Abstract
Protein tyrosine phosphatases (PTPases) have long been thought to be activated by reductants and deactivated by oxidants, owing to the presence of a crucial sulfhydryl group in their catalytic centers. In this article, we report the purification and characterization of Reductant-Inhibited PTPase1 (ZmRIP1) from maize (Zea mays) coleoptiles, and show that this PTPase has a unique mode of redox regulation and signaling. Surprisingly, ZmRIP1 was found to be deactivated by a reductant. A cysteine (Cys) residue (Cys-181) near the active center was found to regulate this unique mode of redox regulation, as mutation of Cys-181 to arginine-181 allowed ZmRIP1 to be activated by a reductant. In response to oxidant treatment, ZmRIP1 was translocated from the chloroplast to the nucleus. Expression of ZmRIP1 in Arabidopsis (Arabidopsis thaliana) plants and maize protoplasts altered the expression of genes encoding enzymes involved in antioxidant catabolism, such as At1g02950, which encodes a glutathione transferase. Thus, the novel PTPase identified in this study is predicted to function in redox signaling in maize.
Plants have evolved a variety of adaptive responses to withstand the environmental stresses encountered during their life cycle. Stress-induced adaptive responses are mediated by cellular signal transduction, which involves complex networks of interconnected signaling pathways. Accordingly, identifying the key signal components will greatly enhance our understanding of the mechanisms that underlie plant stress tolerance and adaptation. As abscisic acid (ABA) is known to be a stress signal (Gomez et al., 1988; Mundy and Chua,1988; Qin and Zeevaart, 2002; Zhu, 2002; Saleh et al., 2005; Verslues and Zhu, 2005), our previous work focused on the mechanisms underlying stress-induced ABA accumulation (Jia and Zhang 2000; Jia et al. 2001, 2002). We revealed that ABA accumulation is controlled by cellular redox status (Jia and Zhang, 2000) and showed that cellular redox status is tightly associated with the expression of a range of crucial stress-responsive genes, such as Responsive to dessication29A (Rd29A), Responsive to dessication29B (Rd29B), early responsive to dehydration10 (ERD10), abscisic aldehyde oxidase (AAO3), and low temperature-induced30 (LTI30). Thus, redox-associated signaling appears to play an important role in plant stress tolerance and adaptation.
Aerobic metabolic processes lead to the production of reactive oxygen species (ROS) in all aerobic organisms (Apel and Hirt, 2004). To withstand the toxicity of excess ROS, plants have evolved a complex array of antioxidant mechanisms that maintain cellular redox homeostasis (Noctor and Foyer, 1998; Buchanan and Balmer, 2005; Xing et al., 2007). As a consequence, the cellular redox status is determined by the integration of ROS activity and the antioxidant system. Emerging evidence suggests that alterations in cellular redox status are exploited for signaling purposes in every type of living organism (Forman et al., 2002; López-Martín et al., 2008; Meyer, 2008). Although both the antioxidant system and redox-associated signaling have been extensively studied, the mechanisms underlying redox regulation in plant signaling, particularly those that relay redox alteration or the redox signal, are poorly understood (Forman et al., 2002; Apel and Hirt, 2004; Oelze et al., 2008). Redox signal relay was thought to reversibly alter the properties of a protein at the posttranslational level by changing its oxidation state. The thiol group of the amino acid Cys is highly sensitive to oxidation, making Cys-containing proteins critical in redox signaling. Protein Tyr phosphatase (PTPase), the activity of which is controlled by the redox state of the cell, is one such protein.
Given the previous finding that stress-induced ABA accumulation can be arrested by reductants (Jia and Zhang, 2000), we examined the function of PTPase in ABA accumulation and stress responses. Interestingly, the PTPase-specific inhibitor, phenylarsine oxide (PAO), can arrest not only ABA accumulation, but also the expression of the stress-responsive genes, suggesting that PTPase plays a critical role in stress-associated redox signaling.
The PTPase family consists of two major groups: the Tyr-specific PTPases, which exclusively dephosphorylate Tyr residues, and the dual-specific protein phosphatases, which dephosphorylate both Tyr and Ser/Thr residues. Both groups of PTPases are characterized by a catalytic domain that contains an active site Cys residue within a conserved diagnostic motif (HCx5R). Oxidants, such as hydrogen peroxide (H2O2), are thought to oxidize the Cys residue in the active center of the PTPase, and thereby inactivate the enzyme. PAO inhibits PTPases by modifying the thiol group of the catalytic Cys residue. In contrast, reductants, such as dithiothreitol (DTT), maintain PTPase in an active state by reducing the reactive Cys residue (Smith and Walker, 1996; Luan et al., 2001; Tonks, 2005). Previous studies showed that stress-induced ABA accumulation and numerous other stress responses are inhibited by both reductants and PAO. This led to the suggestion that plant cells contain a PTPase that can be deactivated rather than activated by reductants. However, to our knowledge, no PTPase that is inhibited by a reductant has been reported from either animal or plant cells. Therefore, identifying a putative reductant-inhibited PTPase in plant cells is of particular interest and will enhance our understanding of the mechanisms involved in redox signaling and plant stress responses.
Using various biochemical techniques, we showed that maize (Zea mays) cells contain several PTPase components that are inactivated rather than activated by reductants. One major component was purified to homogeneity and designated Reductant-Inhibited PTPase1 (ZmRIP1). A Cys residue near the catalytic center of ZmRIP1 was shown to confer the unique mode of redox regulation. Interestingly, ZmRIP1 was found to migrate from chloroplasts to nuclei in response to an oxidative stimulus, implying that it acts as a chloroplast-to-nucleus signaling messenger. This study therefore provides insight into the complex molecular mechanism of redox signaling in maize.
RESULTS
Identification and Purification of Reductant-Inhibited PTPases
Preliminary work demonstrated that both reductants and the PTPase inhibitor, PAO, could block the expression of a series of stress-responsive genes (Supplemental Fig. S1), suggesting that plant cells contain forms of PTPase that can be deactivated rather than activated by reductants. We sought to identify and purify such a PTPase using column chromatography. Total proteins prepared from maize coleoptiles were passed through a DEAE-Sepharose anion-exchange column and PTPase activity was determined based on the dephosphorylation activity toward the [pTyr1018]-epidermal growth factor (EGF) receptor substrate. After elution with a 0 to 0.6 m NaCl gradient, several fractions containing PTPase activity were identified, among which two major fractions (i.e. peak 1 and 2; Fig. 1A) showed the highest activity. Interestingly, peaks 1 and 2 exhibited differed sensitivities to PAO; whereas peak 1 was sensitive to inactivation by PAO, peak 2 showed no sensitivity. Both peaks were inactivated by vanadate, which inhibits phosphatases. In light of previous observations that water-deficit-induced ABA accumulation is blocked by PAO, only peak 1 was pooled and further purified through a SP-Sepharose cation column. Elution with a 0 to 0.5 m NaCl gradient resulted in three peaks (i.e. peaks 1, 2, and 3; Fig. 1B). Peak 2 was not only inactivated by PAO, but was also readily inactivated by DTT, suggesting that maize cells contain reductant-inhibited PTPases, as predicted. Peak 2 was therefore further purified by two successive chromatography columns, i.e. Sephacryl S-200 (Fig. 1C) and Sephacryl S-300 (Fig. 1D) and, after several steps of purification, a sole peak was obtained (Fig. 1D). Biochemical analysis confirmed that the PTPase activity of the purified protein was readily inactivated by both PAO and DTT, whereas it showed no sensitivity to H2O2 and cation regulators normally thought to be important, such as Ca2+, Mg2+, and Mn2+ (Supplemental Fig. S2).
Figure 1.
Column chromatography of crude protein extract from maize coleoptiles. A, The crude protein extract was first desalted on a Sephadex G-25 M column, and then loaded onto a DEAE-Sepharose column. B, The chromatography of step A generated two major peaks; peak 1 was loaded onto a SP Sepharose F.F. column. C, The chromatography of step B generated three major peaks; peak 2 was concentrated and applied to a Sephacryl S-200 column. D, The major peaks obtained in step C were further concentrated and applied to a Sephacryl S-300 column. For A to D, the protein concentration (black circle) in each fraction was monitored by the Bradford Method (Bradford, 1976), and the PTPase activity (white circle) was determined using the [pTyr1018]-EGF receptor as substrate, as described in “Materials and Methods.”
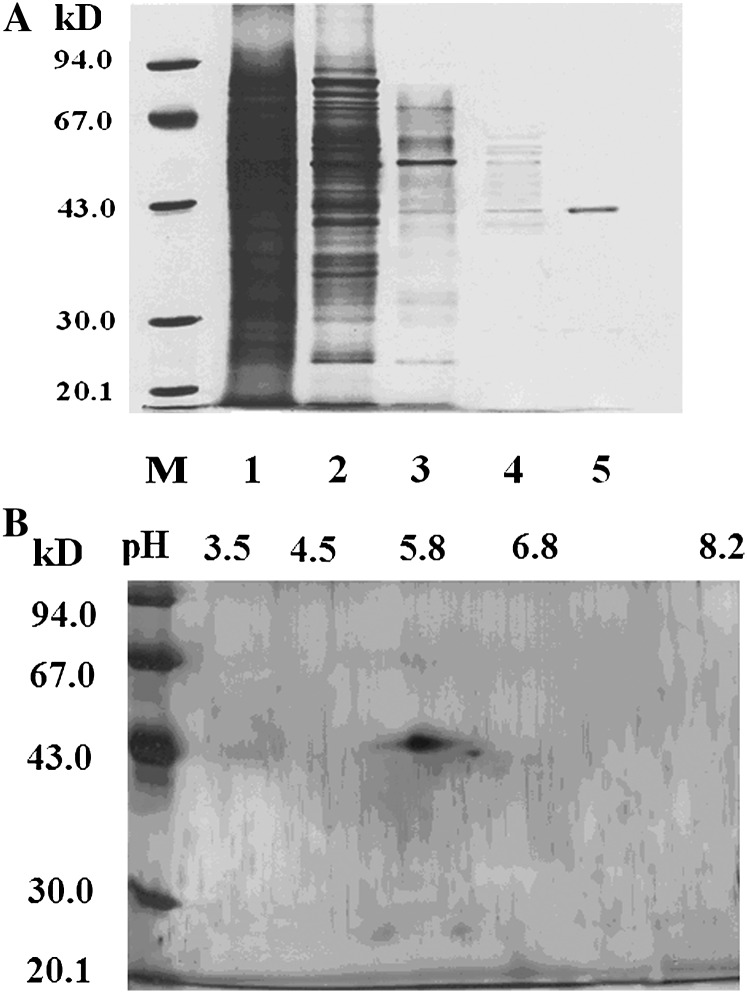
A summary of the purification procedure is shown in Table I. The final specific activity represents a 1,611-fold purification compared with the initial activity in the crude extract. SDS-PAGE showed that the final purification step resulted in a sole band with a Mr of approximately 42 kD (Fig. 2A). The homogeneity of this protein was further evaluated by two-dimensional PAGE with silver staining. Although another faint spot was also observed, only one prominent spot was present, suggesting that the purified protein is almost homogenous (Fig. 2B). We named the protein ZmRIP1, to reflect its unique mode of redox regulation.
Table I. Purification of ZmRIP1 from maize coleoptiles.
Purification was performed as described in “Materials and Methods.” For a single preparation, 150 g of coleoptile was used. Protein was determined by the Bradford Method (Bradford, 1976), and PTPase activity was determined using [pTyr1018]-EGF receptor as substrate.
| Step | Total Protein | Specific Activity | Fold |
|---|---|---|---|
| mg | mmol min−1 mg−1 | ||
| Triton extract | 1,267 | 0.36 | 1 |
| DEAE column | 218 | 2.11 | 6 |
| SP-Sepharose | 17.2 | 63 | 175 |
| Sephacryl S-200 | 0.12 | 325 | 541 |
| Sephacryl S-300 | <0.02 | 580 | 1,611 |
Figure 2.
SDS/PAGE and IEF/SDS-PAGE analysis of the chromatography-purified PTPase. A, The protein fractions from the chromatography column were run with a 12% acrylamide monomer concentration. Key: M, Molecular weight marker; 1, crude extract; 2, DEAE-Sepharose chromatography; 3, SP Sepharose F.F. chromatography; 4, Sephacryl S-200 chromatography; 5, Sephacryl S-300 chromatography. B, The final protein fraction eluted through a Sephacryl S-300 chromatography column was subjected to IEF/SDS-PAGE with a 12% acrylamide monomer concentration and the protein was stained with silver.
Molecular Cloning of ZmRIP1
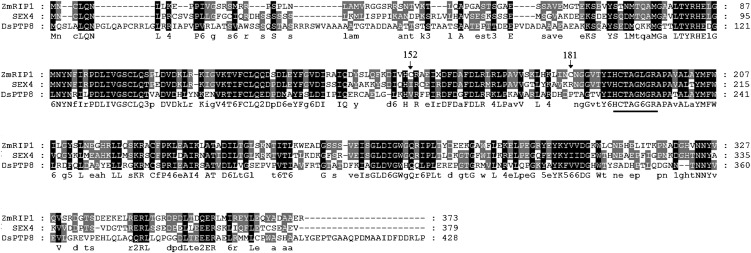
To clone the gene encoding ZmRIP1, N-terminal sequencing of ZmRIP1 was performed; based on the N-terminal sequence, we first searched the National Center for Biotechnology Information (NCBI) database and found that this sequence is almost identical to a hypothetical protein, LOC100216768. The primary structure of LOC100216768 contains a conserved diagnostic motif of PTPase (HCx5R), suggesting that this protein is indeed a PTPase. A search of the maize genetics and genomics database (http://www.maizegdb.org) identified the corresponding gene. The cDNA of this gene is 1,548-bp long and contains 96 bp of the 5′-untranslated region followed by a 1,122-bp open reading frame and a 383-bp 3′-untranslated region. We named the gene ZmRIP1 (accession no. FJ605095) and subsequently cloned it using a primer pair based on its sequence information. Blast analysis of ZmRIP1 in the NCBI database showed major similarities with two proteins; it exhibited 59% identity to starch excess4 (SEX4) from Arabidopsis (Arabidopsis thaliana) and 43% identity to the dual-specificity protein phosphatase 8 from Chlamydomonas reinhardtii (Fig. 3).
Figure 3.
Alignment of ZmRIP1 with SEX4 from Arabidopsis and the dual-specificity protein phosphatase 8 from C. reinhardtii. The amino acid alignments were performed with Clustal X, and the shading of the alignment was generated with GeneDoc software. The conserved diagnostic motif of PTPases is underlined.
In Vitro Expression and Biochemical Characterization
To confirm that ZmRIP1 is the exact gene encoding the purified protein, in vitro expression of ZmRIP1 was carried out. The gene was expressed as a His6-tag fusion protein in yeast (Saccharomyces cerevisiae) cells after induction with Gal. Elution through a nickel affinity column resulted in a major band of approximately 42 kD (Supplemental Fig. S3A). As shown in Supplemental Figure S3B, PTPase activity was detected in the cells grown in Glc, suggesting that the yeast cells themselves contain PTPases. Further analysis showed that this PTPase activity in yeast cells could be activated by the reductant DTT. Induction by Gal led to a significant increase in total PTPase activity and, moreover, this PTPase activity could be inactivated by DTT (Supplemental Fig. S3B). These results confirmed that the protein purified from yeast cells grown in Gal is ZmRIP1, and that ZmRIP1 is the gene that encodes the protein purified from maize.
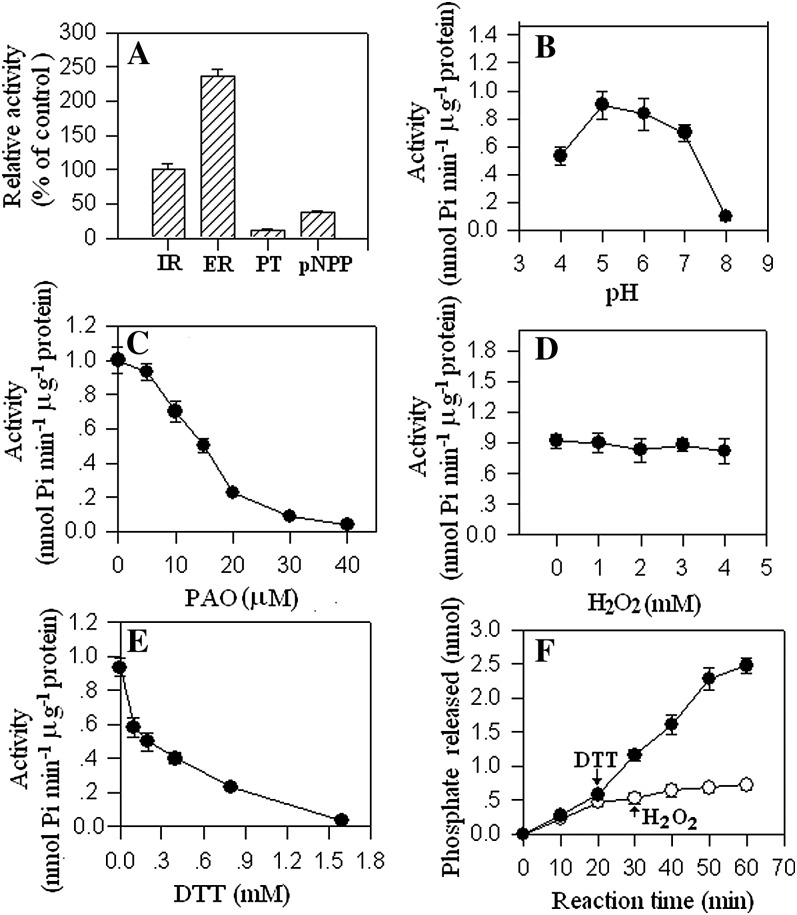
A detailed biochemical characterization is shown in Figure 4. Among several substrates tested, ZmRIP1 showed the highest activity toward [pTyr1018]-EGF receptor (Fig. 4A). PTPase activity peaked at pH 5.0 and decreased significantly at pH 8.0 (Fig. 4B). ZmRIP1 was sensitive to the PTPase-specific inhibitor PAO: 50 μm PAO completely inactivated the enzyme (Fig. 4C). Surprisingly, ZmRIP1 showed no sensitivity to the common inhibitor H2O2. As shown in Figure 4D, 4 mm H2O2 had no effect on the activity of the enzyme. By contrast, ZmRIP1 was readily inhibited by reductants such as DTT; less than 2 mm DTT completely inactivated the activity of the enzyme (Fig. 4E). Further experiments showed that subsequent addition of H2O2 could not relieve the inhibition of ZmRIP1 activity caused by the addition of DTT in the reaction system (Fig. 4F).
Figure 4.
Biochemical characterization of ZmRIP1. A, ZmRIP1 was expressed in and purified from yeast cells and the enzyme activity was assayed with and without the addition of the indicated effectors to the reaction buffer. Key: IR, [pTyr1146]-insulin receptor; ER, [pTyr1018]-EGF receptor; PT, phosphotyrosine; pNPP, p-nitrophenyl phosphate. B to E, ZmRIP1 was expressed in and purified from yeast cells and the enzymatic activity was determined using [pTyr1018]-EGF receptor as substrate. Various effectors were either added or not to the reaction buffer. Values are means ± se of four samples. F, Twenty minutes after the reaction started, DTT was either added (white circle) or not (black circle; i.e. control) to the reaction mixture and, after another 10 min, H2O2 was added to the reaction mixture. Values are means ± se of four samples.
Redox Regulation of ZmRIP1 Activity in Relation to Reactive Cys Residues
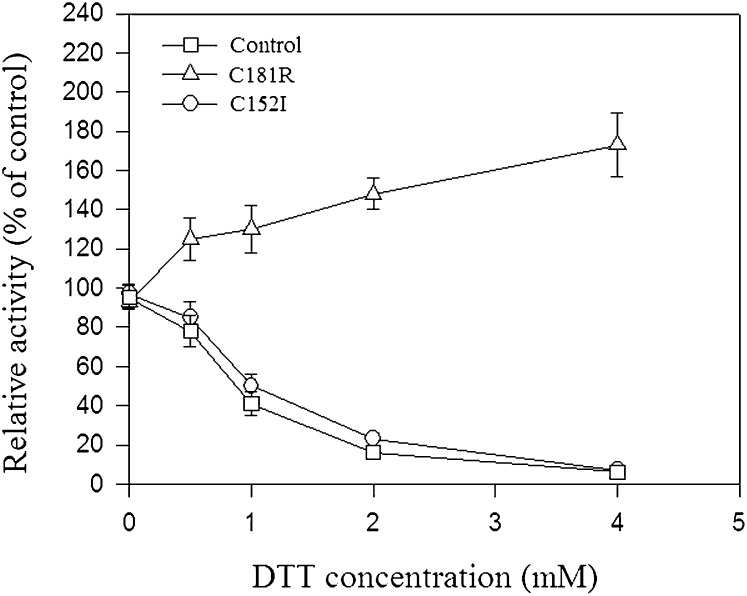
Whereas PTPases are widely known to be activated and inactivated by DTT and H2O2, respectively, ZmRIP1 was found to be inactivated by DTT and unaffected by H2O2. The unique mode of redox regulation of ZmRIP1 prompted us to investigate possible mechanisms underlying ZmRIP1 regulation. In view of the central roles of reactive Cys residues in redox regulation, we investigated the primary structure of ZmRIP1 and found that Cys-152, Cys-181, and the Cys residue in the active center (i.e. Cys-190) are likely to be vicinal cysteines. More importantly, the alignment analysis showed that Cys-152 and Cys-181 are uniquely present in ZmRIP1 (Fig. 3). To examine whether Cys-152 and Cys-181 are involved in the redox regulation of ZmRIP1, the two cysteines were mutated to Ile and Arg residues, respectively, and the effect of this mutation on the redox regulation of ZmRIP1 was evaluated. As shown in Figure 5, mutation of Cys-181 changed the effect of DTT on ZmRIP1 activity from inactivation to activation, whereas mutation of Cys-152 did not affect the redox regulation of ZmRIP1. This suggests that Cys-181, but not Cys-152, is associated with the unique mode of redox regulation of ZmRIP1. A major means by which Cys residues regulate protein activity is by forming disulfide bonds, and thereby altering the protein sulfhydryl content. To determine whether Cys-181 or Cys-152 form disulfides, we evaluated the effect of Cys-181 or Cys-152 mutation on sulfhydryl content. Whereas mutation of Cys-152 resulted in a lower concentration of sulfhydryl than in the wild-type protein, mutation of Cys-181 caused an increase in the level of sulfhydryl (Table II), suggesting that Cys-181 modulates ZmRIP1 activity via a mechanism that regulates disulfide bond formation.
Figure 5.
The effect of point mutations on the biochemical properties of ZmRIP1. Cys-152 and Cys-181 were mutated to Ile-152 and Arg-181, respectively, as described in “Materials and Methods.” Enzymatic activity was determined using [pTyr1018]-EGF receptor as a substrate. DTT was added to the reaction mixture. Values are means ± se of four samples.
Table II. The effect of point mutation on the sulfhydryl content of ZmRIP1.
Cys-152 and Cys-181 were mutated to Ile-152 and Arg-181, respectively, and the sulfhydryl content was determined as described in “Materials and Methods.” Values are means ± se of six samples in three independent experiments. Data were analyzed using Student’s t test. The significance of any differences between wild-type and mutated proteins is indicated by asterisks: *, P < 0.05, where P = 0.0307; **, P < 0.01, where P = 0.0004; n = 6.
| Samples | Experiments | Protein Concentration | A324nm | Sulfhydryl Concentration | Sulfhydryl Content |
|---|---|---|---|---|---|
| μg mL−1 | optical density | μm | pmol ng−1 (Mean ± se) | ||
| Control | Exp. 1 | 31.76 | 1.045 | 6.13 | 192.90 |
| 1.024 | 6.00 | 188.99 | |||
| Exp. 2 | 15.54 | 0.532 | 3.12 | 200.60 | |
| 0.531 | 3.11 | 200.14 | |||
| Exp. 3 | 16.27 | 0.521 | 3.05 | 187.74 | |
| 0.515 | 3.02 | 185.40 | |||
| (192.63 ± 2. 64) | |||||
| C152I | Exp. 1 | 30.22 | 0.879 | 5.15 | 170.53 |
| 0.868 | 5.09 | 168.39 | |||
| Exp. 2 | 21.25 | 0.596 | 3.50 | 164.49 | |
| 0.619 | 3.63 | 170.86 | |||
| Exp. 3 | 28.24 | 0.818 | 4.79 | 169.69 | |
| 0.801 | 4.70 | 166.30 | |||
| (168.38 ± 1.03**) | |||||
| C181R | Exp. 1 | 18.84 | 0.622 | 3.65 | 193.41 |
| 0.632 | 3.70 | 196.55 | |||
| Exp. 2 | 26.10 | 0.954 | 5.59 | 214.26 | |
| 0.952 | 5.58 | 213.77 | |||
| Exp. 3 | 25.62 | 0.988 | 5.79 | 226.05 | |
| 0.942 | 5.52 | 215.48 | |||
| (209.12 ± 5. 09*) |
Chloroplast-to-Nucleus Translocation
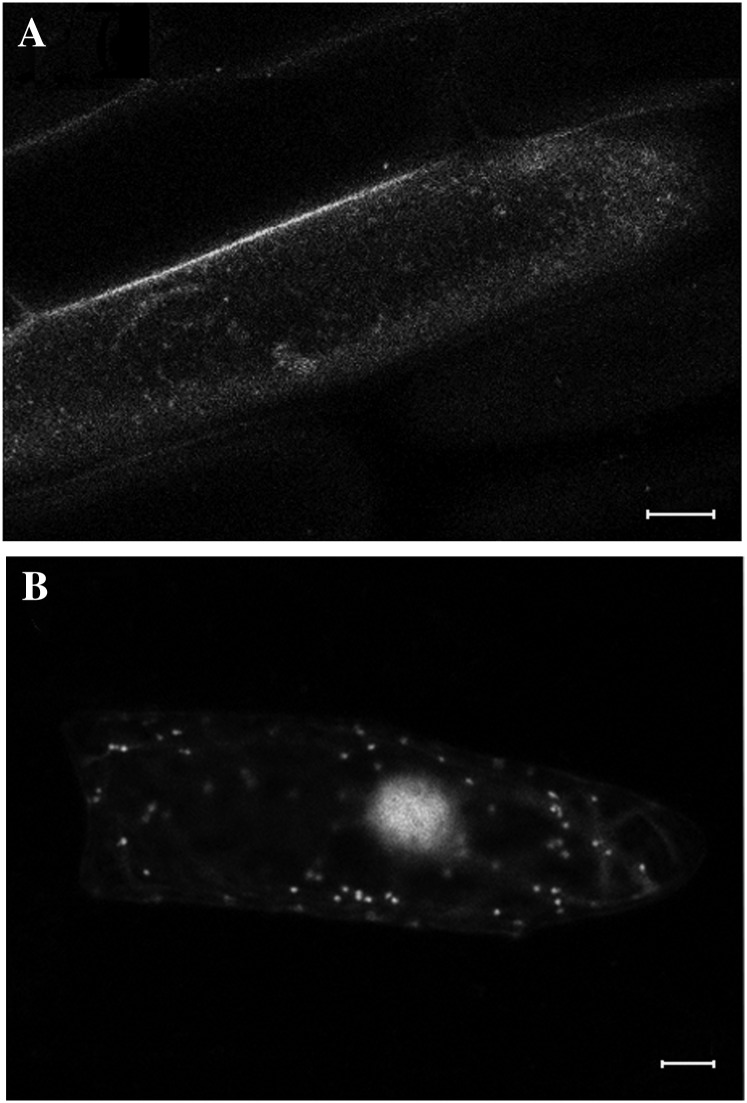
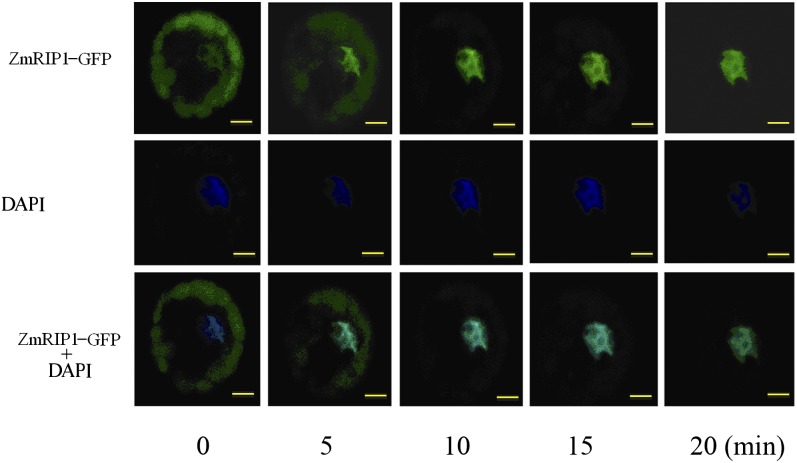
When GFP-ZmRIP1 was introduced into onion (Allium cepa) epidermal cells by particle-mediated DNA delivery, strong fluorescence was observed in nuclei and moderate fluorescence was observed in some small organelles, thought to be plastids (Fig. 6). In maize protoplasts, however, strong fluorescence appeared mainly in chloroplasts in most cells examined. Interestingly, in response to H2O2 treatment, the fluorescence distribution changed and various distinct fluorescence patterns were observed. Four representative fluorescence patterns are shown in Figure 7A: pattern I shows strong fluorescence only in chloroplasts; pattern II shows strong fluorescence in chloroplasts and faint fluorescence in nuclei; pattern III shows fluorescence mainly in nuclei, with faint fluorescence in chloroplasts; and pattern IV shows strong fluorescence only in nuclei. As shown in Figure 7, B and C, under control conditions, >90% of the cells show patterns I and II; by contrast, upon H2O2 treatment, nearly 80% of the cells show patterns III and IV. Furthermore, we noticed that UV excitation during confocal microscopy analysis could trigger the rapid translocation of ZmRIP1 to the nucleus. As shown in Figure 8, the fluorescence of GFP-ZmRIP1 within an individual cell was initially localized to chloroplasts, although faint fluorescence could also be observed in the nucleus. However, soon after UV excitation, stronger fluorescence appeared in the nucleus, whereas the fluorescence intensity in chloroplasts declined. Twenty minutes later, fluorescence was predominantly localized to the nucleus. These observations strongly indicate that ZmRIP1 is capable of translocating from chloroplasts to nuclei.
Figure 6.
Localization of ZmRIP1 in onion epidermal cells. Constructs containing ZmRIP1 fused downstream of GFP were bombarded into onion epidermal cells. GFP fluorescence was then examined by confocal laser-scanning microscopy. A, Construct containing nonfused GFP. B, Construct containing ZmRIP1::GFP. Scale bars = 20 μm.
Figure 7.
Localization and translocation analysis of ZmRIP1 in maize protoplasts. Constructs containing ZmRIP1 fused to GFP were transformed into maize protoplasts and GFP fluorescence was then examined by confocal laser-scanning microscopy. A, Images showing distinct localization patterns of ZmRIP1 in maize protoplasts (scale bars = 5 μm). Pattern I, strong fluorescence is present only in chloroplasts; pattern II, strong fluorescence is present in chloroplasts and faint fluorescence appears in nuclei; pattern III, strong fluorescence is present in nuclei, whereas faint fluorescence appears in chloroplasts; pattern IV, strong fluorescence is present only in nuclei. B, Images showing the effect of H2O2 and DTT treatment on the localization patterns of ZmRIP1 (scale bars = 20 μm). C, Quantitative assessment of ZmRIP1 localization. The number of cells belonging to each of the four different categories described above was determined, and the ZmRIP1 localization was expressed as a percentage of the number of cells of each pattern relative to the total number of cells assessed. Data were analyzed using Student’s t test. Significant differences between control and treated cells belonging to the same category are indicated by asterisks. *, P < 0.01; n = 200.
Figure 8.
Time-course analysis of ZmRIP1 translocation within individual cells. Constructs containing ZmRIP1 fused to GFP were transformed into maize protoplasts and GFP fluorescence was examined by confocal laser-scanning microscopy. The numbers below the figure indicate the time after UV excitation was switched on. Scale bars = 5 μm.
A Role for ZmRIP1 in the Regulation of Genes Encoding Enzymes Involved in Antioxidant Catabolism
Given the potential role of ZmRIP1 in stress-induced ABA accumulation, we further investigated whether ZmRIP1 regulates the expression of a series of genes encoding key enzymes in the ABA biosynthesis pathway. We generated ZmRIP1-expressing transgenic Arabidopsis lines using the LexA-VP16-Estragon Receptor-inducible system. In response to estradiol treatment, ZmRIP1 expression was significantly up-regulated in most of the lines (Supplemental Fig. S4A). To optimize the conditions for the estradiol treatment, line 1 was treated with different concentrations of estradiol for various durations and, based on these results (Supplemental Fig. S4B), the plants were subjected to 10 μm estradiol for 24 h in the following experiment. As shown in Figure 9A, whereas estradiol treatment promoted ZmRIP1 expression, it had no effect on the expression of any of the genes involved in ABA biosynthesis. This suggests that ZmRIP1 is not a mediator of stress-induced ABA accumulation.
Figure 9.
The effect of ZmRIP1 on the expression of genes involved in ABA biosynthesis or the antioxidant system. A, Analysis of gene expression in Arabidopsis transformed with estradiol-inducible ZmRIP1. Wild-type Arabidopsis (ecotype Columbia-0) was transformed with estradiol-inducible pER8-ZmRIP1, and gene expression in transgenic seedlings of line 1 and 7 treated with (+) or without (−) 5 mol/L estradiol for 24 h was evaluated by RT-PCR. B, Analysis of At1g02950 promoter activity in a maize protoplast transient expression system. Protoplasts were prepared from maize leaves. Effector plasmids 35S:ZmRIP1 or empty plasmid pEZS-NL (control) were cotransfected with reporter plasmid pAt1g02950:LUC into maize protoplasts, which were then incubated with or without 200 μm H2O2. Assays were carried out as described in “Materials and Methods.” Values are means ± se of four biological replicates.
To determine the function of ZmRIP1 in cellular redox regulation, we further investigated whether ZmRIP1 is involved in the regulation of a range of genes involved in antioxidant catabolism, such as catalases, superoxide dismutase, ascorbate peroxidase, glutathione reductase, glutathione peroxidase, glutathione transferase, and alternate oxidase (Supplemental Table S1; Supplemental Fig. S5). Among the 25 genes tested by reverse transcription (RT)-PCR, the expression of one gene, At1g02950, which encodes a glutathione transferase, was found to be significantly promoted in response to estradiol treatment (Fig. 9A). To further demonstrate that ZmRIP1 regulates the expression of this gene, we performed a maize protoplast transient expression analysis. As shown in Figure 9B, luciferase activity in protoplasts cotransformed with 35S-ZmRIP1 effector and pAt1g02950-LUC reporter was significantly promoted in response to H2O2 treatment, suggesting that ZmRIP1 mediates the expression of At1g02950.
DISCUSSION
Although the association between cellular redox status regulation and ROS metabolism has been extensively studied for many years, the mechanisms underlying redox-regulated signaling remain largely unknown. In this study, we observed that reductants could arrest the expression of many stress-responsive genes (Supplemental Fig. S1), indicating that an alteration in cellular redox status may play a role in stress signaling. Redox signaling is thought to be mediated by signaling proteins whose activity is regulated by cellular redox status. PTPases are just such proteins, owing to the presence of an active thiol group in their catalytic centers. To test whether PTPases are indeed involved in stress signaling, we examined the effect of PAO (a widely used specific inhibitor of PTPases) on the expression of the water stress-responsive genes. As expected, PAO arrested the expression of a series of crucial stress-responsive genes, including those controlling ABA accumulation. While the pharmacological experiments using both reductants and PAO demonstrated that redox signaling may play an important role in mediating plant responses to water stress, they also raise the possibility that PTPases are involved in stress signaling. PAO and reductants are known to have opposite effects on the regulation of PTPase activity; however, their effects on the expression of many stress-responsive genes were found to be similar (Supplemental Fig. S1). Supposing that the regulatory effects of PAO and reductants on gene expression both resulted from the modulation of PTPases activity, a reasonable explanation for this is that plant cells contain a kind of PTPase that is inactivated rather than activated by reductants. In support of this hypothesis, column chromatography analysis showed that a crude extract of maize coleoptiles indeed contained several PTPase fractions that could be deactivated rather than activated by reductants (Fig. 1).
To our knowledge, this type of PTPase has not been reported before in either animal or plant cells. To further characterize this unique type of PTPase, one reductant-inhibited PTPase protein, designated ZmRIP1, was purified and the encoding gene was cloned. ZmRIP1 showed relatively high similarity with SEX4 from Arabidopsis (Fig. 3). SEX4 is localized to chloroplasts and is involved in starch metabolism (Fordham-Skelton et al., 2002; Kötting et al., 2009). However, unlike SEX4, ZmRIP1 was found to be mainly localized to the nucleus and to small organelles of unknown identity (possibly some type of plastid, e.g. leucoplasts) in onion epidermal cells, or to the nucleus and chloroplasts in maize cells. Further experiments indicated that ZmRIP1 regulates At1g02950, a gene encoding glutathione transferase (Supplemental Fig. S4), whereas it appears to have no role in starch metabolism (data not shown). These results indicate that ZmRIP1 and SEX4 are two functionally different types of PTPase, despite their structural similarities.
Disulfide alteration is a key factor controlling PTPase activity. A change in a single Cys residue is sufficient to have a profound effect on the molecular architecture of PTPases and thus on their enzymatic activity (Mayadas and Wagner, 1992; Chiarugi et al., 2001; Paget and Buttner, 2003; Tonks, 2005; Zimmermann et al., 2007; Winterbourn and Hampton, 2008; Álvarez et al., 2009; Bucciarelli et al., 2009). With this in mind, we searched the protein homology of ZmRIP1 and found that distinct differences between ZmRIP1 and its homologs exist in the Cys residues at two sites (i.e. Cys-152 and Cys-181). Moreover, it was found that these two residues and the catalytic Cys residue (i.e. Cys-190) are close together, and this implies that some critical disulfides might form among these sites. As expected, mutation of Cys-181 altered the redox regulatory property of ZmRIP1 (Fig. 5). The exact mechanism whereby reductants deactivate ZmRIP1 remains unknown. Because mutation of Cys-181 resulted in a decrease in sulfhydryl content, we propose that formation of a Cys-181-based disulfide bond might underlie this mechanism. Reductants might abolish the Cys-181-based disulfide bonds and produce a profound change in the architecture of ZmRIP1 that conceals the catalytic Cys residue. Generally, reductants and oxidants have opposite roles in the regulation of PTPases activity. However, although ZmRIP1 can be readily deactivated by reductants, it shows no sensitivity to H2O2 (Fig. 4D). The reason for this is complex. There is evidence that the redox regulation of PTPases may depend on cofactors; for example, it was reported that inactivation of a PTPase, PTP1B in animal cells, by H2O2 depends on the presence of calcium (Skorey et al., 1997). In this study, the effect of H2O2 on ZmRIP1 was determined in vitro, and it is not known whether and how H2O2 might control ZmRIP1 activity in vivo.
It has been increasingly shown that chloroplast-to-nucleus signaling has a role in the regulation of many cellular biological processes (Nott et al., 2006; Koussevitzky et al., 2007; Ryan and Hoogenraad, 2007; Oelze et al., 2008); however, the exact mechanisms for this are unclear. Plastid metabolites, such as magnesium-protoporphyrin IX and plastoquinone, oxidants (particularly H2O2), and reductants were demonstrated or proposed to be primary signals that mediate nuclear gene expression (Nott et al., 2006; Ryan and Hoogenraad, 2007). However, a direct transfer of a signal protein involved in reversible protein phosphorylation from a plastid to the nucleus has not been reported in plant cells. In animal cells, it was suggested that signaling from the cytoplasm to the nucleus is associated with reversible protein phosphorylation, as evidenced by the transfer of mitogen-activated proteins from the cytoplasm to the nucleus (Kaffman and O’Shea, 1999). In this study, ZmRIP1 was found to move from chloroplasts to the nucleus in response to H2O2. The H2O2-induced ZmRIP1 transfer was relatively slow, typically taking more than 10 h. Interestingly, when ZmRIP1 movement in an individual cell was continuously observed under the UV laser of the confocal microscope, it was found that the UV stimulus could trigger a much quicker transfer of ZmRIP1 from chloroplasts to the nucleus. UV is known to be a strong oxidative stress. Hence, ZmRIP1 appears to be a plastid-to-nucleus transmitted signal and an important player in plant redox signaling.
The exact biological function of ZmRIP1 is of particular interest. Preliminary information provided in this study suggests that ZmRIP1 is involved in the regulation of cellular redox homeostasis, as evidenced by the observation that overexpression of ZmRIP1 resulted in up-regulation of At1g02950, a gene encoding a glutathione transferase that is closely associated with the regulation of redox homeostasis (Supplemental Fig. S5). As fundamental cellular metabolic processes and efficient redox signaling can be guaranteed if redox homeostasis is optimally maintained (Meyer, 2008), there must exist a delicate mechanism that regulates redox homeostasis in plant cells. Given that: (1) ROS metabolism is known to be a major factor in the control of cellular redox homeostasis; (2) excess ROS is produced not only in response to environmental stresses, but also during certain developmental processes under normal conditions; and (3) ROS production is compartmentalized, with some plastids, including chloroplasts, being the major ROS production sites (Hansen et al., 2006; Maulucci et al., 2009; Ushio-Fukai, 2009), the transfer of ZmRIP1 from the chloroplast to the nucleus in response to H2O2 appears to suggest a feedback mechanism for regular regulation of redox homeostasis in maize cells.
The initial objective of this study was to demonstrate the existence of a reductant-inhibited PTPase that has a key role in triggering ABA signal production and the up-regulation of a series of crucial stress-responsive genes in maize. Although a reductant-inhibited PTPase (ZmRIP1) was identified, it was not found to have any relationship with ABA accumulation or the regulation of genes involved in the water-deficit stress response. This implies that, if stress-induced ABA accumulation and other stress responses are indeed associated with reductant-inhibited PTPases, maize cells contain other reductant-inhibited PTPases. Consistent with this, the first chromatography run detected a major PTPase fraction that could readily be deactivated by reductant (Fig. 1A). Further chromatography of this PTPase fraction again revealed several major PTPase fractions that could be readily deactivated by reductant (Fig. 1B). Thus, maize coleoptile cells contain numerous PTPase members that can be deactivated by reductants. However, PTPase activity was examined using a widely used PTPase substrate, the pTyr1018-EGF receptor in this study. It is unclear whether other protein phosphatases show activity toward this substrate. Nonetheless, this study demonstrated the existence of PTPases in plant cells that are deactivated rather than activated by reductants. Pharmacological experiments using reductants (Supplemental Fig. S1; Jia and Zhang, 2000) suggest that the ability of plants to respond to environmental stresses is tightly controlled by cellular redox potential (e.g. it decreases under a low redox potential). This finding that multiple signal proteins are deactivated under a low redox potential contributes to our understanding of the mechanisms involved in the redox regulation of plant stress responses. Since the first ancient life forms evolved under reducing conditions, most fundamental metabolic processes were initially adapted to reducing conditions (Meyer, 2008). It is not known whether this adaptation to reducing conditions is correlated with the tight control of the plant’s responses to changing redox potentials. The biological functions and significance of these reductant-inhibited phosphatases merit further exploration.
MATERIALS AND METHODS
Purification of ZmRIP1
Frozen maize (Zea mays) coleoptiles (150 g) were homogenized with two volumes of buffer A (50 mm MOPS pH 7.4; 10% glycerol; 5 mm EDTA; 1 mm phenylmethylsulfonyl fluoride [PMSF]; 0.5 μg/mL aprotinin; 1.5 μg/mL leupeptin; 0.6 μg/mL pepstain A; 0.5% Triton X-100). The homogenate was filtered through four layers of cheesecloth and centrifuged at 10,000g for 15 min and the supernatant was collected and designated as the crude extract. This was then desalted on a Sephadex G25 M column (5.5 × 20 cm, Pharmacia) and loaded onto a DEAE-Sepharose column (4.5 × 15 cm, Pharmacia) equilibrated with buffer B (25 mm MOPS pH 7.4; 5% glycerol; 5 mm EDTA; 0.5 mm PMSF; 0.5 μg/mL aprotinin; 1.5 μg/mL leupeptin; 0.6 μg/mL pepstain A). The column was washed with buffer B until A280 decreased to <0.05 and was then eluted at 5 mL/min with a linear gradient of 0 to 0.7 NaCl M in buffer B. The eluted solution was collected in 15-mL fractions and the fractions with peak activity were pooled and designated as the DEAE eluate.
Buffer change of the DEAE eluate was carried out on a Sephadex G25 M column (5.5 × 20 cm). The DEAE eluate was applied to a Sephadex G25 M column equilibrated with buffer C (25 mm sodium acetate pH 4.7; 5 mm EDTA; 5% glycerol; 0.5 mm PMSF). The column was eluted with buffer C until A280 reached the baseline. The eluted solution was loaded onto a SP Sepharose F.F. column (2.6 × 18 cm, Pharmacia) equilibrated with buffer C. The column was washed with buffer C until A280 decreased to baseline, and then eluted at 3 mL/min with a linear gradient of 0.1 to 0.5 NaCl in buffer C. The eluted solution was collected in 9-mL fractions and the fractions with peak activity were pooled and designated as the SP eluate.
The SP eluate was concentrated by ultrafiltration to approximately 2 mL and applied to a Sephacryl S200 column (2.6 × 90 cm, Pharmacia) equilibrated with buffer D (i.e. buffer C containing 150 mm NaCl). The column was eluted at 0.5 mL/min with buffer D and the eluted solution was collected in 3-mL fractions. The fractions with peak activity were pooled, concentrated to approximately 300 μL with a 10-kD ultrafiltration apparatus, and applied to a Sephacryl S300 column (1 × 90 cm, Pharmacia). The column was eluted at 0.1 mL/min with buffer D and the eluted solution was collected in 1-mL fractions. The fractions with peak activity were pooled, concentrated, and stored at −70°C.
PTPase Activity Assay
All samples, except those required for chromatography, were first desalted on a Sephadex G25 M column. The PTPase assay was performed according to the method of Ng et al. (1994). Briefly, a 10-μL sample was added to 30-μL reaction buffer (50 mm 3-[N-morpholino] propanesulfonic acid pH 6.0; 5 mm EDTA; 1 mm PMSF). The reaction was initiated by adding 10 μL PTPase substrate (pTyr1018-EGF receptor or other substrates, Sigma). After reacting for 15 min at 30°C, the reaction was stopped by adding 50 μL of malachite green reagent. The absorbance of the released phosphate at 620 nm was measured and compared with a standard curve of inorganic phosphate. To investigate whether H2O2 was able to reverse the effect of DTT on ZmRIP1 activity, a 10-μL enzyme sample was first mixed with 20 μL of reaction buffer and then with 10 μL PTPase substrate to start the reaction. Twenty minutes after the reaction was started, 5 μL DTT was added to the reaction mixture and, after incubation for another 10 min, 5 μL H2O2 was again mixed with the reaction mixture.
Molecular Cloning of ZmRIP1
The protein band on the SDS gel was electrotransferred to a polyvinylidene difluoride membrane and the blot was lightly stained with Coomassie Blue. The N-terminal sequence was determined using an Applied Biosystems 492cLC protein sequencer (Shanghai Genecore BioTechnologies CO., Ltd). The N-terminal sequence of ZmRIP1 was used to query the NCBI database and was found to be almost identical to a hypothetical protein LOC100216768. A search in the maize genetics and genomics database (http://www.maizegdb.org) identified the corresponding gene, which was designated ZmRIP1 (accession no. FJ605095). To clone the cDNA, the following pair of primers was designed: forward, GTCTTGTTTCGTCTCCTCTCGTC; reverse, GCCCGACCTCTCCGTAAAG. Total RNA was prepared from dehydrated maize coleoptiles using TRIzol reagent (Invitrogen). Poly (A+) RNA was isolated using the PolyAtract mRNA isolation system III (Promega) and cDNA was synthesized by RT-PCR using tarnish-reverse transcriptase (Invitrogen).
Expression Analysis of ZmRIP1 in Yeast
The coding sequence of ZmRIP1 was amplified by PCR to introduce a Sami site at the ATG initiation codon and a Clay site fused to the His6 tag at the C terminus. The PCR product was cloned into p426Gal1 and transformed into yeast (Saccharomyces cerevisiae) strain RGY73 (Wu et al., 2002). The RGY73 cells were cultured on a synthetic complete medium lacking uracil (SC-URA)/Glc plate for 4 d at 30°C, and then the culture was induced by transferring the RGY73 cells to SC-URA/Gal for further culture at 30°C until OD600 of the culture medium reached 1.8 to approximately 2.0. As a control, RGY73 was cultured only on SC-URA/Glc. To purify the recombinant protein, yeast cells were centrifuged, and the pellet was resuspended in breaking buffer. After sonication, the solution was centrifuged at 10,000g for 20 min. The protein was purified on a nickel affinity column and eluted with Imidazole Native Elution buffer according to the manufacturer’s instructions (ProBondTM purification system, Invitrogen). The eluate was either collected for a PTPase assay or concentrated for SDS/PAGE as described above.
SDS/PAGE and Isoelectric Focusing/SDS-PAGE of ZmRIP1
SDS/PAGE and isoelectric focusing (IEF)/SDS-PAGE were performed with a Bio-Rad mini-gel apparatus using a discontinuous system. For SDS/PAGE, the final acrylamide monomer concentration was 12% for the separating gel and 4% for the stacking gel. SDS/PAGE was performed according to Laemmli (1970). IEF/SDS-PAGE was performed according to the method of Hochstrasser et al. (1988), with minor modifications. The concentration of ampholytes in the sample buffer and IEF gel monomer solution was 1.6% for pH 5.0 to 7.0 and 0.4% for pH 3.0 to 10.0. IEF was run for 12 h at 750 V, and then SDS/PAGE was performed. The relative Mr and pI were measured using Mr marker proteins (17-0446-01, Pharmacia) and pI marker proteins (17-0471-01, Pharmacia).
Point Mutation of ZmRIP1
To identify the contribution of specific Cys residues to the biochemical characteristics of ZmRIP1, Cys-152 and Cys-181 were mutated to Ile-152 and Arg-181, respectively. The C152I and C181R point mutations were generated using the QuikChange kit (Stratagene) with p426-GAL1-ZmRIDP1-DHA-6His as the expression vector, according to the instructions in the kit’s manual. To generate C152I, the forward primer was TCTACAATTCAAAGATATTGTGCACATTCGTGCGGA-AATTAGGGATTTTGATGC and the reverse primer was GCATCAAAATCCCTAATTTCCG-CACGAATGTGCACAATATCTTTGAATTGAT; to generate C181R, the forward primer was TATATATATTACACCACCATTACGGTTG-ATAAGTTTGTGCAATTTG and the reverse primer was CAAATTGCACAAACTTATCAACCGTAATGGTGGTGT-AACATATATA. The PCR protocol was as follows: 30 s at 95°C, 1 min at 55°C, and 2 min at 68°C (12 cycles). The product was purified using a QIAGEN gel extract kit and transformed into XL1-Blue for sequencing.
Measurement of ZmRIP1 Sulfhydryl Concentration
Measurement of ZmRIP1 sulfhydryls was performed according to the method of Riener et al. (2002), with minor modifications. Briefly, a working solution of 4 mm 4,4′-dithiodipyridine (DTDP) was first prepared by dissolving DTDP in 12 mm HCl solution. For sulfhydryl measurement, 100 μL protein sample was mixed with 20 μL reaction buffer (100 mm NaCl; 50 mm NaH2PO4; 0.5 mm EDTA, adjusted to pH 6.8). The reaction was started by adding 5 μL DTDP working solution and A324 was read against a blank after 5 min incubation at room temperature. The concentration of ZmRIP1 sulfhydryls was calculated based on a calibration curve. For calibration, a solution of 3 mm Cys was first prepared by dissolving Cys in a 12% HCl solution, and the standard samples were prepared by mixing 3 mL reaction buffer with 0, 10, 20, 30, or 40 μL of the 3 mm Cys standard. The reaction was started by adding 125 μL DTDP working solution, and the A324 was read against a blank after 5 min incubation at room temperature.
ZmRIP1 Localization in Onion Epidermal Cells
For subcellular localization of ZmRIP1, the ZmRIP1 cDNA sequence was amplified using the forward primer GGTACCATGAACTGCCTCCAGAACCTG and the reverse primer TCTAGACTAGCGCTCGGCGGCATC; the PCR product was then cloned into a KpnI/XbaI-cleaved pEZS-NL transient expression vector (D. Ehrhardt, Carnegie Institution, Stanford, CA). Onion (Allium cepa) epidermal cells were transformed by particle bombardment using a Bio-Rad PDS1000/He particle gun. Gold particles (1.0-μm diameter) were washed with 100% ethanol and coated with 5 μg of plasmid DNA. After bombardment, the onion cells were incubated in the dark for 20 h. Fluorescence analysis was performed using a Nikon ECLIPSE TE2000-E inverted fluorescence microscope equipped with a Nikon D-ECLIPSE C1 spectral confocal laser-scanning system. GFP fluorescence was examined at 514 nm (excitation) using an argon laser with an emission band of 515 to 530 nm.
Localization and Translocation Assessment of ZmRIP1 in Maize Protoplasts
Maize protoplast isolation and transformation were carried out according to a protocol described by Sheen et al. (1995), with minor modifications. Briefly, leaf strips were digested in digestion solution (0.4 m mannitol; 20 mm MES; pH 5.7, 10 mm CaCl2; 20 mm KCL; 0.1% bovine serum albumin; 1.0% cellulase R10; 0.2% macroenzyme R-10; Yakult Pharmaceuticals) for 3 h. After isolation, the protoplasts were immediately used for the polyethylene glycol transformation. The construct used for the transformation was the same as that used for ZmRIP1 localization in onion epidermal cells (see above). After transformation, the protoplasts were incubated for 16 h, either treated or not with 200 μm H2O2 or 1 mm DTT for 3 h, and then stained with 0.1 mg/mL 4′,6-diamino-phenylindole (DAPI; Sigma-Aldrich) in phosphate-buffered saline for 10 min. ZmRIP1 localization was assessed by visualizing GFP fluorescence with a confocal microscope as described above, and the nuclei were visualized by DAPI fluorescence. GFP and DAPI were excited at 488 nm and a UV laser at 408 nm, respectively. To evaluate the effects of H2O2 and DTT on ZmRIP1 localization more accurately, a statistical analysis of cells with distinct patterns of ZmRIP1 localization was performed. For each assessment, approximately 200 cells were examined.
To observe the effect of UV on ZmRIP1 localization after transformation, the protoplasts were incubated for 12 h without H2O2 or DTT treatment, and then stained with 0.1 mg/mL DAPI (Sigma-Aldrich) in phosphate-buffered saline for 10 min. Localization of ZmRIP1 was observed by visualizing GFP fluorescence with a confocal microscope as described above. For UV treatment, the sample was excited at 408 nm and images were collected with an emission wavelength of 515 to 530 nm. The effect of UV on ZmRIP1 movement was assessed by continuously collecting images of an individual cell while the UV was kept on.
Functional Characterization of ZmRIP1 in Arabidopsis
ZmRIP1 was expressed in Arabidopsis (Arabidopsis thaliana) with an estrogen-receptor-based inducible system. ZmRIP1 cDNA was amplified with primers CTCGAGATGGCTTCTTTCACG-GCAAC and ACTAGTCATAAGAACTCACACGACCTGC; the amplified sequence was then cloned into XhoI/SpeI-cleaved pER8. The pER8 vector was kindly provided by N.-H. Chua and B. Ulker (Zuo et al., 2000). The binary vector was transformed into Agrobacterium tumefaciens strain GV3101 and used to transform wild-type Arabidopsis using the floral-dip method. Expression of ZmRIP1 in the transgenic lines was induced by spraying the seedlings with 5 or 10 μm β-estradiol for different lengths of time. Given that ZmRIP1 is closely correlated with the cellular redox regulation, the expression of genes encoding a range of antioxidant enzymes was extensively analyzed in ZmRIP1-transformed seedlings using RT-PCR (the genes and primer pairs are shown in Supplemental Table S1). Among the genes tested, expression of At1g02950 was found to be significantly promoted in the transgenic seedlings. To further demonstrate the effect of ZmRIP1 on At1g02950 expression, a maize protoplast transient expression analysis was performed. For the reporter construct, the At1g02950 promoter sequence was amplified using the primers CTGCAGTGATGAAAGATCAGAAAACAG and GTCGACA-TATACGATATAGATACACTTG. The PCR product was then cloned into PstI/SalI-cleaved 35S-LUC-NOS transient expression vector (the vector was kindly provided by Jen Sheen). For the effector construct, the ZmRIP1 cDNA sequence was amplified using the primers GGTACCATGAACTGCCTCCAGAACCTG and TCTAGACTAGCGCTCGGCGGC-ATC, and the PCR product was then cloned into KpnI/XbaI-cleaved pEZS-NL transient expression vector. Transient expression analysis in maize protoplasts was performed according to the protocol provided by Jen Sheen’s Lab (http://genetics.mgh.harvard.edu/sheenweb/jen_archive.html).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number FJ605095 (ZmRIP1).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. The effect of a reductant and PAO on the expression of stress-responsive genes.
Supplemental Figure S2. Characterization of the chromatography-purified PTPase.
Supplemental Figure S3. Analysis of ZmRIP1 expression in yeast.
Supplemental Figure S4. Characterization of ZmRIP1-expressing transgenic seedlings.
Supplemental Figure S5. Expression analysis of a range of genes encoding antioxidant enzymes in ZmRIP1 transgenic seedlings.
Supplemental Table S1. Genes encoding the major enzymes involved in antioxidant catabolism and the primers used for RT-PCR.
Supplementary Material
Acknowledgments
We thank N.-H. Chua and B. Ulke for providing pER8, and Jen Sheen for providing the vectors for the maize protoplast transient expression assay.
Glossary
- PTPase
Protein Tyr phosphatase
- ABA
abscisic acid
- ROS
reactive oxygen species
- PAO
phenylarsine oxide
- H2O2
hydrogen peroxide
- EGF
epidermal growth factor
- NCBI
National Center for Biotechnology Information
- RT
reverse transcription
- PMSF
phenylmethylsulfonyl fluoride
- IEF
isoelectric focusing
- DTDP
4,4′-dithiodipyridine
- DAPI
4′,6-diamino-phenylindole
References
- Álvarez R, Vázquez P, Pérez F, Jiménez A, Tirado A, Irles C, González-Serratos H, Ortega A. (2009) Regulation of fast skeletal muscle activity by SERCA1 vicinal-cysteines. J Muscle Res Cell Motil 30: 5–16 [DOI] [PubMed] [Google Scholar]
- Apel K, Hirt H. (2004) Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu Rev Plant Biol 55: 373–399 [DOI] [PubMed] [Google Scholar]
- Bradford MM. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Bucciarelli T, Saliola M, Brisdelli F, Bozzi A, Falcone C, Ilio CD, Martini F. (2009) Oxidation of Cys278 of ADH I isozyme from Kluyveromyces lactis by naturally occurring disulfides causes its reversible inactivation. Biochim Biophys Acta Proteins Proteomics 1794: 563–568 [DOI] [PubMed] [Google Scholar]
- Buchanan BB, Balmer Y. (2005) Redox regulation: a broadening horizon. Annu Rev Plant Biol 56: 187–220 [DOI] [PubMed] [Google Scholar]
- Chiarugi P, Fiaschi T, Taddei ML, Talini D, Giannoni E, Raugei G, Ramponi G. (2001) Two vicinal cysteines confer a peculiar redox regulation to low molecular weight protein tyrosine phosphatase in response to platelet-derived growth factor receptor stimulation. J Biol Chem 276: 33478–33487 [DOI] [PubMed] [Google Scholar]
- Fordham-Skelton AP, Chilley P, Lumbreras V, Reignoux S, Fenton TR, Dahm CC, Pages M, Gatehouse JA. (2002) A novel higher plant protein tyrosine phosphatase interacts with SNF1-related protein kinases via a KIS (kinase interaction sequence) domain. Plant J 29: 705–715 [DOI] [PubMed] [Google Scholar]
- Forman HJ, Torres M, Fukuto J. (2002) Redox signaling. Mol Cell Biochem 234-235: 49–62 [PubMed] [Google Scholar]
- Gomez HJ, Cirillo VJ, Smith SG, Moncloa F. (1988) Lisinopril: efficacy and tolerability in special populations. Curr Opin Cardiol (Suppl 4) 3: S37–S41 [Google Scholar]
- Hansen JM, Go YM, Jones DP. (2006) Nuclear and mitochondrial compartmentation of oxidative stress and redox signaling. Annu Rev Pharmacol Toxicol 46: 215–234 [DOI] [PubMed] [Google Scholar]
- Hochstrasser DF, Harrington MG, Hochstrasser AC, Miller MJ, Merril CR. (1988) Methods for increasing the resolution of two-dimensional protein electrophoresis. Anal Biochem 173: 424–435 [DOI] [PubMed] [Google Scholar]
- Jia W, Xing Y, Lu CM, Zhang J. (2002) Signal transduction from water stress perception to ABA accumulation. Acta Bot Sin 44: 1135–1141 [Google Scholar]
- Jia W, Zhang J. (2000) Water stress-induced abscisic acid accumulation in relation to reducing agents and sulfhydryl modifiers in maize plant. Plant Cell Environ 23: 1389–1395 [Google Scholar]
- Jia WS, Zhang J and, Liang JS. (2001) Initiation and regulation of water deficit-induced abscisic acid accumulation in maize leaves and roots: cellular volume and water relations. J Exp Bot 52: 295–300 [PubMed] [Google Scholar]
- Kaffman A, O’Shea EK. (1999) Regulation of nuclear localization: a key to a door. Annu Rev Cell Dev Biol 15: 291–339 [DOI] [PubMed] [Google Scholar]
- Kötting O, Santelia D, Edner C, Eicke S, Marthaler T, Gentry MS, Comparot-Moss S, Chen J, Smith AM, Steup M, et al. (2009) STARCH-EXCESS4 is a laforin-like phosphoglucan phosphatase required for starch degradation in Arabidopsis thaliana. Plant Cell 21: 334–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koussevitzky S, Nott A, Mockler TC, Hong F, Sachetto-Martins G, Surpin M, Lim J, Mittler R, Chory J. (2007) Signals from chloroplasts converge to regulate nuclear gene expression. Science 316: 715–719 [PubMed] [Google Scholar]
- Laemmli UK. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- López-Martín MC, Romero LC, Gotor C. (2008) Cytosolic cysteine in redox signaling. Plant Signal Behav 3: 880–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan S, Ting J, Gupta R. (2001) Protein tyrosine phosphatases in higher plants. New Phytol 151: 155–164 [DOI] [PubMed] [Google Scholar]
- Maulucci G, Pani G, Labate V, Mele M, Panieri E, Papi M, Arcovito G, Galeotti T, De Spirito M. (2009) Investigation of the spatial distribution of glutathione redox-balance in live cells by using fluorescence ratio imaging microscopy. Biosens Bioelectron 25: 682–687 [DOI] [PubMed] [Google Scholar]
- Mayadas TN, Wagner DD. (1992) Vicinal cysteines in the prosequence play a role in von Willebrand factor multimer assembly. Proc Natl Acad Sci USA 89: 3531–3535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer AJ. (2008) The integration of glutathione homeostasis and redox signaling. Plant Physiol 165: 1390–1403 [DOI] [PubMed] [Google Scholar]
- Mundy J, Chua NH. (1988) Abscisic acid and waterstress induce the expression of a novel rice gene. EM60 J 7: 2279–2286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng DHW, Harder KW, Clark-Lewis I, Jirik F, Johnson P. (1994) Non-radioactive method to measure CD 45 protein tyrosine phosphatase activity isolated directly from cells. J Immunol Methods 179: 177–185 [DOI] [PubMed] [Google Scholar]
- Noctor G, Foyer CH. (1998) Ascorbate and glutathione: keeping active oxygen under control. Annu Rev Plant Physiol Plant Mol Biol 49: 249–279 [DOI] [PubMed] [Google Scholar]
- Nott A, Jung HS, Koussevitzky S, Chory J. (2006) Plastid-to-nucleus retrograde signaling. Annu Rev Plant Biol 57: 739–759 [DOI] [PubMed] [Google Scholar]
- Oelze ML, Kandlbinder A, Dietz KJ. (2008) Redox regulation and overreduction control in the photosynthesizing cell: complexity in redox regulatory networks. Biochim Biophys Acta 1780: 1261–1272 [DOI] [PubMed] [Google Scholar]
- Paget MS, Buttner MJ. (2003) Thiol-based regulatory switches. Annu Rev Genet 37: 91–121 [DOI] [PubMed] [Google Scholar]
- Qin XQ, Zeevaart JAD. (2002) Overexpression of a 9-cis-epoxycarotenoid dioxygenase gene in Nicotiana plumbaginifolia increases abscisic acid and phaseic acid levels and enhances drought tolerance. Plant Physiol 128: 544–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riener CK, Kada G, Gruber HJ. (2002) Quick measurement of protein sulfhydryls with Ellman’s reagent and with 4,4′-dithiodipyridine. Anal Bioanal Chem 373: 266–276 [DOI] [PubMed] [Google Scholar]
- Ryan MT, Hoogenraad NJ. (2007) Mitochondrial-nuclear communications. Annu Rev Biochem 76: 701–722 [DOI] [PubMed] [Google Scholar]
- Saleh OA, Bigot S, Barre FX, Allemand JF. (2005) Analysis of DNA supercoil induction by FtsK indicates translocation without groove-tracking. Nat Struct Mol Biol 12: 436–440 [DOI] [PubMed] [Google Scholar]
- Sheen J, Hwang S, Niwa Y, Kobayashi H, Galbraith DW. (1995) Green-fluorescent protein as a new vital marker in plant cells. Plant J 8: 777–784 [DOI] [PubMed] [Google Scholar]
- Skorey K, Ly HD, Kelly J, Hammond M, Ramachandran C, Huang Z, Gresser MJ, Wang QP. (1997) How does alendronate inhibit protein-tyrosine phosphatases? J Biol Chem 272: 22472–22480 [DOI] [PubMed] [Google Scholar]
- Smith RD, Walker JC. (1996) Plant protein phosphatases. Annu Rev Plant Physiol Plant Mol Biol 47: 101–125 [DOI] [PubMed] [Google Scholar]
- Tonks NK. (2005) Redox redux: revisiting PTPs and the control of cell signaling. Cell 121: 667–670 [DOI] [PubMed] [Google Scholar]
- Ushio-Fukai M. (2009) Compartmentalization of redox signaling through NADPH oxidase-derived ROS. Antioxid Redox Signal 11: 1289–1299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verslues PE, Zhu JK. (2005) Before and beyond ABA: upstream sensing and internal signals that determine ABA accumulation and response under abiotic stress. Biochem Soc Trans 33: 375–379 [DOI] [PubMed] [Google Scholar]
- Winterbourn CC, Hampton MB. (2008) Thiol chemistry and specificity in redox signaling. Free Radic Biol Med 45: 549–561 [DOI] [PubMed] [Google Scholar]
- Wu ZY, Liang F, Hong BM, Young JC, Sussman MR, Harper JF, Sze H. (2002) An endoplasmic reticulum-bound Ca(2+)/Mn(2+) pump, ECA1, supports plant growth and confers tolerance to Mn(2+) stress. Plant Physiol 130: 128–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing KY, Raza A, Löfgren S, Fernando MR, Ho YS, Lou MF. (2007) Low molecular weight protein tyrosine phosphatase (LMW-PTP) and its possible physiological functions of redox signaling in the eye lens. Biochim Biophys Acta 1774: 545–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JK. (2002) Salt and drought stress signal transduction in plants. Annu Rev Plant Biol 53: 247–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann J, Kühne R, Sylvester M, Freund C. (2007) Redox-regulated conformational changes in an SH3 domain. Biochemistry 46: 6971–6977 [DOI] [PubMed] [Google Scholar]
- Zuo J, Niu QW, Chua NH. (2000) Technical advance: an estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J 24: 265–273 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.