Abstract
Tuliposides, the glucose esters of 4-hydroxy-2-methylenebutanoate and 3,4-dihydroxy-2-methylenebutanoate, are major secondary metabolites in tulip (Tulipa gesneriana). Their lactonized aglycons, tulipalins, function as defensive chemicals due to their biological activities. We recently found that tuliposide-converting enzyme (TCE) purified from tulip bulbs catalyzed the conversion of tuliposides to tulipalins, but the possibility of the presence of several TCE isozymes was raised: TCE in tissues other than bulbs is different from bulb TCE. Here, to prove this hypothesis, TCE was purified from petals, which have the second highest TCE activity after bulbs. The purified enzyme, like the bulb enzyme, preferentially accepted tuliposides as substrates, with 6-tuliposide A the best substrate, which allowed naming the enzyme tuliposide A-converting enzyme (TCEA), but specific activity and molecular mass differed between the petal and bulb enzymes. After peptide sequencing, a novel cDNA (TgTCEA) encoding petal TCEA was isolated, and the functional characterization of the recombinant enzyme verified that TgTCEA catalyzes the conversion of 6-tuliposide A to tulipalin A. TgTCEA was transcribed in all tulip tissues but not in bulbs, indicating the presence of a bulb-specific TgTCEA, as suggested by the distinct enzymatic characters between the petal and bulb enzymes. Plastidial localization of TgTCEA enzyme was revealed, which allowed proposing a cytological mechanism of TgTCE-mediated tulipalin formation in the tulip defensive strategy. Site-directed mutagenesis of TgTCEA suggested that the oxyanion hole and catalytic triad characteristic of typical carboxylesterases are essential for the catalytic process of TgTCEA enzyme. To our knowledge, TgTCEA is the first identified member of the lactone-forming carboxylesterases, specifically catalyzing intramolecular transesterification.
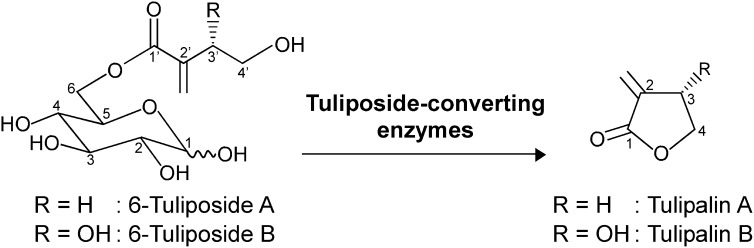
Tuliposides are representative secondary metabolites in the tulip (Tulipa gesneriana; Bergman and Beijersbergen, 1968; Tschesche et al., 1968, 1969; Slob and Varekamp, 1977; Christensen and Kristiansen, 1999). Tuliposides are sugar esters composed of d-Glc and 4′-hydroxy-2′-methylenebutanoyl and/or (3′S)-3′,4′-dihydroxy-2′-methylenebutanoyl side chains (Fig. 1). They have been found in the genera Tulipa, Erythronium, and Gagea in the family Liliaceae and the genera Alstroemeria and Bomarea in Alstroemeriaceae (Slob et al., 1975) as well as in the genus Spiraea in Rosaceae (Kim et al., 1998, 1999). Thus far, seven analogs have been identified: 1-tuliposides A and B, 6-tuliposides A and B, and tuliposides D, E, and F (Tschesche et al., 1968, 1969; Christensen, 1995a, 1995b, 1999; Christensen and Kristiansen, 1995, 1999), with 6-tuliposides A and B the predominant tuliposides produced in tulip cultivars (Christensen and Kristiansen, 1999; Shoji et al., 2005). Because of their chemical lability, 6-tuliposides A and B are spontaneously converted to their lactonized aglycons, tulipalins A and B, respectively, under neutral to basic conditions (Tschesche et al., 1969; Beijersbergen and Lemmers, 1972; van Rossum et al., 1998).
Figure 1.
Reactions catalyzed by tuliposide-converting enzymes. The 1α- and 1β-anomers exist for both 6-tuliposides A and B in tulip plants, and both anomers serve as substrates for tuliposide-converting enzymes.
Tulipalins A and B show antimicrobial activities against a broad range of strains of bacteria and fungi, where tulipalin A functions as an antifungal agent rather than antibacterial and tulipalin B functions in the opposite manner (Bergman, 1966; Bergman et al., 1967; Bergman and Beijersbergen, 1968; Tschesche et al., 1968, 1969; Beijersbergen, 1972; Beijersbergen and Lemmers, 1972; Schönbeck and Schroeder, 1972; Mendgen et al., 2010; Shigetomi et al., 2010, 2011). In addition, tulipalin A has been reported to exhibit high insecticidal activity (Kim et al., 1998, 1999). Thus, tulipalins are considered to serve as a chemical defense for tulip plants against microbial infection and herbivorous insect predation; tuliposides appear to function as storage forms for tulipalins (Beijersbergen and Lemmers, 1972; Schönbeck and Schroeder, 1972). Tuliposides have also been reported to exhibit antimicrobial activities (Tschesche et al., 1968, 1969; Shoji et al., 2005), but it had been controversial whether tuliposide activities originated from tuliposides or from tulipalins formed spontaneously during the course of experiments (Beijersbergen and Lemmers, 1972). Most recently, however, through extensive structure-activity relationship studies using synthetic analogs of tuliposides and tulipalins, it has been clearly demonstrated that the Glc moiety of tuliposides is not indispensable for their inhibitory activities and that the formation of tulipalins plays a key role in antimicrobial action (Shigetomi et al., 2010, 2011).
All parts of tulip plants, such as bulbs, roots, stems, leaves, petals, stamens, and pistils, accumulate large amounts (approximately 0.2%–2% [w/fresh weight]) of tuliposides, whereas tulipalins are far less abundant than tuliposides and sometimes barely detectable (Beijersbergen and Lemmers, 1972; Christensen and Kristiansen, 1999). This fact suggests that inactive tuliposides are stably stored in intact plants and that the conversion of tuliposides into active tulipalins occurs upon infection and wounding. Regardless of the fact that tulipalin-producing conversion reactions occur spontaneously depending on pH, as described above, the possibility of the involvement of a specific enzyme in tulip tissues in such conversion reactions has been suggested (Beijersbergen and Lemmers, 1972). Although no progress has been made for over three decades with respect to this issue, we recently purified and characterized the enzyme tuliposide-converting enzyme (TCE) from tulip bulbs as actively catalyzing the conversion of tuliposides into tulipalins, with 6-tuliposide A the best substrate (Kato et al., 2009a; Fig. 1). Although all parts of tulip possess TCE activity (Kato et al., 2009a, 2009b), specific activities in the crude extracts, as well as the activity ratios toward 6-tuliposides A and B, remarkably differed among the tissues (Kato et al., 2009a). In particular, the enzyme activity in bulbs was markedly higher than those in other tissues. During the course of our preliminary characterization of TCE enzymes in different tulip tissues to reveal the reason for the higher activity in bulbs, we found that the chromatographic behaviors of the TCE activities differ between bulb and petal enzymes, the latter having the second highest TCE activity. These facts led to the hypothesis that the distinct TCE activities with tissues cannot simply be explained by distinct levels of TCE expression but rather by the presence of several TCE isozymes having distinct characteristics: most expressed isozyme differs from tissue to tissue, at least between bulbs and petals.
Here, to prove this hypothesis, as well as to investigate the functional diversity of TCE in tulip tissues, TCE was purified from petals as a representative for tissues other than bulbs, followed by TCE cDNA isolation from petals. Through enzymatic and molecular characterizations, the enzyme was identified as tuliposide A-converting enzyme (TCEA), which catalyzes direct tulipalin A formation from 6-tuliposide A but not the hydrolysis of 6-tuliposide A. To our knowledge, this enzyme is the first identified member of the lactone-forming carboxylesterases, which catalyze an intramolecular transesterification. Moreover, based on subcellular localization of the enzyme, we propose the cytological mechanism of enzyme-mediated tulipalin formation in the tulip defensive strategy.
RESULTS
TCE Activities in Crude Extracts of Tulip Tissues
The TCE activities in the crude tulip extracts were measured using 6-tuliposides A and B as substrate. As shown in Table I, specific activities differed greatly among the tissues. The activity for 6-tuliposide A was highest in bulbs (30 units mg−1 protein) and second highest in petals (7.0 units mg−1 protein). As reported previously by Kato et al. (2009a), the activity ratios toward 6-tuliposides A and B also differed remarkably. The ratios in bulbs, petals, pistils, and stems were over 1.0, suggesting the higher expression of TCE, which favors 6-tuliposide A as substrate, whereas the situation was opposite in leaves and roots, where higher activities for 6-tuliposide B than for 6-tuliposide A were detected.
Table I. TCE activities for 6-tuliposides A and B in the crude extracts of tulip tissues.
Enzyme activity was measured using 4 mm 6-tuliposide A or B as substrate. Data are expressed as means ± sd (n = 3).
| Tissue | Specific Activitya |
Activity Ratio (A/B) | |
|---|---|---|---|
| 6-Tuliposide A | 6-Tuliposide B | ||
| units mg−1 protein | |||
| Bulb | 30 ± 3.2 | 2.9 ± 0.0083 | 10 |
| Petal | 7.0 ± 0.46 | 0.68 ± 0.031 | 10 |
| Pistil | 1.6 ± 0.032 | 0.39 ± 0.043 | 4.1 |
| Stamen | 0.72 ± 0.00058 | 0.71 ± 0.010 | 1.0 |
| Leaf | 0.62 ± 0.019 | 0.87 ± 0.0090 | 0.71 |
| Stem | 0.52 ± 0.0099 | 0.042 ± 0.010 | 12 |
| Root | 0.47 ± 0.0091 | 1.8 ± 0.0054 | 0.26 |
One unit of enzyme activity is defined as the amount of enzyme that catalyzes substrate conversion to product at a rate of 1 μmol min−1.
Purification of TCE from Tulip Petals
Through five steps of column chromatography, the enzyme was purified 254-fold to homogeneity with an overall yield of 17.0% from cell-free extract of tulip petals (Table II). The purified enzyme showed a single 39-kD band on SDS-PAGE (Fig. 2), and gel-filtration analysis on a G3000 SWXL column yielded a single peak, verifying enzyme homogeneity and from which the native enzyme molecular mass was estimated to be 85.4 kD, indicating that the enzyme exists as a dimer. The enzyme had no absorption in the visible light range and a single 280-nm absorption maximum (data not shown), showing that the enzyme was not a chromoprotein.
Table II. Purification of TCE from tulip petals.
Enzyme activity was measured using 4 mm 6-tuliposide A as substrate.
| Step | Total Activitya | Total Protein | Specific Activity | Yield | Purification |
|---|---|---|---|---|---|
| units | mg | units mg−1 | % | -fold | |
| Crude extract | 3,720 | 418 | 8.90 | 100 | 1 |
| DEAE-Toyopearl | 4,110 | 88.3 | 46.6 | 110 | 5 |
| Butyl-Toyopearl | 2,870 | 16.3 | 176 | 77.2 | 20 |
| Gigapite | 1,420 | 2.85 | 498 | 38.2 | 56 |
| Mono-Q HR 5/5 | 1,360 | 0.624 | 2,180 | 36.6 | 245 |
| Superose 12 | 634 | 0.280 | 2,260 | 17.0 | 254 |
One unit of enzyme activity is defined as the amount of enzyme that catalyzes 6-tuliposide A conversion to tulipalin A at a rate of 1 μmol min−1.
Figure 2.
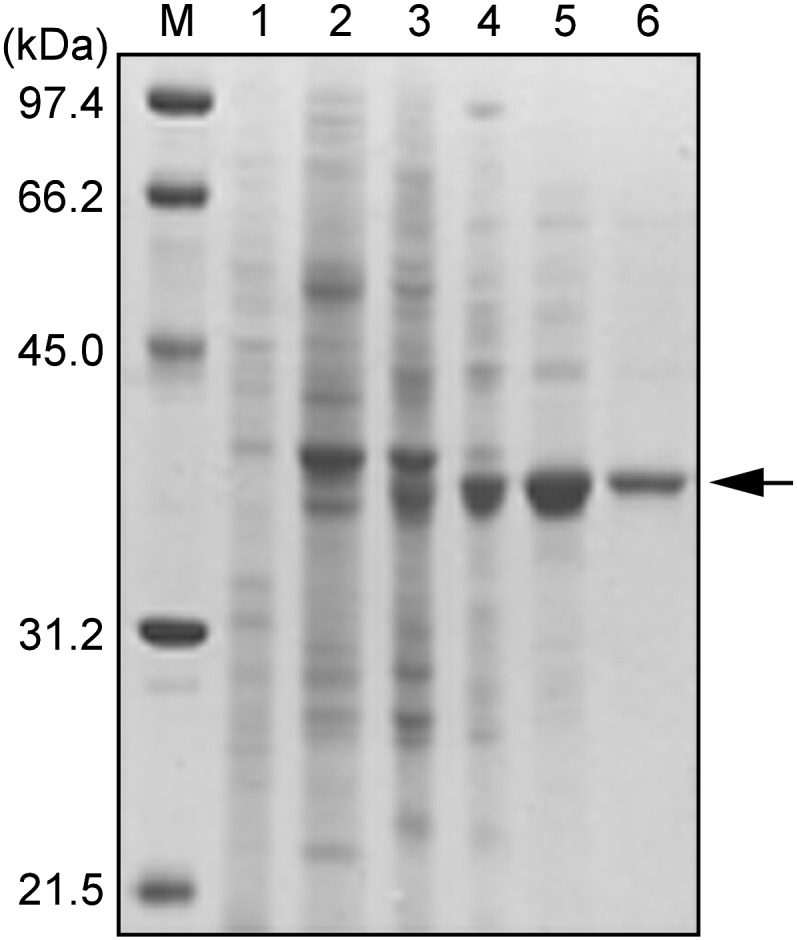
SDS-PAGE of TCE purified from tulip petals. Proteins were separated by 12.5% SDS-PAGE and stained with Coomassie Brilliant Blue R-250. Lane M, Molecular size markers; lane 1, crude extract; lane 2, DEAE-Toyopearl fractions; lane 3, Butyl-Toyopearl fractions; lane 4, Gigapite fractions; lane 5, Mono-Q fractions; lane 6, Superose 12 fractions.
Enzymatic Properties of Petal TCE
The purified enzyme catalyzed the conversion of 6-tuliposide A to tulipalin A with a specific activity of 2,260 units mg−1 at 4 mm 6-tuliposide A, whereas its activity for 6-tuliposide B at 4 mm was only 6.6% (148 units mg−1) of that for 6-tuliposide A, showing that 6-tuliposide A is the natural substrate. As observed for the bulb enzyme (Kato et al., 2009a), stoichiometric analysis of the enzyme reaction for the conversion of 6-tuliposide A clearly showed that petal enzyme catalyzed the formation of tulipalin A but not the ring-opened γ-hydroxy acid, 4-hydroxy-2-methylenebutanoic acid, from 6-tuliposide A. Of the p-nitrophenyl (pNP) derivatives tested for substrates, only pNP acetate was hydrolyzed with 0.44% activity relative to 6-tuliposide A, whereas the enzyme did not react with phosphate and sulfate esters of pNP or glucosides (pNP-α/β-Glcs). The enzyme also showed no activity toward tulipalins A and B to form the corresponding γ-hydroxy acids. Based on these results, the enzyme was designated TCEA; accordingly, the bulb enzyme, which was purified previously (Kato et al., 2009a) and was shown to catalyze tulipalin A formation from 6-tuliposide A, is also hereafter referred to as TCEA.
The temperature and pH optima of the enzyme were determined to be 35°C to 45°C and pH 6.5 to 7.5. The temperature and pH stabilities of the enzyme were also determined by incubating the enzyme for 30 min at various temperatures in 100 mm potassium phosphate (KPi) buffer (pH 6.5) or in 100 mm buffer at various pH levels at room temperature, respectively. Measurement of the remaining activity after the treatments showed that the enzyme was stable up to 45°C for 30 min and in a pH range of 6.0 to 10.0 when maintained at room temperature.
The effects of various additives on enzyme activity were tested with 82 compounds (Supplemental Table S1). Activity was inhibited by approximately 80% and approximately 91% in the presence of NaF (0.2 mm) and phenylmethylsulfonyl fluoride (2 mm), respectively. Among the metal ions, Ag+ (0.2 mm), Hg2+ (0.2 mm), and Cu2+ (2 mm) notably lowered the activity (Supplemental Table S1), and significant effects on enzyme activity were not detected with any other additives.
Isolation of TgTCEA cDNAs from Tulip Petals
N-terminal sequencing of petal enzyme resulted in the detection of multiple amino acids at considerable levels in each Edman degradation cycle regardless of the homogeneity, which was confirmed by SDS-PAGE and gel-filtration analyses. Thus, the enzyme was subjected to internal peptide sequencing. After digestion with lysyl-endopeptidase and peptide mapping, two peptide sequences were determined: sequence 1, SG(R/P)IERFLGTTV, and sequence 2, ?AVVFVAGNDF. As the N-terminal sequence of petal enzyme could not be determined, the enzyme was purified from tulip anthers by procedures similar to those for petals (data not shown), and its N-terminal sequencing was performed. Unlike petal enzyme, anther enzyme gave exclusive, single amino acid signals in each sequencing cycle, and the first 10 amino acids were determined to be ALDDEIVLDL.
To amplify the fragments of cDNA encoding petal TCEA, degenerate primers were designed based on the N-terminal sequence of anther enzyme and the internal peptide sequence 1 of petal enzyme. Degenerate reverse transcription (RT)-PCR from petal mRNA resulted in the isolation of a 74-bp fragment. Its 5′ and 3′ missing parts were amplified by 5′ and 3′ RACE-PCR, and subsequently, RT-PCR was conducted with the primers at the 5′ and 3′ untranslated regions, which resulted in the isolation of two full-length cDNAs, TgTCEA1 (GenBank accession no. AB569208) and TgTCEA2 (AB569209).
TgTCEA1 and TgTCEA2 encoded polypeptides of 385 and 382 amino acids, respectively, and both contained the N-terminal sequence of the mature TCEA from anther (ALDDEIVLDL) and the internal peptide sequences of petal TCEA (SGRIERFLGTTV and RAVVFVAGNDF; Supplemental Fig. S1). These internal sequences were adjacent to Lys on their N-terminal sides, verifying that they arose as a result of lysyl-endopeptidase digestion of purified petal enzyme. TgTCEA1 possessed three amino acid insertions and one amino acid substitution relative to TgTCEA2 in the putative N-terminal signal region and only two amino acid differences in their mature enzyme sequences (Supplemental Fig. S1). A conserved domain search (http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml) suggested that TgTCEA1 and TgTCEA2 possessed sequence motifs for carboxylesterases and were structurally classified to the α/β-hydrolase fold proteins (Ollis et al., 1992). The HGGG motif, constituting an oxyanion hole and the putative catalytic triad residues Ser, Asp, and His, of which the catalytic Ser is present within the conserved pentapeptide sequence Gly-X-Ser-X-Gly, were situated at positions corresponding to those in this protein family (Supplemental Fig. S1; Gershater and Edwards, 2007; Ileperuma et al., 2007). Although most sequences retrieved by BLASTP searches were predicted/putative/hypothetical proteins, of which the hypothetical protein of Vitis vinifera (XP_002285067) exhibited the best score, with 47% identity, the searches clearly showed that TgTCEAs are homologous to the sequences annotated as carboxylesterases and gibberellin receptors, with approximately 40% to 45% identities. Phylogenetic analysis (Supplemental Fig. S2) showed a close relationship of TgTCEAs to the carboxylesterase family proteins, including AeCXE1 of Actinidia eriantha (Ileperuma et al., 2007) and 2-hydroxyisoflavanone dehydratases of Glycyrrhiza echinata and Glycine max (Akashi et al., 2005), rather than to the gibberellin receptor proteins, including rice (Oryza sativa) GID1 (Ueguchi-Tanaka et al., 2005; Shimada et al., 2008).
Genomic PCR with the primers used for the cloning of full-length TgTCEA cDNAs resulted in isolation of the genomic sequences of TgTCEA1 and TgTCEA2. Comparison of cDNA and genomic sequences revealed that the genes contained no intron. In the course of cDNA cloning, the major clones isolated were TgTCEA1 and TgTCEA2, but four additional minor clones designated TgTCEA3 to TgTCEA6 (GenBank accession nos. AB569210–AB569213) were isolated, three of which contained only a few single-nucleotide polymorphisms and the other with a 39-bp deletion relative to TgTCEA1. However, genomic clones corresponding to these minor cDNA clones were not obtained. In addition, other minor sequences in the genomic clones were not isolated by cDNA cloning. Only TgTCEA1 and TgTCEA2 were obtained from both cDNA and genomic DNA.
Enzymatic Characterization of Recombinant TgTCEA
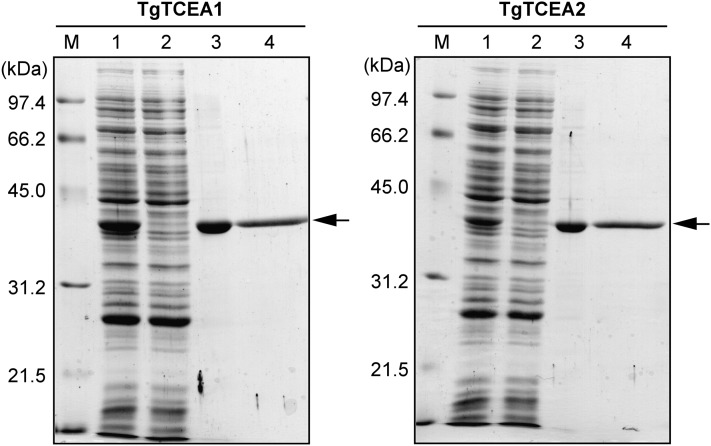
Recombinant TgTCEA1 and TgTCEA2 enzymes were efficiently expressed in a soluble protein fraction in Escherichia coli. N-terminal His-tagged TgTCEA enzymes were purified to homogeneity by metal chelation and gel-filtration chromatography. The purified recombinant enzymes appeared as a 39-kD band on SDS-PAGE (Fig. 3), in agreement with the calculated 37-kD molecular mass, and the native molecular mass was estimated to be 79 kD by gel-filtration analysis, indicating that the recombinant enzymes were expressed as dimers similar to native petal enzyme.
Figure 3.
SDS-PAGE of recombinant TgTCEA enzymes expressed in E. coli. Proteins were separated by 12.5% SDS-PAGE and stained with Coomassie Brilliant Blue R-250. Lane M, Molecular size markers; lane 1, crude extract; lane 2, column run through in TALON affinity chromatography; lane 3, TALON fraction eluted with 200 mm imidazole; lane 4, Superdex 200 fractions.
Purified recombinant enzymes exhibited higher catalytic activities for 6-tuliposide A than for 6-tuliposide B, where the specific activities of TgTCEA1 and TgTCEA2 for 6-tuliposide B were 8.7% and 7.8%, respectively, relative to 6-tuliposide A. This was consistent with the activity ratio of petal enzyme, whose activity for 6-tuliposide B was 6.6% relative to 6-tuliposide A. The kinetic parameters of TgTCEA1 and TgTCEA2 enzymes were determined for 6-tuliposides A and B (Table III). The Km and kcat values of TgTCEA1 for 6-tuliposide A (18 mm and 2,800 s−1, respectively) were remarkably smaller and higher, respectively, than those for 6-tuliposide B (71 mm and 510 s−1). The catalytic efficiency (kcat/Km) of TgTCEA1 for 6-tuliposide A (160 s−1 mm−1) was 22 times higher than for 6-tuliposide B (7.2 s−1 mm−1). The Km and kcat values of TgTCEA2 were comparable to TgTCEA1 for both 6-tuliposides A and B, where TgTCEA2 showed 25 times higher catalytic efficiency for 6-tuliposide A (170 s−1 mm−1) than for 6-tuliposide B (6.7 s−1 mm−1). These results verified that TgTCEA1 and TgTCEA2 encode tuliposide A-converting enzyme.
Table III. Kinetic parameters of TgTCEA enzymes.
Data are expressed as means ± sd (n = 3).
| Enzyme | 6-Tuliposide A |
6-Tuliposide B |
Relative kcat/Km (A/B) | ||||
|---|---|---|---|---|---|---|---|
| Km | kcat | kcat/Km | Km | kcat | kcat/Km | ||
| mm | s−1 | s−1 mm−1 | mm | s−1 | s−1 mm−1 | -fold | |
| TgTCEA1 | 18 ± 1.5 | 2,800 ± 120 | 160 | 71 ± 9.0 | 510 ± 21 | 7.2 | 22 |
| TgTCEA2 | 14 ± 1.3 | 2,400 ± 99 | 170 | 72 ± 6.9 | 480 ± 16 | 6.7 | 25 |
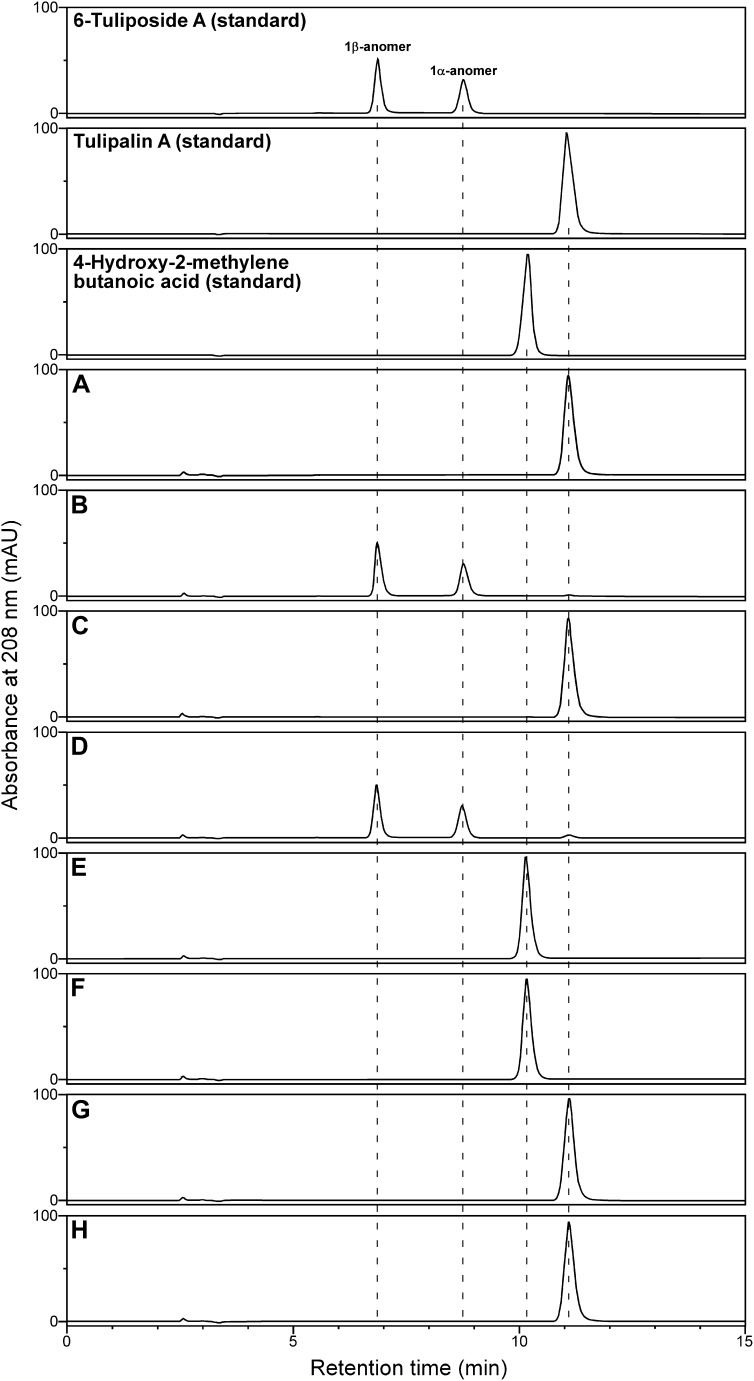
Detailed analysis of the reaction product formed by TgTCEA1 was performed as shown in Figure 4. TgTCEA1 reaction with 6-tuliposide A terminated under acidic conditions formed only tulipalin A as product (Fig. 4, A and B), which was the same as the reaction with purified petal enzyme. Terminating the enzyme reaction with methanol also yielded only tulipalin A as product, but not the hydrolytic product of 6-tuliposide A, 4-hydroxy-2-methylenebutanoic acid (Fig. 4, C and D). In addition, spontaneous conversion of 4-hydroxy-2-methylenebutanoic acid to tulipalin A did not occur during the enzyme reaction and termination (Fig. 4, E and F). These results exclude the possibility that TgTCEA catalyzes the hydrolysis of 6-tuliposide A to form 4-hydroxy-2-methylenebutanoic acid, followed by its spontaneous lactonization to tulipalin A. It was also confirmed that TgTCEA1 does not hydrolyze tulipalin A to form 4-hydroxy-2-methylenebutanoic acid (Fig. 4, G and H), showing that TgTCEA enzyme accepts 6-tuliposide A to catalyze only the intramolecular transesterification but not the hydrolysis.
Figure 4.
HPLC chromatograms of enzyme reaction mixtures of TgTCEA1 with 6-tuliposide A, tulipalin A, and 4-hydroxy-2-methylenebutanoic acid. A and B, Reactions with 6-tuliposide A stopped by phosphoric acid in the presence (A) or absence (B) of TgTCEA1. C and D, Reactions with 6-tuliposide A stopped by methanol in the presence (C) or absence (D) of TgTCEA1. E and F, Reactions with 4-hydroxy-2-methylenebutanoic acid stopped by phosphoric acid in the presence (E) or absence (F) of TgTCEA1. G and H, Reactions with tulipalin A stopped by phosphoric acid in the presence (G) or absence (H) of TgTCEA1. Negative control reactions with heat-inactivated TgTCEA1 also gave the same chromatograms as B, D, F, and H. The dotted lines indicate retention times.
Site-Directed Mutagenesis of TgTCEA1
To examine which two of the three Gly residues in the HGGG motif participate in forming the oxyanion hole, and whether the putative catalytic triad residues, Ser-235, Asp-327, and His-359, indeed play catalytic roles in the TgTCEA reaction, mutations were introduced into these residues (Table IV; Supplemental Fig. S1). In addition, the mutant for His-362, which is three amino acids apart from His-359, was also analyzed. The mutated recombinant enzymes were purified and the enzymatic activities toward 6-tuliposide A were measured. The activity of the G154A mutant was comparable to that of TgTCEA1, but the activities were greatly reduced in the G152A and G153A mutants (Table IV), indicating that Gly-152 and Gly-153 contribute to the oxyanion hole formation. The mutations of each of the putative catalytic triad residues, Ser-235, Asp-327, and His-359, reduced the activity to 0.01%, 0.1%, and 0.0008%, respectively, relative to TgTCEA1 (Table IV). These results strongly suggest that those residues function in a catalytic process in a similar manner to known carboxylesterases catalyzing ester hydrolysis. Although the H362A mutant exhibited the reduced activity (6% of TgTCEA1), the reduction was much more moderate than the mutant for His-359 (0.0008% of TgTCEA1). His-362 seems to contribute to the enzyme conformation rather than to have direct involvement in the catalytic process. It should also be noted that all the mutants tested formed only tulipalin A as product, but not 4-hydroxy-2-methylenebutanoic acid, the hydrolytic product of 6-tuliposide A, showing that the mutations did not affect the product specificity.
Table IV. Specific activity of site-directed mutants of TgTCEA1.
Enzyme activity was measured using 4 mm 6-tuliposide A as substrate. Data are expressed as means ± sd (n = 3).
| Mutant | Specific Activitya | Relative Activity |
|---|---|---|
| units mg−1 protein | % | |
| TgTCEA1 | 326 ± 26.3 | 100 |
| G152A | 0.355 ± 0.0280 | 0.1 |
| G153A | 0.0417 ± 0.00458 | 0.01 |
| G154A | 287 ± 7.78 | 88 |
| S235A | 0.0322 ± 0.00467 | 0.01 |
| D327N | 0.396 ± 0.0231 | 0.1 |
| H359A | 0.00263 ± 0.000215 | 0.0008 |
| H362A | 19.8 ± 0.411 | 6 |
One unit of enzyme activity is defined as the amount of enzyme that catalyzes 6-tuliposide A conversion to tulipalin A at a rate of 1 μmol min−1.
Transcript Profiles of TgTCEA Genes in Tulip Tissues
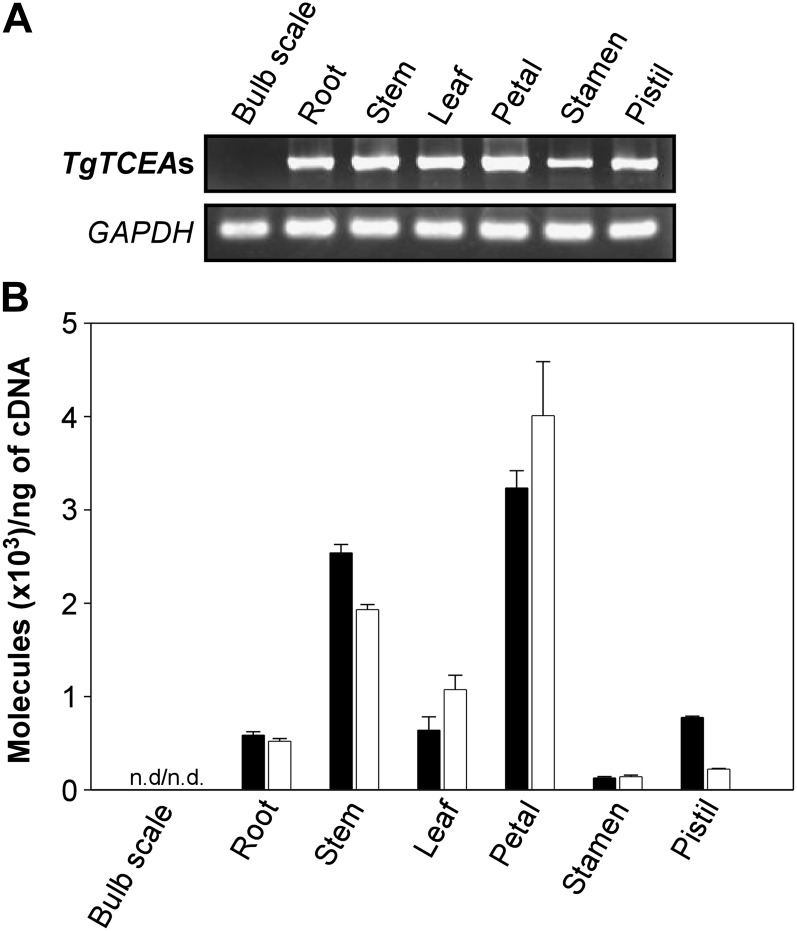
Transcript profiles of TgTCEA genes in tulip tissues were examined by RT-PCR analysis, in which highly identical TgTCEA1 and TgTCEA2 were amplified with primers TCEA-RT-F and TCEA-RT-R common to both sequences (Supplemental Table S2). TgTCEA genes were transcribed in roots, stems, leaves, petals, stamens, and pistils but not in bulb scales (Fig. 5A). Changing the PCR conditions also did not produce any PCR band from bulb scales (data not shown). To further verify the absence of transcripts of TgTCEA genes in bulbs, PCR was performed with six additional primer sets designed from sequences common to both TgTCEA1 and TgTCEA2 genes (Supplemental Table S2), but the PCR products were not detected in bulb scales with any primer combinations (Supplemental Fig. S3), suggesting that the TgTCEA genes identified in this study are not transcribed in bulbs.
Figure 5.
Transcript analysis of TgTCEA genes in tulip tissues. A, RT-PCR with a primer set amplifying both TgTCEA1 and TgTCEA2 genes. B, Transcript levels of TgTCEA1 and TgTCEA2 genes analyzed by quantitative real-time RT-PCR. TgTCEA1 (black bars) and TgTCEA2 (white bars) were amplified with primer sets specific to each of them. Data are expressed as means of triplicate experiments ± sd. n.d., Not detected.
To compare the transcript levels between the TgTCEA1 and TgTCEA2 genes in each tulip tissue, quantitative RT-PCR analysis was performed using gene-specific primers (Supplemental Table S2). The transcript profiles of TgTCEA1 and TgTCEA2 were almost comparable throughout the tissue except for pistils, where 3-fold higher transcription of TgTCEA1 than TgTCEA2 was observed (Fig. 5B). They were transcribed highest in petals, and the absence of transcripts of both genes in bulbs was again confirmed.
Subcellular Localization of TgTCEA1 Enzyme
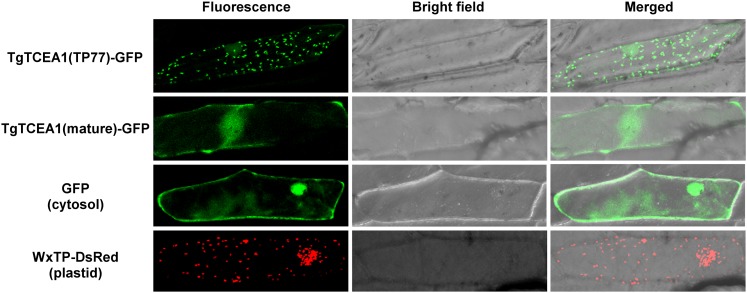
To examine the subcellular localization of TgTCEA1 enzyme, the putative transit peptide of TgTCEA1 (residues 1–77 [TP77]; Supplemental Fig. S1) was fused to GFP and transiently expressed in onion (Allium cepa) epidermal cells. The localization of GFP signals was analyzed based on the confocal images. In the transient expression of the fusion protein TgTCEA1(TP77)-GFP, the green fluorescence was localized to the dotted organelles (Fig. 6), a typical plastidial fluorescence pattern, whose size and pattern were highly similar to those of WxTP-DsRed, a positive control for plastidial localization, which expresses plastid-targeted transit peptide of Waxy protein fused to DsRed (Kitajima et al., 2009). On the other hand, in the expression of transit peptide-truncated mature polypeptides of TgTCEA1 (residues 78–385) fused to GFP, the fluorescence pattern was the same as that of cytosolic GFP (Fig. 6). These results strongly suggest that TgTCEA1 enzyme is localized to plastids.
Figure 6.
Subcellular localization of TgTCEA1(TP77)-GFP and TgTCEA1(mature)-GFP fusion proteins in onion epidermal cells. TgTCEA1(TP77)-GFP expresses the N-terminal 77 amino acids of TgTCEA1, which were predicted as a transit peptide, fused to GFP. TgTCEA1(mature)-GFP expresses mature polypeptides of TgTCEA1 (residues 78–385) fused to GFP. Nontargeted GFP is the control for cytosolic localization. WxTP-DsRed, which expresses the transit peptide of the Waxy protein fused to DsRed, is the control for plastidial localization (Kitajima et al., 2009). Fluorescence signals of TgTCEA1(TP77)-GFP, TgTCEA1(mature)-GFP, and nontargeted GFP in the nucleus are attributable to free diffusion of the protein (Dingwall and Laskey, 1986).
As described above, N-terminal sequencing of purified petal enzyme resulted in the detection of multiple amino acids in each Edman degradation cycle. Because Ser and Pro were detected at considerable levels in each sequencing cycle (data not shown), we hypothesized that this heterologous N terminus of the petal enzyme is due to relaxed recognition of the cleavage site around the repeated Ser and Pro region (residues 64–76 of TgTCEA1; Supplemental Fig. S1). To determine a minimal length of the signal peptide, the subcellular localization of GFPs fused with a deletion series of the TP77 of TgTCEA1 was examined (Supplemental Fig. S4). First, localization of GFPs fused with TP20, TP40, and TP60 was compared, which resulted in the cytosolic localization of TgTCEA1(TP20)-GFP and TgTCEA1(TP40)-GFP and the plastidial localization of TgTCEA1(TP60)-GFP. When TgTCEA1(TP55)-, TgTCEA1(TP50)-, and TgTCEA1(TP45)-GFPs were expressed, all of them exhibited the plastidial localization. TgTCEA1(TP41)-GFP showed clear cytosolic localization, but TgTCEA1(TP42)-, TgTCEA1(TP43)-, and TgTCEA1(TP44)-GFPs showed mixed signals for cytosol and plastids, which were clearly distinct from the TgTCEA1(TP45)-GFP signal, showing exclusive localization in plastids. These results suggest that the minimal length of the plastid-targeted signal of TgTCEA1 is 45 amino acids and that the signal cleavage between residues 45 and 77 is allowed for the TgTCEA1 enzyme to be sorted to the plastids.
DISCUSSION
TCEA Catalyzes the Conversion of 6-Tuliposide A to Tulipalin A
TCEA purified from tulip petals preferentially accepted 6-tuliposide A as substrate, whereas its activity for 6-tuliposide B was 15 times lower, showing that the enzyme is mainly involved in the conversion of 6-tuliposide A to tulipalin A in tulip petals. Although the Km values of TgTCEA1 and TgTCEA2 enzymes for 6-tuliposide A (18 and 14 mm, respectively; Table III) are relatively high, the content of the substrate 6-tuliposide A in petals reaches approximately 20 μmol g−1 fresh weight (Kato et al., 2009b), the equivalent to 20 mm, which still enables TgTCEA1 and TgTCEA2 to exhibit approximately half of their turnover rates (2,800 and 2,400 s−1, respectively; Table III). Such high turnover rates of TgTCEA enzymes appear to compensate for their relatively high Km for 6-tuliposide A. Petal enzyme exhibited similar characters to bulb enzyme (Kato et al., 2009a) with respect to dimerism, substrate specificity, temperature and pH optima, and susceptibility to various inhibitors. However, specific activity differed between petal and bulb enzymes. Petal enzyme activity for 6-tuliposide A was 2,260 units mg−1 (Table II), whereas that of bulb was 42,200 units mg−1 (Kato et al., 2009a). Moreover, molecular mass also differed between petal and bulb enzymes: native and subunit masses of petal enzyme were 85.4 and 39 kD, respectively, and those of bulb enzyme were 70.2 and 35 kD, respectively (Kato et al., 2009a). These results strongly suggested the expression of distinct TCEA isozymes in petals and bulbs, which was supported by the results of RT-PCR analysis of TgTCEA genes isolated from petals in this study (Fig. 5; Supplemental Fig. S3). TgTCEAs were transcribed in petals, roots, stems, leaves, stamens, and pistils but not in bulbs, indicating the presence of a TgTCEA homolog specifically expressed in bulbs. The results also verified the hypothesis proposed here that the remarkably higher enzyme activity in bulb crude extracts than other tissues (Table I) is due to the expression of different isozymes having distinct catalytic properties between bulbs and other tissues, but not due to higher expression of a single enzyme in bulbs than in other tissues. Considering the absence of RT-PCR products in bulbs with any primer combinations (Fig. 5; Supplemental Fig. S3), the TgTCEA isozyme gene in bulbs would be less similar to the petal TgTCEAs. Identification of a gene encoding the bulb-specific isozyme is now being pursued in order to reveal the mechanisms of its bulb-specific expression and higher catalytic activity.
Although all tulip parts possessed TCE activity, the ratios of the specific activities toward 6-tuliposides A and B in crude extracts differed depending on the tissue (Table I; Kato et al., 2009a). If tulip had only TCEs showing higher activity for 6-tuliposide A than for 6-tuliposide B, like the petal and bulb enzymes identified in this and previous studies (Kato et al., 2009a), the 6-tuliposide A/B activity ratios should not fluctuate among the tissues. These facts allowed the prediction of the presence of an enzyme that preferentially catalyzes the conversion of 6-tuliposide B to tulipalin B (i.e. tuliposide B-converting enzyme) in tulip; the purification and characterization of this enzyme are in progress and will be published elsewhere.
TgTCEA Is, to Our Knowledge, the First Identified Member of the Lactone-Forming Carboxylesterases
TgTCEA polypeptides possessed typical sequence motifs for carboxylesterases, including the HGGG motif and the putative catalytic triad residues, Ser-235, Asp-327, and His-359 (Supplemental Fig. S1), of which Ser-235 is situated within the conserved motif (Gly-X-Ser-X-Gly). The involvement of these residues in the catalytic process of TgTCEA enzyme was clearly demonstrated by site-directed mutagenesis (Table IV); the first two Gly residues in the HGGG motif contribute to form the oxyanion hole, and Ser-235, Asp-327, and His-359 function as a catalytic triad. The fact that the Ser enzyme inhibitor phenylmethylsulfonyl fluoride markedly reduced the activity of the purified petal enzyme (Supplemental Table S1) also supports the function of Ser-235 as a catalytic residue. In addition, the pH optimum of the petal enzyme (pH 6.5–7.5) was in a range of the neutral to basic pH optima of carboxylesterase family enzymes (Souleyre et al., 2011), which is reasonable from the viewpoint of the catalytic mechanism; the catalytic triad requires deprotonated His, whose pKa (acidity index) is 6.0, to activate the catalytic Ser.
Despite these features shared with carboxylesterase family enzymes, the reaction catalyzed by TgTCEA is not the hydrolysis of 6-tuliposide A but rather an intramolecular transesterification to form a lactone, tulipalin A. One would expect that TgTCEA hydrolyzes 6-tuliposide A to form γ-hydroxy acid (4-hydroxy-2-methylenebutanoic acid) and that subsequent spontaneous lactonization of the γ-hydroxy acid occurs to form tulipalin A. However, the spontaneous lactonization of γ-hydroxy acid did not occur under the assay conditions used in this study (Fig. 4). In addition, the fact that terminating the enzyme reaction with methanol did not yield a γ-hydroxy acid as a product (Fig. 4) also excludes the possibility that enzymatically formed γ-hydroxy acid is spontaneously lactonized to tulipalin A under acidic conditions of the reaction termination. Moreover, in the course of our studies investigating the resistance mechanism of a tulip pathogenic fungus, Botrytis tulipae, to 6-tuliposide A, it was found that B. tulipae has an enzyme system to convert 6-tuliposide A into γ-hydroxy acid (Y. Kato, S. Ogita, and T. Nomura, unpublished data), where the product γ-hydroxy acid could be detected under the same assay conditions as used in this study. Given all these facts, we can safely conclude that TgTCEA catalyzes the conversion of 6-tuliposide A to tulipalin A without forming γ-hydroxy acid.
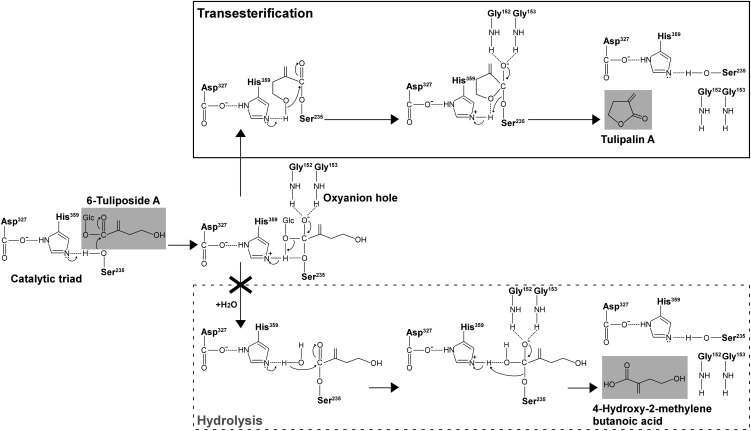
The putative reaction mechanism of TgTCEA enzyme begins with a nucleophilic attack by the catalytic Ser-235 on the carbonyl carbon of 6-tuliposide A, followed by the formation of a tetrahedral intermediate whose structure is stabilized by the oxyanion hole contributed by Gly-152 and Gly-153 (Fig. 7); then, after the elimination of Glc, an intramolecular nucleophilic attack by a terminal hydroxy group of the substrate occurs without incorporating water, leading to the formation of the five-membered ring structure of tulipalin A. If water is incorporated into the acyl-enzyme complex, the hydrolysis reaction that is catalyzed by typical carboxylesterases proceeds to release 4-hydroxy-2-methylenebutanoic acid (Fig. 7). TgTCEA only catalyzes the former intramolecular transesterification, but not the latter hydrolysis reaction, of its natural substrate 6-tuliposide A.
Figure 7.
Putative reaction mechanism of TgTCEA enzyme to convert 6-tuliposide A to tulipalin A. TgTCEA catalyzes the intramolecular transesterification reaction, where intramolecular nucleophilic attack by the terminal hydroxy group of substrate occurs without incorporating water after the elimination of Glc from the substrate. The hydrolysis reaction catalyzed by typical carboxylesterases proceeds via intermolecular nucleophilic attack by activated water, leading to 4-hydroxy-2-methylenebutanoic acid formation. Note that TgTCEA catalyzes a transesterification reaction but not a hydrolysis reaction. For the catalytic triad (Ser, Asp, and His) and oxyanion hole (Gly; Table IV; Supplemental Figure S1), only functional groups involved in catalysis are shown.
Transesterification reactions catalyzed by carboxylesterases have been reported for enzymes in other organisms, such as humans (Imai, 2006). When alcohol is present in abundance, an alcohol, instead of water, can attack the acyl-enzyme complex, generating an ester product. For example, transesterification of cocaine with ethanol and of fatty acyl-CoA with free cholesterol or ethanol have been known to occur (Becker et al., 1994; Brzezinski et al., 1994; Bora et al., 1996; Pennings et al., 2002; Bencharit et al., 2003). However, these reactions are kinetically controlled systems; the reaction equilibrium between hydrolysis and transesterification is affected by the ratio and nucleophilicity of water and alcohol; also, these reactions are intermolecular transesterifications. Considering the above facts, these reactions are different from the TgTCEA reaction that catalyzes the stoichiometric intramolecular transesterification of 6-tuliposide A to tulipalin A in water. To our knowledge, TgTCEA is the first identified member of lactone-forming carboxylesterases that catalyze intramolecular transesterification, and it should also be noted that it is a natural reaction, because TgTCEA was identified based on a functional approach following the activity toward its endogenous substrate, 6-tuliposide A, and not on shunt reactions using artificial substrates.
Carboxylesterases are found across all taxonomic kingdoms and have been well studied, especially in mammals, with respect to catalytic properties toward various ester-containing xenobiotics, fatty acids, and steroids as well as their reaction mechanisms based on crystallography (Imai, 2006; Hosokawa, 2008). However, the role of carboxylesterases in plants has not been well understood, and although many studies have reported enzymatic characterizations, most of them are based on reactions with artificial substrates (Gershater and Edwards, 2007). Therefore, the number of enzymes whose endogenous substrates have been well defined is quite limited, as can be seen from the phylogenetic tree of sequences related to TgTCEA (Supplemental Fig. S2), in which many sequences are for hypothetical proteins and even those annotated are based on sequence similarities. Among them, sequences whose natural functions have been elucidated are for 2-hydroxyisoflavanone dehydratases of G. echinata and G. max (Akashi et al., 2005) and the GID1 gibberellin receptor of rice (Ueguchi-Tanaka et al., 2005; Shimada et al., 2008). Although their sequences are related to carboxylesterases, their functions are not ester hydrolysis; the former enzyme has a Thr instead of Ser in its active site and catalyzes the 1,2-elimination of water from 2-hydroxyisoflavanone, and the latter has lost enzyme activity due to the replacement of an essential His in the catalytic triad with Val and thus functions as a gibberellin receptor. Identification of the natural function of TgTCEA as a lactone-forming carboxylesterase extends the understanding of the functional diversification of plant carboxylesterase family proteins.
Physiological Role of TgTCE-Mediated Tulipalin Formation
There are many examples of phytoanticipins being stored as inactive forms in intact plants and enzymatically converted to active forms upon pathogen infection and/or herbivore predation, as occurs with benzoxazinone-glucosides in gramineous plants (Niemeyer, 1988, 2009; Sicker et al., 2000), the cyanogenic glucoside dhurrin in sorghum (Sorghum bicolor; Poulton, 1990), and glucosinolates in brassicaceous plants (Bones and Rossiter, 2006). Recently, glucosidal intermediates for alkaloid biosynthesis have also been suggested to serve as a defense system (Nomura et al., 2008; Guirimand et al., 2010, 2011). In these systems, toxic aglycons are released from inactive glucosides by corresponding β-glucosidases and myrosinases existing in distinct subcellular compartments. Tuliposides are stored in large quantities in intact tulip tissues, whereas tulipalins are far less abundant than tuliposides and sometimes are barely detectable (Beijersbergen and Lemmers, 1972; Christensen and Kristiansen, 1999). It has been shown that upon infection the tuliposide content decreases whereas the tulipalin content increases (Beijersbergen and Lemmers, 1972; Schönbeck and Schroeder, 1972), indicating that tuliposides function as storage products for the biologically active tulipalins. The identification of TgTCEA here has defined the specific enzyme involved in releasing active tulipalin A. As described above, it is certain that a tuliposide B-converting enzyme also participates in this system to release tulipalin B. The good correlation of the great variation in the content ratio of 6-tuliposides A and B among the tissues and the enzyme activities for 6-tuliposides A and B in the crude extracts (Kato et al., 2009a, 2009b; Table I) suggested that the conversion of 6-tuliposides A and B into tulipalins A and B by enzymes specific to each of them prevails over nonenzymatic tulipalin formation. In this study, TgTCEA1 enzyme was found to be localized in plastids (Fig. 6; Supplemental Fig. S4). Because the amino acid sequence of the plastid-targeted signal of TgTCEA1 (residues 1–77) is highly similar to that of the corresponding region of TgTCEA2 (residues 1–74; Supplemental Fig. S1), TgTCEA2 should also be localized in plastids. It is most likely that Glc ester tuliposides, similar to many glucosidal phytoanticipins, are sequestered in vacuoles, whose acidic pH (Gout et al., 1992; Neuhaus, 2007) enables tuliposides to be stored without chemical decomposition into tulipalins in intact plants. Thus, now we can safely propose the cytological mechanism of the enzyme-mediated tuliposide-tulipalin conversion system: TgTCEA and its substrate 6-tuliposide A, which are compartmentalized in plastids and vacuoles, respectively, in intact cells, are mixed for enzyme reaction to release toxic tulipalin A upon cell disruption by infection and wounding. This must be the case for 6-tuliposide B and tuliposide B-converting enzyme as well. TgTCE-mediated tulipalin formation, which is analogous to the well-known glycoside-aglycon systems in other plants, is considered to be a central strategy of chemical defense in tulip.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
All experiments were performed using tulip (Tulipa gesneriana ‘Murasakizuisho’). The bulbs were purchased from the Toyama Bulb Growers Association in Japan. The plants were grown in a field under natural conditions and from which petals were collected for enzyme purification, preparations of 6-tuliposides A and B and tulipalin B, and cDNA cloning. Crude enzyme solution and RNAs were prepared from bulb scale and tulip plants at flowering stage, cultivated hydroponically in a greenhouse at 20°C/15°C (day/night) with a 12-h photoperiod.
Chemicals
6-Tuliposides A and B were purified from petals collected from 500 plants. After soaking in 10 L of 50% (v/v) methanol at 4°C for 5 d, plant residues were removed by filtration and the resulting extract was concentrated to a small volume by evaporation, resuspended in water, and filtered through filter paper. To the filtrate (500 mL) was added 130 mL of methanol to adjust the methanol concentration to 20% (v/v), and the solution was stirred with 500 mL of DIAION HP20 resin (Mitsubishi Chemical) for 2 h to remove hydrophobic impurities. The resins were then filtered out and rinsed with water, after which the combined filtrate and rinse was concentrated by evaporation, resuspended in a small volume of water, and applied to a charcoal column (charcoal activated for chromatography; 20–150 mesh; Nacalai Tesque) equilibrated with water. The column was eluted sequentially with 0%, 20%, 40%, and 60% (v/v) aqueous methanol. The 40% and 20% methanol fractions were subjected to further purification to yield 6-tuliposides A and B, respectively. Each fraction was evaporated to dryness, resuspended in a small volume of 0.01% (v/v) trifluoroacetic acid (TFA), and individually applied to an ODS column (Cosmosil 75C18-OPN; Nacalai Tesque) equilibrated with 0.01% (v/v) TFA. The column was eluted with 0%, 10%, and 20% (v/v) aqueous methanol containing 0.01% (v/v) TFA. Of the fractions with 0% and 10% methanol, those containing 6-tuliposide A were combined, concentrated by evaporation, and lyophilized to a white powder of pure 6-tuliposide A. Of the fractions with 0% methanol, those containing 6-tuliposide B were combined, concentrated by evaporation, and further subjected to preparative HPLC under the following conditions: column, TSKgel ODS-80Ts, 20 × 250 mm, 5 μm (Tosoh); solvent, 5% (v/v) methanol; flow rate, 5 mL min−1; detection, 255 nm. 6-Tuliposide B was collected, concentrated by evaporation, and lyophilized to a white powder of pure 6-tuliposide B. The purified 6-tuliposide A and B yields amounted to 2.1 and 1.2 g, respectively.
Tulipalin A (α-methylene-γ-butyrolactone) was purchased from Tokyo Chemical Industry. Tulipalin B was prepared from tulip petals via enzymatic conversion and selective extraction as described previously (Kato et al., 2009b). The identities of 6-tuliposides A and B and tulipalin B were confirmed by comparison of 1H- and 13C-NMR spectra recorded with a Bruker Avence II 400 spectrometer with the reported spectra (Tanaka and Yamashita, 1980; Christensen, 1999; Christensen and Kristiansen, 1999; Ohgiya and Nishiyama, 2004; Shigetomi et al., 2008, 2011). 4-Hydroxy-2-methylenebutanoic acid was prepared by alkaline hydrolysis of tulipalin A based on the method described by Morton and Thompson (1978). To an ice-cold stirred solution of tulipalin A (5.0 g, 51 mmol) in methanol (150 mL) was slowly added 38.3 mL of 4 m NaOH (153 mmol), and the mixture was stirred overnight at room temperature. The resulting mixture was concentrated in vacuo to its quarter volume, diluted with 200 mL of brine, and cooled to 0°C. The pH was then adjusted to 4.5 with ice-cold 1 m aqueous KHSO4, and the mixture was quickly extracted with ethyl acetate (5 × 100 mL) saturating with NaCl. The combined extracts were washed with brine, dried over anhydrous Na2SO4, and concentrated in vacuo to give 3.64 g (61.5%) of 4-hydroxy-2-methylenebutanoic acid as a colorless waxy solid. 1H-NMR (400 MHz, CDCl3) δ 7.26 (br, -CH2OH, -CO2H), 6.40 (d, 1H, J = 1.2 Hz, > C = CH2), 5.80 (dd, 1H, J = 1.2, 2.2 Hz, > C = CH2), 3.80 (t, 2H, J = 6.1 Hz, -CH2OH), 2.60 (dt, 2H, J = 0.96, 6.1 Hz, -CH2-C(= CH2) CO2H), 13C-NMR (100 MHz, CDCl3) δ 171.0 (CO2H), 136.7 (> C = CH2), 129.6 (> C = CH2), 61.6 (-CH2OH), 35.1 (-CH2-C(= CH2) CO2H).
Crude Enzyme Extraction
Plant tissues were frozen in liquid nitrogen and ground to a fine powder with mortar and pestle, followed by extraction with 0.1 m KPi buffer (pH 7.0). After centrifugation (15,000g, 10 min), the supernatant was subjected to a PD-10 column (GE Healthcare) equilibrated with the same buffer, and the eluate was used for the enzyme assay for 6-tuliposide A- and 6-tuliposide B-converting activities. Enzyme reaction and detection of reaction products were performed as described below (“Enzyme Assay”).
TCE Purification from Petals
All purification procedures were performed at 4°C, and KPi buffer (pH 6.0) was used throughout the procedure. Freeze-dried tulip petals (10 g) were crushed with mortar and pestle, suspended in 2 L of 10 mm buffer, stirred for 6 h, filtered through cheesecloth, and centrifuged (18,800g, 20 min) to remove plant residues. The supernatant was applied to a DEAE-Toyopearl column (2.5 × 10 cm; Tosoh) equilibrated with 10 mm buffer, and the enzyme was eluted with 100 mm buffer. The active fractions identified by enzyme assay using 6-tuliposide A as substrate as described below were collected and then brought to 30% ammonium sulfate saturation. The solution was applied to a Butyl-Toyopearl column (2.5 × 11 cm; Tosoh) equilibrated with 10 mm buffer containing 30% saturated ammonium sulfate. The active fractions were then eluted with a linear gradient of ammonium sulfate (30%–0% saturation) in 10 mm buffer, combined, and dialyzed. The resulting enzyme solution was loaded on a Gigapite column (1.5 × 11 cm; Seikagaku) equilibrated with 10 mm buffer. The enzyme was eluted with a linear gradient of buffer (10 mm–1 m), and the active fractions were collected and dialyzed. The following two purification steps were carried out using an ÄKTA Explorer System (GE Healthcare). The enzyme solution was applied to a Mono-Q HR5/5 column (GE Healthcare) equilibrated with 10 mm buffer and eluted with a linear gradient of 10 to 50 mm buffer containing 500 mm NaCl at 1 mL min−1. The active fractions were collected and concentrated by ultrafiltration. The enzyme solution was applied to a Superose 12 column (GE Healthcare) equilibrated with 100 mm buffer containing 200 mm NaCl, and the active fractions were eluted with the same buffer.
Protein Analysis
Protein concentrations were determined with a Protein Assay Kit (Bio-Rad) based on the method of Bradford (1976) using a bovine serum albumin standard or by measuring the A280. SDS-PAGE was carried out as described by Laemmli (1970) with an electrophoresis unit (Atto). The molecular mass of the enzyme was estimated as described previously (Kato et al., 2000), with the standards phosphorylase (97.4 kD), bovine serum albumin (66.2 kD), ovalbumin (45.0 kD), carbonic anhydrase (31.2 kD), and soybean trypsin inhibitor (21.5 kD); standards used for gel-filtration analysis on a TSKgel G3000 SWXL (Tosoh) were Glu dehydrogenase (290 kD), lactate dehydrogenase (142 kD), enolase (67.0 kD), adenylate kinase (32.0 kD), and cytochrome c (12.4 kD). The N-terminal amino acid sequence of the purified enzyme was analyzed with a Procise 494 HT System (Applied Biosystems). The internal peptide sequences were determined by digesting the enzyme with lysyl-endopeptidase, and after peptide mapping, two resulting proteolytic peptides were subjected to sequence analysis.
Enzyme Assay
A standard enzyme reaction using 6-tuliposide A or B as a substrate was performed in 50 mm KPi buffer (pH 6.5) containing 4 mm substrate and 5 to 10 μL of appropriately diluted enzyme in a total volume of 50 μL. After incubation at room temperature for 10 min, the reaction was terminated by adding an equal volume of 0.5 m phosphoric acid, followed by dilution with 400 μL of water. Reaction products were detected by HPLC analysis (column, TSKgel ODS-100V, 4.6 × 150 mm, 5 μm [Tosoh]; solvent, 10% [v/v] methanol in 10 mm phosphoric acid; flow rate, 0.65 mL min−1; detection, 208 nm). In assays using pNP-α-Glc, pNP-β-Glc, pNP-acetate, pNP-phosphate, or pNP-sulfate as a substrate, activity was measured by following p-nitrophenol formation at 405 nm.
The effects of pH and temperature on enzyme activity were examined by measuring the activity for 6-tuliposide A under standard assay conditions at various pH levels in several buffers (50 mm) at room temperature or at various temperatures in 50 mm KPi buffer (pH 6.5). The effects of pH and temperature on the enzyme stability were examined by measuring activity for 6-tuliposide A after preincubating the enzyme for 30 min in 100 mm buffer at various pH levels at room temperature or at various temperatures in 100 mm KPi buffer (pH 6.5). The buffers used were Gly-HCl (pH 3.0–4.0), sodium acetate-acetate (pH 3.5–6.0), KPi (pH 6.0–8.0), Tris-HCl (pH 7.5–9.0), and NaHCO3-Na2CO3 (pH 9.0–10.5).
The effects of various compounds on enzyme activity were examined by measuring activity for 6-tuliposide A under standard assay conditions in the presence of various concentrations of inhibitors, coenzymes, and metal ions (Supplemental Table S1).
Cloning of TgTCEA cDNAs
Total RNA was isolated from petals using the RNeasy Plant Mini Kit (Qiagen), followed by DNase I treatment and repurification. Poly(A) RNA was then purified from the total RNA using the MicroPoly(A)Purist Kit (Ambion). Next, cDNA was synthesized from the purified poly(A) RNA, using the SMART RACE cDNA Amplification Kit (Clontech), and subsequently used as a template for PCR with degenerate primers. The primers were designed based on the peptide sequences of N-terminal (TCEA-N-F1 and TCEA-N-F2) and internal (TCEA-Int-R1 and TCEA-Int-R2) regions of purified enzyme (Supplemental Table S2), of which TCEA-N-F2 and TCEA-Int-R2 were nested primers. The first PCR was performed as follows: 2 min at 94°C, followed by 30 cycles of amplification (30 s at 94°C, 1 min at 45°C, and 1 min at 72°C) in a 20-μL reaction mixture containing 2 ng of cDNA, 0.5 μm primers, 0.2 mm deoxyribonucleoside triphosphates (dNTPs), 1× reaction buffer, and 0.5 units of Blend Taq DNA polymerase (Toyobo). The second PCR was performed using 1 μL of diluted (1:50) first-PCR reaction mixture as the template under the same conditions as the first PCR. All possible primer combinations were tested for the first and second PCRs. The second PCR yielded products of around 100 bp with all primer combinations, of which the products amplified by the primer combinations TCEA-N-F1/TCEA-Int-R1 for the first PCR and TCEA-N-F2/TCEA-Int-R2 for the second PCR were cloned into a pT7Blue T-vector (Novagen) and sequenced, resulting in the isolation of a 74-bp fragment. To isolate its 5′ and 3′ missing parts, 5′ and 3′ RACE-PCR was performed using the cDNAs synthesized above as templates. The 5′ RACE was performed with the gene-specific primer TCEA-5RACE-R1 (Supplemental Table S2) and the adaptor primer as follows: 2 min at 94°C, followed by 30 cycles of amplification (30 s at 94°C, 30 s at 55°C, and 1 min at 72°C) in a 20-μL reaction mixture containing 2 ng of cDNA, 1× Universal Primer A Mix, 0.5 μm TCEA-5RACE-R1 primer, 0.2 mm dNTPs, 1× reaction buffer, and 0.5 units of Blend Taq DNA polymerase. The first PCR for the 3′ RACE was performed under the same conditions as for the 5′ RACE except for using the gene-specific primer TCEA-3RACE-F1 (Supplemental Table S2). Then, the second PCR was performed using 1 μL of diluted (1:50) first-PCR reaction mixture as the template in the same reaction mixture as the first PCR except for using 0.5 μm Nested Universal Primer A and 0.5 μm gene-specific primer TCEA-3RACE-F2 (Supplemental Table S2). The PCR conditions were as follows: 2 min at 94°C, followed by 30 cycles of amplification (30 s at 94°C and 1 min at 68°C). The 5′ and 3′ RACE products were then cloned into a pT7Blue T-vector and sequenced.
Finally, the entire coding region of the TgTCEA cDNA was obtained by PCR using petal cDNA (synthesized above for 5′ RACE) as the template with TCEA-RT-F and TCEA-RT-R primers (Supplemental Table S2), which were designed from 5′ and 3′ untranslated regions, respectively. The reaction was performed as follows: 2 min at 94°C, followed by 35 cycles of amplification (10 s at 98°C, 30 s at 55°C, and 1 min at 68°C) in a 50-μL reaction mixture containing 2 ng of cDNA, 0.5 μm primers, 0.2 mm dNTPs, 1.5 mm MgSO4, 1× reaction buffer (version 2), and 1 unit of KOD-Plus DNA polymerase (Toyobo). The PCR products were cloned into the HincII site of a pUC19 vector and sequenced to obtain TgTCEA1 and TgTCEA2.
Cloning of TgTCEA Genomic Sequences
Genomic DNA was isolated from leaves using the DNeasy Plant Mini Kit (Qiagen). Genomic PCR was performed using 50 ng of genomic DNA as the template as follows: 2 min at 94°C, followed by 35 cycles of amplification (10 s at 98°C, 30 s at 60°C, and 2 min at 68°C) in the same reaction mixture as for the RT-PCR to obtain full-length TgTCEA cDNAs. The PCR products were cloned into the HincII site of a pUC19 vector and sequenced.
Expression and Purification of Recombinant TgTCEA Enzymes
The full-length cDNAs (TgTCEA1 and TgTCEA2) were used as templates for PCR to prepare DNA fragments flanked by BamHI and XhoI restriction sites at the 5′ and 3′ ends, respectively; the primer sequences used are shown in Supplemental Table S2 (TCEA-F-BamHI and TCEA-R-XhoI). The amplified DNA fragments corresponding to mature TgTCEA enzymes, whose N termini start from ALDDEIVLDL (Supplemental Fig. S1), were digested with BamHI and XhoI and then cloned into a pET28a vector (Novagen). The resulting plasmids were introduced into the Escherichia coli strain BL21-CodonPlus (DE3)-RIL (Stratagene) for expression of the N-terminal His-tagged enzymes.
Recombinant E. coli was grown in 500 mL of Luria-Bertani medium containing 50 μg mL−1 kanamycin and 35 μg mL−1 chloramphenicol at 37°C until the A600 reached 0.6 to 0.8. After cooling, isopropyl β-d-1-thiogalactopyranoside was added at 1 mm and the culture was incubated at 18°C on an orbital shaker at 200 rpm for 18 h. The following operations were performed at 4°C. The cells were harvested (6,000g, 10 min) and resuspended in 20 mL of 50 mm HEPES buffer (pH 7.5). After sonication (30 pulses per 30 s at 100 W, six times) and centrifugation (15,000g, 30 min), the supernatant containing soluble protein was collected and subjected to purification with a TALON His-Tag Purification Resin (Clontech). The resin (1 mL of bed volume per 20 mL of supernatant), equilibrated with 50 mm HEPES buffer (pH 7.5) containing 200 mm NaCl, was mixed with the supernatant for 1 h, after which the resin was washed with equilibration buffer and the recombinant enzyme was eluted with 5 mL of 50 mm HEPES buffer (pH 7.5) containing 200 mm NaCl and 200 mm imidazole. The eluate was then subjected to gel-filtration chromatography on a Superdex 200 (GE Healthcare) equilibrated with 50 mm HEPES buffer (pH 7.5) containing 150 mm NaCl at 0.5 mL min−1.
Recombinant Enzyme Characterization
Protein quantification and SDS-PAGE analysis were performed as described above. The molecular mass of native recombinant enzyme was estimated by gel-filtration analysis as described above. Enzyme reactions for the determination of kinetic parameters for 6-tuliposides A and B were performed in 50 mm KPi buffer (pH 6.5) at room temperature for 10 min in a total volume of 50 and 25 μL for 6-tuliposides A and B, respectively. The amount of enzyme was adjusted to allow the reaction to proceed linearly, and the reaction termination and quantification of products by HPLC were performed as described above. Kinetic parameters were calculated by nonlinear fitting of the data to the Michaelis-Menten equation using SigmaPlot 2001 (Systat Software).
Site-Directed Mutagenesis of TgTCEA1
Mutated DNA fragments for the expression of TgTCEA1 mutants were prepared by PCR-mediated overlap extension. The restriction sites and the coding region to be expressed were the same as those for the intact TgTCEA1 as described above. The sequences of the mutagenic PCR primers are listed in Supplemental Table S2. Construction of the pET28a expression vector, expression in E. coli, and protein purification were performed as described above.
Transcript Analysis
Total RNA was isolated from bulb scale and tissues (root, stem, leaf, petal, stamen, and pistil) of hydroponically cultivated plants using the RNeasy Plant Mini Kit (except for bulb scale and pistil). RNAs of bulb scale and pistil were isolated by an SDS-phenol method. After the DNase I treatment and repurification, cDNA was synthesized with a SuperScript III reverse transcriptase (1 μg of RNA in 20 μL of reaction mixture; Invitrogen). PCR was performed as follows: 2 min at 94°C, followed by 35 cycles of amplification (30 s at 96°C, 30 s at 60°C, and 1 min at 72°C) in a 20-μL reaction mixture containing 2 ng of cDNA, 0.5 μm primers, 0.2 mm dNTPs, 1× reaction buffer, and 0.5 units of Blend Taq DNA polymerase. TgTCEA cDNAs were amplified with the primers TCEA-RT-F and TCEA-RT-R (Supplemental Table S2), which amplify both TgTCEA1 and TgTCEA2 genes. As an endogenous standard, the tulip GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE (GAPDH; GenBank accession no. AB500108) was amplified with the primers GAPDH-F and GAPDH-R (Supplemental Table S2).
Quantitative RT-PCR for the transcript level of each of the TgTCEA1 and TgTCEA2 genes was performed with the primer sets specific to each gene (Supplemental Table S2). PCR was performed in triplicate on the 7500 Real-Time PCR System (Applied Biosystems) under the following conditions: 30 s at 95°C, followed by 40 cycles of amplification (5 s at 95°C and 34 s at 64°C) in a 10-μL reaction mixture containing 10 ng of template cDNA, 0.2 μm primers, 0.2 μL of ROX Reference Dye II (Takara Bio), and 5 μL of SYBR Premix Ex Taq (Takara Bio). A standard curve was made for every experiment, with the dilution series of the known quantities of a plasmid having each of the target cDNAs as template. The specificity of the amplification was verified by a melt-curve analysis and agarose-gel electrophoresis at the end of each PCR.
Subcellular Localization Analysis of TgTCEA1
The coding sequences for the putative transit peptide at the N terminus of TgTCEA1 (residues 1–77) and for the manure polypeptides of TgTCEA1 (residues 78–385) were amplified by PCR using TgTCEA1/pUC19 plasmid as template with primer sets TCEA-rep-F1/TCEA-rep77-R and TCEA-rep-F2/TCEA-rep-R1 (Supplemental Table S2), respectively. The resulting fragments were inserted into the NcoI site of the GFP expression plasmid PCaMV35SΩ-sGFP(S65T)-NOS3′ (Chiu et al., 1996) with the In-Fusion HD Cloning Kit (Clontech) to express C-terminal GFP-fused proteins, TgTCEA1(TP77)-GFP and TgTCEA1(mature)-GFP. DNA-coated gold particles (0.6 μm; Bio-Rad) were prepared and bombarded into a small piece (approximately 2 × 2 cm) of onion (Allium cepa) with the PDS1000 He Biolistic Particle Delivery System (Bio-Rad) according to the manufacturer’s instructions. The bombarded onion was wrapped with a wet paper towel and incubated at room temperature overnight in a petri dish. PCaMV35SΩ-sGFP(S65T)-NOS3′ without insert and pWxTP-DsRed (Kitajima et al., 2009) were also bombarded as controls for cytosolic and plastidial protein localization, respectively. After the incubation, the peeled epidermis was analyzed using a laser scanning microscope (LSM510 META; Zeiss) for the detection of GFP and DsRed fluorescence.
The nucleotide sequences reported in this paper have been submitted to the GenBank/EMBL/DDBJ databases with accession numbers AB569208 to AB569213.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Alignment of amino acid sequences of TgTCEA1 and TgTCEA2.
Supplemental Figure S2. Unrooted phylogenetic tree of TgTCEA enzymes with selected members of plant α/β-hydrolase fold superfamily proteins.
Supplemental Figure S3. Transcript analysis of TgTCEA genes in tulip tissues by RT-PCR with various primer sets.
Supplemental Figure S4. Subcellular localization of GFPs fused with a deletion series of the plastid-targeted signal of TgTCEA1 in onion epidermal cells.
Supplemental Table S1. Effects of various compounds on the activity of TCE purified from tulip petals.
Supplemental Table S2. Primer sequences used in this study.
Supplementary Material
Acknowledgments
We thank Dr. Kazuaki Shoji (Toyama Prefectural Agricultural, Forestry, and Fisheries Research Center) for the vernalization treatment of tulip bulbs. We are grateful to Dr. Toshiaki Mitsui (Niigata University) for the gift of the WxTP-DsRed plasmid and to Dr. Yasuo Niwa (University of Shizuoka) for providing the PCaMV35SΩ-sGFP(S65T)-NOS3′ plasmid.
Glossary
- TCE
tuliposide-converting enzyme
- pNP
p-nitrophenyl
- KPi
potassium phosphate
- RT
reverse transcription
- TFA
trifluoroacetic acid
- dNTPs
deoxyribonucleotide triphosphate
References
- Akashi T, Aoki T, Ayabe S. (2005) Molecular and biochemical characterization of 2-hydroxyisoflavanone dehydratase: involvement of carboxylesterase-like proteins in leguminous isoflavone biosynthesis. Plant Physiol 137: 882–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker A, Böttcher A, Lackner KJ, Fehringer P, Notka F, Aslanidis C, Schmitz G. (1994) Purification, cloning, and expression of a human enzyme with acyl coenzyme A:cholesterol acyltransferase activity, which is identical to liver carboxylesterase. Arterioscler Thromb 14: 1346–1355 [DOI] [PubMed] [Google Scholar]
- Beijersbergen JCM. (1972) A method for determination of tulipalin A and B concentrations in crude extracts of tulip tissues. Recl Trav Chim Pays Bas 91: 1193–1200 [Google Scholar]
- Beijersbergen JCM, Lemmers CBG. (1972) Enzymic and non-enzymic liberation of tulipalin A (α-methylene butyrolactone) in extracts of tulip. Physiol Plant Pathol 2: 265–270 [Google Scholar]
- Bencharit S, Morton CL, Xue Y, Potter PM, Redinbo MR. (2003) Structural basis of heroin and cocaine metabolism by a promiscuous human drug-processing enzyme. Nat Struct Biol 10: 349–356 [DOI] [PubMed] [Google Scholar]
- Bergman BHH. (1966) Presence of a substance in the white skin of young tulip bulbs which inhibits growth of Fusarium oxysporum. Neth J Plant Pathol 72: 222–230 [Google Scholar]
- Bergman BHH, Beijersbergen JCM. (1968) A fungitoxic substance extracted from tulips and its possible role as a protectant against disease. Neth J Plant Pathol 74: 157–162 [Google Scholar]
- Bergman BHH, Beijersbergen JCM, Overeem JC, Sijpesteijn AK. (1967) Isolation and identification of α-methylenebutyrolactone, a fungitoxic substance from tulips. Recl Trav Chim Pays Bas 86: 709–714 [Google Scholar]
- Bones AM, Rossiter JT. (2006) The enzymic and chemically induced decomposition of glucosinolates. Phytochemistry 67: 1053–1067 [DOI] [PubMed] [Google Scholar]
- Bora PS, Guruge BL, Miller DD, Chaitman BR, Ruyle MS. (1996) Purification and characterization of human heart fatty acid ethyl ester synthase/carboxylesterase. J Mol Cell Cardiol 28: 2027–2032 [DOI] [PubMed] [Google Scholar]
- Bradford MM. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Brzezinski MR, Abraham TL, Stone CL, Dean RA, Bosron WF. (1994) Purification and characterization of a human liver cocaine carboxylesterase that catalyzes the production of benzoylecgonine and the formation of cocaethylene from alcohol and cocaine. Biochem Pharmacol 48: 1747–1755 [DOI] [PubMed] [Google Scholar]
- Chiu W-I, Niwa Y, Zeng W, Hirano T, Kobayashi H, Sheen J. (1996) Engineered GFP as a vital reporter in plants. Curr Biol 6: 325–330 [DOI] [PubMed] [Google Scholar]
- Christensen LP. (1995a) Tuliposides from Alstroemeria revoluta. Phytochemistry 38: 1371–1373 [Google Scholar]
- Christensen LP. (1995b) A further tuliposide from Alstroemeria revoluta. Phytochemistry 40: 49–51 [Google Scholar]
- Christensen LP. (1999) Tuliposides from Tulipa sylvestris and T. turkestanica. Phytochemistry 51: 969–974 [Google Scholar]
- Christensen LP, Kristiansen K. (1995) Isolation and quantification of a new tuliposide (tuliposide D) by HPLC in Alstoroemeria. Contact Dermat 33: 188–192 [DOI] [PubMed] [Google Scholar]
- Christensen LP, Kristiansen K. (1999) Isolation and quantification of tuliposides and tulipalins in tulips (Tulipa) by high-performance liquid chromatography. Contact Dermat 40: 300–309 [DOI] [PubMed] [Google Scholar]
- Dingwall C, Laskey RA. (1986) Protein import into the cell nucleus. Annu Rev Cell Biol 2: 367–390 [DOI] [PubMed] [Google Scholar]
- Gershater MC, Edwards R. (2007) Regulating biological activity in plants with carboxylesterases. Plant Sci 173: 579–588 [Google Scholar]
- Gout E, Bligny R, Douce R. (1992) Regulation of intracellular pH values in higher plant cells: carbon-13 and phosphorus-31 nuclear magnetic resonance studies. J Biol Chem 267: 13903–13909 [PubMed] [Google Scholar]
- Guirimand G, Courdavault V, Lanoue A, Mahroug S, Guihur A, Blanc N, Giglioli-Guivarc’h N, St-Pierre B, Burlat V. (2010) Strictosidine activation in Apocynaceae: towards a “nuclear time bomb”? BMC Plant Biol 10: 182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guirimand G, Guihur A, Ginis O, Poutrain P, Héricourt F, Oudin A, Lanoue A, St-Pierre B, Burlat V, Courdavault V. (2011) The subcellular organization of strictosidine biosynthesis in Catharanthus roseus epidermis highlights several trans-tonoplast translocations of intermediate metabolites. FEBS J 278: 749–763 [DOI] [PubMed] [Google Scholar]
- Hosokawa M. (2008) Structure and catalytic properties of carboxylesterase isozymes involved in metabolic activation of prodrugs. Molecules 13: 412–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ileperuma NR, Marshall SD, Squire CJ, Baker HM, Oakeshott JG, Russell RJ, Plummer KM, Newcomb RD, Baker EN. (2007) High-resolution crystal structure of plant carboxylesterase AeCXE1, from Actinidia eriantha, and its complex with a high-affinity inhibitor paraoxon. Biochemistry 46: 1851–1859 [DOI] [PubMed] [Google Scholar]
- Imai T. (2006) Human carboxylesterase isozymes: catalytic properties and rational drug design. Drug Metab Pharmacokinet 21: 173–185 [DOI] [PubMed] [Google Scholar]
- Kato Y, Nakamura K, Sakiyama H, Mayhew SG, Asano Y. (2000) Novel heme-containing lyase, phenylacetaldoxime dehydratase from Bacillus sp. strain OxB-1: purification, characterization, and molecular cloning of the gene. Biochemistry 39: 800–809 [DOI] [PubMed] [Google Scholar]
- Kato Y, Shoji K, Ubukata M, Shigetomi K, Sato Y, Nakajima N, Ogita S. (2009a) Purification and characterization of a tuliposide-converting enzyme from bulbs of Tulipa gesneriana. Biosci Biotechnol Biochem 73: 1895–1897 [DOI] [PubMed] [Google Scholar]
- Kato Y, Yoshida H, Shoji K, Sato Y, Nakajima N, Ogita S. (2009b) A facile method for the preparation of α-methylene-γ-butyrolactones from tulip tissues by enzyme-mediated conversion. Tetrahedron Lett 50: 4751–4753 [Google Scholar]
- Kim C-S, Datta PK, Hara T, Itoh E, Horiike M. (1999) Precursor of α-methylene-γ-butyrolactone involved in the insecticidal activity of thunberg spiraea, Spiraea thunbergii. Biosci Biotechnol Biochem 63: 152–154 [DOI] [PubMed] [Google Scholar]
- Kim C-S, Hara T, Datta PK, Itoh E, Horiike M. (1998) Insecticidal component in thunberg spiraea, Spiraea thunbergii, against Thrips palmi. Biosci Biotechnol Biochem 62: 1546–1549 [DOI] [PubMed] [Google Scholar]
- Kitajima A, Asatsuma S, Okada H, Hamada Y, Kaneko K, Nanjo Y, Kawagoe Y, Toyooka K, Matsuoka K, Takeuchi M, et al. (2009) The rice α-amylase glycoprotein is targeted from the Golgi apparatus through the secretory pathway to the plastids. Plant Cell 21: 2844–2858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- Mendgen T, Scholz T, Klein CD. (2010) Structure-activity relationships of tulipalines, tuliposides, and related compounds as inhibitors of MurA. Bioorg Med Chem Lett 20: 5757–5762 [DOI] [PubMed] [Google Scholar]
- Morton DR, Thompson JL. (1978) Total synthesis of 3-oxa-4,5,6-trinor-3,7-inter-m-phenylene prostaglandins. 2. Conjugate addition approach. J Org Chem 43: 2102–2106 [Google Scholar]
- Neuhaus HE. (2007) Transport of primary metabolites across the plant vacuolar membrane. FEBS Lett 581: 2223–2226 [DOI] [PubMed] [Google Scholar]
- Niemeyer HM. (1988) Hydroxamic acids (4-hydroxy-1,4-benzoxazin-3-ones): defence chemicals in the Gramineae. Phytochemistry 27: 3349–3358 [Google Scholar]
- Niemeyer HM. (2009) Hydroxamic acids derived from 2-hydroxy-2H-1,4-benzoxazin-3(4H)-one: key defense chemicals of cereals. J Agric Food Chem 57: 1677–1696 [DOI] [PubMed] [Google Scholar]
- Nomura T, Quesada AL, Kutchan TM. (2008) The new β-D-glucosidase in terpenoid-isoquinoline alkaloid biosynthesis in Psychotria ipecacuanha. J Biol Chem 283: 34650–34659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohgiya T, Nishiyama S. (2004) Synthesis of (−)-tulipalin B utilizing 2-bromo-1-alkanes conveniently synthesized from the 2-O-substituted 1,2-dibromoalkane system by regioselective elimination. Heterocycles 63: 2349–2354 [Google Scholar]
- Ollis DL, Cheah E, Cygler M, Dijkstra B, Frolow F, Franken SM, Harel M, Remington SJ, Silman I, Schrag J, et al. (1992) The α/β hydrolase fold. Protein Eng 5: 197–211 [DOI] [PubMed] [Google Scholar]
- Pennings EJ, Leccese AP, Wolff FA. (2002) Effects of concurrent use of alcohol and cocaine. Addiction 97: 773–783 [DOI] [PubMed] [Google Scholar]
- Poulton JE. (1990) Cyanogenesis in plants. Plant Physiol 94: 401–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schönbeck F, Schroeder C. (1972) Role of antimicrobial substances (tuliposides) in tulips attacked by Botrytis spp. Physiol Plant Pathol 2: 91–99 [Google Scholar]
- Shigetomi K, Kishimoto T, Shoji K, Ubukata M. (2008) First total synthesis of 6-tuliposide B. Tetrahedron Asymmetry 19: 1444–1449 [Google Scholar]
- Shigetomi K, Omoto S, Kato Y, Ubukata M. (2011) Asymmetric total synthesis of 6-tuliposide B and its biological activities against tulip pathogenic fungi. Biosci Biotechnol Biochem 75: 718–722 [DOI] [PubMed] [Google Scholar]
- Shigetomi K, Shoji K, Mitsuhashi S, Ubukata M. (2010) The antibacterial properties of 6-tuliposide B: synthesis of 6-tuliposide B analogues and structure-activity relationship. Phytochemistry 71: 312–324 [DOI] [PubMed] [Google Scholar]
- Shimada A, Ueguchi-Tanaka M, Nakatsu T, Nakajima M, Naoe Y, Ohmiya H, Kato H, Matsuoka M. (2008) Structural basis for gibberellin recognition by its receptor GID1. Nature 456: 520–523 [DOI] [PubMed] [Google Scholar]
- Shoji K, Ubukata M, Momonoi K, Tsuji T, Morimatsu T. (2005) Anther-specific production of antimicrobial tuliposide B in tulips. J Jpn Soc Hortic Sci 74: 469–475 [Google Scholar]
- Sicker D, Frey M, Schulz M, Gierl A. (2000) Role of natural benzoxazinones in the survival strategy of plants. Int Rev Cytol 198: 319–346 [DOI] [PubMed] [Google Scholar]
- Slob A, Jekel B, Jong BD, Schlatmann E. (1975) On the occurrence of tuliposides in the Liliiflorae. Phytochemistry 14: 1997–2005 [Google Scholar]
- Slob A, Varekamp HQ. (1977) Tuliposide contents of tulip (Tulipa) species and cultivars during the flowering stage. Proc K Ned Akad Wet C Biol Med Sci 80: 201–211 [Google Scholar]
- Souleyre EJF, Marshall SDG, Oakeshott JG, Russell RJ, Plummer KM, Newcomb RD. (2011) Biochemical characterisation of MdCXE1, a carboxylesterase from apple that is expressed during fruit ripening. Phytochemistry 72: 564–571 [DOI] [PubMed] [Google Scholar]
- Tanaka A, Yamashita K. (1980) Synthesis of (S)-(+)-methyl β,γ-dihydroxy-α-methylenebutyrate and (S)-(-)-tulipalin B. Agric Biol Chem 44: 199–202 [Google Scholar]
- Tschesche R, Kämmerer F-J, Wulff G. (1969) Über die struktur der antibiotisch aktiven substanzen der tulpe (Tulipa gesneriana L.). Chem Ber 102: 2057–2071 [Google Scholar]
- Tschesche R, Kämmerer F-J, Wulff G, Schönbeck F. (1968) [On the antibiotically active substances of the tulip (Tulipa gesneriana)]. Tetrahedron Lett 6: 701–706 [DOI] [PubMed] [Google Scholar]
- Ueguchi-Tanaka M, Ashikari M, Nakajima M, Itoh H, Katoh E, Kobayashi M, Chow TY, Hsing YI, Kitano H, Yamaguchi I, et al (2005) GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature 437: 693–698 [DOI] [PubMed] [Google Scholar]
- van Rossum MWPC, Alberda M, van der Plas LHW. (1998) Tulipaline and tuliposide in cultured explants of tulip bulb scales. Phytochemistry 49: 723–729 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.