMS) determination of protein binding site structure and dynamics: utility in pharmaceutical design. J Cell Biochem. 2001;37S:89–98. doi: 10.1002/jcb.10069. [DOI] [PubMed] [Google Scholar]
- 54.Stephen JC, et al. Epitope mapping by amide hydrogen/deuterium exchange coupled with immobilization of antibody, on-line proteolysis, liquid chromatography and mass spectrometry. Rapid Commun Mass Spectrom. 2009;23:639–647. doi: 10.1002/rcm.3921. [DOI] [PubMed] [Google Scholar]
- 55.Kaltashov IA, Bobst CE, Abzalimov RR. H/D exchange and mass spectrometry in the studies of protein conformation and dynamics: Is there a need for a top-down approach? Anal Chem. 2009;81:7892–7899. doi: 10.1021/ac901366n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Del Mar C, et al. Structure and properties of α-synuclein and other amyloids determined at the amino acid level. Proc Natl Acad Sci U S A. 2005;102:15477–15482. doi: 10.1073/pnas.0507405102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Spada S, Walsh G. Directory of approved biopharmaceutical products. Boca Raton: CRC Press; 2005. p. 317. [Google Scholar]
- 58.Veronese FM, Mero A. The impact of PEGylation on biological therapies. BioDrugs. 2008;22:315. doi: 10.2165/00063030-200822050-00004. [DOI] [PubMed] [Google Scholar]
- 59.Abzalimov RR, Kaltashov IA. Electrospray ionization mass spectrometry of highly heterogeneous protein systems: protein ion charge state assignment via incomplete charge reduction. Anal Chem. 2010;82:7523–7526. doi: 10.1021/ac101848z. [DOI] [PubMed] [Google Scholar]
- 60.Tropak MB, et al. Identification of pharmacological chaperones for Gaucher disease and characterization of their effects on beta-glucocerebrosidase by hydrogen/deuterium exchange mass spectrometry. ChemBioChem. 2008;9:2650–2662. doi: 10.1002/cbic.200800304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tang HY, Speicher DW. Curr Protoc Protein Sci. John Wiley & Sons, Inc.; 2001. Determination of disulfide-bond linkages in proteins. [DOI] [PubMed] [Google Scholar]
- 62.Gau BC, et al. Fast photochemical oxidation of protein footprints faster than protein unfolding. Anal Chem. 2009;81:6563–6571. doi: 10.1021/ac901054w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Maleknia SD, et al. Determination of macromolecular folding and structure by synchrotron x- ray radiolysis techniques. Anal Biochem. 2001;289:103–15. doi: 10.1006/abio.2000.4910. [DOI] [PubMed] [Google Scholar]
- 64.Konermann L, et al. Mass spectrometry combined with oxidative labeling for exploring protein structure and folding. Mass Spectrom Rev. 2010;29:651–667. doi: 10.1002/mas.20256. [DOI] [PubMed] [Google Scholar]
- 65.Mendoza VL, Vachet RW. Probing protein structure by amino acid-specific covalent labeling and mass spectrometry. Mass Spectrom Rev. 2009;28:785–815. doi: 10.1002/mas.20203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Back JW, et al. Chemical cross-linking and mass spectrometry for protein structural modeling. J Mol Biol. 2003;331:303–313. doi: 10.1016/s0022-2836(03)00721-6. [DOI] [PubMed] [Google Scholar]
- 67.Robinson CV, Sali A, Baumeister W. The molecular sociology of the cell. Nature. 2007;450:973–982. doi: 10.1038/nature06523. [DOI] [PubMed] [Google Scholar]
- 68.Abzalimov RR, Frimpong AK, Kaltashov IA. Gas-phase processes and measurements of macromolecular properties in solution: On the possibility of false positive and false negative signals of protein unfolding. Int J Mass Spectrom. 2006;253:207–216. [Google Scholar]
- 69.Cheng Y, et al. Structure of the human transferrin receptor-transferrin complex. Cell. 2004;116:565–76. doi: 10.1016/s0092-8674(04)00130-8. [DOI] [PubMed] [Google Scholar]