Abstract
The plasma membrane H+-ATPase generates an electrochemical gradient of H+ across the plasma membrane that provides the driving force for solute transport and regulates pH homeostasis and membrane potential in plant cells. Recent studies have demonstrated that phosphorylation of the penultimate threonine in H+-ATPase and subsequent binding of a 14-3-3 protein is the major common activation mechanism for H+-ATPase in vascular plants. However, there is very little information on the plasma membrane H+-ATPase in nonvascular plant bryophytes. Here, we show that the liverwort Marchantia polymorpha, which is the most basal lineage of extant land plants, expresses both the penultimate threonine-containing H+-ATPase (pT H+-ATPase) and non-penultimate threonine-containing H+-ATPase (non-pT H+-ATPase) as in the green algae and that pT H+-ATPase is regulated by phosphorylation of its penultimate threonine. A search in the expressed sequence tag database of M. polymorpha revealed eight H+-ATPase genes, designated MpHA (for M. polymorpha H+-ATPase). Four isoforms are the pT H+-ATPase; the remaining isoforms are non-pT H+-ATPase. An apparent 95-kD protein was recognized by anti-H+-ATPase antibodies against an Arabidopsis (Arabidopsis thaliana) isoform and was phosphorylated on the penultimate threonine in response to the fungal toxin fusicoccin in thalli, indicating that the 95-kD protein contains pT H+-ATPase. Furthermore, we found that the pT H+-ATPase in thalli is phosphorylated in response to light, sucrose, and osmotic shock and that light-induced phosphorylation depends on photosynthesis. Our results define physiological signals for the regulation of pT H+-ATPase in the liverwort M. polymorpha, which is one of the earliest plants to acquire pT H+-ATPase.
The plasma membrane H+-ATPase, a member of the superfamily of P-type ATPases, which are characterized by the formation of phosphorylated intermediates during catalysis, has 10 transmembrane segments and N and C termini in the cytosol (Sussman, 1994; Palmgren, 2001). The H+-ATPase is a ubiquitous enzyme from fungi to vascular plants and is a functional monomer with a molecular mass of about 100 kD that can form a dimer or hexamer (Goormaghtigh et al., 1986; Briskin and Reynolds-Niesman, 1989; Kanczewska et al., 2005). The H+-ATPase actively transports H+ out of the cell, coupled with ATP hydrolysis, and creates an electrochemical gradient of H+ across the plasma membrane for energizing substance transport, coupled with many secondary transporters, the maintenance of membrane potential, and pH homeostasis (Duby and Boutry, 2009). Indeed, the H+-ATPase has been shown to be an essential enzyme in yeast and Arabidopsis (Arabidopsis thaliana) plants (Serrano et al., 1986; Haruta et al., 2010).
The structure of the H+-ATPase is highly conserved from fungi to the vascular plants, apart from the C-terminal region. In vascular plants, including one of the most basal of vascular plant lycophytes, Selaginella moellendorffii, the C-terminal region of the H+-ATPase, consisting of around 100 amino acids, is known as an autoinhibitory domain and contains a penultimate Thr (Palmgren et al., 1990, 1991; Palmgren, 2001; Banks et al., 2011). On the other hand, the plasma membrane H+-ATPases in yeast, red algae (Cyanidioschyzon merolae 10D), and green algae (Chlamydomonas reinhardtii, Volvox carteri, and Chlorella variabilis NC64A) lack such a C terminus, and the length of the C terminus varies among species (Portillo, 2000; Matsuzaki et al., 2004; Merchant et al., 2007; Blanc et al., 2010; Prochnik et al., 2010). Here, we define the H+-ATPase having the C-terminal region containing the penultimate Thr as a pT H+-ATPase and others as the non-pT H+-ATPase. Taken together, the pT H+-ATPases probably did exist in the last common ancestor of liverworts and other land plants. However, when pT H+-ATPase appeared in the evolution of plants remains unknown.
The H+-ATPase is known to be regulated by physiological signals at both transcriptional and posttranscriptional levels (Portillo, 2000). Posttranslational regulation of the pT H+-ATPase has been studied extensively. The C-terminal region keeps the H+-ATPase in a low-activity state via an interaction with the catalytic domain under normal conditions, and phosphorylation of the penultimate Thr and subsequent binding of the 14-3-3 protein to the phosphorylated penultimate Thr in response to physiological signals results in activation of the H+-ATPase (Olsson et al., 1998; Fuglsang et al., 1999, 2003; Svennelid et al., 1999; Maudoux et al., 2000; Kinoshita and Shimazaki, 2002). As a physiological signal, blue light is known to activate the H+-ATPase via phosphorylation of the penultimate Thr in stomatal guard cells (Kinoshita and Shimazaki, 1999; Kinoshita et al., 2001; Shimazaki et al., 2007). Moreover, it has been reported that Suc and phytohormones, such as auxin and gibberellic acid, induce phosphorylation of the penultimate Thr in seedlings and culture cells from Arabidopsis (Niittylä et al., 2007; Chen et al., 2010). In addition, osmotic shock is most likely to induce phosphorylation of the penultimate Thr of the H+-ATPase in tomato (Solanum lycopersicum) culture cells (Kerkeb et al., 2002). These results indicate that many physiological signals regulate H+-ATPase activity via regulation of the phosphorylation status of the penultimate Thr of H+-ATPase and that phosphorylation of the penultimate Thr is a major common regulatory mechanism of H+-ATPase in vascular plants (Sze et al., 1999; Kinoshita and Hayashi, 2011). It should be noted that the pT H+-ATPase has been reported to be phosphorylated at multiple sites in addition to the penultimate Thr in vascular plants (Fuglsang et al., 2007; Duby et al., 2009; Rudashevskaya et al., 2012).
The C terminus of non-pT H+-ATPase is also thought to be important for the regulation of its activity. In the yeast Saccharomyces cerevisiae, it has been reported that phosphorylation of two tandemly positioned residues in the C terminus (Ser-911 and Thr-912 in PMA1) activates the H+-ATPase in response to Glc (Lecchi et al., 2007), indicating that the fungal H+-ATPase is regulated in a different manner from the pT H+-ATPase. Posttranslational regulation of the H+-ATPases in red and green algae remains unresolved.
In this study, we performed molecular characterization of plasma membrane H+-ATPase in the liverwort Marchantia polymorpha as a nonvascular plant bryophyte, which represents the most basal lineage of extant land plants. We found that M. polymorpha expresses both pT H+-ATPase and non-pT H+-ATPase. We further provide evidence that the pT H+-ATPase in M. polymorpha is regulated by phosphorylation of its penultimate Thr in response to physiological signals, such as light, Suc, and osmotic shock.
RESULTS
Identification of cDNA Sequences of Plasma Membrane H+-ATPase in M. polymorpha
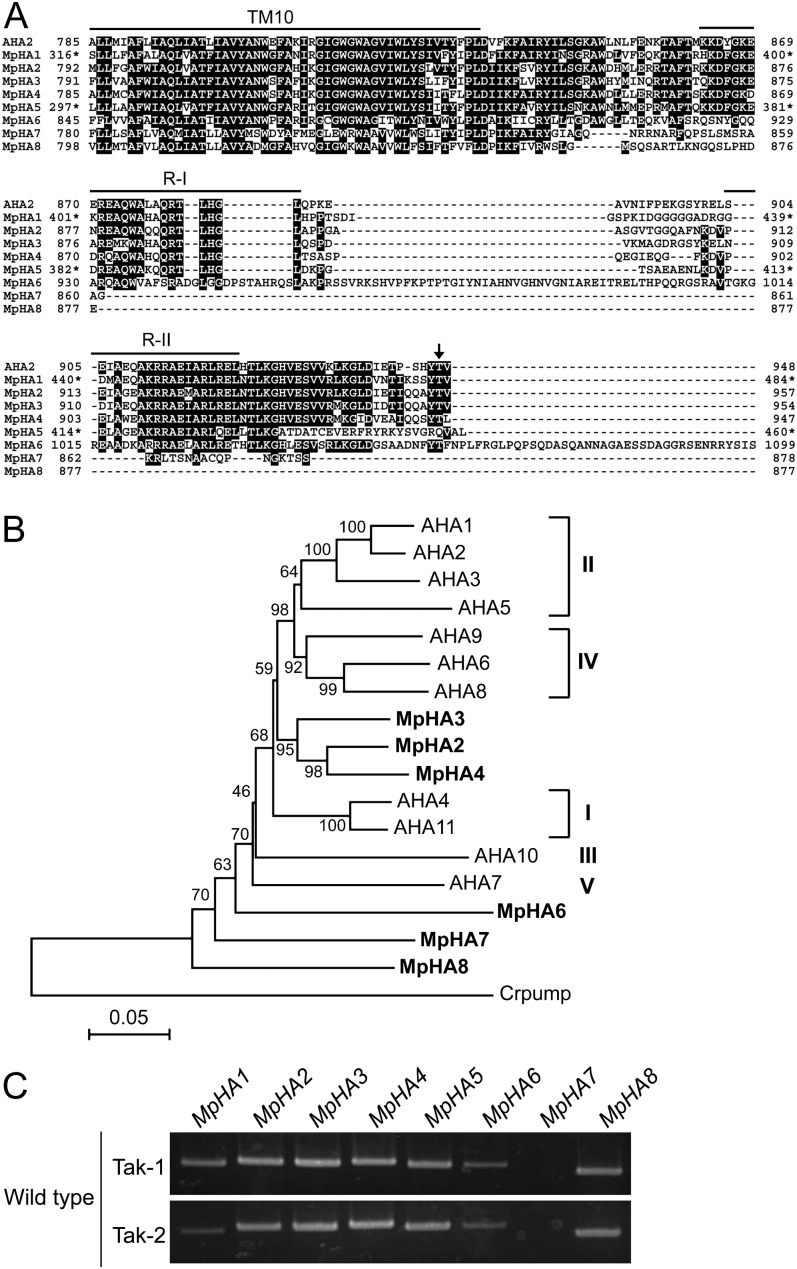
We carried out a BLAST search against M. polymorpha ESTs to find sequences with similarity to the typical plasma membrane H+-ATPase in Arabidopsis, AHA2. Individual ESTs were derived from thalli and protonemata of M. polymorpha, male accession Takaragaike-1 (Tak-1). We found eight H+-ATPase homologs, designated MpHA1 to MpHA8 (Fig. 1A). All isoforms highly conserve a characteristic sequence, GDGVNDAPALKKA, in the catalytic domain of the P-type ATPase (Axelsen and Palmgren, 1998; Supplemental Fig. S1) and show high sequence identity with AHA2 (more than 70%; Supplemental Table S1), providing strong support to our claim that these isoforms are functional homologs as plasma membrane H+-ATPases.
Figure 1.
Molecular characterization of the H+-ATPase in M. polymorpha. A, Alignment of C termini of the H+-ATPases from M. polymorpha (MpHA1–MpHA8) and Arabidopsis (AHA2) with ClustalW (Thompson et al., 1994). Black blocks indicate highly conserved residues. The 10th transmembrane segment (TM10) and region I (R-I) and region II (R-II) within the C-terminal region are indicated by lines. The arrow denotes the penultimate Thr of AHA2, which is the phosphorylated site for activity regulation. Asterisks indicate amino acid numbers of the partial sequences not acquiring N-terminal information for MpHA1 and MpHA5. Dashes indicate gaps introduced to allow for optimal alignment of the sequences. B, Phylogenetic tree of the H+-ATPase proteins from M. polymorpha, Arabidopsis (AHA1–AHA11), and C. reinhardtii (Crpump; XP_001698580). The alignment for the phylogenetic tree was performed with ClustalW using full-length amino acid sequences (Thompson et al., 1994). The phylogenetic tree was created with the MEGA5 software (Tamura et al., 2011) and the neighbor-joining program with 1,000 bootstrap replications. Bootstrap values at the branches represent the percentage obtained in 1,000 replications. The C. reinhardtii pump sequence was used as an outgroup. MpHA1 and MpHA5 were not used to construct the phylogenetic tree, as full-length sequences were not acquired. Roman numerals designate the subfamilies. The scale bar represents 0.05 substitutions per site. C, RT-PCR analysis of isogenes of the H+-ATPase in M. polymorpha. RNA was extracted from each thallus of the male (Tak-1) and female (Tak-2) gametophytes. PCRs were performed with 30 cycles using specific primers for MpHA1 to MpHA8.
Of these, four isoforms (MpHA1, MpHA2, MpHA3, and MpHA4) possess a penultimate Thr and conserve region I and region II, which are important for autoinhibitory effects on the H+-ATPase, in the C-terminal region (Axelsen et al., 1999). In contrast, the remaining isoforms lack such a penultimate Thr in the C terminus and have various C-terminal lengths (Fig. 1A). Phylogenetic analysis using full-length amino acid sequences indicated that MpHA2, MpHA3, and MpHA4 are clustered with Arabidopsis H+-ATPase and that MpHA6, MpHA7, and MpHA8 are close to the non-pT H+-ATPase of Chlamydomonas reinhardtii, which has no penultimate Thr (Fig. 1B). According to the classification of gene families in the pT H+-ATPase, MpHA2, MpHA3, and MpHA4 localize between subfamilies I and IV (Arango et al., 2003). These results suggest that the M. polymorpha genome encodes both pT H+-ATPase and non-pT H+-ATPase genes. Note that MpHA5 has high sequence identity with AHA2 as well as MpHA1 to MpHA4 but no conserved penultimate Thr and that MpHA6 has insertions of over 40 residues in the C-terminal region and a C-terminal extension of 39 residues (Fig. 1A; Supplemental Table S1).
To examine the expression of MpHAs, reverse transcription (RT)-PCR analysis using total RNA from thalli was performed. The results showed that the H+-ATPase isoforms, except for MpHA7, were expressed in thalli (Fig. 1C). All MpHAs showed identical expression properties in both male (Tak-1) and female (Takaragaike-2 [Tak-2]) thalli (Fig. 1C).
Fusicoccin Induces Phosphorylation of the Penultimate Thr of pT H+-ATPases
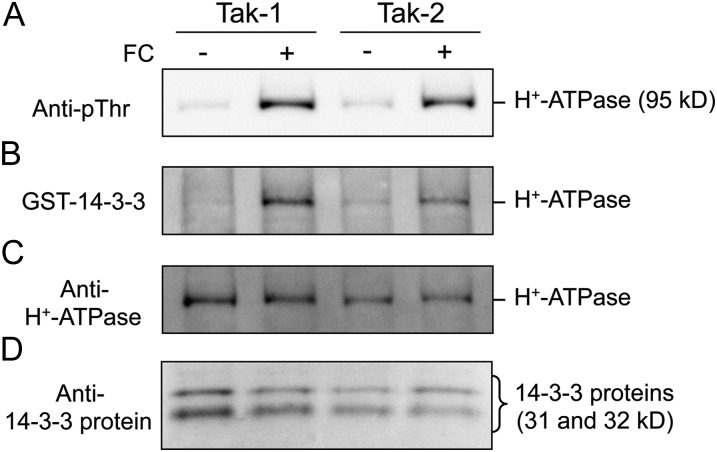
We first performed immunoblot analysis using antibodies raised against the conserved catalytic domain of AHA2 (anti-H+-ATPase; Hayashi et al., 2010) and found that only an apparent 95-kD protein in thalli was recognized (Fig. 2C). This suggests that the 95-kD protein is most likely involved in MpHA1 to MpHA5, because these isoforms show high identity with AHA2 (more than 80%) and have very similar molecular masses to AHA2 (Supplemental Table S1).
Figure 2.
Phosphorylation of the pT H+-ATPase in M. polymorpha. Dark-adapted thalli (male, Tak-1; female, Tak-2) were treated with (+) or without (−) 10 µm FC in the dark for 30 min. Then, thalli were disrupted and the protein extracts subjected to SDS-PAGE. A, FC-induced phosphorylation of the penultimate Thr of H+-ATPase. Phosphorylated H+-ATPase was detected by immunoblot using anti-pThr. B, FC-induced binding of 14-3-3 protein to the H+-ATPase. Protein-blot analysis was performed using GST-14-3-3 protein (Arabidopsis GF14phi) as probe. C, Amounts of the H+-ATPase. The H+-ATPase was detected by immunoblot using antibodies for Arabidopsis AHA2 (anti-H+-ATPase). D, Amounts of 14-3-3 protein. 14-3-3 protein was detected by immunoblot using antibodies for Arabidopsis GF14phi (anti-14-3-3 protein).
To analyze whether the pT H+-ATPase in M. polymorpha (MpHA1–MpHA4) is regulated by phosphorylation of the penultimate Thr, we treated thalli with the fungal toxin fusicoccin (FC), which is an activator of H+-ATPase and accumulates phosphorylated H+-ATPase through inhibition of dephosphorylation of the phosphorylated penultimate Thr in vascular plants (Kinoshita and Shimazaki, 2001; Hayashi et al., 2010). Phosphorylation of the penultimate Thr was detected using antibodies raised against the phosphorylated penultimate Thr-947 of AHA2 (anti-pThr; Hayashi et al., 2010). The results showed that FC at 10 µm induced phosphorylation of the 95-kD protein in thalli without altering the amount of H+-ATPase present in the cells (Fig. 2, A and C). Moreover, protein-blot analysis using 14-3-3 protein (Arabidopsis GF14phi) as a probe revealed that phosphorylated H+-ATPase bound to the 14-3-3 protein. These results indicate that the phosphorylated penultimate Thr creates a binding motif for the 14-3-3 protein, as also seen in vascular plants (Fig. 2B), and that the 95-kD protein contains the pT H+-ATPase in M. polymorpha.
As illustrated in Figure 2A, the H+-ATPases in both male (Tak-1) and female (Tak-2) thalli showed an identical response to FC. We performed further experiments using Tak-1.
Mp14-3-3a Binds to the Phosphorylated H+-ATPase
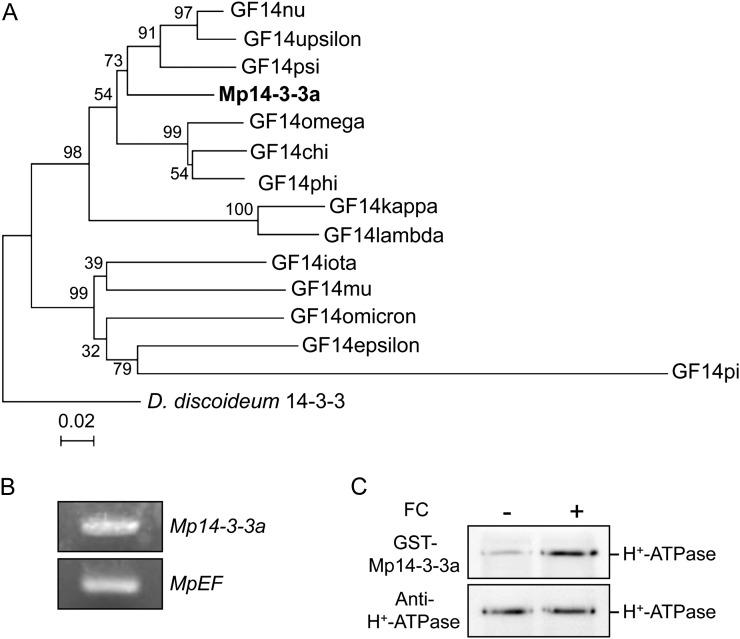
We detected endogenous 14-3-3 proteins in thalli having molecular masses of 31 and 32 kD using antibodies raised against Arabidopsis GF14phi (Fig. 2D; Kinoshita and Shimazaki, 1999). We then performed a BLAST search against M. polymorpha ESTs and found a typical 14-3-3 protein, designated M. polymorpha 14-3-3a (Mp14-3-3a; Fig. 3A; Supplemental Fig. S2). Expression of Mp14-3-3a in thalli was confirmed by RT-PCR (Fig. 3B). Moreover, protein-blot analysis using the recombinant Mp14-3-3a as a probe revealed that it bound to phosphorylated H+-ATPase in thalli (Fig. 3C). These results indicate that the pT H+-ATPases in M. polymorpha might be activated via phosphorylation of the penultimate Thr and subsequent binding of the endogenous 14-3-3 protein, as in vascular plants.
Figure 3.
Molecular characterization of 14-3-3 protein in M. polymorpha. A, Phylogenetic tree of 14-3-3 proteins from M. polymorpha (Mp14-3-3a), Arabidopsis (GF14chi, GF14omega, GF14psi, GF14phi, GF14upsilon, GF14lambda, GF14nu, GF14kappa, GF14mu, GF14epsilon, GF14omicron, GF14iota, GF14pi), and Dictyostelium discoideum 14-3-3 (X95568). The alignment for the phylogenetic tree was performed with ClustalW using full-length amino acid sequences (Thompson et al., 1994). The phylogenetic tree was created with the MEGA5 software (Tamura et al., 2011) and the neighbor-joining program with 1,000 bootstrap replications. Bootstrap values at the branches represent the percentage obtained in 1,000 replications. The D. discoideum 14-3-3 sequence was used as an outgroup. The scale bar represents 0.02 substitutions per site. B, RT-PCR analysis of 14-3-3 protein expression in M. polymorpha thalli. Total RNA was extracted from thalli, and RT-PCR was performed. MpEF was used as a loading control. C, FC-induced binding of 14-3-3 protein of M. polymorpha to the phosphorylated H+-ATPase. Procedures were the same as in Figure 2. Binding of 14-3-3 protein was detected by protein blot using GST-Mp14-3-3a as probe.
Effects of Physiological Signals on the Phosphorylation Status of the pT H+-ATPases in M. polymorpha
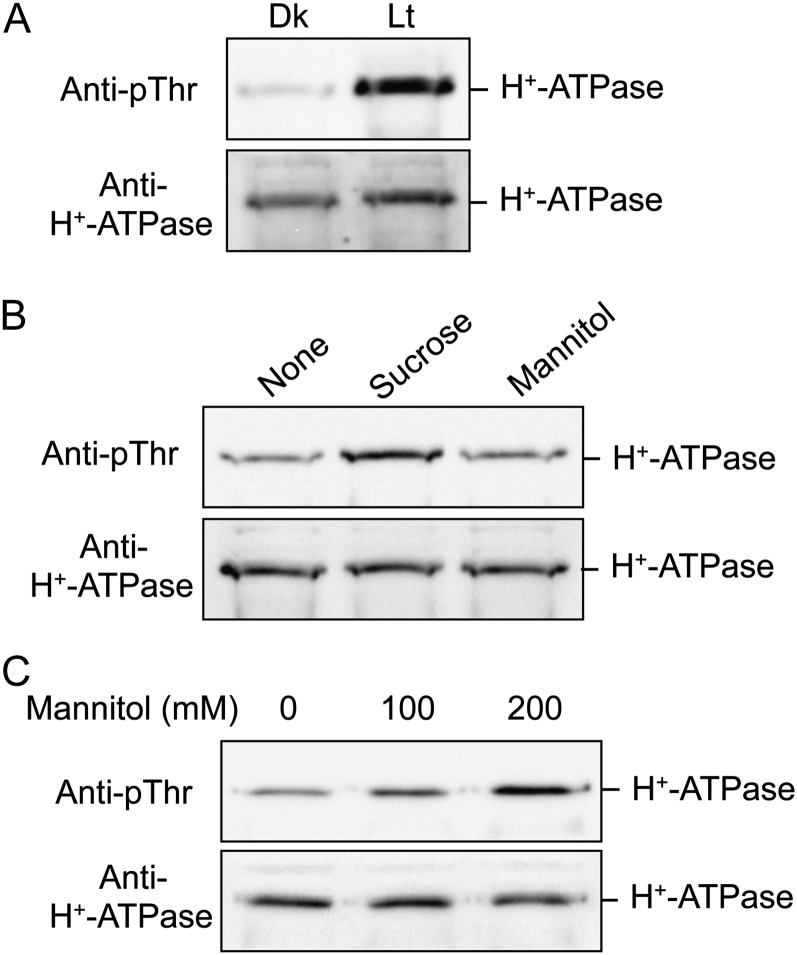
To clarify the physiological signals that regulate the phosphorylation status of the pT H+-ATPase in M. polymorpha, we next examined the effects of putative physiological signals on the pT H+-ATPase in thalli. We treated thalli with white light (50 µmol m−2 s−1 for 30 min; Fig. 4A), Suc as a photosynthesis product (30 mm for 30 min; Fig. 4B), and mannitol as an osmotic agent (100 or 200 mm for 30 min; Fig. 4C). Interestingly, all treatments induced phosphorylation of the H+-ATPase in thalli without altering the H+-ATPase amount. Suc-induced phosphorylation cannot be interpreted as a result of its osmotic pressure, because treatment with the same concentration of mannitol (30 mm for 30 min) had no effect on phosphorylation level (Fig. 4B). Osmotic shock-dependent phosphorylation required over 100 mm mannitol and was concentration dependent between 100 and 200 mm (Fig. 4C).
Figure 4.
Effects of physiological stimuli on phosphorylation level of the pT H+-ATPase in M. polymorpha. A, Light-induced phosphorylation of the H+-ATPase in thalli. Dark-adapted thalli were illuminated with white light for 3 h at 50 µmol m−2 s−1 (Lt) or kept in the dark (Dk). The thalli were then disrupted and the protein extracts subjected to SDS-PAGE. Phosphorylated H+-ATPase and the H+-ATPase were detected by immunoblot using anti-pThr and anti-H+-ATPase, respectively. B, Suc-induced phosphorylation of the H+-ATPase in thalli. Dark-adapted thalli were treated with 30 mm Suc or 30 mm mannitol for 30 min in the dark. Other procedures were the same as in A. Mannitol was used for osmotic control. C, Mannitol-induced phosphorylation of the H+-ATPase in thalli. Dark-adapted thalli were treated with 0, 100, or 200 mm mannitol for 30 min in the dark. Other procedures were the same as in A.
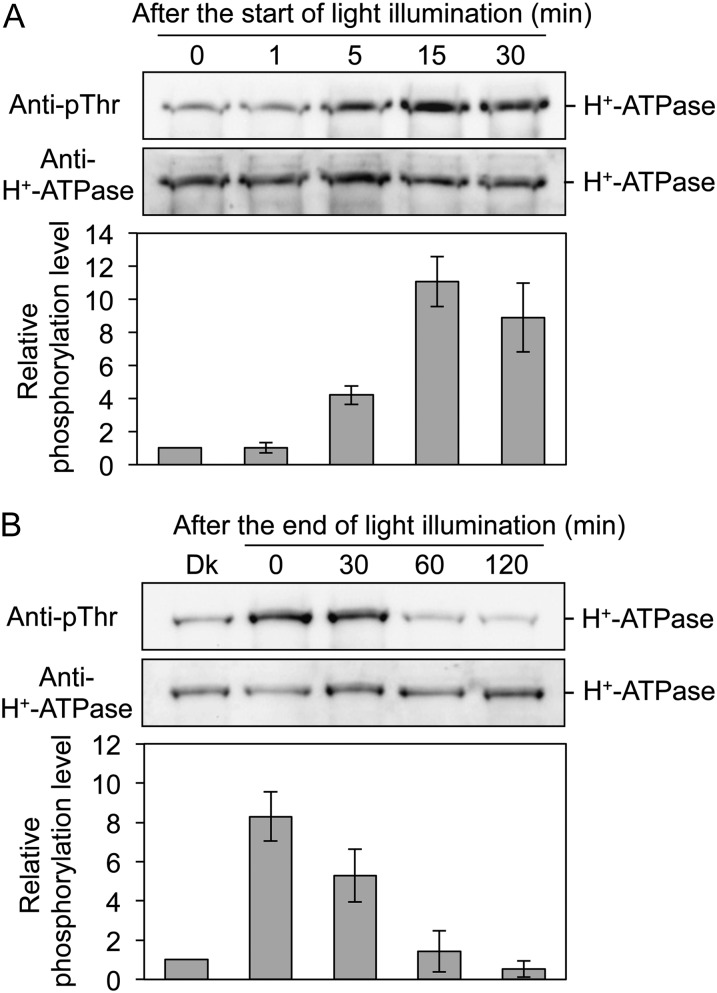
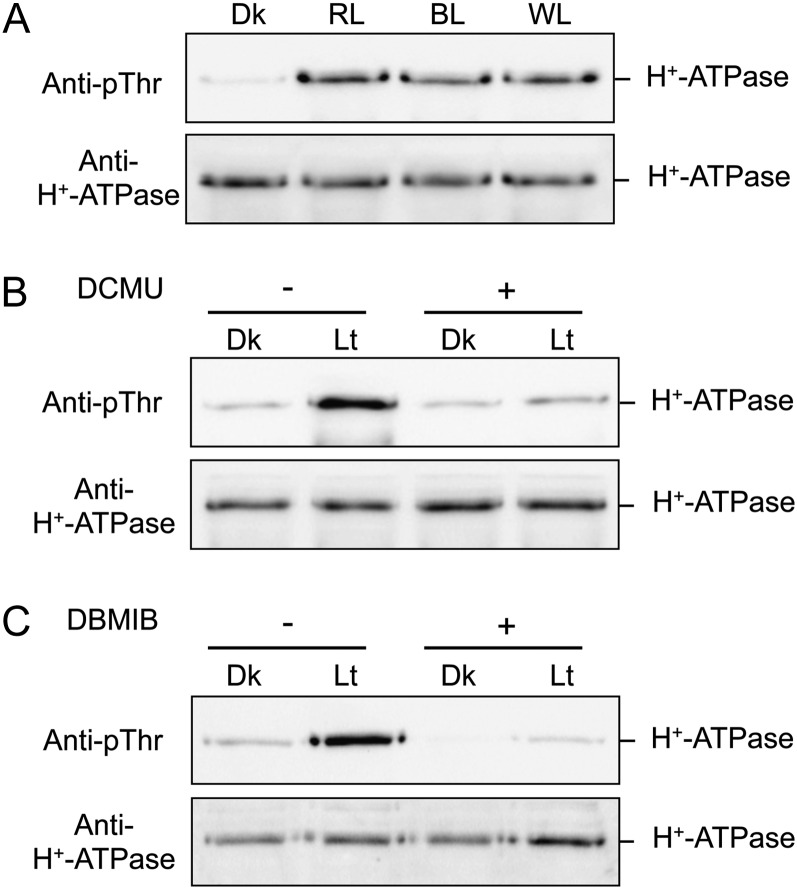
Notably, light illumination had a drastic effect on the phosphorylation level of the H+-ATPase in thalli (Fig. 4A). Therefore, we examined the light-induced phosphorylation of the H+-ATPase in more detail. As shown in Figure 5A, the phosphorylation level of the H+-ATPase reached a maximum within 15 min after the start of illumination. Phosphorylated H+-ATPase was dephosphorylated gradually after the end of light illumination, and phosphorylation level reached the original level in around 60 min (Fig. 5B). We further determined the effects of light quality on phosphorylation and found that red light and blue light (50 µmol m−2 s−1 for 30 min) also induced phosphorylation of the H+-ATPase (Fig. 6A). Interestingly, both 3-(3,4-dichlorophenyl)-1,1-dimethylurea (DCMU) and 2,5-dibromo-3-methyl-6-isopropyl-p-benzoquinone (DBMIB), which are inhibitors of photosynthetic electron transport from PSII to PSI (Trebst, 2007), at 10 µm severely inhibited phosphorylation (Fig. 6, B and C), suggesting that light-induced phosphorylation of the H+-ATPase is regulated by photosynthesis.
Figure 5.
Changes in phosphorylation levels of the H+-ATPase in thalli in response to light. A, Time course of phosphorylation of the H+-ATPase in response to light. Dark-adapted thalli were illuminated with white light at 50 µmol m−2 s−1 and disrupted at 1, 5, 15, and 30 min after the start of illumination. The protein extracts were subjected to SDS-PAGE. Other procedures were the same as in Figure 4A. B, Time course of dephosphorylation of the H+-ATPase at the end of illumination. Dark-adapted thalli (Dk) were illuminated with white light for 30 min at 50 µmol m−2 s−1 and kept in darkness at the end of the illumination. The thalli were disrupted at 0, 30, 60, and 120 min after the end of illumination. The protein extracts were subjected to SDS-PAGE. Other procedures were the same as in Figure 4A. The graphs represent the phosphorylation level of H+-ATPase, as quantified from the ratio of signal intensity from the phosphorylated H+-ATPase to that from the H+-ATPase and expressed relative to the phosphorylation level of the dark-adapted thalli. Values represent means of three independent experiments with sd.
Figure 6.
Analysis of light-induced phosphorylation of the H+-ATPase in thalli. A, Effects of light quality on the phosphorylation of the H+-ATPase. Dark-adapted thalli were illuminated with red light (RL), blue light (BL), or white light (WL) for 30 min at 50 µmol m−2 s−1. The protein extracts were subjected to SDS-PAGE. Other procedures were the same as in Figure 4A. B and C, Effects of DCMU (B) and DBMIB (C) on light-induced phosphorylation of H+-ATPase. Dark-adapted thalli were incubated with (+) or without (−) 10 µm DCMU or 10 µm DBMIB for 30 min in the dark. Thalli were then illuminated with white light (Lt) at 50 µmol m−2 s−1 or kept in the dark (Dk) for 30 min. Other procedures were the same as in Figure 4A.
Analyses of the Signaling Pathway in Light-Induced Phosphorylation of the H+-ATPase in Thalli
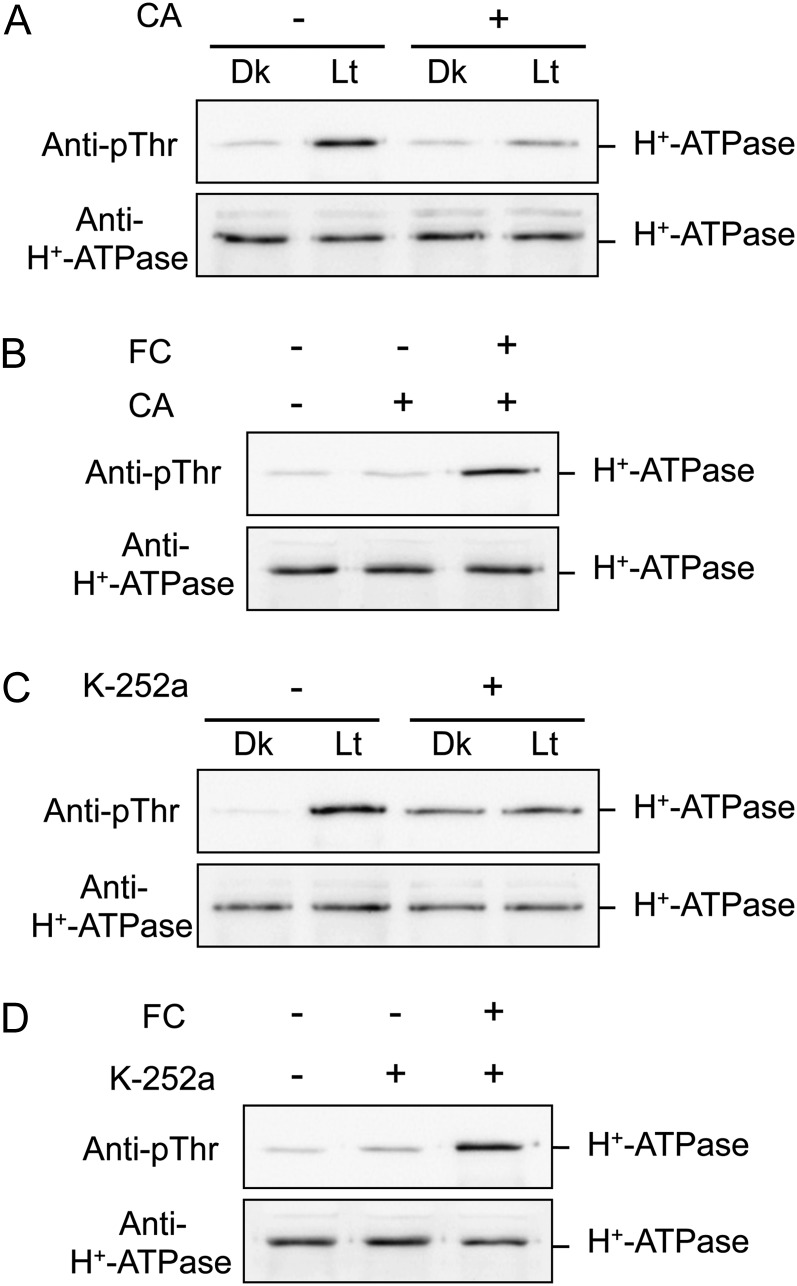
Previous studies have shown that a potent protein kinase inhibitor, K-252a, and a type 1/2A protein phosphatase inhibitor, calyculin A (CA), inhibit blue light-induced phosphorylation/activation of the plasma membrane H+-ATPase in stomatal guard cells (Kinoshita and Shimazaki, 1997, 1999; Hayashi et al., 2011). We tested the effects of K-252a and CA on light-induced phosphorylation of the H+-ATPase in thalli and found that both K-252a at 10 µm \and CA at 0.5 µm severely inhibited phosphorylation (Fig. 7, A and C). In contrast, K-252a and CA had no effect on the FC-induced phosphorylation of the H+-ATPase (Fig. 7, B and D), suggesting that a K-252a-sensitive protein kinase and CA-sensitive protein phosphatase are involved in the signaling pathway of the light-induced phosphorylation of the H+-ATPase in thalli and that a K-252a-insensitive protein kinase catalyzes direct phosphorylation of the H+-ATPase in response to FC. This result is consistent with previous reports showing that direct phosphorylation of the penultimate Thr in H+-ATPase is mediated by a K-252a-insensitive protein kinase (Kinoshita and Shimazaki, 2001; Hayashi et al., 2010).
Figure 7.
Effects of inhibitors on the phosphorylation of H+-ATPase in thalli. A and C, Effects of CA and K-252a on the light-induced phosphorylation of H+-ATPase. Dark-adapted thalli were incubated with (+) or without (−) 0.5 µm CA or 10 µm K-252a for 30 min in the dark. Thalli were then illuminated with white light (Lt) at 50 µmol m−2 s−1 or kept in the dark (Dk) for 30 min. B and D, Effects of CA and K-252a on FC-induced phosphorylation of H+-ATPase. Dark-adapted thalli were incubated with (+) or without (−) 0.5 µm CA or 10 µm K-252a for 30 min, and then FC was added at 10 µm for 30 min in the dark. Other procedures were the same as in Figure 4A.
DISCUSSION
The Plasma Membrane H+-ATPase in M. polymorpha
It has been demonstrated that the H+-ATPase is essential for vascular plants (Zhao et al., 2000; Haruta et al., 2010). Accordingly, treatment with the H+-ATPase inhibitors erythrosine B (Kanczewska et al., 2005) and vanadate (Kinoshita and Shimazaki, 1999) severely inhibited the growth of thalli in the liverwort M. polymorpha (Supplemental Fig. S3), suggesting a critical role of the H+-ATPase in growth and development in M. polymorpha. However, there has not been any study on the plasma membrane H+-ATPase in M. polymorpha. In addition, despite detailed characterization of the pT H+-ATPase in vascular plants, the emergence and evolution of the pT H+-ATPase remain unknown.
In this study, we showed that the liverwort M. polymorpha, which represents the most basal lineage of extant land plants, possesses both the pT H+-ATPase genes (MpHA1–MpHA4), which contain the characteristic penultimate Thr in the C terminus, and the non-pT H+-ATPase genes (MpHA5–MpHA8; Fig. 1). These results strongly suggest that the pT H+-ATPases most likely did exist in the last common ancestor of liverworts. As liverworts represent the most basal lineage of extant land plants, our data suggest that plants had already acquired pT H+-ATPase when they emerged on the terrestrial environment. It should be noted that MpHA6 possesses regions R-I and R-II, which are important for autoinhibitory effects on the H+-ATPase, and YTF in the position of YTV in the pT H+-ATPase. MpHA6, therefore, may be a “transition form” in the evolution to the pT H+-ATPase, because a simple truncation of this gene could create a pT H+-ATPase. A whole-genome sequence has revealed that one of the most basal of vascular plant lycophytes, S. moellendorffii, most likely possesses only the pT H+-ATPase (Banks et al., 2011). In contrast, the bryophyte M. polymorpha possesses both types of H+-ATPase (Fig. 1). These results suggest that non-pT H+-ATPase was lost in the evolutionary transition from bryophytes to vascular plants.
Regulatory Mechanism of the pT H+-ATPase in M. polymorpha
In vascular plants, phosphorylation of the penultimate Thr of the plasma membrane H+-ATPase and subsequent binding of the 14-3-3 protein to the phosphorylated C terminus is the most common activation mechanism for the H+-ATPase (Palmgren, 2001; Duby and Boutry, 2009; Kinoshita and Hayashi, 2011). We found that the pT H+-ATPase in thalli of M. polymorpha is phosphorylated in its penultimate Thr and binds to the 14-3-3 protein in response to FC (Fig. 2). These results clearly indicate that the pT H+-ATPase in M. polymorpha might be activated via an identical mechanism to that in vascular plants. Moreover, we showed that the phosphorylation status of the penultimate Thr of the pT H+-ATPase in thalli is regulated by phosphorylation in response to physiological signals such as light, Suc, and osmotic shock (Fig. 4). Similarly, Suc was reported to induce phosphorylation of the plasma membrane H+-ATPase in Arabidopsis seedlings (Niittylä et al., 2007), and osmotic shock likely induced phosphorylation of the plasma membrane H+-ATPase in tomato culture cells (Kerkeb et al., 2002), suggesting that the phosphorylation status of the pT H+-ATPase in the liverwort M. polymorpha is also regulated by similar physiological signals to those in vascular plants. It should be noted that we measured ATP hydrolytic activity of the H+-ATPase according to a previous method for vascular plants (Kinoshita and Shimazaki, 1999), but we could not detect increased ATP hydrolytic activity of the H+-ATPase in response to physiological signals in cell extracts and microsomes from thalli because of high background noise from nonspecific ATP hydrolytic activity in these samples (data not shown). Further investigations will be needed to establish measurement methods for plasma membrane H+-ATPase activity in the liverwort M. polymorpha and to demonstrate that phosphorylation of the penultimate Thr is correlated with the activation status of the H+-ATPase.
In addition, our results suggest that M. polymorpha possesses the identical/similar protein kinase and protein phosphatase that directly regulate the phosphorylation status of the pT H+-ATPase and might have obtained these components in parallel with the evolution of the pT H+-ATPase. We should note, however, that the protein kinase and phosphatase have not yet been identified in vascular plants, although they have been extensively investigated (Svennelid et al., 1999; Camoni et al., 2000; Hayashi et al., 2010). Identification of protein kinase and phosphatase, including those of M. polymorpha, will provide novel understanding for the regulation of pT H+-ATPase in plants.
Photosynthetic Control of the Phosphorylation Status of the pT H+-ATPase in M. polymorpha
In this study, we found that light induces phosphorylation of the H+-ATPase in thalli and that the photosynthesis inhibitors DCMU and DBMIB inhibit light-induced phosphorylation (Figs. 4A and 6). These results indicate that photosynthesis controls the phosphorylation status of the pT H+-ATPase in thalli. Photosynthetic control of plasma membrane H+-ATPase activity has been reported in vascular plants (Marten et al., 2010). Light-induced cell expansion in Pisum sativum leaves partly depends on a DCMU-sensitive stimulation of the plasma membrane H+-ATPase (Stahlberg and Van Volkenburgh, 1999). In Vallisneria gigantia leaves, photosynthesis-dependent modulation of the enzymatic activity of the plasma membrane H+-ATPase has been shown to be involved in light-induced membrane hyperpolarization, which may have an effect on cytoplasmic streaming (Harada et al., 2002a, 2002b). However, there is no report of photosynthetic control of H+-ATPase phosphorylation. Thus, to our knowledge, the experimental evidence presented here is the first to show photosynthetic control of the phosphorylation status of the penultimate Thr of H+-ATPase in plants. Given that Suc also induced phosphorylation of the pT H+-ATPase in thalli (Fig. 4B), it is possible that light-induced and photosynthesis-dependent phosphorylation of the H+-ATPase is mediated by Suc as a photosynthetic product. Light-induced phosphorylation, however, began within 5 min (Fig. 5A). Further investigation will be needed to clarify the relationship between the time courses of Suc production and H+-ATPase phosphorylation in thalli.
In vascular plants, the H+-ATPase plays a crucial role in the transport of photosynthetic products into sink tissues, such as fruits, tubers, and roots (Palmgren, 2001; Duby and Boutry, 2009). In the case of phloem loading, Suc produced by photosynthesis in mesophyll cells is uploaded into the phloem companion cells by the Suc/H+ symporter utilizing the electrochemical gradient generated by H+-ATPase and is transported to sink tissues via the phloem (Stadler et al., 1995; Burkle et al., 1998; Zhao et al., 2000). However, the liverwort M. polymorpha is a nonvascular plant and has no phloem companion cells. Supplemental Figure S4 presents a cross-section of the thallus, showing distinct layers of tissues: upper epidermis, photosynthetic parenchyma (chlorenchyma), storage parenchyma, lower epidermis, and rhizoids. The chlorenchyma is found just below the upper epidermis. Upper and lower epidermis may also contain chloroplasts. Storage parenchyma occupies the majority of the thallus. Further investigation will be needed to elucidate the localization of the H+-ATPase in the thalli, which is phosphorylated in response to light. This will clarify whether H+-ATPase phosphorylation is mediated by an intracellular or an intercellular signaling pathway and will elucidate the physiological role of the photosynthetic control of H+-ATPase phosphorylation in thalli.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Male and female accessions of Marchantia polymorpha, Tak-1 and Tak-2, respectively (Ishizaki et al., 2008), were grown on one-half-strength Gamborg’s B5 medium containing 1.4% (w/v) agar for 3 weeks under a 16-h-light (50 µmol m−2 s−1)/8-h-dark cycle. Plants were grown at 24°C with a relative humidity of 55% to 75% in growth rooms.
Expression of Transcripts Determined by RT-PCR Analysis
Total RNA was extracted from 3-week-old thalli using the RNeasy Plant Mini Kit (Qiagen). First-strand cDNAs were synthesized from total RNA with the PrimeScript II First Strand cDNA Synthesis Kit using oligo(dT) primer (Takara). The cDNA fragments of the H+-ATPases, Mp14-3-3a, and MpEF were amplified with the following oligonucleotide primers: 5′-GTGTTATTTGGCTGTATAGCATCGTG-3′ and 5′-TCCTCACCTATTCACGTCACC-3′ for MpHA1, 5′-CGGAGTAATCTGGTTGTACTCC-3′ and 5′-CCTAGAACCTTGTGTTCCAAGC-3′ for MpHA2, 5′-GGAGATAGAGGAAGTTACAAGGAG-3′ and 5′-GCTTCTCACATTCACGTTCTCC-3′ for MpHA3, 5′-AAAGGCATGGGATCTTCTTCTG-3′ and 5′-ATGAGCATGCACTCATCAAGTC-3′ for MpHA4, 5′-TCAGCACTCTTCGGCAATCG-3′ and 5′-GAAGAAGTCAGTTCTATCAGCTG-3′ for MpHA5, 5′-GGATGCGATCAAGATCATTTGTC-3′ and 5′-GAGATCGAGTATCTCCTATTCTC-3′ for MpHA6, 5′-GCTCTACTCGAGAACGTTTCG-3′ and 5′-AGCTTGTTGTAGCCAAAGATGTCG-3′ for MpHA7, 5′-ATCATCAGCATCGTTGCCATTCC-3′ and 5′-AATCACCTCGACGACAATGCC-3′ for MpHA8, 5′-TTCGTCGAGGATTAGCGATGG-3′ and 5′-TTAACTGTCCTCGGCATCCTC-3′ for Mp14-3-3a, and 5′-AAGCCGTCGAAAAGAAGGAG-3′ and 5′-TTCAGGATCGTCCGTTATCC-3′ for MpEF. All oligonucleotide primers were designed on the basis of the EST database from M. polymorpha. All PCRs were performed in 30 cycles.
Preparation of Glutathione S-Transferase-Fused Mp14-3-3 Protein
The glutathione S-transferase (GST)-fused Mp14-3-3a (GST-Mp14-3-3a) was expressed in Escherichia coli cells, and the recombinant protein was purified and used as a probe on protein blots. The full-length Mp14-3-3a cDNA was amplified by RT-PCR with two oligonucleotide primers, 5′-CGGGATCCCTTCGTCGAGGATTAGCGATGG-3′ and 5′-CGGGATCCTTAACTGTCCTCGGCATCCTC-3′. The amplified DNA was cloned into the BamHI site of the pGEX-2T vector (GE Healthcare), and the plasmids were transformed into the E. coli BL21 strain. The polypeptide was expressed as a fusion protein with a GST tag and was purified using glutathione-Sepharose 4B beads (GE Healthcare) as described previously (Kinoshita and Shimazaki, 1999). The purified protein was frozen and kept at −25°C until use.
Immunoblot and Protein-Blot Analyses
Immunoblot and protein blot analyses were performed according to previous methods (Kinoshita et al., 2003; Hayashi et al., 2010) with minor modifications. Thalli were homogenized in an ice-cold homogenization buffer (50 mm MOPS-KOH, pH 7.5, 100 mm NaCl, 2.5 mm EDTA, 10 mm NaF, 5 mm dithiothreitol, 1 mm phenylmethylsulfonyl fluoride, and 20 µm leupeptin) using a mortar and pestle. The homogenate was solubilized by adding a half-aliquot of SDS sample buffer (3% [w/v] SDS, 30% [w/v] Suc, 10% [v/v] 2-mercaptoethanol, 0.012% [w/v] Coomassie Brilliant Blue, 1 mm EDTA, and 30 mm Tris-HCl [pH 8.0]). Then, the solubilized sample was centrifuged at 12,000g for 1 min, and the resulting supernatant was subjected to SDS-PAGE.
The polyclonal antibodies raised against the catalytic domain of Arabidopsis (Arabidopsis thaliana) AHA2 (anti-H+-ATPase), phosphorylation of the penultimate Thr-947 of AHA2 (anti-pThr), GST, and Arabidopsis GF14phi (anti-14-3-3 protein) were described previously (Kinoshita and Shimazaki, 1999; Hayashi et al., 2010). Anti-H+-ATPase and anti-pThr recognize not only AHA2 but also other H+-ATPase isoforms in Arabidopsis (Hayashi et al., 2011). For protein blots, we used GF14phi or Mp14-3-3a proteins fused to GST as a probe.
Light Source
White light was obtained from fluorescent lamps (FL 40S N-SDL; National). Both red and blue light were obtained with light-emitting photodiodes (LED-R, maximum intensity at 660 nm; and Stick-B-32, maximum intensity at 470 nm [EYELA]). Photon flux densities were measured with a quantum meter (LI-250; Li-Cor) equipped with a light sensor (LI-190 SA; Li-Cor).
Sequence data from this article can be found in the GenBank/EMBL databases under the following accession numbers: MpHA1 (AB697742), MpHA2 (AB697743), MpHA3 (AB697744), MpHA4 (AB697745), MpHA5 (AB697746), MpHA6 (AB697747), MpHA7 (AB697748), MpHA8 (AB697749), and Mp14-3-3a (AB697750).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Alignment of conserved segments in the catalytic domain of H+-ATPase from M. polymorpha and Arabidopsis with ClustalW (Thompson et al., 1994).
Supplemental Figure S2. Alignment of 14-3-3 proteins from M. polymorpha (Mp14-3-3a) and Arabidopsis (GF14phi) with ClustalW (Thompson et al., 1994).
Supplemental Figure S3. Effects of H+-ATPase inhibitors on growth of the M. polymorpha thalli.
Supplemental Figure S4. Cross-section of M. polymorpha thallus.
Supplemental Table S1. Analysis of MpHAs based on amino acid sequences.
Supplementary Material
Acknowledgments
We are grateful to Dr. A. Harada of Osaka Medical College for helpful discussion.
Glossary
- pT H+-ATPase
penultimate threonine-containing H+-ATPase
- non-pT H+-ATPase
non-penultimate threonine-containing H+-ATPase
- RT
reverse transcription
- FC
fusicoccin
- DCMU
3-(3,4-dichlorophenyl)-1,1-dimethylurea
- DBMIB
2,5-dibromo-3-methyl-6-isopropyl-p-benzoquinone
- CA
calyculin A
- Tak-1
Takaragaike-1
- Tak-2
Takaragaike-2
- GST
glutathione S-transferase
References
- Arango M, Gévaudant F, Oufattole M, Boutry M. (2003) The plasma membrane proton pump ATPase: the significance of gene subfamilies. Planta 216: 355–365 [DOI] [PubMed] [Google Scholar]
- Axelsen KB, Palmgren MG. (1998) Evolution of substrate specificities in the P-type ATPase superfamily. J Mol Evol 46: 84–101 [DOI] [PubMed] [Google Scholar]
- Axelsen KB, Venema K, Jahn T, Baunsgaard L, Palmgren MG. (1999) Molecular dissection of the C-terminal regulatory domain of the plant plasma membrane H+-ATPase AHA2: mapping of residues that when altered give rise to an activated enzyme. Biochemistry 38: 7227–7234 [DOI] [PubMed] [Google Scholar]
- Banks JA, Nishiyama T, Hasebe M, Bowman JL, Gribskov M, dePamphilis C, Albert VA, Aono N, Aoyama T, Ambrose BA, et al. (2011) The Selaginella genome identifies genetic changes associated with the evolution of vascular plants. Science 332: 960–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc G, Duncan G, Agarkova I, Borodovsky M, Gurnon J, Kuo A, Lindquist E, Lucas S, Pangilinan J, Polle J, et al. (2010) The Chlorella variabilis NC64A genome reveals adaptation to photosymbiosis, coevolution with viruses, and cryptic sex. Plant Cell 22: 2943–2955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briskin DP, Reynolds-Niesman I. (1989) Change in target molecular size of the red beet plasma membrane ATPase during solubilization and reconstitution. Plant Physiol 90: 394–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkle L, Hibberd JM, Quick WP, Kuhn C, Hirner B, Frommer WB. (1998) The H+-sucrose cotransporter NtSUT1 is essential for sugar export from tobacco leaves. Plant Physiol 118: 59–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camoni L, Iori V, Marra M, Aducci P. (2000) Phosphorylation-dependent interaction between plant plasma membrane H+-ATPase and 14-3-3 proteins. J Biol Chem 275: 9919–9923 [DOI] [PubMed] [Google Scholar]
- Chen Y, Hoehenwarter W, Weckwerth W. (2010) Comparative analysis of phytohormone-responsive phosphoproteins in Arabidopsis thaliana using TiO2-phosphopeptide enrichment and mass accuracy precursor alignment. Plant J 63: 1–17 [DOI] [PubMed] [Google Scholar]
- Duby G, Boutry M. (2009) The plant plasma membrane proton pump ATPase: a highly regulated P-type ATPase with multiple physiological roles. Pflugers Arch 457: 645–655 [DOI] [PubMed] [Google Scholar]
- Duby G, Poreba W, Piotrowiak D, Bobik K, Derua R, Waelkens E, Boutry M. (2009) Activation of plant plasma membrane H+-ATPase by 14-3-3 proteins is negatively controlled by two phosphorylation sites within the H+-ATPase C-terminal region. J Biol Chem 284: 4213–4221 [DOI] [PubMed] [Google Scholar]
- Fuglsang AT, Borch J, Bych K, Jahn TP, Roepstorff P, Palmgren MG. (2003) The binding site for regulatory 14-3-3 protein in plant plasma membrane H+-ATPase: involvement of a region promoting phosphorylation-independent interaction in addition to the phosphorylation-dependent C-terminal end. J Biol Chem 278: 42266–42272 [DOI] [PubMed] [Google Scholar]
- Fuglsang AT, Guo Y, Cuin TA, Qiu Q, Song C, Kristiansen KA, Bych K, Schulz A, Shabala S, Schumaker KSet al. (2007) Arabidopsis protein kinase PKS5 inhibits the plasma membrane H+-ATPase by preventing interaction with 14-3-3 protein. Plant Cell 19: 1617–1634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuglsang AT, Visconti S, Drumm K, Jahn T, Stensballe A, Mattei B, Jensen ON, Aducci P, Palmgren MG. (1999) Binding of 14-3-3 protein to the plasma membrane H+-ATPase AHA2 involves the three C-terminal residues Tyr946-Thr-Val and requires phosphorylation of Thr947. J Biol Chem 274: 36774–36780 [DOI] [PubMed] [Google Scholar]
- Goormaghtigh E, Chadwick C, Scarborough GA. (1986) Monomers of the Neurospora plasma membrane H+-ATPase catalyze efficient proton translocation. J Biol Chem 261: 7466–7471 [PubMed] [Google Scholar]
- Harada A, Fukuhara T, Takagi S. (2002a) Photosynthetic control of the plasma membrane H+-ATPase in Vallisneria leaves. II. Presence of putative isogenes and a protein equipped with a C-terminal autoinhibitory domain. Planta 214: 870–876 [DOI] [PubMed] [Google Scholar]
- Harada A, Okazaki Y, Takagi S. (2002b) Photosynthetic control of the plasma membrane H+-ATPase in Vallisneria leaves. I. Regulation of activity during light-induced membrane hyperpolarization. Planta 214: 863–869 [DOI] [PubMed] [Google Scholar]
- Haruta M, Burch HL, Nelson RB, Barrett-Wilt G, Kline KG, Mohsin SB, Young JC, Otegui MS, Sussman MR. (2010) Molecular characterization of mutant Arabidopsis plants with reduced plasma membrane proton pump activity. J Biol Chem 285: 17918–17929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi M, Inoue S, Takahashi K, Kinoshita T. (2011) Immunohistochemical detection of blue light-induced phosphorylation of the plasma membrane H+-ATPase in stomatal guard cells. Plant Cell Physiol 52: 1238–1248 [DOI] [PubMed] [Google Scholar]
- Hayashi Y, Nakamura S, Takemiya A, Takahashi Y, Shimazaki K, Kinoshita T. (2010) Biochemical characterization of in vitro phosphorylation and dephosphorylation of the plasma membrane H+-ATPase. Plant Cell Physiol 51: 1186–1196 [DOI] [PubMed] [Google Scholar]
- Ishizaki K, Chiyoda S, Yamato KT, Kohchi T. (2008) Agrobacterium-mediated transformation of the haploid liverwort Marchantia polymorpha L., an emerging model for plant biology. Plant Cell Physiol 49: 1084–1091 [DOI] [PubMed] [Google Scholar]
- Kanczewska J, Marco S, Vandermeeren C, Maudoux O, Rigaud JL, Boutry M. (2005) Activation of the plant plasma membrane H+-ATPase by phosphorylation and binding of 14-3-3 proteins converts a dimer into a hexamer. Proc Natl Acad Sci USA 102: 11675–11680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerkeb L, Venema K, Donaire JP, Rodríguez-Rosales MP. (2002) Enhanced H+/ATP coupling ratio of H+-ATPase and increased 14-3-3 protein content in plasma membrane of tomato cells upon osmotic shock. Physiol Plant 116: 37–41 [DOI] [PubMed] [Google Scholar]
- Kinoshita T, Doi M, Suetsugu N, Kagawa T, Wada M, Shimazaki K. (2001) Phot1 and phot2 mediate blue light regulation of stomatal opening. Nature 414: 656–660 [DOI] [PubMed] [Google Scholar]
- Kinoshita T, Emi T, Tominaga M, Sakamoto K, Shigenaga A, Doi M, Shimazaki K. (2003) Blue-light- and phosphorylation-dependent binding of a 14-3-3 protein to phototropins in stomatal guard cells of broad bean. Plant Physiol 133: 1453–1463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita T, Hayashi Y. (2011) New insights into the regulation of stomatal opening by blue light and plasma membrane H+-ATPase. Int Rev Cell Mol Biol 289: 89–115 [DOI] [PubMed] [Google Scholar]
- Kinoshita T, Shimazaki K. (1997) Involvement of calyculin A- and okadaic acid- sensitive protein phosphatase in the blue light response of stomatal guard cells. Plant Cell Physiol 38: 1281–1285 [Google Scholar]
- Kinoshita T, Shimazaki K. (1999) Blue light activates the plasma membrane H+-ATPase by phosphorylation of the C-terminus in stomatal guard cells. EMBO J 18: 5548–5558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita T, Shimazaki K. (2001) Analysis of the phosphorylation level in guard-cell plasma membrane H+-ATPase in response to fusicoccin. Plant Cell Physiol 42: 424–432 [DOI] [PubMed] [Google Scholar]
- Kinoshita T, Shimazaki K. (2002) Biochemical evidence for the requirement of 14-3-3 protein binding in activation of the guard-cell plasma membrane H+-ATPase by blue light. Plant Cell Physiol 43: 1359–1365 [DOI] [PubMed] [Google Scholar]
- Lecchi S, Nelson CJ, Allen KE, Swaney DL, Thompson KL, Coon JJ, Sussman MR, Slayman CW. (2007) Tandem phosphorylation of Ser-911 and Thr-912 at the C terminus of yeast plasma membrane H+-ATPase leads to glucose-dependent activation. J Biol Chem 282: 35471–35481 [DOI] [PubMed] [Google Scholar]
- Marten I, Deeken R, Hedrich R, Roelfsema MRG. (2010) Light-induced modification of plant plasma membrane ion transport. Plant Biol (Stuttg) (Suppl 1) 12: 64–79 [DOI] [PubMed] [Google Scholar]
- Matsuzaki M, Misumi O, Shin-I T, Maruyama S, Takahara M, Miyagishima SY, Mori T, Nishida K, Yagisawa F, Nishida K, et al. (2004) Genome sequence of the ultrasmall unicellular red alga Cyanidioschyzon merolae 10D. Nature 428: 653–657 [DOI] [PubMed] [Google Scholar]
- Maudoux O, Batoko H, Oecking C, Gevaert K, Vandekerckhove J, Boutry M, Morsomme P. (2000) A plant plasma membrane H+-ATPase expressed in yeast is activated by phosphorylation at its penultimate residue and binding of 14-3-3 regulatory proteins in the absence of fusicoccin. J Biol Chem 275: 17762–17770 [DOI] [PubMed] [Google Scholar]
- Merchant SS, Prochnik SE, Vallon O, Harris EH, Karpowicz SJ, Witman GB, Terry A, Salamov A, Fritz-Laylin LK, Maréchal-Drouard L, et al. (2007) The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318: 245–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niittylä T, Fuglsang AT, Palmgren MG, Frommer WB, Schulze WX. (2007) Temporal analysis of sucrose-induced phosphorylation changes in plasma membrane proteins of Arabidopsis. Mol Cell Proteomics 6: 1711–1726 [DOI] [PubMed] [Google Scholar]
- Olsson A, Svennelid F, Ek B, Sommarin M, Larsson C. (1998) A phosphothreonine residue at the C-terminal end of the plasma membrane H+-ATPase is protected by fusicoccin-induced 14-3-3 binding. Plant Physiol 118: 551–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmgren MG. (2001) Plant plasma membrane H+-ATPases: powerhouses for nutrient uptake. Annu Rev Plant Physiol Plant Mol Biol 52: 817–845 [DOI] [PubMed] [Google Scholar]
- Palmgren MG, Larsson C, Sommarin M. (1990) Proteolytic activation of the plant plasma membrane H+-ATPase by removal of a terminal segment. J Biol Chem 265: 13423–13426 [PubMed] [Google Scholar]
- Palmgren MG, Sommarin M, Serrano R, Larsson C. (1991) Identification of an autoinhibitory domain in the C-terminal region of the plant plasma membrane H+-ATPase. J Biol Chem 266: 20470–20475 [PubMed] [Google Scholar]
- Portillo F. (2000) Regulation of plasma membrane H+-ATPase in fungi and plants. Biochim Biophys Acta 1469: 31–42 [DOI] [PubMed] [Google Scholar]
- Prochnik SE, Umen J, Nedelcu AM, Hallmann A, Miller SM, Nishii I, Ferris P, Kuo A, Mitros T, Fritz-Laylin LK, et al. (2010) Genomic analysis of organismal complexity in the multicellular green alga Volvox carteri. Science 329: 223–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudashevskaya EL, Ye J, Jensen ON, Fuglsang AT, Palmgren MG. (2012) Phosphosite mapping of P-type plasma membrane H+-ATPase in homologous and heterologous environments. J Biol Chem 287: 4904–4913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano R, Kielland-Brandt MC, Fink GR. (1986) Yeast plasma membrane ATPase is essential for growth and has homology with (Na+ + K+), K+- and Ca2+-ATPases. Nature 319: 689–693 [DOI] [PubMed] [Google Scholar]
- Shimazaki K, Doi M, Assmann SM, Kinoshita T. (2007) Light regulation of stomatal movement. Annu Rev Plant Biol 58: 219–247 [DOI] [PubMed] [Google Scholar]
- Stadler R, Brandner J, Schulz A, Gahrtz M, Sauer N. (1995) Phloem loading by the PmSUC2 sucrose carrier from Plantago major occurs into companion cells. Plant Cell 7: 1545–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahlberg R, Van Volkenburgh E. (1999) The effect of light on membrane potential, apoplastic pH and cell expansion in leaves of Pisum sativum L. var. Argenteum: role of the plasma-membrane H+-ATPase and photosynthesis. Planta 208: 188–195 [Google Scholar]
- Sussman MR. (1994) Molecular analysis of proteins in the plant plasma membrane. Annu Rev Plant Physiol Plant Mol Biol 45: 211–234 [Google Scholar]
- Svennelid F, Olsson A, Piotrowski M, Rosenquist M, Ottman C, Larsson C, Oecking C, Sommarin M. (1999) Phosphorylation of Thr-948 at the C terminus of the plasma membrane H+-ATPase creates a binding site for the regulatory 14-3-3 protein. Plant Cell 11: 2379–2391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sze H, Li X, Palmgren MG. (1999) Energization of plant cell membranes by H+-pumping ATPases: regulation and biosynthesis. Plant Cell 11: 677–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trebst A. (2007) Inhibitors in the functional dissection of the photosynthetic electron transport system. Photosynth Res 92: 217–224 [DOI] [PubMed] [Google Scholar]
- Zhao R, Dielen V, Kinet JM, Boutry M. (2000) Cosuppression of a plasma membrane H+-ATPase isoform impairs sucrose translocation, stomatal opening, plant growth, and male fertility. Plant Cell 12: 535–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.