Abstract
WRKY transcription factors are encoded by a large gene superfamily with a broad range of roles in plants. Recently, several groups have reported that proteins containing a short VQ (FxxxVQxLTG) motif interact with WRKY proteins. We have recently discovered that two VQ proteins from Arabidopsis (Arabidopsis thaliana), SIGMA FACTOR-INTERACTING PROTEIN1 and SIGMA FACTOR-INTERACTING PROTEIN2, act as coactivators of WRKY33 in plant defense by specifically recognizing the C-terminal WRKY domain and stimulating the DNA-binding activity of WRKY33. In this study, we have analyzed the entire family of 34 structurally divergent VQ proteins from Arabidopsis. Yeast (Saccharomyces cerevisiae) two-hybrid assays showed that Arabidopsis VQ proteins interacted specifically with the C-terminal WRKY domains of group I and the sole WRKY domains of group IIc WRKY proteins. Using site-directed mutagenesis, we identified structural features of these two closely related groups of WRKY domains that are critical for interaction with VQ proteins. Quantitative reverse transcription polymerase chain reaction revealed that expression of a majority of Arabidopsis VQ genes was responsive to pathogen infection and salicylic acid treatment. Functional analysis using both knockout mutants and overexpression lines revealed strong phenotypes in growth, development, and susceptibility to pathogen infection. Altered phenotypes were substantially enhanced through cooverexpression of genes encoding interacting VQ and WRKY proteins. These findings indicate that VQ proteins play an important role in plant growth, development, and response to environmental conditions, most likely by acting as cofactors of group I and IIc WRKY transcription factors.
WRKY proteins are a relatively recently identified class of sequence-specific DNA-binding transcription factors found almost exclusively in plants (Rushton et al., 2010). The characteristic structural feature of WRKY proteins is the highly conserved WRKY domain, which contains the almost invariant WRKYGQK sequence at the N terminus followed by a Cx4-5Cx22-23HxH or Cx7Cx23HxC zinc-finger motif (Rushton et al., 2010). Genes encoding WRKY proteins have been identified in low photosynthetic and nonphotosynthetic eukaryotes, but they have greatly proliferated and form large superfamilies only in higher plants with more than 70 members in Arabidopsis (Arabidopsis thaliana; Zhang and Wang, 2005). Based on the number and structures of the conserved WRKY zinc-finger motifs, WRKY proteins were initially classified into three groups (Eulgem et al., 2000). The first group contains two Cx4Cx22-23HxH zinc-finger motifs, the second group contains one Cx4-5Cx23HxH zinc-finger motif, and the third group contains one Cx7Cx23HxC zinc-finger motif. More recent analyses, however, have shown that group II WRKY proteins can be further divided into IIa, IIb, IIc, IId, and IIe subgroups (Zhang and Wang, 2005; Rushton et al., 2010). In the green alga Chlamydomonas reinhardtii as well as in the nonphotosynthetic slime mold Dictyostelium discoideum and unicellular protist Giardia lamblia, there is only a single group I WRKY gene (Zhang and Wang, 2005), suggesting that group I WRKY proteins with two WRKY domains are the ancestors to the other groups of WRKY proteins.
Since their initial identification (Ishiguro and Nakamura, 1994), plant WRKY transcription factors have been subjected to intensive analysis for their biological functions. Earlier studies from analysis of the expression patterns of isolated WRKY genes and the cis-acting elements in defense gene promoters suggested an important role of plant WRKY transcription factors in plant defense responses. For example, we have previously analyzed the expression profile of the Arabidopsis WRKY gene superfamily and discovered that more than 70% of the gene family members are responsive to pathogen infection and salicylic acid (SA) treatment (Dong et al., 2003). The indirect evidence for the involvement of WRKY proteins in plant defense responses has been confirmed by studies using loss-of-function knockout/down mutants or overexpression lines of WRKY genes. These studies have shown that WRKY transcription factors can act as positive or negative regulators of plant defense responses (Li et al., 2004, 2006; Journot-Catalino et al., 2006; Kim et al., 2006, 2008; Xu et al., 2006; Zheng et al., 2006; Lai et al., 2008). Other studies have found that WRKY proteins play critical roles in plant hormone signaling (Chen et al., 2010; Shang et al., 2010), secondary metabolism (Wang et al., 2010b; Suttipanta et al., 2011), and plant responses to abiotic stresses (Jiang and Deyholos, 2009; Li et al., 2009, 2011). In addition, several WRKY proteins regulate plant growth and developmental processes including trichome (Johnson et al., 2002) and seed (Luo et al., 2005) development, germination (Jiang and Yu, 2009), and leaf senescence (Robatzek and Somssich, 2002; Miao et al., 2004). Thus, WRKY proteins have important roles in a broad range of biological processes in plants.
Despite the wide range of biological functions, almost all analyzed WRKY proteins recognize the TTGACC/T W-box sequences (de Pater et al., 1996; Rushton et al., 1996; Wang et al., 1998; Chen and Chen, 2000; Cormack et al., 2002). This raises the question of how the functional specificity of different members of the transcription factor superfamily is achieved. One potentially important specificity determinant could be the nucleotides adjacent to the TTGACC/T core sequence, which may influence both the binding specificity and binding affinity of WRKY proteins to the core W boxes (Ciolkowski et al., 2008). Other mechanisms may include differential expression of WRKY genes and difference in transcription-activating or -repressing activities of different WRKY proteins. As found with other transcription factors, differential DNA-binding and transcription-regulatory activities of many WRKY transcription factors could well be regulated by their interacting partners such as coactivators, chromatin remodelers, and enzymes that modify histone. For example, we have previously reported both physical and functional interactions between structurally related Arabidopsis WRKY18, WRKY40, and WRKY60 (Xu et al., 2006). The three pathogen-induced WRKY proteins formed both homocomplexes and heterocomplexes with significantly different DNA-binding activity (Xu et al., 2006). More recently, we have shown that Arabidopsis WRKY38 and WRKY62 interact with HISTONE DEACETYLASE19 (HDA19) in plant basal defense (Kim et al., 2008). Both WRKY38 and WRKY62 were transcriptional activators in plant cells but the activating activities were largely abolished by overexpressed HDA19. Functional analysis using knockout mutants indicated that HDA19 had a positive role whereas WRKY38 and WRKY62 had negative roles in plant basal defense. The physical interaction of the functionally opposing WRKY proteins with HDA19 may, therefore, act to fine tune plant basal defense responses (Kim et al., 2008).
Over the past several years, several groups including ours have reported physical interactions of WRKY transcription factors with proteins containing a conserved FxxxVQxLTG or VQ motif. Arabidopsis WRKY25 and WRKY33 interact with MKS1, a VQ protein that also acts as a substrate of MAPK4 (MPK4) from Arabidopsis (Andreasson et al., 2005; Qiu et al., 2008). HAIKU1 (IKU1), another VQ protein important for endosperm growth and seed size, interacts with WRKY10, which also regulates Arabidopsis seed size (Wang et al., 2010a). Very recently, we have shown that WRKY33 interacts with two other VQ proteins, SIGMA FACTOR-INTERACTING PROTEIN1 (SIB1) and SIB2 (Lai et al., 2011). MKS1, SIB1, and SIB2 recognize the C-terminal WRKY domain and stimulate the DNA-binding activity of WRKY33 (Lai et al., 2011). Further analysis indicated that the conserved V and Q residues in the VQ motif of SIB1 are important for its interaction with WRKY33 (Lai et al., 2011). Based on these and additional analyses of the interacting WRKY and VQ proteins, we have proposed that the short VQ motif represents the core of a WRKY-interacting domain and the VQ proteins are important regulators of DNA binding and other molecular activities of WRKY transcription factors (Lai et al., 2011). Here we report our expanded structural and functional analysis of the 34 Arabidopsis VQ genes and their gene products. We have discovered that the structurally diverse VQ proteins interact with the C-terminal WRKY domains of group I WRKY proteins and the sole WRKY domains of group IIc WRKY proteins, which share a number of amino acid residues critical for interaction with VQ proteins. Additional analysis using gene expression, knockout, and overexpression lines strongly suggest that plant VQ genes play important roles in plant growth, development, and defense responses.
RESULTS
Identification of VQ Proteins from Arabidopsis
Five VQ motif-containing proteins from Arabidopsis have been previously reported and functionally analyzed. These known VQ proteins are MKS1 (Andreasson et al., 2005), IKU1 (Wang et al., 2010a), CALMODULIN-BINDING PROTEIN25 (CAMBP25; Perruc et al., 2004), SIB1, and SIB2 (Xie et al., 2010; Lai et al., 2011). Loss-of-function mutants and/or overexpression lines for the VQ genes are altered in seed size, tolerance to abiotic stress, or resistance to pathogen infection, indicating that members of this protein family play important roles in plant growth, development, and responses to environmental conditions. To understand the extensiveness of VQ proteins in Arabidopsis, we analyzed the entire family of the 34 genes encoding proteins containing the FxxxVQxxLTG motif in Arabidopsis (Xie et al., 2010; Table I). A majority of the Arabidopsis VQ genes are intronless and encode relatively small proteins with fewer than 300 amino acid residues. VQ2 is the largest VQ protein with 430 amino acid residues and is predicted to be encoded by a gene with four introns. The N-terminal region of approximately 240 amino acids of VQ2 is highly homologous with VQ3, which is encoded by an intronless gene adjacent to VQ2 on chromosome I (Fig. 1). The remaining, C-terminal region of VQ2, which are encoded by the other three exons, contains a RNA-recognition motif that is absent in VQ3. It is likely that VQ2 and VQ3 were resulted from relatively recent gene duplication, after which VQ2 has acquired additional functions through fusion with other gene(s).
Table I. Properties of the Arabidopsis VQ proteins.
AGI ID, Arabidopsis Genome Initiative Identification Number; CDS, coding DNA sequence; n, Nucleus; p, plastid; m, mitochondria.
| Name | AGI ID | Other Name | No. of Amino Acids | Subcellular Localizationa | Intron within CDS |
|---|---|---|---|---|---|
| AtVQ1 | AT1G17147 | 99 | p/n | 0 | |
| AtVQ2 | AT1G21320 | 430 | n | 4 | |
| AtVQ3 | AT1G21326 | 239 | n | 0 | |
| AtVQ4 | AT1G28280 | 247 | n/m | 0-1 | |
| AtVQ5 | AT1G32585 | 220 | n | 0 | |
| AtVQ6 | At1G32610 | 291 | n | 0-1 | |
| AtVQ7 | AT1G35830 | 302 | n/m | 0 | |
| AtVQ8 | AT1G68450 | 152 | n/p | 0 | |
| AtVQ9 | AT1G78310 | 311 | n/p | 0 | |
| AtVQ10 | AT1G78410 | 104 | p/n | 0 | |
| AtVQ11 | AT1G80450 | 177 | n/p | 0 | |
| AtVQ12 | AT2G22880 | 114 | p/m | 0 | |
| AtVQ13 | AT2G33780 | 204 | n/m | 0 | |
| AtVQ14 | AT2G35230 | IKU1 | 402 | n | 0-1 |
| AtVQ15 | AT2G41010 | CAMBP25 | 238 | n | 0 |
| AtVQ16 | AT2G41180 | SIB2 | 141 | n/p | 0 |
| AtVQ17 | AT2G42140 | 172 | p | 0 | |
| AtVQ18 | AT2G44340 | 188 | n/p | 0 | |
| AtVQ19 | AT3G15300 | 219 | n/p | 0 | |
| AtVQ20 | AT3G18360 | 285 | n/p | 0 | |
| AtVQ21 | AT3G18690 | MKS1 | 222 | n | 0 |
| AtVQ22 | AT3G22160 | 192 | n/p | 0 | |
| AtVQ23 | AT3G56710 | SIB1 | 151 | p/n | 0 |
| AtVQ24 | AT3G56880 | 245 | n | 0 | |
| AtVQ25 | AT3G58000 | 175 | n/m | 0 | |
| AtVQ26 | AT3G60090 | 157 | n | 0 | |
| AtVQ27 | AT4G15120 | 193 | n/p | 0 | |
| AtVQ28 | AT4G20000 | 208 | n | 0 | |
| AtVQ29 | AT4G37710 | 117 | p/n | 0 | |
| AtVQ30 | AT4G39720 | 290 | n | 0 | |
| AtVQ31 | AT5G08480 | 174 | p/n | 0-1 | |
| AtVQ32 | AT5G46780 | 237 | n | 0-1 | |
| AtVQ33 | AT5G53830 | 243 | p/n | 0 | |
| AtVQ34 | AT5G65170 | 363 | n | 0 |
Based on prediction in the Subcellular Proteomic Database (http://suba.plantenergy.uwa.edu.au).
Figure 1.
Alignment of VQ motif sequences of Arabidopsis proteins. The highly conserved residues in the VQ motif are indicated in red.
The previously characterized IKU1 (VQ14), CAMBP25 (VQ15), and MKS1 (VQ21) are localized exclusively in the nucleus. On the other hand, both SIB1 (VQ23) and SIB2 (VQ16) are dual targeted, interacting with both plastic SIG1 and nuclear WRKY proteins. Through search of the Subcellular Proteomic Database, which contains large proteomic and GFP localization sets and precompiled bioinformatics predictions for subcellular localization of proteins from Arabidopsis, we found that a majority of Arabidopsis VQ proteins are also localized in the nucleus (Table I). Interestingly, a substantial number of Arabidopsis VQ proteins also contain a putative chloroplast or mitochondrial targeting signal at their N terminus, in addition to the nuclear localization signal (Table I). Thus, like SIB1 and SIB2, some members of the Arabidopsis VQ protein family appear to be dual targeted to both the nucleus and chloroplasts or mitochondria.
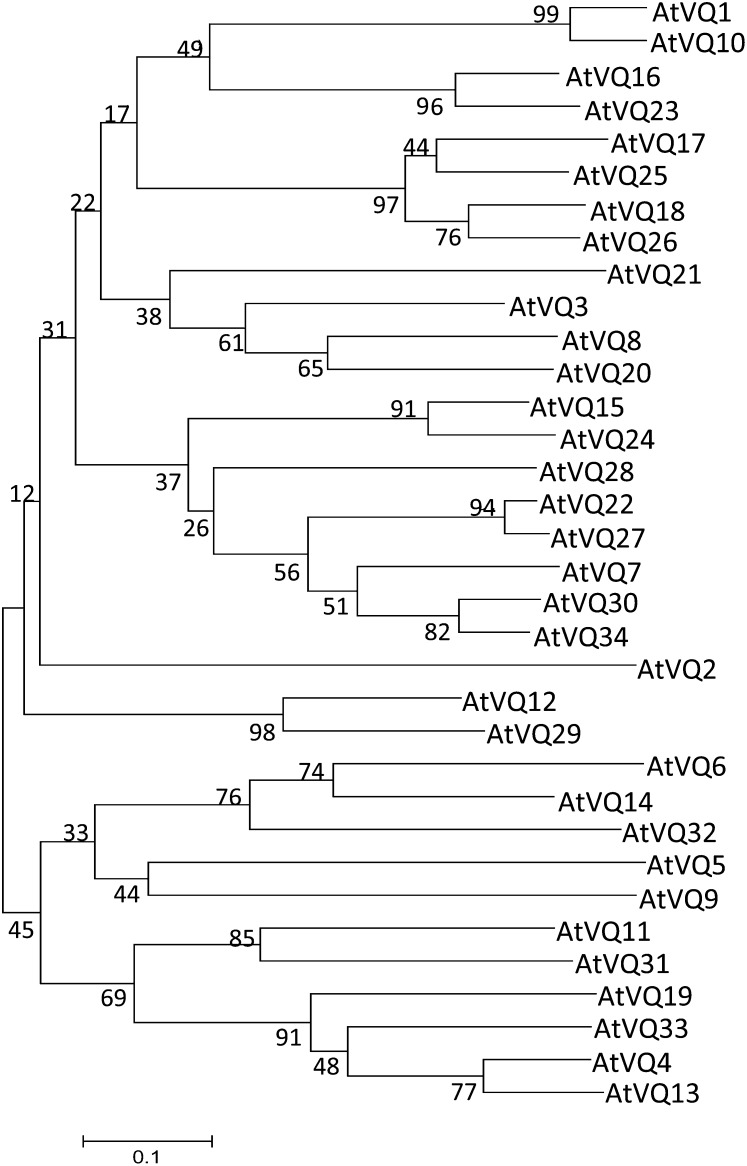
Apart from the short VQ motif, there is relatively low sequence homology among the Arabidopsis VQ proteins (Fig. 1). When the full-length protein sequences of the 34 VQ proteins were used to construct the phylogenetic tree, there were only four branches with bootstrap values larger than 95% (Fig. 2). A majority of the branches, particularly those deep ones had low confidence values (Fig. 2), likely due to the divergent sequences of the protein family.
Figure 2.
Phylogenetic trees of the VQ proteins from Arabidopsis. The tree was inferred using the neighbor-joining method. Phylogenetic analyses were conducted in MEGA5. Bootstrap values from 1,000 replicates were used to assess the robustness of the tree.
Interacting Partners of VQ Proteins from Arabidopsis
Among the five Arabidopsis VQ proteins functionally analyzed, four have been shown to interact with WRKY proteins. In addition, we have previously reported that Arabidopsis VQ1 interacts with WRKY33 (Lai et al., 2011). Subsequent screens also identified VQ10 interacted with WRKY33 and closely related WRKY25 and WRKY26 (Table II). In this study, we first analyzed using yeast (Saccharomyces cerevisiae) two-hybrid assays whether CAMBP25/VQ15, one of the five functionally characterized VQ proteins, also interacted with WRKY proteins. For this purpose, we fused the CAMBP25 gene with the AD domain of the Gal4 transcription factor in the yeast two-hybrid prey vector pAD. We also cloned cDNA fragments corresponding to the DNA-binding domains for WRKY25, WRKY33, WRKY51, WRKY70, and WRKY38 into the pBD bait vector. The fused pAD and pBD vectors were then cotransformed into yeast cells and tested for LacZ reporter gene expression through assays of β-galactosidase activity. Both assays showed that CAMBP25/VQ15 was capable of interacting with both WRKY25 and WRKY51 (Table II).
Table II. Proteins that interact with VQ proteins from Arabidopsis.
AGI ID, Arabidopsis Genome Initiative Identification Number; Last column: 1, This study; 2, Perruc et al. (2004); 3, Andreasson et al. (2005); 4, Morikawa et al. (2002); 5, Lai et al. (2011); 6, Wang et al. (2010a); 7, Arabidopsis Interactome Mapping Consortium (2011).
| VQ-Interacting Protein | Source | |||||
|---|---|---|---|---|---|---|
| VQ Protein |
WRKY Protein |
Non-WRKY Protein |
||||
| Name | AGI ID | Name | AGI ID | Name | AGI ID | |
| AtVQ1 | AT1G17147 | WRKY33(I) | AT2G38470 | 1 | ||
| AtVQ8 | AT1G68450 | WRKY20(I) | AT4G26640 | RACK1C | At3g18130 | 7 |
| WRKY24(IIc) | AT5G41570 | |||||
| WRKY34(I) | AT4G26440 | |||||
| AtVQ9 | AT1G78310 | WRKY20(I) | AT4G26640 | MPK3 | At3g45640 | |
| AtVQ10 | AT1G78410 | WRKY25(I) | AT2G30250 | 1 | ||
| WRKY26(I) | AT5G07100 | |||||
| WRKY33(I) | AT2G38470 | |||||
| AtVQ12 | AT2G22880 | WRKY20(I) | AT4G26640 | CSN5A | AT1G22920 | 7 |
| WRKY23(I) | AT2G47260 | |||||
| WRKY24(IIc) | AT5G41570 | |||||
| AtVQ14 | AT2G35230 | WRKY10(IIc) | AT1G55610 | 6 | ||
| AtVQ15 | AT2G41010 | WRKY25(I) | AT2G30250 | CaM1 | AT5G37780 | 1, 2 |
| WRKY51(IIc) | AT5G64810 | |||||
| AtVQ16 | AT2G41180 | WRKY25(I) | AT2G30250 | 4, 5 | ||
| WRKY33(I) | AT2G38470 | |||||
| AtVQ20 | AT3G18360 | WRKY20(I) | AT4G26640 | 7 | ||
| WRKY75(IIc) | AT5G13080 | |||||
| AtVQ21 | AT3G18690 | WRKY25(I) | AT2G30250 | MPK4 | At4g01370 | 3 |
| WRKY33(I) | AT2G38470 | |||||
| AtVQ22 | AT3G22160 | WRKY51(IIc) | AT5G64810 | |||
| AtVQ23 | AT3G56710 | WRKY3(I) | AT4G26640 | SIGA | At1g64860 | 4, 5 |
| WRKY4(I) | AT1G13960 | |||||
| WRKY20(I) | AT4G26640 | |||||
| WRKY25(I) | AT2G30250 | |||||
| WRKY33(I) | AT5G64810 | |||||
| AtVQ24 | AT3G56880 | WRKY20(I) | AT4G26640 | 7 | ||
| WRKY75(IIc) | AT5G13080 | |||||
| AtVQ32 | AT5G46780 | WRKY20(I) | AT4G26640 | NDL1 | AT5G56750 | 7 |
To determine whether other Arabidopsis VQ proteins are capable of interacting with WRKY proteins, we utilized a recently published proteome-wide binary protein-protein-interaction map of Arabidopsis (Arabidopsis Interactome Mapping Consortium, 2011). The map was generated by testing all pairwise combinations of a collection of approximately 8,000 Arabidopsis open reading frames with an improved high-throughput binary interactome mapping pipeline based on the yeast two-hybrid system. The interaction map contains approximately 6,200 highly reliable interactions between approximately 2,700 proteins. Through searching the Arabidopsis interactome database, we found eight VQ proteins (VQ8–10, VQ12, VQ16, VQ20, VQ24, and VQ32) in the generated interactome database with interacting partners. Significantly, all these eight VQ proteins had WRKY proteins as interacting partners (Table II). These interacting WRKY proteins all belong to either group I or group IIc WRKY proteins (Table II). Several of these eight VQ proteins also interacted with other non-WRKY proteins. For example, VQ9 interacted with MPK3 (Table II), a key signaling regulator involved in a variety of biological processes including defense responses (Asai et al., 2002; Pitzschke et al., 2009; Mao et al., 2011). VQ32 also interacted with N-MYC DOWN-REGULATED-LIKE1 (NDL1; Table II), a positive regulator of auxin transport in a G-protein-mediated pathway (Mudgil et al., 2009).
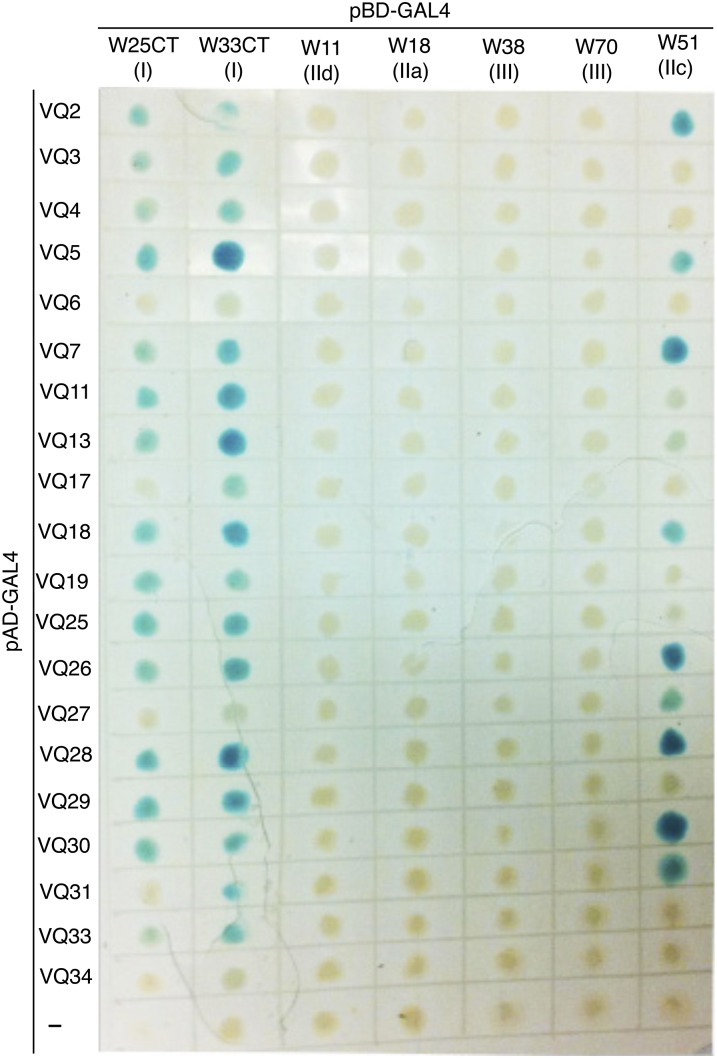
To further examine the partnership between the WRKY and VQ proteins, we analyzed the remaining 20 VQ proteins from the protein family for interaction with the WRKY domains from various groups of the WRKY superfamily. These tested WRKY domains included the C-terminal WRKY domains of WRKY25 and WRKY33 (both group I) and the WRKY domains of WRKY11 (group IId), WRKY18 (group IIa), WRKY38 (group III), WRKY70 (group III), and WRKY51 (group IIc; Fig. 3). Again, we fused these VQ genes with the AD domain of the Gal4 transcription factor in the yeast two-hybrid vector pAD and cDNA fragments corresponding to the DNA-binding domains for the WRKY genes into the pBD vector. The fused pAD and pBD recombinant vectors were then cotransformed into yeast cells and tested for LacZ reporter gene expression through assays of β-galactosidase activity. As shown in Figure 3, a majority of the 20 VQ proteins interacted with the C-terminal WRKY domains of both WRKY25 and WRKY33, albeit with varying intensities based on the β-galactosidase activity. VQ6, VQ17, and VQ34 failed to interact with WRKY25 and only interacted weakly with WRKY33 based on the very low β-galactosidase activities detected (Fig. 3). About 50% of the 20 VQ proteins also interacted with group IIc WRKY51 based on the positive β-galactosidase activity detected in the cotransformed yeast cells. On the other hand, none of the 20 VQ proteins interacted with the DNA-binding WRKY domains of group IId (WRKY11), group IIa (WRKY18), or group III (WRKY38 and WRKY70) WRKY proteins (Fig. 3). To determine whether VQ proteins recognized the other two groups of WRKY domains, we assayed the interactions of VQ5, VQ7, and VQ10 with the WRKY domains of WRKY6 (group IIb) and WRKY22 (group IIe) but failed to detect any significant β-galactosidase activity in yeast cells cotransformed with the fused bait and prey vectors (see Supplemental Fig. S1).
Figure 3.
Interaction of Arabidopsis VQ proteins with WRKY proteins in yeast cells. The Gal4 DNA BD-WRKY domain fusion bait vectors were contransformed with the activation domain (AD)-VQ fusion prey vectors into yeast cells and the transformant cells were assayed for LacZ reporter gene expression. The empty pAD prey vector was used as negative control (−).
Structural Features of WRKY Domains Critical for Interaction with VQ Proteins
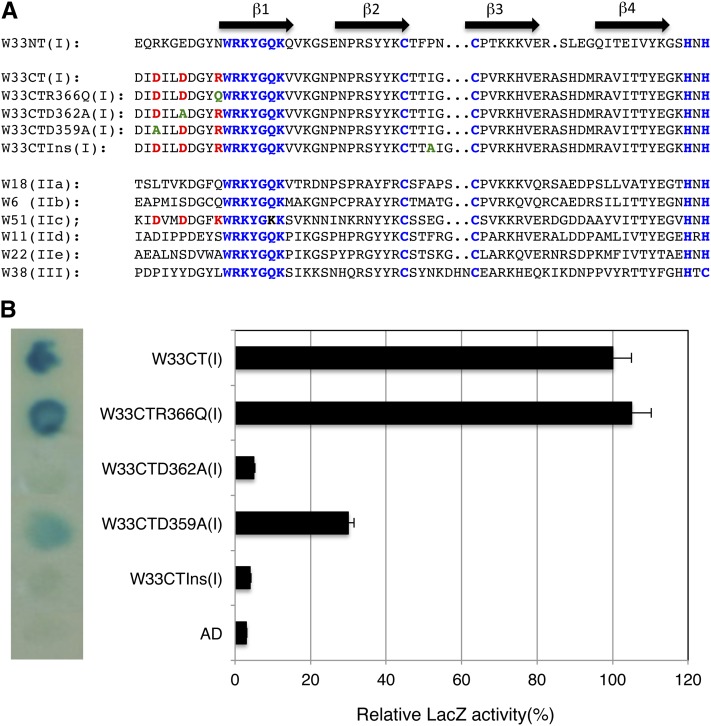
Assays of the 34 Arabidopsis VQ proteins in yeast cells revealed a clear pattern: They interacted with the C-terminal DNA-binding WRKY domains of group I and the sole WRKY domains of group IIc WRKY proteins, but not with the N-terminal WRKY domain of group I or the WRKY domain of group IIa, IIb, IId, IIe, or III WRKY proteins (Table II; Fig. 3; Supplemental Fig. S1). The consensus sequences of WRKY domains for each WRKY group in higher plants have been generated (Rushton et al., 2010). Comparison of the consensus sequences revealed that a basic amino acid (Arg or Lys) residue in the region immediately preceding the WRKYGQK signature sequence is unique to the C-terminal WRKY domain of group I WRKY proteins and the sole WRKY domain of group IIc WRKY proteins (Rushton et al., 2010; Fig. 4A). There are also two Asp residues in the region preceding the WRKYGQK signature sequence in the C-terminal WRKY domain of group I WRKY proteins and the sole WRKY domain of group IIc and III WRKY proteins (Rushton et al., 2010). For the C-terminal WRKY domain of WRKY33 (W33CT), these three amino acid residues correspond to Asp-359 (D359), Asp-362 (D362), and Arg-366 (R366; Fig. 4A). To test their role in interaction with VQ proteins, we changed Arg-366, which immediately precedes the WRKYGQK signature sequence of W33CT, to a Gln residue (W33CTR366Q), a consensus residue at the same position in group IIa and IIb WRKY proteins (Fig. 4A). We also changed Asp-359 and Asp-362 of W33CT to an Ala residue to generate the W33CTD359A and W33CTD362A mutant WRKY domains, respectively (Fig. 4A). The mutant C-terminal WRKY domains of WRKY33 were then tested for interacting with VQ10. As shown in Figure 4B, W33CTR366Q interacted with the VQ protein as strongly as wild-type C-terminal WRKY domain of WRKY33 (W33CT), indicating that the basic amino acid residue immediately preceding the WRKYGQK sequence is not critical for interaction with the VQ protein. By contrast, the W33CTD362A mutant WRKY domain failed to interact with the same VQ protein based on the β-galactosidase activity assays (Fig. 4B). W33CTD359A interacted positively but weakly with VQ10, based on the relatively low β-galactosidase activities (Fig. 4B). Thus, Asp-362 is a critical residue for interaction with VQ proteins.
Figure 4.
Identification of critical amino acid residues of C-terminal WRKY domain of WRKY33 for interaction with VQ10. A, Sequence comparison of WRKY domains of WRKY33 (W33), WRKY18 (W18), WRKY6 (W6), WRKY51 (W51), WRKY11 (W11), WRKY22 (W22), WRKY38 (W38), which belong to group I, IIa, IIb, IIc, IId, IIe, and III WRKY proteins, respectively. Mutant C-terminal WRKY domains of WRKY33 (W33CT) with D395A, D362A, or R366Q substitution or with an Ala inserted between the two zinc-finger Cys residues (W33CTIns) are also shown. The WRKYGQK sequences and the Cys (C) and His (H) residues involved in zinc coordination are indicated in blue. The two Asp (D) residues and one basic residue in the region preceding the WRKYGQK sequence shared by the W33CT and W51 WRKY domains are in red. Resulted amino acid residues in the mutant W33CT proteins are in green. B, Yeast two-hybrid assays of interactions of VQ10 with wild-type and mutant forms of the C-terminal WRKY domains of WRKY33. pAD-VQ10 fusion vector was cotransformed with various pBD-W33CT constructs into yeast cells. Yeast transformants were analyzed for the LacZ reporter gene expression through assays of β-galactosidase activity on membrane using 5-bromo-4-chloro-3-indoly1-β-d-galactopyranoside (left section) or o-nitrophenyl-β-d-galactopyranose (right section) as substrate.
Comparison of the consensus sequences also revealed that there are only four amino acid residues between the two conserved Cys residues (Cx4C) in the zinc-finger structure of the C- and N-terminal WRKY domains of group I WRKY proteins and the WRKY domains of group IIc WRKY domains (Rushton et al., 2010; Fig. 4A). On the other hand, there are five or seven residues between the conserved Cys residues in the zinc-finger structure of the WRKY domains of the other five groups of WRKY proteins (group IIa, IIb, IId, IIe, and III; Rushton et al., 2010; Fig. 4A). We inserted an Ala residue in the middle of the four residues between the conserved Cys residues in the zinc-finger structure of the C-terminal WRKY domain of WRKY33 (W33CTIns), as found in group IIa and IIb WRKY proteins (Fig. 4A). As shown in Figure 4B, the W33CTIns mutant WRKY domain failed to interact with VQ10 based on the β-galactosidase activity assays. This result indicated that the number of amino acid residues between the two conserved Cys residues involved in zinc coordination is also critical for interaction with VQ proteins.
Quantitative Real-Time-PCR Analysis of VQ Genes Expression Profiles
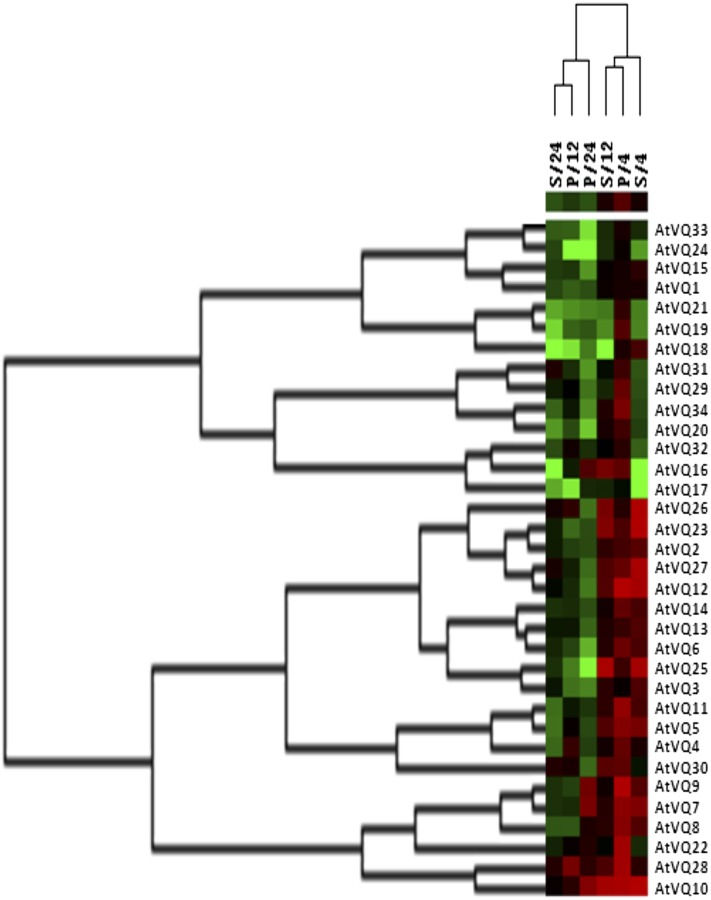
We have previously analyzed the expression profile of the Arabidopsis WRKY gene family and showed that almost 70% of the WRKY genes in Arabidopsis were differentially regulated, mostly up-regulated, in response to pathogen infection or SA treatment (Dong et al., 2003). This observation strongly suggested that a major function of the WRKY gene superfamily is the regulation of genes associated with plant defense and stress responses. To examine their roles in plant defense, we also analyzed the expression levels of the 34 VQ genes from Arabidopsis. Arabidopsis plants were inoculated with a virulent strain of Pseudomonas syringae or treated with SA (1 mm). Leaf samples were collected at 0, 4, 12, and 24 h post pathogen inoculation (hpi) or SA treatment and the transcript levels of the VQ genes were quantified using quantitative real-time (qRT)-PCR. Cluster analysis of the expression profiles divided the VQ genes into two groups (Fig. 5). In the first group, the levels of transcript of the VQ genes including VQ19 and VQ21 were overall reduced after SA treatment and at later hours after pathogen infection (12 and 24 hpi). In the other group, the transcript levels were up-regulated at 4 and 12 h post SA treatment (Fig. 4). Many of these SA-induced VQ genes were also up-regulated at earlier hpi (4 hpi). At later hours, particularly at 24 h post pathogen infection or SA treatment, the transcript levels of many of the induced VQ genes were not further increased and some of them even declined (Fig. 5). Thus, induction of many of the VQ genes by pathogen infection and SA was rapid but transient. Generally speaking, the transcript levels of Arabidopsis VQ genes changed quite dynamically upon pathogen infection or SA treatment.
Figure 5.
Cluster analysis of the expression profiles of Arabidopsis VQ genes. VQ gene expression was determined by qRT-PCR. VQ genes are represented as rows and treatment/time point as columns in the matrix. Red, black, and green elements indicate up-regulated, no change, and down-regulated VQ genes, respectively, in the matrix. Cluster analysis was performed on expression profiles of 34 VQ genes across six transients treatments (P, P. syringae infection; S, SA treatment) and time points (4, 12, and 24 h after pathogen inoculation or SA treatment). The horizontal and vertical dendrograms indicate the degree of similarity between expression profiles for VQ genes and conditions tested, respectively.
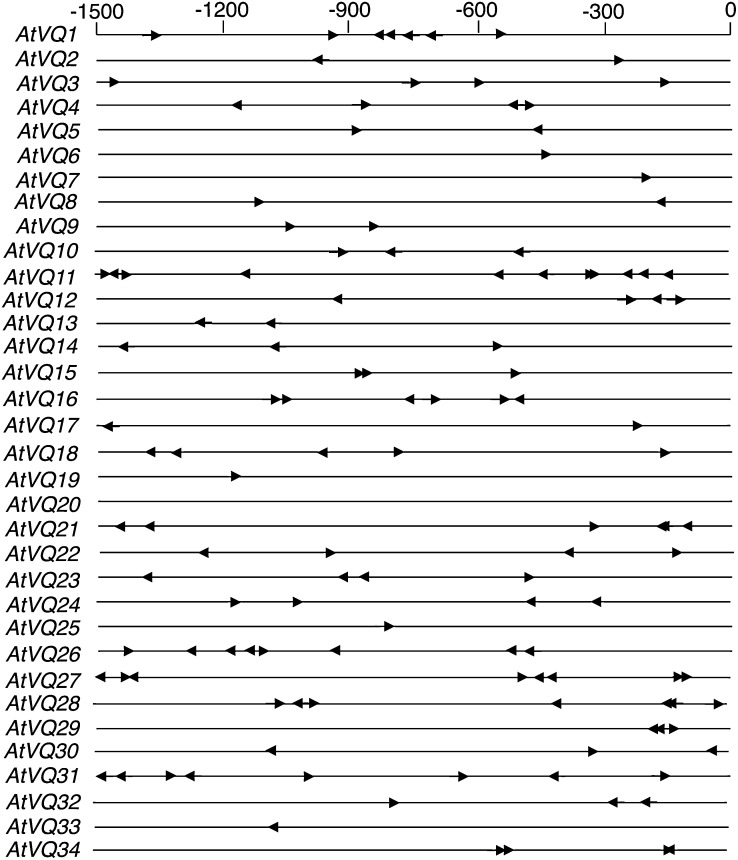
Analysis of VQ Gene Promoters from Arabidopsis
We have previously shown that the average frequencies of the W boxes in pathogen- and SA-regulated WRKY gene promoters from Arabidopsis are substantially higher than the statistically expected frequencies (Dong et al., 2003). The overrepresentation of the W boxes suggests extensive self and cross regulation among members of the WRKY gene family. To examine possible regulation of Arabidopsis VQ genes by WRKY transcription factors, we inspected 1.5-kb sequences upstream of the predicted translational start sites of the Arabidopsis VQ genes for the core W-box sequence (TTGAC). The average frequencies of the W boxes in the 1.5-kb promoters of the 34 VQ genes from Arabidopsis are 3.8 (Fig. 6), which is higher than the statistically expected frequency of 3.04. Among the 34 VQ genes, 16 (47%) contain four or more copies of the TTGAC pentamer sequence in their promoters (Fig. 6). VQ11 contains the highest number of WRKY-binding sites (11 copies of TTGAC), whereas VQ20 does not contain even a single WRKY-binding site in its promoter (Fig. 6). However, neither the induction/repression by pathogen or SA, nor the degree of either response, appeared to correlate well with the number of W boxes in the VQ gene promoters. A similar trend has been previously found with Arabidopsis WRKY genes (Dong et al., 2003).
Figure 6.
W-box elements in the promoter of Arabidopsis VQ genes. Numbering is from predicted translation start codons.
Functional Analysis of VQ Genes from Arabidopsis
To analyze directly the biological functions of Arabidopsis VQ genes, we first sought a loss-of-function approach through characterization of T-DNA or transposon insertion mutants. Through screening of insertion lines obtained from various stock centers, we isolated T-DNA or transposon insertion mutants for eight new VQ genes (VQ4, VQ5, VQ7, VQ8, VQ10, VQ20, VQ24, and VQ32). Mutants for four additional VQ genes have been previously reported and characterized (Andreasson et al., 2005; Wang et al., 2010a; Lai et al., 2011). Thus, T-DNA or transposon insertion mutants exist only for less than 50% of VQ genes from Arabidopsis. This low frequency is likely due to the small gene sizes of VQ genes. For the mutants of the eight VQ genes, we examined their overall phenotypes in growth, development, and resistance to a virulent strain of the bacterial pathogen P. syringae and the necrotrophic fungal pathogen Botrytis cinerea. Except for the mutant for VQ8, we observed no major alteration in growth, development, or disease resistance in the mutants for the VQ genes.
VQ8 encodes a VQ protein with an N-terminal signal peptide predicted to be chloroplast targeting (Table I). A transposon-tagged line was identified and verified by PCR using primers flanking the insertion site in the gene (Fig. 7, A and B; Kuromori et al., 2006). The vq8-1 mutant displayed pale-green and stunted-growth phenotypes throughout the entire life cycle (Fig. 7C). As expected for a loss-of-function mutant, the F1 progeny of the cross between vq8-1 and No-0 wild-type plants were normal in growth and development. Furthermore, survey of F2 progeny showed that the mutant phenotypes were observed only in vq8-1/vq8-1 homozygous mutant plants, indicating that the vq8-1 mutant is a recessive loss-of-function allele (Fig. 7B).
Figure 7.
Identification and characterization of Arabidopsis vq8-1 mutant. A, The structure of the intronless VQ8 gene. The transposon insertion site of the vq8-1 mutant is indicated. P1 and P2 are the two primers used for identification of homozygous vq8-1 mutant plants. B, Genetic analysis of vq8-1. Col-0 wild type (WT) was crossed with the vq8-1 and the F2 progeny were screened for homozygous vq8 progeny and scored for the absence (−) or presence (+) of its mutant phenotypes. C, The phenotypes of the vq8-1 mutant. The picture was taken at 7 weeks after germination.
As a complementing approach for functional analysis of the VQ genes, we generated transgenic plants that constitutively overexpressed VQ genes. The coding sequences of the VQ genes were subcloned behind the cauliflower mosaic virus (CaMV) 35S promoter in a binary plant transformation vector and transformed into Arabidopsis plants. RNA blotting was used to identify F0 transformant plants with elevated levels of transcripts for the transformed VQ gene (see Supplemental Fig. S2). Those transgenic F1 lines with increased VQ gene expression were analyzed for alterations in growth, development, and disease resistance. Due to the large number of transgenic lines for almost 30 VQ genes, our analysis of the transgenic lines was primarily focused on highly altered phenotypes that could be readily detected and verified. Generally speaking, overexpression of a majority of the VQ genes had no major effects on growth or development in transgenic plants. However, grossly altered growth was observed in transgenic plants overexpressing some of the VQ genes. Specifically, transgenic plants with elevated transcript levels for VQ17, VQ18, or VQ22 were highly stunted in growth (Fig. 8A). Overexpression of VQ29 didn’t alter growth but substantially delayed the flowering time of the transgenic plants (Fig. 8B).
Figure 1.
Growth and developmental phenotypes of transgenic VQ-overexpressing Arabidopsis plants. A, Stunted growth of transgenic plants overexpressing VQ17, VQ18, or VQ22. The picture of Col-0 wild type (WT) and two lines (L) of transgenic overexpression plants for each VQ gene was taken 7 weeks after germination. B, Delayed flowering of transgenic plants overexpressing VQ29. The picture of Col-0 wild type (WT) and two lines (L) of transgenic VQ29 overexpression plants was taken 10 weeks after germination. The RNA-blotting analysis of the overexpressed VQ genes in the transgenic lines is shown in Supplemental Figure S2S.
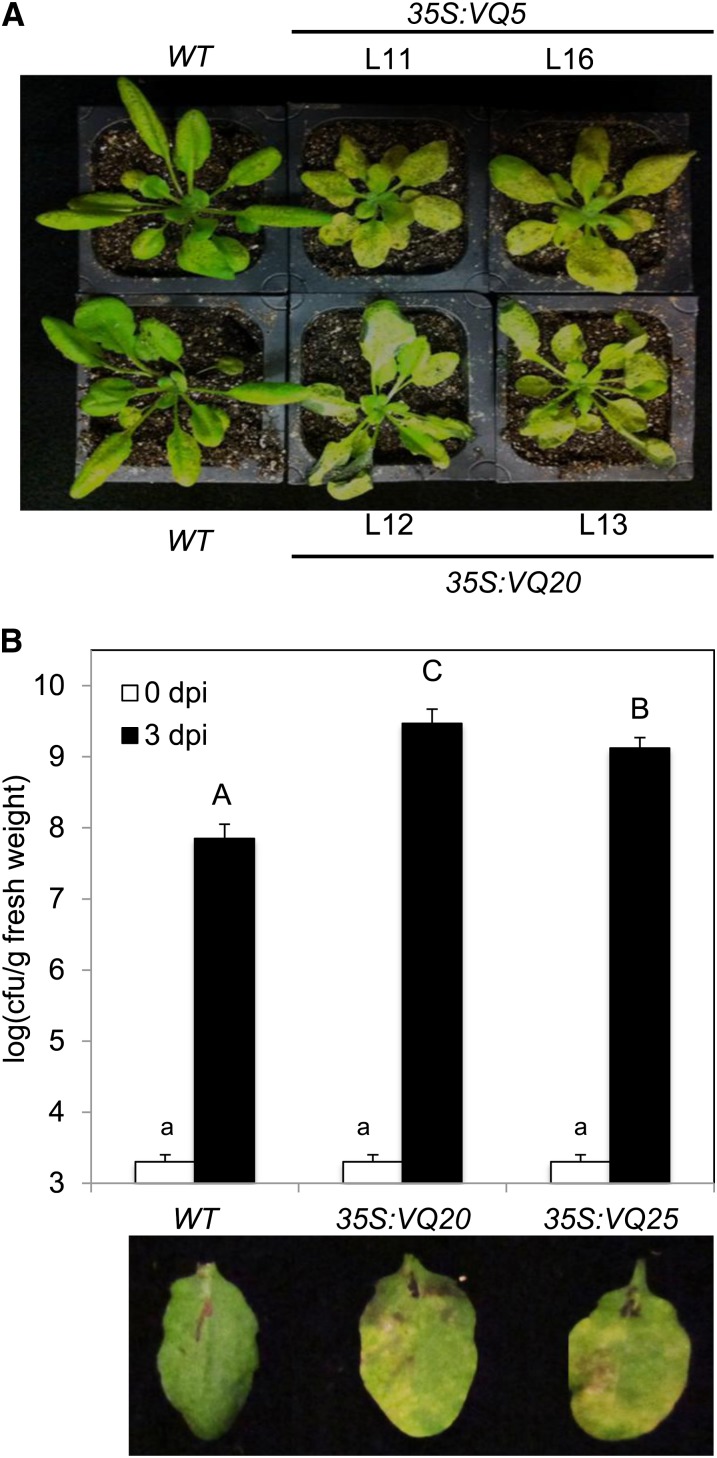
We also examined the responses of the VQ overexpression lines to P. syringae and B. cinerea and found relatively minor alteration in responses to the pathogens for a number of VQ genes. Overexpression of three VQ genes, however, had major effects on plant resistance to the two pathogens. Transgenic plants overexpressing VQ5 displayed increased chlorosis after infection of B. cinerea (Fig. 9A), indicating enhanced susceptibility to the necrotrophic fungal pathogen. Transgenic plants overexpressing VQ20 displayed enhanced disease symptoms after infection of either B. cinerea or P. syringae (Fig. 9), suggesting that VQ20 is a negative regulator in plant defense responses. In addition, transgenic plants overexpressing VQ25 were also highly susceptible to P. syringae but were normal in resistance to B. cinerea (Fig. 9).
Figure 9.
Altered disease resistance of transgenic VQ-overexpressing Arabidopsis plants. A, Enhanced susceptibility to B. cinerea. Col-0 wild type (WT) and two independent lines (L) of transgenic plants overexpressing VQ5 or VQ20 were spray inoculated with Botrytis and the picture was taken at the third d post inoculation (dpi). B, Enhanced susceptibility to P. syringae. Col-0 wild type (WT) and transgenic plants overexpressing VQ20 (lines 12 and 13) or VQ25 (lines 7 and 8) were inoculated with a virulent strain of P. syringae pv tomato DC3000 (OD600= 0.0002 in 10 mm MgCl2). Samples were taken at 0 and 3 dpi to determine the growth of the bacterial pathogen (top section). According to Duncan’s multiple range test (P= 0.01), means of colony-forming units (cfu) do not differ significantly at 0 dpi if they are indicated with the same lowercase letter and do not differ significantly at 2 dpi if they are indicated with the same uppercase letter. Images from representative inoculated leaves taken at 3 dpi are also shown (bottom section). The RNA-blotting analysis of the overexpressed VQ genes in the transgenic lines is shown in Supplemental Figure S2S.
Functional Interactions of Interacting WRKY and VQ Proteins
VQ proteins interact physically with group I and IIc WRKY proteins and may regulate DNA binding and other molecular activities of the two groups of WRKY transcription factors. To determine possible functional interaction between VQ proteins and their interacting WRKY partners, we compared the phenotypes of transgenic plants overexpressing a single VQ or an interacting WRKY gene with those of transgenic plants cooverexpressing both the interacting VQ and WRKY genes. For this purpose, we chose genes encoding VQ10 and the WRKY25, WRKY26, and WRKY33 interacting partners. These genes were selected for analysis of functional interactions because they were among the earliest identified interacting partners during our study of VQ proteins (Table II). As shown in Figure 10, transgenic plants overexpressing VQ10, WRKY25, WRKY26, or WRKY33 were largely normal in growth as judged from the sizes of plants at mature stages. However, when a VQ10-overxpressing line was crossed with lines overexpressing the three WRKY genes, the F1 progeny were substantially altered in growth. Specifically, those F1 progeny cooverexpressing VQ10 and either WRKY25 or WRKY33 were greatly stunted in growth as indicated by the drastically reduced plant sizes at mature stages (Fig. 10). The transgenic plants cooverexpressing VQ10 and WRKY26, on the other hand, were not significantly reduced in sizes at mature stage when compared to those expressing either VQ10 or WRKY26 (Fig. 10). However, the leaves of the transgenic plants coexpressing the interacting VQ10 and WRKY26 genes were substantially darker and rounder than those of wild-type and transgenic plants overexpressing a single VQ10 or WRKY26 gene (Fig. 10).
Figure 10.
Enhanced phenotypes in transgenic plants cooverexpressing interacting VQ10 and WRKY genes. Transgenic plants overexpressing VQ10 (line 1) were crossed with transgenic plants overexpressing WRKY25 (line 1), WRKY26 (line 1), or WRKY33 (line 1). The F1 progeny of the crossing were compared with Col-0 and their parental lines for growth alteration. The images for the plants were taken 6 weeks after germination.
DISCUSSION
As a large protein superfamily, WRKY transcription factors are diverse in both structures and functions. As transcription factors, the diverse biological functions of WRKY proteins are due to their ability in regulating expression of a wide range of target genes through coordination with other DNA-binding or non-DNA-binding interacting proteins. Four VQ motif-containing proteins from Arabidopsis have been previously reported to interact with WRKY proteins and play critical roles in the biological processes in which their respective interacting WRKY proteins are involved (Andreasson et al., 2005; Qiu et al., 2008; Wang et al., 2010a; Xie et al., 2010; Lai et al., 2011). In this study, we expanded the study to the entire family of 34 VQ proteins in Arabidopsis and provided new important insights into the structures, interaction partnership with WRKY proteins, and biological functions of the new protein family.
VQ Proteins as Group I and IIc WRKY-Interacting Proteins
We have recently shown that Arabidopsis VQ proteins SIB1 (VQ23), SIB2 (VQ16), and MKS1 (VQ21) specifically recognize the C-terminal WRKY domain and stimulate the DNA-binding activity of WRKY33 (Lai et al., 2011). We have further revealed that the conserved V and Q residues in the short VQ motif of SIB1 are important for its physical interactions with WRKY33 and proposed that the short VQ motif is the core of the WRKY-interacting domain (Lai et al., 2011). To extend the analysis, we examined the entire family of 34 VQ proteins in Arabidopsis for interaction with WRKY proteins. Apart from the short VQ motif, there is relatively low sequence homology among the 34 Arabidopsis VQ proteins (Fig. 1). Despite the structural diversity, all 34 VQ proteins from Arabidopsis were capable of interacting with various WRKY proteins (Table II; Fig. 3). The WRKY proteins identified or assayed for interactions with the 34 VQ proteins included WRKY3, WRKY4, WRKY20, WRKY23, WRKY25, WRKY33, and WRKY34 (Table II; Fig. 3), which all belong to group I WRKY proteins with two WRKY domains (Eulgem et al., 2000). The interactions of these group I WRKY proteins with VQ proteins are likely through their C-terminal WRKY domains based on the inability of the N-terminal WRKY domain of WRKY33 to interact with VQ proteins (Lai et al., 2011; Supplemental Fig. S1). Additional WRKY proteins identified to interact with VQ proteins include WRKY10, WRKY24, WRKY51, and WRKY75, which belong to group IIc WRKY proteins (Table II; Fig. 3). By contrast, the WRKY domains from group IIa (WRKY18), IIb (WRKY6), IId (WRKY11), IIe (WRKY22), and III (WRKY38 and WRKY70) failed to interact with VQ proteins (Fig. 3; Supplemental Fig. S1). Thus, VQ proteins appeared to interact only with group I and IIc WRKY transcription factors.
Using WRKY33 as a model, we have identified structural features unique to the two groups of WRKY proteins that are critical for their interaction with VQ proteins. These structural features include the presence of four amino acid residues between the two conserved Cys residues involved in zinc coordination and two charged amino acid residues in the region that precede the WRKYGQK signature sequence (D359 and D362 for WRKY33). However, the N-terminal WRKY domains of group I also contain these similar structural features but failed to interact with VQ proteins (Supplemental Fig. S1). The N-terminal WRKY domains of group I WRKY proteins also have very low DNA-binding activity and, therefore, may lack certain unknown structural features necessary for strong DNA binding and interaction with VQ proteins.
According to the structure solved by NMR, the C-terminal WRKY domain of WRKY4 is an antiparallel β-sheet forming from four β-strands (Yamasaki et al., 2012). For the longer C-terminal WRKY domain of WRKY4 or WRKY1 with more residues at the N terminus, there is actually another β-strand that is linked through a long bridging loop with the β-strand in which the WRKYGQK sequence is located (Duan et al., 2007). Asp-262 of the C-terminal WRKY domain of WRKY33 would be expected to be located in the C-terminal end of the long bridging loop (Duan et al., 2007). Asp-262 of WRKY33 is also adjacent to the highly conserved Asp-263, which forms a salt bridge with the Trp residue in the invariant WRKYGQK sequence (Duan et al., 2007), which, in turn, is adjacently connected with the first Cys residue of the zinc-finger motif (Yamasaki et al., 2012). Therefore, Asp-262 and those residues located between the two Cys residues involved in zinc coordination, although well separated in the primary sequence, are in close proximity at one of the ends of the β-sheet, which may act as the interface for interaction with the small VQ motif. The recently solved solution structure of the C-terminal DNA-binding WRKY domain of WRKY4 in complex with the W-box DNA has further revealed that the β-sheet of the WRKY domain enters the major groove of DNA, nearly perpendicular to the DNA helical axis (Yamasaki et al., 2012). Residues located in the β-strand containing the conserved WRKYGQK motif are in direct contact with DNA bases (Yamasaki et al., 2012). Therefore, VQ proteins could potentially regulate the DNA binding of group I and IIc WRKY proteins by specifically recognizing the part of the β-sheets of their DNA-binding WRKY domains that directly contacts DNA molecules.
Biological Functions of VQ Genes
Due to the relatively large number of genes and the lack of knockout mutants for many of the gene family members, our functional analysis of the VQ gene family was by no means comprehensive but did provide a glimpse into the potential biological functions of the gene family in plant growth, development, and defense responses. We showed that overexpressing of some VQ genes alone or in combination with their interacting WRKY genes, led to altered phenotypes in growth and flowering (Figs. 8 and 10). Because overexpression could lead to physiologically irrelevant phenotypes, the inferred biological functions in plant growth and development should been considered with caution. On the other hand, WRKY genes with important roles in growth and development including trichome and seed development have been identified (Johnson et al., 2002; Wang et al., 2010a). In addition, many WRKY and VQ genes are highly responsive to pathogen infection and some interacting VQ and WRKY genes could be induced at sufficiently high levels locally, potentially to impact the expression of not only defense-related genes but also genes associated with growth and development. There is increasing body of evidence that plant growth is intrinsically linked with plant stress and defense responses (Kazan and Manners, 2009). Plant hormones including auxin and brassinosteroids, which are primarily associated with growth and development, have now been directly linked or implicated in plant defense responses to a variety of microbial pathogens (Remans et al., 2006; Stotz et al., 2011; Belkhadir et al., 2012). Plant VQ and their interacting WRKY partners could play roles in interplay of plant growth, development, and stress responses.
Using qRT-PCR we showed that many of the VQ genes from Arabidopsis were responsive to pathogen infection and SA treatment (Fig. 5), suggesting possible roles in plant defense. In Arabidopsis, SA-mediated defense is important for resistance to biotrophic pathogens such as P. syringae and Hyaloperonospora arabidopsidis, whereas jasmonic acid (JA) often mediates plant defense against necrotrophic pathogens such as B. cinerea (Glazebrook, 2005). A number of studies have shown that the SA and JA signaling pathways are mutually antagonistic and, as a result, many genes involved in plant defense often play opposite roles in resistance to the two distinct types of plant pathogens (Kunkel and Brooks, 2002). Overexpression of MKS1 led to increased expression of SA-regulated PR1 gene expression but compromised plant resistance to necrotrophic B. cinerea (Andreasson et al., 2005; Petersen et al., 2010). In addition, the mks1 mutants were compromised in resistance to biotrophic pathogens P. syringae and H. arabidopsidis (Petersen et al., 2010). These results indicated that MKS1 plays a positive role in SA-mediated defense against biotrophic pathogens but a negative role in JA-regulated defense against necrotrophic pathogens. On the other hand, the closely related SIB1 and SIB2 proteins act as coactivators of WRKY33 in JA-mediated plant defense against necrotrophic pathogens (Lai et al., 2011). Interestingly, overexpression of SIB1 and SIB2 also increased resistance to hemibiotrophic pathogen P. syringae (Xie et al., 2010). By contrast, overexpression of VQ20 compromised resistance to both biotrophic and necrotrophic pathogens (Fig. 9). Thus, the impacts of mutations or overexpression of VQ genes on plant resistance to biotrophic pathogens are not always opposite to those on resistance to necrotrophic pathogens. Therefore, VQ genes have complex roles in plant defense responses, which could, in part, be due to their ability to interact with multiple WRKY proteins.
The Modes of Action of VQ Proteins
Out of the 72 members of the WRKY gene superfamily in Arabidopsis, there are a total of 32 group I and IIc WRKY genes (13 group I and 19 group IIc). This number is similar to the 34 VQ genes in the plant. Despite the similar numbers, the interaction partnership between the WRKY and VQ proteins is not highly specific. A single VQ protein interacted with multiple WRKY proteins and different VQ proteins had partially overlapping pools of interacting WRKY partners, which raised the question of how VQ genes achieve their functional specificity. In this regard, it is useful to consider several factors that could influence the formation and properties of the WRKY/VQ protein complexes in plant cells. First, members within the VQ and WRKY gene families differ in their spatial and temporal expression patterns. Because only coexpressed WRKY and VQ proteins can interact in vivo, the number of major WRKY/VQ complexes for a specific VQ protein in a given type of cell or tissue under a given condition may not be as large as indicated from the yeast two-hybrid assays. Identification of the major WRKY interaction partners for a VQ protein in the physiological context would, therefore, be key to understanding the biological functions of the VQ gene. Second, apart from the highly conserved short VQ motif, VQ proteins are highly divergent in flanking amino acid sequences, which could influence the specificity of interaction of a VQ protein with WRKY partners. Indeed, although a majority of the VQ proteins interacted with group I WRKY25 and WRKY33, only about half interacted with group IIc WRKY51 in yeast cells (Fig. 3). Thus, structurally divergent VQ proteins may be quite selective in interactions with group II WRKY proteins. Moreover, as VQ proteins appear to recognize the part of the WRKY domains that contacts DNA, they could differ substantially in their effects on DNA binding of their interacting WRKY partners. Unlike SIB1 and SIB2, which stimulate the DNA-binding activity of WRKY33, other WRKY33-interacting VQ proteins might inhibit DNA binding of WRKY33, thereby antagonizing the positive roles of WRKY33, SIB1, and SIB2 in plant defense against necrotrophic pathogens. Another possibility is the potential varying effects of different VQ proteins on the DNA-binding specificity of their interacting WRKY partners, particularly to the nucleotides neighboring the conserved W boxes, which may play a critical role in determining target gene specificity of WRKY transcription factors (Ciolkowski et al., 2008). Third, there are other proteins besides WRKY proteins that interact with VQ proteins (Table I), some of which such as MPK3, MPK4, and CaM1 are regulatory proteins in signal transduction. MPK4 interacts with VQ21 (MKS1) and regulates the activity of the VQ21/WRKY33 complex (Qiu et al., 2008). Likewise, interaction of VQ9 and VQ15 with MPK3 and CaM1, respectively, might also alter the interactions of the VQ proteins with WRKY proteins and, as a consequence, WRKY-regulated gene expression.
Both SIB1 and SIB2 are targeted to chloroplasts and interact with a σ factor of chloroplast RNA polymerase (Morikawa et al., 2002). Interestingly, a large percentage of the VQ proteins from Arabidopsis contain chloroplast- or mitochondria-targeting signal peptides at their N terminus (Table I), raising the possibility that many of the VQ proteins might function in the two organelles. The vq8 mutant plants were pale green (Fig. 8), likely due to defects in chloroplast development. Genetic and molecular analyses have identified genes that encode chloroplast (plastid)-localized proteins involved in the synthesis and signaling of plant defense signal molecules, such as SA and JA. Chloroplasts are also a major source of reactive oxygen species and redox-active soluble molecules that affect the expression of both chloroplast and nuclear genes associated with defense responses. In addition, it is known that plant disease resistance is strongly influenced by environmental factors such as light (Chandra-Shekara et al., 2006; Griebel and Zeier, 2008). The fact that many members of a WRKY-interacting protein family are targeted to chloroplasts or mitochondria raised the possibility that WRKY/VQ-regulated gene expression may be coordinated with certain processes in the two organelles during plant defense and stress responses.
In summary, we found that structurally divergent VQ proteins from Arabidopsis were all capable of forming complexes with group I and IIc WRKY proteins through physical interaction of conserved VQ and WRKY domains. Several highly conserved amino acid residues located at the end of the β-sheets of the DNA-binding domains of the two groups of WRKY proteins with direct contacts with DNA are critical for interaction with VQ proteins. VQ genes exhibited dynamic expression patterns in response to pathogen infection and SA treatment. Functional analysis of the VQ genes indicated that they play important roles in plant growth, development, and defense responses. These results provided strong evidence that plant VQ proteins have evolved primarily as cofactors of WRKY proteins and play critical roles in the network of WRKY-mediated gene expression.
MATERIALS AND METHODS
Plant Growth Conditions
Arabidopsis (Arabidopsis thaliana) plants were grown in growth chambers or rooms at 22°C, 120 μE m−2 light with 12 h of light/12 h of darkness.
Yeast Two-Hybrid Assays
Interactions between WRKY domains and VQ proteins were assayed using Gal4-based yeast (Saccharomyces cerevisiae) two-hybrid system as described (Lai et al., 2011). pAD-Gal-VQ and pBD-Gal-WRKY fusion constructs were generated from PCR-amplified DNA fragments using the gene-specific primers listed in Supplemental Tables S1 and S2, respectively. The prey and bait plasmids were transformed to yeast strain YRG-2. After overnight culture, the transformants were plated on membrane and assayed for β-galactosidase activity using 5-bromo-4-chloro-3-indoly1-β-d-galactopyranoside as substrate. Quantitative assays of β-galactosidase activity of yeast transformants were performed using o-nitrophenyl-β-d-galactopyranose as substrate.
Site-Directed Mutagenesis
Changes of amino acid residues in the region preceding the WRKYGQK sequence of the C-terminal WRKY domain of WRKY33 (W33CT) were achieved through site-directed mutagenesis using either the QuikChange kit from Agilent or overlapping PCR with the following primers: W33CTD359A (gtagtgcagacaacgagtgatattgccattct), W33CTD362A (gtagtgcagacaacgagtgatattgccattct), and W33CTR366Q (cttgacgacggttaccaatggagaaaatacggccaga). Insertion of an Ala residue in the region between the two Cys residues in the zinc finger of the C-terminal WRKY domain of WRKY33 (W33CTIns) was performed using the QuikChange kit (primer: actacaagtgcacaaccgctatcggttgtccagtgagga).
qRT-PCR
Total RNA was isolated from 5- to 6-week-old pathogen-infected or SA-treated Arabidopsis Columbia-0 (Col-0) wild-type plants using the Trizol reagent according to the supplier’s instruction. Extracted RNA was treated with DNase (remove contaminating DNA) and reverse transcribed using the SuperScript III first-strand synthesis system for reverse transcriptase-PCR (Invitrogen). qRT-PCR was performed with an iCycler iQ multicolor real-time PCR detection system (Bio-Rad). PCRs were performed using the SYBR Green PCR master mix (Applied Biosystems) and gene-specific primers listed in Supplemental Table S3. The PCR conditions consisted of denaturation at 95°C for 3 min, followed by 40 cycles of denaturation at 95°C for 30 s, annealing at 58°C for 30 s, and extension at 72°C for 30 s. Relative gene expression was calculated as previously described (Livak and Schmittgen, 2001). The Arabidopsis ACT2 gene was used as internal control as previously described (Huang et al., 2010).
Expression patterns of the 34 VQ genes were used for cluster analysis. To determine the degree of regulation of each VQ gene after pathogen infection or SA treatment, we calculated the induction/repression fold by determining the relative expression levels compared to those in uninfected/untreated plants. The degree-of-regulation data across six transients (three time points × two treatments) produced a matrix from which a gene expression similarity metric (the Peterson correlation coefficient) was calculated using the implementation as previously reported (Eisen et al., 1998). The similarity metrics were analyzed to determine clusters of the VQ genes with similar expression profiles. The cluster analysis was performed using the software tools written by Eisen (Eisen et al., 1998; http://rana.lbl.gov/EisenSoftware.htm).
Identification of T-DNA/Transposon Insertion Mutants
T-DNA/transposon insertion mutants for the following eight new VQ genes were identified: VQ4 (Sail_589_A07), VQ5 (GABI_063E09), VQ7 (Salk_099709), VQ8 (Pst20793), VQ10 (SK6114), VQ20 (Salk_002300), VQ24 (Salk_151487), and VQ32 (Salk_035635). PCR for identification of homozygous mutants were performed using gene-specific primers listed in Supplemental Table S1.
Generation of Transgenic VQ and WRKY Lines
For generating transgenic VQ overexpression lines, the full-length coding sequences for VQ genes were first excised from the corresponding pAD-Gal-VQ recombinant vectors using appropriate restriction enzymes and inserted behind the CaMV 35S promoter in the plant transformation vector PFGC5941. The resulted plasmids were transformed into Col-0 wild-type plants using the Agrobacterium-mediated floral-dip procedure (Clough and Bent, 1998). Transformants were identified for resistance to Basta. Transgenic plants overexpressing the transformed VQ transgene were identified by northern blotting (Supplemental Fig. S2).
For generating transgenic WRKY overexpression lines, WRKY25, WRKY26, and WRKY33 full-length cDNAs were inserted behind CaMV 35S promoter in the pOCA30 plant transformation vector and transformed into Arabidopsis Col-0 plants. Transformants were identified for resistance to kanamycin and transgenic lines overexpressing the transformed WRKY genes were identified by northern blotting (Supplemental Fig. S2).
RNA Gel-Blot Analysis
Total RNA was isolated from leaves and separated on 1.2% agarose-formaldehyde gels and blotted to nylon membranes according to standard procedures. Blots were hybridized with 32P-dATP-labeled gene-specific probes. Hybridization was performed in PerfectHyb plus hybridization buffer (Sigma) overnight at 68°C. The membrane was then washed for 15 min twice with 2× SSC (1× SSC is 0.15 m NaCl and 0.015 m sodium citrate) and 1% SDS and for 10 min with 0.1× SSC and 1% SDS at 68°C.
Pathogen Inoculation and SA Treatment
The bacterial strain Pseudomonas syringae DC3000 were grown overnight in King’s B medium containing rifampicin (50 μg/mL) and kanamycin (25 μg/mL). The bacterial cells were harvested and suspended in 10 mm MgCl2. The cells were then diluted to OD600= 0.0001 and infiltrated into the abaxial surface of the leaves. Growth and spray inoculation of Botrytis cinerea were performed as previously described (Zheng et al., 2006).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Interaction of Arabidopsis VQ5, VQ7, and VQ10 with WRKY proteins in yeast cells.
Supplemental Figure S2. VQ and WRKY gene overexpression in transgenic plants.
Supplemental Table S1. Primers for pAD-Gal fusion and overexpression constructs.
Supplemental Table S2. Primers for pBD-Gal fusion constructs.
Supplemental Table S3. Primers for quantitative reverse transcriptase-PCR.
Supplementary Material
Acknowledgments
We thank the Arabidopsis Resource Center at Ohio State University, the Nottingham Arabidopsis Stock Centre, and RIKEN Bioresource Center for providing the Arabidopsis mutants.
Glossary
- hpi
h post pathogen inoculation
- qRT
quantitative real-time
- CaMV
cauliflower mosaic virus
- JA
jasmonic acid
- Col-0
Columbia-0
References
- Andreasson E, Jenkins T, Brodersen P, Thorgrimsen S, Petersen NH, Zhu S, Qiu JL, Micheelsen P, Rocher A, Petersen M, et al. (2005) The MAP kinase substrate MKS1 is a regulator of plant defense responses. EMBO J 24: 2579–2589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Interactome Mapping Consortium (2011) Evidence for network evolution in an Arabidopsis interactome map. Science 333: 601–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai T, Tena G, Plotnikova J, Willmann MR, Chiu WL, Gomez-Gomez L, Boller T, Ausubel FM, Sheen J. (2002) MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415: 977–983 [DOI] [PubMed] [Google Scholar]
- Belkhadir Y, Jaillais Y, Epple P, Balsemão-Pires E, Dangl JL, Chory J. (2012) Brassinosteroids modulate the efficiency of plant immune responses to microbe-associated molecular patterns. Proc Natl Acad Sci USA 109: 297–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra-Shekara AC, Gupte M, Navarre D, Raina S, Raina R, Klessig D, Kachroo P. (2006) Light-dependent hypersensitive response and resistance signaling against Turnip Crinkle Virus in Arabidopsis. Plant J 45: 320–334 [DOI] [PubMed] [Google Scholar]
- Chen C, Chen Z. (2000) Isolation and characterization of two pathogen- and salicylic acid-induced genes encoding WRKY DNA-binding proteins from tobacco. Plant Mol Biol 42: 387–396 [DOI] [PubMed] [Google Scholar]
- Chen H, Lai Z, Shi J, Xiao Y, Chen Z, Xu X. (2010) Roles of Arabidopsis WRKY18, WRKY40 and WRKY60 transcription factors in plant responses to abscisic acid and abiotic stress. BMC Plant Biol 10: 281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciolkowski I, Wanke D, Birkenbihl RP, Somssich IE. (2008) Studies on DNA-binding selectivity of WRKY transcription factors lend structural clues into WRKY-domain function. Plant Mol Biol 68: 81–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cormack RS, Eulgem T, Rushton PJ, Köchner P, Hahlbrock K, Somssich IE. (2002) Leucine zipper-containing WRKY proteins widen the spectrum of immediate early elicitor-induced WRKY transcription factors in parsley. Biochim Biophys Acta 1576: 92–100 [DOI] [PubMed] [Google Scholar]
- de Pater S, Greco V, Pham K, Memelink J, Kijne J. (1996) Characterization of a zinc-dependent transcriptional activator from Arabidopsis. Nucleic Acids Res 24: 4624–4631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J, Chen C, Chen Z. (2003) Expression profiles of the Arabidopsis WRKY gene superfamily during plant defense response. Plant Mol Biol 51: 21–37 [DOI] [PubMed] [Google Scholar]
- Duan MR, Nan J, Liang YH, Mao P, Lu L, Li L, Wei C, Lai L, Li Y, Su XD. (2007) DNA binding mechanism revealed by high resolution crystal structure of Arabidopsis thaliana WRKY1 protein. Nucleic Acids Res 35: 1145–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. (1998) Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA 95: 14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulgem T, Rushton PJ, Robatzek S, Somssich IE. (2000) The WRKY superfamily of plant transcription factors. Trends Plant Sci 5: 199–206 [DOI] [PubMed] [Google Scholar]
- Glazebrook J. (2005) Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu Rev Phytopathol 43: 205–227 [DOI] [PubMed] [Google Scholar]
- Griebel T, Zeier J. (2008) Light regulation and daytime dependency of inducible plant defenses in Arabidopsis: phytochrome signaling controls systemic acquired resistance rather than local defense. Plant Physiol 147: 790–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Gu M, Lai Z, Fan B, Shi K, Zhou YH, Yu JQ, Chen Z. (2010) Functional analysis of the Arabidopsis PAL gene family in plant growth, development, and response to environmental stress. Plant Physiol 153: 1526–1538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiguro S, Nakamura K. (1994) Characterization of a cDNA encoding a novel DNA-binding protein, SPF1, that recognizes SP8 sequences in the 5′ upstream regions of genes coding for sporamin and beta-amylase from sweet potato. Mol Gen Genet 244: 563–571 [DOI] [PubMed] [Google Scholar]
- Jiang W, Yu D. (2009) Arabidopsis WRKY2 transcription factor mediates seed germination and postgermination arrest of development by abscisic acid. BMC Plant Biol 9: 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Deyholos MK. (2009) Functional characterization of Arabidopsis NaCl-inducible WRKY25 and WRKY33 transcription factors in abiotic stresses. Plant Mol Biol 69: 91–105 [DOI] [PubMed] [Google Scholar]
- Johnson CS, Kolevski B, Smyth DR. (2002) TRANSPARENT TESTA GLABRA2, a trichome and seed coat development gene of Arabidopsis, encodes a WRKY transcription factor. Plant Cell 14: 1359–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Journot-Catalino N, Somssich IE, Roby D, Kroj T. (2006) The transcription factors WRKY11 and WRKY17 act as negative regulators of basal resistance in Arabidopsis thaliana. Plant Cell 18: 3289–3302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazan K, Manners JM. (2009) Linking development to defense: auxin in plant-pathogen interactions. Trends Plant Sci 14: 373–382 [DOI] [PubMed] [Google Scholar]
- Kim KC, Fan B, Chen Z. (2006) Pathogen-induced Arabidopsis WRKY7 is a transcriptional repressor and enhances plant susceptibility to Pseudomonas syringae. Plant Physiol 142: 1180–1192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KC, Lai Z, Fan B, Chen Z. (2008) Arabidopsis WRKY38 and WRKY62 transcription factors interact with histone deacetylase 19 in basal defense. Plant Cell 20: 2357–2371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel BN, Brooks DM. (2002) Cross talk between signaling pathways in pathogen defense. Curr Opin Plant Biol 5: 325–331 [DOI] [PubMed] [Google Scholar]
- Kuromori T, Wada T, Kamiya A, Yuguchi M, Yokouchi T, Imura Y, Takabe H, Sakurai T, Akiyama K, Hirayama T, et al. (2006) A trial of phenome analysis using 4000 Ds-insertional mutants in gene-coding regions of Arabidopsis. Plant J 47: 640–651 [DOI] [PubMed] [Google Scholar]
- Lai Z, Li Y, Wang F, Cheng Y, Fan B, Yu JQ, Chen Z. (2011) Arabidopsis sigma factor binding proteins are activators of the WRKY33 transcription factor in plant defense. Plant Cell 23: 3824–3841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Z, Vinod KM, Zheng Z, Fan B, Chen Z. (2008) Roles of Arabidopsis WRKY3 and WRKY4 transcription factors in plant responses to pathogens. BMC Plant Biol 8: 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Brader G, Kariola T, Palva ET. (2006) WRKY70 modulates the selection of signaling pathways in plant defense. Plant J 46: 477–491 [DOI] [PubMed] [Google Scholar]
- Li J, Brader G, Palva ET. (2004) The WRKY70 transcription factor: a node of convergence for jasmonate-mediated and salicylate-mediated signals in plant defense. Plant Cell 16: 319–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Fu Q, Chen L, Huang W, Yu D. (2011) Arabidopsis thaliana WRKY25, WRKY26, and WRKY33 coordinate induction of plant thermotolerance. Planta 233: 1237–1252 [DOI] [PubMed] [Google Scholar]
- Li S, Fu Q, Huang W, Yu D. (2009) Functional analysis of an Arabidopsis transcription factor WRKY25 in heat stress. Plant Cell Rep 28: 683–693 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Luo M, Dennis ES, Berger F, Peacock WJ, Chaudhury A. (2005) MINISEED3 (MINI3), a WRKY family gene, and HAIKU2 (IKU2), a leucine-rich repeat (LRR) KINASE gene, are regulators of seed size in Arabidopsis. Proc Natl Acad Sci USA 102: 17531–17536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao G, Meng X, Liu Y, Zheng Z, Chen Z, Zhang S. (2011) Phosphorylation of a WRKY transcription factor by two pathogen-responsive MAPKs drives phytoalexin biosynthesis in Arabidopsis. Plant Cell 23: 1639–1653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Y, Laun T, Zimmermann P, Zentgraf U. (2004) Targets of the WRKY53 transcription factor and its role during leaf senescence in Arabidopsis. Plant Mol Biol 55: 853–867 [DOI] [PubMed] [Google Scholar]
- Morikawa K, Shiina T, Murakami S, Toyoshima Y. (2002) Novel nuclear-encoded proteins interacting with a plastid sigma factor, Sig1, in Arabidopsis thaliana. FEBS Lett 514: 300–304 [DOI] [PubMed] [Google Scholar]
- Mudgil Y, Uhrig JF, Zhou J, Temple B, Jiang K, Jones AM. (2009) Arabidopsis N-MYC DOWNREGULATED-LIKE1, a positive regulator of auxin transport in a G protein-mediated pathway. Plant Cell 21: 3591–3609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perruc E, Charpenteau M, Ramirez BC, Jauneau A, Galaud JP, Ranjeva R, Ranty B. (2004) A novel calmodulin-binding protein functions as a negative regulator of osmotic stress tolerance in Arabidopsis thaliana seedlings. Plant J 38: 410–420 [DOI] [PubMed] [Google Scholar]
- Petersen K, Qiu JL, Lütje J, Fiil BK, Hansen S, Mundy J, Petersen M. (2010) Arabidopsis MKS1 is involved in basal immunity and requires an intact N-terminal domain for proper function. PLoS ONE 5: e14364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitzschke A, Schikora A, Hirt H. (2009) MAPK cascade signalling networks in plant defence. Curr Opin Plant Biol 12: 421–426 [DOI] [PubMed] [Google Scholar]
- Qiu JL, Fiil BK, Petersen K, Nielsen HB, Botanga CJ, Thorgrimsen S, Palma K, Suarez-Rodriguez MC, Sandbech-Clausen S, Lichota J, et al. (2008) Arabidopsis MAP kinase 4 regulates gene expression through transcription factor release in the nucleus. EMBO J 27: 2214–2221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remans R, Spaepen S, Vanderleyden J. (2006) Auxin signaling in plant defense. Science 313: 171. [DOI] [PubMed] [Google Scholar]
- Robatzek S, Somssich IE. (2002) Targets of AtWRKY6 regulation during plant senescence and pathogen defense. Genes Dev 16: 1139–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushton PJ, Somssich IE, Ringler P, Shen QJ. (2010) WRKY transcription factors. Trends Plant Sci 15: 247–258 [DOI] [PubMed] [Google Scholar]
- Rushton PJ, Torres JT, Parniske M, Wernert P, Hahlbrock K, Somssich IE. (1996) Interaction of elicitor-induced DNA-binding proteins with elicitor response elements in the promoters of parsley PR1 genes. EMBO J 15: 5690–5700 [PMC free article] [PubMed] [Google Scholar]
- Shang Y, Yan L, Liu ZQ, Cao Z, Mei C, Xin Q, Wu FQ, Wang XF, Du SY, Jiang T, et al. (2010) The Mg-chelatase H subunit of Arabidopsis antagonizes a group of WRKY transcription repressors to relieve ABA-responsive genes of inhibition. Plant Cell 22: 1909–1935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stotz HU, Jikumaru Y, Shimada Y, Sasaki E, Stingl N, Mueller MJ, Kamiya Y. (2011) Jasmonate-dependent and COI1-independent defense responses against Sclerotinia sclerotiorum in Arabidopsis thaliana: auxin is part of COI1-independent defense signaling. Plant Cell Physiol 52: 1941–1956 [DOI] [PubMed] [Google Scholar]
- Suttipanta N, Pattanaik S, Kulshrestha M, Patra B, Singh SK, Yuan L. (2011) The transcription factor CrWRKY1 positively regulates the terpenoid indole alkaloid biosynthesis in Catharanthus roseus. Plant Physiol 157: 2081–2093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A, Garcia D, Zhang H, Feng K, Chaudhury A, Berger F, Peacock WJ, Dennis ES, Luo M. (2010a) The VQ motif protein IKU1 regulates endosperm growth and seed size in Arabidopsis. Plant J 63: 670–679 [DOI] [PubMed] [Google Scholar]
- Wang H, Avci U, Nakashima J, Hahn MG, Chen F, Dixon RA. (2010b) Mutation of WRKY transcription factors initiates pith secondary wall formation and increases stem biomass in dicotyledonous plants. Proc Natl Acad Sci USA 107: 22338–22343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Yang P, Fan B, Chen Z. (1998) An oligo selection procedure for identification of sequence-specific DNA-binding activities associated with the plant defence response. Plant J 16: 515–522 [DOI] [PubMed] [Google Scholar]
- Xie YD, Li W, Guo D, Dong J, Zhang Q, Fu Y, Ren D, Peng M, Xia Y. (2010) The Arabidopsis gene SIGMA FACTOR-BINDING PROTEIN 1 plays a role in the salicylate- and jasmonate-mediated defence responses. Plant Cell Environ 33: 828–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Chen C, Fan B, Chen Z. (2006) Physical and functional interactions between pathogen-induced Arabidopsis WRKY18, WRKY40, and WRKY60 transcription factors. Plant Cell 18: 1310–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki K, Kigawa T, Watanabe S, Inoue M, Yamasaki T, Seki M, Shinozaki K, Yokoyama S. (2012) Structural basis for sequence-specific DNA recognition by an Arabidopsis WRKY transcription factor. J Biol Chem 287: 7683–7691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wang L. (2005) The WRKY transcription factor superfamily: its origin in eukaryotes and expansion in plants. BMC Evol Biol 5: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z, Qamar SA, Chen Z, Mengiste T. (2006) Arabidopsis WRKY33 transcription factor is required for resistance to necrotrophic fungal pathogens. Plant J 48: 592–605 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.