Abstract
Isoflavonoids are a class of phenylpropanoids made by legumes, and consumption of dietary isoflavonoids confers benefits to human health. Our aim is to understand the regulation of isoflavonoid biosynthesis. Many studies have shown the importance of transcription factors in regulating the transcription of one or more genes encoding enzymes in phenylpropanoid metabolism. In this study, we coupled bioinformatics and coexpression analysis to identify candidate genes encoding transcription factors involved in regulating isoflavonoid biosynthesis in Lotus (Lotus japonicus). Genes encoding proteins belonging to 39 of the main transcription factor families were examined by microarray analysis of RNA from leaf tissue that had been elicited with glutathione. Phylogenetic analyses of each transcription factor family were used to identify subgroups of proteins that were specific to L. japonicus or closely related to known regulators of the phenylpropanoid pathway in other species. R2R3MYB subgroup 2 genes showed increased expression after treatment with glutathione. One member of this subgroup, LjMYB14, was constitutively overexpressed in L. japonicus and induced the expression of at least 12 genes that encoded enzymes in the general phenylpropanoid and isoflavonoid pathways. A distinct set of six R2R3MYB subgroup 2-like genes was identified. We suggest that these subgroup 2 sister group proteins and those belonging to the main subgroup 2 have roles in inducing isoflavonoid biosynthesis. The induction of isoflavonoid production in L. japonicus also involves the coordinated down-regulation of competing biosynthetic pathways by changing the expression of other transcription factors.
The phenylpropanoid pathway in higher plants produces a range of phenolic metabolites derived from the aromatic amino acids Phe and Tyr. These metabolites protect plants during biotic and abiotic challenges such as pathogen attack or exposure to UV light (Landry et al., 1995; Jin et al., 2000; Kliebenstein et al., 2002; Winkel-Shirley, 2002). They also serve as signaling molecules, such as salicylic acid and nodulation factors (Subramanian et al., 2007).
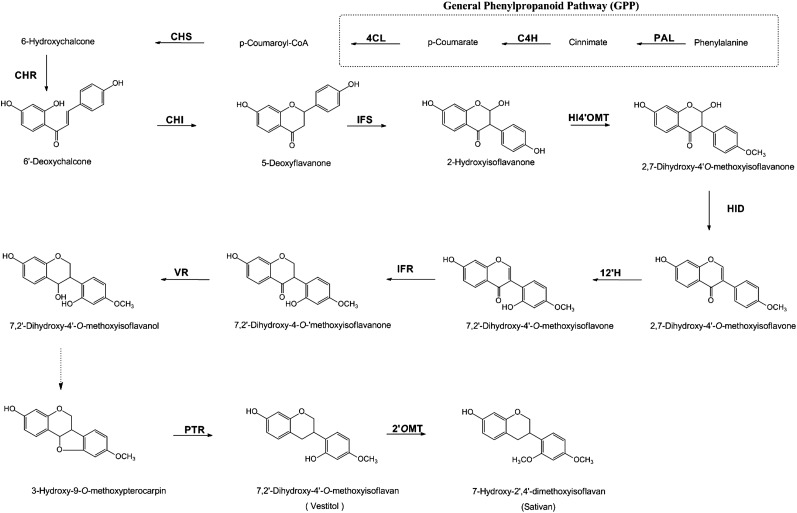
The first three enzymatic steps in phenylpropanoid biosynthesis, catalyzed by phenylalanine ammonia lyase (PAL), cinnamate 4-hydroxylase (C4H), and p-coumaroyl coenzyme A ligase (4CL; Vogt, 2010), are usually referred to as the general phenylpropanoid pathway (GPP), and their activities provide precursors for the synthesis of most phenylpropanoids. Their product, p-coumaroyl-CoA, can either enter the flavonoid pathway or be used for the synthesis of hydroxycinnamic acids and monolignols. The first committed step in flavonoid metabolism is the condensation of three molecules of malonyl-CoA and one molecule of 4-courmaroyl-CoA, catalyzed by chalcone synthase (CHS) to produce chalcones that can be elaborated into aurones, flavones, flavonols, isoflavonoids, phlobaphenes, 3-deoxyanthocyanidins, tannins, and anthocyanins. In legume species, chalcones and flavanones provide precursors for isoflavonoid biosynthesis (Fig. 1).
Figure 1.
Enzymatic steps in the isoflavonoid pathway. C4H, Cinnamate 4-hydroxylase; CHI, chalcone isomerase; CHR, chalcone reductase; CHS, chalcone synthase; 4CL, p-coumaroyl CoA-ligase; HID, 2-hydroxyisoflavanone dehydratase; HI4′OMT, S-adenosyl-l-Met:2,7,4′-trihydroxyisoflavanone 4′-O-methyltransferase; IFR, isoflavone reductase; IFS, isoflavone synthase; I2′H, isoflavone 2′-hydroxylase; 2′OMT, 2′-O-methyltransferase; PAL, Phe ammonia lyase; PTR, pterocarpan reductase; VR, vestitone reductase.
R2R3MYB transcription factors (TFs) regulate the activity of some branches of phenylpropanoid metabolism. This plant-specific TF family is defined by a common DNA-binding domain of two repeats of about 50 amino acids. Examination of R2R3MYB TFs by phylogenetic analysis has revealed functionally distinct subgroups (Stracke et al., 2001; Jiang et al., 2004; Dubos et al., 2010), of which several are involved in the regulation of particular branches of phenylpropanoid metabolism; for example, anthocyanin production (Paz-Ares et al., 1987; Quattrocchio et al., 1998; Schwinn et al., 2006), phlobaphene biosynthesis (Grotewold et al., 1994), flavonol biosynthesis (Mehrtens et al., 2005), hydroxycinnamic acid biosynthesis (Tamagnone et al., 1998; Jin et al., 2000), and monolignol biosynthesis (Zhou et al., 2009; Zhong et al., 2010). In legumes, there is an extra dimension to the regulatory control of phenylpropanoid metabolism because they produce isoflavonoids that serve as phytoalexins and as signaling molecules for nodulation (Subramanian et al., 2007).
Isoflavonoids in the diet have been linked to anticancer and antiaging health benefits that are associated with their phytoestrogenic and antioxidant properties (Dixon and Steele, 1999). As a consequence, there is interest in understanding how isoflavonoid metabolism can be engineered in tissues where high levels might be beneficial. One approach to engineering synthesis is to identify TFs that bind to specific sequences in the promoters of target genes and to overexpress these TFs in target tissues (Martin, 1996; Dixon and Steele, 1999). However, TFs that regulate isoflavonoid biosynthesis in legumes have yet to be identified.
We have mined the genome sequence of the model legume Lotus (Lotus japonicus) for genes encoding TFs to identify those strongly correlated in their expression with the induction of isoflavonoids. We were interested in TFs that are induced by elicitation as candidates for specific regulators of isoflavonoid biosynthesis.
RESULTS
Identification of Members of 39 TF Families in Lotus
Searches of the genome of Lotus (Sato et al., 2008) were performed for each TF family using a hidden Markov model (HMM) profile corresponding to the conserved DNA-binding domain. A summary of the number of proteins discovered for each family is shown in Table I, organized by pFAM clans to reflect those families that have a discernible single evolutionary origin or organized by noncognate TF domain fold types with similar structural characteristics.
Table I. Summary of the TF families showing the number of genes that were identified in the Lotus genome for each family and printed to the TF microarray.
The number of genes from Arabidopsis and rice are indicated when the genes from these species were used in phylogenetic analysis. –, Data not available. Small TF families not studied (from table 1 of Mitsuda and Ohme-Takagi, 2009) are as follows: Alfin, AT-hook, AS2, bHSH, C2C2 (Zn)-CO, C3H-type 1 (Zn), CAMTA, CBF5, CCAAT, CPP (Zn), CSD, DBP, DTT, GIF, GRF, HRT, LUG, Nin-like, NOZZLE (NZZ), SET (PcG), RB, SAP, Sir2, Sigma70-like, SNF, SW13, Swi, TUB, ULT, VOZ, VIP3, Whirly, and ZIM.
| Protein Category | TF Family | pFAM Clan Identifier (Description) | pFAM Identifier | HMM Profile Used | No. of Proteins | |||
|---|---|---|---|---|---|---|---|---|
| Arabidopsis | Rice | Lotus (This Study) | Lotus (LegumeTFDB) | |||||
| Helix-turn-helix motifs | CL0123 (helix-turn-helix) | |||||||
| MYB | PF00249 | |||||||
| (R1)R2R3MYBa | In house | (5) 127 | 106 | (4) 135 | 109 | |||
| SHAQKY and GARPa | In house | 88 | 90 | 57 | 12 + 25 | |||
| MYB (other) | 46 | – | ||||||
| Homeodomain | ||||||||
| HD | PF00046 | pFAM seed | 97 | 97 | 81 | 73 | ||
| ZF-HDa | PF04770 | pFAM seed | 17 | 19 | – | |||
| HSF | PF00447 | In house | 24 | 25 | 18 | 18 | ||
| E2F-DP | PF02319 | pFAM seed | 8 | 7 | 7 | |||
| Trihelix | PF10545 | pFAM seed | 26 | 20 | 13 | |||
| Helix-loop-helix motifs | ||||||||
| bHLH | PF00010 | In house | 171 | 181 | 122 | 102 | ||
| TCPa | PF03634 | In house | 24 | 22 | 22 | 22 | ||
| Zinc finger(-like) motifs | ||||||||
| C2H2 | CL0361 (classical C2H2 and C2HC) | PF00096 | pFAM seed | 120 | 128 | 140 | 102 | |
| DOF (C2C2 type)a | PF02701 | pFAM seed | 36 | 31 | 22 | 22 | ||
| GATA (C2C2 type) | CL0167 (zinc β-ribbon) | PF00320 | pFAM seed | 26 | 17 | 15 | ||
| LIM | PF00412 | pFAM seed | 13 | 12 | 12 | |||
| HD-PHD | PF00628 | pFAM seed | 86 | 63 | 63 | |||
| TAZ (putative zinc finger) | PF02135 | pFAM seed | 9 | 3 | 3 | |||
| WRKY (zinc finger-like)a | CL0274 (WRKY-GCM1) | PF03106 | In house | 73 | 103 | 70 | 72 | |
| YABBY (C2C2 type with an HMG box)a | CL0114 (HMG box) | PF04690 | pFAM seed | 5 | 6 | 5 | ||
| Other large TF families | ||||||||
| AP2/ERF familya | CL0081 (MBD-like) | PF00847 | 163 | |||||
| AP2-like | In house | 18 | 26 | 17 | ||||
| ERF-like | In house | 128 | 143 | 130 | ||||
| B3 (VP1/ABI3-like)a | CL0405 (pseudo-barrel domain) | PF02362 | In house | 98 | 92 | 53 | 40 | |
| bZIP | CL0018 (bZIP-like Leu zipper) | PF07716 | In house | 77 | 98 | 46 | 47 | |
| MADS | PF00319 | pFAM seed | 105 | 74 | 88 | 88 | ||
| NACa | PF02365 | pFAM seed | 108 | 139 | 110 | 105 | ||
| Other small TF families | ||||||||
| Auxin response factor (ARF)a | PF06507 | pFAM seed | 23 | 20 | 18 | |||
| ARID | PF01388 | pFAM seed | 24 | 10 | 9 | |||
| AUX-IAAa | PF02309 | pFAM seed | 28 | 30 | 20 | |||
| BBR-BPCa | PF06217 | pFAM seed | 7 | 4 | 4 | |||
| BES1a | PF05687 | pFAM seed | 8 | 4 | 4 | |||
| EIL | PF04873 | pFAM seed | 6 | 7 | 7 | |||
| FHA | CL0357 (SMAD/FHA) | PF00498 | pFAM seed | 16 | 14 | – | ||
| GeBPa | PF04504 | pFAM seed | 21 | 6 | 3 | |||
| GRASa | PF03514 | pFAM seed | 33 | 42 | 45 | |||
| JumonC | CL0029 (cupin) | PF02373 | pFAM seed | 17 | 16 | 15 (JUMONJI) | ||
| LEAFY | PF01698 | pFAM seed | 1 | 2 | 2 | |||
| MBF1 | PF08523 | pFAM seed | 3 | 5 | – | |||
| PLATZa | PF04640 | pFAM seed | 10 | 10 | 10 | |||
| S1Faa | PF04689 | pFAM seed | 4 | 1 | 2 | |||
| SBPa | PF03110 | pFAM seed | 17 | 16 | 16 | |||
| SRSa | PF05142 | pFAM seed | 10 | 9 | 9 | |||
One of the 20 families and subfamilies that are confined to the plant kingdom.
For the R2R3MYB family only, sequences were also mined from EST collections (The Lotus japonicus Gene Index at the Dana-Farber Cancer Institute; Lee et al., 2005), and unique gene fragments were isolated by reverse transcription (RT)-PCR. This resulted in the identification of 16 additional R2R3MYB genes not represented in the genome sequence (build 1.0), of which nine were not represented in GenBank (see “Materials and Methods”).
Recently, Mochida et al. (2010) estimated the number of proteins in different TF families in Lotus, Medicago (Medicago truncatula), and soybean (Glycine max). The number of proteins we identified in different TF families in Lotus was similar to theirs for each TF family. Based on several collections of plant TF family data sets (Mitsuda and Ohme-Takagi, 2009), a current maximum of 72 families are recognized to contain genes that encode transcriptional regulators. We studied 39 of these families in Lotus, including all the largest families. Some smaller families of TFs were not included (Table I) because no members have been shown to regulate phenylpropanoid metabolism in other plant species.
We gave each Lotus protein a name based on the TF acronym (e.g. LjMYB1) to convey membership in particular TF families. For the R2R3MYB family only, each Lotus protein was assigned a name based on the protein in Arabidopsis (Arabidopsis thaliana) with which it shared the highest sequence similarity, deduced by observing an initial phylogenetic tree. For 31 genes, only a tentative name assignment (denoted by “t”) could be made because of incomplete sequence data, although this could be replaced in the future to correspond to a numbered Arabidopsis name. Recently, Gray et al. (2009) recommended a randomized naming scheme for TFs that we adopted for the other TF families studied.
We used the unique regions of each gene to design 60-mer oligonucleotides for a custom Agilent microarray to follow changes in gene expression during isoflavonoid elicitation; a total of 1,456 TF genes were included on the microarray. The sequences were also used for phylogenetic analyses, using an amino acid alignment of the DNA-binding domains, to define subgroups of related protein sequences in each TF family. Genes from the microarray experiment that showed significant increases or decreases in expression were then compared with the complete set of proteins for each family from Arabidopsis, rice (Oryza sativa), and proteins from other species with known functions. These analyses placed the genes from Lotus in the context of existing functional information available for similar genes from other species.
All protein sequences and accession numbers are available online at the IT3F Web site (Bailey et al., 2008; http://jicbio.nbi.ac.uk/IT3F/), in addition to the phylogenetic trees for each family and a breakdown of the families into their main subgroups. It is also possible to interrogate the trees with new sequences and locate the subgroup to which the query sequences belong.
Changes in Gene Expression Associated with Isoflavonoid Elicitation
We focused on identifying genes encoding TFs that were induced or repressed after elicitation of young leaf explants with reduced glutathione (GSH), which induces isoflavonoid production (Robbins et al., 1991), to define potential regulators of the GPP and the isoflavonoid pathway. GSH treatment may signal additional changes in gene expression, particularly those associated with biotic challenges (Foyer and Noctor, 2005), although these are often slower than isoflavonoid elicitation and may involve reductions in gene expression (Hérouart et al., 1993; Wingsle and Karpinski, 1996; Baier and Dietz, 1997). To enable the association of TFs with these pathways, a comprehensive set of genes encoding the enzymes in these pathways was compiled, and gene-specific oligonucleotides were added to the microarray slides to act as control genes for the induction of the pathway (Supplemental Table S1). For some genes, phylogenetic analysis was carried out to identify the Lotus gene that most likely encoded the correct enzyme (Supplemental Figs. S1–S4).
Induction of Isoflavonoid Pathway Genes after Elicitation
After 3 h of elicitation, all the genes encoding enzymes in the isoflavonoid pathway already identified in Lotus (Shimada et al., 2007) showed between 6- and 127-fold increases in the steady-state levels of their mRNA (ISOFLAVONE SYNTHASE [IFS], S-ADENOSYL-l-MET:2,7,4'-TRIHYDROXYISOFLAVANONE 4'-O-METHYLTRANSFERASE [HI4'OMT], ISOFLAVONE 2'-HYDROXYLASE [I2'H], ISOFLAVONE REDUCTASE [IFR], VESTITONE REDUCTASE [VR], and PTEROCARPAN REDUCTASE [PTR]). After 6 h, the steady-state levels of all these transcripts started to decrease (Supplemental Table S2). We also identified genes potentially encoding new isoforms of enzymes in the isoflavonoid pathway by observing their induction by elicitation. The I2′H enzyme has been reported to be encoded by a single-copy gene, CYP81E6 (Shimada et al., 2000). However, two additional CYP81E genes (CYP81E40 and CYP81E41) were discovered in close proximity to the gene encoding CYP81E6 on chromosome 2. CYP81E41 was induced by elicitation, indicating a role during the GSH-induced elicitation of isoflavonoid biosynthesis, but CYP81E40 decreased in expression.
2-Hydroxyisoflavanone dehydratase (HID) is a member of the carboxylesterase family; in Lotus, carboxylesterase-like proteins are encoded by a small cluster of genes. Only one of these genes (HID1) has been suggested to encode HID, due to its high level of identity (75%) to HID of Glycyrrhiza echinata (Shimada et al., 2007). Elicitation experiments showed that another carboxylesterase-like gene (HID2) was induced along with HID1 (Supplemental Table S2). HID2 shows lower identity (52% and 51%, respectively) to GeHID, but the HID2 protein is predicted to contain the oxyanion hole and the catalytic triad characteristic of the active site of members of the carboxylesterase family (Akashi et al., 2005), indicating that it is a catalytically active enzyme.
After elicitation by GSH, another putative HID (HID4) and a VR-like protein also showed decreased gene expression (Supplemental Table S3), suggesting that they are unlikely to be involved directly in isoflavonoid production after elicitation, even though they may have the expected catalytic activity.
PTR catalyzes the final step in vestitol biosynthesis by converting the pterocarpan into an isoflavan (Fig. 1). In Lotus, four PTRs have been characterized biochemically and shown to catalyze this reaction. However, only two of these (PTR1 and PTR2) display enantiospecificity for (−)-medicarpin in vitro, their naturally occurring substrate (Akashi et al., 2006). GSH elicitation induced the expression of PTR1, PTR2, and PTR3 but not of the phylogenetically most distinct gene, PTR4 (Supplemental Fig. S2).
Changes in Expression of Other Phenylpropanoid Pathway Genes
After 3 h of elicitation, genes encoding enzymes in the GPP (PAL, C4H, and 4CL) showed between 8- and 4,376-fold increases in the steady-state levels of their mRNAs. Most of the genes in the core flavonoid pathway (CHS, CHALCONE REDUCTASE [CHR], and CHALCONE ISOMERASE [CHI]) showed between 12- and 189-fold increases in the steady-state levels of their mRNAs (Supplemental Table S2). Based on sequence similarity with known CHR genes, an additional gene encoding CHR (LjSGA_019910.1) was identified from the Lotus genome sequence that showed a similar level of induction of gene expression to the CHR genes identified by Shimada et al. (2007).
Not all genes encoding isoforms of the enzymes of the phenylpropanoid or flavonoid pathways showed increases in the steady-state levels of their mRNA after elicitation. Individual genes encoding isoforms of PAL and 4CL displayed marked down-regulation after treatment with GSH, as did single genes encoding isoforms of the flavonoid enzymes CHS, CHI, and FLAVANOL SYNTHASE (FLS) (Supplemental Table S3).
R2R3MYB TFs
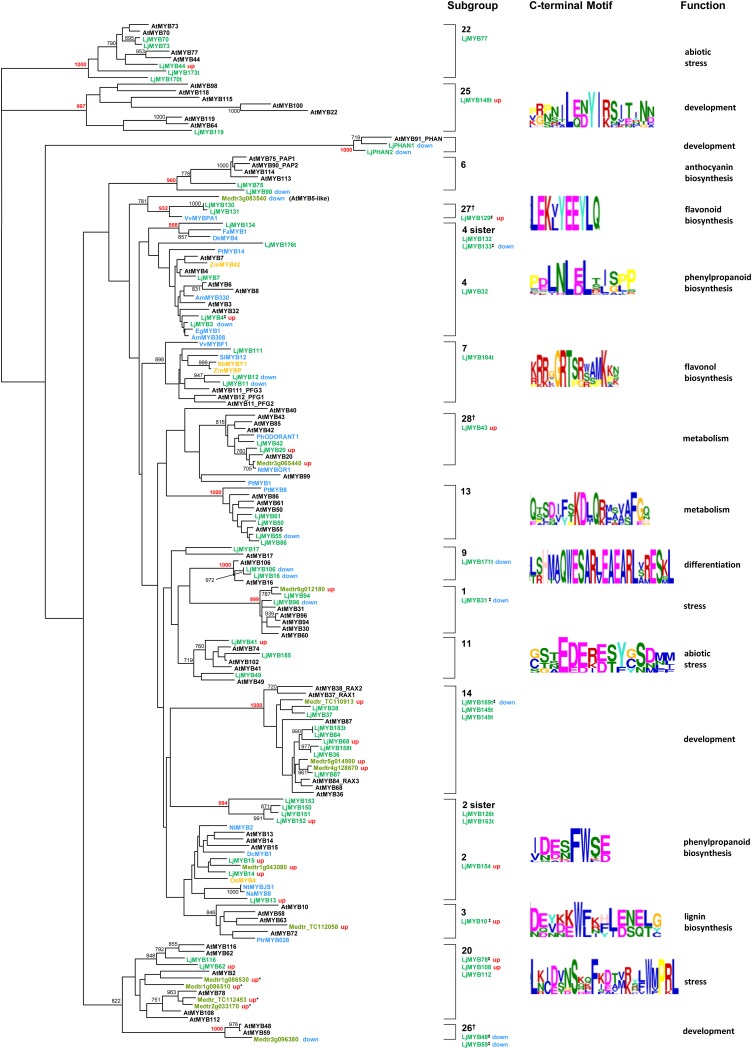
After 3 h of elicitation, a total of 20 R2R3MYB genes showed greater than 2-fold increases in the steady-state levels of their mRNA (Table II). Phylogenetic analysis performed with the full complement of Arabidopsis, rice, and Lotus R2R3MYB proteins showed that each of the induced Lotus genes fitted well into one of 10 subgroups defined by Stracke et al. (2001): 2, 3, 4, 11, 14, 20, 22, and 25, plus two new subgroups numbered in this study as 27 and 28. Subgroup 28 includes PhODORANT1, which regulates Phe biosynthesis in solanaceous plants (Verdonk et al., 2005; Dal Cin et al., 2011). The structural relationships between these genes and other genes belonging to each subgroup are shown in Figure 2. Of these 10 subgroups, five are known to contain proteins that regulate particular parts of the phenylpropanoid pathway in other plant species (subgroups 2, 3, 4, 27, and 28). Subgroups 11, 20, and 22 are linked to abiotic stress responses, and subgroups 14 and 25 are linked to developmental processes (Dubos et al., 2010).
Table II. List of Lotus R2R3MYB genes whose steady-state expression levels were induced or repressed more than 2-fold after GSH elicitation, arranged by the family subgroup to which they belong.
| Genes Induced or Represssed | TF | Subgroup | Kazusa Identifier | 0 to 3 h | 0 to 6 h |
|---|---|---|---|---|---|
| Induced | Fold Increase | ||||
| MYB13 | 2 | chr1.CM0295.20.nd | 951.8 | 393.0 | |
| MYB14 | 2 | chr5.CM0071.410.nd | 18.2 | 20.2 | |
| MYB15 | 2 | chr6.CM1613.30.nc | 168.0 | 128.5 | |
| MYB152 (sister clade) | 2 | LjSGA_148662.1 | 16.4 | 58.2 | |
| MYB154 | 2 | chr1.CM0600.130.nd | 27.6 | 30.9 | |
| MYB10a | 3 | 22.8 | 24.6 | ||
| MYB4a | 4 | 6.9 | 5.1 | ||
| MYB41 | 11 | chr3.CM0711.160.nd | 19.9 | 15.6 | |
| MYB68 | 14 | LjT05A08.60.nd | 18.9 | 10.6 | |
| MYB62 | 20 | chr4.CM0042.760.nc | 86.3 | 106.1 | |
| MYB78a | 20 | 34.3 | 97.1 | ||
| MYB108 | 20 | LjSGA_070863.1 | 61.7 | 327.8 | |
| MYB44 | 22 | chr5.CM0096.100.nc | 3.5 | 2.8 | |
| MYB148t | 25 | LjSGA_039113.1 | 4.3 | 14.9 | |
| MYB129a | 27b | 23.4 | 31.8 | ||
| MYB43 | 28b | LjSGA_050296.1 | 60.3 | 42.4 | |
| MYB20 | 28b | LjSGA_014958.1 | 2.9 | 7.8 | |
| Repressed | |||||
| MYB31a | 1 | −3.13 | −7.82 | ||
| MYB96 | 1 | chr1.CM0023.70.nc | −16.08 | −22.49 | |
| MYB3 | 4 | LjT15F17.90.nd | −6.18 | −8.59 | |
| MYB133 (sister clade)a | 4 | −2.74 | −3.05 | ||
| MYB11 | 7 | chr5.CM0239.580.nc | −76.13 | −68.23 | |
| MYB12 | 7 | LjSGA_016141.1 | −24.55 | −21.55 | |
| MYB16 | 9 | chr1.CM0141.320.nc | −7.22 | −7.14 | |
| MYB106 | 9 | LjSGA_020545.1 | −30.81 | −52.24 | |
| MYB171t | 9 | LjSGA_057038.1 | −5.57 | −5.09 | |
| MYB55 | 13 | chr5.CM0048.320.nd | −35.24 | −22.37 | |
| MYB169ta | 14 | −4.87 | −5.09 | ||
| MYB59a | 26b | −10.49 | −24.76 | ||
| MYB48a | 26b | −54.63 | −59.44 | ||
| PHAN1 | Orthologous to AtPHANTASTICA | chr3.LjT11G09.160.nc | −10.85 | −7.10 | |
| PHAN2 | Orthologous to AtPHANTASTICA | chr3.LjT11G09.150.nc | −52.68 | −38.50 | |
| MYB144t | Subgroup membership unclear | LjSGA_068650.1 | na | −4.29 |
A gene not present in the published genome sequence (Sato et al., 2008). bNew subgroups given numbers in this study. na, Not assessed.
Figure 2.
Phylogenetic tree showing 16 subgroups of the R2R3MYB family. Lotus proteins corresponding to genes that increased (up; red) or decreased (down; blue) in expression after elicitation are shown together with other R2R3MYBs from Lotus (Lj; green) and Arabidopsis (At; black) and R2R3MYBs from other species known to regulate phenylpropanoid metabolism as follows: blue, dicots and gymnosperms (Pinus taeda [Pt], tobacco [Nt], Petunia hybrida [Ph], Solanum lycopersicum [Sl], Vitis vinifera [Vv], Fragaria × ananassa [Fa], Diospyros kaki [Dk], Antirrhinum majus [Am], Eucalyptus gunnii [Eg], P. trichocarpa [Ptr], Nicotiana attenuata [Na], and Daucus carota [Dc]); and yellow, monocot species (rice [Os], Sorghum bicolor [Sb], and Zea mays [Zm]). Due to space constraints, only Arabidopsis and Lotus sequences are included for subgroup 6. Lotus sequences insufficiently complete over the DNA-binding domain were excluded from the final tree but are indicated alongside the subgroup they belong to. Bootstrap values (greater than 70%) are indicated and are shown in red if they are greater than 90%. Numerical identifiers for each subgroup (Stracke et al., 2001) are indicated to the right of the tree. Subgroup clades with low bootstrap support were verified by detecting whether the majority of subgroup members shared a distinct motif in the C-terminal region of the protein. R2R3MYB proteins from Medicago are shown (Medtr; green) that correspond to genes that were induced or repressed in cells treated with yeast elicitor in microarray experiments by Naoumkina et al. (2007). * Genes induced in both yeast elicitor-treated cells and cells treated with methyl jasmonate; methyl jasmonate does not induce early isoflavonoid pathway gene transcripts de novo. † New subgroups given numbers in this study (subgroups 26, 27, and 28). ‡ Genes not present in the genome sequence (build 1.0).
Two sets of closely related genes in Lotus fell into R2R3MYB subgroup 2 and encoded the subgroup 2 motif in their C-terminal domains: [IV][DN][ED][SN]FWS[ED] (Stracke et al., 2001). The genes in one of the sets (LjMYB13, LjMYB14, LjMYB15, and LjMYB154) are orthologous to the Arabidopsis genes AtMYB13, AtMYB14, and AtMYB15. The steady-state levels of the transcripts of these four genes increased between 18- and 951-fold after elicitation. The other set of genes (LjMYB150, LjMYB151, LjMYB152, LjMYB153, and the gene fragments MYB126t and MYB163t) encode a sister clade to subgroup 2, having clear differences in the amino acid sequences of their DNA-binding domains compared with LjMYB13, LjMYB14, LjMYB15, and LjMYB154 but still comprising part of subgroup 2 because they also encode the C-terminal, subgroup 2 motif. A TBLASTN search of all plant sequences in the Gene Indices at the Dana-Farber Cancer Institute and GenBank with these proteins (July 2011) revealed genes from 13 other dicot species that also belong to the subgroup 2 sister clade. In this clade, there is a legume-specific cluster of genes supported by a significant bootstrap value within which resides the LjMYB152 gene (Supplemental Fig. S5). Only this gene in the subgroup 2 sister clade showed an increase in steady-state transcript levels (50-fold) at 6 h after elicitation.
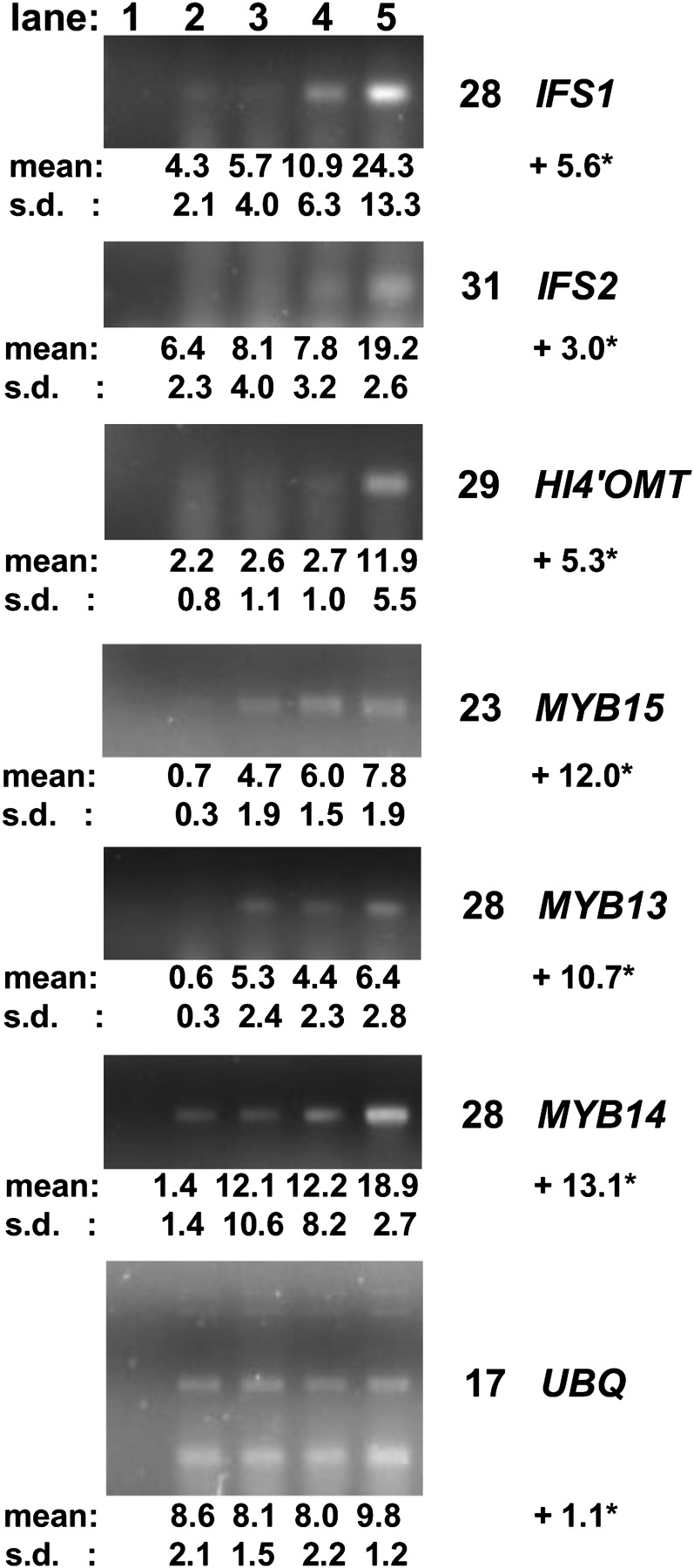
Changes in transcript levels after elicitation were confirmed using the SYBR-Gold gel stain method for LjMYB13, LjMYB14, LjMYB15, IFS1, IFS2, and HI4′OMT in young leaves elicited with GSH for 7 h (Fig. 3). Unlike IFS and HI4′OMT transcripts, transcripts for LjMYB13, LjMYB14, and LjMYB15 also showed significant increases (6.7- to 8.8-fold) in leaves in buffered solution (pH 5.8) without GSH. It is possible that these genes respond to stress associated with submerging leaves in water but that this stress response is insufficient to trigger the expression of the IFS and HI4′OMT genes after 6 h.
Figure 3.
Quantitative RT-PCR experiments to confirm the induction of IFS, HI4′OMT, LjMYB13, LjMYB14, and LjMYB15 gene transcripts during a 7-h elicitation experiment using young leaf explants from Lotus (var Gifu). A gel is shown for the PCR band generated for each gene (the minimum number of cycles required is indicated to the right). Mean signal intensity values (intensity units × mm2 × 102) ± sd (s.d.) from three independent experiments are shown below each PCR band. Lane 1, No-DNA PCR control; lane 2, untreated leaves, 0-h control; lane 3, leaves in MES-buffered solution (pH 5.8); lane 4, leaves in MES-buffered solution containing 10 mm GSH (pH 5.8); lane 5, leaves in unbuffered solution containing 10 mm GSH (pH 3.1). * Fold change in the amount of transcript between uninduced tissue (lane 2) and induced tissue (lane 5) is indicated below the gene name; a positive value indicates higher gene expression in the induced state, and a negative value indicates lower gene expression in the induced state. A t test performed on the band signal intensity for these samples showed significant changes (at a confidence level of greater than 95%), except for MYB5 (at a confidence level of greater than 90%) and the ubiquitin (UBQ) control gene, which showed no significant changes.
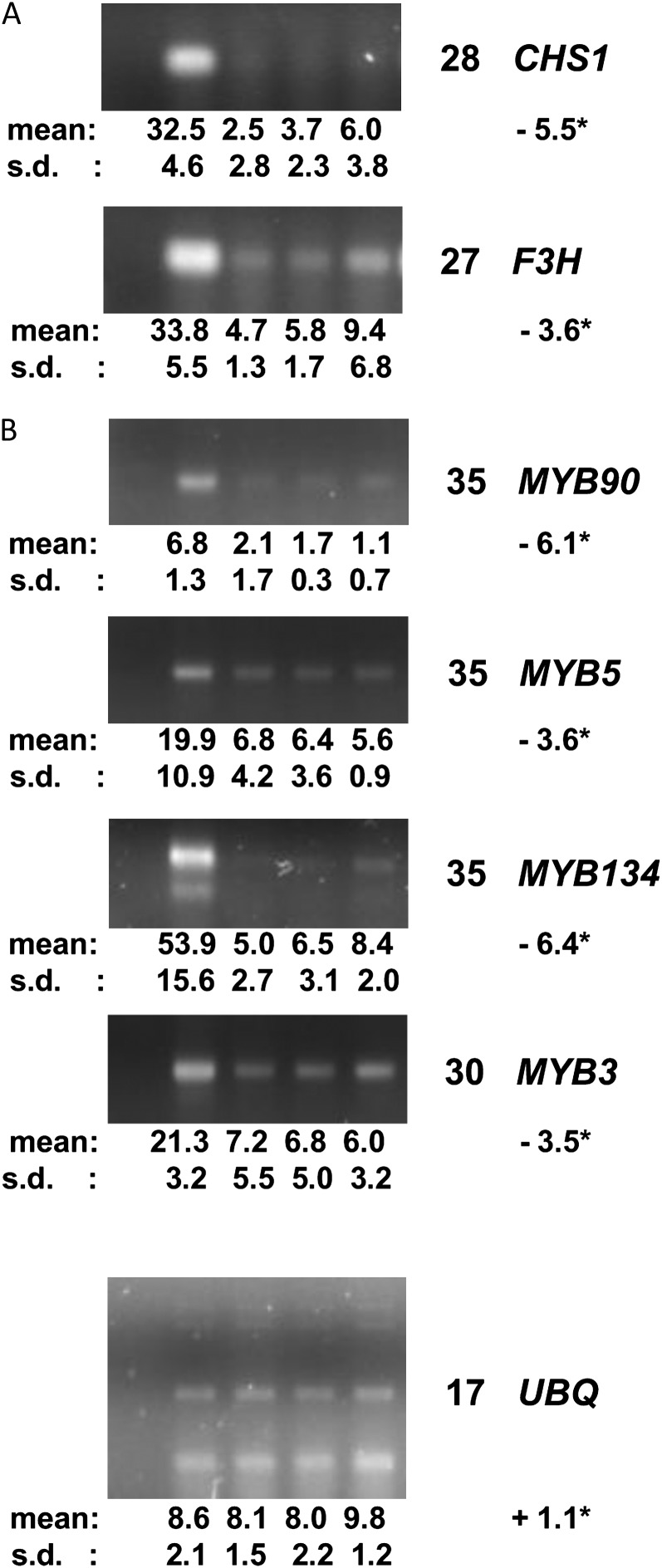
After 3 h of elicitation, a total of 16 R2R3MYB genes showed greater than 2-fold decreases in steady-state levels of their mRNA (Table II). Phylogenetic analysis (Fig. 2) showed that each of the down-regulated Lotus genes fell into one of eight R2R3MYB subgroups: 1, 4, 7, 9, 13, and 14 (Stracke et al., 2001), the clade of genes related to the Arabidopsis PHANTASTICA-like genes, and a new subgroup we have numbered 26. Of these subgroups, three are known to include TFs that regulate particular parts of the phenylpropanoid pathway (subgroups 4, 7, and 13). Other R2R3MYB genes orthologous to known phenylpropanoid-related genes that were expressed at low levels in the microarray experiments were assayed using the SYBR-Gold gel stain method to quantify their expression dynamics after elicitation. LjMYB90 (subgroup 6), a likely regulator of anthocyanin biosynthesis, LjMYB5 (the ortholog of AtMYB5), and LjMYB134 (encoding a member of the subgroup 4 sister group) all decreased in their transcript levels (Fig. 4). The ortholog of the Lotus MYB5 gene in Medicago, Medtr3g083540, also showed reduced transcript levels after elicitation (Naoumkina et al., 2007). The abundance of LjMYB136, LjMYB137, and LjMYB139 (subgroup 5) transcripts was very low in young leaves prior to elicitation, consistent with their proposed roles in positively regulating condensed tannin biosynthesis (Yoshida et al., 2008), and were undetectable in elicited samples after 40 PCR cycles. Yoshida et al. (2008) reported the same trend for LjMYB136 and LjMYB139 gene expression after GSH treatment.
Figure 4.
Quantitative RT-PCR experiments showing the changes in gene expression of genes encoding CHS1 (Kazusa identifier chr1.CM0593.380.nc) and F3H (Kazusa identifier chr4.CM0119.240.nc; A) and a survey of further phenylpropanoid-related R2R3MYB genes (B). For descriptions of the experiments and gel lanes, see Figure 3.
Changes in Other TFs after Elicitation
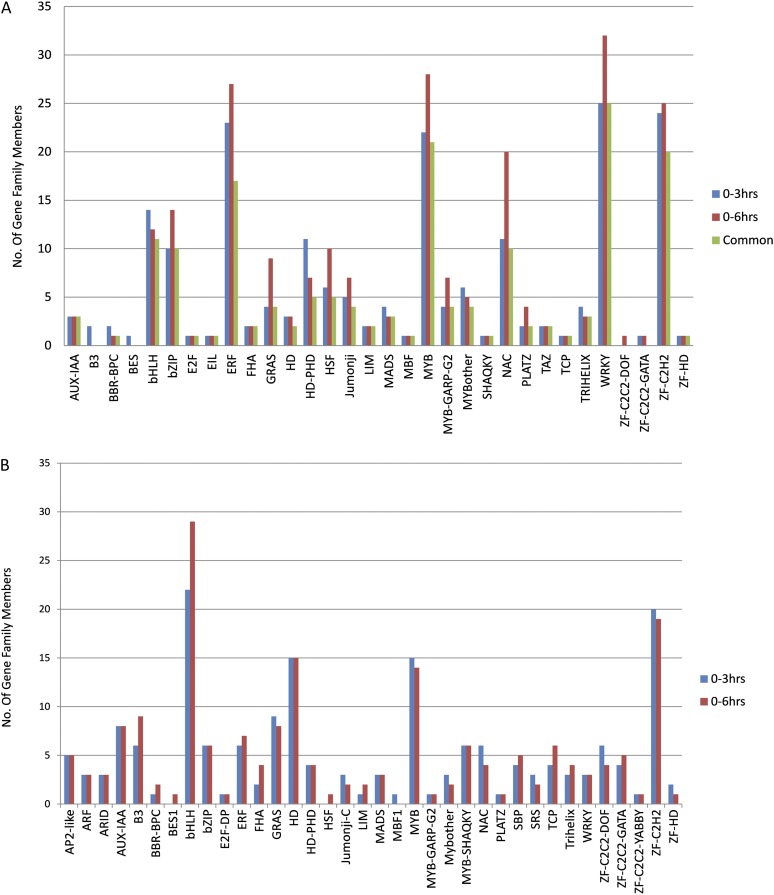
Many genes encoding members of other TF families showed a greater than 2-fold change in their steady-state transcript levels after elicitation with GSH (Fig. 5). The top six TF families with the largest number of genes that were induced in their expression by elicitation were ETHYLENE RESPONSE FACTOR (ERF), ZINC FINGER CYS2HIS2 (ZF-C2H2), BASIC HELIX-LOOP-HELIX (bHLH), and BASIC LEUCINE ZIPPER (bZIP). These families contain gene members that are known for their roles in inducing the expression of genes that protect the plant against abiotic and biotic stresses, such as WRKY (Rushton et al., 2010), MYB (Dubos et al., 2010), and C2H2 (Ciftci-Yilmaz and Mittler, 2008) TFs. The most predominant families of TFs down-regulated in Lotus and Medicago were the bHLH, ZF-C2H2, MYB, and HD families. The effects of GSH elicitation of TF gene expression in Lotus leaves were comparable to elicitation by a yeast elicitor in Medicago protoplasts (Naoumkina et al., 2008); the seven TF families with the most abundant transcript levels in the initial hours after elicitation were the same. Lotus TF genes showed three different patterns of up-regulated expression after elicitation: (1) those that were rapidly induced and whose transcript levels were subsequently sustained; (2) those with a delayed response in transcript levels; and (3) those that were rapidly but only transiently induced. These different kinetics may reflect different functional roles in response to GSH treatment and the effects of GSH on biotic stress responses, in addition to isoflavonoid elicitation (Foyer and Noctor, 2005).
Figure 5.
Histogram showing the number of genes in different TF families that showed a greater than 2-fold increase (A) or decrease (B) in the steady-state transcript levels after elicitation.
Overexpression of LjMYB14 in Lotus Induces Genes Encoding Enzymes in the GPP and Some Specific for Isoflavonoid Biosynthesis
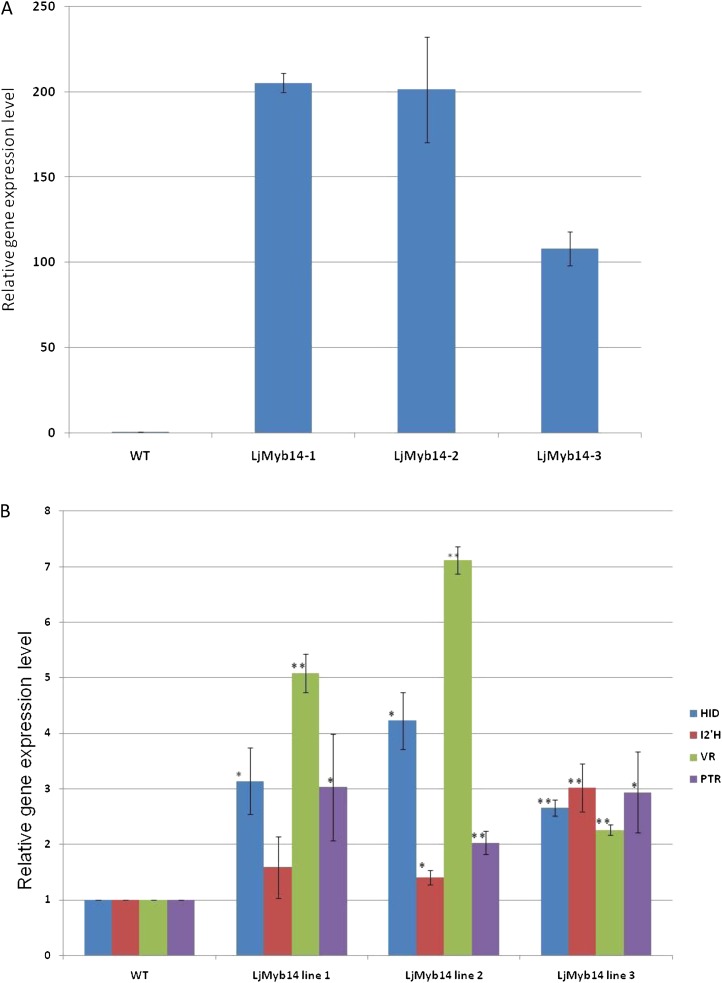
To test the role of selected TFs in isoflavonoid metabolism, a construct for constitutive overexpression of LjMYB14 using the Lotus ubiquitin promoter was assembled and transformed into Lotus (var. MG-20). Microarray analysis of transcript levels in leaves from three independent transformants (without elicitation) was performed, and only those genes for which there was a greater than 2-fold increase in expression in all three transgenic lines compared with controls were analyzed further. The transgenic plants showed increased expression both of genes encoding specific isoforms of the GPP enzymes (PAL, C4H, 4CL) and genes encoding enzymes of the isoflavonoid pathway (HID, I2′H, VR, and PTR; Table III; Supplemental Table S5); the latter set of genes were verified by quantitative RT-PCR analysis (Fig. 6). Not all of the genes encoding enzymes required for vestitol biosynthesis were up-regulated by overexpression of LjMYB14, most notably, the IFS and IFR genes. This confirmed the microarray data, which showed that subgroup 2 R2R3MYB genes (LjMYB13, LjMYB14, and LjMYB15) were moderately induced by treatment of leaf discs with water alone (possibly a response to wounding), whereas IFS and H14′OMT were induced only by GSH treatment.
Table III. Genes associated with general phenylpropanoid and isoflavonoid metabolism and UDP-glycosyltransferases that displayed more than 2-fold increases in gene expression in Lotus plants overexpressing LjMYB14.
| Pathway or Enzyme Class | Gene Name | Kazusa Identifier | Line LjMYB14-1 | Line LjMYB14-2 | Line LjMYB14-3 |
|---|---|---|---|---|---|
| General phenylpropanoid | Fold Increase | ||||
| PAL4 | chr1.CM0033.830 | 2.4 | 2.1 | 2.5 | |
| PAL6 | chr1.CM0033.960 | 2.5 | 2.2 | 2.7 | |
| CYP73A99(b)a (C4H3) | LjSGA_082453.1 | 9.2 | 11.2 | 9.3 | |
| CYP73A99(a)a (C4H4) | LjSGA_029001.1 | 8.1 | 10.0 | 4.6 | |
| CYP73A98 (C4H5) | LjSGA_031602.1 | 3.2 | 3.2 | 3.3 | |
| 4CL1/16 | chr2.CM0018.1240/ | 2.8 | 2.7 | 2.9 | |
| chr6.CM0057.160 | |||||
| 4CL20 | 3.0 | 3.3 | 2.8 | ||
| Isoflavonoid | |||||
| PKR5/6b,c | CM1092.70/CM1092.80 | 9.1 | 10.2 | 13.7 | |
| HI4′ OMT | LjT24P23.70 | 2.5 | 2.3 | 3.0 | |
| HID2 | LjB01D01.120 | 3.2 | 3.6 | 4.3 | |
| CYP81E40 (I2′H) | chr2.CM0250.70 | 4.4 | 4.5 | 3.2 | |
| VR | chr1.CM1255.320 | 6.8 | 8.0 | 7.0 | |
| PTR3b (AB265591) | chr3.CM0216.50 | 3.2 | 3.0 | 3.3 | |
| UDP-glycosyltransferase | |||||
| UGT71B-l.ike | LjSGA_025093.1 | 8.1 | 8.1 | 6.9 | |
| UGT71B-l.ike | LjSGA_066183.1 | 4.9 | 5.7 | 4.0 | |
| UGT71B-l.ike | LjSGA_084705.1 | 5.3 | 4.5 | 4.3 | |
| UGT71B-l.ike | LjSGA_128268.1 | 9.8 | 9.3 | 8.6 | |
| UGT72B4 | chr1.CM0579.30 | 2.7 | 3.4 | 3.0 | |
| UGT72B5 | chr3.LjT41F16 | 9.0 | 8.7 | 8.7 | |
| UGT73B-like | chr4.CM0739.130 | 11.8 | 4.4 | 4.1 | |
| UGT73B-like | chr6.LjT16B20.10 | 2.00 | 2.2 | 2.7 | |
| UGT79B13 | chr3.LjT14J20.160 | 3.3 | 3.4 | 4.5 | |
| UGT87E1 | chr3.LjT09A09.120 | 7.0 | 4.8 | 5.6 | |
| UGT90A-like | chr1.LjT14B18.140 | 27.6 | 31.5 | 22.9 | |
| UGT92A-like | chr1.LjT14B18.160 | 26.7 | 26.4 | 23.6 | |
| UGT92A-like | LjT30N24.300 | 15.6 | 35.1 | 34.4 |
Presumed to be a single gene represented by two nonoverlapping gene fragments. bShimada et al. (2007). cSequence identity too high to discriminate individual genes.
Figure 6.
Quantitative RT-PCR results of relative gene expression normalized to wild-type (WT) expression levels for the LjMYB14 transgene (A) and genes specific for isoflavonoid biosynthesis up-regulated in transgenic lines (B). Expression levels of all genes were calculated relative to actin, and the increase in gene expression in the transgenic plants is expressed as fold change normalized to relative expression in wild-type MG-20. * Significant at P < 0.05 and ** significant at P < 0.01 compared with wild-type levels.
Despite the up-regulation of some genes of isoflavonoid biosynthesis by overexpression of LjMYB14, no increase in isoflavonoid levels was observed in leaves from the overexpression lines compared with leaves from control (MG-20) plants either unelicited or after elicitation (Table IV).
Table IV. Amount of vestitol in leaves of control plants (MG-20) and plants overexpressing LjMYB14 before elicitation or after elicitation by GSH.
| Plant Line | Vestitol in Unelicited Leaves | Vestitol in Elicited Leaves | Vestitol in Elicitation Medium | Total Vestitol after Elicitation | Fold Elicitation |
|---|---|---|---|---|---|
| µg g−1 dry wt | |||||
| MG-20 | 1.83 | 58.95 | 11.06 | 70.01 | 38.3 |
| LjMYB14-1 | 9.02 | 44.90 | 8.93 | 53.83 | 6.0 |
| LjMYB14-2a | 1.14 | 49.73 | 12.66 | 62.39 | 54.7 |
| LjMYB14-2b | 0.61 | 46.18 | 10.47 | 56.65 | 92.9 |
| LjMYB14-3 | 1.11 | 60.13 | 9.87 | 70.00 | 63.1 |
DISCUSSION
One approach to engineering the synthesis of important natural products from plants is to identify TFs that bind to specific sequences in the promoters of genes encoding the enzymes of biosynthesis and overexpress these TFs in target tissues (Cone et al., 1993; Martin, 1996; Grotewold et al., 1998; Dixon and Steele, 1999; Borevitz et al., 2000; Butelli et al., 2008; Cutanda-Perez et al., 2009). TFs, therefore, have the potential to act as molecular switches to induce or repress the accumulation of specific metabolites and can overcome “bottlenecks” that limit the amounts of metabolites that accumulate when a single enzyme activity in a metabolic pathway is engineered.
Our aim was to mine all the genes encoding TFs in Lotus and, from the analysis of gene expression dynamics after elicitation, to discover good candidates for regulators of isoflavonoid biosynthesis. Our global approach identified 1,456 genes encoding TFs that belong to 39 TF families. Phylogenetic trees were constructed for comparative analyses to related proteins from Arabidopsis. We wished to see whether there might be novel, Lotus-specific clades of particular TFs or whether there had been any expansions of particular subclades of TF genes in legumes that might have diverged to regulate isoflavonoid metabolism in a similar way to the clade of R2R3MYB proteins (subgroup 12) that regulate glucosinolate metabolism in the Brassicaceae (Sønderby et al., 2007).
Identification of TF Genes Specific to Lotus and Their Expression after Elicitation
The numbers of examples of species-specific gene duplications and retention events were broadly comparable to Arabidopsis but were more modest in Lotus than in other dicot species. This might have been predicted from the small genome size of Lotus. For example, Wilkins et al. (2009) reported 192 R2R3MYB genes in the diploid tree species Populus trichocarpa (compared with an estimated 135 for Lotus). However, by viewing TF family trees containing Arabidopsis, rice, and Lotus proteins and a global protein alignment, clades of proteins specific to Lotus were observed in the TF families as follows: B3 (one clade containing LjB3-11); C2H2 (three clades containing LjC2H2-20, -50, or -27); ERF (one clade containing LjERF72); MYB1R (one clade containing GARP27 and GARP32); NAC (one clade containing LjNAC57); and MADS (four clades containing LjMADS26, -32, -29, or -47). In the MADS family, there were many cases of gene duplication events having occurred relatively recently in Arabidopsis, Lotus, and the monocot lineage, probably by tandem duplication. Clades of proteins containing significantly more Lotus proteins than the orthologous clades in Arabidopsis were apparent for the R2R3MYB family (subgroup 2 and its sister clade [three Arabidopsis proteins, 10 Lotus proteins] and subgroup 5 [one Arabidopsis protein, five Lotus proteins]) and the bHLH family (subgroup IVa [four Arabidopsis proteins, 12 Lotus proteins]). Both the R2R3MYB subgroups have been linked previously to secondary metabolism, and LjMYB152, which belongs to the subgroup 2 sister clade, showed a large increase in gene expression upon elicitation in the microarray experiments. Although we found proteins from nonlegume species in this clade, LjMYB152 belongs to a phylogenetically distinct inner clade comprising legume-only genes, together with LjMYB150 and LjMYB151 but not LjMYB153. The presence of this inner clade and the fact that LjMYB153 and a Medicago protein fall outside it suggest that the inner clade cannot be a reflection of speciation only but that these genes have arisen after gene duplication and diversified in leguminous species, possibly adopting a role in regulating isoflavonoid biosynthesis. One other legume-specific clade showed changes in gene expression after elicitation: LjMADS47 and a related protein, LjMADS82, increased in expression by a modest 2.5- and 4.3-fold, respectively.
The expression of R2R3MYB subgroup 2 genes in other species is known to be induced in response to biotic and abiotic stress, and some subgroup 2 TFs have been shown to bind to the promoters of genes encoding enzymes of the GPP (PAL [Sugimoto et al., 2000; Maeda et al., 2005] and 4CL [Gális et al., 2006]). Generally, these TFs are thought to induce shikimate and phenylpropanoid metabolism in response to stress (Chen et al., 2006; Ding et al., 2009). In Lotus, increased activity of these proteins likely increases the flux through the GPP, providing substrates for the isoflavonoid pathway. All the Lotus genes encoding proteins belonging to subgroup 2 (LjMYB13, LjMYB14, LjMYB15, LjMYB154) showed increases in their steady-state transcript levels in response to GSH elicitation.
Overexpression of LjMYB14 Increases the Expression of Isoflavonoid Biosynthetic Genes
Microarray analysis of LjMYB14 transgenic lines revealed increased expression of PAL, C4H, and 4CL genes in leaves without elicitation. Overexpression of LjMYB14 also induced the expression of some of the genes involved specifically in isoflavonoid biosynthesis (HID, I2′H, VR, PTR). Although the activity of LjMYB14 was not sufficient to increase isoflavonoid production, our data support the view that subgroup 2 R2R3MYB proteins contribute to the control of isoflavonoid biosynthesis by enhancing the supply of coumaroyl-CoA precursors from the GPP and by inducing some of the genes encoding enzymes specific for isoflavonoid biosynthesis. This suggests that inducers of the shikimate pathway and general phenylpropanoid metabolism may have evolved a more extended role in legumes, assuming roles in regulating the production of the isoflavonoid phytoallexins. We are currently investigating the activity of the subgroup 2 sister group of proteins to determine whether they, in combination with subgroup 2 proteins (LjMYB13, LjMYB14, LjMYB15, LjMYB154), activate isoflavonoid biosynthesis in response to biotic and abiotic stresses.
Expression of Other TF Genes Related to Secondary Metabolism after Elicitation
Apart from R2R3MYB TFs, few genes from other classes of TF have been shown to have a role in regulating secondary metabolism. However, a role in anthocyanin biosynthesis has been demonstrated for bHLH TFs that belong to subgroup 3f (Ludwig et al., 1989; Goodrich et al., 1992). A Lotus gene belonging to this subgroup, bHLH114, decreased 60-fold in expression after elicitation. Members of this subgroup from other species encode proteins that interact with R2R3MYB TFs to coregulate the anthocyanin pathway (Goff et al., 1992) or the proanthocyanidin pathway (Nesi et al., 2001; Paolocci et al., 2007). The Lotus bHLHs belonging to subgroup 3f likely interact with MYB proteins regulating anthocyanin/proanthocyanin biosynthesis, some of which were also observed to decrease in expression in microarray and quantitative RT-PCR experiments: LjMYB5 (AtMYB5-like), LjMYB90 (subgroup 6), LjMYB133, and LjMYB134 (subgroup 4 sister). Thus, the expression of a MYB-bHLH-WD40 complex may be attenuated to allow metabolites from the flavonoid pathway to be diverted to isoflavonoid biosynthesis.
Lotus genes orthologous to the WRKY1 gene controlling sesquiterpene gene expression in cotton (Gossypium hirsutum; Xu et al., 2004) were induced (LjWRKY39, -44, -46; Supplemental Fig. S6). Overexpression of four Medicago WRKY genes in tobacco (Nicotiana tabacum) resulted in increased accumulation of flavonoids and other phenolic compounds (Naoumkina et al., 2008). These genes belong to four distinct subgroups of the WRKY family (Supplemental Fig. S6; accession nos. EU526033–EU526036), so it is possible that WRKY TFs have a general ability to induce phenylpropanoids when expressed at high levels or in response to stress. These four Medicago genes were induced by yeast elicitor together with 23 other WRKY genes (Naoumkina et al., 2008). The same number of Lotus genes were induced in our elicitation experiments, 10 of which are orthologous to the genes induced in Medicago (Supplemental Fig. S6) and represent genes worthy of further investigation of their roles in isoflavonoid biosynthesis.
Evidence of Altered Metabolite Flux through the Phenylpropanoid Pathway after Elicitation
The induction of isoflavonoid biosynthesis by elicitation was accompanied by significant decreases in transcript levels of genes encoding members of other R2R3MYB subgroups, including subgroups 4, 7, and 13, which are known to regulate branches of phenylpropanoid metabolism (Jin et al., 2000; Newman et al., 2004; Mehrtens et al., 2005). Members of subgroup 4 are repressors of targeting genes of the GPP (Jin et al., 2000) or monolignol biosynthesis (Legay et al., 2007). By analogy, LjMYB3 and LjMYB133 (subgroup 4 and subgroup 4 sister) could contribute to isoflavonoid biosynthesis by derepressing target genes in the GPP or specific targets in isoflavonoid metabolism. Interestingly, an ortholog of AtMYB4, LjMYB4, was up-regulated by elicitation. Its activity could contribute to enhancing isoflavonoid production by inhibiting the expression of genes encoding specific isoforms of GPP enzymes in the same way that AtMYB4 negatively regulates 4CL-1 (which is specific for hydroxycinnamate biosynthesis) but not 4CL-3 (which is involved in flavonoid biosynthesis; Jin et al., 2000). Down-regulation of members of subgroups 7 and 13 reflects their roles in positively regulating competing pathway branches. The primary role of genes belonging to subgroup 7 is the control of the genes encoding the enzymes in the flavonoid pathway, including CHS and CHI, required for flavonol biosynthesis in dicot plants and 3-deoxy-flavonoid biosynthesis in monocot plants (Grotewold et al., 1994; Mehrtens et al., 2005). The fact that two genes from this subgroup showed a large decrease in steady-state transcript levels upon elicitation in our experiments and that this was mirrored by down-regulation of CHS1, FLAVANONE 3-HYDROXYLASE (F3H), and FLS suggested that the flavonol branch of the flavonoid pathway competes for the precursors required for isoflavonoid biosynthesis and that the reduced expression of the subgroup 7 TFs likely plays a role in directing the flux of metabolites into the isoflavonoid pathway. This confirms reports that GSH elicitation of Lotus leaves results in reduced levels of kaempferol and quercetin flavonols (Lanot and Morris, 2005).
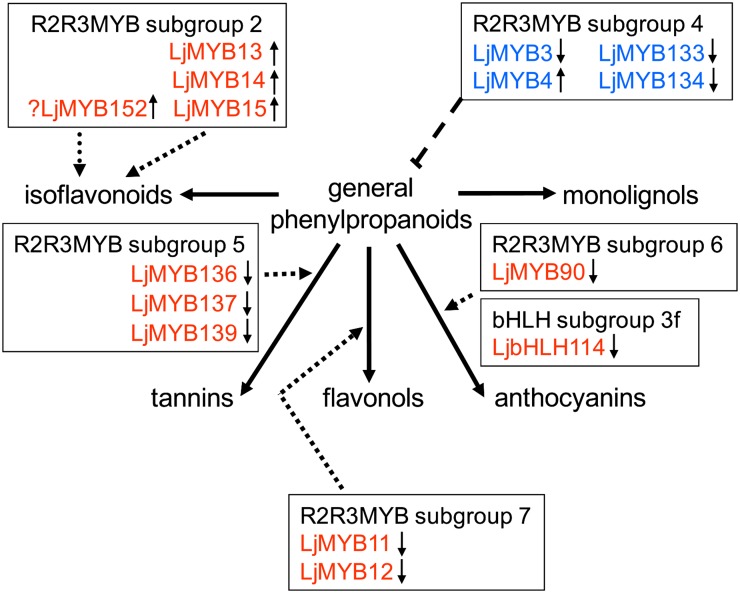
CONCLUSION
Based on the known roles of R2R3MYB TFs in regulating the GPP and flavonoid pathways and the Lotus genes that were up-regulated (Table II; subgroups 2, 3, 4, and 28) and down-regulated (Table II; subgroups 4, 7, and 13; Fig. 4; subgroup 4 and 6 and AtMYB5-like), we propose that several members from these subgroups act coordinately to induce the flux to isoflavonoids and/or reduce the flux of metabolites through competing branches of phenylpropanoid metabolism, such as those leading to flavonols and anthocyanins, so that the precursor metabolite pool can be channeled effectively into isoflavonoids (Fig. 7). The most likely regulators of isoflavonoid biosynthesis are members of R2R3MYB subgroup 2, whose genes show rapid and sustained induction after elicitation, a view supported by our data on the effects of the overexpression of LjMYB14, which enhanced the expression of genes of GPP metabolism and some of the genes specific for isoflavonoid metabolism. The activity of LjMYB14 was not sufficient to induce isoflavonoid accumulation, however, suggesting that additional TFs are required for the induction of key genes (IFS, IFR) in isoflavonoid biosynthesis. A prime candidate for such an additional regulator of isoflavonoid biosynthesis is LjMYB152, belonging to the legume-specific sister group to subgroup 2, which shows similar kinetics in its transcript levels after elicitation to the induction of expression of isoflavonoid biosynthetic genes.
Figure 7.
Proposed TF involvement in the regulation of the phenylpropanoid pathway in Lotus based on this and previous studies. Indicated are TFs corresponding to genes that increase (↑) or decrease (↓) in expression after elicitation that may activate (red) or repress (blue) the different biosynthetic pathways. ?, A potential regulator of isoflavonoid biosynthesis.
MATERIALS AND METHODS
Data Mining for Genes in the Lotus Genome
The Lotus (Lotus japonicus) genome (Sato et al., 2008) was searched for each TF family with the HMMER software suite (Eddy, 1998) using the full set of Lotus predicted protein sequences (file: protein_sequence.gz, July 2008, from the Kazusa DNA Research Institute) and an HMM profile corresponding to each conserved DNA-binding domain. HMM profiles (http://pfam.sanger.ac.uk/) were, in some instances, optimized using the full set of the DNA-binding domains from the Arabidopsis (Arabidopsis thaliana) and rice (Oryza sativa) proteins. Inclusion or exclusion of a gene from the family was assessed by observing the alignment of the gene to the HMM profile and using an alignment of all hits generated by the HMMALIGN program.
The Lotus japonicus Gene Index at the Dana-Farber Cancer Institute was searched for tentative consensus sequences using the TBLASTN program (Altschul et al., 1997) with known R2R3MYB protein query sequences. The corresponding EST clones were obtained from the Plant cDNA Bank Section, Kazusa DNA Research Institute, and sequenced with the BigDye Terminator version 3.1 Cycle Sequencing Kit (Applied Biosystems; http://www.appliedbiosystems.com).
The Lotus genome was mined for genes encoding the enzymes of the isoflavonoid pathway using the BLASTX program with sequences reported by Shimada et al. (2007). Where genes belonged to large families (365 cytochrome P450s, 191 glucosyl transferases, and 64 β-glucosidases), all genes in the family were analyzed.
Cloning of R2R3MYB Genes by RT-PCR and 3′ RACE Methods
RNA was extracted from GSH-treated and control leaf tissue using TRI reagent (Sigma-Aldrich; http://www.sigmaaldrich.com), precipitated with 2 m LiCl, and then cleaned on a Qiagen (http://www.qiagen.com) RNA column. First-strand cDNA was synthesized from 5 μg of RNA using SuperScript II RNase-H reverse transcriptase (Invitrogen; http://www.invitrogen.com) in a total volume of 20 µL. To amplify gene-specific fragments, PCR was performed using 0.2 μL of the first-strand reaction, 1.25 units of AmpliTaq polymerase (Applied Biosystems), and 0.5 μm each of forward and reverse degenerate primers, designed by Romero et al. (1998) to recognize the C-terminal ends of the R2 and R3 MYB domain repeats, respectively. The PCR program was as follows: 95°C for 2 min, and 40 cycles of 94°C for 45 s, 55°C for 2 min, and 72°C for 3 min. The resulting 180-bp fragments were cloned into the pGEM-T Easy vector (Promega; http://www.promega.com). MYB genes were also isolated and cloned from RNA extracted using the CONCERT plant RNA reagent (Invitrogen) from flower, root, silique, seed, seedling, stem, and whole drought-stressed plants (at the point of wilting).
To obtain the unconserved C-terminal portion of each MYB gene for subsequent gene expression experiments, 3′ RACE was performed with the SMART RACE cDNA Amplification Kit (Clontech; http://www.clonetech.com), primed with a 5′ gene-specific primer designed to the 180-bp R3 MYB fragment and a 3′ oligo(dT)18 primer (Frohman et al., 1988), with 5 μg of RNA as the template for the cDNA reaction.
Phylogenetic Analysis
To compare proteins identified in Lotus with proteins in the same family in other model species, data sets containing the full set of predicted protein sequences in the Arabidopsis genome (file: TAIR9_pep_20090619, June 2009) and, for some families, the rice genome (file: all.pep [Rice MSU Osa1 Release 6.1], June 2009) were searched using the HMMER program for significant hits. The hit sequences were aligned to the corresponding HMM profile using the HMMALIGN program with the “-m” option. This alignment was used to generate a neighbor-joining tree with the PHYLIP software package (Felsenstein et al., 1994). To provide statistical support for each node on the tree, a consensus tree was generated from 1,000 bootstrap data sets. Motif logos were prepared using MEME software running on the MEME suite Web server (Bailey et al., 2009).
Isoflavonoid Elicitation
Isoflavonoid elicitation was performed with young leaves of Lotus (ecotype Gifu for the R2R3MYB gene cloning, 3′ RACE, and quantitative RT-PCR, and ecotype MG20 for the microarray experiments). In both cases, the method by Robbins et al. (1991) for leaf material was used except that plants were grown from seed for 4 to 7 weeks in an isolated growth cabinet prior to the elicitation experiments to avoid biotic stress (18/6-h day/night cycle and 24°C/18°C day/night temperature regime), the elicitation procedure with 10 mm GSH was performed in the dark at room temperature (23°C–25°C) for 6 to 8 h with gentle shaking (80–100 rpm), and all solutions contained 0.005% Silwet (Sigma) to reduce leaf surface tension.
Microarray Design and Hybridization Experiments
Microarray slides were designed and produced using Agilent eArray (Agilent Technologies; http://www.agilent.com) using the following parameters: two probes per target, base composition methodology, best distribution, and 3′ bias using the Agilent Lotus transcriptome as the reference database.
Total RNA was extracted from approximately 100 mg of prepared leaf tissue using the RNeasy Plant Mini kit (Qiagen) and then treated with DNase I, and RNA integrity was assessed using the 2100 BioAnalyzer (Agilent Technologies). RNA labeling was performed using kits from Agilent Technologies: the Quick-Amp labeling kit (for the GSH elicitation experiment) or the Low-Input Quick-Amp labeling kit (for transgenic plant analysis). The labeled RNA was purified using RNeasy mini spin columns (Qiagen). For GSH elicitation experiments and transgenic plant analysis, 600 and 200 ng of Cy3-labeled RNA, respectively, was hybridized overnight at 65°C.
The microarrays were scanned using Microarray Scanner G2505B, the raw image files were processed using Feature Extraction software version 10.7.3.1, and data analysis was performed using GeneSpring GX software (all from Agilent Technologies). Differentially expressed genes were determined as genes showing greater than 2-fold absolute change in expression for two independent probes in three biological replicates and passing an unpaired t test (P < 0.05) and the Benjamini-Hochberg multiple testing correction.
Cloning of LjMYB14
cDNA was generated from Lotus plants (ecotype MG20) elicited for 10 h with 10 mm GSH. PCR amplification of LjMYB14 was performed using the primers 5′-ATGGTAAGAGCTCCTTGTTGT-3′ (forward) and 5′-TCAGAACTCGGGCAGTTCTACTG-3′ (reverse), then cloned into pCR-BLUNTII-TOPO (Invitrogen) and sequenced to confirm sequence fidelity. LjMYB14 was then transferred to a plant expression vector, pUB-GW-Hyg (Maekawa et al., 2008), via pDONR207 (Invitrogen) using the primers attBMyb14fwd (5′-GGGACAAGTTTGTACAAAAAAGCAGGCTTAATGGTAAGAGCTCCTTGT-3′) and attBMyb14rev (5′-GGGGACCACTTTGTACAAGAAAGCTGGGTATCAGAACTCGGGCAGTTC-3′) to generate the plant expression vector pUBI-MYB14. pUBI-MYB14 was transformed into Agrobacterium tumefaciens GV3101 for transformation of Lotus (Stiller et al., 1997), ecotype MG20.
Analysis of Transgenic Plants
T1 transgenic plants were germinated on one-half-strength Murashige and Skoog medium containing 15 mg L−1 hygromycin to select for segregating transformants. RNA was extracted from approximately 100 mg of plant material using the RNeasy Plant Mini kit (Qiagen) and treated with DNase I. For each plant, cDNA was generated using the SuperScript III first-strand cDNA synthesis kit (Invitrogen), and LjMYB14 expression was assessed using the LjMyb14fwd and LjMyb14rev primers to confirm transformants.
Metabolite Profiling
Analytical liquid chromatography-mass spectrometry was carried out using an Agilent 1100 Series apparatus (Agilent Technologies) according to the method of Robbins et al. (1991). A 4.6- × 250-mm Spherisorb C18 ODS2 5-µm column (Agilent Technologies) was used at a flow rate of 0.7 mL min−1. The mobile phases and gradient used were the same as those of Robbins et al. (1991). The mass spectrometer was run in positive electrospray mode.
Gene Expression Analysis Using SYBR-Gold Gel Stain
For Figures 3 and 4, PCR was prepared as described for degenerate PCR but contained 5 μL of a 1:50 dilution of cDNA and 0.1 μm each of forward and reverse gene-specific primers (Supplemental Table S4). The PCR products were separated by gel electrophoresis, and the gel was incubated in a solution containing SYBR-Gold nucleic acid gel stain (Sigma-Aldrich) for 30 min and then visualized using an ImageQuant imaging device (Bio-Rad; http://www.bio-rad.com). The corresponding Quantity One software package was used to calculate the fluorescence intensity of the PCR bands. Detection of the PCR products by SYBR-Gold was at least 4-fold more sensitive than ethidium bromide and was shown to detect the concentration of PCR products in a linear manner (Supplemental Fig. S7).
Quantitative RT-PCR
For Figure 6, quantitative RT-PCR was performed using DyNAmo Flash SYBR master mix (Finnzymes; http://www.finnzymes.com) and run with the Rotor-Gene 6000 cycler (Corbett; http://www.corbettlifescience.com). Samples contained 100 ng of cDNA and 0.5 µm each of forward and reverse gene-specific primers (Supplemental Table S4). The PCR program was as follows: 95°C for 7 min, and 45 cycles of 95°C for 15 s, 57°C for 30 s, and 65°C for 30 s. Dissociation curves were run on all samples to ensure that only a single PCR product was produced. The default settings of the Rotor-Gene 1.7 software were used to quantify cycle threshold values. Relative mRNA levels were determined using the ΔΔCT method with actin as the reference gene (Livak and Schmittgen, 2001). The results represent averages of a minimum of three replicates per gene for wild-type plants and each independent LjMYB14 constitutive overexpressing line.
Novel sequence data from this article can be found in the GenBank and EMBL databases under accession numbers JN863531 (LjMYB10), JN863532 (LjMYB48), JN863533 (LjMYB31), JN863534 (LjMYB132), JN863535 (LjMYB59), JN863536 (LjMYB112), JN863537 (LjMYB32), JN863538 (LjMYB77), and JN863539 (LjMYB169t). Microarray data from this article can be found in the Gene Expression Omnibus database under accession numbers GSE31240 (GSH elicitation experiment) and GSE31739 (transgenic plant analysis).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Phylogenetic tree of CHI-like proteins.
Supplemental Figure S2. Phylogenetic tree of the Nmr family containing proteins that encode enzymes for IFR, LAR, PTR, and PLR (for pinoresinol-lariciresinol reductase).
Supplemental Figure S3. Phylogenetic tree of the epimerase family containing proteins that encode enzymes for DFR and VR.
Supplemental Figure S4. Phylogenetic tree of the 2-oxoglutarate and Fe(II)-dependent oxygenase family containing proteins that encode enzymes for F3H, ANS/LDOX, and FLS.
Supplemental Figure S5. Phylogenetic tree of subgroup 2 proteins from Arabidopsis, Lotus, rice, and Brachypodium distachyon and a related but distinct set of proteins that were found to date (July 2011) to be present in 13 dicot species, including legume species.
Supplemental Figure S6. Phylogenetic tree of the WRKY family showing proteins from Lotus and Medicago (Medicago truncatula).
Supplemental Figure S7. Graph showing that SYBR-Gold nucleic acid gel stain detects DNA in a linear manner.
Supplemental Table S1. Summary of Lotus genes encoding enzymes in the phenylpropanoid pathway used in the microarray experiments.
Supplemental Table S2. Genes encoding phenylpropanoid biosynthetic enzymes that increased in expression 3 or 6 h after elicitation.
Supplemental Table S3. Genes encoding phenylpropanoid biosynthetic enzymes that decreased in expression 3 or 6 h after elicitation.
Supplemental Table S4. Sequences of primers used to confirm gene expression profiles.
Supplemental Table S5. Raw intensity signal for isoflavonoid-related genes with increased expression levels in all three independent transgenic lines constitutively expressing LjMYB14 compared with wild-type MG20.
Supplemental File S1. Protein sequences with appropriate accession identifiers of all the Lotus TFs reported in this study and all other R2R3MYB and WRKY proteins used in the phylogenetic analyses.
Supplementary Material
Acknowledgments
We thank Baldeep Kular for support with metabolic profiling.
Glossary
- GPP
general phenylpropanoid pathway
- TF
transcription factor
- HMM
hidden Markov model
- RT
reverse transcription
- GSH
reduced glutathione
References
- Akashi T, Aoki T, Ayabe S-i. (2005) Molecular and biochemical characterization of 2-hydroxyisoflavanone dehydratase: involvement of carboxylesterase-like proteins in leguminous isoflavone biosynthesis. Plant Physiol 137: 882–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akashi T, Koshimizu S, Aoki T, Ayabe S-i. (2006) Identification of cDNAs encoding pterocarpan reductase involved in isoflavan phytoalexin biosynthesis in Lotus japonicus by EST mining. FEBS Lett 580: 5666–5670 [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baier M, Dietz KJ. (1997) The plant 2-Cys peroxiredoxin BAS1 is a nuclear-encoded chloroplast protein: its expressional regulation, phylogenetic origin, and implications for its specific physiological function in plants. Plant J 12: 179–190 [DOI] [PubMed] [Google Scholar]
- Bailey PC, Dicks J, Wang TL, Martin C. (2008) IT3F: a Web-based tool for functional analysis of transcription factors in plants. Phytochemistry 69: 2417–2425 [DOI] [PubMed] [Google Scholar]
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. (2009) MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 37: W202–W208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borevitz JO, Xia Y, Blount J, Dixon RA, Lamb C. (2000) Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell 12: 2383–2394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butelli E, Titta L, Giorgio M, Mock H-P, Matros A, Peterek S, Schijlen EGWM, Hall RD, Bovy AG, Luo J, et al. (2008) Enrichment of tomato fruit with health-promoting anthocyanins by expression of select transcription factors. Nat Biotechnol 26: 1301–1308 [DOI] [PubMed] [Google Scholar]
- Chen YH, Zhang XB, Wu W, Chen ZL, Gu HY, Qu LJ. (2006) Overexpression of the wounding-responsive gene AtMYB15 activates the shikimate pathway in Arabidopsis. J Int Plant Biol 48: 1084–1095 [Google Scholar]
- Ciftci-Yilmaz S, Mittler R. (2008) The zinc finger network of plants. Cell Mol Life Sci 65: 1150–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cone KC, Cocciolone SM, Burr FA, Burr B. (1993) Maize anthocyanin regulatory gene pl is a duplicate of c1 that functions in the plant. Plant Cell 5: 1795–1805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutanda-Perez M-C, Ageorges A, Gomez C, Vialet S, Terrier N, Romieu C, Torregrosa L. (2009) Ectopic expression of VlmybA1 in grapevine activates a narrow set of genes involved in anthocyanin synthesis and transport. Plant Mol Biol 69: 633–648 [DOI] [PubMed] [Google Scholar]
- Dal Cin V, Tieman DM, Tohge T, McQuinn R, de Vos RC, Osorio S, Schmelz EA, Taylor MG, Smits-Kroon MT, Schuurink RC, et al. (2011) Identification of genes in the phenylalanine metabolic pathway by ectopic expression of a MYB transcription factor in tomato fruit. Plant Cell 23: 2738–2753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding ZH, Li SM, An XL, Liu XJ, Qin HM, Wang D. (2009) Transgenic expression of MYB15 confers enhanced sensitivity to abscisic acid and improved drought tolerance in Arabidopsis thaliana. J Genet Genomics 36: 17–29 [DOI] [PubMed] [Google Scholar]
- Dixon RA, Steele CL. (1999) Flavonoids and isoflavonoids: a gold mine for metabolic engineering. Trends Plant Sci 4: 394–400 [DOI] [PubMed] [Google Scholar]
- Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C, Lepiniec L. (2010) MYB transcription factors in Arabidopsis. Trends Plant Sci 15: 573–581 [DOI] [PubMed] [Google Scholar]
- Eddy SR. (1998) Profile hidden Markov models. Bioinformatics 14: 755–763 [DOI] [PubMed] [Google Scholar]
- Felsenstein J. (2004) PHYLIP (Phylogeny Inference Package) Version 3.6. J. Felsenstein, Department of Genome Sciences, University of Washington, Seattle
- Foyer CH, Noctor G. (2005) Oxidant and antioxidant signalling in plants: a re-evaluation of the concept of oxidative stress in a physiological context. Plant Cell Environ 28: 1056–1071 [Google Scholar]
- Frohman MA, Dush MK, Martin GR. (1988) Rapid production of full-length cDNAs from rare transcripts: amplification using a single gene-specific oligonucleotide primer. Proc Natl Acad Sci USA 85: 8998–9002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gális I, Šimek P, Narisawa T, Sasaki M, Horiguchi T, Fukuda H, Matsuoka K. (2006) A novel R2R3 MYB transcription factor NtMYBJS1 is a methyl jasmonate-dependent regulator of phenylpropanoid-conjugate biosynthesis in tobacco. Plant J 46: 573–592 [DOI] [PubMed] [Google Scholar]
- Goff SA, Cone KC, Chandler VL. (1992) Functional analysis of the transcriptional activator encoded by the maize B gene: evidence for a direct functional interaction between two classes of regulatory proteins. Genes Dev 6: 864–875 [DOI] [PubMed] [Google Scholar]
- Goodrich J, Carpenter R, Coen ES. (1992) A common gene regulates pigmentation pattern in diverse plant species. Cell 68: 955–964 [DOI] [PubMed] [Google Scholar]
- Gray J, Bevan M, Brutnell T, Buell CR, Cone K, Hake S, Jackson D, Kellogg E, Lawrence C, McCouch S, et al. (2009) A recommendation for naming transcription factor proteins in the grasses. Plant Physiol 149: 4–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grotewold E, Chamberlin M, Snook M, Siame B, Butler L, Swenson J, Maddock S, St Clair G, Bowen B. (1998) Engineering secondary metabolism in maize cells by ectopic expression of transcription factors. Plant Cell 10: 721–740 [PMC free article] [PubMed] [Google Scholar]
- Grotewold E, Drummond BJ, Bowen B, Peterson T. (1994) The myb-homologous P gene controls phlobaphene pigmentation in maize floral organs by directly activating a flavonoid biosynthetic gene subset. Cell 76: 543–553 [DOI] [PubMed] [Google Scholar]
- Hérouart D, Van Montagu M, Inzé D. (1993) Redox-activated expression of the cytosolic copper/zinc superoxide dismutase gene in Nicotiana. Proc Natl Acad Sci USA 90: 3108–3112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang C, Gu X, Peterson T. (2004) Identification of conserved gene structures and carboxy-terminal motifs in the Myb gene family of Arabidopsis and Oryza sativa L. ssp. indica. Genome Biol 5: R46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, Cominelli E, Bailey P, Parr A, Mehrtens F, Jones J, Tonelli C, Weisshaar B, Martin C. (2000) Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. EMBO J 19: 6150–6161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein DJ, Lim JE, Landry LG, Last RL. (2002) Arabidopsis UVR8 regulates ultraviolet-B signal transduction and tolerance and contains sequence similarity to human regulator of chromatin condensation 1. Plant Physiol 130: 234–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry LG, Chapple CC, Last RL. (1995) Arabidopsis mutants lacking phenolic sunscreens exhibit enhanced ultraviolet-B injury and oxidative damage. Plant Physiol 109: 1159–1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanot A, Morris P. (2005). Elicitation of isoflavan phytoalexins in Lotus japonicus. In A Marquez, ed, Lotus japonicus Handbook. Springer, Dordrecht, The Netherlands, pp 355–361.
- Lee Y, Tsai J, Sunkara S, Karamycheva S, Pertea G, Sultana R, Antonescu V, Chan A, Cheung F, Quackenbush J. (2005) The TIGR Gene Indices: clustering and assembling EST and known genes and integration with eukaryotic genomes. Nucleic Acids Res 33: D71–D74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legay S, Lacombe E, Goicoechea M, Brière C, Séguin A, Mackay J, Grima-Pettenati J. (2007) Molecular characterization of EgMYB1, a putative transcriptional repressor of the lignin biosynthetic pathway. Plant Sci 173: 542–549 [Google Scholar]
- Livak KJ, Schmittgen TD. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Δ Δ C(T)) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Ludwig SR, Habera LF, Dellaporta SL, Wessler SR. (1989) Lc, a member of the maize R gene family responsible for tissue-specific anthocyanin production, encodes a protein similar to transcriptional activators and contains the myc-homology region. Proc Natl Acad Sci USA 86: 7092–7096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda K, Kimura S, Demura T, Takeda J, Ozeki Y. (2005) DcMYB1 acts as a transcriptional activator of the carrot phenylalanine ammonia-lyase gene (DcPAL1) in response to elicitor treatment, UV-B irradiation and the dilution effect. Plant Mol Biol 59: 739–752 [DOI] [PubMed] [Google Scholar]
- Maekawa T, Kusakabe M, Shimoda Y, Sato S, Tabata S, Murooka Y, Hayashi M. (2008) Polyubiquitin promoter-based binary vectors for overexpression and gene silencing in Lotus japonicus. Mol Plant Microbe Interact 21: 375–382 [DOI] [PubMed] [Google Scholar]
- Martin C. (1996) Transcription factors and the manipulation of plant traits. Curr Opin Biotechnol 7: 130–138 [Google Scholar]
- Mehrtens F, Kranz H, Bednarek P, Weisshaar B. (2005) The Arabidopsis transcription factor MYB12 is a flavonol-specific regulator of phenylpropanoid biosynthesis. Plant Physiol 138: 1083–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuda N, Ohme-Takagi M. (2009) Functional analysis of transcription factors in Arabidopsis. Plant Cell Physiol 50: 1232–1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochida K, Yoshida T, Sakurai T, Yamaguchi-Shinozaki K, Shinozaki K, Tran L-SP. (2010) LegumeTFDB: an integrative database of Glycine max, Lotus japonicus and Medicago truncatula transcription factors. Bioinformatics 26: 290–291 [DOI] [PubMed] [Google Scholar]
- Naoumkina M, Farag MA, Sumner LW, Tang Y, Liu C-J, Dixon RA. (2007) Different mechanisms for phytoalexin induction by pathogen and wound signals in Medicago truncatula. Proc Natl Acad Sci USA 104: 17909–17915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naoumkina MA, He X, Dixon RA. (2008) Elicitor-induced transcription factors for metabolic reprogramming of secondary metabolism in Medicago truncatula. BMC Plant Biol 8: 132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesi N, Jond C, Debeaujon I, Caboche M, Lepiniec L. (2001) The Arabidopsis TT2 gene encodes an R2R3 MYB domain protein that acts as a key determinant for proanthocyanidin accumulation in developing seed. Plant Cell 13: 2099–2114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman L, Perazza D, Juda L, Campbell M. (2004) Involvement of the R2R3-MYB, AtMYB61, in the ectopic lignification and dark-photomorphogenic components of the det3 mutant phenotype. Plant J 37: 239–250 [DOI] [PubMed] [Google Scholar]
- Paolocci F, Robbins MP, Madeo L, Arcioni S, Martens S, Damiani F. (2007) Ectopic expression of a basic helix-loop-helix gene transactivates parallel pathways of proanthocyanidin biosynthesis: structure, expression analysis, and genetic control of leucoanthocyanidin 4-reductase and anthocyanidin reductase genes in Lotus corniculatus. Plant Physiol 143: 504–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paz-Ares J, Ghosal D, Wienand U, Peterson PA, Saedler H. (1987) The regulatory c1 locus of Zea mays encodes a protein with homology to myb proto-oncogene products and with structural similarities to transcriptional activators. EMBO J 6: 3553–3558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quattrocchio F, Wing JF, van der Woude K, Mol JN, Koes R. (1998) Analysis of bHLH and MYB domain proteins: species-specific regulatory differences are caused by divergent evolution of target anthocyanin genes. Plant J 13: 475–488 [DOI] [PubMed] [Google Scholar]
- Robbins M, Hartnoll J, Morris P. (1991) Phenylpropanoid defence responses in transgenic Lotus corniculatus. 1. Glutathione elicitation of isoflavan phytoalexins in transformed root cultures. Plant Cell Rep 10: 59–62 [DOI] [PubMed] [Google Scholar]
- Romero I, Fuertes A, Benito MJ, Malpica JM, Leyva A, Paz-Ares J. (1998) More than 80 R2R3-MYB regulatory genes in the genome of Arabidopsis thaliana. Plant J 14: 273–284 [DOI] [PubMed] [Google Scholar]
- Rushton PJ, Somssich IE, Ringler P, Shen QJ. (2010) WRKY transcription factors. Trends Plant Sci 15: 247–258 [DOI] [PubMed] [Google Scholar]
- Sato S, Nakamura Y, Kaneko T, Asamizu E, Kato T, Nakao M, Sasamoto S, Watanabe A, Ono A, Kawashima K, et al. (2008) Genome structure of the legume, Lotus japonicus. DNA Res 15: 227–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwinn K, Venail J, Shang Y, Mackay S, Alm V, Butelli E, Oyama R, Bailey P, Davies K, Martin C. (2006) A small family of MYB-regulatory genes controls floral pigmentation intensity and patterning in the genus Antirrhinum. Plant Cell 18: 831–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada N, Akashi T, Aoki T, Ayabe S-i. (2000) Induction of isoflavonoid pathway in the model legume Lotus japonicus: molecular characterization of enzymes involved in phytoalexin biosynthesis. Plant Sci 160: 37–47 [DOI] [PubMed] [Google Scholar]
- Shimada N, Sato S, Akashi T, Nakamura Y, Tabata S, Ayabe S-i, Aoki T. (2007) Genome-wide analyses of the structural gene families involved in the legume-specific 5-deoxyisoflavonoid biosynthesis of Lotus japonicus. DNA Res 14: 25–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sønderby IE, Hansen BG, Bjarnholt N, Ticconi C, Halkier BA, Kliebenstein DJ. (2007) A systems biology approach identifies a R2R3 MYB gene subfamily with distinct and overlapping functions in regulation of aliphatic glucosinolates. PLoS ONE 2: e1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiller J, Martirani L, Tuppale S, Chian R-J, Chiurazzi M, Gresshoff PM. (1997) High frequency transformation and regeneration of transgenic plants in the model legume Lotus japonicus. J Exp Bot 48: 1357–1365 [Google Scholar]
- Stracke R, Werber M, Weisshaar B. (2001) The R2R3-MYB gene family in Arabidopsis thaliana. Curr Opin Plant Biol 4: 447–456 [DOI] [PubMed] [Google Scholar]
- Subramanian S, Stacey G, Yu O. (2007) Distinct, crucial roles of flavonoids during legume nodulation. Trends Plant Sci 12: 282–285 [DOI] [PubMed] [Google Scholar]
- Sugimoto K, Takeda S, Hirochika H. (2000) MYB-related transcription factor NtMYB2 induced by wounding and elicitors is a regulator of the tobacco retrotransposon Tto1 and defense-related genes. Plant Cell 12: 2511–2528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamagnone L, Merida A, Parr A, Mackay S, Culianez-Macia FA, Roberts K, Martin C. (1998) The AmMYB308 and AmMYB330 transcription factors from Antirrhinum regulate phenylpropanoid and lignin biosynthesis in transgenic tobacco. Plant Cell 10: 135–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdonk JC, Haring MA, van Tunen AJ, Schuurink RC. (2005) ODORANT1 regulates fragrance biosynthesis in petunia flowers. Plant Cell 17: 1612–1624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt T. (2010) Phenylpropanoid biosynthesis. Mol Plant 3: 2–20 [DOI] [PubMed] [Google Scholar]
- Wilkins O, Nahal H, Foong J, Provart NJ, Campbell MM. (2009) Expansion and diversification of the Populus R2R3-MYB family of transcription factors. Plant Physiol 149: 981–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingsle G, Karpinski S. (1996) Differential redox regulation by glutathione of glutathione reductase and CuZn-superoxide dismutase gene expression in Pinus sylvestris L. needles. Planta 198: 151–157 [DOI] [PubMed] [Google Scholar]
- Winkel-Shirley B. (2002) Biosynthesis of flavonoids and effects of stress. Curr Opin Plant Biol 5: 218–223 [DOI] [PubMed] [Google Scholar]
- Xu Y-H, Wang J-W, Wang S, Wang J-Y, Chen X-Y. (2004) Characterization of GaWRKY1, a cotton transcription factor that regulates the sesquiterpene synthase gene (+)-δ-cadinene synthase-A. Plant Physiol 135: 507–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K, Iwasaka R, Kaneko T, Sato S, Tabata S, Sakuta M. (2008) Functional differentiation of Lotus japonicus TT2s, R2R3-MYB transcription factors comprising a multigene family. Plant Cell Physiol 49: 157–169 [DOI] [PubMed] [Google Scholar]
- Zhong R, Lee C, Ye Z-H. (2010) Evolutionary conservation of the transcriptional network regulating secondary cell wall biosynthesis. Trends Plant Sci 15: 625–632 [DOI] [PubMed] [Google Scholar]
- Zhou J, Lee C, Zhong R, Ye Z-H. (2009) MYB58 and MYB63 are transcriptional activators of the lignin biosynthetic pathway during secondary cell wall formation in Arabidopsis. Plant Cell 21: 248–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.