Abstract
Sulfoquinovosyldiacylglycerol (SQDG) lipids, found in plants and photosynthetic bacteria, can substitute for phospholipids under phosphate limiting conditions. Here, various low-energy ion activation strategies have been evaluated for the identification and characterization of deprotonated SQDG lipids from a crude membrane lipid extract of Rhodobacter sphaeroides, using collision- induced dissociation - tandem mass spectrometry (CID-MS/MS) in either a triple quadrupole mass spectrometer or in a hybrid quadrupole ion trap-multipole mass spectrometer coupled with high resolution / accurate mass analysis capabilities. In the triple quadrupole instrument, using energy resolved CID-MS/MS experiments, the SQDG head group specific product ion at m/z 225 (C6H9O7S−), rather than m/z 81 (SO3H−), was determined to provide the greatest sensitivity for SQDG lipid detection, and is therefore the preferred `fingerprint' ion for the identification of this lipid class from within complex lipid mixtures when using precursor ion scan mode MS/MS experiments. A comparison of conventional ion trap CID-MS/MS and -MSn, with `low Q' CID-MS/MS, pulsed Q dissociation (PQD)-MS/MS and higher energy collision induced dissociation (HCD)-MS/MS performed in an LTQ Orbitrap Velos mass spectrometer, revealed that HCD-MS/MS coupled with high resolution/accurate mass analysis represents the most sensitive, and perhaps most importantly the most specific strategy, for ion trap based identification and characterization of SQDG lipids, due to the ability to readily distinguish the SQDG head group specific product ion at m/z 225.0069 from other products that may be present at the same nominal m/z value. Finally, the mechanisms responsible for formation of each of the major product ions observed by low-energy CID-MS/MS of deprotonated SQDG lipids were elucidated using uniform H/D exchange, HCD-MS/MS and high resolution mass analysis. Formation of the m/z 225 `fingerprint' ion occurs via a charge-remote cis-elimination reaction, likely involving transfer of a hydrogen from the hydroxyl group located on the C2 position of the sugar ring.
Introduction
The non-phosphorous containing sulfoquinovosyldiacylglycerol (SQDG) lipids (Scheme 1) are preferentially, though not exclusively, found in the membranes of most photosynthetic bacteria, higher plants, mosses, ferns and algae [1–3]. Although SQDG lipids are not essential for anoxygenic photosynthesis, and no evidence has suggested that SQDG plays an essential role in oxygenic photosynthesis [1–5], they do demonstrate conditional importance under phosphate-limiting conditions [6–8], and become physiologically essential when phosphatidylglycerol (PG) lipids are depleted through genetic mutation [1–3]. SQDG lipids have also been found to inhibit DNA polymerases and display antitumor and antiviral effects, probably due to their anionic detergent-like structure [9,10].
Scheme 1.

General structure of sulfoquinovosyldiacylglyceride (SQDG) lipids under physiological pH.
Under normal phosphate growth conditions, SQDG lipids are relatively low in abundance in the membranes of Rhodobacter sphaeroides (R. sphaeroides), compared to phospholipids, but have been demonstrated to undergo a significant increase in abundance under phosphate-limiting anaerobic or aerobic growth conditions [4–7,11,12]. Despite the drastically altered lipid profiles observed under these conditions, the bacteria are still viable and exhibit unhampered expression of a fully active four-subunit membrane protein complex of the terminal enzyme of the respiratory electron transport chain, cytochrome c oxidase (CcO) [11]. Notably, a number of lipid molecules have previously been demonstrated to be co-purified with CcO, and highly conserved lipid binding sites have been identified crystallographically in the core subunits of CcO isolated from R. sphaeroides and bovine heart [13–17], suggesting an important functional role for lipids in CcO [18,19], as well as a requirement for lipids for achieving high-resolution CcO crystal structures. Importantly, abundant SQDG lipids have recently been found within the CcO enzymes purified from both the wild type R. sphaeroides as well as a cardiolipin-deficient mutant of R. sphaeroides (formed by disruption of the cardiolipin synthase (cls) gene), under normal and phosphate deficient growth conditions, and were identified for the first time in CcO crystals grown from the wild type and cardiolipin-deficient R. sphaeroides grown under normal phosphate conditions [11,12]. Depletion of cardiolipin and/or phosphate deficiency did not impact any aspect of CcO structure or behavior, which was rationalized as being due to a tolerance for quantitative substitution of cardiolipin or other phospholipids with non-phosphorous containing negatively charged lipids (such as SQDG) in this bacterial system.
Previous studies to identify and/or characterize SQDG lipids have involved the use of thin layer chromatography (TLC), combined with a series of functional group-selective reaction sprays, including a positive reaction with the sugar group-specific α-naphthol and negative results with phosphate and amino group-specific reactions [5,20]. Quantification of SQDG lipids has also been performed by TLC, in combination with metabolic radioactive labeling, or by gas chromatography-mass spectrometry (GC-MS) following plate scraping, re-extraction, saponification and chemical derivatization [5,11,21,22]. Alternative techniques capable of providing more detailed structural information on SQDG lipids have included (i) nuclear magnetic resonance (NMR, 1H and 13C) [7,10,23–25] (note however, that NMR typically requires a relatively large amount of rigorously purified lipid for analysis), and (iii) tandem mass spectrometry (MS/MS) methods, including fast atom bombardment (FAB)-sector high energy collision-induced dissociation (CID)-MS/MS [7,26–28], matrix-assisted laser desorption/ionization-time of flight (MALDI-TOF) post-source decay (PSD) [21], MALDI- ion trap TOF CID-MSn [29], electrospray ionization (ESI)- low-energy triple quadrupole CID-MS/MS in mass spectrometer [25,30–32], and ESI- low-energy ion trap CID-MSn [7], in the negative ionization mode. Using these MS/MS approaches, the presence of SQDG lipids have been determined by the observation of characteristic product ions at m/z 80 (SO3−) under high energy CID-MS/MS conditions [26,27], at m/z 225 under both high and low energy CID conditions [25–32], and at m/z 81 (SO3H−) under low energy CID conditions [25,30].
Unfortunately, as most lipid classes form precursor ions at m/z 600–1500, and their structural identification often relies on the observation of characteristic low m/z product ions formed in the tandem mass spectra [33,34], the activation q value (typically 0.25) associated with performing conventional CID in quadrupole ion trap mass spectrometers imposes a low-mass cutoff (LMCO) on the m/z range. This can result in an inability to detect product ions that fall below this LMCO, including the characteristic ions indicated above for SQDG lipids. To overcome this limitation, multistage CID-MSn (e.g., MS3 or MS4) may be employed, whereby product ions initially formed at higher m/z (i.e., above the LMCO) are subjected to further dissociation to yield the desired low mass products [29]. Alternatively, decreasing the activation q value of CID in the ion trap can decrease the LMCO for MS/MS, thereby allowing the observation of low m/z product ions [11]. However, this approach also places the precursor ions into a shallower trapping potential, potentially leading to decreased ion stability and undesired ion losses during ion activation. Pulsed Q collision induced dissociation (PQD) in the ion trap, which is achieved through initially applying a pulse at high q for precursor ion activation, followed by a rapid drop of the q value down to 0.05 to trap the fragment ions for detection [35], can also be used to extend the low mass limit to a much lower value during CID-MS/MS, thereby providing a more complete complement of product ions in a single spectrum, thereby providing information that is similar to that observed by performing CID-MSn in ion traps, or CID-MS/MS in triple quadrupole mass spectrometers. However, the sensitivity of PQD is potentially limited due to its typically lower fragmentation efficiency compared to conventional CID. Finally, the use of higher energy collisionally activated dissociation (HCD) in the multipole collision cell of a hybrid linear quadrupole ion trap - Orbitrap mass spectrometer allows the efficient capture of low m/z product ions, with the resultant MS/MS spectra being similar to those observed in a triple quadrupole instrument, but with the advantage of mass analysis being performed at high resolution and mass accuracy to facilitate unambiguous product ion assignment [36].
Given the physiological significance of SQDG lipids as noted above, there remains a continued need to develop effective modern tandem mass spectrometry approaches to examine and understand their fragmentation characteristics in order to accurately identify their molecular structures, and to achieve sensitive quantification of each molecular species using a minimum amount of sample, especially for the analysis of trace abundant lipids that remain bound in the purified CcO protein complex or in CcO protein crystals. Furthermore, although the possible channels for formation of the characteristic MS/MS product ions from SQDG lipids have previously been rationalized [25,26], the mechanisms associated with the formation of these ions have not been systematically investigated to date. The alternative mass spectrometry and ion activation methods described above have particular promise for lipid identification and characterization, as well as for understanding their associated fragmentation mechanisms. However, the formation and relative abundances of the various product ions that are observed using these strategies, and a demonstration of their applicability for sensitive and specific lipid identification, characterization or quantitative analysis, including determination of which product ion(s) provide the greatest sensitivity and specificity for use as characteristic `fingerprint' ions of SQDG lipids from within complex lipid extracts when using these techniques, have not previously been reported.
Materials and Methods
Materials
Cell membrane pellets from R. sphaeroides strains were obtained as described previously [11,12]. Methanol (HPLC grade) was purchased from Sigma-Aldrich (St. Louis, MO). Isopropanol was obtained from Jade Scientific (Canton, MI). Chloroform (HPLC grade) and ammonium acetate was from Mallinckrodt Baker (Phillipsburg, NJ). CD3OD (D, 99.8%) and CDCl3 (D, 99.8%) were purchased from Cambridge Isotope Laboratories (Andover, MA). Glass vials used for lipid extraction were purchased from either Fisher Scientific (Waltham, MA) or from SUN-Sri Chromatography Accessories (Rockwood, TN), and were washed prior to use.
Extraction of lipids from isolated membranes
To 10 μL of suspended membrane from R. sphaeroides cells (diluted with water to approximately 1.4 mg/mL protein concentration determined by BCA assay), 50 μL of 50% methanol was added followed by 100 μL of chloroform:methanol (2:1, v/v), with vigorous vortexing and centrifugation. The organic layer was removed then 100 μL of chloroform was added to the aqueous layer, followed by vortexing and centrifugation. The organic layers were combined, dried under a stream of Nitrogen and re-dissolved in 100 μL isopropanol:methanol:chloroform (4:2:1, v/v/v) containing 20mM ammonium acetate, immediately prior to use.
Triple quadruple mass spectrometer product ion scan, collision energy-resolved product ion scan and precursor ion scan mode CID-MS/MS analysis
Negative ionization mode ESI-MS, product ion scan mode CID-MS/MS, collision energy-resolved product ion scan mode CID-MS/MS and precursor ion scan mode CID-MS/MS analysis of total membrane lipid extracts was performed using a Thermo Scientific model TSQ Quantum Ultra triple quadrupole (QqQ) mass spectrometer (San Jose, CA). Total membrane lipid extracts were introduced to the mass spectrometer by direct infusion using an Advion BioSciences Triversa Nanomate nano-electrospray ionization (nESI) source (Ithaca, NY). nESI conditions were optimized to maximize the sensitivity and stability of the precursor ions of interest while minimizing `in-source' fragmentation. Product ion scan mode CID-MS/MS experiments were performed on monoisotopically isolated precursor ions using Q1 and Q3 peak widths set at 0.5 Da, Argon collision gas pressure of 1.5 mTorr, a laboratory collision energy of 50 eV (optimized in order to maximize the intensity of the most abundant product ion within the observable m/z range), and a scan rate of 250 m/z sec−1. Collision-energy-resolved product ion scan mode CID-MS/MS experiments were obtained by varying the laboratory collision energy from 20 to 70 eV using a step size of 5 eV. Precursor ion scan mode CID-MS/MS experiments to monitor for the characteristic m/z 225 `fingerprint' ion of SQDG lipids was performed using the same conditions as described above for the precursor ion scan MS/MS experiments. Each spectrum shown was the average of 50–100 scans.
High resolution ion trap ESI-MS, CID-MS/MS, -MSn and HCD-MS/MS analysis
High resolution negative ionization mode ESI-MS, CID-MS/MS and -MSn analyses were performed using a high resolution / accurate mass Thermo Scientific model LTQ Orbitrap Velos mass spectrometer equipped with a dual pressure ion trap and a HCD multipole collision cell (San Jose, CA). Samples were introduced to the mass spectrometer by direct infusion using an Advion BioSciences Triversa Nanomate nano-electrospray ionization (nESI) source (Ithaca, NY). nESI conditions were optimized to maximize the sensitivity and stability of the precursor ions of interest while minimizing `in-source' fragmentation. Orbitrap MS spectra were acquired with a mass analyzer resolution of 100,000, while all MS/MS and MSn spectra were acquired using the Orbitrap with a mass analyzer resolution of 7,500. External calibration of the instrument was performed using the standard LTQ calibration mixture. Conventional CID-MS/MS and -MSn experiments on monoisotopically isolated precursor ions were performed using an activation time of 10 ms and an activation q value of 0.25. `Low q' CID-MS/MS spectra were performed using an activation q value of 0.20. PQD-MS/MS and HCD-MS/MS spectra were both acquired using default activation times and a low mass limit of m/z 50. For each experiment, the normalized collision energies were optimized in order to maximize the intensity of the most abundant product ion within the observable m/z range. Automated Gain Control (AGC) target numbers were maintained at the same values (default settings) for all MS/MS experiments to allow a comparison of the relative sensitivity of the various ion activation methods for product ion formation. Each spectrum shown is the average of 100–200 scans. Theoretical m/z values were calculated using the tool found at: http://www.cem.msu.edu/~reusch/OrgPage/mass.htm.
Hydrogen/Deuterium exchange
H/D exchange of lipid extracts was achieved by drying the original lipid extracts under vacuum then redissolving in CDCl3:CD3OD (1:1, v/v) solvent. This process was repeated twice, then the samples were dissolved in CDCl3:CD3OD (1:1, v/v) containing 20 mM ammonium acetate immediately prior to mass spectrometry analysis. The H/D-exchanged lipid extracts were then infused into the LTQ Orbitrap Velos mass spectrometer using a conventional ESI source at a flow rate of 1 μL/min, under conditions optimized to minimize back exchange. Typical ESI conditions were: spray voltage 3.0 kV, heated capillary temperature 200 °C, S-lens 60%, sheath gas flow rate 10 arbitrary units, sweep gas flow rate 30 arbitrary units. Fully H/D exchanged precursor ions of the SQDG lipids were monoisotopically isolated then subjected to HCD-MS/MS using the optimized conditions as described above.
Results and Discussion
Utilization of product ion scan mode and energy resolved CID-MS/MS, and precursor ion scan mode CID-MS/MS in a triple quadrupole mass spectrometer to examine the fragmentation behavior and selectively identify SQDG lipids from a crude total lipid extract of R. sphaeroides
In order to elucidate the detailed mechanisms and product ion structures associated with the gas-phase dissociation reactions of SQDG lipids, as well as to demonstrate which product(s) provide the greatest sensitivity and specificity for use as characteristic `fingerprint' ions for their detection from within complex lipid extracts when using a triple quadrupole mass spectrometer, a crude lipid membrane extract from R. sphaeroides was subjected to analysis. Figure 1A shows the spectrum obtained by negative ion mode ESI-MS of the crude membrane lipid extract of R. sphaeroides obtained as described in the Methods section above. The identities of each of the ions in this spectrum have recently been reported in the literature [11,12]. CID-MS/MS of the abundant precursor ion at m/z 819.5 in Figure 1A resulted in the product ion spectrum shown in Figure 1B, while CID-MS/MS product ion spectra for the m/z 793.4, m/z 821.4 and m/z 845.5 and m/z 847.5 precursor ions are shown in Supplemental Figures S1A-D, respectively. The very similar fragmentation behavior of each of these precursor ions containing different fatty acyl chains (i.e., combinations of 16:0, 18:1 and 18:0) indicate that the fatty acid compositions have little influence on the observed fragmentation pathways.
Figure 1.
Negative ionization mode triple quadrupole ESI-mass spectrometry and tandem mass spectrometry analysis of a total membrane lipid extract from R. sphaeroides. (A) ESI-MS spectrum, (B) CID-MS/MS product ion spectra of m/z 819.5 (SQDG 18:1/16:0) acquired at a laboratory collision energy of 47 eV, and (C) energy resolved CID-MS/MS of m/z 819.5. The yield of the precursor and product ions at each collision energy value are shown as their observed ion counts. The inset to panel C shows an expanded region of the collision energy breakdown curve in order to observe the individual fragmentation channels at higher collision energies. (D) CID-MS/MS precursor ion scan (laboratory collision energy of 50 eV) to monitor for the characteristic m/z 225 ion in order to selectively identify all SQDG lipid species in the total membrane lipid extract from R. sphaeroides.
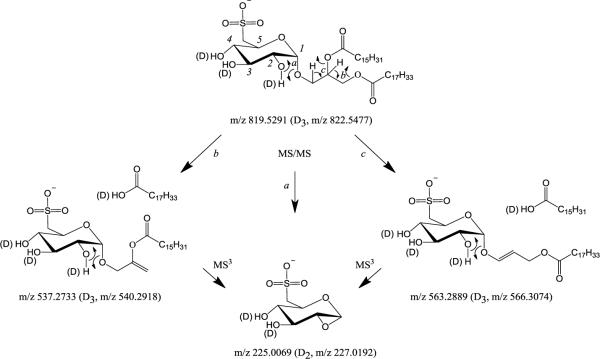
The most abundant product ion in Figure 1B is observed at m/z 225, corresponding to the dehydrated sulfoquinovosyl head group of an SQDG lipid (pathway a in Scheme 2). In addition, a series of other product ions that likely originate from the sulfoquinovosyl head group (m/z 207, 165, 153, 149, 125, 95, and 81) are also observed, albeit at much lower abundance than m/z 225. Formation of m/z 225 as the most abundant product upon performing low energy CID-MS/MS of SQDG lipids has previously been reported by others [25,30–32], and has been employed by Welti et al. in precursor ion scan MS/MS experiments as a characteristic `signature' to selectively identify the presence of SQDG lipids from within complex lipid extracts [31,32]. However, the presence of a product at a nominal m/z of 225 could also correspond to the deprotonated fatty acid anion of a 14:1 fatty acyl moiety. Furthermore, de Souza et al. previously reported a triple quadrupole product ion scan MS/MS experiment that yielded the product ion at m/z 81 as the most abundant species [30]. Note, however, that CID in the triple quadrupole can result in the formation of product ions from both primary and secondary fragmentation reactions, depending on the collision energy and gas pressure employed during the fragmentation. Thus, the differences between the results observed here and those reported previously may arise due to different CID conditions applied between the two instruments.
Scheme 2.
Proposed pathways for the multistage gas-phase dissociation reactions of the deprotonated SQDG 18:1/16:0 lipid from a total membrane lipid extract from R. sphaeroides.
The m/z 563.3 and 537.4 product ions in Figure 1B correspond to the neutral losses of 16:0 and 18:1 RCOOH fatty acid chains from the sn-1 or sn-2 positions of the glycerol backbone, respectively, while the product ions at m/z 255.3 and m/z 281.3 correspond to deprotonated 16:0 RCOO− 18:1 RCOO− fatty acid anions, respectively. Based on the results obtained from NMR, and data from regioselective enzymatic hydrolysis, Keusgen et al. have previously reported that the relative intensity of the fragment ion resulting from loss of the sn-1 fatty acyl moiety from deprotonated SQDG lipids upon low energy CID-MS/MS in a triple quadruple mass spectrometer is significantly higher than the relative intensity of the fragment ion resulting from loss of the sn-2 fatty acyl group, thereby allowing occupancy of the sn-1 and sn-2 positions of the glycerol backbone to be assigned based on the relative abundances of the respective fatty acyl chain neutral loss or anionic product ions [25]. This is in contrast to previous reports for the dissociation of glycerophospholipid anions, where loss of the sn-2 acyl group is favored, thereby yielding more abundant [M-H-R2COOH]− product ions [37]. Another study reported a collision energy dependence to the ratio of sn-1/sn-2 carboxylate anions from deprotonated glycerophospholipids, whereby the sn-2 carboxylate anion is observed as the dominant product at low collision energy and the sn-1 carboxylate anion is preferentially observed at higher energy [38]. Here, however, collision energy-resolved MS/MS (Figure 1C) revealed no significant change in sn-1/sn-2 product ion abundances as a function of collision energy. Thus, given the greater abundance of the 18:1 acyl chain neutral loss and anionic product ions observed here compared to the 16:0 acyl chain losses, the 18:1 and 16:0 fatty acyl chains within the m/z 819.5 precursor ion have been assigned to the sn-1 and sn-2 positions of the glycerol backbone, respectively (pathways b and c in Scheme 2). However, given that the abundances of the 537.3/563.4 and 281.3/255.3 product ions are quite similar, it is likely that the precursor ion at m/z 819.5 contains a mixture of SQDG 18:1/16:0 and SQDG 16:0/18:1 structural isomers. Note that structural information to allow characterization of the specific position of the double bond within the 18:1 fatty acyl chain is not obtained under the low energy CID-MS/MS conditions employed here.
To further understand the origin of the various acyl chain and head group specific product ions from the deprotonated SQDG lipids, and to evaluate which head group specific product ions (e.g., m/z 225 vs. m/z 81) provide the greatest sensitivity for detection of this lipid class from within a complex lipid extract using precursor ion scan mode MS/MS experiments on the triple quadruple mass spectrometer, collision-energy-resolved CID-MS/MS experiments were performed for the m/z 819.5 (SQDG 18:1/16:0) (Figure 1C and Supplemental Figure S2) and m/z 845.5 (SQDG 18:1/18:1) (Supplemental Figure S3) precursor ions. For both precursors, when the laboratory collision energy was raised above the threshold for fragmentation (25–30 eV), the abundance of the precursor ion gradually decreased, while that of first-generation product ions started to increase. For the m/z 819.5 precursor ion in Figure 1C, these included m/z 563 and m/z 537 (i.e., the product ions formed via neutral losses of the 18:1 and 16:0 acyl chains, and m/z 225 (i.e., the SQDG head group product ion). Above a laboratory collision energy of 45 eV, the abundance of the m/z 563 and 537 product ions started to decrease, whereas the abundance of m/z 225 kept increasing, suggesting that m/z 225 could also be generated via secondary fragmentation of the m/z 563 and 537 ions at higher collision energies, in addition to being generated directly from the precursor ion as a first-generation product. The abundance of the m/z 281 and 255 product ions (i.e., the 18:1 and 16:0 fatty acid anions) were also observed to increase above a laboratory collision energy of 45 eV, reaching a maximum abundance around 55 eV, indicating that these products could also be formed via secondary fragmentation of the m/z 563 and 537 product ions. Above a laboratory collision energy of 45–50 eV, m/z 225 decreased in abundance, while m/z 165, 95 and 81 started to increase. Notably, however, the absolute abundance of m/z 81 was not observed to exceed the maximum product ion abundance of m/z 225 at any of the collision energies examined, even though its relative abundance (expressed as a percentage of the total precursor and product ion abundance, see Supplemental Figure S2) reached a greater value than m/z 225 ion at the highest collision energy examined. Thus m/z 225, rather than m/z 81, would appear to be a more sensitive `fingerprint' ion for SQDG identification, albeit subject to a potential lack of selectively due to its potential for overlap with a 14:1 fatty acyl anion, if present.
Having confirmed that the product ion at m/z 225 is the most abundant fingerprint ion for SQDG lipid identification in the triple quadrupole instrument, it was then applied for the selective identification of SQDG lipids from within the total membrane lipid extract of R. sphaeroides, using a precursor ion scan mode CID-MS/MS experiment at the optimal laboratory collision energy of 50 eV as determined in Figure 1C (see Figure 1D). Compared to the ESI-MS spectrum, this scan resulted in higher sensitivity (via the enhancement in signal-to-noise provided through the use of MS/MS compared to MS), allowing the identification of numerous SQDG lipids in addition to those described in Figure 1 and Supplemental Figure S1, including many at low relative abundance. (e.g., m/z 805.6, m/z 817.5, m/z 833.6, m/z 835.6, m/z 837.6, m/z 861.5 and m/z 863.5). In contrast, a precursor ion scan mode CID-MS/MS experiment to monitor for the m/z 81 product ion, using an optimized laboratory collision energy of 70 eV as determined in Figure 1C, resulted in significantly lower signal-to-noise that limited the observation of these low abundance species (data not shown).
High resolution CID-MS/MS and -MSn fragmentation of SQDG lipids in a hybrid quadrupole linear ion trap mass spectrometer
Further insights into the identities, fragmentation lineage and relative detection sensitivity and specificity of SQDG lipids was then afforded by the use of high resolution ESI-MS, conventional ion trap CID-MS/MS and MSn, `low Q' CID-MS/MS, pulsed Q dissociation (PQD)-MS/MS and HCD-MS/MS performed in an LTQ Orbitrap Velos mass spectrometer. High resolution negative ion mode ESI-MS of the same lipid extract examined using the triple quadrupole mass spectrometer (Figure 1A) resulted in the spectrum shown in Figure 2A. As expected, the spectra were very similar, albeit with the accurate mass capabilities of this instrument providing valuable information regarding the elemental compositions of the various ions that were observed. Conventional negative ion mode CID-MS/MS (i.e., using an activation q value of 0.25) on the m/z 819.5 precursor ion in Figure 2A, acquired at high resolution in the Orbitrap, yielded the spectrum shown in Figure 2B. In contrast to the triple quadrupole CIDMS/MS data, the low mass cut-off associated with the conventional ion trap CID-MS/MS experiment precluded observation of the m/z 225 product ion. Instead, product ions corresponding to neutral losses of the 18:1 (m/z 537.2725) and 16:0 (m/z 563.2879) fatty acids were observed as the dominant species. Notably, the accurate mass information provided by this instrument could be used to unambiguously confirm the identities of these neutral losses. The significantly lower abundance of the m/z 255.2323 (16:0 RCOO−, m/z 255.2325) and m/z 281.2479 (18:1 RCOO−, m/z 281.2482) product ions in the CID-MS/MS spectrum is consistent with the energy resolved triple quadrupole data in Figure 1C that indicated these ions as being formed via secondary fragmentation of the initial neutral loss products, or are formed as competitive products only at higher collision energies. CID-MS3 of the m/z 563.2879 and m/z 537.2725 product ions resulted in the spectra shown in Figure 2C and Figure 2D, respectively. Both ions fragmented to form m/z 225.0067 as the dominant product ion, whose mass is consistent with a composition of C6H9O7S− (calculated m/z of 225.0069) corresponding to the SQDG head group, and ruling out the possibility of it being a 14:1 fatty acid RCOO− anion (C14H25O2−, m/z 225.1856). CID-MS4 of m/z 225 ion from these spectra (the spectrum obtained by CID-MS4 of the m/z 225.0067 ion in Figure 2C is shown in Figure 2E), resulted in the formation of products at m/z 206.9962 (C6H7O6S−, m/z 206.9963), 164.9855 (C4H5O5S−, m/z 164.9858), 152.9855 (C3H5O5S−, m/z 152.9858), 148.9905 (C4H5O4S−, m/z 148.9909), 125.0236 (C6H5O3−, m/z 125.0239), 94.9808 (CH3O3S−, m/z 94.9803) and 80.9644 (SO3H−, m/z 80.9646), that could be used to further confirm its identify. Notably, the order of ion generation in these CID-MSn spectra acquired using the quadrupole linear ion trap is very consistent with the ion hierarchy obtained from the product ion scan mode and collision-energy-resolved CID-MS/MS experiments performed using the triple quadrupole shown earlier. Furthermore, the product ion at m/z 225 is again observed as the more abundant product, compared to m/z 81, confirming this product as the preferred `fingerprint ion' for SQDG lipid identification.
Figure 2.
High resolution negative ionization mode ESI-ion trap mass spectrometry and multistage tandem mass spectrometry analysis of a total membrane lipid extract from R. sphaeroides. (A) ESI-MS spectrum, (B) CID-MS/MS product ion spectra of m/z 819.5286 (SQDG 18:1/16:0), (C) CID-MS3 product ion spectra of m/z 563.2879 from panel B, (D) CID-MS3 product ion spectra of m/z 537.2725 from panel B, and (E) CID-MS4 product ion spectra of m/z 225.0067 from panel C.
Next, we examined the potential utility of alternate methods for ion activation (namely `low Q' ion trap CID-MS/MS (Figure 3A), PQD ion trap CID-MS/MS (Figure 3B) and multipole HCD-MS/MS (Figure 3C)), as a means to improve the sensitivity for detection of the characteristic SQDG head group specific C6H9O7S− product ion at m/z 225.0067, compared to the use of conventional ion trap CID-MS/MS and -MS3. In each experiment, the normalized collision energy used was optimized in order to observe the m/z 225.0067 product at the greatest abundance. In Figure 3A, the use of a lower activation `q' value of 0.2 resulted in a spectrum similar to that observed by conventional CID-MS/MS, but with the LMCO achieved under these conditions (m/z 180) allowing observation of the characteristic m/z 225 product ion at an abundance of 3.23E4. Notably, this was approximately 2.5 times the abundance of the m/z 225 product ion observed by using the conventional CID-MS3 experiment, as seen in Figures 2C and 2D. In contrast, the use of PQD-MS/MS and HCD-MS/MS (Figures 3B and 3C, respectively) yielded spectra that were more similar to that observed using the triple quadrupole mass spectrometer, with m/z 225 being observed as the most abundant product ion (abundances of 4.71E4 and 2.06E5, respectively) in each case. In addition to the ability of HCD-MS/MS to yield the m/z 225 product ion with significantly greatest sensitivity compared to the other methods, the observation of other product ions formed via neutral losses of the 18:1 and 16:0 acyl chains (m/z 537.2737 and 563.2897), and their corresponding fatty acid anions (m/z 281.2485 and 255.2330), as well as a variety of abundant low m/z product ions formed via further dissociation of the m/z 225 product ion, enabled the identity of the SQDG 18:1/16:0 lipid to be readily assigned. Thus, from these experiments, we conclude that HCD-MS/MS coupled with the use of high resolution/accurate mass analysis (that allows the characteristic m/z 225.0069 product ion to be identified and differentiated from other product ions with the same nominal m/z value), represents the most sensitive, and perhaps most importantly, a specific strategy, for the identification and characterization of SQDG lipids from within complex mixtures.
Figure 3.
High resolution negative ionization mode ESI-tandem mass spectrometry analysis of m/z 819.5 from a total membrane lipid extract from R. sphaeroides using alternative ion activation methods. (A) `Low Q' ion trap CID-MS/MS (activation q = 0.2), (B) PQD ion trap CID-MS/MS, and (C) HCD multipole CID-MS/MS. The insets to panels B and C show expanded regions of the low m/z range in each spectrum.
Elucidation of the gas-phase fragmentation mechanisms of SQDG lipids
In order to elucidate the mechanisms associated with the fragmentation pathways observed above, high resolution HCD-MS/MS experiments were performed after subjecting the crude lipid membrane extract from R. sphaeroides to uniform H/D exchange in solution, prior to introduction to the mass spectrometer by ESI. SQDG lipids contain three H/D-exchangeable OH groups in their sugar ring; thus H/D exchange experiments can be used to distinguish between fragmentation reactions involving hydroxyl OH protons and C–H protons. After incubating the total membrane lipid extract with CD3OD (99.9%), over 90% of the total precursor ions were fully exchanged by the addition of 3 deuteriums, and displayed a 3 Da shift upon ESI-MS, as expected (data not shown). The spectrum obtained by HCD-MS/MS of the 822.5 precursor ion, formed by H/D exchange of the 819.5 precursor ion of the SQDG 18:1/16:0 lipid, is shown in Figure 4A, while Figures 4B–D show expanded regions of the product ion spectrum in order to more readily observe the various product ions that were formed, and their deuterium content. The numbers indicated in parenthesis for each product ion indicate the number of deuterium atoms that are present in each product ion.
Figure 4.
High resolution negative ionization mode ESI HCD-MS/MS analysis of m/z 822.5 from a total membrane lipid extract from R. sphaeroides following in-solution H/D exchange using CD3OD. Panel (A) shows the full m/z range, while panels (B, C and D) show expanded regions of the spectrum from panel A. The numbers indicated in parenthesis for each product ion indicate the number of deuterium atoms that are present in each case.
In the spectrum shown in Figure 4, the RCOOH neutral loss product ions originally observed at m/z 537.2737 and m/z 563.2897 in Figure 3C each display a 3 Da mass shift to m/z 540.2911 and m/z 566.3063, respectively, indicating that no deuterium atoms were incorporated within the neutral fatty acid species that were lost. Based on this, loss of the neutral RCOOH groups likely occur by deprotonation of the sn-2 C–H proton for loss of the R1COOH acyl chain, and deprotonation of one of the four sn-1 and sn-3 glycerol backbone CH-protons for loss of the R2COOH acyl chain, via the charge-remote cis-elimination processes shown in Scheme 2, pathways a and b. Formation of the deprotonated fatty acid anions at m/z 255.2322 and m/z 281.2477 could occur either by an intermolecular proton transfer reaction prior to separation of the ion-molecule complex of the m/z 540.2911 or m/z 566.3063 product ions and their corresponding neutral fatty acids that were initially formed in Scheme 2, or by a charge-directed intramolecular proton transfer from the sn-1, sn-2 or sn-3 positions of the glycerol backbone to the negative charge of the sulfite moiety, resulting in direct anion formation (pathway not shown).
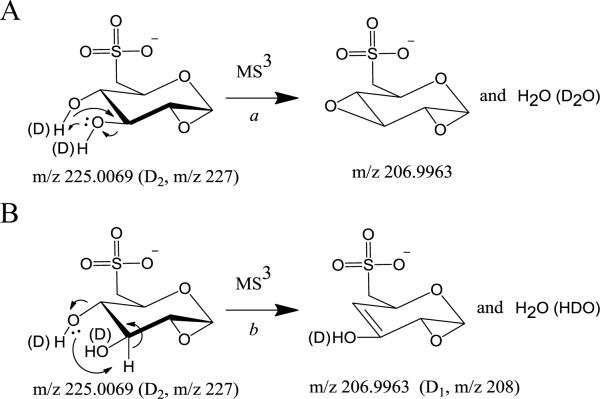
The head group specific product ion originally observed at m/z 225.0071 in Figure 3C was observed at m/z 227.0188, indicating the incorporation of two deuteriums within this product ion. A likely mechanism for this process is shown in Scheme 2, pathway c, involving a charge-remote cis-elimination reaction with transfer of the deuteron from the hydroxyl group located on the C2 position of the sugar ring. A comparison between the secondary product ions formed from the m/z 225 ion (Figure 3C) with those from the H/D exchanged version at m/z 227 (Figure 4) provides interesting clues on how several of these ions are formed. m/z 80.9642 (HSO3−) in Figure 4 is proposed to form from m/z 227.0188 through a charge-directed 5-member ring proton transfer process as shown in Scheme 3A, involving the negative charge on the sulfite moiety and the C5-H of the sugar ring. The lower abundance ion at m/z 81.9708 (DSO3−) in Figure 4 is likely formed via intermolecular H/D transfer prior to separation of the ion-molecule complex of the HSO3− product ion and its corresponding neutral that is initially formed from fragmentation of the m/z 227.0188 ion (Scheme 3A). Formation of m/z 94.9799 (containing no deuterium) from m/z 227.0188 is also likely to occur through a charge-directed process by attacking the C4-H of the sugar ring, as shown in Scheme 3B, while the m/z 95.9861 product ion containing one deuterium is likely formed by either a more favorable intermolecular H/D transfer prior to separation of the ion-molecule complex of the CH3SO3− product ion and its corresponding neutral, or by initial intramolecular nucleophilic attack from the negative charge of the sulfite moiety at the C3-hydroxyl group of the sugar ring, resulting in transfer of a deuteron and subsequent bond cleavage. Based on the observation of products at m/z 206.9959, m/z 208.0021 and 209.0085 in Figure 4, the ion at m/z 206.9965 in Figure 3C corresponding to the loss of H2O from the m/z 225 ion, seems to be able to occur via several mechanisms; the most likely involving the charge-remote pathways shown in Scheme 4, because both D2O (Scheme 4A) and HDO (Scheme 4B) losses are observed. Notably, and consistent with the result that the fragmentations of SQDG require much higher collision energies than typical non-glyco phospholipids, the major fragmentation pathways of deprotonated SQDG lipids by CID-MS/MS are all charge-remote.
Scheme 3.
Proposed pathways for the formation of m/z 81 (A) and m/z 95 (B) from m/z 225 (D2, m/z 227).
Scheme 4.
Proposed pathways for the formation of m/z 207 (A) and m/z 208 (B) from m/z 225 (D2, m/z 227).
Conclusions
The results from this study demonstrate the utility of advanced tandem mass spectrometry based ion activation strategies, particularly when coupled with high resolution MS/MS analysis, as well as associated mechanistic studies, to obtain insights into the gas-phase fragmentation reactions and facilitate the selective identification of individual species such as SQDG lipids, from within complex lipid extracts. The results from this work are expected to facilitate future efforts directed toward the development and application of optimized strategies for the sensitive identification, characterization and quantitative analysis of trace abundant lipids within complex membrane lipid extracts, or from within purified protein complexes or protein crystals.
Supplementary Material
Acknowledgements
This article is dedicated to Alex Harrison on the occasion of his 80th birthday, and in recognition of his seminal contributions to our understanding of biomolecular gas-phase ion chemistry, structure and fragmentation. This work was supported by NIH GM26916 (S.F.-M.) and a MSU Research Excellence Fund grant (S.F.-M., G.E.R.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Sato N. Roles of the acidic lipids sulfoquinovosyl diacylglycerol and phosphatidylglycerol in photosynthesis: their specificity and evolution. J. Plant. Res. 2004;117:495–505. doi: 10.1007/s10265-004-0183-1. [DOI] [PubMed] [Google Scholar]
- 2.Benning C. Biosynthesis and function of the sulfolipid sulfoquinovosyl diacylglycerol. Ann. Rev. Plant. Physiol. Plant. Mol. Biol. 1998;49:53–75. doi: 10.1146/annurev.arplant.49.1.53. [DOI] [PubMed] [Google Scholar]
- 3.Benning C. Questions remaining in sulfolipid biosynthesis: A historical perspective. Photosynth Res. 2007;92:199–203. doi: 10.1007/s11120-007-9144-6. [DOI] [PubMed] [Google Scholar]
- 4.Gueler S, Seeliger A, Haertel H, Renger G, Benning C. A null mutant of Synechococcus sp. PCC7942 deficient in the sulfolipid sulfoquinovosyl diacylglycerol. J. Biol. Chem. 1996;271:7501–7507. doi: 10.1074/jbc.271.13.7501. [DOI] [PubMed] [Google Scholar]
- 5.Benning C, Somerville CR. Isolation and genetic complementation of a sulfolipid-deficient mutant of Rhodobacter sphaeroides. J. Bacteriol. 1992;174:2352–2360. doi: 10.1128/jb.174.7.2352-2360.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benning C, Beatty JT, Prince RC, Somerville CR. The sulfolipid sulfoquinovosyldiacylglycerol is not required for photosynthetic electron transport in Rhodobacter sphaeroides but enhances growth under phosphate limitation. Proc. Natl. Acad. Sci. U.S.A. 1993;90:1561–1565. doi: 10.1073/pnas.90.4.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Benning C, Huang Z-H, Gage DA. Accumulation of a novel glycolipid and a betaine lipid in cells of Rhodobacter sphaeroides grown under phosphate limitation. Arch. Biochem. Biophys. 1995;317:103–111. doi: 10.1006/abbi.1995.1141. [DOI] [PubMed] [Google Scholar]
- 8.van Mooy BAS, Fredricks HF, Evans CT, Devol AH. Sulfolipids dramatically decrease phosphorus demand by picocyanobacteria in oligotrophic marine environments. Proc. Natl. Acad. Sci. U.S.A. 2006;103:8607–8612. doi: 10.1073/pnas.0600540103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ohta K, Mizushina Y, Hirata N, Takemura M, Sugawara F, Matsukage A, Yoshida S, Sakaguchi K. Sulfoquinovosyldiacylglycerol, KM043, a new potent inhibitor of eukaryotic DNA polymerases and HIV-reverse transcriptase type 1 from a marine red alga, Gigartina tenella. Chem. Pham. Bull. 1998;46:684–686. doi: 10.1248/cpb.46.684. [DOI] [PubMed] [Google Scholar]
- 10.Hanashima S, Mizushina Y, Yamazaki T, Ohta K, Takahashi S, Sahara H, Sakaguchi K, Sugawara F. Synthesis of sulfoquinovosylacylglycerols, inhibitors of Eukaryotic DNA Polymerase a and b. Bioorg. Med. Chem. 2001;9:367–376. doi: 10.1016/s0968-0896(00)00252-2. [DOI] [PubMed] [Google Scholar]
- 11.Zhang X, Tamot B, Hiser C, Reid GE, Benning C, Ferguson-Miller SM. Cardiolipin-Deficiency in Rhodobacter sphaeroides Alters the Lipid Profile of Membranes and of Crystallized Cytochrome Oxidase, but Structure and Function are Maintained. Biochemistry. 2011;50:3879–3890. doi: 10.1021/bi101702c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang X, Hiser C, Tamot B, Benning C, Reid GE, Ferguson-Miller SM. Combined Genetic and Metabolic Manipulation of Lipids in Rhodobacter sphaeroides Reveals Nonphospholipid Substitutions in Fully Active Cytochrome c Oxidase. Biochemistry. 2011;50:3891–3902. doi: 10.1021/bi1017039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qin L, Sharpe MA, Garavito RM, Ferguson-Miller S. Conserved lipid-binding sites in membrane proteins: a focus on cytochrome c oxidase. Curr. Opin. Struct. Biol. 2007;17:444–450. doi: 10.1016/j.sbi.2007.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Awasthi YC, Chuang TF, Keenan TW, Crane FL. Tightly bound cardiolipin in cytochrome oxidase. Biochim Biophys Acta. 1971;226:42–52. doi: 10.1016/0005-2728(71)90176-9. [DOI] [PubMed] [Google Scholar]
- 15.Shinzawa-Itoh K, Aoyama H, Muramoto K, Terada H, Kurauchi T, Tadehara Y, Yamasaki A, Sugimura T, Kurono S, Tsujimoto K, Mizushima T, Yamashita E, Tsukihara T, Yoshikawa S. Structures and physiological roles of 13 integral lipids of bovine heart cytochrome c oxidase. EMBO J. 2007;26:1713–1725. doi: 10.1038/sj.emboj.7601618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qin L, Hiser C, Mulichak A, Garavito RM, Ferguson-Miller S. Identification of conserved lipid/detergent-binding sites in a high-resolution structure of the membrane protein cytochrome c oxidase. Proc. Natl. Acad. Sci. U.S.A. 2006;103:16117–16122. doi: 10.1073/pnas.0606149103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Distler AM, Allison J, Hiser C, Qin L, Hilmi Y, Ferguson-Miller SM. Mass spectrometric detection of protein, lipid and heme components of cytochrome c oxidase from R. sphaeroides and the stabilization of non-covalent complexes from the enzyme. Eur. Mass. Spectrom. 2004;10:295–308. doi: 10.1255/ejms.594. [DOI] [PubMed] [Google Scholar]
- 18.Yamaoka-Koseki S, Urade R, Kito M. Cardiolipins from rats fed different dietary lipids affect bovine heart cytochrome c oxidase activity. J. Nutr. 1991;121:956–958. doi: 10.1093/jn/121.7.956. [DOI] [PubMed] [Google Scholar]
- 19.Zhang M, Mileykovskaya E, Dowhan W. Cardiolipin is essential for organization of complexes III and IV into a supercomplex in intact yeast mitochondria. J. Biol. Chem. 2005;280:29403–29408. doi: 10.1074/jbc.M504955200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vieler A, Wilhelm C, Goss R, Suß R, Schiller J. The lipid composition of the unicellular green alga Chlamydomonas reinhardtii and the diatom Cyclotella meneghiniana investigated by MALDI-TOF MS and TLC. Chem. Phys. Lipids. 2007;150:143–155. doi: 10.1016/j.chemphyslip.2007.06.224. [DOI] [PubMed] [Google Scholar]
- 21.Rossak M, Schafer A, Xu N, Gage DA, Benning C. Accumulation of sulfoquinovosyl-1-o-dihydroxyacetone in a sulfolipid-deficient mutant of rhodobacter sphaeroides inactivated in sqdC. Arch. Biochem. Biophys. 1997;340:219–230. doi: 10.1006/abbi.1997.9931. [DOI] [PubMed] [Google Scholar]
- 22.Gorchein A. The separation and identification of the lipids of rhodopseudomonas spheroides. Proc. Royal. Soc., Ser. B. 1968;170:279–297. doi: 10.1098/rspb.1968.0039. [DOI] [PubMed] [Google Scholar]
- 23.Cedergren RA, Hollingsworth RI. Occurrence of sulfoquinovosyl diacylglycerol in some members of the family rhizobiaceae. J. Lipid Res. 1994;35:1452–1461. [PubMed] [Google Scholar]
- 24.Golik J, Dickey JK, Todderud G, Lee D, Alford J, Huang S, Klohr S, Eustice D, Aruffo A, Agler ML. Isolation and Structure Determination of Sulfonoquinovosyl Dipalmitoyl Glyceride, a P-Selectin Receptor Inhibitor from the Alga Dictyochloris fragrans. J. Nat. Prod. 1997;60:387–389. doi: 10.1021/np9606761. [DOI] [PubMed] [Google Scholar]
- 25.Keusgen M, Curtis JM, Thibault P, Walter JA, Windust A, Ayer SW. Sulfoquinovosyl diacylglycerols from the alga heterosigma carterae. Lipids. 1997;32:1101–1112. doi: 10.1007/s11745-997-0142-9. [DOI] [PubMed] [Google Scholar]
- 26.Gage DA, Huang Z-H, Benning C. Comparison of sulfoquinovosyl diacylglycerol from spinach and the purple bacterium Rhodobacter sphaeroides by fast atom bombardment tandem mass spectrometry. Lipids. 1992;27:632–636. doi: 10.1007/BF02536123. [DOI] [PubMed] [Google Scholar]
- 27.Sprott GD, Larocque S, Cadotte N, Dicaire CJ, McGee M, Brisson JR. Novel polar lipids of halophilic eubacterium Planococcus H8 and archaeon Haloferax volcanii. Biochim. Biophys. Acta. Mol. Cell. Biol. Lipids. 2003;1633:179–188. doi: 10.1016/j.bbalip.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 28.Kim YH, Yoo JS, Kim MS. Structural characterization of sulfoquinovosyl, monogalactosyl and digalactosyl diacylglycerols by FAB-CID-MS/MS. J. Mass Spectrom. 1997;32:968–977. [Google Scholar]
- 29.Naumann I, Darsow KH, Walter C, Lange HA, Buchholz R. Identification of sulfoglycolipids from the alga Porphyridium purpureum by matrix-assisted laser desorption/ionisation quadrupole ion trap time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 2007;21:3185–3192. doi: 10.1002/rcm.3190. [DOI] [PubMed] [Google Scholar]
- 30.de Souza LM, Iacomini M, Gorin PAJ, Sari RS, Haddad MA, Sassaki GL. Glyco- and sphingophosphonolipids from the medusa Phyllorhiza punctata: NMR and ESI-MS/MS fingerprints. Chem. Phys. Lipids. 2007;145:85–96. doi: 10.1016/j.chemphyslip.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 31.Welti R, Wang X, Williams TD. Electrospray ionization tandem mass spectrometry scan modes for plant chloroplast lipids. Anal. Biochem. 2003;314:149–152. doi: 10.1016/s0003-2697(02)00623-1. [DOI] [PubMed] [Google Scholar]
- 32.Welti R, Wang X. Lipid species profiling: a high-throughput approach to identify lipid compositional changes and determine the function of genes involved in lipid metabolism and signaling. Curr. Opin. Plant Biol. 2004;7:337–344. doi: 10.1016/j.pbi.2004.03.011. [DOI] [PubMed] [Google Scholar]
- 33.Pulfer M, Murphy RC. Electrospray mass spectrometry of phospholipids. Mass Spectrom. Rev. 2003;22:332–364. doi: 10.1002/mas.10061. [DOI] [PubMed] [Google Scholar]
- 34.Han X, Yang K, Gross RW. Multi-dimensional mass spectrometry-based shotgun lipidomics and novel strategies for lipidomic analyses. Mass Spectrom Rev. 2011 doi: 10.1002/mas.20342. DOI: 10.1002/mas.20342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.ThermoScientific Pulsed q collision induced dissociation (pqd) on linear ion trap mass spectrometers. Thermo Scientific Product Support Bulletin. 2006;124:62577–62578. [Google Scholar]
- 36.Schuhmann K, Herzog R, Schwudke D, Metelmann-Strupat W, Bornstein SR, Shevchenko A. Bottom-Up Shotgun Lipidomics by Higher Energy Collisional Dissociation on LTQ Orbitrap Mass Spectrometers. Anal. Chem. 2011;83:5480–5487. doi: 10.1021/ac102505f. [DOI] [PubMed] [Google Scholar]
- 37.Huang Z-H, Gage DA, Sweeley CC. Characterization of diacylglycerylphosphocholine molecular species by FAB-CAD-MS/MS: A general method not sensitive to the nature of the fatty acyl groups. J. Am. Soc. Mass Spectrom. 1992;3:71–78. doi: 10.1016/1044-0305(92)85020-K. [DOI] [PubMed] [Google Scholar]
- 38.Hvattum E, Hagelin G, Larsen A. Study of mechanisms involved in the collision-induced dissociation of carboxylate anions from glycerophospholipids using negative ion electrospray tandem quadrupole mass spectrometry. Rapid Commun. Mass Spectrom. 1998;12:1405–1409. doi: 10.1002/(SICI)1097-0231(19981015)12:19<1405::AID-RCM338>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.