Abstract
The cytoskeletal protein talin activates integrin receptors by binding of its FERM domain to the cytoplasmic tail of β-integrin. Talin also couples integrins to the actin cytoskeleton, largely by binding to and activating the cytoskeletal protein vinculin, which binds to F-actin through the agency of its five-helix bundle tail (Vt) domain. Talin activates vinculin by means of buried amphipathic α-helices coined vinculin binding sites (VBSs) that reside within numerous four- and five-helix bundle domains that comprise the central talin rod, which are released from their buried locales by means of mechanical tension on the integrin:talin complex. In turn, these VBSs bind to the N-terminal seven-helix bundle (Vh1) domain of vinculin, creating an entirely new helix bundle that severs its head-tail interactions. Interestingly, talin harbors a second integrin binding site coined IBS2 that consists of two five-helix bundle domains that also contain a VBS (VBS50). Here we report the crystal structure of VBS50 in complex with vinculin at 2.3 Å resolution and show that intramolecular interactions of VBS50 within IBS2 are much more extensive versus its interactions with vinculin. Indeed, the IBS2-vinculin interaction only occurs at physiological temperature and the affinity of VBS50 for vinculin is about 30 times less than other VBSs. The data support a model where integrin binding destabilizes IBS2 to allow it to bind to vinculin.
Keywords: cytoskeleton, crystallography, focal adhesion, protein-protein interaction
Introduction
Talin is a large cytoskeletal protein that couples integrin receptors to the actin cytoskeleton and directs the necessary final step for inside-out integrin activation. Talin harbors a FERM domain within its N-terminus and has a large rod domain comprised of over sixty α-helices organized into four- or five-helix bundle domains.1 The talin-integrin interaction occurs by binding of the talin F3 FERM subdomain with the cytoplasmic tail of the β-subunit of the integrin. This triggers the separation of the α- and β-integrin tails and conformational changes in the ectodomains that promote high affinity binding to ligands of the extracellular matrix.2, 3 The talin rod domain contains at least 11 vinculin binding site (VBSs), two actin binding sites, a dimerization domain, and a second integrin binding site (IBS2),4 although the affinity for the IBS2-integrin interaction is much weaker.5
Structure-function analyses have suggested that the talin FERM domain converts integrins into a high affinity state whereas the talin IBS2 domain directs the clustering of integrin receptors.6 The crystal structure of the IBS2 domain has shown that it is comprised of two five-helix bundles coined IBS2-A and IBS2-B, and both bundles directly contribute to integrin binding and are required for localization of talin to focal adhesions and for binding to acidic phospholipids.5 Finally, differential scanning calorimetry experiments have suggested that the binding of integrin to IBS2 destabilizes both five-helix bundle domains.
Vinculin is a key regulator of cell–cell (adherens junctions) and cell–matrix (focal adhesions) contacts, and also controls integrin clustering, force generation, and the strength of adhesion. The structure of this 1066 residue polypeptide chain from homo sapiens at 2.85 Å resolution showed that this cytoskeletal protein harbors three seven-helix bundle subdomains, Vh1, Vh2, and Vh3, which together with one four-helix bundle, Vt2, comprise the vinculin head domain, VH, that is linked to the five-helix bundle tail domain, Vt, by means of a proline-rich region.7 The muscle specific metavinculin isoform is very similar, where the first Vt α-helix is replaced by another from the 68 residue metavinculin specific insert.8 Vinculin is held in its closed inactive conformation by extensive hydrophobic VH-Vt interactions, which can be disrupted by the vinculin binding sites (VBSs) of talin9–14 and α-actinin,15, 16 and by the VBSs of the Shigella invasin IpaA17–19 and the Rickettsial sca4 antigen.20 These VBSs activate vinculin by burying within and inducing helix bundle conversion of the N-terminal four-helix subdomain of Vh1, which displace Vt from a distance and expose its cryptic F-actin binding sites.
The talin rod domain harbors eleven VBSs14 and α-helix α4 of the first IBS2 five-helix bundle domain contains such a vinculin binding site, VBS50.14 Here we report the 2.3 Å crystal structure of the vinculin head domain Vh1 in complex with talin-VBS50. While VBS50 is part of a five-helix bundle when either in IBS2 or when bound to vinculin, it engages in fewer interactions when bound to vinculin and, accordingly, it has a reduced affinity for vinculin versus other VBSs of talin. Collectively, these data support a model where integrin binding to IBS2 unfurls this five-helix bundle to allow talin-VBS50 to interact with vinculin.
Results
The crystal structure of vinculin in complex with talin-VBS50
The fact that IBS2 of talin can bind to both integrin and to vinculin suggested that this domain might coordinate the interactions of integrin receptors with vinculin. To gain insights into the role of the IBS2 domain of talin in the interplay of integrins and vinculin, we solved the structure of vinculin binding site 50 (VBS50) that is present within the IBS2 domain in complex with the Vh1 domain of vinculin at 2.3 Å resolution. This structure revealed that VBS50 binds to vinculin in a mode very similar to other Vh1:talin-VBS crystal structures [Fig. 1(A)]. All Vh1:VBS crystal structures10–12, 14 have the VBS inserted into the N-terminal four-helix Vh1 subdomain by helix bundle conversion.9 Further, in all Vh1:talin-VBS crystal structures, the VBS α-helix is parallel to Vh1 α-helix α1 and antiparallel to α-helix α2. The 21 core residues of VBS1, VBS2, VBS11, VBS33, VBS36, VBS50, and VBS58 align with those of VBS3 with r.m.s.d. ranging from 0.5 Å to 1.1 Å.
Figure 1.
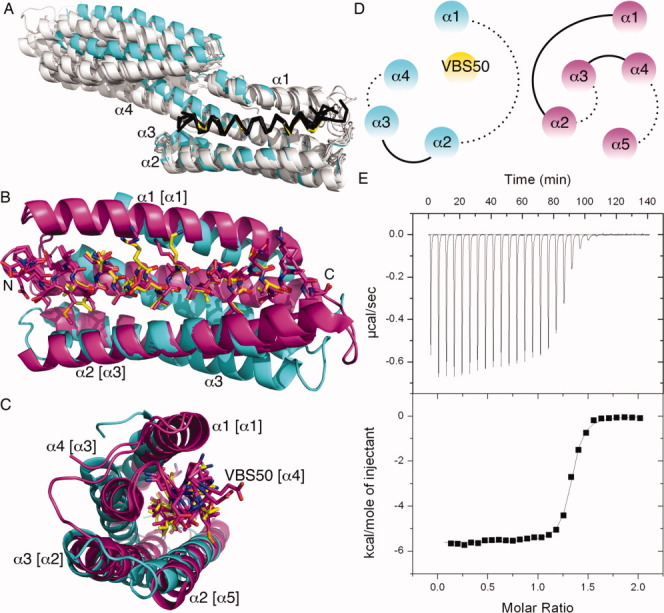
Talin-IBS2 unfurls upon binding to vinculin. A: Superposition of the crystal structures of all talin-VBSs (shown as Cα traces) in their Vh1-bound states (shown as a cartoon) onto our Vh1:VBS50 (Vh1, cyan; VBS50, yellow). Vh1 α-helices α1 through α4 are labeled. VBS1 (residues 608-627), VBS11 (residues 822–841), VBS2 (residues 855–874), VBS33 (residues 1525–1544), VBS36 (residues 1633–1652), VBS3 (residues 1949–1968), and VBS58 (residues 2345–2364) are shown in black bound to Vh1, which is shown in white, and they were aligned to VBS50 (residues 2080–2099). The N-terminal Vh1-bound subdomain structures are almost identical while the C-terminal four-helix Vh1 subdomain that does not bind to these VBSs is found in several relative orientations relative to the N-terminal four-helix bundle subdomain. B: Cartoon drawing of human vinculin Vh1 domain (cyan) in complex with VBS50 of the talin IBS2 (shown in yellow) ball-and-stick representation. Human VBS50 (residues 2079–2097) in their Vh1-bound state are superimposed onto VBS50 as seen in the IBS2 crystal structures from mouse (magenta, PDB entry 3dyj; only one subunit of the asymmetric unit is shown) and Drosophila melanogaster (magenta, PDB entry 3fyq) with r.m.s.d. of 0.4 Å and 0.28 Å for 133 or 90 atoms, respectively. This superposition places α1 (labeled, residues 7–26) of Vh1 in a similar position as α1 of IBS2 (residues 1976–2001; bracketed α1) but with opposite directionality; α2 of Vh1 (residues 40–62) similarly to α5 of IBS2 (residues 2108–2138); α3 (Vh1 residues 68–95) similarly to α2 of IBS2 (residues 2011–2037); and α4 of Vh1 (residues 102–129) similarly to α3 of IBS2 (residues 2040–2068) with a relative tilt of about 45°. The termini of VBS50 are labeled (N and C, respectively). C: Perpendicular view, along the horizontal axis of panel B. α-Helices α1-α4 of Vh1 are labeled, which are structurally similar to α-helices [bracketed] α1 (residues 1975–2001), α5 (residues 2108–2139), α2 (residues 2011–2037), and α3 (residues 2040–2069) of IBS2, respectively, while VBS50 (residues 2078–2099) is α-helix α4 in the mouse and Drosophila IBS2 crystal structures (residues 2073–2102). D: Topologies of Vh1 in complex with VBS50 (left) and VBS50 within the IBS2 domain (right) in the same orientation and color coding as in panel C. Dotted lines indicate connectivity of the respective α-helices, which are on the bottom of the α-helices in this view. E: ITC titration of VBS50 to vinculin. The total heat exchanged during each injection of VBS50 to Vh1 is fit to a single-site binding model with stoichiometry, KD and ΔH° as independent parameters, where N = 1.3, KD = 99 nM, ΔH° = −5544 ± 24 cal mol−1, and TΔS = 3.98 kcal mol−1 or N = 1.3, KD = 97 nM, ΔH° = −5673 ± 23 cal mol−1, and TΔS = 3.71 kcal mol−1 for a second independent experiment (not shown). No significant heat exchange was observed for VBS50 titration into the reaction buffer alone.
The final refined model has good stereochemistry as evidenced by the MolProbity21 score of 1.3 corresponding to 100th percentile in comparison to similar resolution structures and an all atom clash score of 5.59 (equivalent to 99th percentile in comparison to similar resolution structures). Structural analysis by MolProbity21 revealed no outlier residues with 99.6% falling in favored regions of the Ramachandran plot.
Comparison of intra- versus intermolecular VBS50 interactions
The crystal structure of the talin IBS2 domain containing the VBS50 has been determined for both mouse5 and Drosophila22 talin. The VBS50 α-helix is the fourth α-helix of a five-helix bundle of the IBS2 domain [Fig. 1(B–D)], and the VBS50 residues involved in vinculin binding are buried in the talin IBS2 structure; thus, talin needs to be activated to expose this cryptic VBS50 binding site to allow binding to vinculin. Nevertheless, binding to vinculin does not alter the conformation of VBS50, as this α-helix in complex with vinculin can be superimposed with r.m.s.d. of 0.3–0.4 Å in its unbound state. Further, while the environment of VBS50 within IBS2 is different compared to its vinculin bound state, where VBS50 is parallel to Vh1 α-helices α1 and α3 but is rather antiparallel to α1, α3, and α5 within IBS2, these helices fulfill similar positions in binding to VBS50 that are not affected by the distinct topologies [Fig. 1(D)].
Interestingly, comparison of the structures revealed that VBS50 (residues, 2078–2099) engages in several additional intramolecular interactions versus those seen when bound to vinculin (Supporting Information Figs. S1–S3). Specifically, 29 talin IBS2 residues are in contact with eighteen VBS50 residues, while only 21 vinculin Vh1 residues interact with 15 VBS50 residues. Indeed, with the exception of talin residues Leu-2080, Asp-2086, Ala-2090, and Gly-2092, all other VBS50 residues are in contact with more talin residues in IBS2 than with vinculin residues in the Vh1:VBS50 structure. Further, this feature is seen for both hydrophobic and polar interactions, where there is only one polar interaction in the Vh1:VBS50 structure versus two electrostatic interactions for VBS50 within the IBS2 five-helix bundle structure. This is consistent with an increase of about 20% in buried solvent accessible surface area of VBS50 in the IBS2 structure compared to its interaction with the Vh1. The additional buried surface area is accounted by VBS50 and other IBS2 residues. However, the shape complementarity is similar in both cases (0.75 for VBS50 within IBS2 and 0.78 for the Vh1:VBS50 complex).
Talin-VBS50 binding to Vh1 in solution
The affinity for binding of VBS1, -2, and -3 to Vh1 was determined (binding constants of 15, 33, and 3 nM, respectively) and three residues were found to be responsible for the weaker binding of VBS2.16 In particular, the smaller side chain of residue Ala-868 (corresponding to VBS50 residue Gly-2092) was not able to interact with Vh1 versus the corresponding valine residue of VBS1 and VBS3. Furthermore, inspection of our Vh1:VBS50 structure suggests that Lys-2099 (which is also a lysine in VBS2) introduces a positive charge near Vh1 residues His-22 and Arg-105 (Supporting Information Fig. S3), which seems unfavorable compared to the Gln residue in this position in VBS1 and VBS3 or to Asn, Asp, or Gly residues in the three bacterial IpaA-VBSs, which bind Vh1 with 110 pM, 6 nM, and 54 pM affinities, respectively.17, 18 To address the binding properties of VBS50 we performed isothermal titration calorimetry (ITC). As predicted, we observed weaker binding (less than 100 nM) [Fig. 1(E)] compared with all other VBSs.
Discussion
The talin VBSs are found buried within four- or five-helix bundle domains within the large C-terminal talin rod domain. The recent structures of Vh1:VBS3311 and the four-helix VBS33 bundle domain23 showed that VBS33 engages in fewer intramolecular interactions than intermolecular interactions with Vh1. In contrast, the Vh1:VBS50 structure reported here together with the five-helix IBS2 bundle harboring VBS505, 22 shows that VBS50 engages in significantly more intramolecular interactions within the IBS2 five-helix bundle, consistent with the finding that the VBSs of talin need to be activated by force to unfurl the rod and expose these VBSs.24, 25 Indeed, the binding of VBS50 with vinculin is 3- to 33-fold weaker than that of other determined talin-VBSs and 16- to 540-fold weaker than that of the VBSs of the Shigella invasin IpaA. In accord with these findings, IBS2 binding to Vh1 was temperature dependent (more was bound at 37 °C versus ambient temperature) and the 58 °C melting temperature of IBS2 suggests that the physiological temperature might destabilize the IBS2 bundle to allowing it to bind to vinculin.5 Collectively, the data support a model whereby local force transmitted by binding of integrin receptors unfurls the IBS2 domain to allow VBS50 to bind and activate vinculin.
Materials and Methods
Vh1:VBS50 crystallization
The human vinculin Vh1 domain (residues 1–258) was generated as described.9 Crystallization screens of Vh1 protein and VBS50 mixed in 1:5 molar ratio were performed at two temperatures. Cocrystals were obtained from Hampton crystal screen I with a reservoir of 0.1M Hepes pH 7.5, 10% w/v polyethylene glycol 6000, and 5% 2-Methyl-2,4-pentanediol. Crystals were transferred directly from the initial 96-well screening plate into Paratone-N oil and flash-frozen in liquid nitrogen.
X-ray diffraction data collection, reduction, structure determination, and crystallographic refinement
X-ray diffraction data of Vh1:VBS50 crystals were obtained to 2.3 Å Bragg spacings at the Advanced Photon Source (APS), Argonne National Laboratory (ANL), SER-CAT ID-22 beam line and integrated and scaled using autoPROC26 which uses XDS27 and SCALA28 as the data reduction engine. The crystals belong to space group P212121 (a = 34 Å, b = 68 Å, c = 138 Å) with one Vh1:VBS50 heterodimer in the asymmetric unit resulting in a solvent content of 0.49 and a crystal volume per unit of protein molecular weight, VM,29 of 2.4 Å3/Da. The data collection statistics are summarized in Table I.
Table I.
X-ray Data Reduction and Crystallographic Refinement Statistics
| (A) X-ray data reduction statistics | |
| Space group | P212121 |
| Unit cell dimensions | |
| a, b, c | 33.5 Å, 68.2 Å, 137.6 Å |
| Resolution (last shell) | 68.8 Å–2.25 Å (2.26 Å–2.25 Å) |
| Total measurements (last shell) | 109,559 (1,168) |
| Number of unique reflections (last shell) | 15,637 (163) |
| Wavelength | 1 Å |
| R-mergea (last shell) | 0.065 (0.592) |
| I/σ(I) (last shell) | 16.4 (3.3) |
| Completeness (last shell) | 0.997 (1) |
| Redundancy (last shell) | 7 (7.2) |
| (B) Crystallographic refinement statistics | |
| Resolution (last shell) | 48.8 Å–2.25 Å (2.4 Å–2.25 Å) |
| No. of reflections (working set) | 14,799 |
| No. of reflections (test set) | 787 |
| R-factorb (last shell) | 0.208 (0.235) |
| R-freec (last shell) | 0.243 (0.320) |
| No. of amino acid residues | 263 |
| No. of atoms | 2,117 |
| No. of solvent molecules | 69 |
| Average B-factor | |
| Protein | 70.7 Å2 |
| Solvent | 64.3 Å2 |
| R.m.s.d. from ideal geometry | |
| Bond lengths | 0.01 Å |
| Bond angles | 1.1° |
R-merge = 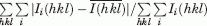
R-factor = 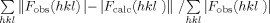 where <|Fcalc|> denotes the expectation of |Fcalc(hkl)| used in defining the likelihood refinement target.
where <|Fcalc|> denotes the expectation of |Fcalc(hkl)| used in defining the likelihood refinement target.
The free R-factor is a cross-validation residual calculated by using about 5% reflections, which were randomly chosen and excluded from the refinement.
Phases for the Vh1:VBS50 structure were obtained by molecular replacement using the Vh1 domain of the Vh1:IpaA-VBS structure (PDB entry 2ibf)19 as the search model and the CCP4 program MOLREP.30 Crystallographic refinement of Vh1:VBS50 was performed with BUSTER31 together with extensive manual model building using COOT.32
Isothermal titration calorimetry
ITC experiments were carried out at 22 °C using a VP-ITC calorimeter (Microcal). Both human Vh1 and VBS50 were kept in same buffer (20 mM Tris-HCl, 150 mM NaCl, pH 8) and were filtered and degassed along with the buffer for each experiment. The sample cell was filled with 30 μM Vh1 protein solution and the syringe was filled with a 300 μM VBS50 solution while the reference cell contained just the buffer. Each titration consisted of 28 injections, each of 10 μl volumes except the first injection (2 μl), and 20 s duration with a 5 min interval between additions. The binding isotherms were fitted using a single-site model as implemented in the Microcal Origin software, utilizing the nonlinear least-squares minimization routine to derive the binding stoichiometry (n), the equilibrium binding constant (KD), and changes in entropy (ΔS) and enthalpy (ΔH) occurring due to the binding reaction.
PDB Coordinates
The coordinates have been deposited with the Protein Data Bank (PDB entry 4dj9).
Acknowledgments
The authors are indebted to their colleagues at Scripps Florida: John Cleveland for critical review of the manuscript and many fruitful discussions, Philippe Bois and Zhen Wu for sequencing, and Philippe Bois for many helpful discussions. We are also grateful to the staff at the Advanced Photon Source, SER-CAT, for synchrotron support. TI is supported by grants from the National Institutes of Health and by start-up funds provided to Scripps Florida from the State of Florida. SDY is a fellow of the Department of Defense. This is publication no. 21482 from the Scripps Research Institute.
Glossary
Abbreviations
- ANL
Argonne National Laboratory
- APS
Advanced Photon Source
- CCP4
Collaborative Computational Project No. 4
- COOT
Crystallographic Object-Oriented Toolkit
- FERM
4.1 protein/ezrin/radixin/moesin
- IBS2
integrin binding site 2
- ITC
isothermal titration calorimetry
- PDB
protein data bank
- r.m.s.d.
root mean squares deviation
- r.p.m.
revolutions per minute
- SER-CAT
South Eastern Regional Collaborative Access Team
- VBS
vinculin binding site
- VH
vinculin head domain
- Vh1
N-terminal domain of vinculin
- Vh2
2nd domain of vinculin
- VM
crystal volume per unit of protein molecular weight
- Vt2
4th domain of vinculin
- Vt
vinculin tail domain
- XDS
X-ray Detector Software.
Supplementary material
Additional Supporting Information may be found in the online version of this article.
References
- 1.Critchley DR. Biochemical and structural properties of the integrin-associated cytoskeletal protein talin. Annual Rev Biophys. 2009;38:235–254. doi: 10.1146/annurev.biophys.050708.133744. [DOI] [PubMed] [Google Scholar]
- 2.Calderwood DA. Integrin activation. J Cell Sci. 2004;117:657–666. doi: 10.1242/jcs.01014. [DOI] [PubMed] [Google Scholar]
- 3.Calderwood DA, Zent R, Grant R, Rees DJ, Hynes RO, Ginsberg MH. The Talin head domain binds to integrin β subunit cytoplasmic tails and regulates integrin activation. J Biol Chem. 1999;274:28071–28074. doi: 10.1074/jbc.274.40.28071. [DOI] [PubMed] [Google Scholar]
- 4.Moes M, Rodius S, Coleman SJ, Monkley SJ, Goormaghtigh E, Tremuth L, Kox C, van der Holst PP, Critchley DR, Kieffer N. The integrin binding site 2 (IBS2) in the talin rod domain is essential for linking integrin β subunits to the cytoskeleton. J Biol Chem. 2007;282:17280–17288. doi: 10.1074/jbc.M611846200. [DOI] [PubMed] [Google Scholar]
- 5.Gingras AR, Ziegler WH, Bobkov AA, Joyce MG, Fasci D, Himmel M, Rothemund S, Ritter A, Grossmann JG, Patel B, Bate N, Goult BT, Emsley J, Barsukov IL, Roberts GC, Liddington RC, Ginsberg MH, Critchley DR. Structural determinants of integrin binding to the talin rod. J Biol Chem. 2009;284:8866–8876. doi: 10.1074/jbc.M805937200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tanentzapf G, Brown NH. An interaction between integrin and the talin FERM domain mediates integrin activation but not linkage to the cytoskeleton. Nat Cell Biol. 2006;8:601–606. doi: 10.1038/ncb1411. [DOI] [PubMed] [Google Scholar]
- 7.Borgon RA, Vonrhein C, Bricogne G, Bois PR, Izard T. Crystal structure of human vinculin. Structure. 2004;12:1189–1197. doi: 10.1016/j.str.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 8.Rangarajan ES, Lee JH, Yogesha SD, Izard T. A helix replacement mechanism directs metavinculin functions. PLoS ONE. 2010;5:e10679. doi: 10.1371/journal.pone.0010679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Izard T, Evans G, Borgon RA, Rush CL, Bricogne G, Bois PR. Vinculin activation by talin through helical bundle conversion. Nature. 2004;427:171–175. doi: 10.1038/nature02281. [DOI] [PubMed] [Google Scholar]
- 10.Izard T, Vonrhein C. Structural basis for amplifying vinculin activation by talin. J Biol Chem. 2004;279:27667–27678. doi: 10.1074/jbc.M403076200. [DOI] [PubMed] [Google Scholar]
- 11.Yogesha SD, Sharff A, Bricogne G, Izard T. Intermolecular versus intramolecular interactions of the vinculin binding site 33 of talin. Protein Sci. 2011;20:1471–1476. doi: 10.1002/pro.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fillingham I, Gingras AR, Papagrigoriou E, Patel B, Emsley J, Critchley DR, Roberts GC, Barsukov IL. A vinculin binding domain from the talin rod unfolds to form a complex with the vinculin head. Structure. 2005;13:65–74. doi: 10.1016/j.str.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 13.Gingras AR, Vogel KP, Steinhoff HJ, Ziegler WH, Patel B, Emsley J, Critchley DR, Roberts GC, Barsukov IL. Structural and dynamic characterization of a vinculin binding site in the talin rod. Biochemistry. 2006;45:1805–1817. doi: 10.1021/bi052136l. [DOI] [PubMed] [Google Scholar]
- 14.Gingras AR, Ziegler WH, Frank R, Roberts GC, Critchley DR, Emsley J. Mapping and consensus sequence identification for multiple vinculin binding sites within the talin rod. J Biol Chem. 2005;280:37217–37224. doi: 10.1074/jbc.M508060200. [DOI] [PubMed] [Google Scholar]
- 15.Bois PR, Borgon RA, Vonrhein C, Izard T. Structural dynamics of α-actinin-vinculin interactions. Mol Cell Biol. 2005;25:6112–6122. doi: 10.1128/MCB.25.14.6112-6122.2005. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 16.Bois PR, O'Hara BP, Nietlispach D, Kirkpatrick J, Izard T. The vinculin binding sites of talin and α-actinin are sufficient to activate vinculin. J Biol Chem. 2006;281:7228–7236. doi: 10.1074/jbc.M510397200. [DOI] [PubMed] [Google Scholar]
- 17.Izard T, Tran Van Nhieu G, Bois PR. Shigella applies molecular mimicry to subvert vinculin and invade host cells. J Cell Biol. 2006;175:465–475. doi: 10.1083/jcb.200605091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Park H, Valencia-Gallardo C, Sharff A, Tran Van Nhieu G, Izard T. A novel Vinculin binding site of the IpaA invasin of Shigella. J Biol Chem. 2011;286:23214–23224. doi: 10.1074/jbc.M110.184283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tran Van Nhieu G, Izard T. Vinculin binding in its closed conformation by a helix addition mechanism. Embo J. 2007;26:4588–4596. doi: 10.1038/sj.emboj.7601863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park H, Lee JH, Cossart P, Gouin E, Izard T. The Rickettsia surface cell antigen 4 applies mimicry to bind to and activate vinculin. J Biol Chem. 2011;286:35096–35103. doi: 10.1074/jbc.M111.263855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen VB, Arendall WB, III, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cheung TY, Fairchild MJ, Zarivach R, Tanentzapf G, Van Petegem F. Crystal structure of the talin integrin binding domain 2. J Mol Biol. 2009;387:787–793. doi: 10.1016/j.jmb.2009.01.053. [DOI] [PubMed] [Google Scholar]
- 23.Gingras AR, Bate N, Goult BT, Patel B, Kopp PM, Emsley J, Barsukov IL, Roberts GC, Critchley DR. Central region of talin has a unique fold that binds vinculin and actin. J Biol Chem. 2010;285:29577–29587. doi: 10.1074/jbc.M109.095455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee SE, Kamm RD, Mofrad MR. Force-induced activation of talin and its possible role in focal adhesion mechanotransduction. J Biomech. 2007;40:2096–2106. doi: 10.1016/j.jbiomech.2007.04.006. [DOI] [PubMed] [Google Scholar]
- 25.del Rio A, Perez-Jimenez R, Liu R, Roca-Cusachs P, Fernandez JM, Sheetz MP. Stretching single talin rod molecules activates vinculin binding. Science. 2009;323:638–641. doi: 10.1126/science.1162912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vonrhein C, Flensburg C, Keller P, Sharff A, Smart O, Paciorek W, Womack T, Bricogne G. Data processing and analysis with the autoPROC toolbox. Acta Crystallogr D Biol Crystallogr. 2011;67:293–302. doi: 10.1107/S0907444911007773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J Appl Crystallogr. 1993;26:795–800. [Google Scholar]
- 28.Evans P. Scaling and assessment of data quality. Acta Crystallogr D Biol Crystallogr. 2006;62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 29.Matthews BW. Solvent content of protein crystals. J Mol Biol. 1968;33:491–497. doi: 10.1016/0022-2836(68)90205-2. [DOI] [PubMed] [Google Scholar]
- 30.Vagin A, Teplyakov A. MOLREP: an automated program for molecular replacement. J Appl Cryst. 1997;30:1022–1025. [Google Scholar]
- 31.Bricogne G, Blanc E, Brandl M, Flensburg C, Keller P, Paciorek P, Roversi P, Sharff A, Smart OS, Vonrhein C, Womack TO. BUSTER version 2.9. Cambridge. United Kingdom: Global Phasing Ltd; 2011. [Google Scholar]
- 32.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


