Abstract
In Arabidopsis (Arabidopsis thaliana), ethylene is perceived by a receptor family consisting of five members. Subfamily 1 members ETHYLENE RESPONSE1 (ETR1) and ETHYLENE RESPONSE SENSOR1 (ERS1) have histidine kinase activity, unlike the subfamily 2 members ETR2, ERS2, and ETHYLENE INSENSITIVE4 (EIN4), which lack amino acid residues critical for this enzymatic activity. To resolve the role of histidine kinase activity in signaling by the receptors, we transformed an etr1-9;ers1-3 double mutant with wild-type and kinase-inactive versions of the receptor ETR1. Both wild-type and kinase-inactive ETR1 rescue the constitutive ethylene-response phenotype of etr1-9;ers1-3, restoring normal growth to the mutant in air. However, the lines carrying kinase-inactive ETR1 exhibit reduced sensitivity to ethylene based on several growth response assays. Microarray and real-time polymerase chain reaction analyses of gene expression support a role for histidine kinase activity in eliciting the ethylene response. In addition, protein levels of the Raf-like kinase CONSTITUTIVE TRIPLE RESPONSE1 (CTR1), which physically associates with the ethylene receptor ETR1, are less responsive to ethylene in lines containing kinase-inactive ETR1. These data indicate that the histidine kinase activity of ETR1 is not required for but plays a modulating role in the regulation of ethylene responses. Models for how enzymatic and nonenzymatic regulation may facilitate signaling from the ethylene receptors are discussed.
The gaseous hormone ethylene plays roles throughout the plant life cycle (Mattoo and Suttle, 1991; Abeles et al., 1992). Ethylene regulates seed germination, seedling growth, leaf and petal abscission, fruit ripening, organ senescence, as well as stress and pathogen responses. In Arabidopsis (Arabidopsis thaliana), ethylene is perceived by a five-member family of receptors composed of ETHYLENE RESPONSE1 (ETR1), ETR2, ETHYLENE RESPONSE SENSOR1 (ERS1), ERS2, and ETHYLENE INSENSITIVE4 (EIN4) (Schaller and Kieber, 2002; Chen et al., 2005; O’Malley et al., 2005; Kendrick and Chang, 2008). The ethylene receptors can be divided into two subfamilies based on phylogenetic analysis and some shared structural features, subfamily 1 being composed of ETR1 and ERS1 and subfamily 2 being composed of ETR2, ERS2, and EIN4 (Chang and Stadler, 2001; Schaller and Kieber, 2002; Chen et al., 2005). Genetic analysis indicates that the receptors serve as negative regulators of the ethylene response, that there is functional overlap among the receptors, and that the subfamily 1 receptors generally play the predominant role in ethylene signaling (Hua and Meyerowitz, 1998; Wang et al., 2003; Qu et al., 2007).
The ethylene receptors have a similar overall modular structure, each containing three conserved transmembrane domains near the N terminus, followed by a GAF domain, and then signal output motifs in the C-terminal half. The transmembrane domains contain the ethylene-binding site (Schaller and Bleecker, 1995; Hall et al., 1999; Rodríguez et al., 1999) and also serve to localize the receptor to the endoplasmic reticulum (ER) and possibly to the Golgi apparatus (Chen et al., 2002; Dong et al., 2008; Grefen et al., 2008). The GAF domain has been implicated in protein-protein interactions among the receptors and may help mediate the formation of higher order receptor clusters (Gao et al., 2008; Grefen et al., 2008). In their C-terminal halves, all five receptors contain His kinase-like domains and, excepting ERS1 and ERS2, also receiver domains. His kinase and receiver domains are signaling elements originally identified as components in bacterial phosphorelays and are now known to be present in plants, fungi, and slime molds (Schaller et al., 2008, 2011). In two-component systems, His kinases autophosphorylate on a conserved His residue, often in response to an environmental stimulus (Mizuno, 1997; Stock et al., 2000; Gao and Stock, 2009); this phosphate is then transferred to a conserved Asp residue within a receiver domain. Receiver domains are sometimes found joined to the His kinases (as occurs with ETR1, ETR2, and EIN4) and sometimes in separate proteins referred to as response regulators. The subfamily 1 receptors ETR1 and ERS1 have functional His kinase domains based on in vitro analysis, suggesting that they could function like canonical His kinases in a two-component signaling pathway (Gamble et al., 1998; Moussatche and Klee, 2004). However, the subfamily 2 receptors ETR2, ERS2, and EIN4 lack the necessary residues for His kinase activity and, based on in vitro analysis, are now thought to function as Ser/Thr kinases (Moussatche and Klee, 2004).
Truncation studies using ETR1 demonstrate the importance of the C-terminal half of the protein for signal output, but this importance appears to be largely independent of the enzymatic activity contained in the His kinase domain (Qu and Schaller, 2004; Xie et al., 2006). Instead, the key role for the His kinase domain appears to be as a docking site for the downstream Raf-like kinase CONSTITUTIVE TRIPLE RESPONSE1 (CTR1), mutations of which have substantial effects on ethylene signaling (Kieber et al., 1993; Huang et al., 2003). Nevertheless, several studies suggest that the His kinase activity of ETR1 may modulate aspects of ethylene signaling. In one study, when a kinase-inactive mutant of ETR1 was introduced into the triple mutant etr1;etr2;ein4, it was found to rescue the mutant phenotype but showed increased sensitivity to ethylene (Qu and Schaller, 2004). A second study suggests that kinase activity regulates the ability of seedlings to recover normal growth after ethylene treatment (Binder et al., 2004). In a third study, the response regulator ARABIDOPSIS RESPONSE REGULATOR2 (ARR2) was implicated as playing a positive role in ethylene signaling as part of a phosphorelay involving ETR1 (Hass et al., 2004).
Although suggestive, none of these studies was performed in a genetic background that eliminated the endogenous His kinase activity of ETR1 and ERS1. This was due to the fact that, although null mutations were initially isolated for four of the five ethylene receptor family members (Hua and Meyerowitz, 1998), only a partial loss-of-function mutation was available for the subfamily 1 member ERS1 (Zhao et al., 2002; Hall and Bleecker, 2003; Wang et al., 2003; Qu et al., 2007). Of particular significance to the study of His kinase activity, Wang et al. (2003) reported that canonical His kinase activity was not required for signaling by ETR1; however, interpretation of this result is complicated by residual levels of ERS1 activity later identified in the background line used for the study (Xie et al., 2006; Qu et al., 2007). Thus, the degree to which His kinase activity contributes to the signal output of the ethylene receptors has not been resolved. We recently isolated a null mutation in ERS1 as well as an additional null mutation in ETR1 (Qu et al., 2007). The resulting constitutive ethylene-response phenotype of the etr1-9;ers1-3 double mutant was more pronounced than that identified in any previous combination of receptor mutations, pointing to the major role of subfamily 1 in mediating ethylene signal transduction in Arabidopsis and raising the question of how much of this role might be due to their His kinase activity (Qu et al., 2007). Here, we address the role of receptor His kinase activity by examining the rescue of the etr1-9;ers1-3 double mutant by kinase-inactive versions of ETR1.
RESULTS
Kinase-Inactive ETR1 Rescues the Constitutive Ethylene-Response Phenotype of etr1-9;ers1-3
We addressed the role of receptor His kinase activity in signaling by examining the ability of kinase-inactive versions of ETR1 to rescue the constitutive ethylene-response phenotype found in the etr1-9;ers1-3 double mutant, following the general strategy illustrated in Figure 1A. The His kinase domain of ETR1 contains conserved residues essential for activity based on the well-characterized His kinases of bacteria and prior characterization of ETR1 (Gamble et al., 1998, 2002; Stock et al., 2000; Moussatche and Klee, 2004). These include a His residue (His-353) that serves as the autophosphorylation site and a catalytic domain with two groups of conserved Gly residues referred to as the G1 and G2 boxes (Fig. 1B). Mutations in these conserved residues eliminate the autophosphorylation of ETR1 when examined in vitro (Gamble et al., 1998, 2002; Moussatche and Klee, 2004). Based on this information, we generated three kinase-inactive versions of ETR1 for analysis in plants. The mutant ETR1-G2 contains a mutated G2 box (G545A and G547A) predicted to interfere with catalysis by disrupting ATP binding to the catalytic domain (Gamble et al., 2002). The ETR1-G2 mutant, in addition to interfering with autophosphorylation activity, should be incapable of transphosphorylating other receptors, a consideration because ethylene receptors form higher order clusters (Gao et al., 2008; Grefen et al., 2008). For a second kinase-inactive mutant (ETR1-H/G2), we combined the G2 mutation of ETR1 with a mutation of the His (His-353Gln) that serves as the phosphor-accepting site (Gamble et al., 1998; Moussatche and Klee, 2004). Inclusion of the His mutation should eliminate the autophosphorylation of ETR1, either by other His kinases of Arabidopsis or by any residual kinase activity remaining in the G2 box mutant of ETR1. We also generated a third mutant, in which we combined a mutation (Asp-659Asn) of the putatively phosphorylated Asp of the receiver domain with the prior two mutations to create ETR1-H/G2/D. The rationale for the Asp-659Asn mutation was that, although the receiver domain would not be phosphorylated by kinase-inactive ETR1, it could potentially serve as a target for other Arabidopsis His kinases such as the cytokinin receptors, which like ethylene receptors localize to the ER membrane (Chen et al., 2002; Dong et al., 2008; Caesar et al., 2011; Wulfetange et al., 2011).
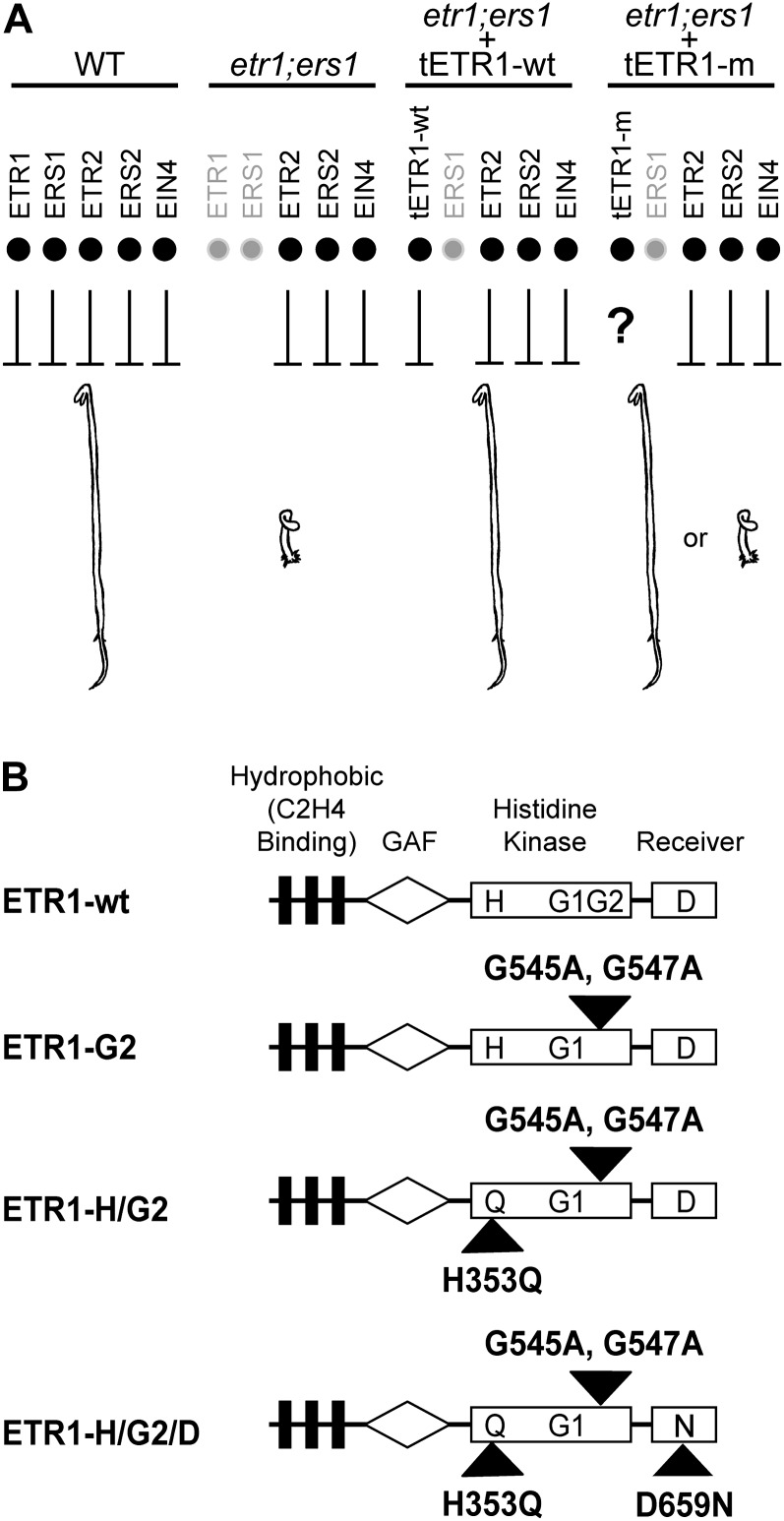
Figure 1.
Experimental strategy and constructs used for analysis. A, Effect of subfamily 1 receptors on the repression of ethylene responses. In wild-type (WT) plants, all five ethylene receptors serve to repress ethylene responses. In the etr1-9;ers1-3 double mutant (etr1;ers1), the remaining subfamily 2 receptors (ETR2, ERS2, and EIN4) are not sufficient to repress ethylene responses, and dark-grown seedlings show a constitutive ethylene-response phenotype. Transgenic expression of ETR1 (tETR1-wt) in the etr1-9;ers1-3 background rescues the mutant phenotype. Other modified versions of ETR1 (tETR1-m) can then be tested to determine if they rescue the mutant etr1-9;ers1-3 phenotype. B, Structure of ETR1 and constructs used for analysis. The hydrophobic ethylene-sensing domain, the GAF domain, the His kinase domain, and the receiver domain are indicated. The predicted phosphorylation sites are indicated by H for His-353 and D for Asp-659. G1 and G2 indicate the positions of the G1 and G2 boxes within the kinase domain. Black triangles indicate the positions of site-directed mutations introduced to eliminate His kinase activity.
All ETR1 constructs were derived from a genomic fragment that contains both promoter and coding regions of ETR1 (Chang et al., 1993). Wild-type and kinase-inactive versions of ETR1 were transformed into the etr1-9;ers1-3 double mutant, which lacks the His kinase-containing receptors of subfamily 1 and exhibits a strong constitutive ethylene-response phenotype (Qu et al., 2007). Because the homozygous double mutant is sterile, constructs were initially transformed into etr1-9/etr1-9;ers1-3/ERS1 plants, with plants homozygous for etr1-9, ers1-3, and the transgene identified in subsequent generations. Several independent lines were isolated for each construct and characterized for their ability to rescue dark-grown and light-grown phenotypes of etr1-9;ers1-3.
Dark-grown seedlings of etr1-9;ers1-3 exhibit a constitutive ethylene-response phenotype when grown in the absence of ethylene (in air; Qu et al., 2007). As shown in Figure 2A, this mutant phenotype is characterized by the inhibition of root and hypocotyl elongation, an exaggerated apical hook, and a thickening of the hypocotyl. These features contrast sharply with the etiolated phenotype observed in wild-type seedlings as well as in the single etr1-9 and ers1-3 mutants (Fig. 2A). As expected, transgenic expression of wild-type ETR1 (tETR1-wt) rescues the constitutive ethylene-response phenotype of dark-grown etr1-9;ers1-3 seedlings (Fig. 2A; Qu et al., 2007). The kinase-inactive versions of ETR1 (tETR1-G2, tETR1-H/G2, and tETR1-H/G2/D) also rescue the constitutive ethylene-response phenotype of etr1-9;ers1-3, the transgenic seedlings proving phenotypically indistinguishable from seedlings rescued by the expression of wild-type ETR1 (Fig. 2A).
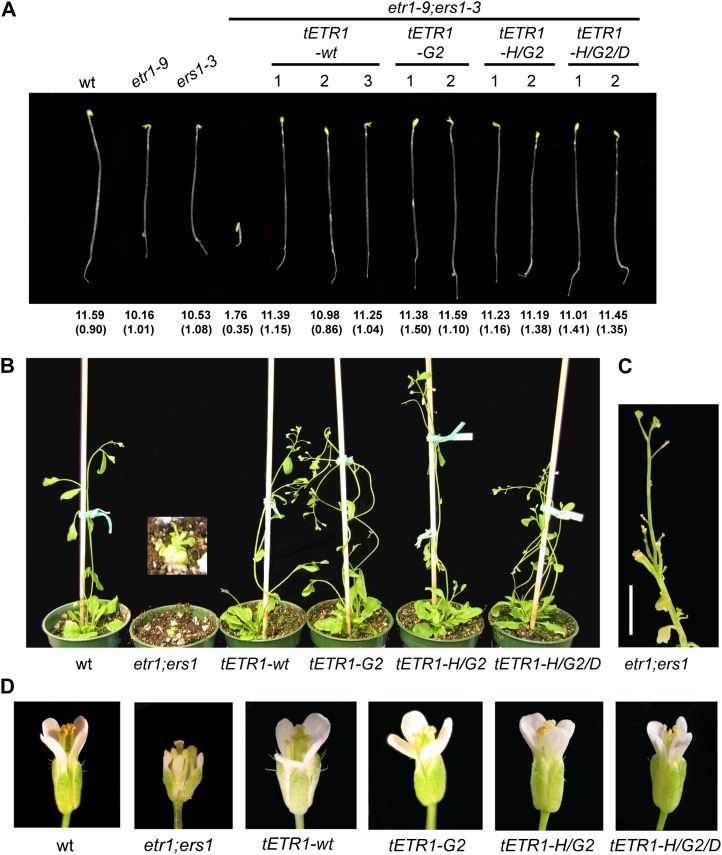
Figure 2.
Kinase-inactive versions of ETR1 rescue the constitutive ethylene-response phenotypes of etr1-9;ers1-3 plants. Constructs for tETR1-wt, tETR1-G2, tETR1-H/G2, and tETR1-H/G2/D were transformed into the etr1-9;ers1-3 background, and multiple lines homozygous for the transgene were isolated. A, Phenotypes of dark-grown seedlings. Comparison of transgenic lines with Ws wild-type (wt), etr1-9, ers1-3, and etr1-9;ers1-3 seedlings grown in the absence of ethylene (air). Representative 4-d-old seedlings are shown. Mean hypocotyl length is given in mm based on measurement of at least 20 seedlings with sd in parentheses. B, Phenotypes of 5-week-old plants. The inset shows a closeup of etr1-9;ers1-3. C, Inflorescence of a 7-week-old etr1-9;ers1-3 mutant. Bar = 5 mm. D, Floral phenotypes of adult plants. Flowers of equivalent age are shown. Note early developmental arrest of the etr1-9;ers1-3 mutant, which is rescued by introduction of the kinase-deficient versions of ETR1.
The etr1-9;ers1-3 mutant also exhibits a pronounced constitutive ethylene-response phenotype when grown in the light. Compared with the wild type, the mutant plants are dwarfed, late flowering, sterile, exhibit premature leaf senescence, and have altered floral morphology and development (Fig. 2, B–D; Qu et al., 2007). Expression of the wild-type transgene tETR1-wt in the etr1-9;ers1-3 background rescues all these phenotypes, indicating that they originate from a lack of the ethylene receptors (Fig. 2, B–D; Qu et al., 2007). The kinase-inactive versions of ETR1 also rescue these light-grown phenotypes of etr1-9;ers1-3 (Fig. 2, B–D), the transgenic plants proving indistinguishable from plants rescued by the expression of wild-type ETR1. Overall, these data indicate that the kinase-inactive versions of ETR1 can functionally replace a wild-type version of ETR1 in terms of their ability to rescue the constitutive ethylene-response phenotypes observed in the etr1-9;ers1-3 double mutant.
Plants with Kinase-Inactive ETR1 Exhibit Reduced Ethylene Responsiveness
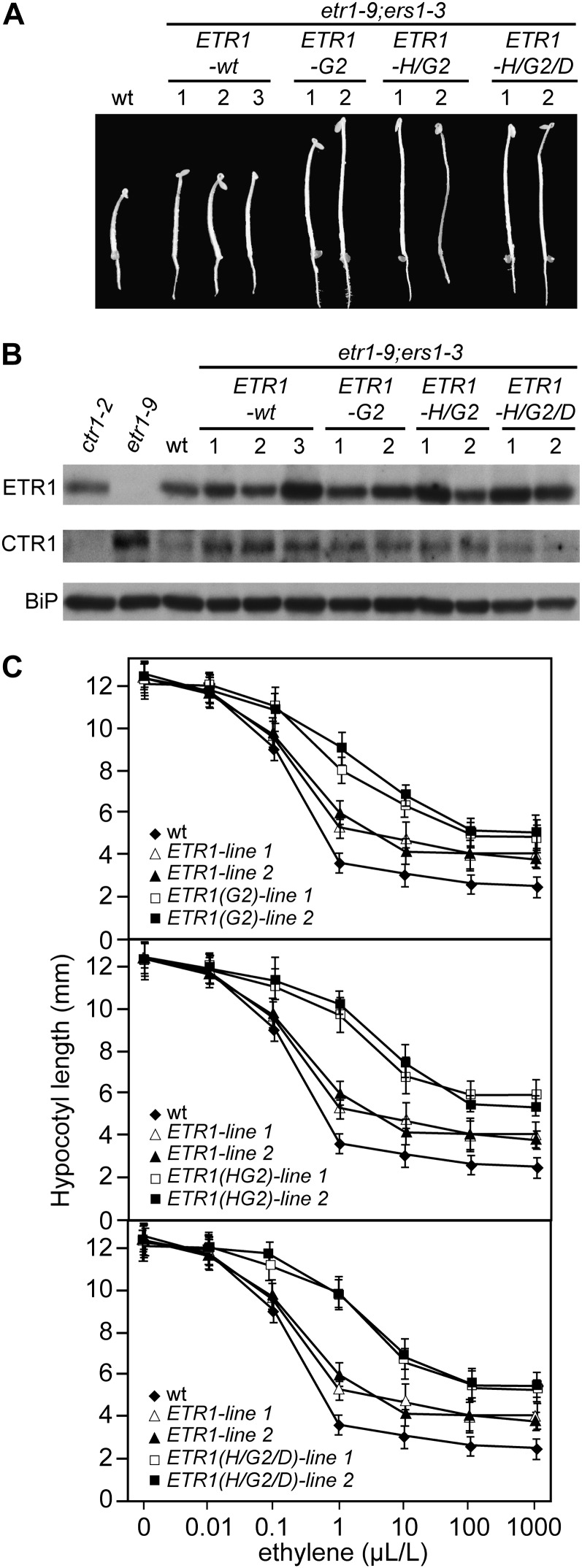
The etr1-9;ers1-3 lines containing kinase-inactive ETR1 were indistinguishable from those containing wild-type ETR1 or from wild-type plants themselves when examined in the absence of ethylene (in air). This raises the question of whether there might be a difference among the lines in terms of their response to ethylene. Growth of dark-grown seedlings in the presence of 1 µL L−1 ethylene suggested that this might be the case (Fig. 3A). All the transgenic lines exhibited the triple response to ethylene, characterized by reductions in hypocotyl and root length, formation of an apical hook, and thickening of the hypocotyl, but this response was less pronounced in the kinase-inactive lines (Fig. 3A). In particular, the hypocotyls of the kinase-inactive transgenic lines were longer than those containing wild-type ETR1. Immunoblot analysis, performed with membrane proteins from dark-grown seedlings, confirmed the expression of ETR1 in the transgenic lines and revealed a range of ETR1 protein levels (Fig. 3B). Significantly, levels of the kinase-inactive ETR1 fell within the expression range exhibited in the wild-type ETR1 lines, indicating that phenotypic differences were not due to differences in the protein levels.
Figure 3.
Altered ethylene response in dark-grown seedlings containing kinase-inactive versions of ETR1. A, Phenotypes of 4-d-old seedlings grown in the presence of 1 µL L−1 ethylene. B, Protein expression of ETR1 and CTR1 in dark-grown seedlings based on immunoblot analysis using anti-ETR1 and anti-CTR1 antibodies. BiP was used as a loading control. C, Ethylene dose-response curves of hypocotyl growth for the kinase-deficient transgenic lines. Two independent lines each for ETR1-G2, ETR1-H/G2, and ETR1-H/G2/D are shown (white and black squares). For comparison in each case, ethylene dose-response curves are shown for the wild type (wt; black diamonds) and two tETR1-wt transgenic lines (white and black triangles). Values represent means ± sd (n = 20).
We confirmed the apparent difference in ethylene responsiveness by performing a quantitative ethylene dose-response analysis of growth in the transgenic lines (Fig. 3C). Seedlings were grown in the dark in ethylene at concentrations ranging from 0 to 1,000 µL L−1 ethylene, and the hypocotyl lengths were measured after 4 d of growth. The ethylene biosynthesis inhibitor aminoethylvinylglycine (AVG) was included in the growth medium to inhibit endogenous ethylene production by the seedlings. In the absence of ethylene, both wild-type and transgenic lines exhibited a similar hypocotyl length, consistent with our earlier observation that both wild-type and kinase-inactive versions of ETR1 rescue the constitutive ethylene-response phenotype of etr1-9;ers1-3 (Fig. 2A). The two transgenic lines of tETR1-wt exhibited ethylene responsiveness, indicating that the addition of the wild-type ETR1 restored ethylene responsiveness to the etr1-9;ers1-3 mutant line (Fig. 3C). The ethylene responsiveness of tETR1-wt was slightly less than that observed in native wild-type ecotype Wassilewskija (Ws) seedlings, which could be due to additional sequence not contained on the 7.3-kb genomic fragment used for transformation, the chromosome location of the transgenes, or the fact that the genetic background still lacks ERS1 and thus is not identical to wild-type Ws. In contrast, transgenic lines containing kinase-inactive tETR1-G2, tETR1-H/G2, or tETR1-H/G2/D exhibited a substantially different ethylene dose response from those containing tETR1-wt (Fig. 3C). The kinase-inactive lines demonstrated a partial ethylene-insensitive phenotype observable at all concentrations from 0.1 to 1,000 µL L−1 ethylene, with this response being most pronounced at 1 to 10 µL L−1. Several additional points can be made from analysis of the dose-response curves. First, the dose-response curves are quite similar for the two independent lines examined for each transgene, in spite of differences in ETR1 protein levels, indicating that the effects of the mutations on the ethylene-response phenotype outweigh any potential effects of ETR1 expression. Second, the ethylene insensitivity exhibited by the tETR1-H/G2 and tETR1-H/G2/D lines was slightly greater than that of the tETR1-G2 lines (e.g. at 1 µL L−1 ethylene, the tETR1-wt lines averaged 46.2%, the tETR1-G2 lines 68.6%, the tETR1-H/G2 lines 78.7%, and the tETR1-H/G2/D lines 80.6% hypocotyl length compared with their untreated controls), suggesting that His-353 may serve as a phosphorylation site in tETR1-G2, albeit at a reduced level compared with tETR1-wt. Third, the ethylene insensitivity exhibited by the tETR1-H/G2/D lines was similar to that observed in the tETR1-H/G2 lines (e.g. at 1 µL L−1 ethylene, the tETR1-H/G2/D lines averaged 80.6% and the tETR1 lines averaged 78.7% hypocotyl length compared with their untreated controls), suggesting that the maximal contribution of kinase activity to ETR1 signaling has been reached.
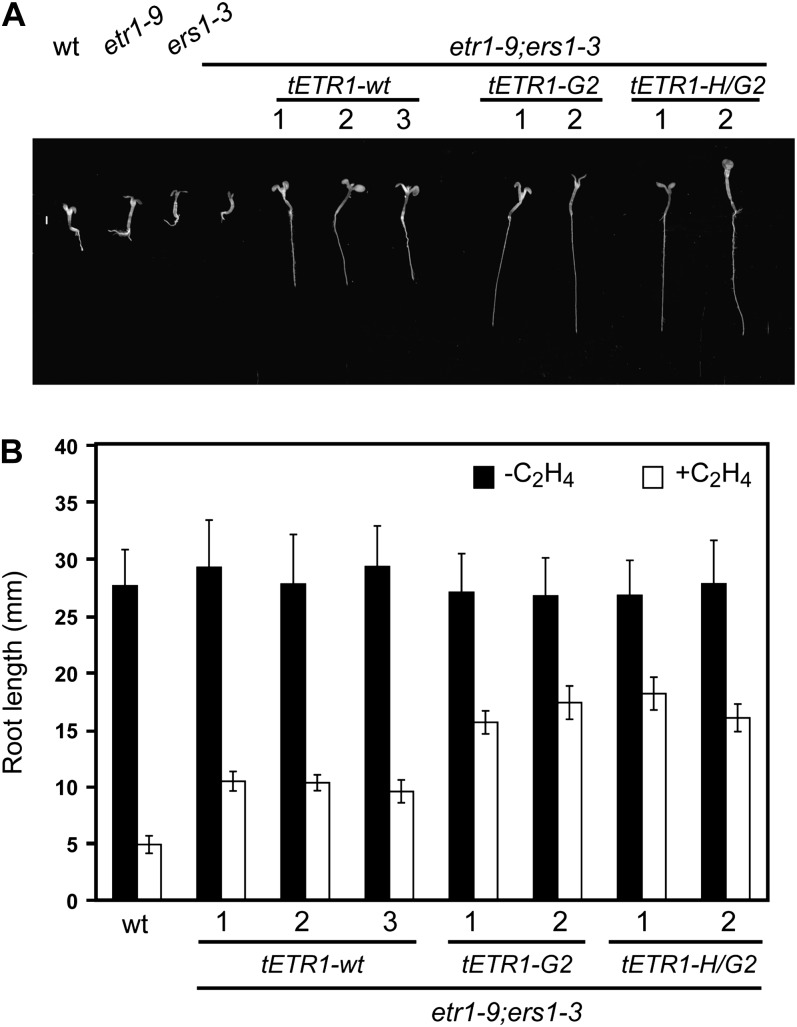
We also examined the ethylene responsiveness of seedlings grown in the light (Fig. 4). For this purpose, seedlings were grown under continuous light for 7 d in the absence or presence of 10 µL L−1 ethylene. We did not observe any difference in shoot growth between the tETR1-wt and the kinase-inactive lines, whether grown in the absence or presence of ethylene (Fig. 4A). However, the root-growth response of the kinase-inactive lines to ethylene was reduced compared with that of the tETR1-wt lines, the kinase-inactive lines thus exhibiting reduced ethylene sensitivity for root growth (Fig. 4B). Overall, these phenotypic data support a role for His kinase activity in the establishment of ethylene responses, because kinase-inactive versions of ETR1 exhibit reduced ethylene sensitivity compared with wild-type ETR1.
Figure 4.
Altered root growth response to ethylene of light-grown seedlings containing kinase-inactive versions of ETR1. A, Phenotypes of 7-d-old seedlings grown in the presence of 10 µL L−1 ethylene under continuous light. B, Root lengths of seedlings grown in the absence (black bars) or presence (white bars) of 10 µL L−1 ethylene. Values represent means ± sd (n > 12). wt, Wild type.
Gene Expression Analysis of Seedlings with Kinase-Inactive ETR1
To gain information at the molecular level on how the ethylene response differed between kinase-active and kinase-inactive ethylene receptor lines, we performed a microarray analysis. For this purpose, we used the tETR1-wt #2 and tETR1-H/G2 #2 lines, choosing these lines for comparison because they exhibit similar levels of ETR1 protein to each other as well as to the native level of ETR1 found in wild-type seedlings (Fig. 3B). RNA was prepared from seedlings grown in the dark in the presence or absence of 1 µL L−1 ethylene, because we have observed maximal differences in the hypocotyl growth response under these growth conditions (Fig. 3, A and C). Samples were prepared in triplicate for analysis.
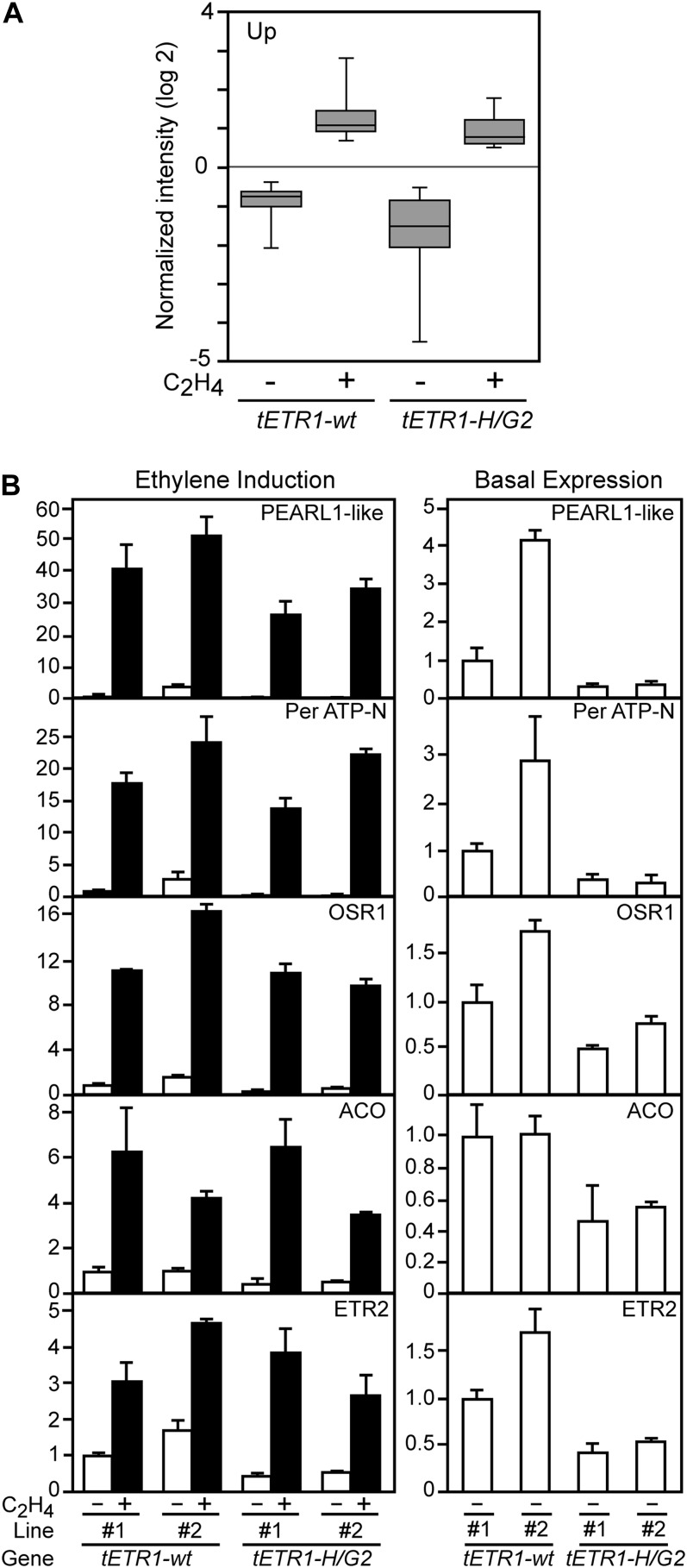
From the microarray analysis, we identified a group of 40 genes whose expression is induced 3-fold or more by ethylene in the tETR1-wt sample (Supplemental Table S1). So as to work with a robust set of ethylene-induced genes, independent of effects due to transgenes and experimental variation, we compared the genes identified in this data set with those previously identified in wild-type seedlings grown under similar conditions for ethylene treatment (Alonso et al., 2003). Almost half (19) of the ethylene-induced genes from our microarray met the criterion of also being induced in this independent experiment (Table I). We used box-plot analysis to visualize how the expression of this group of ethylene-induced genes compares between tETR1-wt and tETR1-H/G2 (Fig. 5A). In both cases, we observe an induction of this gene set by ethylene. However, we observe differences in the expression levels of the genes that relate to the kinase activity. Most pronounced is a decrease in the basal-level expression (minus exogenous ethylene) for the genes in the tETR1-H/G2 line compared with the tETR1-wt line. There is also a decrease under ethylene induction conditions (plus exogenous ethylene) for the genes in the tETR1-H/G2 background compared with tETR1-wt, although this difference is not as pronounced as that observed under basal conditions.
Table I. Ethylene up-regulated genes.
Genes with expression 3-fold up-regulated by 1 µL L−1 ethylene in tETR1-wt were identified, and those common to the up-regulated gene set of Alonso et al. (2003) are shown. C2H4, Ethylene.
| Gene No. | Description | Fold Up-Regulation | Normalized Expression (log2 Transformed) |
|||
|---|---|---|---|---|---|---|
| tETR1-wt |
tETR1-H/G2 |
|||||
| −C2H4 | +C2H4 | −C2H4 | +C2H4 | |||
| At1g72290 | Drought-induced protein | 19.01 | −2.07 | 2.18 | −2.64 | 1.78 |
| At2g41230 | ORGAN SIZE RELATED1 (OSR1) | 11.64 | −0.77 | 2.78 | −2.22 | 1.19 |
| At1g23730 | Carbonic anhydrase | 8.00 | −0.55 | 2.45 | −0.83 | 1.32 |
| At4g12470 | pEARL1-like | 6.31 | −0.79 | 1.86 | −2.09 | 1.63 |
| At1g06080 | DELTA 9 DESATURASE1 (ADS1) | 6.11 | −1.18 | 1.43 | −2.13 | 1.04 |
| At4g35150 | O-Methyltransferase | 6.08 | −1.26 | 1.35 | −1.64 | 1.01 |
| At5g19890 | Peroxidase ATP-N | 6.02 | −1.31 | 1.28 | −4.49 | 1.25 |
| At5g20820 | Putative protein | 4.78 | −1.02 | 1.24 | −1.31 | 1.35 |
| At5g33340 | CONSTITUTIVE DISEASE RESISTANCE1 (CDR1) | 3.53 | −0.37 | 1.45 | −0.91 | 0.53 |
| At3g17680 | Putative protein | 3.39 | −0.78 | 0.98 | −1.40 | 0.84 |
| At1g33790 | Myrosinase-binding protein | 3.18 | −0.63 | 1.04 | −0.62 | 0.60 |
| At4g12410 | Auxin-induced protein | 3.18 | −0.95 | 0.72 | −1.46 | 0.71 |
| At1g73830 | BR ENHANCED EXPRESSION3 (BEE3) | 3.12 | −0.80 | 0.84 | −2.04 | 0.80 |
| At5g09970 | CYTOCHROME P450, FAMILY 78, SUBFAMILY A, POLYPEPTIDE7 (CYP78A7) | 3.09 | −0.75 | 0.88 | −0.67 | 0.56 |
| At3g15370 | ALPHA EXPANSIN12 (ATEXPA12) | 3.09 | −0.75 | 0.87 | −0.58 | 0.57 |
| At5g15720 | GDSL-MOTIF LIPASE7 (GLIP7) | 3.05 | −0.66 | 0.95 | −2.02 | 0.63 |
| At3g21510 | HISTIDINE-CONTAINING PHOSPHOTRANSMITTER1 (AHP1) | 3.03 | −0.64 | 0.96 | −1.51 | 0.72 |
| At5g63660 | LOW-MOLECULAR-WEIGHT CYSTEINE-RICH74/PLANT DEFENSIN2.5 (LCR74/PDF2.5) | 3.02 | −0.60 | 1.00 | −0.82 | 0.64 |
| At3g23150 | ETR2 | 3.01 | −0.50 | 1.09 | −1.85 | 0.61 |
Figure 5.
Expression analysis of ethylene-induced genes. A, Box-plot analysis of microarray expression data for the genes shown in Table I. The bottom and top of each box represent the 25th and 75th percentile for data expression values, the band in the middle of the box represents the median expression value, and the ends of the box whiskers represent the minimum and maximum expression values for the data. Expression values for the box-plot analysis are log2 transformed. B, Real-time RT-PCR analysis for the expression of selected genes. Two independent lines for each of the transgenes tETR1-wt and tETR1-H/G2 were analyzed, tETR1-wt-line 2 and tETR1-H/G2-line 2 being those used for microarray analysis. The expression level of tETR1-wt-line 1 in the absence of ethylene is set to 1. The left panel for each gene shows expression in the absence and presence of 1 µL L−1 ethylene. The right panel shows just the basal level of expression in the absence of ethylene treatment, at a scale to allow for better comparison of expression.
We performed quantitative reverse transcription (qRT)-PCR on a subset of the induced genes, examining expression in two independent lines each for tETR1-wt and tETR1-H/G2 (Fig. 5B). Some differences in ethylene-regulated gene expression are predicted between these lines due to the differing protein levels for ETR1 (Fig. 3B). tETR1-wt-line 1 and tETR1-H/G2-line 1 exhibit higher protein levels, whereas tETR1-wt-line 2 and tETR1-H/G2-line 2 exhibit lower protein levels, being similar to the native ETR1 protein level (Fig. 3B). Because ETR1 serves as a negative regulator of ethylene responses, the lines with higher ETR1 protein levels are predicted to more strongly suppress ethylene-responsive gene expression. In general, this prediction is born out by the qRT-PCR analysis. For example, the basal expression level for ethylene-regulated genes is generally lower in tETR1-wt-line 1 than in tETR1-wt-line 2 (four of the five genes exhibiting lower expression and one exhibiting similar expression). In addition, the expression level after ethylene induction is lower in tETR1-wt-line 1 compared with tETR1-wt-line 2 for four of the five genes examined.
Several points can be made based on comparison of the tETR1-wt and the kinase-inactive tETR1-H/G2 lines. First, the qRT-PCR analysis (Fig. 5B) confirms the microarray analysis for tETR1-wt-line 2 and tETR1-H/G2-line 2 (Table I; Fig. 5A). Second, there is a clear difference in basal gene expression levels for the ethylene-induced genes that extends across both lines for each transgene, in spite of the differences in ETR1 protein levels; lines 1 and 2 of tETR1-H/G2 demonstrate a lower basal gene expression level than lines 1 and 2 of tETR1-wt. Third, we also observed that, when protein levels of ETR1 are taken into account (e.g. comparing tETR1-wt-line 1 with tETR1-H/G2-line 1 and comparing tETR1-wt-line 2 with tETR1-H/G2-line 2), the two genes with the highest levels of induction (EARLY ARABIDOPSIS ALUMINUM INDUCED1-like [pEARL1-like] and Per ATP-N) exhibit reduced ethylene induction in the kinase-inactive lines. We thus observe reduced expression of ethylene-induced genes in the tETR1-H/G2 lines, particularly under basal expression conditions, providing a potential molecular basis for the reduced phenotypic response to ethylene observed in these kinase-inactive lines.
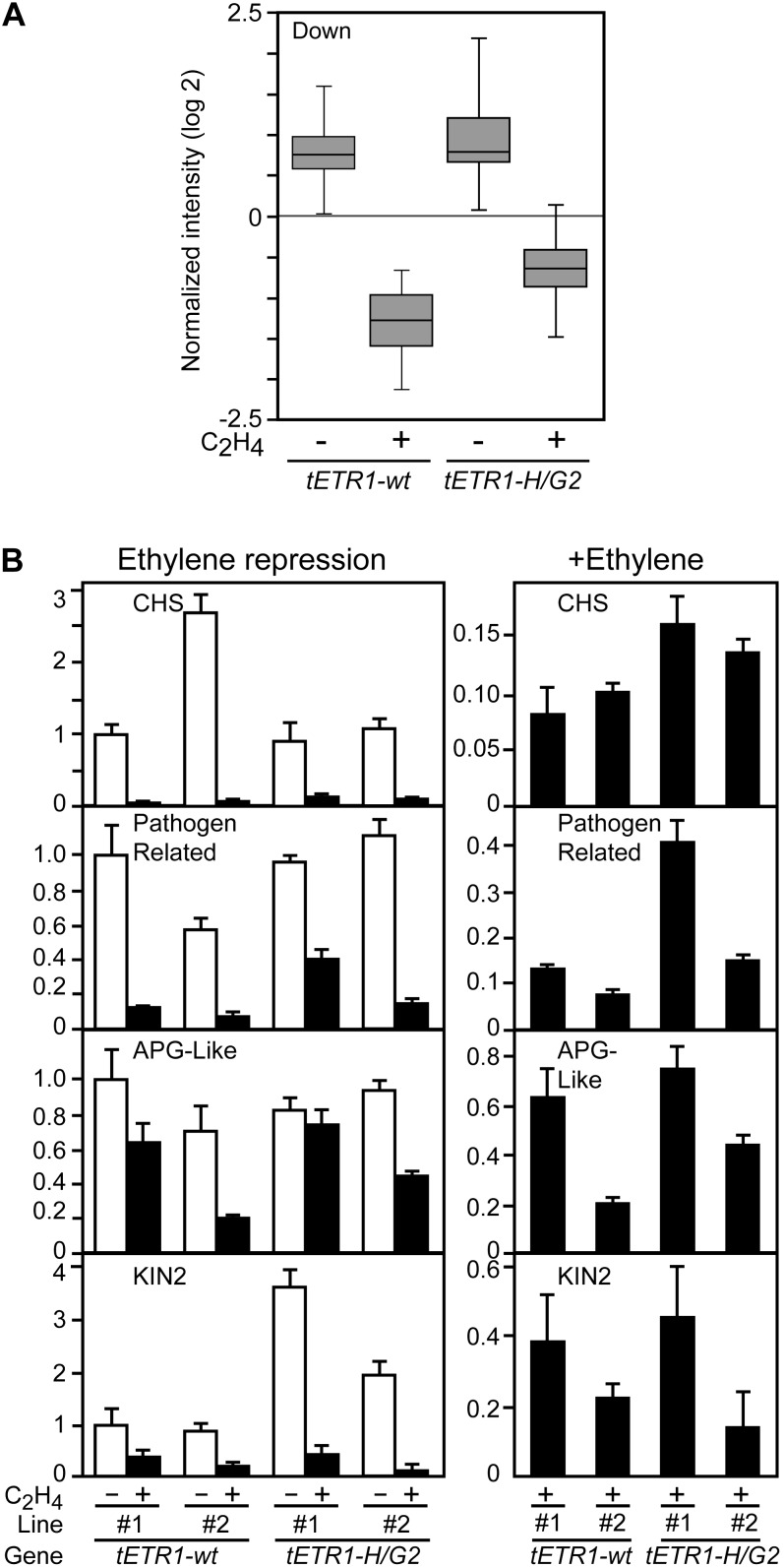
Using a similar strategy employed for analyzing ethylene-induced genes, we also examined genes whose expression is repressed by ethylene. In this case, of the 151 genes whose expression was repressed 3-fold or more by ethylene in the tETR1-wt sample (Supplemental Table S2), only 14 were robustly confirmed by the microarray analysis of Alonso et al. (2003; Table II). Box-plot analysis for these repressed genes indicated a similar basal level of expression for tETR1-wt and tETR1-H/G2 but a differing level of expression in the presence of exogenous ethylene, with gene expression not being suppressed as effectively in the kinase-inactive tETR1-H/GH2 line (Fig. 6A). Follow-up analysis on a subset of these genes by qRT-PCR supports this effect on treatment with exogenous ethylene (Fig. 6B). Three of the four genes (CHALCONE SYNTHASE [CHS], Pathogen-related, and Anther-specific proline-rich protein-like [APG-like]) examined exhibit reduced suppression when protein levels of ETR1 are taken into account, with two of these (CHS and Pathogen-related) exhibiting reduced suppression in the kinase-inactive lines regardless of the ETR1 protein level. The fourth gene examined (COLD-RESPONSIVE 6.6 [COR6.6/KIN2]) does not show a difference between the kinase-active and -inactive lines upon ethylene treatment but does show a difference in the basal levels, with substantially higher basal levels of expression in both kinase-inactive tETR1-H/G2 lines compared with the tETR1-wt lines. Thus, overall, analysis of the ethylene-repressed genes is consistent with our analysis of the ethylene-induced genes, with various genes in the kinase-inactive tETR1-H/G2 lines demonstrating a decreased ability to induce the ethylene response at the molecular level.
Table II. Ethylene down-regulated genes.
Genes with expression 3-fold down-regulated by 1 µL L−1 ethylene in tETR1-wt were identified, and those common to the down-regulated gene set of Alonso et al. (2003) are shown. C2H4, Ethylene.
| Gene No. | Description | Fold Down-Regulation | Normalized Expression (log2 Transformed) |
|||
|---|---|---|---|---|---|---|
| tETR1-wt |
tETR1-H/G2 |
|||||
| −C2H4 | +C2H4 | −C2H4 | +C2H4 | |||
| At4g37410 | CYTOCHROME P450, FAMILY 81, SUBFAMILY F, POLYPEPTIDE 4 (CYP81F4) | 5.81 | 0.91 | −1.63 | 1.65 | −1.48 |
| At5g15960 | COR6.6/KIN2 | 5.51 | 0.78 | −1.68 | 2.19 | −0.82 |
| At5g09530 | Periaxin-like protein | 5.23 | 0.28 | −2.11 | 0.22 | −1.38 |
| At5g13930 | CHS | 4.83 | 1.60 | −0.67 | 0.89 | −0.64 |
| At5g48000 | CYTOCHROME P450, FAMILY 708, SUBFAMILY A, POLYPEPTIDE 2 (CYP708A2) | 4.17 | 1.10 | −0.96 | 0.71 | −0.69 |
| At4g25780 | Pathogenesis-related protein | 4.10 | 0.62 | −1.42 | 1.90 | −0.43 |
| At1g80160 | Putative protein | 4.01 | 1.05 | −0.96 | 0.10 | 0.15 |
| At3g29250 | SHORT-CHAIN DEHYDROGENASE REDUCTASE4 (SDR4) | 3.90 | 0.59 | −1.37 | 1.09 | −0.57 |
| At1g66800 | Cinnamyl ADH | 3.83 | 0.77 | −1.17 | 1.27 | −0.87 |
| At5g48010 | Cycloartenol synthase | 3.48 | 0.88 | −0.92 | 0.78 | −0.67 |
| At1g75900 | APG-like | 3.25 | 0.05 | −1.65 | 0.82 | −0.08 |
| At2g38760 | ANNEXIN3 | 3.20 | 1.01 | −0.67 | 0.66 | −0.40 |
| At5g41080 | Putative protein | 3.17 | 0.62 | −1.04 | 0.74 | −0.88 |
| At1g29660 | Lipase/hydrolase | 3.06 | 0.25 | −1.36 | 0.35 | −0.19 |
Figure 6.
Expression analysis of ethylene-repressed genes. A, Box-plot analysis of microarray expression data for the ethylene-repressed genes shown in Table II. Symbols are as in Figure 5A. B, Real-time RT-PCR analysis for the expression of selected genes. The expression level of tETR1-wt-line 1 in the absence of ethylene is set to 1. The left panel for each gene shows expression in the absence and presence of 1 µL L−1 ethylene. The right panel shows just the level of expression in the ethylene-treated samples, at a scale to allow for better comparison of expression.
Analysis of CTR1 Protein Levels
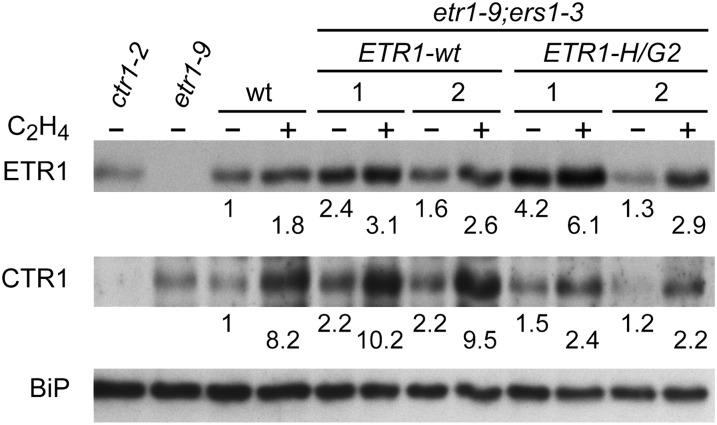
The Raf-like kinase CTR1 serves as a negative regulator of ethylene responses and physically associates with the ethylene receptors to suppress ethylene signal transduction (Clark et al., 1998; Cancel and Larsen, 2002; Gao et al., 2003). The subfamily 1 mutant etr1-9;ers1-3 employed as the genetic background for our studies results in a substantial loss of the membrane-associated CTR1 (maximal CTR1 protein level at the membrane is approximately 35% of that found in the wild type; Qu et al., 2007), accounting in part for the mutant’s constitutive ethylene-response phenotype. Based on the role of CTR1 in suppressing ethylene responses, one potential explanation for the reduced ethylene sensitivity observed in the kinase-inactive ETR1 lines would be if kinase-inactive ETR1 localized greater levels of CTR1 to the membrane than wild-type ETR1. Therefore, we immunologically characterized the levels of membrane-associated CTR1 in the different lines (Fig. 3B). The null mutant ctr1-2 served as a negative control for the expression of CTR1 (Kieber et al., 1993; Gao et al., 2003). All the transgenic lines exhibited CTR1 protein levels similar to or greater than those found in the wild type, consistent with the ability of these lines to suppress the constitutive ethylene-response phenotype observed in the etr1-9;ers1-3 background. Significantly, the lines containing kinase-inactive versions of ETR1 did not exhibit any more CTR1 protein than tETR1-wt lines.
Previous work has demonstrated that CTR1 protein levels are not constant in wild-type Arabidopsis membranes but increase in response to ethylene (Gao et al., 2003), potentially due to short-term transcriptional induction of CTR1 (Winter et al., 2007; S.N. Shakeel and G.E. Schaller, unpublished data). Therefore, we followed up on our initial analysis of CTR1 protein by quantitatively characterizing the levels of membrane-associated CTR1 in etiolated seedlings of tETR1-wt and tETR1-H/G2 grown in the absence or presence of 1 µL L−1 ethylene (Fig. 7), the same ethylene treatment conditions used for gene expression analysis. In both the ETR1-wt and ETR1-H/G2 lines, CTR1 is present and, as in the wild type, exhibits an ethylene-induced increase in protein levels. However, similar to what we observed in our analysis of ethylene-induced gene expression (Table I; Fig. 5), we observed a slightly decreased level of CTR1 protein in the ETR1-H/G2 lines compared with ETR1-wt in the absence of ethylene. Perhaps most significantly, the degree to which CTR1 levels were induced by exogenous ethylene was substantially less in the tETR1-H/G2 lines than in the wild-type or the tETR1-wt lines. Immunological examination of ETR1 in the lines reveals a small ethylene-dependent increase in protein levels, but this does not correlate with the differences in CTR1 induction (Fig. 7). Thus, differences in CTR1 protein levels in the ETR1-H/G2 lines compared with ETR1-wt lines cannot directly account for the difference in phenotypes but rather seem to reflect the differences in ethylene sensitivity.
Figure 7.
Effect of His kinase activity of the receptors on the level of membrane-associated CTR1. Membrane proteins were isolated from dark-grown seedlings grown in the absence or presence of 1 µL L−1 ethylene. Levels of ETR1 and CTR1 were determined by immunoblot analysis using anti-ETR1 and anti-CTR1 antibodies. BiP was used as a loading control. The ctr1-2 and etr1-9 null mutants served as negative controls for the expression of CTR1 and ETR1, respectively. Protein levels of ETR1 and CTR1 are shown below each blot; these are normalized to the loading control and are expressed relative to the level found in wild-type (wt) seedlings grown in the absence of ethylene.
DISCUSSION
The role that ethylene receptors play in ethylene signal transduction involves their ability to (1) repress ethylene responses in the absence of ethylene (in air) and (2) establish ethylene responses in the presence of ethylene. The ethylene receptors function in concert with the physically associated Raf-like kinase CTR1 to accomplish this purpose (Clark et al., 1998; Cancel and Larsen, 2002; Gao et al., 2003). CTR1 is a negative regulator of ethylene signaling, and loss-of-function mutations in CTR1 result in a constitutive ethylene response (Kieber et al., 1993; Huang et al., 2003). Similarly, higher order loss-of-function mutations in the ethylene receptors also result in a constitutive ethylene-response phenotype (Hua and Meyerowitz, 1998; Qu et al., 2007), apparently due to the loss of CTR1 from the ER membrane location of the ethylene receptor/CTR1 complex (Gao et al., 2003; Qu et al., 2007). Thus, the role of ethylene receptors in air involves maintaining both the kinase activity and the correct location for action of CTR1, resulting in the repression of the ethylene response. The role of the ethylene receptors upon binding ethylene is then achieved by transmitting information, whether enzymatic or conformational, to CTR1 to reduce its kinase activity, thereby derepressing the ethylene response.
A characteristic feature of the subfamily 1 ethylene receptors is their conserved His kinase domain, a feature noted after the initial cloning of ETR1 and one that immediately raised the question of what role His kinase activity might play in signaling (Chang and Meyerowitz, 1995). Resolution of this question optimally requires a genetic background that eliminates the endogenous His kinase activity of ETR1 and ERS1 and was initially addressed after the isolation of null etr1 alleles (Hua and Meyerowitz, 1998) and the T-DNA insertion allele ers1-2 (Zhao et al., 2002; Wang et al., 2003). The constitutive ethylene-response phenotype of an etr1-7;ers1-2 double mutant was rescued by subfamily 1 receptors but not by subfamily 2 receptors, indicating that a subfamily 1-specific feature was required for functional complementation, potentially His kinase activity (Wang et al., 2003). However, a kinase-inactive version of ETR1 also rescued the mutant phenotype, indicating that canonical His kinase activity was not required for signaling (Wang et al., 2003). This might have been the end of the story, but isolation of a new T-DNA insertion allele of ERS1 (ers1-3), coupled with molecular and genetic analyses, indicated that the ers1-2 allele is not a complete null (the ers1-2 T-DNA is inserted into the 5′ untranslated region and the ERS1 transcript is still produced, whereas the ers1-3 T-DNA is inserted into the second exon and disrupts the production of the ERS1 transcript; Xie et al., 2006; Qu et al., 2007). Thus, the etr1-7;ers1-2 background used by Wang et al. (2003) contains residual activity from ERS1, and the degree to which His kinase activity contributes to the signal output of the ethylene receptors could not be fully ascertained, prompting models for how His kinase activity might function in concert with CTR1 to mediate signaling (Lin et al., 2009). To resolve the role of His kinase activity in ethylene signaling, we used the double mutant etr1-9;ers1-3, which is null for both subfamily 1 ethylene receptors, and analyzed the ability of kinase-inactive forms of ETR1 to rescue the constitutive ethylene-response phenotype found in the mutant.
A key conclusion from our study, consistent with the earlier study by Wang et al. (2003), is that His kinase activity is not absolutely required for signaling by the receptors, either for repressing ethylene responses in air or for establishing ethylene responses upon ethylene binding. The etr1-9;ers1-3 double mutant exhibits substantially reduced levels of membrane-associated CTR1, accounting for its strong constitutive ethylene-response phenotype (Qu et al., 2007). Both wild-type and kinase-inactive versions of ETR1 are capable of binding to CTR1, based on analysis in a yeast model system (Gao et al., 2003) and our immunoblot analysis of the transgenic lines reported here. Thus, the rescue of the double-mutant phenotype is consistent with both wild-type and kinase-inactive versions of ETR1 reforming a functional signaling complex with CTR1, which is then able to repress the constitutive ethylene-response phenotype of the etr1-9;ers1-3 mutant in air. Significantly, kinase-inactive ETR1 can also induce an ethylene response, based on phenotypic and gene expression analyses. This indicates that ETR1 has an alternative mechanism, not involving its His kinase activity, by which to regulate the kinase activity of CTR1. The physical interaction of CTR1 with the receptors (Clark et al., 1998; Cancel and Larsen, 2002; Gao et al., 2003) suggests that its regulation could be accomplished due to conformational changes in the receptors being transmitted to the associated CTR1.
Although the His kinase is not absolutely required for signaling by ETR1, our study does demonstrate that this enzymatic activity modulates signaling. Kinase-inactive ETR1 was less effective than wild-type ETR1 at inducing an ethylene response, based on phenotypic analysis of dark- and light-grown seedlings as well as the molecular analysis of ethylene-regulated gene expression. A concern with any genetic study involving site-directed mutations is whether the phenotype is due to the known effect of the site-directed mutations or to an unintended side effect. To minimize the possibility of side effects, we made relatively conserved changes in the amino acids in which we incorporated established modifications based on work with bacterial and fungal two-component signaling elements (Iuchi, 1993; Pan et al., 1993; Yang and Inouye, 1993; Posas et al., 1996). In addition, we made use of three mutant versions of ETR1, the effects of these mutations being consistent with a shared mechanistic role in a His-Asp phosphorelay. Specifically, we found that the reduced ethylene sensitivity observed in ETR1-G2 is further accentuated in the ETR1-H/G2 mutant, suggesting that His-353 and the G2 box, although separate in the primary sequence, play a role in the same signaling mechanism. Furthermore, the Asp mutation had no additive effect, consistent with its role being downstream and dependent on the His kinase activity of ETR1.
The decreased ethylene sensitivity of kinase-inactive ETR1 expressed in the etr1-9;ers1-3 background contrasts with the results obtained from expression in the etr1-7;ers1-2 or the etr1-7;etr2-3;ein4-4 background (Wang et al., 2003; Qu and Schaller, 2004). No difference was observed between wild-type ETR1 and kinase-inactive ETR1 in their ability to rescue the constitutive ethylene-response phenotype of etr1-7;ers1-2 or in their ability to mediate ethylene responses in the transgenic seedlings (Wang et al., 2003). Our ability to detect differences between wild-type ETR1 and kinase-inactive ETR1 when expressed in the etr1-9;ers1-3 background is likely due to this background being null for both subfamily 1 receptors, whereas the etr1-7;ers1-2 background contains residual levels of ERS1 (Xie et al., 2006; Qu et al., 2007). When expressed in the etr1-7;etr2-3;ein4-4 background, kinase-inactive ETR1 was found to be slightly less effective than wild-type ETR1 in rescuing the constitutive ethylene-response phenotype of the background (Qu and Schaller, 2004). Additionally, the transgenic etr1-7;etr2-3;ein4-4 lines containing kinase-inactive ETR1 also exhibited slightly increased ethylene sensitivity compared with lines containing wild-type ETR1 (Qu and Schaller, 2004), an interesting result because it is the opposite of what is found in the etr1-9;ers1-3 background. This difference in ethylene responsiveness likely arises from the substantive differences in receptor makeup of the etr1-7;etr2-3;ein4-4 and etr1-9;ers1-3 backgrounds (O’Malley et al., 2005; Qu et al., 2007), although the specific basis is unknown. It is notable, however, that the phenotype of decreased ethylene sensitivity we associate here with kinase-inactive ETR1 is only uncovered when the His kinase activity of both subfamily 1 receptors ETR1 and ERS1 is eliminated from the background.
Two possibilities, not mutually exclusive, could account for phosphorylation playing a modulating role in ethylene signal transduction (Mason and Schaller, 2005). First, ETR1 could transmit a signal through a multistep phosphorelay involving downstream phosphotransfer proteins (AHPs) and response regulators (ARRs); this alternative and secondary pathway could augment output from the primary CTR1-dependent pathway. Support for such an alternative ethylene-signaling pathway comes from evidence that the ethylene receptors interact with AHP proteins (Urao et al., 2000; Scharein et al., 2008), that the response regulator ARR2 may modulate ethylene signaling (Hass et al., 2004; Mason et al., 2005), and that mutants of CTR1 still demonstrate a residual ethylene response (Larsen and Chang, 2001; Hall and Bleecker, 2003).
A second possibility is that the phosphorylation of ETR1 affects signaling through well-established components of the pathway, a possibility consistent with the broad effect on transcription of ethylene-regulated genes in the kinase-inactive mutant. Phosphorylation is a commonly used mechanism to elicit conformational changes in proteins as well as to modulate interactions between proteins; therefore, phosphorylation could, for instance, modulate the conformational information passed between ETR1 and the physically associated CTR1. Consistent with such a possibility is the finding that both the His kinase and receiver domains of the ethylene receptors associate with CTR1 (Clark et al., 1998; Cancel and Larsen, 2002; Gao et al., 2003; Zhong et al., 2008). Based on our data, the kinase-inactive ETR1 is more effective at suppressing the ethylene response than wild-type ETR1; thus, according to this model, autophosphorylation of ETR1 would serve as a part of the means by which CTR1 is inactivated (i.e. in the kinase-inactive ETR1 lines, CTR1 is more active; thus, it is more difficult to induce an ethylene response). Interestingly, we observed lower levels of CTR1 associated with the kinase-inactive ETR1 lines than with the wild-type ETR1 lines, even though these lines were more effective at suppressing the ethylene response. The reduced levels of CTR1 in the kinase-inactive lines may reflect their reduced ethylene responsiveness, because one of the ethylene responses is to induce increased levels of membrane-associated CTR1 (Gao et al., 2003). Alternatively, the reduced CTR1 levels may reflect part of a feedback mechanism to modulate output from the receptors, we having observed that CTR1 levels do not always directly correlate with the total receptor levels, actually increasing in the etr1-7 and etr1-9 null mutants (Gao et al., 2003; Qu et al., 2007).
Other elements of the ethylene signal transduction pathway, including other ethylene receptors, the essential downstream signaling component EIN2, and the signaling modulator RTE1, also physically associate with ETR1 (Gao et al., 2008; Grefen et al., 2008; Bisson et al., 2009; Chen et al., 2010) and could have their activities potentially modified through interactions with phospho-ETR1. Interestingly, in vitro analysis suggests that the phosphorylation of ETR1 may reduce its affinity for EIN2, supporting the possibility that His phosphorylation may affect and modulate interactions among components of the ethylene receptor signaling complex (Bisson et al., 2009).
The simplest mechanistic model, consistent with our data indicating that autophosphorylation serves a role in establishing an ethylene response, is that ethylene stimulates the His kinase activity and autophosphorylation of ETR1. The kinase-inactive ETR1would thus be less effective at initiating an ethylene response, resulting in the reduced sensitivity to ethylene in our growth response assays. Similarly, we also observed a reduced ability by the kinase-inactive ETR1 to regulate gene expression in response to ethylene, particularly when considered on a per protein basis with wild-type ETR1. In some cases, the effects on gene expression were strong enough that the kinase-inactive ETR1 lines differed from the wild-type ETR1 lines, regardless of the ETR1 protein level. This effect was most apparent when examining the basal expression levels of ethylene-induced genes, which were consistently lower in the kinase-inactive ETR1 lines. Only endogenous ethylene is present under basal expression conditions, some ethylene being produced by seedlings even when the ethylene biosynthesis inhibitor AVG is incorporated into the growth medium (Sanders et al., 1991). This low level of ethylene is apparently sufficient to induce expression of the genes in the wild-type ETR1 lines to a greater extent than in the kinase-inactive ETR1 lines. A more substantial role for autophosphorylation at low ethylene levels is consistent with our growth response curves (Fig. 3), where growth differences between wild-type ETR1 and kinase-inactive ETR1 lines decreased at the higher ethylene concentrations (e.g. 100 and 1,000 µL L−1). A role of phosphorylation in establishing an ethylene response is consistent with several previous genetic analyses in which phenotypes of kinase-deficient forms of ETR1 were chiefly observed after ethylene treatment (Binder et al., 2004; Qu and Schaller, 2004).
On the other hand, this model for the ethylene regulation of kinase activity is not consistent with the in vitro analysis of a bacterially expressed version of ETR1, in which ethylene is reported to repress autophosphorylation of the receptor (Voet-van-Vormizeele and Groth, 2008). If ethylene binding indeed suppresses the kinase activity of ETR1, more complicated models are suggested for how phosphorylation might serve to facilitate the ethylene response. Perhaps phospho-dependent signaling in the absence of ethylene induces the expression of a positive regulator of the ethylene response, with this positive regulator persisting during the time period of the ethylene response. However, it is not certain how well the in vitro bacterially based system mimics native ETR1, as preparation of the recombinant protein involved denaturation and renaturation (Voet-van-Vormizeele and Groth, 2008), potentially separating or perturbing the dimeric structure necessary for signal perception (Schaller et al., 1995; Rodríguez et al., 1999). The apparent inconsistency between the genetic and in vitro results suggests that it will be important to biochemically analyze the kinase activity of ETR1 using systems that more closely emulate signaling in plants.
MATERIALS AND METHODS
Plasmid Construction and Plant Transformation
All ETR1 constructs were derived from a 7.3-kb genomic ETR1 fragment containing the full-length ETR1 coding sequence and native genomic promoter (Chang et al., 1993). Construction of the wild-type ETR1-expressing plasmid has been described previously (Qu and Schaller, 2004). The ETR1-G2 mutant was generated as described previously (Gamble et al., 2002) and cloned into the binary vector pCAMBIA2380 (Gao et al., 2003). For the construction of ETR1-H/G2, the ETR1-G2 genomic clone was subcloned into pALTERII, the site-directed mutation made using the Altered Site Mutagenesis System (Promega), and the mutant product was then cloned into pCAMBIA2380. The primer used for mutagenesis (5′-CTAGCGGTTATGAACCAAGAAATGCGAACACC-3′) results in mutation of the conserved autophosphorylated His residue to a Gln residue. The ETR1-H/G2/D mutant was similarly constructed from ETR1-H/G2, the primer used for mutagenesis (5′-CAAAGTGGTCTTCATGAAGGTGTGCATGCCC-3′) resulting in mutation of the conserved Asp residue of the receiver domain to an Asn residue. The constructs were introduced into Agrobacterium tumefaciens strain GV3101 and used to transform an etr1-9/ers1-9;ers1-3/ERS1 line (Qu et al., 2007) by the floral dip method (Clough and Bent, 1998). Lines homozygous for both etr1-9 and ers1-3 were identified by PCR-based genotyping, and those homozygous for the transgene were based on the segregation of antibiotic resistance and by PCR-based genotyping (Qu et al., 2007).
Plant Growth Conditions and Ethylene Response Assays
Treatment and analysis of the triple response of dark-grown Arabidopsis (Arabidopsis thaliana) seedlings to ethylene (Chen and Bleecker, 1995; Roman and Ecker, 1995) were performed as described (Qu and Schaller, 2004). AVG, an inhibitor of ethylene biosynthesis, was included in growth medium at a concentration of 5 µm. Plates were placed in sealed containers with 0 to 1,000 µL L−1 ethylene as indicated. To examine seedlings growing in the absence of ethylene, hydrocarbon-free air was passed to remove ethylene synthesized by the seedlings. Seedlings were examined after 4 d of growth in the dark at 22°C, time 0 corresponding to the time when the plates were removed from 4°C (for stratification) and brought to 22°C for germination and growth. To measure hypocotyl length, seedlings were grown on vertically oriented square plates, the plates were scanned, and hypocotyl length was measured using ImageJ software (version 1.32; National Institutes of Health). To examine growth in the light, the plants were grown at 22°C under constant light on Murashige and Skoog basal medium with Gamborg’s vitamins (pH 5.75; Sigma), 1% (w/v) Suc, and 8% (w/v) agar (Qu et al., 2007).
Immunoblot Analysis
Immunoblot analysis was performed using microsomal fractions isolated from dark-grown Arabidopsis seedlings essentially as described (Gao et al., 2008). Briefly, plant material was homogenized in a buffer containing 30 mm Tris (pH 8.3 at 4°C), 150 mm NaCl, 1 mm EDTA, and 20% (v/v) glycerol with protease inhibitors (Sigma-Aldrich; P9599) and then centrifuged at 8,000g for 15 min (Chen et al., 2002; Zhao et al., 2002). The supernatant was then centrifuged at 100,000g for 30 min, and the resulting membrane pellet was resuspended in 10 mm Tris (pH 7.6 at 22°C), 150 mm NaCl, 0.1 mm EDTA, and 10% (v/v) glycerol with protease inhibitors. Protein concentration was determined by use of the bicinchoninic acid reagent (Pierce) according to the manufacturer after first adding 0.1 mL of 0.5% (w/v) SDS to solubilize membrane proteins. Bovine serum albumin was used as a standard for protein assays. ETR1 was identified by use of a polyclonal anti-ETR1 antibody generated against amino acids 401 to 738 of ETR1 (Chen et al., 2002). CTR1 was identified by use of a polyclonal anti-CTR1 antibody (Gao et al., 2003). An anti-BiP antibody was used as a loading control (Stressgen Biotech; SPA-818E). Relative expression levels for ETR1 and CTR1 were determined using the program ImageJ version 1.38x (http://rsbweb.nih.gov/ij/), with quantification of the scanned exposed film from the immunoblots being made by comparison with a dilution series (Zhao et al., 2002).
Gene Expression Analysis
Total RNA was isolated using the RNeasy plant kit according to the manufacturer’s instructions (Qiagen). The nucleic acid integrity was assessed by evaluating rRNA bands on agarose gels, and its quality was determined by calculating the 260:280 and 260:230 spectrophotometric ratios. When necessary, the RNA was cleaned further by phenol:chloroform:isoamyl alcohol (25:24:1) extraction, followed by precipitation with 0.15 m sodium acetate and 50% (v/v) isopropanol.
Microarray analysis was performed using GeneChip ATH1 Arabidopsis genome arrays (Affymetrix) on 10 µg of total RNA isolated from 4-d-old dark-grown seedlings, grown in the presence or absence of 1 µL L−1 ethylene. Three independent biological replicates were performed for each experimental treatment. Because we were examining the gene expression basis for a subtle phenotypic difference, transgenic lines carrying wild-type and kinase-inactive versions of ETR1 were grown together on the same petri plates, so as to minimize any potential effects of plate-to-plate variation. Samples were independently labeled (Genechip IVT labeling kit), hybridized, washed (Fluidics Station 450), and scanned by the Dartmouth Genomics and Microarray Laboratory (http://geiselmed.dartmouth.edu/dgml/) according to the manufacturers’ recommended procedures. Affymetrix GeneChip data files (CEL files) were imported into GeneSpring GX 11.5 (Agilent Technologies) for data analysis. The data were normalized by baseline transformation, which is equivalent to per chip to the 50th percentile and per gene to the median. Analysis of these data indicates that only 1.64% of the probe sets vary in expression by 2-fold or greater when comparing tETR1-wt with tETR1-H/G2 and that ethylene treatment of tETR1-wt results in a 2-fold or greater change in expression for 3.00% of the 22,810 probe sets present on the array, consistent with the majority of the genes not being differentially expressed under the experimental conditions. The raw data were filtered by expression, with the requirement that at least one sample out of six for a line exceed a lower cutoff percentile of 20%. The data were statistically analyzed using an unpaired t test, and data were accepted with P values less than or equal to 0.05. For interpretation of the data, the GeneSpring fold-change mode was used, and putative ethylene-regulated genes were identified based on their exhibiting a 3-fold change in the ethylene-treated sample compared with the control sample. The complete data set is deposited in Array-Express (http://www.ebi.ac.uk/arrayexpress) with accession number E-MEXP-3574 and is also available as Supplemental Table S3.
Real-time PCR was performed as described (Argyros et al., 2008) using primer sets specific to genes for pEARL1-like (At4g12470; 5′-AGTCCTAAACCAAAGCCAGTCCCA-3′ and 5′-CGATATTGTGCACTGGCATCGCAT-3′), Peroxidase ATP-N (At5g19890; 5′-CGCCATTATCGCCAAATTTGTAGCCG-3′ and 5′-TCATCGCACATGTGAAGTCCCTGA-3′), OSR1 (At2g41230; 5′-ATGAGGGTTCATGATCAACGGCTG-3′ and 5′-GGCTGGGCTCATTAGAAGGAGAAA-3′), ACC oxidase (At1g77330; 5′-GTGATGGATGAGAATTTGGGTTTGCC-3′ and 5′-ATCGATCCACTCGCCGTCTTTCAA-3′), ETR2 (At3g23150; 5′-AGAGAAACTCGGGTGCGATGT-3′ and 5′-TCACTGTCGTCGCCACAATC-3′), CHS (At5g13930; 5′-TGCTTACATGGCTCCTTCTCTGGA-3′ and 5′-ATCTCAGAGCAGACAACGAGGACA-3′), a pathogen-related gene (At4g25780; 5′-TGACCACGACTCCTTGCAGTTCTT-3′ and 5′-ATGAAGATCCCACCATTGTCGCAC-3′), APG-like (At1g75900; 5′-TTTGCGTCCGGAGGTTCTGGTTAT-3′ and 5′-CTGAGGCAGAGTCAGACATAAGAG-3′), COR6.6/KIN2 (At5g15960; 5′-TGTATCGGATGCGGCAGCG-3′ and 5′-TTTGAATATAAGTTTGGCTCGTCT-3′), and the control β-tubulin (At5g62700; 5′-CGTAAGCTTGCTGTGAATCTCATC-3′ and 5′-CTGCTCGTCAACTTCCTTTGTG-3′).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table S1. Ethylene up-regulated genes identified by microarray analysis.
Supplemental Table S2. Ethylene down-regulated genes identified by microarray analysis.
Supplemental Table S3. Complete microarray data set.
Supplementary Material
Glossary
- ER
endoplasmic reticulum
- AVG
aminoethylvinylglycine
- Ws
ecotype Wassilewskija
- qRT
quantitative reverse transcription
- RT
reverse transcription
References
- Abeles FB, Morgan PW, Saltveit ME., Jr (1992) Ethylene in Plant Biology, Ed 2. Academic Press, San Diego.
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Argyros RD, Mathews DE, Chiang Y-H, Palmer CM, Thibault DM, Etheridge N, Argyros DA, Mason MG, Kieber JJ, Schaller GE. (2008) Type B response regulators of Arabidopsis play key roles in cytokinin signaling and plant development. Plant Cell 20: 2102–2116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder BM, O’Malley RC, Wang W, Moore JM, Parks BM, Spalding EP, Bleecker AB. (2004) Arabidopsis seedling growth response and recovery to ethylene: a kinetic analysis. Plant Physiol 136: 2913–2920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisson MM, Bleckmann A, Allekotte S, Groth G. (2009) EIN2, the central regulator of ethylene signalling, is localized at the ER membrane where it interacts with the ethylene receptor ETR1. Biochem J 424: 1–6 [DOI] [PubMed] [Google Scholar]
- Caesar K, Thamm AM, Witthöft J, Elgass K, Huppenberger P, Grefen C, Horak J, Harter K. (2011) Evidence for the localization of the Arabidopsis cytokinin receptors AHK3 and AHK4 in the endoplasmic reticulum. J Exp Bot 62: 5571–5580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancel JD, Larsen PB. (2002) Loss-of-function mutations in the ethylene receptor ETR1 cause enhanced sensitivity and exaggerated response to ethylene in Arabidopsis. Plant Physiol 129: 1557–1567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Kwok SF, Bleecker AB, Meyerowitz EM. (1993) Arabidopsis ethylene-response gene ETR1: similarity of product to two-component regulators. Science 262: 539–544 [DOI] [PubMed] [Google Scholar]
- Chang C, Meyerowitz EM. (1995) The ethylene hormone response in Arabidopsis: a eukaryotic two-component signaling system. Proc Natl Acad Sci USA 92: 4129–4133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Stadler R. (2001) Ethylene hormone receptor action in Arabidopsis. Bioessays 23: 619–627 [DOI] [PubMed] [Google Scholar]
- Chen QG, Bleecker AB. (1995) Analysis of ethylene signal-transduction kinetics associated with seedling-growth response and chitinase induction in wild-type and mutant Arabidopsis. Plant Physiol 108: 597–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YF, Etheridge N, Schaller GE. (2005) Ethylene signal transduction. Ann Bot (Lond) 95: 901–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YF, Gao Z, Kerris RJ, III, Wang W, Binder BM, Schaller GE. (2010) Ethylene receptors function as components of high-molecular-mass protein complexes in Arabidopsis. PLoS ONE 5: e8640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YF, Randlett MD, Findell JL, Schaller GE. (2002) Localization of the ethylene receptor ETR1 to the endoplasmic reticulum of Arabidopsis. J Biol Chem 277: 19861–19866 [DOI] [PubMed] [Google Scholar]
- Clark KL, Larsen PB, Wang X, Chang C. (1998) Association of the Arabidopsis CTR1 Raf-like kinase with the ETR1 and ERS ethylene receptors. Proc Natl Acad Sci USA 95: 5401–5406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Dong C-H, Rivarola M, Resnick JS, Maggin BD, Chang C. (2008) Subcellular co-localization of Arabidopsis RTE1 and ETR1 supports a regulatory role for RTE1 in ETR1 ethylene signaling. Plant J 53: 275–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamble RL, Coonfield ML, Schaller GE. (1998) Histidine kinase activity of the ETR1 ethylene receptor from Arabidopsis. Proc Natl Acad Sci USA 95: 7825–7829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamble RL, Qu X, Schaller GE. (2002) Mutational analysis of the ethylene receptor ETR1: role of the histidine kinase domain in dominant ethylene insensitivity. Plant Physiol 128: 1428–1438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao R, Stock AM. (2009) Biological insights from structures of two-component proteins. Annu Rev Microbiol 63: 133–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z, Chen YF, Randlett MD, Zhao XC, Findell JL, Kieber JJ, Schaller GE. (2003) Localization of the Raf-like kinase CTR1 to the endoplasmic reticulum of Arabidopsis through participation in ethylene receptor signaling complexes. J Biol Chem 278: 34725–34732 [DOI] [PubMed] [Google Scholar]
- Gao Z, Wen C-K, Binder BM, Chen Y-F, Chang J, Chiang Y-H, Kerris RJ, III, Chang C, Schaller GE. (2008) Heteromeric interactions among ethylene receptors mediate signaling in Arabidopsis. J Biol Chem 283: 23801–23810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grefen C, Städele K, Růzicka K, Obrdlik P, Harter K, Horák J. (2008) Subcellular localization and in vivo interactions of the Arabidopsis thaliana ethylene receptor family members. Mol Plant 1: 308–320 [DOI] [PubMed] [Google Scholar]
- Hall AE, Bleecker AB. (2003) Analysis of combinatorial loss-of-function mutants in the Arabidopsis ethylene receptors reveals that the ers1 etr1 double mutant has severe developmental defects that are EIN2 dependent. Plant Cell 15: 2032–2041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall AE, Chen QG, Findell JL, Schaller GE, Bleecker AB. (1999) The relationship between ethylene binding and dominant insensitivity conferred by mutant forms of the ETR1 ethylene receptor. Plant Physiol 121: 291–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hass C, Lohrmann J, Albrecht V, Sweere U, Hummel F, Yoo SD, Hwang I, Zhu T, Schäfer E, Kudla Jet al. (2004) The response regulator 2 mediates ethylene signalling and hormone signal integration in Arabidopsis. EMBO J 23: 3290–3302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua J, Meyerowitz EM. (1998) Ethylene responses are negatively regulated by a receptor gene family in Arabidopsis thaliana. Cell 94: 261–271 [DOI] [PubMed] [Google Scholar]
- Huang Y, Li H, Hutchison CE, Laskey J, Kieber JJ. (2003) Biochemical and functional analysis of CTR1, a protein kinase that negatively regulates ethylene signaling in Arabidopsis. Plant J 33: 221–233 [DOI] [PubMed] [Google Scholar]
- Iuchi S. (1993) Phosphorylation/dephosphorylation of the receiver module at the conserved aspartate residue controls transphosphorylation activity of histidine kinase in sensor protein ArcB of Escherichia coli. J Biol Chem 268: 23972–23980 [PubMed] [Google Scholar]
- Kendrick MD, Chang C. (2008) Ethylene signaling: new levels of complexity and regulation. Curr Opin Plant Biol 11: 479–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieber JJ, Rothenberg M, Roman G, Feldmann KA, Ecker JR. (1993) CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the raf family of protein kinases. Cell 72: 427–441 [DOI] [PubMed] [Google Scholar]
- Larsen PB, Chang C. (2001) The Arabidopsis eer1 mutant has enhanced ethylene responses in the hypocotyl and stem. Plant Physiol 125: 1061–1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z, Zhong S, Grierson D. (2009) Recent advances in ethylene research. J Exp Bot 60: 3311–3336 [DOI] [PubMed] [Google Scholar]
- Mason MG, Mathews DE, Argyros DA, Maxwell BB, Kieber JJ, Alonso JM, Ecker JR, Schaller GE. (2005) Multiple type-B response regulators mediate cytokinin signal transduction in Arabidopsis. Plant Cell 17: 3007–3018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason MG, Schaller GE. (2005) Histidine kinase activity and the regulation of ethylene signal transduction. Can J Bot 83: 563–570 [Google Scholar]
- Mattoo AK, Suttle JC, editors (1991) The Plant Hormone Ethylene. CRC Press, Boca Raton, FL
- Mizuno T. (1997) Compilation of all genes encoding two-component phosphotransfer signal transducers in the genome of Escherichia coli. DNA Res 4: 161–168 [DOI] [PubMed] [Google Scholar]
- Moussatche P, Klee HJ. (2004) Autophosphorylation activity of the Arabidopsis ethylene receptor multigene family. J Biol Chem 279: 48734–48741 [DOI] [PubMed] [Google Scholar]
- O’Malley RC, Rodriguez FI, Esch JJ, Binder BM, O’Donnell P, Klee HJ, Bleecker AB. (2005) Ethylene-binding activity, gene expression levels, and receptor system output for ethylene receptor family members from Arabidopsis and tomato. Plant J 41: 651–659 [DOI] [PubMed] [Google Scholar]
- Pan SQ, Charles T, Jin S, Wu Z-L, Nester EW. (1993) Preformed dimeric state of the sensor protein VirA is involved in plant-Agrobacterium signal transduction. Proc Natl Acad Sci USA 90: 9939–9943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posas F, Wurgler-Murphy SM, Maeda T, Witten EA, Thai TC, Saito H. (1996) Yeast HOG1 MAP kinase cascade is regulated by a multistep phosphorelay mechanism in the SLN1-YPD1-SSK1 “two-component” osmosensor. Cell 86: 865–875 [DOI] [PubMed] [Google Scholar]
- Qu X, Hall BP, Gao Z, Schaller GE. (2007) A strong constitutive ethylene-response phenotype conferred on Arabidopsis plants containing null mutations in the ethylene receptors ETR1 and ERS1. BMC Plant Biol 7: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu X, Schaller GE. (2004) Requirement of the histidine kinase domain for signal transduction by the ethylene receptor ETR1. Plant Physiol 136: 2961–2970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez FI, Esch JJ, Hall AE, Binder BM, Schaller GE, Bleecker AB. (1999) A copper cofactor for the ethylene receptor ETR1 from Arabidopsis. Science 283: 996–998 [DOI] [PubMed] [Google Scholar]
- Roman G, Ecker JR. (1995) Genetic analysis of a seedling stress response to ethylene in Arabidopsis. Philos Trans R Soc Lond B Biol Sci 350: 75–81 [DOI] [PubMed] [Google Scholar]
- Sanders IO, Harpham NVJ, Raskin I, Smith AR, Hall MA. (1991) Ethylene binding in wild type and mutant Arabidopsis thaliana (L.) Heynth. Ann Bot (Lond) 68: 97–103 [Google Scholar]
- Schaller GE, Bleecker AB. (1995) Ethylene-binding sites generated in yeast expressing the Arabidopsis ETR1 gene. Science 270: 1809–1811 [DOI] [PubMed] [Google Scholar]
- Schaller GE, Kieber JJ. (2002) Ethylene. The Arabidopsis Book 1: e0071, doi/10.1199/tab.0071. [DOI] [PMC free article] [PubMed]
- Schaller GE, Kieber JJ, Shiu S-H. (2008) Two-component signaling elements and histidyl-aspartyl phosphorelays. The Arabidopsis Book 6: e0112, doi/10.1199/tab.0112. [DOI] [PMC free article] [PubMed]
- Schaller GE, Ladd AN, Lanahan MB, Spanbauer JM, Bleecker AB. (1995) The ethylene response mediator ETR1 from Arabidopsis forms a disulfide-linked dimer. J Biol Chem 270: 12526–12530 [DOI] [PubMed] [Google Scholar]
- Schaller GE, Shiu SH, Armitage JP. (2011) Two-component systems and their co-option for eukaryotic signal transduction. Curr Biol 21: R320–R330 [DOI] [PubMed] [Google Scholar]
- Scharein B, Voet-van-Vormizeele J, Harter K, Groth G. (2008) Ethylene signaling: identification of a putative ETR1-AHP1 phosphorelay complex by fluorescence spectroscopy. Anal Biochem 377: 72–76 [DOI] [PubMed] [Google Scholar]
- Stock AM, Robinson VL, Goudreau PN. (2000) Two-component signal transduction. Annu Rev Biochem 69: 183–215 [DOI] [PubMed] [Google Scholar]
- Urao T, Miyata S, Yamaguchi-Shinozaki K, Shinozaki K. (2000) Possible His to Asp phosphorelay signaling in an Arabidopsis two-component system. FEBS Lett 478: 227–232 [DOI] [PubMed] [Google Scholar]
- Voet-van-Vormizeele J, Groth G. (2008) Ethylene controls autophosphorylation of the histidine kinase domain in ethylene receptor ETR1. Mol Plant 1: 380–387 [DOI] [PubMed] [Google Scholar]
- Wang W, Hall AE, O’Malley R, Bleecker AB. (2003) Canonical histidine kinase activity of the transmitter domain of the ETR1 ethylene receptor from Arabidopsis is not required for signal transmission. Proc Natl Acad Sci USA 100: 352–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ. (2007) An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wulfetange K, Lomin SN, Romanov GA, Stolz A, Heyl A, Schmülling T. (2011) The cytokinin receptors of Arabidopsis are located mainly to the endoplasmic reticulum. Plant Physiol 156: 1808–1818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie F, Liu Q, Wen C-K. (2006) Receptor signal output mediated by the ETR1 N terminus is primarily subfamily I receptor dependent. Plant Physiol 142: 492–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Inouye M. (1993) Requirement of both kinase and phosphatase activities of an Escherichia coli receptor (Taz1) for ligand-dependent signal transduction. J Mol Biol 231: 335–342 [DOI] [PubMed] [Google Scholar]
- Zhao XC, Qu X, Mathews DE, Schaller GE. (2002) Effect of ethylene pathway mutations upon expression of the ethylene receptor ETR1 from Arabidopsis. Plant Physiol 130: 1983–1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S, Lin Z, Grierson D. (2008) Tomato ethylene receptor-CTR interactions: visualization of NEVER-RIPE interactions with multiple CTRs at the endoplasmic reticulum. J Exp Bot 59: 965–972 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.