Abstract
Plant thioredoxins (Trxs) constitute a complex family of thiol oxidoreductases generally sharing a WCGPC active site sequence. Some recently identified plant Trxs (Clot, Trx-like1 and -2, Trx-lilium1, -2, and -3) display atypical active site sequences with altered residues between the two conserved cysteines. The transcript expression patterns, subcellular localizations, and biochemical properties of some representative poplar (Populus spp.) isoforms were investigated. Measurements of transcript levels for the 10 members in poplar organs indicate that most genes are constitutively expressed. Using transient expression of green fluorescent protein fusions, Clot and Trx-like1 were found to be mainly cytosolic, whereas Trx-like2.1 was located in plastids. All soluble recombinant proteins, except Clot, exhibited insulin reductase activity, although with variable efficiencies. Whereas Trx-like2.1 and Trx-lilium2.2 were efficiently regenerated both by NADPH-Trx reductase and glutathione, none of the proteins were reduced by the ferredoxin-Trx reductase. Only Trx-like2.1 supports the activity of plastidial thiol peroxidases and methionine sulfoxide reductases employing a single cysteine residue for catalysis and using a glutathione recycling system. The second active site cysteine of Trx-like2.1 is dispensable for this reaction, indicating that the protein possesses a glutaredoxin-like activity. Interestingly, the Trx-like2.1 active site replacement, from WCRKC to WCGPC, suppresses its capacity to use glutathione as a reductant but is sufficient to allow the regeneration of target proteins employing two cysteines for catalysis, indicating that the nature of the residues composing the active site sequence is crucial for substrate selectivity/recognition. This study provides another example of the cross talk existing between the glutathione/glutaredoxin and Trx-dependent pathways.
In proteins, Cys residues are prone to oxidation, leading, for example, to the formation of sulfenic acids or nitrosothiols upon reaction with reactive oxygen or nitrogen species. Overoxidation can be prevented through the formation of disulfide bridges either internally or between two distinct peptides or with the Cys of glutathione (GSH), a process named glutathionylation (Zaffagnini et al., 2012b). All these oxidized forms are reversible and reduced back to Cys residues by thioredoxins (Trxs) or glutaredoxins (Grxs), two types of thiol oxidoreductases displaying redox active Cys residues in a CxxC or CxxS active site motif (Rouhier et al., 2008; Zaffagnini et al., 2012b). These dithiol-disulfide exchange reactions are thus essential for many processes, regulating or regenerating the activity of many proteins. In most living organisms, Trxs are thought to be the major disulfide reductases, whereas Grxs preferentially catalyze deglutathionylation reactions. The reduction of disulfide bonds is a two-step process where the first Cys of the active site, named catalytic Cys, performs a nucleophilic attack on the target disulfide, forming a transient heterodisulfide, and the second Cys, also referred to as resolving Cys, then reduces this intermolecular disulfide (Collet and Messens, 2010). A few years ago, Trxs were defined as small proteins of 10 to 14 kD with a specific WCGPC active site signature. However, the recent sequencing of many genomes, in particular from plant species, helped identify many variations in the size of the proteins, in their domain organization, and/or in their active site sequence (Chibani et al., 2009). The conventional Trxs have generally a low midpoint redox potential compared with other redoxins, between −270 and −330 mV at pH 7.0, and an N-terminal Cys with a pKa around 7 (Collin et al., 2003; Ren et al., 2009; Collet and Messens, 2010). Site-directed mutagenesis established that the active site Pro is a key residue determining the reducing property of Trx and that the Gly maintains the conformation of the active site (Holmgren, 1985; Eklund et al., 1991; Roos et al., 2007). Besides, it was shown that the Trp preceding the catalytic Cys contributes to Trx stability. Indeed, the altered redox potential and Cys pKa value observed for a W28A variant of a Staphylococcus aureus Trx was attributed to partial protein unfolding (Roos et al., 2010). This Trp is also part of the contact area involved in substrate recognition (Menchise et al., 2000). A few other residues have also been demonstrated to be important both for the reactivity and redox properties of Trxs. In Escherichia coli Trx1, Asp-27 and Lys-57 contribute to the lowering of the Cys thiol group pKa (Dyson et al., 1997). In addition, the residue (usually an Ile) positioned just before the conserved cis-Pro found in all members of the Trx superfamily downstream of the active site and facing it in three-dimensional structures is also very important, because its mutation strongly modifies the redox potential, the Cys pKa, and substrate recognition (Ren et al., 2009).
Photosynthetic organisms contain a large number of Trx isoforms (approximately 40), as compared with nonphotosynthetic organisms such as E. coli, Saccharomyces cerevisiae, and humans. The classical Trxs with WC[G/P]PC active sites, formerly studied, are located in several subcellular compartments including the cytosol (h-type Trxs, tetratricopeptide domain-containing Trxs), the nucleus (h-type Trxs and nucleoredoxin), mitochondria (h- and o-type Trxs), and plastids (m-, f-, x- y-, and z-type Trxs, NADPH thioredoxin reductase chloroplastic (NTRc); Meyer et al., 2005; Chibani et al., 2009; Meng et al., 2010). The subcellular localizations of several Trxs, especially of members of the nucleoredoxin and Trx h classes, are only based on predictions and have not yet been experimentally confirmed. The plastids constitute the subcellular compartment possessing the highest number of Trxs. Trxs m and f are primarily involved in the light-dependent regulation of key carbon metabolism enzymes (Lemaire et al., 2007). Trxs x, y, NTRc, and CDSP32 (for chloroplast drought-induced protein of 32 kD) are likely involved in stress responses serving as reductants for antioxidant enzymes such as thiol peroxidases (Tpxs) and methionine sulfoxide reductases (MSRs; Vieira Dos Santos and Rey, 2006). Trx z is implicated in plastid development (Arsova et al., 2010). Most plastidial Trxs with a single domain are likely maintained reduced by light through the ferredoxin (Fdx)/ferredoxin-thioredoxin reductase (FTR) system, whereas Trxs h and o, usually found in the cytosol and in mitochondria, are reduced by NADPH via NADPH-thioredoxin reductase (NTR; Schürmann and Jacquot, 2000; Laloi et al., 2001; Gelhaye et al., 2004; Balmer et al., 2006; Reichheld et al., 2007; Schürmann and Buchanan, 2008; Chibani et al., 2011). The peculiar plastidial NTRc, formed by a fusion between an NTR and a Trx module, is dependent on NADPH (Pérez-Ruiz et al., 2006). It is also worth mentioning that a plant Trx h isoform is specifically reduced by a glutathione reductase (GR)/GSH/Grx system instead of the classical Trx reductase pathway (Gelhaye et al., 2004; Koh et al., 2008).
More recently, new classes of atypical Trxs named Clot, Trx-like, and Trx-lilium have been identified (Meyer et al., 2005). Higher plants generally contain one Clot isoform, two to four Trx-like isoforms, and four to five isoforms of Trx-lilium (Chibani et al., 2009). These proteins are specific to photosynthetic eukaryotes, as these classes are also present in some algae but not in cyanobacteria. Although sharing a clear homology with other Trxs, they possess very different active site sequences: Clot exhibits a WCPDC active site, Trx-like proteins have WCRVC or WCRKC active sites, whereas Trxs-lilium most often possess GCGGC, SCGSC, or WCASC active sites. The Trxs-lilium and the so-called Trx-like2 (WCRKC active site) from Arabidopsis (Arabidopsis thaliana) are localized in chloroplasts, most likely in the stromal fraction (Cain et al., 2009; Dangoor et al., 2009). However, little biochemical information is available on these atypical Trxs. Some Trxs-lilium, although having a less negative redox potential compared with other plastidial Trxs, could constitute good reductants of 2-Cys PrxA but are poor activators of NADP-malate dehydrogenase (MDH; Dangoor et al., 2009).
In this study, we have investigated the transcript expression patterns of the poplar (Populus trichocarpa) genes encoding Clot, Trxs-like, and Trxs-lilium in various plant organs and have experimentally determined the subcellular localization of the isoforms, which has not been previously performed. On the other hand, selected members, as well as protein variants constructed by site-directed mutagenesis, were produced as recombinant proteins and purified. Using a combination of biochemical approaches and activity assays, the specificity of these Trxs versus different known target proteins (NADP-MDH, Tpxs [either glutathione peroxidases {Gpxs} or peroxiredoxins {Prxs}], and MSR) or versus the possible reducing systems (NADPH/NTR, NADPH/NTRc, NADPH/FNR/Fdx/FTR, or NADPH/GR/GSH) was tested.
RESULTS
Sequence Characteristics and Subcellular Localization of Clot, Trx-like1, and Trx-like2.1
Previous phylogenetic studies on photosynthetic and nonphotosynthetic organisms revealed the presence of a large number of Trxs with undefined roles. For instance, the poplar genome contains 45 Trx-related sequences and four Trx reductases (Chibani et al., 2009). Besides classical Trxs, poplar contains one, four, and five isoforms of Clot, Trx-like, and Trx-lilium, respectively. Clot proteins (WCPDC active site) constitute a single phylogenetic subgroup, whereas Trx-like proteins are distributed in two distinct subgroups called Trx-like1 (WCRVC active site) and Trx-like2 (WCRKC active site). Trx-lilium proteins are split in three subgroups named Trx-lilium1, Trx-lilium2, and Trx-lilium3 (usually GCGGC, WCASC and SCGSC active sites; respectively; Supplemental Fig. S1; Chibani et al., 2009).
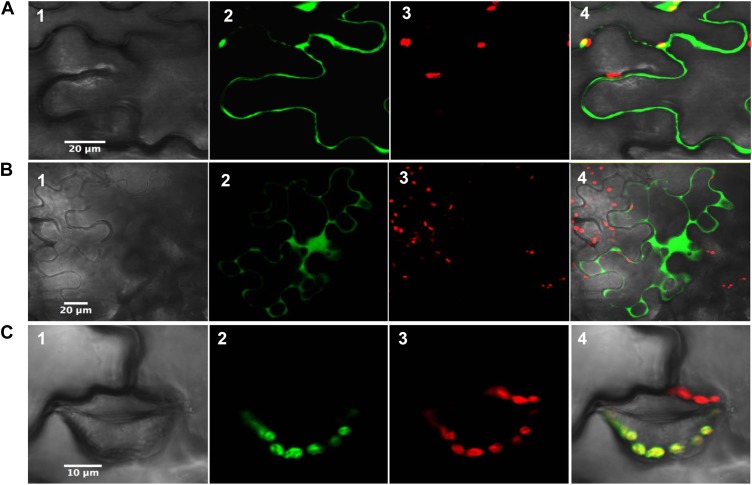
Although the subcellular localization has been addressed previously for most Arabidopsis orthologs, some doubt persisted for a few isoforms (Cain et al., 2009; Dangoor et al., 2009). The analysis of protein targeting by different prediction programs suggested a cytosolic localization for poplar Clot, Trx-like1, and also Trx-like2.1, despite the presence of an N-terminal extension in Trx-like2.1. Their subcellular localization was assessed by fusing the whole protein to the N terminus of GFP. As expected from the lack of an N-terminal transit peptide in their sequences, Clot and Trx-like1 were found in the cytosol (Fig. 1, A and B). In addition, fluorescence was also observed in the nucleus. Whether this signal was due to transient overexpression or whether it has a physiological significance will have to be further investigated. On the other hand, Trx-like2.1 was clearly targeted to the chloroplast (Fig. 1C).
Figure 1.
Subcellular localization of poplar Clot (A), Trx-like1 (B), and Trx-like2.1 (C) in tobacco cells. The entire open reading frames were fused upstream of the 5′ end of a GFP coding sequence. Panels 1, Cells under visible light; panels 2, fluorescence of the GFP construct; panels 3, fluorescence of chlorophyll (red); panels 4, merged images.
Expression Analysis of Atypical Trx Genes in Different Poplar Organs
In order to gain information about the transcript expression patterns, reverse transcription (RT)-PCR experiments have been performed using RNA extracted from different poplar organs (roots, stems, young leaves, mature leaves, petioles, stamens, female catkins, and fruits; Fig. 2). With the exceptions of Trx-like1 and Trx-lilium1.2, which were not expressed or were below the detection level in roots and stamens and in stems and stamens, respectively, transcripts have been detected in all organs for all other Trxs tested. The transcripts of Clot, Trx-lilium1.1, Trx-lilium2.1, Trx-lilium2.2, and Trx-lilium3 were present in all tissues, indicating that they are constitutively expressed (Fig. 2). On the other hand, some transcript variations were observed for members of the Trx-like subgroup. For example, Trx-like2.1 and Trx-like2.3 transcripts were only barely detected in fruits and petioles, Trx-like2.1 being also weakly expressed in roots and young leaves. Trx-like2.2 was also less expressed in roots, petioles, and stems. The fact that Trx-like2.1 was more expressed in photosynthetic organs than in nonphotosynthetic organs is consistent with its plastidial localization.
Figure 2.
Transcript expression profiles in poplar organs. RT-PCR was performed from total RNAs extracted from roots (R), stems (S), young leaves (Yl), mature leaves (Ml), petioles (P), stamens (St), female catkins (Fc), and fruits (Fr). Trx h1 was used as a control, as it has been shown to be constitutively expressed in all organs tested (Rouhier et al., 2006). For these experiments, 35 PCR cycles were used to amplify each Trx transcript from cDNA obtained from 1 µg of total RNA.
Reductase Activity of Atypical Trxs
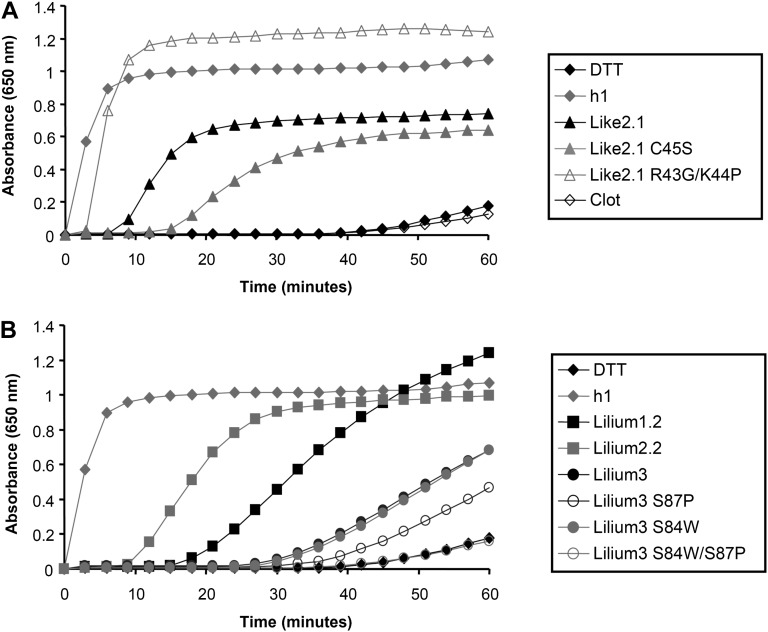
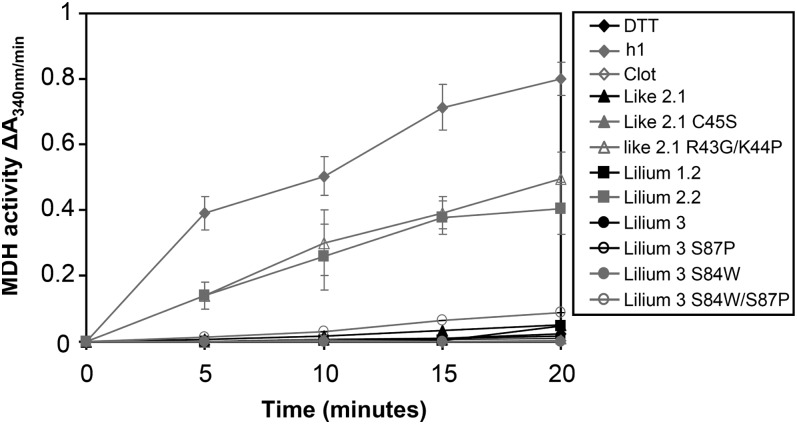
To characterize their biochemical properties, the mature forms (i.e. devoid of N-terminal targeting sequences when present) of all these proteins were expressed in E. coli and purified to homogeneity by conventional purification techniques and in an untagged form unless otherwise indicated. All the recombinant proteins were produced at high levels in the soluble fraction, except Trx-like1. For the latter, all attempts to solubilize the protein by adding an N-terminal His tag, by changing culture conditions, or by coexpressing chaperones were unsuccessful (data not shown). In addition, to explore the importance of the active site for protein reactivity, some amino acids constituting the active site of two representatives, Trx-like2.1 and Trx-lilium3, have been mutated. For Trx-like2.1, the WCRKC active site has been changed into WCRKS (C45S variant) or into WCGPC (R43G/K44P variant). For Trx-lilium3, the SCGSC active site was mutated into WCGSC (S84W variant), SCGPC (S87P variant), and WCGPC (S84W/S87P variant). The reductase activity was first tested by measuring in vitro their ability to reduce insulin disulfide bridges using dithiothreitol (DTT) as a reductant in comparison with poplar Trx h1 used as a positive control (Fig. 3; Behm and Jacquot, 2000). Although less efficient than Trx h1, Trx-like2.1 was able to reduce insulin. As expected, the activity drops down with the monocysteinic mutant Trx-like2.1 C45S, indicating that the resolving Cys is important for disulfide reduction. The remaining activity might be attributed to a reaction involving the successive nucleophilic attack of two Trx molecules. Accordingly, protein dimers were observed on nonreducing SDS gels (data not shown). Interestingly, the Trx-like2.1 R43G/K44P variant (WCGPC active site) has an efficiency very comparable to Trx h1, showing that the dipeptide motif separating the two catalytic Cys residues is very important for protein reactivity and/or substrate recognition. Among Trx-lilium isoforms, Trx-lilium2.2 was more efficient than Trx-lilium1.2 and Trx-lilium3. Because the major difference is the presence of a Trp in the active site of Trx-lilium2.2, which is absent in the two other isoforms, we tested Trx-lilium3 variants with WCGSC, SCGPC, and WCGPC active sites. None of these mutated proteins became more efficient in insulin reduction. In marked contrast with Trx-like2.1, changing the SCGSC active site into WCGPC did not improve the capacity of this protein to reduce insulin. Clot was not active at all in this classical activity assay, raising the question of its reductase activity.
Figure 3.
Reduction of insulin by Trx-like and Trx-lilium proteins. Insulin reduction was measured using a DTT-based assay and 10 µm Clot and Trx-like2.1 (A) or Trxs-lilium (B) by measuring the turbidity at 650 nm caused by the precipitation of reduced insulin. Each trace is a representative experiment from two to three repetitions.
Regeneration of Oxidized Trxs
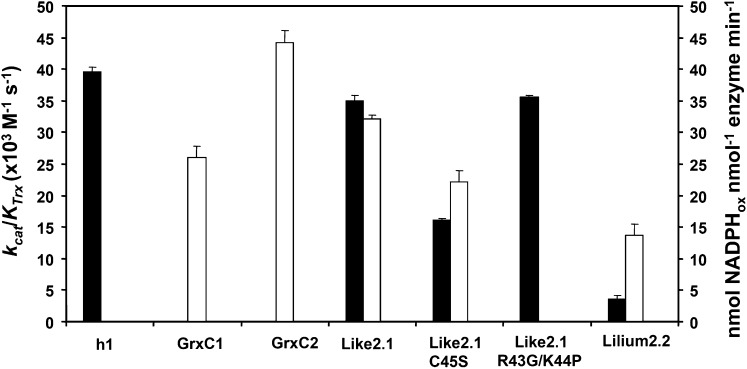
We further characterized the redox properties of these proteins by examining their possible regeneration pathways. We first tested their ability to reduce 5,5′-dithio-bis-2-nitrobenzoic acid (DTNB), an artificial substrate containing a disulfide, in the presence of an NADPH/NTR system that allows investigation of the capacity of Trxs to be regenerated by NTR. Only Trx-like2.1 and to a lesser extent Trx-lilium2.2 efficiently reduced DTNB, with catalytic efficiencies (kcat/KTrx) of 35 and 3.5 × 103 m−1 s−1, respectively, comparable, in the case of Trx-like2.1, to the value obtained with Trx h1 (39 × 103 m−1 s−1; Fig. 4). The catalytic efficiency of the reaction catalyzed by the R43G/K44P variant was also similar (around 35 × 103 m−1 s−1). Surprisingly, the Trx-like2.1 C45S variant conserved the capacity to reduce DTNB, although the catalytic efficiency was decreased by a factor of 2 (16 versus 35 × 103 m−1 s−1). The fact that Trx-lilium1.2 and Trx-lilium3 mutants were able to reduce insulin but not DTNB indicates that they are not reduced by NTR.
Figure 4.
Measurement of typical Trx or Grx activity. Black bars represent the catalytic efficiency (kcat/KTrx) of the tested proteins in a typical Trx assay measuring the capacity to reduce DTNB at the expense of AtNTRB. White bars represent the activity (expressed as nmol oxidized NADPH nmol−1 enzyme min−1) of the tested proteins in a typical Grx assay measuring the capacity to reduce a β-mercaptoethanol-GSH adduct using a GSH recycling system. Only the proteins for which an appreciable activity was detected are represented. The data are represented as means ± sd of at least two separate experiments.
Because Trx-like2.1 is plastidial, we have tested the physiological FTR and NTRc reducing systems for its regeneration. The reduction by FTR was measured using a recently developed procedure employing a four-component electron transfer system, NADPH/FNR/Fdx/FTR (Chibani et al., 2011). After incubation of oxidized Trx with this reducing system, the redox state was assessed after alkylation of free thiol groups with 2-kD methoxyl polyethylene glycol (mPEG) maleimide and subsequent separation on nonreducing SDS gels. Contrary to Trx z, systematically used as a control, and although they contain an intramolecular disulfide, none of these atypical Trxs were reduced by the Fdx/FTR system, as illustrated for Trx-like2.1 (Fig. 5; data not shown). The reduction of Trx-like2.1 by NTRc was then investigated by taking advantage of the fact that Trx-like2.1, unlike NTRc, directly reduces PrxIIE (see below). Although NTRc was functional (as tested in the DTNB reduction assay; data not shown), it was not able to support Trx-like2.1 activity, indicating that, in the case of the full-length NTRc, neither its NTR domain nor its Trx domain was able to reduce Trx-like2.1 (Supplemental Fig. S2).
Figure 5.
Assessment of poplar Trx-like2.1 reduction by Synechocystis Fdx-Trx reductase. The redox state of Trx-like2.1 (A) or Trx z (B) was analyzed by nonreducing SDS-PAGE after oxidizing or reducing treatments and alkylation. Lanes 1, Trxs untreated and not alkylated. Lanes 2 to 5 represent mPEG maleimide-alkylated proteins: lanes 2, untreated Trxs; lanes 3, Trxs incubated with 500 µm oxidized DTT; lanes 4, Trxs incubated with 100 µm reduced DTT; lanes M, molecular mass markers (from top to bottom, 75, 50, 37, 25, 20, and 15 kD); lanes 5, Trxs incubated for 15 min with 20 µm NADPH, 20 nm FNR, 0.5 µm Fdx, and 0.5 µm FTR. Note that FNR, Fdx, and FTR were only slightly detectable by Coomassie blue staining due to the low concentrations used. As previously observed, the shift arising from the alkylation of the thiol groups of the two active site Cys residues of Trx (lanes 4) was larger than expected (approximately 12 kD instead of 4 kD; Chibani et al., 2011).
The fact that some Trxs exhibited no or poor reductase activity in the presence of the various Trx reductases prompted us to investigate whether they possess Grx-like activity using GSH as a reductant. The typical 2-hydroxyethyl disulfide (HED) assay was used to measure the ability of all these Trxs to catalyze the reduction of a mixed disulfide formed between GSH and HED (Fig. 4). Trx-like2.1 and Trx-lilium2.2 reduced the β-mercaptoethanol-GSH adduct with an efficiency comparable to poplar GrxC1 (CGYC active site) and GrxC2 (CPFC active site) used as controls. Interestingly, the monocysteinic Trx-like2.1 C45S variant was almost as efficient as the wild-type form, supporting the fact that, in Trx-like2.1, the second Cys is dispensable, as for most dithiol Grxs active in this assay (Bushweller et al., 1992). Other Trxs (Trx h1, Trx-lilium1.2, wild-type and mutated Trxs-lilium3) did not display any recordable activity, indicating that they either cannot reduce glutathionylated molecules or use GSH for their regeneration.
Midpoint Redox Potentials of Atypical Trxs
Next, we investigated whether differences in the midpoint redox potentials could explain the different capacities of these atypical Trxs to reduce insulin disulfide bonds and their ability to be regenerated or not by GSH or Trx reductases. The midpoint redox potentials of these Trxs were found to be slightly different, ranging from −239 to −265 mV at pH 7.0 (Table I). Despite having an adequate redox potential of −255 mV, the inability of Clot to reduce insulin indicates that other factors, such as the presence of an acidic residue in the active site (WCPDC), might explain the absence of reductase activity. Trx-like2.1 has the lowest redox potential, with a value of −265 mV. The mutations achieved in the Trx-like2.1 R43G/K44P variant, which improved its capacity to reduce insulin, also affected its redox potential (−250 mV), but in an opposite way compared with our expectation. Poplar Trxs-lilium had redox potentials ranging from −239 to −247 mV, very similar to those determined for their Arabidopsis orthologs (Dangoor et al., 2009). In the case of poplar Trx-lilium3, the mutations introduced into the active site strongly affected its redox potential in the expected way, with decreases to −266 and −264 mV for the S84W and S87P single mutants, respectively, and to −268 mV for the S84W/S87P double mutant. Nevertheless, although the redox potential of Trx-lilium3 S84W/S87P became more electronegative and similar to conventional plant Trx h, this change did not improve its capacity to reduce insulin.
Table I. Redox midpoint potentials of wild-type Trx and mutated Trx variants.
For mutated Trxs, the mutation appears in boldface. The values represent means ± sd of three separate experiments. Individual titration representations are provided as Supplemental Figure S3.
| Proteins | Active Site Sequences | Redox Potentials |
|---|---|---|
| mV | ||
| Clot | WCPDC | −255 ± 2 |
| Trx-like2.1 | WCRKC | −265 ± 1 |
| Trx-like2.1R43G/K44P | WCGPC | −250 ± 2 |
| Trx-lilium1.2 | GCGGC | −247 ± 2 |
| Trx-lilium2.2 | WCASC | −239 ± 2 |
| Trx-lilium3 | SCGSC | −242 ± 1 |
| Trx-lilium3 S87P | SCGPC | −263 ± 1 |
| Trx-lilium3 S84W | WCGSC | −266 ± 2 |
| Trx-lilium3 S84W/S87P | WCGPC | −268 ± 1 |
Regeneration of Physiological Target Proteins
As some Arabidopsis Trx-like and Trx-lilium orthologs were shown to weakly activate Arabidopsis NADP-MDH (Dangoor et al., 2009), an enzyme regulated by the formation of intramolecular disulfides, the capacity of these wild-type and mutated poplar plastidial Trxs to reduce the Sorghum bicolor NADP-MDH protein was investigated. Among wild-type Trxs, only Trx-lilium2.2 was able to activate this enzyme, but with a lower efficiency as compared with Trx h1 (Fig. 6). Interestingly, the Trx-like2.1 R43G/K44P mutant acquired the capacity to activate SbNADP-MDH with an efficiency comparable to Trx-lilium2.2, further showing the importance of the active site sequence for protein partner recognition.
Figure 6.
Activation of SbNADP-MDH by wild-type and mutated Trxs-like and Trx-lilium. NADP-MDH activation was achieved in the presence of 500 µm DTT and 10 µm wild-type and mutated Trxs. Every 5 min over a 20-min period, an aliquot was used to measure NADP-MDH activity by following NADPH oxidation at 340 nm. The data are represented as means ± sd of at least two separate experiments.
Next, to assess the specificity of these atypical Trxs toward other putative physiological target proteins, their ability to regenerate various Tpxs or MSRs was measured in coupled assays in the presence of an NADPH/NTR or an NADPH/GR/GSH reduction system (Table II). We selected a cytosolic and two plastidial Prxs named PtPrxIIB, PtPrxIIE, and PtPrxQ, respectively, as well as two plastidial Gpxs, PtGpx1 and PtGpx3, and three plastidial MSRs, PtMSRA4, AtMSRB2, and AtMSRB1. The catalytic and recycling mechanisms of these enzymes are well characterized. PrxIIB, PrxIIE, and MSRB1 are enzymes that use a single Cys residue in their catalytic cycle (Rouhier et al., 2002; Tarrago et al., 2009). The sulfenic acid formed upon catalysis is either reduced by GSH or directly by Trxs (Tarrago et al., 2009, 2010). PtPrxQ, PtGpx1, PtGpx3, AtMSRB2, and PtMSRA4 are enzymes that form an intramolecular disulfide in the course of their catalysis (Rouhier et al., 2004, 2007; Navrot et al., 2006; Tarrago et al., 2009). In this case, the disulfide is usually uniquely reduced by Trxs but not by GSH or Grxs. Trx h1, which is able to reduce all target proteins except AtMSRB1, was used as a control. Whereas all other Trxs were unable to reduce the tested target proteins, wild-type Trx-like2.1 as well as the C45S variant supported the activity of PtPrxIIE, PtPrxIIB, and AtMSRB1 in the presence of a GSH reducing system (Table II). The presence of Trx-like2.1 was crucial, because GSH alone was not able to regenerate the target proteins, except for PtPrxIIE, which was slightly reduced by GSH. Remarkably, the activities measured with Trx-like2.1 C45S were in the same range as those obtained with the wild-type form (even two and three times better for AtMSRB1 and PtPrxIIE, respectively), indicating that the second Cys of the Trx-like2.1 active site is not required for this GSH-dependent reaction. Although Trx-like2.1 was reduced by NTR, it did not provide electrons to PrxIIB and PrxIIE using this regeneration system, whereas Trx h1 did. Regarding the Trx-like2.1 R43G/K44P variant (WCGPC active site), the two substitutions in the active site were sufficient for the protein to acquire, together with the NADPH/NTR system, the capacity to reduce most enzymes whose catalytic mechanism involves the formation of an intramolecular disulfide (PtPrxQ, PtGpx1 and -3, and PtMSRA but not AtMSRB2). Interestingly, this variant conserved the capacity to reduce both PtPrxIIB and PtPrxIIE, but in the presence of the NADPH/NTR reducing system instead of the NADPH/GR/GSH one. Note also that it has lost the capacity to regenerate AtMSRB1 regardless of the reducing system. These results suggest that, in the case of the two Prxs, the sulfenic acid can be directly reduced by the catalytic Cys of the Trx-like2.1 R43G/K44P variant but not in the case of AtMSRB1, the reduction of which requires the presence of GSH.
Table II. Regeneration of Tpx or MSR family members by wild-type and mutated Trx-like2.1 in the presence of an NADPH/NTR system (NTS) or an NADPH/GR/GSH system (NGS).
Data are expressed as nmol oxidized NADPH min−1 nmol−1 enzyme. Dashes indicate that no significant activity was detected.
| Targets | Trx h1 |
Trx-like2.1 |
Trx-like2.1 C45S |
Trx like2.1 R43G/K44P |
||||
| NTS | NGS | NTS | NGS | NTS | NGS | NTS | NGS | |
| PtPrxIIB | 7.7 ± 0.5 | –- | –- | 16.3 ± 1.1 | –- | 15.7 ± 0.1 | 5.8 ± 0.2 | –- |
| PtPrxIIE | 11.6 ± 1.5 | –- | –- | 2.4 ± 0.4 | –- | 7.8 ± 0.3 | 8.4 ± 0.3 | –- |
| PtPrxQ | 10.7 ± 0.5 | –- | –- | –- | –- | –- | 2.4 ± 0.4 | –- |
| PtGpx1 | 10.2 ± 0.8 | –- | –- | –- | –- | –- | 4.8 ± 0.7 | –- |
| PtGpx3 | 10.2 ± 0.2 | –- | –- | –- | –- | –- | 5.6 ± 0.1 | –- |
| PtMSRA4 | 2.6 ± 0.2 | –- | –- | –- | –- | –- | 1.6 ± 0.3 | –- |
| AtMSRB2 | 16.2 ± 1 | – | –- | –- | –- | –- | –- | –- |
| AtMSRB1 | –- | –- | –- | 1.9 ± 0.1 | –- | 4.4 ± 0.1 | –- | –- |
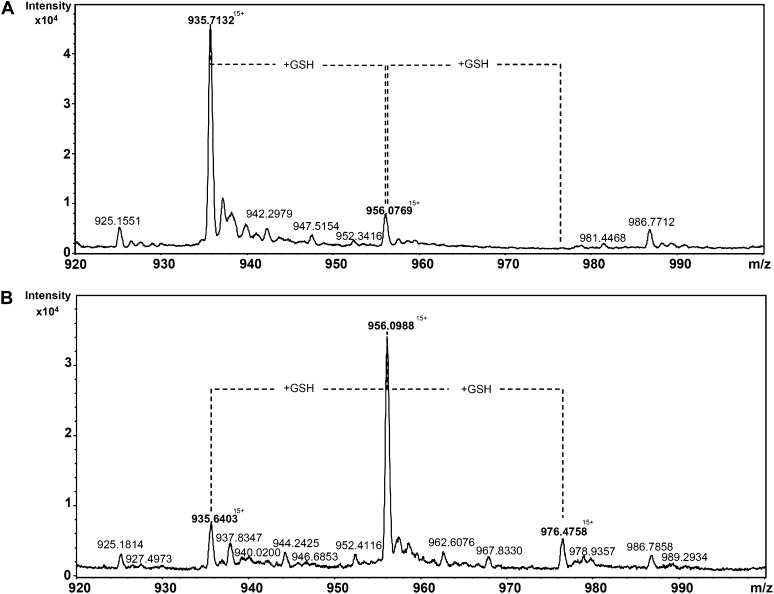
The specific reduction of proteins using for catalysis a single Cys, which is glutathionylated in the course of their regeneration mechanism, suggests that Trx-like2.1 should itself become transiently glutathionylated. To confirm the possibility of forming a GSH adduct, in vitro glutathionylation assays have been performed with the Trx-like2.1 C45S variant, and the redox state of the protein was subsequently analyzed by mass spectrometry analyses. An untreated Trx-like2.1 C45S had the expected mass of 14,020 D, if we consider that the N-terminal Met has been cleaved, which is most likely the case, because the second residue is an Ala (Fig. 7). A small additional peak corresponding to a molecular mass of 14,326 D and consistent with the presence of a GSH adduct was also detected, indicating that a small part of the protein was glutathionylated in E. coli. An oxidation treatment with S-nitrosoglutathione (GSNO), performed on a prereduced Trx-like2.1 C45S, led to the formation of three protein species, a major form with a molecular mass of 14,326 D and two minor forms of 14,020 and 14,632 D, compatible with the presence of either an unmodified protein or of a protein with one (+306 D) or two (+712 D) GSH molecules. As expected, the two protein species with GSH adducts disappeared by incubating the protein with DTT (data not shown). Based on the presence of two Cys residues in the Trx-like2.1 C45S variant (the catalytic Cys and the additional nonconserved Cys-67), trypsic digestion helped us establish that the peptide containing the catalytic Cys (ELSQPIIIDWMASWCR) was predominantly glutathionylated (data not shown).
Figure 7.
Electrospray ionization (ESI)-time-of-flight mass spectra of glutathionylated Trx-like2.1 C45S. ESI mass-to-charge ratio (m/z) spectra of an untreated (A) or GSNO-treated (B) Trx-like2.1 C45S samples were obtained using a Bruker microTOF-Q spectrometer in denaturing conditions. From the multiply charged ion spectra initially obtained, we focused on the peak with 15 charges on the ion. The deconvolution of the ESI spectra indicated that the ion with a m/z of approximately 935 atomic mass units (amu) corresponded to a molecular mass of 14,020 D, the one with a m/z of approximately 956 amu corresponded to a molecular mass of 14,326 D, and the one with a m/z of approximately 976 amu corresponded to a molecular mass of 14,632 D. The intensity of the signal is represented as arbitrary units. However, assuming that the different forms of the protein were identically ionized, the abundance of each species in the sample is well correlated to the intensity of each species on the ESI mass spectra.
DISCUSSION
The Plastidial Trx Equipment of Photosynthetic Organisms
Clot, Trx-like, and Trx-lilium isoforms belong to atypical Trx classes found in all higher plants and contain nonconventional active site signatures (Chibani et al., 2009). At the beginning of this work, limited information was available for these proteins regarding their localization, expression, reduction systems, and possible partners. Arabidopsis Trxs-lilium, also referred to as ACHT (for atypical Cys- and His-rich Trxs), and the two Trx-like2 members were known to be located in plastids (Cain et al., 2009; Dangoor et al., 2009). The chloroplastic localization of poplar Trx-like2.1, established here using a translational GFP fusion approach, is in accordance with the finding that the Arabidopsis orthologs are imported in the stroma in in vitro chloroplast import assays (Cain et al., 2009). Contrary to poplar Trx-like2.1, the predictions for poplar Trx-like2.2 and Trx-like2.3 were less ambiguous and indicated the presence of a plastidial targeting sequence, but this will have to be experimentally proven. With two members in Arabidopsis and three in poplar, Trxs-like2 extend the already very large number of plastidial Trxs. Overall, there are at least 20 and 24 potential plastidial Trxs in Arabidopsis and poplar, respectively, all types (Trxs f, x, m, y, and z, CDSP32, HCF164, Trxs-lilium, Trxs-like2, and NTRc) being present in both species (Lennartz et al., 2001; Collin et al., 2003, 2004; Cain et al., 2009; Chibani et al., 2009; Dangoor et al., 2009; Meng et al., 2010). Trx m and Trxs-lilium display the highest numbers of isoforms, four and eight for Trx m in Arabidopsis and poplar, respectively, and five for Trxs-lilium in both species. All these proteins, with the exception of HCF164, are assumed to be in the stromal fraction. This multiplicity of plastidial Trxs is likely related to different spatiotemporal expression patterns, to the reduction of specific target proteins, or to the use of specific reducing pathways.
Transcript Expression Patterns of Plastidial Trxs
Transcripts coding for Trxs-like and Trxs-lilium have been detected in all poplar organs tested (Fig. 2), suggesting that the diversity of plastidial Trxs is probably not explained by differential expression at the organ level, although, for example, transcripts of Trx-like2.1 and Trx-like2.2 seem to be mainly present in photosynthetic organs and female flowers. In Arabidopsis, Dangoor et al. (2009) reported a preferential expression of Trx-lilium2.2 in green tissues. In addition, a more detailed analysis using the Genevestigator tool (Hruz et al., 2008) revealed that, in Arabidopsis, some atypical Trxs are expressed in a very specific manner as a function of tissue age or cell type. Indeed, Arabidopsis Trx-lilium1.1, -1.2, and -1.3 are expressed at a very high level in apical meristem, seed suspensor, and anther, respectively, the transcript abundance of these three Trxs being lower and comparable in all other tissues. Regarding AtTrx-like1, a high and specific expression is detected in the abscission zone of anthers and in imbibed seeds. Note also that most typical plastidial Trxs (m, f, x, and y) are specifically expressed in photosynthetic tissues except Trx y1, the transcript level of which is much higher in seeds and roots (Collin et al., 2004). Finally, some Trx genes display substantial variations in their expression in relation to the environment. This is the case of CDSP32 and of the cytosolic Trx h5, the expression of which is triggered by environmental conditions leading to oxidative stress (Broin et al., 2000; Laloi et al., 2004). Altogether, these data argue in favor of a specialization of plant Trxs as a function of the organ type, developmental stage, and/or environmental conditions, but this can vary depending on the species considered.
One Trx-like and One Trx-lilium Use GSH for Their Regeneration
Although belonging to the same protein superfamily, Grxs, which possess YC[P/G/S]Y[C/S] active site sequences, are usually reduced by GSH whereas Trxs are reduced by Fdx- or NADPH-dependent Trx reductases. In addition, it is commonly accepted that Grxs preferentially reduce protein-GSH adducts, whereas Trxs are more specific for the reduction of disulfide bonds formed within a single polypeptide or between two polypeptides. However, in the set of atypical Trxs tested here, two proteins, Trx-like2.1 and Trx-lilium2.2, are able to reduce a β-mercaptoethanol-GSH mixed disulfide, a preferential Grx substrate, with an efficiency comparable (around 0.58 s−1) to the poplar Grxs tested here but lower when compared with previous results obtained with two plastidial Grxs (1.21 s−1 for AtGrxC5 and 23.10 s−1 for poplar GrxS12; Couturier et al., 2011; Zaffagnini et al., 2012a). Strikingly, both proteins can also be reduced by Arabidopsis NTRB. This is not unprecedented, because a Trx-like protein from the parasitic nematode Haemonchus contortus, which displays a CRSC active site sequence, possesses a similar property (Sotirchos et al., 2009). However, considering the plastidial localization of the proteins, their reduction by NTR should not be of physiological importance, unless protein dual targeting exists according to environmental conditions or developmental stage (Dangoor et al., 2009; this work). On the contrary, the fact that they are reduced neither by NTRc nor by FTR makes these enzymes strictly dependent on the NADPH/GR/GSH system for their regeneration in plastids.
Previous studies already illustrated the cross-reactivity existing between the Trx and GSH/Grx reducing pathways, some Grxs being regenerated by Trx reductases (Johansson et al., 2004; Zaffagnini et al., 2008). In addition, some Trxs are reduced by GSH and/or Grx but not by NTR. As an example, the presence of an additional Cys residue at position 4 in poplar Trx h4 (WCGPC active site) prevents its recycling by NTR but renders this enzyme GSH and Grx dependent (Koh et al., 2008). Besides, the poplar TrxCxxS3, harboring an unusual WCMPS active site motif, is also recycled by GSH, but the activity is extremely low, with a difference of approximately 3 orders of magnitude with this study (Gelhaye et al., 2003). In addition, some genetic and biochemical evidence indicates that the two yeast cytosolic Trxs, although reduced by Trx reductases, are apparently able to catalyze the deglutathionylation of some protein substrates (Greetham et al., 2010). On the other hand, the findings that, in knockout plants for the cytosolic and mitochondrial NTRs, the cytosolic Trx h3 is only partially oxidized, and that buthionine sulfoximine, a specific inhibitor of GSH biosynthesis, leads to full oxidation, indicate a direct or indirect reduction of the Trx by GSH in vivo (Reichheld et al., 2007).
Based on sequence similarity with Grxs, Clot proteins, first identified in Drosophila as required for the biosynthesis of drosopterin, an eye pigment, were assumed to be GSH-dependent enzymes (Giordano et al., 2003). Surprisingly, no significant activity was detected for the Clot proteins, either from poplar or Arabidopsis (data not shown). The orthologs from S. cerevisiae (also referred to as ScGrx8) or from mammals (referred to as TRP14), which possess the same active site, are also not able or are very poorly able to reduce insulin, and a faint activity in the HED assay was detected for ScGrx8 (Jeong et al., 2004; Eckers et al., 2009). However, it has been shown that TRP14 can reduce a disulfide in the dynein light chain LC8, contributing to the inhibitory activity of LC8 toward the nuclear factor κB (Jung et al., 2008). Similarly, plant Clot proteins might have very specific protein partners.
Putative Physiological Targets of Atypical Trxs
For the two proteins exhibiting a GSH- and/or NTR-dependent activity, Trx-like2.1 and Trx-lilium2.2, we have investigated their capacity to regenerate known plastidial members of the Tpx and MSR families. Whereas Trx-lilium2.2 did not exhibit any significant activity, Trx-like2.1 was able to specifically regenerate the established GSH/Grx-dependent proteins (PrxIIE and MSRB1), whose catalytic mechanism relies on a single redox active Cys but not the strictly Trx-dependent targets (PrxQ, Gpxs, MSRA4, and MSRB2), whose catalytic mechanism involves two or three Cys residues (Fig. 8). The fact that the second active site Cys of Trx-like2.1 is not essential for the reaction led us to propose a regeneration mechanism similar to the one employed by Grxs. Indeed, they generally use a monothiol mechanism where the second active site Cys is not required, although it can modulate (either increase or decrease) protein reactivity (Bushweller et al., 1992; Couturier et al., 2009, 2011). Thus, from a biochemical point of view, Trx-like2.1 constitutes a redundant system with plastidial Grxs. Nevertheless, if we compare the turnover numbers of the reactions, it is generally five to 16 times less efficient. The kcat values of the reactions catalyzed by PrxIIE and MSRB1 were 0.04 and 0.03 s−1, respectively, with Trx-like2.1, whereas the values obtained for the same enzymes with poplar GrxS12 or AtGrxC5 were 0.66 and 0.17 s−1 or 0.5 and 0.35 s−1, respectively (Vieira Dos Santos et al., 2007; Gama et al., 2008; Couturier et al., 2011).
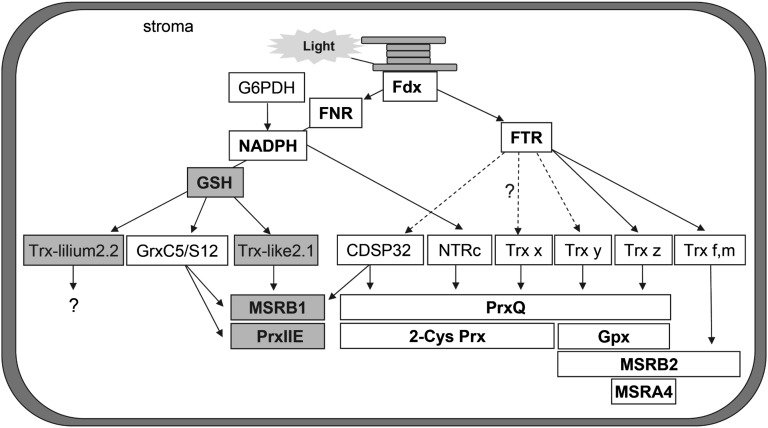
Figure 8.
Model depicting the reducing pathways for plastidial Trxs and their specificity toward known physiological targets of the Tpx and MSR families. Except for MSRA, which was only tested with Trx z, all other reductant/target protein couples have generally been exhaustively tested in previous studies. The conclusions represented here summarize results presented previously (Broin et al., 2002; Collin et al., 2003, 2004; Rouhier et al., 2004; Rey et al., 2005; Moon et al., 2006; Navrot et al., 2006; Pérez-Ruiz et al., 2006; Vieira Dos Santos et al., 2007; Couturier et al., 2009, 2011; Pulido et al., 2010; Tarrago et al., 2010; Chibani et al., 2011). New insights provided by this study (i.e. reduction of Trx-lilium2 and Trx-like2.1 by GSH and regeneration of PrxIIE and MSRB1 by Trx-like2.1) are highlighted by gray boxes. Dotted arrows indicate that reduction of these Trxs by FTR is possible but not experimentally determined.
Regarding the catalytic mechanism, the sulfenic acid initially formed on the Prx or MSR proteins probably reacts with GSH, forming a glutathionylated intermediate, which is subsequently attacked by the catalytic Cys of Trx-like2.1 (Supplemental Fig. S4; Vieira Dos Santos et al., 2007; Tarrago et al., 2009). The latter becomes itself glutathionylated, a possibility demonstrated by the in vitro glutathionylation treatment of the Trx-like2.1 C45S. In the presence of GSH, it is most likely that the glutathionylated Trx form is resolved by another GSH molecule. This is different from the peculiar CDSP32, which can support the activity of AtMSRB1 without GSH by directly reducing the sulfenic acid formed on the catalytic Cys (Tarrago et al., 2010).
However, it is also clear from our redox titration of Trx-like2.1 that an intramolecular disulfide can be formed. The midpoint redox potential determined for this disulfide was −265 mV at pH 7.0, a value comparable to certain values obtained with other cytosolic or plastidial Trxs and that should be adequate for the reduction of Prxs and MSRs forming an intramolecular disulfide. Because Trxs known to reduce these proteins usually exhibit a WCGPC active site, the reactivity of a Trx-like2.1 variant was examined. Contrary to our expectation, the redox midpoint potential is increased in this variant (−250 mV). However, this mutated Trx was able to reduce most of these target proteins in the presence of an NTR reducing system. This result suggests that, at least for Trx-like2.1, the presence of two basic residues in the active site sequence, replacing the usual hydrophobic residues, is of high importance for target protein recognition. However, this is not true for the interaction with NTR, because wild-type and mutated Trx-like2.1 proteins displayed similar catalytic efficiencies. A mutational study performed with E. coli Trx1 showed that the presence of a positive charge decreases its catalytic efficiency, most likely by affecting protein-protein interactions, although in this case the mutation did not change the midpoint redox potential (Lin and Chen, 2004). The fact that the redox potential is probably not the major determinant for the interaction of these atypical Trxs with their protein partners is also illustrated by the following results. Although having a redox potential of −255 mV, Clot is not reduced by AtNTRB. Thus, it might be that the presence of an acidic residue in the active site motif (WCPDC) prevents such interaction, whereas it could favor protein-protein interactions with other yet unknown targets. This observation also raises the question of the in vivo reduction of this cytosolic protein. In the case of Trx-lilium3, we have observed that changing the active site from SCGSC to WCGPC decreased the midpoint redox potential from −242 to −268 mV. This result is consistent with a study conducted on a Staphylococcus aureus Trx, showing that replacement of the active site Pro-31 by a Ser or a Thr changed the redox potential from −268 mV for the wild-type protein to −236 and −244 mV for the two mutated proteins (Roos et al., 2007). Nevertheless, although the redox potential of Trx-lilium3 S84W/S87P became more electronegative and similar to conventional plant Trx h, this change did not improve its capacity to reduce insulin.
All Trxs-lilium, although forming three independent subgroups, have very similar active site sequences (formed by small and uncharged amino acids) and midpoint redox potentials, and they all have the capacity to reduce disulfide bonds, as indicated by their insulin reductase activity. However, Trx-lilium2.2 was the only one able to activate SbNADP-MDH (less efficiently than Trx h1) and was unable to support the activity of the tested Prxs and MSRs. In addition, only Trx-lilium2.2 exhibited GSH- and NTR-dependent activities, suggesting that some specificity exists among Trx-lilium isoforms. Some differences might arise from the presence of an N-terminal extension of about 25 amino acids in Trx-lilium1.2 and a C-terminal extension of about 50 amino acids in Trx-lilium3.
CONCLUSION
This study reports the biochemical characterization of poplar isoforms belonging to three atypical Trx classes, namely Clot, Trx-like, and Trx-lilium. The mutational analysis of the active site sequences of these atypical Trxs indicates that the dipeptide separating the two Cys residues of the active site is an important factor for providing particular redox properties and substrate specificities toward both the reducing systems and the target proteins. In particular, the positively charged nature of the Trx-like2.1 active site is a major determinant for its capacity to reduce specific MSR and Tpx at the expense of GSH. Although all proteins, except Clot, exhibit disulfide reductase activity with insulin, only two proteins (Trx-lilium2.2 and Trx-like2.1) exhibited a detectable activity with the potential plastidial partners that were tested. Hence, the question of the nature of the physiological targets and roles of all these proteins should now be addressed using other approaches, such as the study of knockout and knockdown plants and in vivo or in vitro methods to identify protein partners. For instance, besides their usual disulfide reductase activity, it has recently been shown that some plastidial Trxs f and m have chaperone properties (Sanz-Barrio et al., 2012).
MATERIALS AND METHODS
Intracellular Localization via GFP Fusion
The full-length coding sequences of poplar (Populus spp.) PtClot, PtTrx-like1, and PtTrx-like2.1 were cloned into the NcoI and BamHI sites of pCK-GFP S65C using the primers detailed in Supplemental Table S1. The sequences, fused to the N-terminal part of GFP, are under the control of a double 35S promoter (Menand et al., 1998). Nicotiana benthamiana cells were then transfected by bombardment of leaves with tungsten particles coated with plasmid DNA, and images were obtained with a Zeiss LSM510 confocal microscope. Chloroplasts were visualized by the natural fluorescence of chlorophyll.
RT-PCR
Semiquantitative RT-PCR was used to estimate the transcript expression in various poplar organs (i.e. roots, young and mature leaves, stems, petioles, fruits, stamens, and female catkins). Total RNA was isolated with the RNeasy Plant Mini kit (Qiagen) from 100 mg of frozen tissue. To remove contaminating DNA, the samples were treated with DNase I (Qiagen). A total of 1 µg of RNA was converted to cDNA using reverse transcriptase (Qiagen). The PCR program used was as follows: 94°C for 3 min and 35 cycles of 94°C for 30s, 52°C for 45s, and 72°C for 90s. The primers used for these RT-PCR experiments are those designed for cloning the sequence of the mature forms of the proteins (Supplemental Table S1).
Cloning and Site-Directed Mutagenesis
The sequences encoding the mature forms of Trx-like2.1, Trx-lilium2.2, and Trx-lilium3 were amplified from a Populus tremula × Populus tremuloides leaf cDNA library, those for Clot and Trx-lilium1.2 were amplified from a P. trichocarpa × Populus deltoides root cDNA library, and those for Trx-like1 were amplified from a P. tremula × P. tremuloides flower cDNA library, using the primers described in Supplemental Table S1. In the case of Trx-like1 and Clot, the amplified sequence corresponded to the full-length protein. For Trx-like2.1, Trx-lilium2.2, and Trx-lilium3, the putative plastidial targeting sequences were removed and the amplified fragment was coded for proteins devoid of the first 73, 82, 79, and 69 amino acids, respectively. After digestion with NcoI and BamHI, PCR fragments were inserted into the pET-3d expression plasmid. For this cloning, owing to the use of an NcoI restriction site, a codon for Ala was added after the ATG start codon in the case of Clot, Trx-like1, Trx-like2.1, and Trx-lilium3. Site-directed mutagenesis was used to create variants for Trx-like2.1 (R43G/K44P and C45S) and Trx-lilium3 (S84W, S87P, and S84W/S87P) using the two complementary mutagenic primers described in Supplemental Table S1. The amino acid numbering is based on the sequences of the mature forms expressed as recombinant proteins. The introduction of the mutation in the DNA sequence was verified by sequencing.
Production and Purification of Recombinant Trxs
The recombinant plasmids obtained were used to transform the BL21(DE3) pSBET strain of Escherichia coli. The bacteria were grown to a final volume of approximately 2.4 L at 37°C, and protein production was induced during the exponential growth phase by adding 100 µm isopropyl-β-thiogalactoside (Euromedex). The bacteria were harvested by centrifugation at 4,400g for 20 min and then resuspended in buffer A (30 mm Tris-HCl, pH 8.0, 1 mm EDTA, and 200 mm NaCl). Cells were disrupted by sonication, and all Trxs were found in the soluble fraction after centrifugation at 20,000g for 30 min, except Trx-like1, which was completely insoluble. The soluble fraction was then precipitated with ammonium sulfate successively up to 40% and 80% of the saturation, and the precipitated proteins were collected by centrifugation (20,000g for 15 min). The protein pellet was redissolved in buffer A, and the sample was loaded onto an ACA 44 gel filtration column (Biosepra) equilibrated with buffer A. The proteins of interest were identified using SDS-PAGE and Coomassie blue staining. The fractions containing the protein were pooled, dialyzed, and loaded on a DEAE-Sepharose column (Sigma) equilibrated with buffer A without NaCl. The proteins were eluted using a 0 to 0.4 m NaCl gradient, selected based on the highest purity, dialyzed, concentrated, and finally stored in buffer A at −20°C until further use. Final purity was checked by 15% SDS-PAGE.
Determination of the Midpoint Redox Potentials
Oxidation-reduction titrations were carried out at ambient temperature by measuring fluorescence resulting from the reaction between protein thiol groups and monobromobimane (Sigma) as described previously (Hirasawa et al., 1999). The reaction mixtures contained 50 µg of protein in 100 mm HEPES buffer, pH 7.0, containing defined mixtures of oxidized and reduced DTT to set the ambient potential. Equilibration times of either 2 or 3 h were used. The redox potentials were calculated by fitting the curve to the Nernst equation for a single two-electron redox couple using GraphPad Prism version 4.0 and using a redox potential value of −327 mV for DTT at pH 7.0.
Reductase Activity
The insulin reduction assay was carried out in a 500-µL reaction mixture containing 100 mm phosphate, pH 7.9, 2 mm EDTA, 0.75 mg mL−1 bovine insulin, 500 µm DTT, and 10 µm Trx at 30°C. The reduction of insulin was monitored as the increase in turbidity at 650 nm due to insulin precipitation. Nonenzymatic reduction of insulin by DTT was monitored in the absence of Trx.
The ability of Trxs to catalyze the reduction of DTNB was measured at 25°C by monitoring the increase in A412 caused by the release of TNB−. The reaction medium contained 30 mm Tris-HCl, pH 8.0, 2 mm EDTA, 200 µm NADPH, 0.5 µm Arabidopsis (Arabidopsis thaliana) NTRB, 100 µm DTNB, and varying concentrations of Trxs ranging from 0.25 to 20 µm. Control experiments were performed under the same conditions but in the absence of Trx.
Reduction of Trxs by the Fdx/FTR System
The reduction of oxidized plastidial Trxs by an NADPH/FNR/Fdx/FTR system was assessed by SDS-PAGE separation after alkylation with mPEG maleimide (Laysan Bio) as described earlier (Chibani et al., 2011).
HED Activity Assay
A 500-µL mixture containing 200 µm NADPH, 0.5 unit of GR, 500 µm GSH, and 700 µm HED was prepared in 100 mm Tris-HCl, pH 8.0, and 1 mm EDTA. After a 2-min incubation, 250 nm Trx or Grx was added. For all proteins, this concentration is within the linear response range of the enzyme. The decrease in A340 was followed using a Cary 50 spectrophotometer (Agilent Technologies). HED activity was determined after subtracting the spontaneous reduction rate observed in the absence of Trx or Grx and expressed as nmol NADPH oxidized min−1 nmol−1 enzymes.
Reduction or Regeneration of Target Proteins
The peroxidase activities of poplar PrxIIE, PrxIIB, PrxQ, Gpx1, and Gpx3 were measured spectrophotometrically by following NADPH oxidation at 340 nm in the presence of 10 µm Trxs and either the NADPH/NTR system or the NADPH/GR/GSH system. The 500-µL reaction mixture contained 30 mm Tris-HCl, pH 8.0, 1 mm EDTA, 200 µm NADPH, 250 µm hydrogen peroxide, 300 nm PrxIIE, PrxIIB, PrxQ, Gpx1, or Gpx3, and either 0.8 µm AtNTRB or 0.5 unit of GR and 500 µm GSH. The reaction was started by adding the Tpxs. The MSR activities of poplar MSRA4, AtMSRB1, and AtMSRB2 were measured in the same conditions except that MSRs (5 µm PtMSRA and 2.5 µm AtMSRB1) replaced Tpxs and the substrate was 2 mm N-acetyl-Met sulfoxide instead of hydrogen peroxide. The rate of NADPH oxidation was determined after subtracting the spontaneous rate in the absence of Prx, Gpx, or MSR.
The activation of the Sorghum bicolor NADP-MDH was carried out as described (Jacquot et al., 1981). The incubation mixture contained 500 µm DTT, 10 µm Trxs, and 10 µg of recombinant SbMDH (0.75 mg mL−1) in 100 mm Tris-HCl, pH 8.0, buffer. Every 5 min, an aliquot of 20 µL was added to a standard assay mixture containing 100 mm Tris-HCl, pH 8.0, 189 μm oxaloacetate, and 800 μm NADPH. The activity was measured by following the decrease in A340.
Glutathionylation of Trx-like2.1 C45S and Mass Spectrometry Analysis
For glutathionylation experiments, 100 µm prereduced Trx-like2.1 C45S was incubated with 5 mm GSNO for 30 min at 25°C. Prereduction was performed by incubating the protein with an excess of DTT (usually 10 mm) for 30 min and subsequent desalting on G-25 columns. Three samples (untreated, GSNO treated, and GSNO treated subsequently reduced by DTT) were then analyzed using a Bruker microTOF-Q spectrometer (Bruker Daltonics) equipped with an Apollo II electrospray ionization source and a procedure similar to that described by Couturier et al. (2011). Trypsin digestion and subsequent analyses by reverse-phase liquid chromatography-electrospray ionization-tandem mass spectrometry using a capillary HPLC system coupled to a quadrupole time-of-flight mass spectrometer (CapLC Q-TOF Ultima; Waters) were performed as described (Bäckström et al., 2007).
DNA sequences for these poplar atypical Trxs have been deposited in GenBank under the following accession numbers: JQ407766 for Trx-like1, JQ407767 for Trx-like2.1, JQ407770 for Trx-lilium1.2, JQ407768 for Trx-lilium2.2, JQ407769 for Trx-lilium3, and JQ407771 for Clot.
Supplemental Data
The following materials are available in the online version of this article.
Supplementary Material
Acknowledgments
Technical support from François Dupire for mass analyses is gratefully acknowledged.
Glossary
- Trx
thioredoxin
- Grx
glutaredoxin
- Tpx
thiol peroxidase
- MSR
methionine sulfoxide reductase
- Fdx
ferredoxin
- FTR
ferredoxin-Trx reductase
- GR
glutathione reductase
- GSH
glutathione
- MDH
malate dehydrogenase
- Gpx
glutathione peroxidase
- Prx
peroxiredoxin
- RT
reverse transcription
- DTT
dithiothreitol
- HED
2-hydroxyethyl disulfide
- DTNB
5,5′-dithio-bis-2-nitrobenzoic acid
References
- Arsova B, Hoja U, Wimmelbacher M, Greiner E, Ustün S, Melzer M, Petersen K, Lein W, Börnke F. (2010) Plastidial thioredoxin z interacts with two fructokinase-like proteins in a thiol-dependent manner: evidence for an essential role in chloroplast development in Arabidopsis and Nicotiana benthamiana. Plant Cell 22: 1498–1515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bäckström S, Elfving N, Nilsson R, Wingsle G, Björklund S. (2007) Purification of a plant mediator from Arabidopsis thaliana identifies PFT1 as the Med25 subunit. Mol Cell 26: 717–729 [DOI] [PubMed] [Google Scholar]
- Balmer Y, Vensel WH, Cai N, Manieri W, Schürmann P, Hurkman WJ, Buchanan BB. (2006) A complete ferredoxin/thioredoxin system regulates fundamental processes in amyloplasts. Proc Natl Acad Sci USA 103: 2988–2993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behm M, Jacquot JP. (2000) Isolation and characterization of thioredoxin h from poplar xylem. Plant Physiol Biochem 38: 363–369 [Google Scholar]
- Broin M, Cuiné S, Eymery F, Rey P. (2002) The plastidic 2-cysteine peroxiredoxin is a target for a thioredoxin involved in the protection of the photosynthetic apparatus against oxidative damage. Plant Cell 14: 1417–1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broin M, Cuiné S, Peltier G, Rey P. (2000) Involvement of CDSP 32, a drought-induced thioredoxin, in the response to oxidative stress in potato plants. FEBS Lett 467: 245–248 [DOI] [PubMed] [Google Scholar]
- Bushweller JH, Aslund F, Wüthrich K, Holmgren A. (1992) Structural and functional characterization of the mutant Escherichia coli glutaredoxin (C14→S) and its mixed disulfide with glutathione. Biochemistry 31: 9288–9293 [DOI] [PubMed] [Google Scholar]
- Cain P, Hall M, Schröder WP, Kieselbach T, Robinson C. (2009) A novel extended family of stromal thioredoxins. Plant Mol Biol 70: 273–281 [DOI] [PubMed] [Google Scholar]
- Chibani K, Tarrago L, Schürmann P, Jacquot JP, Rouhier N. (2011) Biochemical properties of poplar thioredoxin z. FEBS Lett 585: 1077–1081 [DOI] [PubMed] [Google Scholar]
- Chibani K, Wingsle G, Jacquot JP, Gelhaye E, Rouhier N. (2009) Comparative genomic study of the thioredoxin family in photosynthetic organisms with emphasis on Populus trichocarpa. Mol Plant 2: 308–322 [DOI] [PubMed] [Google Scholar]
- Collet JF, Messens J. (2010) Structure, function, and mechanism of thioredoxin proteins. Antioxid Redox Signal 13: 1205–1216 [DOI] [PubMed] [Google Scholar]
- Collin V, Issakidis-Bourguet E, Marchand C, Hirasawa M, Lancelin JM, Knaff DB, Miginiac-Maslow M. (2003) The Arabidopsis plastidial thioredoxins: new functions and new insights into specificity. J Biol Chem 278: 23747–23752 [DOI] [PubMed] [Google Scholar]
- Collin V, Lamkemeyer P, Miginiac-Maslow M, Hirasawa M, Knaff DB, Dietz KJ, Issakidis-Bourguet E. (2004) Characterization of plastidial thioredoxins from Arabidopsis belonging to the new y-type. Plant Physiol 136: 4088–4095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couturier J, Koh CS, Zaffagnini M, Winger AM, Gualberto JM, Corbier C, Decottignies P, Jacquot JP, Lemaire SD, Didierjean Cet al. (2009) Structure-function relationship of the chloroplastic glutaredoxin S12 with an atypical WCSYS active site. J Biol Chem 284: 9299–9310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couturier J, Ströher E, Albetel AN, Roret T, Muthuramalingam M, Tarrago L, Seidel T, Tsan P, Jacquot JP, Johnson MK, et al. (2011) Arabidopsis chloroplastic glutaredoxin C5 as a model to explore molecular determinants for iron-sulfur cluster binding into glutaredoxins. J Biol Chem 286: 27515–27527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangoor I, Peled-Zehavi H, Levitan A, Pasand O, Danon A. (2009) A small family of chloroplast atypical thioredoxins. Plant Physiol 149: 1240–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyson HJ, Jeng MF, Tennant LL, Slaby I, Lindell M, Cui DS, Kuprin S, Holmgren A. (1997) Effects of buried charged groups on cysteine thiol ionization and reactivity in Escherichia coli thioredoxin: structural and functional characterization of mutants of Asp 26 and Lys 57. Biochemistry 36: 2622–2636 [DOI] [PubMed] [Google Scholar]
- Eckers E, Bien M, Stroobant V, Herrmann JM, Deponte M. (2009) Biochemical characterization of dithiol glutaredoxin 8 from Saccharomyces cerevisiae: the catalytic redox mechanism redux. Biochemistry 48: 1410–1423 [DOI] [PubMed] [Google Scholar]
- Eklund H, Gleason FK, Holmgren A. (1991) Structural and functional relations among thioredoxins of different species. Proteins 11: 13–28 [DOI] [PubMed] [Google Scholar]
- Gama F, Bréhélin C, Gelhaye E, Meyer Y, Jacquot JP, Rey P, Rouhier N. (2008) Functional analysis and expression characteristics of chloroplastic Prx IIE. Physiol Plant 133: 599–610 [DOI] [PubMed] [Google Scholar]
- Gelhaye E, Rouhier N, Gérard J, Jolivet Y, Gualberto J, Navrot N, Ohlsson PI, Wingsle G, Hirasawa M, Knaff DB, et al. (2004) A specific form of thioredoxin h occurs in plant mitochondria and regulates the alternative oxidase. Proc Natl Acad Sci USA 101: 14545–14550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelhaye E, Rouhier N, Jacquot JP. (2003) Evidence for a subgroup of thioredoxin h that requires GSH/Grx for its reduction. FEBS Lett 555: 443–448 [DOI] [PubMed] [Google Scholar]
- Giordano E, Peluso I, Rendina R, Digilio A, Furia M. (2003) The clot gene of Drosophila melanogaster encodes a conserved member of the thioredoxin-like protein superfamily. Mol Genet Genomics 268: 692–697 [DOI] [PubMed] [Google Scholar]
- Greetham D, Vickerstaff J, Shenton D, Perrone GG, Dawes IW, Grant CM. (2010) Thioredoxins function as deglutathionylase enzymes in the yeast Saccharomyces cerevisiae. BMC Biochem 11: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirasawa M, Schürmann P, Jacquot JP, Manieri W, Jacquot P, Keryer E, Hartman FC, Knaff DB. (1999) Oxidation-reduction properties of chloroplast thioredoxins, ferredoxin:thioredoxin reductase, and thioredoxin f-regulated enzymes. Biochemistry 38: 5200–5205 [DOI] [PubMed] [Google Scholar]
- Holmgren A. (1985) Thioredoxin. Annu Rev Biochem 54: 237–271 [DOI] [PubMed] [Google Scholar]
- Hruz T, Laule O, Szabo G, Wessendorp F, Bleuler S, Oertle L, Widmayer P, Gruissem W, Zimmermann P. (2008) Genevestigator v3: a reference expression database for the meta-analysis of transcriptomes. Adv Bioinformat 2008: 420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquot JP, Buchanan BB, Martin F, Vidal J. (1981) Enzyme regulation in C(4) photosynthesis: purification and properties of thioredoxin-linked NADP-malate dehydrogenase from corn leaves. Plant Physiol 68: 300–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong W, Yoon HW, Lee SR, Rhee SG. (2004) Identification and characterization of TRP14, a thioredoxin-related protein of 14 kDa: new insights into the specificity of thioredoxin function. J Biol Chem 279: 3142–3150 [DOI] [PubMed] [Google Scholar]
- Johansson C, Lillig CH, Holmgren A. (2004) Human mitochondrial glutaredoxin reduces S-glutathionylated proteins with high affinity accepting electrons from either glutathione or thioredoxin reductase. J Biol Chem 279: 7537–7543 [DOI] [PubMed] [Google Scholar]
- Jung Y, Kim H, Min SH, Rhee SG, Jeong W. (2008) Dynein light chain LC8 negatively regulates NF-κB through the redox-dependent interaction with IκBα. J Biol Chem 283: 23863–23871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh CS, Navrot N, Didierjean C, Rouhier N, Hirasawa M, Knaff DB, Wingsle G, Samian R, Jacquot JP, Corbier Cet al. (2008) An atypical catalytic mechanism involving three cysteines of thioredoxin. J Biol Chem 283: 23062–23072 [DOI] [PubMed] [Google Scholar]
- Laloi C, Mestres-Ortega D, Marco Y, Meyer Y, Reichheld JP. (2004) The Arabidopsis cytosolic thioredoxin h5 gene induction by oxidative stress and its W-box-mediated response to pathogen elicitor. Plant Physiol 134: 1006–1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laloi C, Rayapuram N, Chartier Y, Grienenberger JM, Bonnard G, Meyer Y. (2001) Identification and characterization of a mitochondrial thioredoxin system in plants. Proc Natl Acad Sci USA 98: 14144–14149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaire SD, Michelet L, Zaffagnini M, Massot V, Issakidis-Bourguet E. (2007) Thioredoxins in chloroplasts. Curr Genet 51: 343–365 [DOI] [PubMed] [Google Scholar]
- Lennartz K, Plücken H, Seidler A, Westhoff P, Bechtold N, Meierhoff K. (2001) HCF164 encodes a thioredoxin-like protein involved in the biogenesis of the cytochrome b(6)f complex in Arabidopsis. Plant Cell 13: 2539–2551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin TY, Chen TS. (2004) A positive charge at position 33 of thioredoxin primarily affects its interaction with other proteins but not redox potential. Biochemistry 43: 945–952 [DOI] [PubMed] [Google Scholar]
- Menand B, Maréchal-Drouard L, Sakamoto W, Dietrich A, Wintz H. (1998) A single gene of chloroplast origin codes for mitochondrial and chloroplastic methionyl-tRNA synthetase in Arabidopsis thaliana. Proc Natl Acad Sci USA 95: 11014–11019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menchise V, Corbier C, Didierjean C, Jacquot JP, Benedetti E, Saviano M, Aubry A. (2000) Crystal structure of the W35A mutant thioredoxin h from Chlamydomonas reinhardtii: the substitution of the conserved active site Trp leads to modifications in the environment of the two catalytic cysteines. Biopolymers 56: 1–7 [DOI] [PubMed] [Google Scholar]
- Meng L, Wong JH, Feldman LJ, Lemaux PG, Buchanan BB. (2010) A membrane-associated thioredoxin required for plant growth moves from cell to cell, suggestive of a role in intercellular communication. Proc Natl Acad Sci USA 107: 3900–3905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer Y, Reichheld JP, Vignols F. (2005) Thioredoxins in Arabidopsis and other plants. Photosynth Res 86: 419–433 [DOI] [PubMed] [Google Scholar]
- Moon JC, Jang HH, Chae HB, Lee JR, Lee SY, Jung YJ, Shin MR, Lim HS, Chung WS, Yun DJ, et al (2006) The C-type Arabidopsis thioredoxin reductase ANTR-C acts as an electron donor to 2-Cys peroxiredoxins in chloroplasts. Biochem Biophys Res Commun 348: 478–484 [DOI] [PubMed] [Google Scholar]
- Navrot N, Collin V, Gualberto J, Gelhaye E, Hirasawa M, Rey P, Knaff DB, Issakidis E, Jacquot JP, Rouhier N. (2006) Plant glutathione peroxidases are functional peroxiredoxins distributed in several subcellular compartments and regulated during biotic and abiotic stresses. Plant Physiol 142: 1364–1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Ruiz JM, Spínola MC, Kirchsteiger K, Moreno J, Sahrawy M, Cejudo FJ. (2006) Rice NTRC is a high-efficiency redox system for chloroplast protection against oxidative damage. Plant Cell 18: 2356–2368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulido P, Spínola MC, Kirchsteiger K, Guinea M, Pascual MB, Sahrawy M, Sandalio LM, Dietz KJ, González M, Cejudo FJ. (2010) Functional analysis of the pathways for 2-Cys peroxiredoxin reduction in Arabidopsis thaliana chloroplasts. J Exp Bot 61: 4043–4054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichheld JP, Khafif M, Riondet C, Droux M, Bonnard G, Meyer Y. (2007) Inactivation of thioredoxin reductases reveals a complex interplay between thioredoxin and glutathione pathways in Arabidopsis development. Plant Cell 19: 1851–1865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren G, Stephan D, Xu Z, Zheng Y, Tang D, Harrison RS, Kurz M, Jarrott R, Shouldice SR, Hiniker A, et al. (2009) Properties of the thioredoxin fold superfamily are modulated by a single amino acid residue. J Biol Chem 284: 10150–10159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rey P, Cuiné S, Eymery F, Garin J, Court M, Jacquot JP, Rouhier N, Broin M. (2005) Analysis of the proteins targeted by CDSP32, a plastidic thioredoxin participating in oxidative stress responses. Plant J 41: 31–42 [DOI] [PubMed] [Google Scholar]
- Roos G, Garcia-Pino A, Van Belle K, Brosens E, Wahni K, Vandenbussche G, Wyns L, Loris R, Messens J. (2007) The conserved active site proline determines the reducing power of Staphylococcus aureus thioredoxin. J Mol Biol 368: 800–811 [DOI] [PubMed] [Google Scholar]
- Roos G, Geerlings P, Messens J. (2010) The conserved active site tryptophan of thioredoxin has no effect on its redox properties. Protein Sci 19: 190–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouhier N, Couturier J, Jacquot JP. (2006) Genome-wide analysis of plant glutaredoxin systems. J Exp Bot 57: 1685–1696 [DOI] [PubMed] [Google Scholar]
- Rouhier N, Gelhaye E, Gualberto JM, Jordy MN, De Fay E, Hirasawa M, Duplessis S, Lemaire SD, Frey P, Martin F, et al. (2004) Poplar peroxiredoxin Q: a thioredoxin-linked chloroplast antioxidant functional in pathogen defense. Plant Physiol 134: 1027–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouhier N, Gelhaye E, Jacquot JP. (2002) Glutaredoxin-dependent peroxiredoxin from poplar: protein-protein interaction and catalytic mechanism. J Biol Chem 277: 13609–13614 [DOI] [PubMed] [Google Scholar]
- Rouhier N, Kauffmann B, Tete-Favier F, Palladino P, Gans P, Branlant G, Jacquot JP, Boschi-Muller S. (2007) Functional and structural aspects of poplar cytosolic and plastidial type a methionine sulfoxide reductases. J Biol Chem 282: 3367–3378 [DOI] [PubMed] [Google Scholar]
- Rouhier N, Lemaire SD, Jacquot JP. (2008) The role of glutathione in photosynthetic organisms: emerging functions for glutaredoxins and glutathionylation. Annu Rev Plant Biol 59: 143–166 [DOI] [PubMed] [Google Scholar]
- Sanz-Barrio R, Fernández-San Millán A, Carballeda J, Corral-Martínez P, Seguí-Simarro JM, Farran I. (2012) Chaperone-like properties of tobacco plastid thioredoxins f and m. J Exp Bot 63: 365–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schürmann P, Buchanan BB. (2008) The ferredoxin/thioredoxin system of oxygenic photosynthesis. Antioxid Redox Signal 10: 1235–1274 [DOI] [PubMed] [Google Scholar]
- Schürmann P, Jacquot JP. (2000) Plant thioredoxin systems revisited. Annu Rev Plant Physiol Plant Mol Biol 51: 371–400 [DOI] [PubMed] [Google Scholar]
- Sotirchos IM, Hudson AL, Ellis J, Davey MW. (2009) A unique thioredoxin of the parasitic nematode Haemonchus contortus with glutaredoxin activity. Free Radic Biol Med 46: 579–585 [DOI] [PubMed] [Google Scholar]
- Tarrago L, Laugier E, Zaffagnini M, Marchand C, Le Maréchal P, Rouhier N, Lemaire SD, Rey P. (2009) Regeneration mechanisms of Arabidopsis thaliana methionine sulfoxide reductases B by glutaredoxins and thioredoxins. J Biol Chem 284: 18963–18971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarrago L, Laugier E, Zaffagnini M, Marchand CH, Le Maréchal P, Lemaire SD, Rey P. (2010) Plant thioredoxin CDSP32 regenerates 1-cys methionine sulfoxide reductase B activity through the direct reduction of sulfenic acid. J Biol Chem 285: 14964–14972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira Dos Santos C, Laugier E, Tarrago L, Massot V, Issakidis-Bourguet E, Rouhier N, Rey P. (2007) Specificity of thioredoxins and glutaredoxins as electron donors to two distinct classes of Arabidopsis plastidial methionine sulfoxide reductases B. FEBS Lett 581: 4371–4376 [DOI] [PubMed] [Google Scholar]
- Vieira Dos Santos C, Rey P. (2006) Plant thioredoxins are key actors in the oxidative stress response. Trends Plant Sci 11: 329–334 [DOI] [PubMed] [Google Scholar]
- Zaffagnini M, Bedhomme M, Marchand CH, Couturier JR, Gao XH, Rouhier N, Trost P, Lemaire SP. (2012a) Glutaredoxin s12: unique properties for redox signaling. Antioxid Redox Signal 16: 17–32 [DOI] [PubMed] [Google Scholar]
- Zaffagnini M, Bedhomme M, Marchand CH, Morisse S, Trost P, Lemaire SD. (2012b) Redox regulation in photosynthetic organisms: focus on glutathionylation. Antioxid Redox Signal 16: 567–586 [DOI] [PubMed] [Google Scholar]
- Zaffagnini M, Michelet L, Massot V, Trost P, Lemaire SD. (2008) Biochemical characterization of glutaredoxins from Chlamydomonas reinhardtii reveals the unique properties of a chloroplastic CGFS-type glutaredoxin. J Biol Chem 283: 8868–8876 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.