Molecular characterization and subtyping show genetic diversities within clonal complexes.
Keywords: Enterohemorrhagic E. coli O157:H7, clonal complexes, genetic diversity, molecular evolution, research
Abstract
Escherichia coli O157:H7 variants were examined for trait mutations and by molecular subtyping to better define clonal complexes postulated on the O157:H7 evolution model. Strains of β-glucuronidase–positive, sorbitol-negative O157:H7 isolated in United States and Japan were identical to A5 clonal strain and shared sequence type (ST)–65 by multilocus sequence typing (MLST); thus, they belong in A5. However, these strains exhibited pulsed-field gel electrophoresis (PFGE) profile differences that suggested genomic divergence between populations. Sorbitol-fermenting O157 (SFO157) strains from Finland, Scotland, and Germany were identical to A4 clonal strain and belong in A4. Some SFO157 strains, isolated years apart and from different countries, had identical PFGE profiles, suggesting a common origin. Despite similarities, some Finnish and Scottish and all of the German strains have ST-75 (“German clone”), whereas others have ST-76, a new variant (“Scottish clone”). MLST of strains in other clonal complexes also discriminated strains thought to be identical and showed that genetic differences will further distinguish clonal populations into subclones.
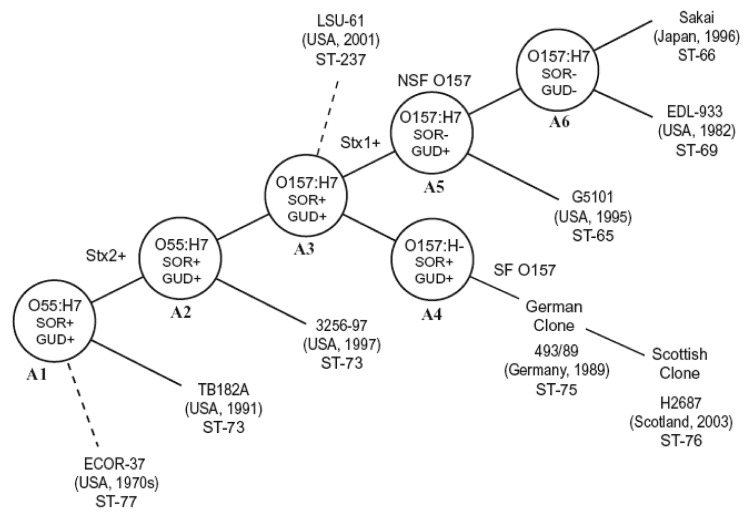
An evolutionary model postulates that Escherichia coli O157:H7 evolved from ancestral E. coli by stepwise acquisition or loss of virulence and phenotypic traits (1). At the center of the model is a hypothetical “intermediate” strain (ancestor A3), which evolved from the A2 clonal complex of O55:H7 strains that are closely related but ancestral to O157:H7 (2). It is hypothesized that the A3 intermediate strain is a missing link, a primitive O157:H7 that ferments sorbitol (SOR), is β-glucuronidase positive (GUD+), and has the Shiga toxin 2 gene (stx2), and from which evolved 2 distinct pathways. The loss of SOR phenotype and the acquisition of stx1 gene led to the emergence of the A5 clonal complex of SOR–, GUD+ O157:H7 strains, which then lost GUD expression, resulting in the prototypic O157:H7 clonal complex (A6). Also from A3 emerged a divergent lineage caused in part by the loss of motility giving rise to the A4 clonal complex of SOR+, GUD+, nonmotile strains that are designated as SFO157. These clonal complexes on the model were predicted on the basis of phenotypes, multilocus enzyme electrophoresis (MLEE), and the presence of the +93 single nucleotide polymorphisms (SNP) in the uidA gene that encode for GUD (1). However, except for the A6 clonal complex (O157:H7), from which strains were readily available, only a few strains existed for the other clonal complexes. As a result, these clonal complexes were not well defined because of the limited characterization criteria used and the lack of strains.
Molecular typing methods have improved the ability to characterize and discriminate closely related strains. Genetic studies have also elucidated some of the mutations that occurred in the stepwise emergence of clonal complexes. For example, in the transition from A5 to A6, the loss of GUD expression was found to be due to a frame-shift mutation caused by G-G insertion at +686 in the uidA gene (3). Similarly, in the divergence of A4 from A3, the loss of motility was caused by a 12-bp deletion in the flhC flagella master regulon gene (4). These mutations provide unique markers for tracing the model’s evolutionary events and, coupled with better typing methods, have provided more discriminatory means to reexamine genetic relatedness among O157:H7 clonal complexes.
In addition, new or previously rare strains have been isolated more frequently. The A5 clonal complex of GUD+ O157:H7 was represented solely by strain G5101, isolated from a hemorrhagic colitis patient in the United States in 1994 (5). Similarly, the A4 clone complex of SFO157 was represented by 493–89 and a few other German strains isolated from hemolytic uremic syndrome (HUS) patients in Bavaria in 1988 (6). Recently, however, GUD+ O157:H7 strains have been isolated in the United States and from outbreaks and sporadic infections in Hokkaido, Japan (7). Likewise, SFO157 strains, which have also been isolated from cattle (8), are increasingly causing sporadic infections and outbreaks of HUS (9,10) and have been isolated in the Czech Republic (11), Finland (12,13), Scotland (14), and other countries (6).
In this study, we examined strains from various clonal complexes, including GUD+ O157:H7 strains from the United States and Japan and SFO157 strains from Finland, Scotland, and Germany, for unique mutational markers. We also used molecular typing to better define the various clonal complexes in the O157:H7 evolution model.
Materials and Methods
Bacterial Strains
The 3 GUD+ O157:H7 strains from the United States were originally isolated from clinical samples, and the 22 strains from Japan were isolated from outbreak patients with diarrhea or no symptoms and sporadic cases in Hokkaido (7). The SFO157 strains included 8 from Germany (15), 8 from Finland (13), and 5 from Scotland (14). Most of these were isolated from patients with symptoms of hemorrhagic colitis or HUS. However, 3 Scottish strains (designated H1085 and variations thereof) were isolated from cat feces at a farm where a hemorrhagic colitis infection had occurred. The other strains examined are from the Food and Drug Administration (FDA) and the Shiga Toxin-Producing Escherichia coli Center (Michigan State University, East Lansing, MI, USA), except for LSU-61, an O157:H7 strain isolated from deer (16).
Characterization
To reconfirm their phenotypic traits, strains were plated on cefixime-tellurite sorbitol MacConkey agar (Bacteriological Analytical Manual, www.cfsan.fda.gov/~ebam/bam-4a.html) to test for SOR fermentation and resistance to 2.5 μg/mL potassium tellurite. The plates also had a ColiComplete disc (BioControl, Bellevue, WA, USA) to test for GUD activity. All isolates were serotyped for O157 and H7 antigens (RIM O157:H7, Remel, Lenexa, KS, USA) and tested by several PCR assays for trait virulence genes, mutational markers, and SNPs. A multiplex PCR (17) was used to test for stx1, stx2, γ-eae for intimin, ehxA for enterohemolysin, and the +93 SNP in the uidA. The GUD+ O157:H7 strains were tested for the +776 SNP and for the +686 G-G insertion in uidA (3); the SFO157 strains were tested for the presence of the H7 fliC gene and for the 12-bp flhC deletion (4). Shiga toxin (Stx) production was verified serologically with the Verotoxin-producing E. coli–Reversed Passive Latex Agglutination Test (Denka Seiken, Japan), and enterohemolysin activity was tested on Ca++ blood agar plates (18).
Pulsed-Field Gel Electrophoresis (PFGE)
XbaI-digested genomic DNA was analyzed in 1% agarose gel in 0.5× Tris-boric acid-EDTA TBE buffer at 14°C by using CHEF MAPPER (BioRad, Hercules, CA, USA) (19). The runtime was 18 h at 6V/cm, with initial and final switch times of 2.16 and 54.17 s, respectively. The gel was stained with ethidium bromide (1 μg/mL), observed on the Gel Doc 2000 system (BioRad), and analyzed with the BioNumerics fingerprinting software (Applied Maths, St-Martens-Latem, Belgium).
Multilocus Sequence Typing (MLST)
The MLST protocol (www.shigatox.net/cgi-bin/mlst7/index) uses PCR primers to amplify internal segments of 7 specific housekeeping genes (aspartate aminotransferase [aspC], caseinolytic protease [clpX], acyl-CoA synthetase [fadD], isocitrate dehydrogenase [icdA], lysine permease [lysP], malate dehydrogenase [mdh] and uidA), which are purified and sequenced. Each unique sequence is given an allele number, and the combinations of alleles from the 7 genes are used to compile the organism’s allelic profile. Each unique profile is designated as a sequence type (ST), which is then compared with those of other pathogenic E. coli strains in the EcMLST database (20).
Results
Strain Characterizations
All the GUD+ O157:H7 strains examined had traits identical to the A5 type strain (G5101); were SOR–, GUD+, and tellurite resistant; and produced both O157 and H7 antigens (Table 1). Except for 2 US strains (TW06289 and TW06290) that did not have stx1, all carried stx1, stx2, γ-eae, ehxA, and the +93 uidA SNP. Serologic analysis confirmed the production of both Stx, and all strains also had enterohemolysin activity. All GUD+ O157:H7 strains had the +776 SNP and, consistent with their GUD+ phenotype, did not have the +686 uidA G-G insertion. Because these strains expressed H7, they were not tested for the H7 fliC gene or for the flhC deletion.
Table 1. Characteristics of Escherichia coli O157 strains*.
| Source | No. | SOR | GUD | Τe | O157 | H7 | fliC | stx1 | stx2 | γeae | ehxA | +93 | +776 | +686 | BAP | ΔflhC |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| G5101 | – | + | R | + | + | + | + | + | + | + | + | + | – | + | – | |
| USA | 1 | – | + | R | + | + | ND | + | + | + | + | + | + | – | + | NA |
| 2 | – | + | R | + | + | ND | – | + | + | + | + | + | – | + | NA | |
| Japan | 22 | – | + | R | + | + | ND | + | + | + | + | + | + | – | + | NA |
| 498/89 | + | + | S | + | – | + | – | + | + | + | + | – | NA | – | + | |
| Germany | 5 | + | + | S | + | – | + | – | + | + | + | + | – | NA | – | + |
| 2 | + | + | S | + | – | + | – | – | + | + | + | – | NA | – | + | |
| Scotland | 4 | + | + | S | + | – | + | – | + | + | + | + | – | NA | – | + |
| 1 | + | + | S | + | – | + | – | + | + | – | + | – | NA | – | + | |
| Finland | 6 | + | + | S | + | – | + | – | + | + | + | + | – | NA | – | + |
| 2 | + | + | S | + | – | + | – | – | + | + | + | – | NA | – | + | |
| LSU-61 | + | + | R | + | + | + | – | – | + | + | + | – | NA | + | – |
*SOR, sorbitol fermentation; GUD, β-glucuronidase activity; Te, tellurite resistant (R) or sensitive (S); O157, antigen; H7, antigen; fliC, H7 flagella gene; stx1, shiga toxin 1 gene; stx2, shiga toxin 2 gene; γeae, γ-intimin; ehxA, enterohemolysin gene; +93, uidA single nucleotide polymorphisms (SNP); +776, uidA SNP; +686, uidA G-G insertion; BAP, enterohemolysin activity on blood agar plates; ΔflhC, 12-bp deletion; ND, not done; NA, not applicable.
All SFO157 strains examined were SOR+, GUD+, and tellurite sensitive, and expressed the O157 but not the H7 antigen (Table 1). Despite the absence of H7, all strains carried the H7 fliC gene. Except for 2 German (210–89, CB1009) and 2 Finnish (IH56929, IH56776) strains that did not have stx, all SFO157 carried only stx2, γ-eae, +93 uidA SNP, and the 12-bp flhC deletion, but not the +776 uidA SNP. Since SFO157 strains are GUD+, they were not tested for the +686 insertion in uidA. Also, except for strain H1085 1a/1, all SFO157 strains carried ehxA, but none had enterohemolysin activity. All these traits are consistent with those of the A4 type strain (493–89).
Analysis of LSU-61 showed that it had a mix of traits from various clonal complexes. It is SOR+, GUD+, tellurite resistant, and had both O157 and H7 antigens. It had the +93 SNP but not the +776 SNP in uidA; had no stx; had γ-eae and ehxA; and showed enterohemolytic activity on blood agar plates (Table 1).
Molecular Subtyping
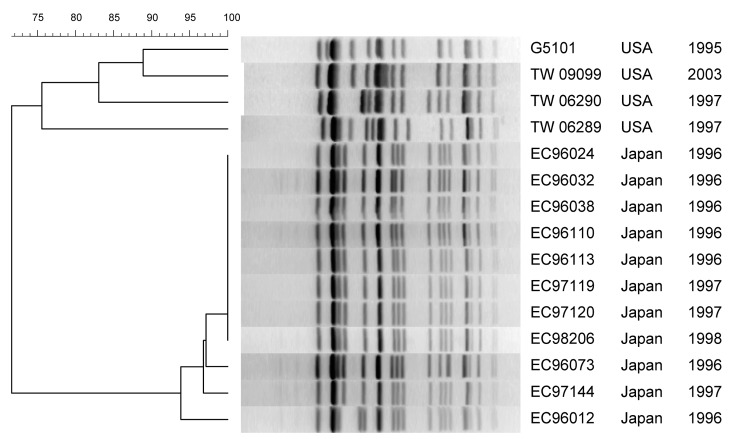
PFGE profiles of the US and 11/22 Japanese GUD+ O157:H7 strains are shown in Figure 1. The Japanese strains had nearly identical profiles, showing >95% similarity. Among the US strains, TW09099 and G5101 shared ≈90% similarity, but the other 2 strains were only 75% similar to G5101. The profiles of the US and Japanese strains shared only ≈70% similarity overall. MLST showed that all GUD+ O157:H7, including G5101, had ST-65 (Table 2).
Figure 1.
Pulsed-field gel electrophoresis of XbaI-digested DNA from GUD+ O157:H7 strains. Strain designation, source and year of isolation are shown at right. This unweighted pair-group method with arithmetic mean dendrogram was generated in BioNumerics software (Applied Maths, St-Martens-Latem, Belgium) by using Die coefficient with a 1.0% lane optimization and 1.0% band position tolerance. The scale above the dendrogram indicates percent similarity.
Table 2. Sequence type, serotype, and isolation information of strains of Escherichia coli.
| Sequence type | Strain | Source | Year | Serotype |
|---|---|---|---|---|
| 65* | G5101 | USA | 1995 | O157:H7 |
| EC96038 | Japan | 1996 | O157:H7 | |
| EC96024 | Japan | 1996 | O157:H7 | |
| EC97144 | Japan | 1997 | O157:H7 | |
| TW09099 | USA | 2003 | O157:H7 | |
| TW06290 | USA | 1997 | O157:H7 | |
| TW06289 | USA | 1997 | O157:H7 | |
| 66 | 86–24 | USA | 1986 | O157:H7 |
| 93–111 | USA | 1993 | O157:H7 | |
| Sakai | Japan | 1996 | O157:H7 | |
| 69 | EDL-933 | USA | 1982 | O157:H7 |
| 73 | C586–65 | Sri Lanka | 1965 | O55:H7 |
| TB182A | USA | 1991 | O55:H7 | |
| 5905 | USA | 1994 | O55:H7 | |
| 3256–97 | USA | 1997 | O55:H7 | |
| 75 | 493/89 | Germany | 1989 | O157:H– |
| 1782/88 | Germany | 1988 | O157:H– | |
| 5412/89 | Germany | 1989 | O157:H– | |
| CB569 | Germany | 1987 | O157:H– | |
| 210/89 | Germany | 1989 | O157:H– | |
| CB1009 | Germany | 1990 | O157:H– | |
| 514/91 | Germany | 1991 | O157:H– | |
| 4326/93 | Germany | 1993 | O157:H– | |
| IH 53440 | Finland | 1997 | O157:H– | |
| IH 56776 | Finland | 1998 | O157:H– | |
| IH 57086 | Finland | 1999 | O157:H– | |
| H1410 | Scotland | 2002 | O157:H– | |
| IH 57225 | Finland | 1990s | O157:H– | |
| 76 | IH 57201 | Finland | 1999 | O157:H– |
| IH 56909 | Finland | 1999 | O157:H– | |
| IH 56969 | Finland | 1999 | O157:H– | |
| IH 56929 | Finland | 1999 | O157:H– | |
| H2687 | Scotland | 2003 | O157:H– | |
| H1085C | Scotland | 2003 | O157:H– | |
| H1085 3a | Scotland | 2003 | O157:H– | |
| H1085 1a/1 | Scotland | 2003 | O157:H– | |
| 77 | ECOR-37† | USA | 1970s | Ont:Hnt |
| 237 | LSU-61 | USA | 2001 | O157:H7 |
*Only selected β-glucuronidase positive O157:H7 strains from Japan are shown. †Not typeable.
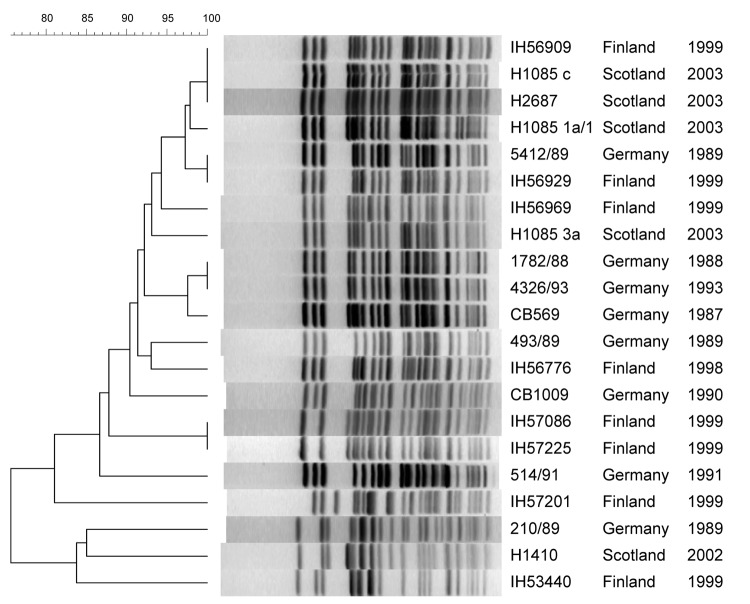
The PFGE profiles of some SFO157 strains isolated within Germany (1782/88 and 4326/93) and Finland (IH57086 and IH57225) were identical (Figure 2). But profile identity was also observed among strains from Finland (IH56906) and Scotland (H1085c and H2687) and strains from Finland (IH56929) and Germany (5412/89) (Figure 2). MLST analysis showed that all the German, 4 Finnish, and 1 Scottish strain had ST-75 but that the other 4 Scottish strains and the rest of the Finnish strains had a distinct mdh allele and were genotyped as ST-76 (Table 2).
Figure 2.
Pulsed-field gel electrophoresis of XbaI-digested DNA from SFO157 strains. Strain designation, source, and year of isolation are shown at right. This unweighted pair-group method with arithmetic mean dendrogram was generated in BioNumerics software (Applied Maths, St-Martens-Latem, Belgium) by using Die coefficient with a 1.0% lane optimization and 1.0% band position tolerance. The scale above the dendrogram indicates percent similarity.
Discussion
The stepwise evolutionary model postulates that ancestral O157 clonal group (A3) split into 1 lineage, leading to the common GUD–, SOR– O157:H7 (NSF O157) clonal complex (A6) and a second branch of SFO157 that retained many primitive traits (A4) (Figure 3). The A4 and A5 clonal complexes on the evolution model are closely related to O157:H7 (A6 clonal complex) (Figure 3) and share many traits, including the +93 uidA SNP, which is found only in O157:H7 and its nonmotile variants (1). Another common trait is the γ-eae allele, which is also found in few other serotypes (21), including the O55:H7 strains in the A2 clonal complex that is ancestral to O157:H7 (2) (Figure 3). The A5 clonal complex of GUD+ O157:H7 strains is postulated to have emerged from the A3 intermediate strain. Analysis of A5 type strain (G5101) showed that it carried a +776 uidA SNP, which appears to have been acquired before the emergence of A5, because it is found only in A5 strains and A6 clonal complex of O157:H7 strains (3). All the GUD+ O157:H7 strains tested had identical traits as G5101, including the unique uidA markers (+93 and +776 SNP and absence of +686 G-G insertion) that are consistent with the mutational events postulated for the emergence of A5 and confirm that these strains also belong in A5. The fact that all these GUD+ O157:H7 strains have ST-65 supports that conclusion (Table 2). Despite similarities, however, there were differences in PFGE profiles. The Japanese strains had nearly identical PFGE profiles, which is consistent with the results by Nagano et al. (7), who also observed profile identity among clinical and environmental GUD+ O157:H7 isolates in Japan (22). In contrast, the US strains showed more diversity and shared only 70% similarity with the Japanese strains, which suggests the occurrence of recent genomic divergences among the US populations of GUD+ O157:H7 strains.
Figure 3.
Evolution model for Escherichia coli O157:H7. Figure modified and updated from (1) to include the sequence type (ST) data showing subclones within clonal complexes. Some strains, whose position on the model remains to be determined, are shown with dashed lines.
The SFO157 strains of A4 clonal complex were also postulated to have evolved from the A3 intermediate strain (Figure 3). Analysis of the A4 type strain 493–89 showed that a key mutation that led to the emergence of A4 was the loss of motility caused by the 12-bp flhC deletion (4). Other A4 traits included tellurite sensitivity and the carriage of both ehxA and the H7 fliC, neither of which is expressed. All the SFO157 strains tested had these markers, which were postulated to have been acquired in the emergence of A4, which confirmed that these SFO157 strains also belong in the A4 clonal complex. The genomic similarities of the SFO157 strains are also reflected in their PFGE profiles, as 2 German strains (1782/88 and 4326/93) isolated 5 years apart had identical profiles; a Finnish strain (IH56909) isolated in 1999 was identical to 2 Scottish strains (H1085c, H2687) that were isolated in 2003; and a Finnish strain (IH56929) was identical to a German strain (5412/89) that was isolated 10 years earlier. These results are consistent with similarities observed among SFO157 strains from Germany (15), Finland (12,13), and the Czech Republic (11). The fact that there are profile identities among strains isolated years apart and from different geographic areas suggests that these SFO157 strains isolated from various European countries may have a common origin. Despite PFGE profile similarities, MLST showed genetic differences within the SFO157 populations (Table 2). The German, 4 Finnish, and 1 Scottish strain (H1410) had ST-75, which we designated as the “German clone,” while the remaining Scottish and Finnish strains had ST-76 (“Scottish clone”) (Figure 3). This genetic difference existed among strains with identical PFGE profiles, as German strain 5412/89 and Finnish strain IH56929 had ST-75 and ST-76, respectively.
Preliminary studies show that MLST (Table 2) may also better define other clonal complexes. In the A6 complex of O157:H7 strains, the Sakai strain that caused outbreaks in Japan and 2 US strains had identical lysP alleles (ST-66) but were distinct from the commonly used EDL933 reference strain (ST-69), a finding that suggests that genetic differences may also further distinguish the A6 clonal complex into subclones (Figure 3).
The A1 clonal complex, which consists of O55:H7 strains that are SOR+, GUD+, and carry γ-eae, was postulated to have given rise to A2 by the acquisition of the stx2 phage (1). Based on phenotypes, 2 O55:H7 strains (C586–65 and TB182A) that did not have stx were previously thought to be A1 strains, but MLST showed both strains to have ST-73, identical to the Stx2-producing O55:H7 strains (5905 and 3256–97) in the A2 clonal complex (Table 2). Analogous to the data obtained with the GUD+ O157:H7 strains, these O55:H7 strains had the same ST, but the PFGE profiles of C586–65 and TB182A shared only 60% similarity to 5905 and 3256–97 (data not shown). The genetic data on the transition from A1 to A2 is limited, so it is uncertain whether C586–65 and TB182A are derivatives of A1 or, perhaps, are A2 strains that have lost the stx2 phage (see below).
Similarly, a marmoset isolate of E. coli (ECOR37) was previously shown to be closely related to A2 by MLEE. This strain had A2-like traits, including γ-eae (23), but its O and H antigens were serologically untypeable. PCR testing showed that ECOR37 had both the O55 wzx gene, required for export of O lipopolysaccharide, and the H7 fliC gene (data not shown) and, therefore, is an O55:H7 strain. However, ECOR37 shared only 60% similarity in PFGE profiles with A2 strains (data not shown). MLST also showed ECOR37 to have distinct mdh and clpX alleles (ST-77) (Table 2) and so this strain does not appear to belong in the A2 clonal complex (ST-73). We can only speculate that ECOR37 is an ancestral strain of A1 and A2, but its position in the model is uncertain (Figure 3).
LSU-61 is an O157:H7 that is SOR+; GUD+; tellurite resistant; carried γ-eae, ehxA, the +93, but not the +776 uidA SNP; and had no stx. Except for the absence of stx2, LSU-61 had many of the traits proposed for the A3 intermediate, which has not yet been isolated. If LSU-61 is the A3 intermediate strain, we would expect it to be genetically related to the other clones because A3 is thought to have evolved from A2 and to be the intermediate for both A4 and A5 (Figure 3). However, the PFGE profile of LSU-61 showed only 60% similarity to stains in the A2, A4, A5, and A6 clonal complexes (data not shown), and it had a distinct fadD allele (ST-237) (Table 2). Despite these dissimilarities, the fact that LSU-61 has both O157 and H7 antigens and carries traits of neighboring clonal complexes (especially +93 uidA SNP and γ-eae) is compelling evidence that it belongs in the O157:H7 complex. The absence of the +776 uidA SNP in LSU-61 indicates that it is ancestral to A5; however, the exact position of LSU-61 on the evolutionary model remains to be determined (Figure 3).
In our study, we encountered various strains that had identical STs and traits as the clonal type strains, except for stx. Both stx1 and stx2 are phage encoded, and there is great diversity in stx-phage insertion sites among strains (24). Sometimes, these phages may be induced, resulting in strains that have lost the ability to produce Stx (25) or conversely, strains may acquire the ability to produce Stx by phage infection. This acquisition or loss of stx phages shows that this trait may not be a stable marker. Thus, the association of stx genotypes with clonal complexes should be interpreted with caution.
In summary, the use of unique trait markers and molecular typing methods better defined some of the clonal complexes postulated on the O157:H7 evolution model. The GUD+ O157:H7 and the SFO157 strains obtained worldwide had the unique mutation markers postulated for the emergence of the A5 and A4 clonal types and, therefore, belonged in these respective clonal complexes. Molecular subtyping showed genetic similarities and identities among strains within clonal complexes, but MLST identified genetic differences that further segregated these strains into subclones within a clonal complex.
Acknowledgments
Sequence typing was conducted by the STEC Center, part of the Food and Waterborne Diseases Integrated Research Network supported with federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health (NIH), Department of Health and Human Services, under NIH research contract no. N01-AI-30058.
Biography
Dr Feng is a research microbiologist with the Center for Food Safety and Applied Nutrition, FDA. His research interests are molecular characterization and evolutionary analysis of enterohemorrhagic E. coli O157:H7 and its atypical variants as well as detection of these pathogens in foods.
Footnotes
Suggested citation for this article: Feng PCH, Monday SR, Lacher DW, Allison L, Siitonen A, Keys C, et al. Genetic diversity among clonal lineages within Escherichia coli O157:H7 stepwise evolutionary model. Emerg Infect Dis [serial on the Internet]. 2007 Nov [date cited]. Available from http://www.cdc.gov/EID/content/13/11/1701.htm
References
- 1.Feng P, Lampel K, Karch H, Whittam T. Genotypic and phenotypic changes in the emergence of E. coli O157:H7. J Infect Dis. 1998;177:1750–3. 10.1086/517438 [DOI] [PubMed] [Google Scholar]
- 2.Whittam TS, Wolfe M, Wachsmuth I, Orskov F, Orskov I, Wilson R. Clonal relationships among Escherichia coli strains that cause hemorrhagic colitis and infantile diarrhea. Infect Immun. 1993;61:1619–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Monday SR, Whittam T, Feng P. Genetic and evolutionary analysis of mutations in the gusA gene that caused the absence of β-glucuronidase activity in Escherichia coli O157:H7. J Infect Dis. 2001;184:918–21. 10.1086/323154 [DOI] [PubMed] [Google Scholar]
- 4.Monday SR, Minnich S, Feng P. A 12 base-pair deletion in the flagellar master control gene flhC causes non-motility of the pathogenic German sorbitol-fermenting Escherichia coli O157:H- strains. J Bacteriol. 2004;186:2319–27. 10.1128/JB.186.8.2319-2327.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hayes PS, Blom K, Feng P, Lewis J, Strockbine N, Swaminathan B. Isolation and characterization of a β-D-glucuronidase-producing strain of Escherichia coli O157:H7 in the United States. J Clin Microbiol. 1995;33:3347–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karch H, Bielaszewska M. Sorbitol-fermenting Shiga toxin-producing Escherichia coli O157:H- strains: epidemiology, phenotypic and molecular characteristics and microbiological diagnosis. J Clin Microbiol. 2001;39:2043–9. 10.1128/JCM.39.6.2043-2049.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nagano H, Okui T, Fujiwara O, Uchiyama Y, Tamate N, Kumada H, et al. Clonal structure of Shiga toxin (Stx)-producing and β-D-glucuronidase-positive Escherichia coli O157:H7 strains isolated from outbreaks and sporadic cases in Hokkaido, Japan. J Med Microbiol. 2002;51:405–16. [DOI] [PubMed] [Google Scholar]
- 8.Bielaszewska M, Schmidt H, Liesgang A, Prager R, Rabach W, Tschape H, et al. Cattle can be a reservoir of sorbitol-fermenting shiga toxin–producing Escherichia coli O157:H- strains and a source of human disease. J Clin Microbiol. 2000;38:3470–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ammon A, Petersen LR, Karch H. A large outbreak of hemolytic uremic syndrome caused by an unusual sorbitol-fermenting strain of Escherichia coli O157:H-. J Infect Dis. 1999;179:1274–7. 10.1086/314715 [DOI] [PubMed] [Google Scholar]
- 10.Gerber A, Karch H, Allerberger F, Verweyen H, Zimmerhackl L. Clinical course and the role of shiga toxin-producing Escherichia coli infection in the hemolytic-uremic syndrome in pediatric patients, 1997–2000, in Germany and Austria: a prospective study. J Infect Dis. 2002;186:493–500. 10.1086/341940 [DOI] [PubMed] [Google Scholar]
- 11.Bielaszewska M, Schmidt H, Karmali M, Khakhria R, Janda J, Bláhová K, et al. Isolation and characterization of sorbitol-fermenting Shiga toxin (verocytotoxin)–producing Escherichia coli O157:H- strains in the Czech Republic. J Clin Microbiol. 1998;36:2135–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eklund M, Bielaszewska M, Nakari U, Karch H, Siitonen A. Molecular and phenotypic profiling of sorbitol-fermenting Escherichia coli O157:H- human isolates from Finland. Clin Microbiol Infect. 2006;12:634–41. 10.1111/j.1469-0691.2006.01478.x [DOI] [PubMed] [Google Scholar]
- 13.Saari M, Cherasty T, Leino K, Siitonen A. Phage types and genotypes of shiga toxin–producing Escherichia coli O157 in Finland. J Clin Microbiol. 2001;39:1140–3. 10.1128/JCM.39.3.1140-1143.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taylor P, Allison L, Willshaw G, Cheasty T, Hanson M. Sorbitol-fermenting Escherichia coli O157 in Scotland. 5th International Symposium on Shiga Toxin (Verotoxin)–producing E. coli Infections. Edinburgh, Scotland, 2003. Jun. Abstract no. P280. [Google Scholar]
- 15.Karch H, Bohm H, Schmidt H, Gunzer F, Aleksic S, Heesemann J. Clonal structure and pathogenicity of Shiga-like toxin-producing, sorbitol-fermenting Escherichia coli O157:H-. J Clin Microbiol. 1993;31:1200–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dunn JR, Keen JE, Moreland D, Thompson R. Prevalence of Escherichia coli O157:H7 in white-tailed deer from Louisiana. J Wildl Dis. 2004;40:361–8. [DOI] [PubMed] [Google Scholar]
- 17.Feng P, Monday S. Multiplex PCR for detection of trait and virulence factors in enterohemorrhagic Escherichia coli serotypes. Mol Cell Probes. 2000;14:333–7. 10.1006/mcpr.2000.0323 [DOI] [PubMed] [Google Scholar]
- 18.Bettelheim KA. Identification of enterohemorrhagic Escherichia coli by means of their production of enterohemolysin. J Appl Bacteriol. 1995;79:178–80. [DOI] [PubMed] [Google Scholar]
- 19.Centers for Disease Control and Prevention. Standardized molecular subtyping of foodborne bacterial pathogens by pulsed-field gel electrophoresis: CDC training manual. Atlanta: The Centers; 2003. [Google Scholar]
- 20.Qi W, Lacher D, Bumbaugh A, Hyma K, Ouellette L, Large T, et al. EcMLST: an online database for multi locus sequence typing of pathogenic Escherichia coli. Proceedings of the Institute of Electrical and Electronics Engineers (IEEE) Computational Systems Bioinformatics Conference, 2004. Washington: IEEE Computer Society; 2004. [Google Scholar]
- 21.Zhang WL, Kohler B, Oswald E, Beutin L, Karch H, Morabito S, et al. Genetic diversity of intimin genes of attaching effacing Escherichia coli strains. J Clin Microbiol. 2002;40:4486–92. 10.1128/JCM.40.12.4486-4492.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nagano H, Hirochi T, Fujita K, Wakamori Y, Takeshi K, Yano S. Phenotypic and genotypic characterization of β-D-glucuronidase–positive Shiga toxin–producing Escherichia coli O157:H7 isolated from deer. J Med Microbiol. 2004;53:1037–43. 10.1099/jmm.0.05381-0 [DOI] [PubMed] [Google Scholar]
- 23.McGraw EA, Li J, Selander R, Whittam T. Molecular evolution and mosaic structure of α, β, and γ intimins of pathogenic Escherichia coli. Mol Biol Evol. 1999;16:12–22. [DOI] [PubMed] [Google Scholar]
- 24.Karch H, Meyer T, Russmann H, Heesemann J. Frequent loss of Shiga-like toxin genes in clinical isolates of Escherichia coli upon subcultivation. Infect Immun. 1992;60:3464–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Besser TE, Shaikh N, Holt N, Tarr P, Konkel M, Malik-Kale P, et al. Greater diversity of shiga toxin-encoding bacteriophage insertion sites among Escherichia coli O157:H7 isolates from cattle than in those from humans. Appl Environ Microbiol. 2007;73:671–9. 10.1128/AEM.01035-06 [DOI] [PMC free article] [PubMed] [Google Scholar]