Abstract
Improvement of strawberry (Fragaria × ananassa) fruit flavor is an important goal in breeding programs. To investigate genetic factors controlling this complex trait, a strawberry mapping population derived from genotype ‘1392’, selected for its superior flavor, and ‘232’ was profiled for volatile compounds over 4 years by headspace solid phase microextraction coupled to gas chromatography and mass spectrometry. More than 300 volatile compounds were detected, of which 87 were identified by comparison of mass spectrum and retention time to those of pure standards. Parental line ‘1392’ displayed higher volatile levels than ‘232’, and these and many other compounds with similar levels in both parents segregated in the progeny. Cluster analysis grouped the volatiles into distinct chemically related families and revealed a complex metabolic network underlying volatile production in strawberry fruit. Quantitative trait loci (QTL) detection was carried out over 3 years based on a double pseudo-testcross strategy. Seventy QTLs covering 48 different volatiles were detected, with several of them being stable over time and mapped as major QTLs. Loci controlling γ-decalactone and mesifurane content were mapped as qualitative traits. Using a candidate gene approach we have assigned genes that are likely responsible for several of the QTLs. As a proof of concept we show that one homoeolog of the O-methyltransferase gene (FaOMT) is the locus responsible for the natural variation of mesifurane content. Sequence analysis identified 30 bp in the promoter of this FaOMT homoeolog containing putative binding sites for basic/helix-loop-helix, MYB, and BZIP transcription factors. This polymorphism fully cosegregates with both the presence of mesifurane and the high expression of FaOMT during ripening.
Fruit flavor is a key characteristic for consumer acceptability and is therefore not surprising that its improvement is receiving increasing importance in strawberry (Fragaria × ananassa) breeding programs. Aroma compounds are key contributors to fruit flavor perception, which relies in a combination of taste, smell, appearance, and texture (Taylor and Hort, 2004). Thus, the volatile composition of strawberry fruits has been extensively studied and more than 360 constituents have been reported, including esters, aldehydes, ketones, alcohols, terpenes, furanones, and sulfur compounds (Latrasse, 1991; Zabetakis and Holden, 1997; Ménager et al., 2004; Jetti et al., 2007). Strawberry aroma increases rapidly as fruit ripens and differs both between species and cultivars due to different quantities and/or combination of many of this complex mixture of compounds (Ulrich et al., 2007). In addition, growing practices, seasonal variations, and storage conditions also affect fruit volatile profile (Zabetakis and Holden, 1997; Forney et al., 2000).
Analysis of the aroma value (the ratio of compound concentration to odor threshold) has indicated that less than 20 compounds contribute significantly to strawberry flavor (Schieberle and Hofmann, 1997; Ulrich et al., 1997; Jetti et al., 2007). Esters, formed by esterification of alcohols and acyl-CoA, constitute the largest and one of the most important groups contributing to the aroma of strawberry fruit (Pérez et al., 1992, 2002). Among them, ethyl 2-methylbutanoate, ethyl and methyl butanoates, ethyl and methyl hexanoates, and hexyl and (E)-2-hexenyl acetates have been reported as key aroma compounds for strawberry fruit, providing green and sweet fruity notes (Schieberle and Hofmann, 1997; Pérez et al., 2002; Ménager et al., 2004). Terpenes, which include linalool, nerolidol, terpineol, or α-pinene, constitute the other important group, providing pleasant citrus and spicy notes and reaching up to 20% of total fruit volatiles in some cultivars of strawberry (Loughrin and Kasperbauer, 2002). Other compounds considered important for strawberry flavor are hexanal, (Z)-3-hexenal, 2-heptanone, and γ-decalactone, the latter being a noteworthy cultivar-specific compound conferring peach (Prunus persica)-like flavor to strawberry fruit (Larsen and Poll, 1992; Larsen et al., 1992; Schieberle and Hofmann, 1997; Ulrich et al., 1997). Yet the two most important contributors to strawberry aroma are furanones and most specifically 2,5-dimethyl-4-hydroxy-3(2H)-furanone (DMHF or furaneol) and 2,5-dimethyl-4-methoxy-3(2H)-furanone (DMMF or mesifurane; Pyysalo et al., 1979; Larsen and Poll, 1992; Pérez et al., 1996; Raab et al., 2006). Furaneol imparts caramel and sweet notes at high concentrations and fruity notes at lower concentrations, whereas mesifurane has been described as having a more burnt, sherry-like, or fusty aroma (Larsen and Poll, 1992; Pérez et al., 1996).
Major progress has been achieved in defining the pathways for volatile biosynthesis in plants and this has resulted in the identification of many genes encoding biosynthetic enzymes (Yamashita et al., 1976; Schwab et al., 2008; Klee, 2010; Osorio et al., 2010; Pérez and Sanz, 2010). A number of these loci have been used in transgenic approaches to engineer volatile production (Beekwilder et al., 2004; Lunkenbein et al., 2006a, 2006b). However, the desired effects have not always been obtained, as regulation of metabolic flux or substrate availability might be important parameters influencing their biosynthetic pathway. The main advantage of quantitative trait loci (QTL) analysis compared with the former approaches is that it allows the identification of loci that, by definition, alter the target trait in natural (and mapping) populations. In addition, QTL analyses can contribute to increase our knowledge of the molecular mechanisms by which aroma compounds are regulated in fruits, which remain largely unknown (Aharoni et al., 2004; Klee, 2010).
High-throughput assays are expensive and technically challenging and, furthermore, volatiles are environmentally influenced to a large extent. Despite existing challenges, a number of studies has addressed the analysis of QTLs affecting fruit volatiles. Tomato (Solanum lycopersicum) is by far the most studied fruit and a number of QTLs have been identified in intraspecific crosses (Causse et al., 2001; Saliba-Colombani et al., 2001; Zanor et al., 2009) and introgression lines generated with crosses with wild relatives such as Solanum pennellii (Schauer et al., 2006; Tieman et al., 2006) and Solanum habrochaites (Mathieu et al., 2009). Other studies have investigated the genetic basis of aroma compounds by a QTL-based approach in apple (Malus domestica; Zini et al., 2005; Dunemann et al., 2009), grape (Vitis vinifera; Doligez et al., 2006; Battilana et al., 2009), rose (Rosa × hybrida) (Spiller et al., 2010; Spiller et al., 2011) or Eucalyptus (O’Reilly-Wapstra et al., 2011). In strawberry, the inheritance patterns of key aroma compounds were examined in segregating populations of strawberry and Fragaria virginiana showing quantitative inheritance, typical of polygenic traits (Carrasco et al., 2005; Olbricht et al., 2008). However, to our knowledge, no loci controlling strawberry volatile compounds have been mapped to date, probably due to its octoploid constitution as well as its susceptibility to inbreeding depression.
The cultivated strawberry is an allo-octoploid species (2n = 8x = 56) originated from the hybridization between two wild octoploid species, Fragaria chiloensis and F. virginiana (Darrow, 1966). A number of cytological genome models have been proposed for the octoploid species, but the most widely accepted to date is that of Bringhurst (1990), who proposed the genomic conformation AAA’A’BBB’B’, which assumes a diploidization of the octoploid Fragaria genomes and disomic inheritance. Up to four diploid ancestors have contributed to the genomes of octoploid strawberries, with an ancestor of Fragaria vesca (2n = 2x = 14) being the maternal donor of the A subgenomes (Rousseau-Gueutin et al., 2009). A high colinearity between the genomes of strawberry and F. vesca has been reported (Rousseau-Gueutin et al., 2008; Sargent et al., 2009; Zorrilla-Fontanesi et al., 2011a). This, together with the availability of a comprehensive genome sequence and annotated gene predictions for the diploid species (Shulaev et al., 2011) will greatly facilitate genetic investigations in the cultivated strawberry.
Mapping of QTLs controlling fruit aroma and volatile levels and subsequent identification of linked molecular markers is an important goal for future marker-assisted selection (MAS) in strawberry. To accelerate the process of QTL identification, and gain insight into the biological mechanism, the candidate gene (CG) approach can be used to identify genes governing the amount of volatile compounds including those contributing to aroma (Pflieger et al., 2001). The aim of this study was to use the linkage maps of cultivated strawberry to locate selected CGs involved in aroma biosynthesis and to identify genomic regions controlling volatile compounds through QTL detection. Because strawberry is a highly heterozygous species, we used a F1 population and a pseudo-backcross strategy to create separate parental maps (Grattapaglia and Sederoff, 1994). The parental lines of the mapping population ‘232’ and ‘1392’ differed, among other traits (Zorrilla-Fontanesi et al., 2011a), in the overall fruit flavor scores annotated during the breeding program of these two selections. In this study, the parental and the 95 progeny lines were phenotypically evaluated for the content of individual volatile compounds in fruit purees using headspace solid phase microextraction coupled to gas chromatography and mass spectrometry (HS-SPME-GC-MS) over four successive years. A high number of major and stable QTLs were detected and cosegregation with CG playing a potential role in the variation of volatile compounds were also found. One of these associations was studied in more detail and expression studies as well as promoter sequence analysis in contrasting lines resulted in the identification of FaOMT as the gene responsible for the variation in mesifurane content, a key compound for strawberry flavor. Overall, this study gives important clues for understanding the genetic basis of aroma/flavor regulation in strawberry fruit.
RESULTS
Volatile Profiling and Analysis of the Variation in the ‘232’ × ‘1392’ Mapping Population
Automated HS-SPME sampling coupled to gas chromatographic separation produced chromatograms with more than 300 distinct peaks for each of the four assessed years. Among them, 87 volatiles including the majority of those previously shown to contribute to the aroma of strawberry could be identified using gas chromatography (GC)-mass spectrometry (MS). Two compounds that have been reported as key for strawberry flavor, ethyl 2-methylbutanoate and furaneol, were not detected. For furaneol, this was due to its water-soluble nature and thermal instability (Pérez et al., 1996). The identified substances included 43 (49.4%) esters, 16 (18.3%) aldehydes, eight (9.2%) alcohols, nine (10.3%) ketones and alkanes, six (6.9%) terpenes, and five (5.7%) furans (Table I). The relative content of these 87 volatiles in fruits of the parents and F1 progeny over 3 of the 4 years (2007–2009), along with their corresponding identification codes and descriptive statistics, are shown in Table I. The parental lines ‘232’ and ‘1392’ displayed similar relative content for several volatile compounds, such as the majority of alcohols and esters, but line ‘1392’ (selected for good flavor) displayed higher relative concentration of aldehydes, ketones, furans, and terpenes. These differences were significant in the 3 years for 15 compounds (Table I). The segregating progeny displayed even higher variation for most of the volatiles, with the exception of compounds 44, 53, 83, and 86 that showed little variation or very low content in the majority of individuals and therefore were not used for QTL analysis.
Table I.
Relative content of the 87 identified volatiles in ripe fruits of the ‘232’ × ‘1392’ population in 3 years
For the F1 progeny, the range of variation is shown. Data are normalized to the concentration of each metabolite in a reference sample containing a mix of all assessed individuals for each year. Volatiles significantly different between the parental lines over the three years (P < 0.05; Student’s t test) are labeled in bold in the parental columns. Volatiles with normal distributions in the progeny are labeled in bold in the corresponding minimum and maximum columns (P ≥ 0.05; Shapiro-Wilks test).
| Volatile Compound | Code | Clustera | Parental Lines |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 232 |
1392 |
F1 Progeny |
|||||||||
| 2007 | 2008 | 2009 | 2007 | 2008 | 2009 | 2007 | 2008 | 2009 | |||
| Alcohols | |||||||||||
| 1-penten-3-ol | 4 | A | 0.622 | 0.710 | 0.398 | 1.863 | 1.209 | 2.579 | 0.072–2.500 | 0.039–3.643 | 0.295–2.528 |
| (E)-2-hexen-1-ol | 17 | A | 0.374 | 0.611 | 0.684 | 0.579 | 0.497 | 1.339 | 0.237–2.777 | 0.030–3.050 | 0.310–4.180 |
| 1-hexanol | 18 | A | 0.049 | 0.635 | 0.737 | 0.617 | 0.784 | 1.231 | 0.195–2.813 | 0.121–3.747 | 0.278–4.693 |
| 2-heptanol | 24 | C | 0.524 | 0.370 | 0.789 | 1.458 | 2.401 | 0.945 | 0.152–8.941 | 0.066–13.739 | 0.080–12.575 |
| 1-octanol | 50 | D | 1.070 | 0.609 | 1.234 | 1.265 | 0.912 | 0.444 | 0.122–3.884 | 0.281–5.067 | 0.304–2.983 |
| 2-nonanolb | 53 | C | 1.000 | 1.000 | 1.000 | 1.000 | 20.029 | 1.000 | 1.000–1.000 | 0.020–12.851 | 0.359–11.302 |
| 1-decanol | 70 | C | 3.071 | 1.000 | 0.887 | 8.486 | 1.000 | 0.359 | 0.602–21.860 | 0.048–8.095 | 0.131–23.992 |
| Eugenol | 74 | D | 0.761 | 1.359 | 1.197 | 0.824 | 1.000 | 0.669 | 0.583–13.480 | 0.195–7.127 | 0.115–6.346 |
| Aldehydes | |||||||||||
| Pentanal | 7 | B | 0.603 | 0.691 | 0.701 | 0.905 | 1.332 | 1.814 | 0.281–1.596 | 0.217–2.081 | 0.343–1.997 |
| (E)-2-pentenal | 9 | B | 0.298 | 0.848 | 0.563 | 0.937 | 1.696 | 2.814 | 0.097–3.621 | 0.070–18.026 | 0.127–3.059 |
| (Z)-3-hexenal | 11 | B | 0.924 | 0.957 | 0.634 | 0.594 | 1.246 | 1.599 | 0.264–1.397 | 0.016–2.702 | 0.596–2.552 |
| Hexanal | 12 | B | 0.907 | 0.988 | 0.865 | 0.954 | 1.181 | 1.077 | 0.448–1.567 | 0.044–1.656 | 0.569–2.068 |
| (E)-2-hexenal | 16 | B | 0.882 | 0.991 | 0.871 | 1.093 | 1.109 | 1.358 | 0.403–1.533 | 0.299–1.625 | 0.574–1.794 |
| Heptanal | 25 | B | 0.918 | 0.995 | 0.948 | 0.681 | 0.852 | 0.985 | 0.223–1.576 | 0.058–1.880 | 0.500–2.098 |
| (E)-2-heptenal | 29 | B | 0.750 | 0.796 | 0.736 | 1.139 | 1.296 | 1.476 | 0.272–1.555 | 0.017–2.091 | 0.562–1.816 |
| Benzaldehyde | 31 | C | 0.569 | 0.601 | 0.710 | 0.593 | 1.252 | 0.999 | 0.104–1.618 | 0.357–2.858 | 0.349–1.985 |
| (E, Z)-2,4-heptadienal | 37 | A | 0.537 | 0.769 | 0.620 | 0.107 | 1.263 | 1.526 | 0.107–1.897 | 0.060–13.740 | 0.353–1.837 |
| Octanal | 39 | B | 0.895 | 0.979 | 1.057 | 0.767 | 0.752 | 0.811 | 0.233–1.977 | 0.244–2.100 | 0.302–2.696 |
| (E)-2-octenal | 49 | B | 0.775 | 0.710 | 0.819 | 1.292 | 1.249 | 1.366 | 0.187–1.474 | 0.304–1.759 | 0.606–1.915 |
| Nonanal | 55 | B | 0.932 | 0.767 | 1.191 | 0.819 | 0.647 | 0.615 | 0.159–1.651 | 0.247–1.370 | 0.514–2.399 |
| (E)-2-nonenal | 58 | D | 0.652 | 0.407 | 1.082 | 1.275 | 0.481 | 0.687 | 0.159–1.874 | 0.239–1.211 | 0.402–2.491 |
| Decanal | 64 | B | 0.823 | 0.655 | 1.208 | 0.982 | 0.630 | 0.901 | 0.135–1.274 | 0.222–1.372 | 0.551–2.490 |
| 3,4-dimethylbenzaldehyde | 67 | B | 0.302 | 1.358 | 1.074 | 0.418 | 0.612 | 1.033 | 0.209–1.189 | 0.057–4.166 | 0.226–2.188 |
| (E)-2-decenal | 69 | B | 1.345 | 1.349 | 1.587 | 1.524 | 0.816 | 0.777 | 0.284–3.154 | 0.261–2.562 | 0.350–2.879 |
| Esters | |||||||||||
| Ethyl acetate | 2 | C | 12.236 | 1.000 | 0.163 | 0.145 | 0.821 | 0.447 | 0.203–16.028 | 0.428–9.797 | 0.092–29.278 |
| 1-methylethyl acetate | 3 | A | 0.837 | 1.464 | 0.701 | 1.601 | 1.008 | 0.968 | 0.080–7.372 | 0.023–4.579 | 0.093–29.858 |
| Methyl butanoate | 8 | C | 3.690 | 1.257 | 0.522 | 1.829 | 1.308 | 0.841 | 0.058–5.423 | 0.001–4.222 | 0.186–10.643 |
| Ethyl butanoate | 10 | D | 7.746 | 0.338 | 0.363 | 0.227 | 1.073 | 0.928 | 0.041–22.575 | 0.110–4.667 | 0.116–7.559 |
| Butyl acetate | 13 | D | 2.049 | 0.914 | 0.479 | 0.840 | 1.229 | 0.385 | 0.018–13.605 | 0.058–7.321 | 0.044–5.833 |
| Methyl pentanoate | 14 | C | 1.454 | 1.176 | 0.754 | 0.994 | 1.064 | 1.105 | 0.281–4.385 | 0.262–3.536 | 0.296–5.426 |
| 1-methylethyl butanoate | 15 | C | 2.473 | 0.926 | 0.594 | 1.837 | 3.808 | 0.962 | 0.019–8.425 | 0.114–5.180 | 0.194–7.043 |
| 3-methylbutyl acetate | 19 | A | 2.321 | 1.949 | 0.547 | 0.397 | 0.374 | 0.324 | 0.051–7.114 | 0.136–5.864 | 0.074–8.688 |
| 2-methylbutyl acetate | 20 | A | 3.935 | 2.211 | 1.131 | 0.441 | 0.354 | 0.379 | 0.085–5.991 | 0.100–5.590 | 0.118–4.294 |
| S-methyl thiobutanoate | 22 | D | 3.057 | 1.757 | 0.983 | 1.402 | 1.722 | 0.703 | 0.049–4.121 | 0.049–8.568 | 0.172–4.143 |
| Propyl butanoate | 23 | D | 1.441 | 0.499 | 0.343 | 1.144 | 1.964 | 0.771 | 0.171–8.500 | 0.136–5.227 | 0.078–6.757 |
| Pentyl acetate | 26 | A | 1.324 | 0.740 | 0.551 | 0.737 | 0.944 | 0.868 | 0.257–2.937 | 0.131–10.126 | 0.091–2.873 |
| 3-methyl-2-butenyl acetate | 27 | A | 3.597 | 3.297 | 1.358 | 0.329 | 0.257 | 0.349 | 0.168–5.764 | 0.174–6.340 | 0.158–3.395 |
| Methyl hexanoate | 28 | C | 1.656 | 0.930 | 0.687 | 1.556 | 1.230 | 0.846 | 0.058–3.771 | 0.218–2.925 | 0.264–2.408 |
| Methyl 2-hexenoate | 30 | C | 0.937 | 0.954 | 1.805 | 1.918 | 2.077 | 0.821 | 0.103–4.634 | 0.090–6.064 | 0.125–6.660 |
| Butyl butanoate | 33 | D | 1.403 | 0.541 | 1.517 | 0.818 | 2.236 | 0.880 | 0.003–7.280 | 0.008–7.608 | 0.020–6.414 |
| Ethyl hexanoate | 35 | C | 2.429 | 0.241 | 0.460 | 0.298 | 0.950 | 0.796 | 0.021–5.776 | 0.112–3.134 | 0.104–4.187 |
| (Z)-3-hexenyl acetate | 38 | A | 0.608 | 0.568 | 0.696 | 0.508 | 0.439 | 1.435 | 0.198–4.270 | 0.069–3.240 | 0.240–2.620 |
| Hexyl acetate | 40 | A | 0.761 | 0.610 | 0.661 | 0.854 | 0.986 | 1.071 | 0.262–3.202 | 0.019–3.071 | 0.418–2.137 |
| (E)-2-hexenyl acetate | 41 | A | 0.733 | 0.544 | 0.643 | 0.607 | 0.540 | 1.274 | 0.321–2.719 | 0.121–3.249 | 0.195–2.511 |
| 1-methylbutyl butanoate | 42 | C | 0.754 | 0.225 | 0.807 | 1.141 | 6.856 | 0.880 | 0.267–9.140 | 0.088–12.325 | 0.097–10.870 |
| 1-methylethyl hexanoate | 43 | C | 0.836 | 0.576 | 0.660 | 2.927 | 3.368 | 1.081 | 0.033–6.602 | 0.117–5.168 | 0.115–3.795 |
| 1-methylhexyl acetate | 44 | C | 1.000 | 1.000 | 1.000 | 1.591 | 1.000 | 1.000 | 0.909–4.822 | 1.000–1.000 | 0.712–11.688 |
| Ethyl 2-hexenoate | 45 | C | 1.520 | 1.000 | 0.214 | 0.851 | 1.000 | 1.000 | 0.814–14.064 | 1.000–18.834 | 0.177–13.146 |
| Methyl benzoate | 56 | A | 0.285 | 0.829 | 0.592 | 1.279 | 1.336 | 1.361 | 0.199–6.001 | 0.200–7.413 | 0.274–5.795 |
| Methyl octanoate | 57 | C | 0.588 | 0.798 | 0.790 | 2.109 | 0.870 | 0.603 | 0.035–5.370 | 0.147–2.421 | 0.151–3.333 |
| Benzyl acetate | 59 | A | 0.657 | 0.887 | 0.546 | 1.206 | 0.963 | 1.726 | 0.056–3.728 | 0.209–4.430 | 0.172–3.746 |
| Butyl hexanoate | 60 | D | 0.791 | 0.373 | 1.387 | 1.122 | 1.482 | 0.506 | 0.006–8.239 | 0.006–6.859 | 0.028–4.461 |
| Hexyl butanoate | 61 | D | 0.584 | 0.454 | 1.371 | 1.030 | 2.354 | 0.684 | 0.012–3.956 | 0.015–3.591 | 0.034–5.575 |
| Ethyl octanoate | 62 | C | 1.150 | 0.210 | 0.456 | 0.295 | 0.685 | 0.596 | 0.068–4.628 | 0.037–3.511 | 0.055–4.607 |
| Octyl acetate | 63 | D | 1.029 | 1.010 | 1.021 | 1.464 | 0.709 | 0.340 | 0.018–6.691 | 0.012–4.535 | 0.023–3.974 |
| Nonyl acetate | 71 | D | 0.869 | 0.815 | 1.268 | 1.418 | 0.905 | 0.609 | 0.227–3.932 | 0.179–3.453 | 0.172–3.294 |
| Methyl decanoate | 72 | D | 0.345 | 1.000 | 0.868 | 1.788 | 1.069 | 0.786 | 0.127–2.908 | 0.029–3.022 | 0.120–2.804 |
| (−)-myrtenyl acetate | 73 | B | 0.665 | 0.417 | 0.306 | 0.652 | 0.608 | 2.706 | 0.515–4.887 | 0.093–3.723 | 0.203–6.252 |
| Hexyl hexanoate | 75 | D | 0.323 | 0.366 | 1.087 | 0.835 | 1.736 | 0.629 | 0.008–8.606 | 0.013–3.479 | 0.039–5.973 |
| Octyl butanoate | 76 | D | 0.965 | 0.538 | 2.210 | 1.041 | 1.331 | 0.568 | 0.001–5.399 | 0.003–3.403 | 0.009–4.541 |
| Ethyl decanoate | 77 | D | 0.662 | 0.126 | 0.471 | 0.686 | 0.431 | 0.760 | 0.507–17.314 | 0.116–21.490 | 0.103–10.533 |
| 1-methyloctyl butanoate | 78 | D | 0.431 | 0.750 | 2.280 | 0.407 | 1.413 | 0.966 | 0.051–15.578 | 0.007–7.257 | 0.128–17.682 |
| Decyl acetate | 79 | D | 0.501 | 0.913 | 0.956 | 1.359 | 0.798 | 0.419 | 0.088–11.174 | 0.029–4.815 | 0.046–8.255 |
| Cinnamyl acetate | 81 | D | 0.963 | 1.200 | 1.669 | 0.655 | 3.967 | 0.103 | 0.251–20.895 | 0.041–11.486 | 0.052–4.706 |
| Methyl dodecanoateb | 83 | D | 1.227 | 0.268 | 1.000 | 1.227 | 1.912 | 1.000 | 0.831–2.711 | 0.060–6.497 | 1.000–10.216 |
| Octyl hexanoate | 85 | D | 0.473 | 0.317 | 1.042 | 1.136 | 1.170 | 0.332 | 0.003–8.965 | 0.002–5.185 | 0.008–6.097 |
| Ethyl dodecanoateb | 86 | B | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 0.659 | 1.000–1.000 | 1.000–1.704 | 0.498–11.232 |
| Ketones and alkane | |||||||||||
| Acetone | 1 | D | 2.036 | 0.343 | 0.720 | 1.155 | 1.366 | 0.898 | 0.041–7.729 | 0.014–5.294 | 0.120–8.821 |
| 1-penten-3-one | 5 | B | 0.256 | 0.674 | 0.410 | 1.059 | 1.906 | 2.760 | 0.055–2.309 | 0.032–2.231 | 0.260–2.822 |
| 2-pentanone | 6 | C | 0.492 | 0.457 | 0.530 | 1.530 | 1.568 | 0.991 | 0.250–3.919 | 0.106–9.864 | 0.173–4.278 |
| 2-heptanone | 21 | D | 0.459 | 0.366 | 0.904 | 1.061 | 1.510 | 0.987 | 0.042–4.084 | 0.030–3.487 | 0.132–3.233 |
| 6-methyl-5-hepten-2-one | 32 | C | 0.546 | 0.453 | 0.966 | 0.888 | 0.746 | 1.207 | 0.202–1.546 | 0.350–1.581 | 0.523–1.825 |
| Acetophenone | 51 | B | 1.420 | 0.825 | 0.487 | 1.579 | 1.154 | 0.472 | 0.393–5.538 | 0.213–2.401 | 0.259–1.235 |
| 2-nonanone | 52 | D | 0.758 | 0.280 | 0.862 | 0.711 | 3.173 | 0.732 | 0.218–3.805 | 0.060–4.283 | 0.182–2.679 |
| (Z)-geranylacetone | 80 | D | 0.569 | 0.746 | 1.850 | 0.994 | 0.776 | 1.884 | 0.213–2.130 | 0.261–2.304 | 0.407–4.058 |
| Decane | 36 | B | 0.624 | 2.101 | 3.733 | 0.214 | 0.579 | 1.000 | 0.128–2.085 | 0.047–3.091 | 0.263–10.157 |
| Furans | |||||||||||
| 2-pentylfuran | 34 | B | 0.809 | 0.732 | 0.798 | 1.021 | 1.113 | 1.108 | 0.377–1.710 | 0.269–2.078 | 0.464–1.811 |
| 2-(1-pentenyl)furan | 47 | B | 0.531 | 0.791 | 0.760 | 1.045 | 0.851 | 1.177 | 0.166–2.299 | 0.086–1.951 | 0.357–2.213 |
| Mesifurane | 48 | C | 0.375 | 0.368 | 0.830 | 0.609 | 1.986 | 0.768 | 0.003–4.815 | 0.001–4.486 | 0.002–4.728 |
| γ-decalactone | 82 | D | 0.007 | 0.011 | 0.027 | 2.131 | 2.059 | 1.167 | 0.000–4.456 | 0.000–4.300 | 0.002–4.028 |
| γ-dodecalactone | 87 | D | 0.452 | 0.563 | 1.512 | 0.842 | 1.142 | 0.493 | 0.003–7.154 | 0.008–5.380 | 0.001–4.095 |
| Terpenes | |||||||||||
| Limonene | 46 | B | 1.671 | 0.269 | 0.449 | 0.485 | 0.587 | 1.806 | 0.116–3.604 | 0.075–5.797 | 0.298–2.073 |
| Linalool | 54 | C | 0.625 | 0.414 | 0.399 | 2.242 | 1.704 | 1.910 | 0.189–2.357 | 0.355–1.879 | 0.394–2.072 |
| Terpineol | 65 | C | 0.336 | 0.404 | 0.599 | 2.137 | 2.449 | 1.825 | 0.069–3.177 | 0.125–2.913 | 0.376–2.717 |
| Myrtenol | 66 | D | 0.529 | 0.081 | 0.155 | 4.850 | 3.662 | 2.413 | 0.381–10.168 | 0.081–13.460 | 0.133–17.472 |
| Nerol | 68 | D | 0.544 | 0.079 | 0.563 | 4.349 | 6.667 | 1.309 | 0.412–6.530 | 0.079–15.292 | 0.160–3.662 |
| Nerolidol | 84 | C | 0.260 | 0.111 | 0.933 | 1.493 | 1.598 | 0.641 | 0.019–4.488 | 0.080–2.532 | 0.077–3.415 |
Clusters according to Supplemental Figure S3. bThese volatiles showed little variation in the progeny in most of the analyzed years and were not used in the QTL analysis.
Most of the identified compounds showed continuous variation, typical of polygenic inheritance, although their distributions were generally skewed toward low values and/or were significantly deviated from normality (α = 5%; Table I). Transgressive segregation occurred in both directions and volatiles 2, 24, 42, 45, 66, and 70 showed the most transgressive behavior, with a relative range of variation between approximately 0.05 and 29.30 along the 3 years (Table I). The Shapiro-Wilk test (Shapiro and Wilk, 1965) was used to verify the normality of trait distributions and several transformations were applied to those traits nonnormally distributed (see “Materials and Methods”). The transformation that best fitted to normality was employed in the subsequent QTL analysis.
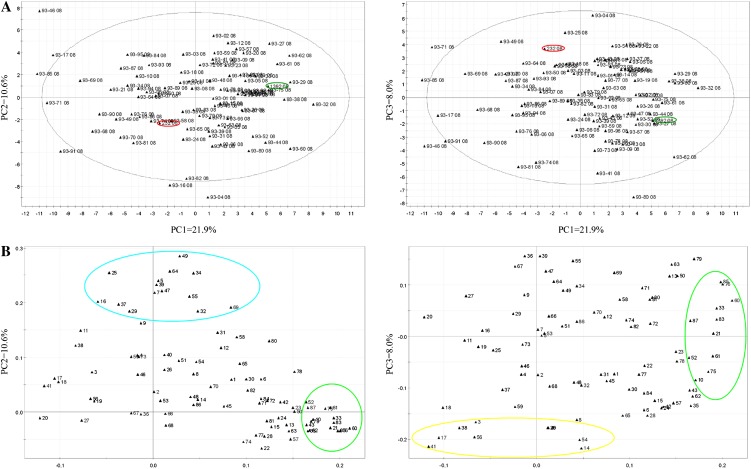
Principal component analysis (PCA) was used to analyze the differences in volatile profiles in the population during the four seasons under study. Although the distribution of individuals and the principal component weights varied for each season, similar conclusions could be obtained for each year and therefore only the results for the chosen vectors, displayed in two-dimensional plots for the year 2008 are shown (Fig. 1). As observed in the other years, individual lines were distributed evenly without forming clear clusters. The parental lines are well separated in both biplots, indicating that they contain relatively different volatile profiles (Fig. 1A). Principal component representations of loadings indicated that ethyl, butyl, hexyl, and octyl esters of hexanoic and butanoic acids (compounds 35, 60, 75, 85, 10, 33, 61, and 76), along with 2-nonanone, 2-heptanone, and γ-dodecalactone were important for the separation of lines across PC1 (Fig. 1B). The second PC separated the lines primarily on the basis of the concentration of most aldehydes (7, 9, 16, 25, 29, 37, 39, 49, 55, 64, and 69), two furans (34 and 47), and two ketones (5 and 32). Finally, concentrations of 1-methylethyl, pentyl, and hexyl acetates (3, 26, and 40), both (E)-2- and (Z)-3-hexenyl acetates (38 and 41), methyl pentanoate (14), methyl benzoate (56), and (E)-2-hexenol (17) accounted in the variation of volatile profiles across the PC3.
Figure 1.
PCA of the ‘232’ × ‘1392’ volatile profiles in year 2008. A, Points show the PCA scores of each line. ‘232’ and ‘1392’ parental lines are indicated in red and green, respectively. B, Loading plots of PC1 and PC2 (left) and PC1 and PC3 (right). Volatile compounds that accounted most for the variability of aroma profiles across PC1, PC2, and PC3 are highlighted in green, blue, and yellow circles, respectively. For volatile codes see Table I. PC1, PC2, and PC3: First, second, and third principal components, respectively. [See online article for color version of this figure.]
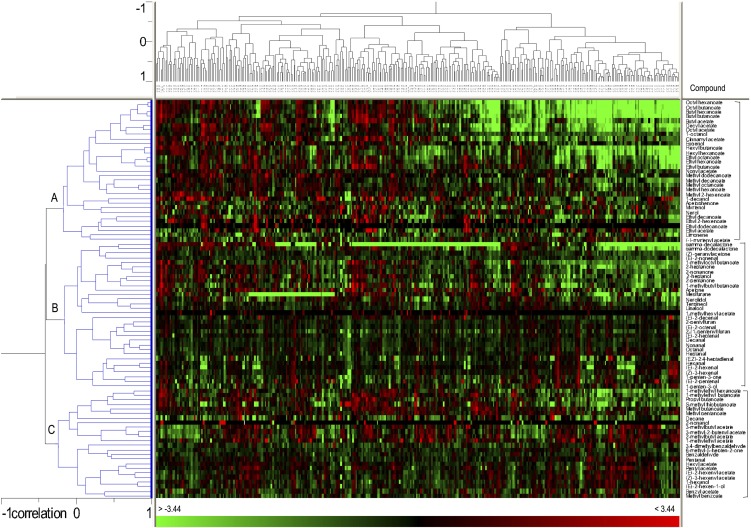
Hierarchical cluster analysis (HCA) using the results from the four seasons of volatile profiles of parental lines as well as those of the F1 progeny was used to further investigate the relationship between both compounds and individuals of the population (Fig. 2). This analysis grouped the volatiles into three distinct clusters (A–C), each of them containing biosynthetically related aroma compounds. This result validates our analysis and further reveals the complex metabolic network underlying volatile production in strawberry fruit. Cluster A grouped approximately 36% of the identified volatiles and was enriched in esters, which are quantitatively the main contributors to the aroma of strawberry fruit. Cluster A also included three alcohols; 1-decanol (70), 1-octanol (50), and eugenol (74) and the terpene alcohols myrtenol (66) and nerol (68; Fig. 2). Cluster B included approximately 36% of the identified volatiles and it contained a subcluster with the majority of aldehydes, such as (E)-2-heptenal (29), (E)-2-octenal (49), nonanal (55), decanal (64), and (E)-2-decenal (69; Fig. 2). The other subcluster B contained a more diverse set of compounds including three monoterpene alcohols, linalool, terpineol, and nerolidol (54, 65, and 84); furans, such as mesifurane, γ-decalactone, and γ-dodecalactone (48, 82, and 87), and ketones, such as acetone, 2-pentanone, 2-heptanone, or 2-nonanone (1, 6, 21, and 52; Fig. 2). Cluster C grouped about 28% of the identified volatiles and included mainly esters of acetic acid, such as 1-methylethyl, pentyl, hexyl, and benzyl acetates (3, 26, 40, and 59), 2-methylbutyl acetate (20), (Z)-3-hexenyl acetate (38) or (E)-2-hexenyl acetate (41), 1-hexanol (18), and four esters of butanoic acid (Fig. 2). Clustering of esters and alcohols is an expected result, because esters are enzymatically synthesized by coupling the respective acids and alcohols (Yamashita et al., 1977). Volatile profiles displayed considerable variation between different years (Table I), suggesting an important influence of environmental factors. However, in the HCA samples from the same line in the four different years were in general closely associated, indicating that, despite a clear environmental influence, the genotypic variation may be sufficient for QTL detection (Fig. 2; Supplemental Fig. S2).
Figure 2.
HCA and heat map representation of volatile profiles in ‘232’ × ‘1392’ over four successive years (2006–2009). Individuals with a relative content for a given compound similar, lower, or higher than that of the reference sample are shown in black, green, or red, respectively. Clusters of volatiles are indicated by different letters. [See online article for color version of this figure.]
Cluster Validation Using Correlation Analyses
For a deeper understanding of the production of volatile compounds in strawberry, a correlation-based approach was adopted, which has been shown as a useful tool to gain insight into metabolic pathways and networks (Raamsdonk et al., 2001; Weckwerth et al., 2004). To identify coregulated compounds in the population, the pairwise correlation for each volatile was analyzed against every other volatile. Pearson correlation coefficients were calculated for year 2008 (Supplemental Table S1) and the corresponding heat map representation and HCA is shown in Supplemental Figure S3. Of the 7,569 possible pairs analyzed, 2,558 resulted in significant correlations (P < 0.05). Of these pairs, most of them (2,176) showed positive correlation coefficients and only 382 showed negative correlation coefficients. The highest negative correlations were found between 2-methylbutyl acetate (20) and other three volatiles: hexyl hexanoate (75; r = −0.52), ethyl butanoate (10; r = −0.39), and nerolidol (84; r = −0.38). Negative correlations were also found between alcohols and esters, as for example between 1-hexanol (18) and butyl hexanoate (60; r = −0.30) or octyl hexanoate (85; r = −0.29). By contrast, high positive correlations were found between the alcohols (E)-2-hexen-1-ol (17) and 1-hexanol (18; r = 0.86), between the esters (Z)-3-hexenyl acetate (38) and (E)-2-hexenyl acetate (41; r = 0.78), between the aldehydes (E)-2-heptenal (29) and (E)-2-octenal (49; r = 0.87) or nonanal (55) and decanal (64; r = 0.81) and between a group of 12 esters, whose correlation coefficients ranged between 0.65 and 0.85. As previously reported in tomato (Zanor et al., 2009), a high positive correlation was found between the terpenes linalool (54) and terpineol (65; r = 0.77). Because all these strong pairwise correlations involve volatiles that share a common structure and belong to the same family, a likely explanation is that they are in the same biochemical (biosynthetic) pathway and/or display mutual control by a single enzyme. However, it should be noted that nonneighboring metabolites may be highly correlated and, by contrast, metabolites that participate in common reactions may not always exhibit significant correlation (Steuer et al., 2003; Camacho et al., 2005).
HCA based on pairwise correlations distinguished four major clusters of biosynthetically related compounds, which were highly similar to those found in the HCA of lines and metabolites. Some of them could be divided into minor subclusters containing volatiles with remarkable associations from a metabolic point of view (Supplemental Fig. S3). The first cluster (A) contained mainly esters of acetic acid and alcohols, such as (E)-2-hexen-1-ol (17) or 1-hexanol (18), and comprised approximately 16% of the identified volatiles (Supplemental Fig. S3). The second large group (cluster B) contained approximately 24% of the identified compounds and it essentially included aldehydes, such as (E)-2-heptenal (29), (E)-2-octenal (49), nonanal (55), decanal (64), or (E)-2-decenal (69; Supplemental Fig. S3). Cluster C comprised about 26% of the volatiles, with a subcluster including the high and positively correlated terpenes linalool (54) and terpineol (65; Supplemental Fig. S3). Finally, the other subcluster C and cluster D contained ketones, furans, and the majority of esters (Supplemental Fig. S3). In theory, compounds derived from the same precursor or synthesized by the same enzyme should cluster together and therefore, QTLs controlling the variation of one of them may also control the variation of the others.
Genetic Mapping of QTLs Controlling Aroma Compounds in Strawberry Fruits
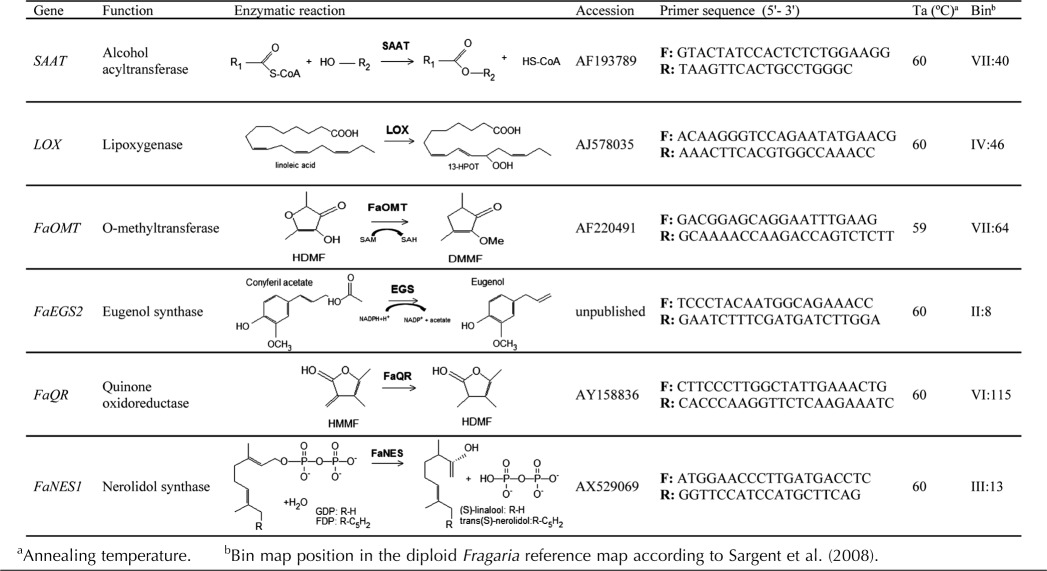
For the QTL analyses, additional markers, including a number derived from aroma CGs (Table II), were added to the established maps of the ‘232’ × ‘1392’ mapping population (Zorrilla-Fontanesi et al., 2011a). The ‘232’ map had 253 markers that were distributed in 40 linkage groups (LGs) with a cumulative length of 910 centimorgans (cM; Table III). The ‘1392’ map consisted of 227 markers distributed in 36 LGs with a cumulative length of 869 cM (Table III). The integrated map included 363 markers distributed in 39 LGs and spanned a cumulative length of 1,400 cM (Table III; Supplemental Fig. S1). Combined, the ‘232’ × ‘1392’ map spanned 63.8% of he published octoploid reference map (Rousseau-Gueutin et al., 2008). The markers mapped in this study, including CG markers, displayed complete synteny with the diploid F. vesca reference map (Supplemental Fig. S1).
Table II.
List of the CGs positioned onto the ‘232’ × ‘1392’ and diploid Fragaria reference maps
Table III.
Description of parental and integrated linkage maps of octoploid strawberry using single-dose markers coded as CP (for cross pollinated) in JoinMap 4
| Description of the Linkage Maps | Female Map | Male Map | Integrated Map |
|---|---|---|---|
| SSR | 184 | 157 | 255 |
| Amplified fragment length polymorphism | 53 | 57 | 83 |
| Sequence-tagged site and SSCP | 16 | 13 | 25 |
| Total | 253 | 227 | 363 |
| Total no. of LGs | 40 | 36 | 39 |
| Mean no. of markers per LG (±sd) | 6.3 (±3.8) | 6.3 (±4.5) | 7.9 (±5.3) |
| Range of marker no. per LG | 2–16 | 2-17 | 2-20 |
| Average marker spacing (cM) | 4.3 | 4.6 | 4.4 |
| Mean size per LG (cM ± sd) | 22.7 (±20.1) | 24.1 (±18.5) | 30.4 (±21.4) |
| Range size per LG (cM) | 0.0–84.5 | 0.0–65.1 | 2.4–84.5 |
| Largest gap (cM) | 31.7 | 33.7 | 33.7 |
| Cumulative genome length (cM) | 909.5 | 868.5 | 1,400.1 |
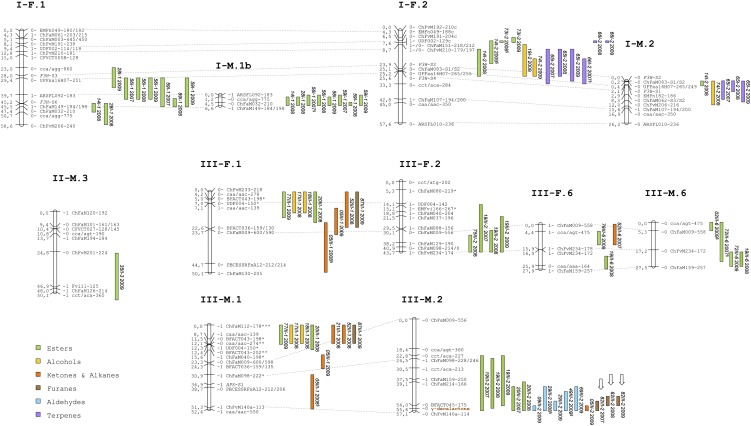
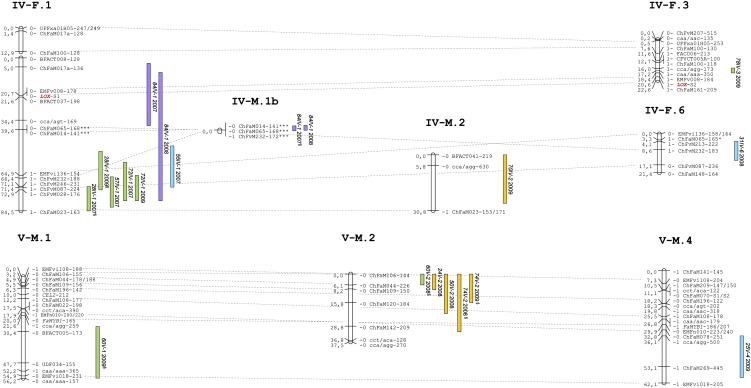
A total of 194 significant associations were found between markers and phenotypes (Supplemental Tables S2–S4; Fig. 3). Because QTLs for each trait detected in the same chromosomal regions (with overlapping confidence intervals) in different years or in both parental maps were considered to be the same, the 194 significant associations can be summarized into 70 QTLs for 48 different volatile compounds. Among them, 35 (50.0%) QTLs were stable over two or all three assessed years and are indicated in bold in Supplemental Tables S2 to S4.
Figure 3.
Location of QTLs controlling aroma compounds analyzed in three consecutive years (2007–2009) using IM. Only LGs including QTLs are shown. Color bars represent 1-LOD support intervals. Location of loci controlling γ-decalactone and mesifurane are highlighted in brown and the QTLs associated to them are indicated by arrows. Segregation distortion is indicated by *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001. §, QTLs detected below the threshold but significant in other years. [See online article for color version of this figure.]
QTLs were identified across the seven homoeology groups (HGs) of the ‘232’ × ‘1392’ maps, ranging from one QTL for ester 35 in HG II to 22 QTLs for different esters, alcohols, aldehydes, ketones, and furans in HG III. With the exception of HG II, clusters of QTLs were detected in all HGs, indicating linkage or pleiotropic effects of loci. The largest cluster was found in female and male LG VI-1, which comprised 16 QTLs for 11 esters, two alcohols, two terpenes, and one ketone. Several clusters of QTLs involved volatiles that were grouped in the HCA and/or were significantly correlated, i.e. QTLs for esters in LG I-1 (14 and 28) and in LG VI-1 (13, 33, 60, 63, 76, 79, and 85), QTLs for alcohols in LG III-1 (17, 18) and in LG V-2 (50 and 74), QTLs for aldehydes in LG III-2 (9, 29, 49, and 69), and QTLs for the terpenes linalool (54) and terpineol (65) in LG VI-1 and VI-2 (Supplemental Tables S1–S4; Figs. 2 and 3). Another cluster of QTLs controlling terpenes was found in LG I-F.2 comprising terpineol (65) and myrtenol (66), although they were not significantly correlated (Fig. 3; Supplemental Tables S1 and S3; Supplemental Fig. S3). QTLs for γ-decalactone (82III-2) and γ-dodecalactone (87III-1), two volatiles significantly correlated (r = 0.42) and clustered in the HCA, did not colocate in the same LG and were mapped to different homoeologous LGs of HG III (Supplemental Tables S1 and S4; Figs. 2–3). In agreement with their common enzymatic reaction, QTLs for different esters frequently colocated with QTLs controlling alcohols. They clustered in HG I, HG III, HG V, and HG VI (Fig. 3). As an example, a QTL for 1-octanol in HG VI (50VI-1) stable in all three years colocated with QTLs for octyl acetate (63VI-1), octyl butanoate (76VI-1), and octyl hexanoate (85VI-1), also stable in all three years. These volatiles grouped together in the HCA and were significantly correlated (Fig. 2; Supplemental Fig. S3; Supplemental Table S1).
Between one and three QTLs have been identified per volatile trait, with the percentage of phenotypic variation (R2) explained by each QTL ranging from 14.2% to 92.8% (Supplemental Tables S2–S4). One major QTL was detected for 31 volatiles and between two and three QTLs were detected for the remaining 17 compounds. This high proportion of major QTLs suggests that variation in strawberry fruit aroma is regulated by a limited set of loci with a high effect rather than multiple loci with reduced effects, in contrast to the regulation of other agronomic and fruit quality traits (Zorrilla-Fontanesi et al., 2011a). Mesifurane (48) and γ-decalactone (82) displayed qualitative variation suggestive of single locus inheritance. Mesifurane was detected in fruits of both parental lines and about 25% of the progeny presented concentrations of this compound close to the detection limit in the four seasons (expected 3:1 ratio; P = 0.36). In contrast to mesifurane, γ-decalactone was present in fruits of only one parental line, ‘1392’, and the observed segregation pattern in the progeny matched the expected 1:1 ratio (P = 0.76). The two traits were scored as presence or absence in the segregating population and mapped accordingly. Mesifurane and γ-decalactone were also mapped as QTL and shown to colocalize to LG VII-F/M.2 and LG III-M.2, respectively (Fig. 3; Supplemental Fig. S1). For γ-decalactone, the QTL explained about 90% of the total variance, corroborating our single gene hypothesis (Supplemental Table S4). The detected QTL for mesifurane explained from 42% up to 67.3% of the phenotypic variance, indicating a strong effect of this locus in the control of total variation (Supplemental Table S4).
Association of CGs with Aroma QTLs
With the exception of FaOMT (see below), none of the aroma CGs mapped in this study (Table II) located within the QTL intervals, or if they colocated they were not functionally related to the corresponding volatiles. However, most of the markers used in the elaboration of these maps were derived from ESTs obtained from strawberry fruit. This opens the possibility of identifying CGs that account for the QTL. Among those markers, and hence genes that could be responsible for the variation in related compounds, we found marker ChFaM149, located in the ripening up-regulated cinnamyl alcohol dehydrogenase (CAD) gene (Blanco-Portales et al., 2002). This gene mapped to the log of the odds (LOD) peak or within the confidence interval of QTLs for methyl hexanoate, pentanoate, benzoate, and benzyl acetate (14I-1, 28I-1, 56I-1, and 59I-1; Supplemental Table S2; Fig. 3). Two other ESTs within the confidence intervals of QTLs have homology to transcription factors with possible regulatory effects. One of these markers was ChFaM083 (in a putative zinc-binding transcription factor) mapped to LG I-2, inside the confidence interval of QTLs controlling esters (14I-2, 19I-2, and 73I-2), eugenol (74I-2), and terpenes (65I-2 and 66I-2), all of them stable in two or all three years (Supplemental Tables S2 and S3; Fig. 3). The second marker was ChFaM109 (in a putative transcriptional activator), mapped within the confidence interval of QTLs controlling butyl hexanoate (60V-2) and alcohols 2-heptanol, 1-hexanol, and eugenol (24V-2, 50V-2, and 74V-2; Supplemental Tables S2 and S3; Fig. 3).
One Homoeolog of FaOMT Controls the Production of Mesifurane
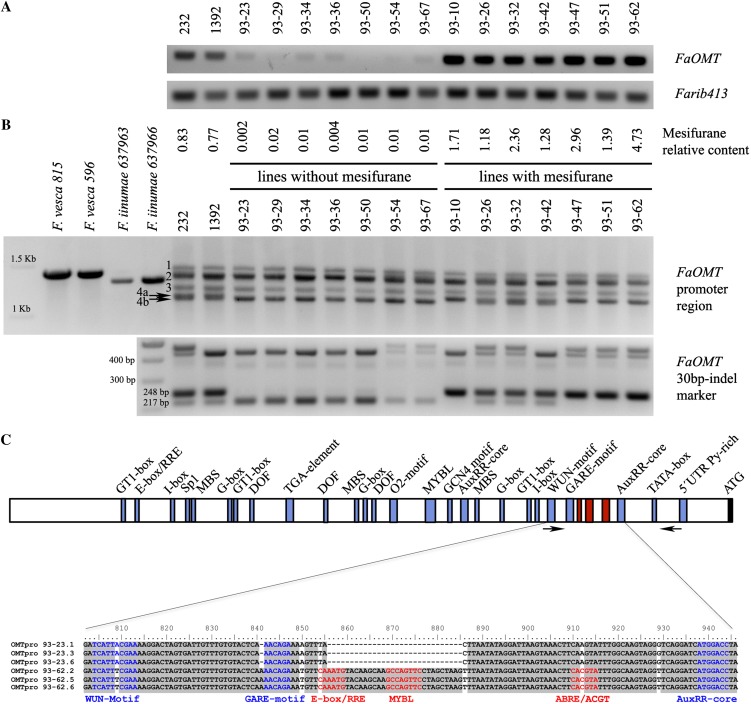
The strawberry protein O-methyltransferase (FaOMT) controls variation in mesifurane content via the methylation of furaneol using S-adenosyl-l-Met as methyl donor (Wein et al., 2002; Lunkenbein et al., 2006b). The gene FaOMT was mapped to the bottom of LG VII-F.1, linked to marker ChFaM160. The QTL 48VII-2 controlling mesifurane content was mapped at approximately the same position in another LG of the same HG VII (Supplemental Table S4; Fig. 3). Therefore, it is possible that one homoeolog of FaOMT (up to four homoeologous genes can be present in an octoploid genome) is the gene controlling the variation in mesifurane content. To determine if a FaOMT homoeolog located in LGVII-2 is the gene underlying the QTL controlling mesifurane content, we analyzed the expression level in ripe fruit showing contrasting mesifurane content, including the parental lines, both heterozygous for the QTL, and seven progeny lines with and without mesifurane using semiquantitative reverse transcription (RT)-PCR (Fig. 4A). This analysis showed high expression of FaOMT in those lines containing mesifurane (the parental and the seven selected lines) but barely detectable expression in those lines scored as not producing mesifurane. This result supports that FaOMT is the gene responsible for the natural variation observed in mesifurane content in cultivated strawberry and that this variation is dependent on FaOMT expression in ripe fruits.
Figure 4.
Characterization of lines with contrasting mesifurane content in the population ‘232’ × ‘1392’. A, Analysis of FaOMT transcripts in fruits of parental lines, seven lines without significant amount of mesifurane in fruits, and seven lines with mesifurane. B, Amplification of FaOMT promoter regions in selected lines and species. Bands associated to mesifurane content are indicated by arrows. C, Schematic representation of the FaOMT promoter region. Common cis-acting elements are indicated in blue and motifs specific to the functional allele are indicated in red. Arrows indicate the position of primers of FaOMT 30-bp indel marker. [See online article for color version of this figure.]
To investigate whether differences in FaOMT expression are based on sequence polymorphisms in the promoter, primers flanking a fragment of 1,417 bp were designed using the genome sequence of F. vesca (http://www.strawberrygenome.org). These primers amplified a fragment of the expected size in two different accessions of F. vesca and a fragment of a slightly smaller size in two accessions of Fragaria iinumae, considered as other putative donor to the strawberry octoploid genome (Rousseau-Gueutin et al., 2009; Fig. 4B). Amplification in octoploid strawberry showed five distinct bands on agarose gel (Fig. 4B). Whereas the three larger bands were monomorphic, the two smaller bands varied between the lines (Fig. 4B, bands 4a and 4b). All seven individuals that lacked mesifurane always contained band 4b, whereas the individuals with mesifurane contained 4a.
To further investigate the polymorphisms of bands 4a and 4b, the fragments were gel isolated, cloned, and sequenced. DNA alignment between each of the strawberry clones and F. vesca promoter identified a number of single nucleotide polymorphisms (SNPs), insertion/deletions (indels), and rearrangements in the promoter (Supplemental Fig. S4). The most consistent difference between the functional and nonfunctional alleles was a 30-bp indel at −275 bp from the ATG (Fig. 4C; Supplemental Fig. S4). Primers flanking the 30-bp indel were used to genotype the entire population (arrows in Fig. 4C). As an example, the amplification products in the previously selected individuals using the newly developed primers are shown in Figure 4B. The genotyping confirmed previous findings showing that individuals lacking mesifurane only contained the 4b allele (217-bp band; Fig. 4B) presumably being a nonfunctional allele that is not or very lowly expressed. The fact that half of the lines in the population produced mesifurane and presented both bands indicates the dominance of the functional allele, consistent with being an expression QTL.
Analysis of Potential Cis-Regulatory Elements in the FaOMT Promoter Sequence
Databases of known position-specific scoring matrices were used to search for putative transcription factor binding motifs present in the FaOMT promoter sequences (detailed in “Materials and Methods”). Then we focused in those putative binding motifs present in the promoter of the active alleles but missing in the promoters of the inactive alleles. The most relevant cis-regulatory elements and their position relative to the promoter sequence of the active 93-62 allele are listed in Table IV. The analysis detected a putative TATA box at −131 bp from the ATG start codon and potential cis-regulatory elements associated with hormone-, light-, and stress-related responses in all promoters. These cis-regulatory elements of the 4a and 4b promoters are depicted in Figure 4C. The promoters were particularly enriched in light-responsive elements such as Sp1, G-Box, I-box, and GT-1, suggesting that FaOMT could be tightly regulated by light (Terzaghi and Cashmore, 1995; Toledo-Ortiz et al., 2003). In addition, a number of hormone-responsive motifs were identified, with three motifs related to auxin regulation (one TGA element and two AuxRR-core motifs) and one GARE motif implicated in gibberellin responsiveness.
Table IV.
Main cis-regulatory motifs identified in the FaOMT promoter sequence
Motifs exclusive of active promoter allele of 93-62 are labeled in bold.
| Motif | Function | Strand | Distance from ATGa | Sequence |
|---|---|---|---|---|
| GT1-box | Light-responsive element | − | −1,034 | GGTTAA |
| − | −845 | |||
| + | −353 | |||
| E-box/RRE | Myc-like bHLH binding factors | − | −1,004 | CANNTG |
| I-box/GATA | Cis-regulatory element involved in light responsiveness | − | −946 | GATAA |
| − | −343 | |||
| ACGT | Cis-acting regulatory element involved in light responsiveness, bZIP binding factors | − | −850 | ACGTca |
| + | −622 | ACGTaa | ||
| − | −422 | ACGTaa | ||
| Sp1 | Light-responsive element | + | −922 | CC(G/A)CCC |
| MBS | MYB binding site involved in drought inducibility | + | −917 | YAACKG |
| MBS | MYB binding site involved in drought inducibility | + | −634 | YAACKG |
| + | −458 | |||
| MYBL | MYB-like proteins | + | −518 | tatTAGTta |
| TGA element | Auxin-responsive element | + | −754 | AACGAC |
| AuxRR core | Cis-regulatory element involved in auxin responsiveness | + | −475 | GGTCCAT |
| − | −192 | |||
| P-box/DOF | DNA binding with one finger (DOF) | + | −815 | AAAG |
| + | −694 | |||
| − | −613 | |||
| O2 motif | Opaque-2 like (bZIP) transcriptional activators | − | −584 | tCCACttc |
| GCN4 like | Cis-regulatory element involved in endosperm expression | + | −494 | TGTGTCA |
| WUN motif | Wound-responsive element | + | −326 | TCATTACGAA |
| GARE motif | Gibberellin-responsive element | + | −288 | AAACAGA |
| E-box/RRE | Myc-likebHLHbinding factors | + | −276 | CANNTG |
| MYBL | MYB-like proteins | + | −263 | gcCAGTtc |
| ACGT | Cis-acting regulatory element involved in light responsiveness,bZIPbinding factors | + | −220 | cACGTa |
| ABRE | ABAresponse element | − | −220 | TACGTG |
| CAAT-box | Common cis-acting element in promoters and enhancers | ± | 30 boxes in 93-23 and 31 in 93-62 | |
| TATA-box | Core promoter element at about −30 of transcription start | + | −131 | ATATAT |
| 5′-UTR Py-rich stretch | Element in 5′-UTR conferring high transcription levels | + | −83 | tTTCTTCTCT |
Distance (in bp) from ATG in allele 93-62. For common motifs, distance may vary in other promoter alleles.
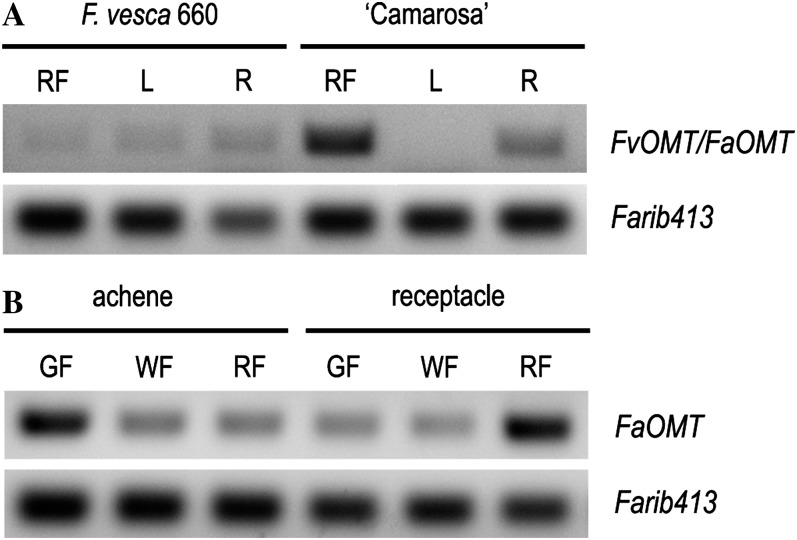
The potential motifs that were found in the functional allele but missing in the rest of the sequences concentrated in the 30-bp indel region (from −276 to −220 bp from the ATG) and included (1) an E-box/RRE motif, (2) a potential MYBL motif, and (3) a sequence with high homology to an ABRE motif and to an ACGT-containing element (Table IV; Fig. 4C; Supplemental Fig. S4). Therefore, it is likely that these specific motifs are responsible for driving high FaOMT expression in strawberry fruit. The region around the 30-bp indel was not conserved between the FvOMT promoter and the functional FaOMT allele (Supplemental Fig. S4), suggesting that this allele might not be expressed in F. vesca red ripe fruits. Therefore, we analyzed the expression of FvOMT in red fruits, leaves, and roots of F. vesca and the commercial strawberry cv Camarosa (Fig. 5A). Whereas only high expression of FaOMT was found in red fruits of Camarosa, low expression of FvOMT was found in all tissues of F. vesca, including red fruits. In agreement, comparison of volatile profiles in red fruits of F. vesca and Camarosa indicated that the content of mesifurane was 100-fold higher in the cultivated species (data not shown). Similarly, other researchers have reported lower content of mesifurane and high furaneol/mesifurane ratios in other accessions of F. vesca (Zabetakis and Holden, 1997; Ulrich et al., 2007).
Figure 5.
Analysis of FvOMT and FaOMT transcript accumulation by semiquantitative RT-PCR in different tissues of F. vesca and strawberry (A) and separately in achene and receptacle along strawberry fruit ripening (B). RF, Red fruit; L, leaf; R, root; GF, green fruit; WF, white fruit.
To further investigate the function of FaOMT, we analyzed separately the expression in the receptacle and the achene at different stages of fruit ripening in the strawberry cultivar Camarosa (Fig. 5B). This analysis showed different expression patterns in each organ. FaOMT expression increased during ripening in the receptacle tissue while in the achene, the highest expression was observed in green fruit, decreasing later during ripening. This expression pattern is consistent with the role of FaOMT in the biosynthesis of mesifurane in the receptacle but also with the additional role that has been proposed in lignin biosynthesis (Lunkenbein et al., 2006b), which might be more relevant in the achene.
DISCUSSION
Variation in Volatile Compounds in ‘232’ × ‘1392’
The relative content of 87 volatiles identified by GC and MS was analyzed in the parental lines and F1 progeny over four successive years. In agreement with their quantitative contribution to the aroma of strawberry fruit, the majority of the volatiles were esters (49.4%), aldehydes and alcohols (27.6%), followed by several ketones, terpenes, and furans (23.0%). All these compounds are known to occur in strawberry fruit and many are shown to contribute to its aroma (Pérez and Sanz, 2010). Differences in their relative concentration in each line in the four assessed years are most probably due to environmental factors. Nevertheless, remarkable differences between genotypes exist, which is in accordance with previous studies performed in apple (Malus domestica) and strawberry, where volatile profiles were more dependent on genotype than on environmental conditions (Forney et al., 2000; Rowan et al., 2009a). Most of the volatiles showed a distribution typical of polygenic inheritance and, generally, levels of compounds in the F1 individuals showed transgressive behavior. HCA based on both volatile profiles and correlation data grouped aroma compounds in similar clusters. Thus, volatiles belonging to the same biochemical pathway were normally grouped in the same cluster, suggesting a coregulation of these metabolites.
Mapping volatile compound content as single Mendelian traits have also been used for the genetic dissection of scent metabolites in diploid roses, where nerol and neryl acetate were mapped as single traits in the rose genome and geranyl acetate was mapped as an oligogenic trait controlled by two independent loci (Spiller et al., 2010). In agreement with our data, qualitative differences in mesifurane content have been reported in an analysis of five strawberry cultivars (Larsen et al., 1992). Similarly, γ-decalactone has been described as an important cultivar-specific volatile in strawberry (Larsen and Poll, 1992; Larsen et al., 1992; Schieberle and Hofmann, 1997; Ulrich et al., 1997) and was detected in 44% of the progeny generated by crossing two strawberry cultivars that strongly differed in flavor (Olbricht et al., 2008). In accordance, γ-decalactone was only detected in one parental line (‘1392’) and in approximately half of the progeny. Therefore, markers linked to the locus controlling γ-decalactone in LG III-2 might be useful tools for future MAS, because it explained up to approximately 93.3% of the phenotypic variation and this compound confers a pleasant peach-like flavor note to strawberry fruit (Larsen and Poll, 1992).
QTLs Controlling the Aroma of Strawberry and Associated CGs
Two different QTL detection methods, the nonparametric Kruskal-Wallis test and interval mapping (IM), were employed to map a large number of loci controlling aroma compounds in the strawberry population ‘232’ × ‘1392’. Although the first method is less powerful than IM, it allowed the detection of significant associations between marker genotypes and raw phenotypic data, confirming most of the QTLs detected using IM. QTLs for approximately 55% of the identified volatiles were detected in this study and 50% of them were stable in two or all three analyzed years. Most of the QTLs (50.3%) controlled ester production, which are quantitatively the main contributors to the aroma of strawberry fruit.
An unexpectedly high proportion of detected QTLs explained a large proportion of the phenotypic variation (from approximately 30% to up to 93%) and were also stable in all assessed years, being potential candidates for future MAS in strawberry. They included QTLs for butyl acetate (13VI-1), benzyl acetate (59VII-1), octyl acetate (63VI-1), decyl acetate (79VI-1), 3-methylbutyl acetate (19III-2), terpineol (65VI-2), myrtenol (66I-2), 6-methyl-5-hepten-2-one (32VI-1), mesifurane (48VII-2), γ-decalactone (82III-2), and γ-dodecalactone (87VII-1). The identification of underlying genes would be of great interest to elucidate the control of aroma volatile levels in strawberry fruit.
QTLs for diverse volatiles belonging to the same or different biosynthetic pathways clustered in similar genomic regions. Clusters of esters and alcohols were the most commonly found, indicating a possible pleiotropic effect of loci, because the biosynthesis of alcohols could affect the content of related esters. In addition, homoeoQTLs (QTLs for the same compounds at similar positions on different homoeologous LGs) for several compounds were detected in four HGs (i.e. QTLs 19III-2 and 19III-6; Fig. 3), suggesting that more than one homoeologous gene is controlling a fraction of the variation of each of those compounds. QTLs for terpenes linalool (54) and terpineol (65) clustered in LG VI-1 and LG VI-2 close to markers ChFaM164, Fvi020, and/or ChFvM244, which are tightly linked to marker BFACT047 in other homoeologous LGs of the integrated octoploid map and in the diploid Fragaria reference map (Supplemental Fig. S1). Interestingly, terpenes nerol and citronellol have been mapped as a single Mendelian trait and a QTL, respectively, at the same distance to marker BFACT047 in Rosa LG III (Spiller et al., 2010, 2011). Recent comparative studies between these two species have shown synteny and high colinearity of rose LG III and Fragaria LG VI (Gar et al., 2011; Iwata et al., 2012). Taken together, these results indicate that orthologous loci could control the content of different terpenes in these two closely related genera within the Rosaceae. If this is the case, the gene responsible would most probably be acting early in the terpene biosynthetic pathway or may display a broad substrate affinity.
In this study, several CGs involved in strawberry aroma biosynthesis were mapped and some found associated with QTLs for specific volatiles. Interestingly, a cluster of QTLs controlling four phenyl-derived esters, eugenol, and terpineol colocated in LGs I-1 and I-2 with CAD homoeologs. The enzyme CAD catalyzes the reversible conversion of cinnamyl aldehydes to the corresponding alcohols, the last step in the biosynthesis of monolignols (Singh et al., 2010). Strikingly, CAD has been reported to be expressed even in cells that do not make lignin (Singh et al., 2010). Although CAD expression pattern was related to vascular bundle formation in strawberry fruit (Aharoni et al., 2002), it has been suggested that cinnamyl alcohol derivatives produced by CAD activity may be implicated in fruit flavor and aroma (Mitchell and Jelenkovic, 1995; Singh et al., 2010). Thus, an association between CAD and phenylpropanoid-derived volatile compounds could be explained by a higher CAD activity redirecting and increasing the substrates for production of phenylpropene compounds such as eugenol. However, it cannot be excluded that in addition to cinnamyl alcohol, CAD may have affinity for other substrates in strawberry receptacle, as has been shown for aromatic and terpene alcohols (Mitchell and Jelenkovic, 1995). This possibility deserves further investigation to determine whether CAD plays a role in the production of diverse volatile compounds.
The largest cluster of aroma QTLs, involving 16 different volatiles, was mapped to LG VI-1. Among them, five acetate esters colocated with QTLs for other related esters, such as butyl and octyl hexanoates and butanoates, and two alcohols, 1-octanol and 1-decanol. The majority of these compounds was grouped in cluster D and were also highly correlated between each other (Fig. 2; Supplemental Fig. S3). We could speculate that the locus underlying this common QTL might be controlling the content of a common precursor in their biosynthesis. In agreement, substrate availability is a key limiting factor in the synthesis of volatiles and the role of substrates in their regulation is currently under study (Dudareva et al., 2004). Similarly to our results, the content of a number of acetate esters correlated in apple and a number of them have been mapped to MG9, which is syntenic to a region of FGVI (Dunemann et al., 2009; Rowan et al., 2009a, 2009b; Illa et al., 2011a, 2011b). Increasing the resolution of the ‘232’ × ‘1392’ maps and a search of CGs in the confidence interval of the QTL in the F. vesca genome is under way. Because the QTL could be conserved between strawberry and apple, the identification of common CGs in the intervals of these QTLs in F. vesca and apple genomes would reduce the number of potential candidates and might hasten the identification of the underlying gene.
The Gene FaOMT Controls the Variation in Mesifurane Content
We have shown that a QTL controlling mesifurane content (48VII-2) occurs at the same location as one of the four homoeologs of the FaOMT gene in LG VII-2. The substrate specificity of FaOMT indicates that it is involved in the methylation of furaneol to mesifurane (Wein et al., 2002). Furthermore, the inhibition of FaOMT in transgenic strawberry plants resulted in a near total loss of mesifurane (Lunkenbein et al., 2006b). This data together with the dramatic reduction of FaOMT expression in lines with trace content of mesifurane provide strong evidence indicating that FaOMT is the locus controlling mesifurane content in strawberry fruit and is responsible for its natural variation.
The observed size and sequence variability in the examined OMT (FaOMT and FvOMT) promoter region between different species, as well as between different homoeologs in the octoploid population indicates that a large number of SNPs and indels have shaped different alleles of this promoter. It is believed that the abscisic acid (ABA)/auxin ratio could be part of the signal that triggers fruit ripening in strawberry (Perkins-Veazie, 1995). Auxin stimulates receptacle expansion during early fruit development and later inhibits fruit ripening (Given et al., 1988), whereas exogenous ABA markedly promotes fruit maturation (Manning, 1994; Perkins-Veazie, 1995; Jia et al., 2011). Thus, many ripening up-regulated genes are negatively regulated by auxin but positively by ABA in strawberry fruit. Although they need to be experimentally verified, the studied FaOMT promoter region contains cis-regulatory elements involved in auxin, ABA, and gibberellin regulation. Interestingly, equivalent motifs have been found in the promoter regions of other ripening regulated genes in strawberry (Bustamante et al., 2009; Tisza et al., 2010). However, the most abundant motifs in the FaOMT promoter were related to light regulation, with a much higher number of them compared with those found in the light-inducible promoter of the strawberry gene FaGaLUR (Agius et al., 2005).
Despite the identification of the motifs above commented, only three potential cis-regulatory elements (E-box/RRE motif, MYBL motif, and an ABRE/ACGT motif) located in the proximal 30-bp indel region were specific to the functional allele, indicating that the rest of them are not sufficient for high expression in strawberry fruit. The E-box/RRE motif is recognized by the basic/helix-loop-helix (bHLH) transcription factors and has been characterized in diverse biological processes of different eukaryotes, including yeasts, animals, and plants (Toledo-Ortiz et al., 2003). In plants, light regulatory units sufficient for light responsiveness have been identified in the promoters of four coordinately expressed genes encoding enzymes that catalyze successive steps in the flavonol biosynthetic pathway (Hartmann et al., 2005). Each unit consists of two cis-elements, namely a MYB-recognition element (MRE) and an ACGT-containing element. The MRE element is recognized by R2R3-MYB transcription factors whereas several basic region/Leu zipper (BZIP) factors can bind to the ACGT-containing element (Weisshaar et al., 1991). An additional third element, an E-box/RRE motif, was identified in the promoter of one of the genes, in the chalcone synthase, and shown necessary for tissue-specific production of flavonoids (Hartmann et al., 2005). Interestingly, the three specific motifs identified in the FaOMT functional allele resemble the light regulatory units identified in the AtCHS promoter, with the E-box/RRE and the ABRE/ACGT corresponding to the binding sites of bHLH and BZIP transcription factors, respectively. Although the MRE motif was not identified, a putative MYBL motif was detected, which could be recognized by a MYB protein.
FaOMT down-regulation also affected the concentration of feruloyl 1-O-β-d-Glc and caffeoyl 1-O-β-d-Glc, therefore in addition to its role in the methylation of furaneol, it has also been suggested a role in the methylation of lignin precursors (Lunkenbein et al., 2006b). Because of the expression pattern of FaOMT during fruit ripening, it was assumed that in the beginning of fruit development FaOMT could be involved in the lignification of the vascular bundles in the expanding fruit while in the red stage could be involved in the methylation of furaneol (Wein et al., 2002). Our expression analysis of achenes and receptacle support a dual role of FaOMT in strawberry fruit. Whereas at early stages it would provide precursors for achene lignification, at later stages it could be involved in the conversion of furaneol into mesifurane in ripe receptacles.
CONCLUSION
This study provides a genetic map of QTLs that represent a useful resource for the identification of the loci responsible for the variation of a number of volatiles in strawberry fruit. Some QTLs control the variation of volatiles that contribute significantly to the aroma/flavor of fruits but others may control volatile compounds relevant to plant survival, defense against pathogens, or to plant-plant interaction. Some of them were mapped to well-defined regions and the availability of the genome sequence of F. vesca as well as the future sequencing of octoploid strawberry will allow the identification of many of the genes underlying these QTLs. Many of the QTLs identified explained a large proportion of the phenotypic variation and were stable over different years. Thus, associated molecular markers will represent useful tools for the selection of genotypes with enhanced concentrations of important aroma volatiles using molecular breeding approaches. QTLs identified in the genetic background of this strawberry population will gain from further studies involving other strawberry cultivars or even other Fragaria species to better understand the genetic architecture of strawberry aroma. Furthermore, because many volatile compounds are common to different important crops and ornamental species, QTLs identified in strawberry will facilitate advances in other species.
Using genetic, metabolomic, and molecular approaches we have identified functional and inactive alleles of the gene FaOMT and shown that the expression of this gene is responsible for the natural variation in mesifurane content in strawberry fruit. Because the substrate of this enzyme, furaneol, is considered among the most important compounds influencing strawberry aroma, the selection of nonfunctional alleles of FaOMT using the marker here developed will be desirable in new elite cultivars. From a biotechnological perspective, the 30-bp indel might provide an important tool to engineer promoters able to drive high and specific expression in the receptacle during fruit ripening of selected genes.
MATERIALS AND METHODS
Plant Material
The F1 mapping population, comprising 95 progeny lines, was raised from the cross between two Instituto Andaluz de Investigación y Formación Agraria y Pesquera selection lines, ‘232’ and ‘1392’, with contrasting agronomical and fruit quality traits (Zorrilla-Fontanesi et al., 2011a). ‘232’ is a very productive strawberry (Fragaria × ananassa) line, whereas ‘1392’ has firmer and tastier fruits. The mapping population was grown in the strawberry-producing area of Huelva (Spain) under commercial conditions during four successive years (2006–2009). In the first growing season (2006), only one plant from each progeny line was available and grown. For the three subsequent years, four plants of each line were vegetatively propagated and grown. The diploid Fragaria bin mapping population FV × FB used in this study has been described in Sargent et al. (2008).
Volatile Compound Analysis
Sample Preparation
To analyze the aroma profiles of fruit purees of the octoploid strawberry population, 10 to 15 fully ripe fruits were harvested the same day (at the middle of the season) from the parental and each of the 95 F1 lines in each of four successive years (2006, 2007, 2008, and 2009). Fruits were immediately cut, frozen in liquid nitrogen, and stored at −80°C. Later, fruits were powdered in liquid nitrogen using a coffee grinder and stored at −80°C until GC-MS analyses. Prior to the analysis of volatile compounds, frozen fruit powder (1 g fresh weight) of each sample was weighed in a 7-mL vial, closed, and incubated at 30°C for 5 min. Then 300 μL of a NaCl saturated solution were added. A total of 900 μL of the homogenized mixture were then transferred to a 10-mL screw cap headspace vial, from where the volatiles were immediately collected.
Automated HS-SPME-GC-MS
The volatiles were sampled by HS-SPME (Pawliszyn, 1997) with a 65-μm polydimethylsiloxane/divinyl-benzene fiber (Supelco). Initially, vials were tempered at 50°C for 10 min. Then the volatiles were extracted by exposing the fiber to the vial headspace for 30 min under continuous agitation and heating at 50°C. The extracted volatiles were desorbed in the GC injection port for 1 min at 250°C in splitless mode. Incubation of the vials, extraction, and desorption of the volatiles were performed automatically by a CombiPAL autosampler (CTC Analytics). Chromatography was performed on a DB-5ms (60 m × 0.25 mm × 1 μm) column (J&W Scientific) with helium as carrier gas at a constant flow of 1.2 mL/min. GC interface and MS source temperatures were 260°C and 230°C, respectively. Oven temperature conditions were 40°C for 3 min, 5°C/min ramp until 250°C, and then held at 250°C for 5 min. Mass spectra were recorded in scan mode in the 35 to 220 mass-to-charge ratio range by a 5975B mass spectrometer (Agilent Technologies) at an ionization energy of 70 eV and a scanning speed of 7 scans/s. Chromatograms and spectra were recorded and processed using the Enhanced ChemStation software (Agilent Technologies).
Compound Identification and Relative Quantification
Compounds were unequivocally identified by comparison of both mass spectrum and retention time to those of pure standards (SIGMA-Aldrich) except 2-(1-pentenyl)furan, which was tentatively identified by comparison of its mass spectrum with those in the NIST05 library. Peak areas of selected specific ions were integrated for each compound. Then, they were normalized by comparing with the peak area of the same compound in a reference sample injected regularly (a mixture of all the samples from the mapping population for each year) to correct for variations in detector sensitivity and fiber aging. Data were expressed as the relative content of each metabolite compared with the reference sample.
Statistical Analysis
Descriptive statistical analysis of the identified compounds was performed using different modules of the STATISTICA 7.0 software package (StatSoft, Inc., 2007). Range of variation in the F1 progeny, skewness, and kurtosis were calculated only for the last three years (2007–2009), and the Shapiro-Wilk test (Shapiro and Wilk, 1965) was applied to test normality of trait distributions. For those volatiles deviating from normality, several transformations (Ln, Log10, Log2, inverse of square root, square root, square, cube, reciprocal, and arcsine in degrees or radians) were tested and the transformation that gave the least-skewed result was used in the subsequent QTL analysis.
Two of the identified compounds (γ-decalactone and mesifurane) were resolved into single Mendelian traits and analyzed as both a single gene and a quantitative trait. To analyze γ-decalactone as a major gene, genotypes with relative values higher or lower than 0.05 (meaning 20 times less than the reference sample) were considered as producing or not producing γ-decalactone, respectively. For mesifurane, although lines considered not producing mesifurane contained in general less than one-twentieth the content in the reference sample, the limit for scoring the lines as producing or not producing was established at 0.1 (meaning 10 times less than the reference sample).
For PCA, Log2-transformed relative values and the program SIMCA-P version 11 (Umetrics) were employed, with the variables centered and scaled to unit of variance. Then, Acuity 4.0 software (Axon Instruments) was used to examine volatiles that significantly varied between individuals by ANOVA and also to represent those volatiles in the HCA and the corresponding heat map, using Log2-transformed relative values from data of all four years (2006–2009). In the heat map representation, green and red regions indicate levels 3.44-fold higher or lower than those of the reference sample, respectively, as depicted in the reference color bar. Finally, Pearson correlation coefficients between the identified volatiles were calculated with SPSS 15.0 software for year 2008 and the corresponding heat map representation of pairwise correlations was performed with Acuity 4.0 software. Green and red regions indicate negative or positive correlation between traits, respectively.
CG Analysis
DNA for molecular markers and CG analyses was isolated from young leaves of the parents and F1 mapping population using a modified CTAB method based on that of Doyle and Doyle (1990).
Six strawberry CGs involved in volatile formation, alcohol acyltransferase (SAAT), lipoxygenase (LOX), O-methyltransferase (FaOMT), eugenol synthase2 (FaEGS2), quinone oxidoreductase (FaQR), and nerolidol synthase1 (FaNES1), were amplified in the ‘232’ × ‘1392’ mapping population and in the FV × FB bins using the single-strand conformational polymorphisms (SSCPs) technique (Table II). In addition, the strawberry homolog of the elongated hypocotyl5 from Arabidopsis (Arabidopsis thaliana; FaHY5; K. Merchante and V. Valpuesta, unpublished data) along with the strawberry cytosolic ascorbate peroxidase (Sargent et al., 2007) were also analyzed by SSCPs in the octoploid mapping population (ascorbate peroxidase) or in both populations (FaHY5). Primers used to amplify FaHY5 were: FaHY5-F: 5′-GTCTTCCAGCTCTGCATTCC and FaHY5-R: 5′-CCCTCTTTCTCTGGCTCTCA. For SSCP analysis, PCR reactions were carried out as previously described (Zorrilla-Fontanesi et al., 2011a) using the corresponding annealing temperature for each primer pair (Table II). PCR products, which ranged between 167 and 402 bp in length, were loaded and electrophoresed in agarose and nondenaturing gels, as described (Zorrilla-Fontanesi et al., 2011a, 2011b). At least two different SSCP bands from each gene were picked from the gels, amplified, and directly sequenced to verify their identity. The microsatellite EMFv010 (James et al., 2003) was also amplified in the octoploid mapping population to increase saturation in HG VI.
Linkage Mapping
Polymorphic SSR and SSCP bands plus the two volatile compounds resolved as Mendelian loci were scored by two different observers. Then, the χ2 analysis for goodness of fit was performed to test the segregation ratios obtained to those expected for single and multiple dose markers under disomic or octosomic inheritance (Lerceteau-Köhler et al., 2003). Markers were considered to have significantly skewed ratios at P ≤ 0.05. Linkage analyses and map construction were performed using JoinMap 4 (van Ooijen, 2006) and the population coded as CP (for cross pollinated). Two independent parental maps and the integrated map were constructed using the novel molecular markers and those previously located in the ‘232’ × ‘1392’ map (Zorrilla-Fontanesi et al., 2011a). To generate the maps, a double pseudo-testcross strategy was employed, including 1:1, 3:1, and codominant markers segregating from each parental line (Grattapaglia and Sederoff, 1994). Grouping was performed using independence LOD and the default settings in JoinMap. Groups were generally chosen from a LOD of 5.0 to 8.0, although for some groups this value was decreased to 4.0 or 3.0 if markers comprising the groups showed linkage in previous maps.
The strongest cross link parameter was used to assign ungrouped markers to already established LGs or to join different LGs belonging to the same chromosome based in previous versions of octoploid strawberry maps (Rousseau-Gueutin et al., 2008; Sargent et al., 2009; Zorrilla-Fontanesi et al., 2011a). Map construction was performed using the regression mapping algorithm (Stam, 1993) and the following JoinMap parameters: Rec = 0.40, LOD = 1, Jump = 5.0, and ripple = 1. The Kosambi mapping function was used to convert recombination frequencies into map distances and markers showing distorted segregation were included in the maps when they did not disrupt previous marker order. The seven HGs were named I to VII, as the corresponding LGs in the diploid Fragaria reference map, followed by an F (for female LGs), an M (for male LGs), or F/M (for integrated LGs). Linkage maps were drawn using MapChart 2.2 for Windows (Voorrips, 2002).
QTL Analysis
Because of the high number of traits and the different origin (seed in the first year and vegetatively propagated plants in the second, third, and fourth years), we decided to exclude the data obtained in the first year (2006). QTL analyses were performed using MapQTL 5 (van Ooijen, 2004) on data from the last three years (2007–2009). Due to nonnormality for most of the metabolites, the raw relative data for a total of 83 volatile compounds (44, 53, 83, and 86 were excluded) were analyzed first by the nonparametric Kruskal-Wallis rank-sum test. A stringent significance level of P = 0.005 was used as threshold, as suggested by van Ooijen (2004). Second, different data transformations were tested with the aim of identifying the most appropriate to achieve normality (see statistical analysis section). This is preferred for subsequent QTL analysis based on IM. Lastly, the genetic linkage map of each parental line and transformed data sets for most traits were used to identify and locate QTLs using IM (Lander and Botstein, 1989). Identified QTLs were described by the marker with the highest significance level in the corresponding QTL region. For IM, the all-markers mapping approach was used to upgrade marker information (Knott and Haley, 1992; Maliepaard and van Ooijen, 1994). This method employs not only the flanking markers but also markers from neighboring intervals to calculate the probabilities of a QTL. Five neighboring intervals and a step size of 3 cM were used. Significance LOD thresholds were estimated with a 1,000-permutation test (Churchill and Doerge, 1994) for each volatile and year on each map and QTLs with LOD scores greater than the genome-wide threshold at P ≤ 0.05 were declared significant. For each LOD peak, the 1-LOD support interval was determined (van Ooijen, 1992). The percentage of variance explained by each QTL and the genotypic information coefficient (which ranges from 0 to 1, with 0 indicating no marker information and 1 complete or maximum marker information) were also calculated. QTLs were named in italics using the volatile code followed by the name of the LG in which the QTL was located. QTL positions and 1-LOD confidence intervals were drawn using MapChart 2.2 for Windows.
Gene Expression Analysis
Total RNA was extracted from different tissues of strawberry and Fragaria vesca Reine des vallées (accession no. IFAPA660) as previously described (Manning, 1991). Prior to RT, RNA was treated with DNase I (Fermentas) to remove any residual contaminating genomic DNA. First-strand cDNA was synthesized using the i-script kit (Bio-Rad) and following the procedure described by the manufacturer. Two microliters of a dilution of the reaction product was subjected to subsequent semiquantitative RT-PCR in a 20-μL reaction volume and 25 PCR cycles. Primers used to amplify the Farib413 (18S-26S interspacer ribosomal gene) constitutive control were described elsewhere (Muñoz et al., 2010). Sequences of primers used to analyze FaOMT expression are shown in Table II and they amplify a fragment of the 3′ untranslated region (UTR) of the cDNA. This primer pair amplified with approximately the same efficiency a product of the same size using F. vesca or strawberry genomic DNA as template (data not shown). The amplification products were separated on 1.5% agarose gel stained with ethidium bromide and visualized with UV light. For analysis of FaOMT expression in contrasting lines of the mapping population, RNA was extracted from the same pool of fruit tissue used for volatile profiling in season 2009. For the rest of expression studies, two independent biological replicates were assessed. The same results were obtained and only one is shown.
Promoter Isolation and Analysis
For characterization of the FaOMT promoter region from strawberry lines, we sequenced three independent clones obtained from (1) band 4a of line 93-62 (this line produces mesifurane and seems to be homozygous for the functional allele), (2) band 4b of line 93-23 (this line does not produce mesifurane), and (3) band 2 from the parental line ‘1392’ (this line produces mesifurane). Further, the fragment size for band 2 was most similar in size to the F. vesca allele of FvOMT. PCR was carried out using genomic DNA of the selected mapping lines and primers OMT-Pro-F (5′-TGGTTGTGCAATTTTCTCCA-3′) and OMT-Pro-R (5′-ATGGGTCGGAGTCATCTGAG-3′). Forward primer was designed according to F. vesca accession HI 4 and reverse primer according to strawberry sequences such as that of the original clone AF220491. PCR were performed using an annealing temperature of 59°C and amplicons were separated in agarose electrophoresis. Selected bands were isolated and purified from the agarose gel using the FavorPrep GEL/PCR purification kit (Favorgen) and cloned into the pGEM-T Easy vector (Promega). Three independent clones per band were sequenced. In agreement with their similar size, band 2 from line ‘1392’ was the most similar to the FvOMT promoter sequence. However, the three strawberry sequences, in addition to different lengths (1,362, 1,368, and 1,376 bp) presented several SNPs and indels among them, thus representing three different FaOMT alleles. The clones sequenced from band 4a of line 93-62, thus representing active alleles, were 1,165-bp long and two alleles were identified differing in only two SNPs. Sequences from band 4b of 93-23 were 1,133-bp long and two different alleles, differing in eight SNPs, were identified. It is important to note that strawberry is an octoploid; therefore up to 16 different alleles (four for each homoeologous gene) can be segregating in the progeny derived from a cross between two different heterozygous genotypes. The nucleotide sequences of the different promoter regions have been submitted to the EMBL/GenBank under the accession numbers JQ322651 to JQ322659.
To develop a PCR marker for discrimination between functional and nonfunctional FaOMT alleles, novel primers were designed flanking the 30-bp indel in the promoter sequences: FaOMT-SI/NO-F (5′-CGATCATTTCGAAAAGGACTA-3′) and FaOMT-SI/NO-R (5′-AAGCAGGGTTAGTTGTGGAGA-3′). PCRs were performed in a final reaction volume of 15 μL comprising 2 μL template DNA, 1× PCR buffer, 2 mm MgCl2, 200 μm dNTPs, 0.2 μm each primer, and 0.5 units Taq polymerase (GeneCraft) following the touchdown protocol described by Sargent et al. (2003).
Sequence analyses and comparisons were carried out using the BioEdit software. Identification of putative cis-acting elements was performed using the following softwares: PLANTCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html; Lescot et al., 2002), PLACE (http://www.dna.affrc.go.jp/PLACE; Higo et al., 1999), and MatInspector (http://www.genomatix.de/online_help/help_matinspector/matinspector_help.html; Cartharius et al., 2005).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers JQ322651 to JQ322659.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Integrated linkage map of the cultivated strawberry.
Supplemental Figure S2. HCA and heat map representation of volatile profiles of each individual in the population during the four assessed years.
Supplemental Figure S3. HCA and heat map representation of pairwise correlations between volatile compounds in 2008.
Supplemental Figure S4. Sequence alignment of FaOMT and FvOMT promoter sequences.
Supplemental Table S1. Pairwise correlations among the 87 volatiles identified in the ‘232’ × ‘1392’ mapping population for year 2008.
Supplemental Table S2. QTLs controlling the content of esters.
Supplemental Table S3. QTLs controlling the content of alcohols and terpene alcohols.
Supplemental Table S4. QTLs controlling the content of aldehydes, ketones, and furans.
Supplementary Material
Acknowledgments
HS-SPME-GC-MS was performed at the Metabolomics laboratory in Instituto de Biología Molecular y Celular de Plantas, Consejo Superior de Investigaciones Científicas-Universidad Politécnica de Valencia. We thank Teresa Caballero for her excellent technical assistance, Maria Pilar López and Nuria Cabedo for valuable help in compound identification, and Asunción Fernández and Antonio Monforte for helpful discussions on QTL analysis.
Glossary
- QTL
quantitative trait loci
- MAS
marker-assisted selection
- CG
candidate gene
- HS-SPME-GC-MS
headspace solid phase microextraction coupled to gas chromatography and mass spectrometry
- HS-SPME
headspace solid phase microextraction
- GC
gas chromatography
- MS
mass spectrometry
- PCA
principal component analysis
- HCA
hierarchical cluster analysis
- LGs
linkage groups
- HGs
homology groups
- RT
reverse transcription
- SNPs
single nucleotide polymorphisms
- IM
interval mapping
- ABA
abscisic acid
- bHLH
basic/helix-loop-helix
- MRE
MYB-recognition element
- BZIP
basic region/Leu zipper
- SSCPs
single-strand conformational polymorphisms
- UTR
untranslated region
References
- Agius F, Amaya I, Botella MA, Valpuesta V. (2005) Functional analysis of homologous and heterologous promoters in strawberry fruits using transient expression. J Exp Bot 56: 37–46 [DOI] [PubMed] [Google Scholar]
- Aharoni A, Giri AP, Verstappen FW, Bertea CM, Sevenier R, Sun Z, Jongsma MA, Schwab W, Bouwmeester HJ. (2004) Gain and loss of fruit flavor compounds produced by wild and cultivated strawberry species. Plant Cell 16: 3110–3131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aharoni A, Keizer LC, Van Den Broeck HC, Blanco-Portales R, Muñoz-Blanco J, Bois G, Smit P, De Vos RC, O’Connell AP. (2002) Novel insight into vascular, stress, and auxin-dependent and -independent gene expression programs in strawberry, a non-climacteric fruit. Plant Physiol 129: 1019–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battilana J, Costantini L, Emanuelli F, Sevini F, Segala C, Moser S, Velasco R, Versini G, Stella Grando M. (2009) The 1-deoxy-D:-xylulose 5-phosphate synthase gene co-localizes with a major QTL affecting monoterpene content in grapevine. Theor Appl Genet 118: 653–669 [DOI] [PubMed] [Google Scholar]
- Beekwilder J, Alvarez-Huerta M, Neef E, Verstappen FWA, Bouwmeester HJ, Aharoni A. (2004) Functional characterization of enzymes forming volatile esters from strawberry and banana. Plant Physiol 135: 1865–1878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco-Portales R, Medina-Escobar N, López-Ráez JA, González-Reyes JA, Villalba JM, Moyano E, Caballero JL, Muñoz-Blanco J. (2002) Cloning, expression and immunolocalization pattern of a cinnamyl alcohol dehydrogenase gene from strawberry (Fragaria × ananassa cv. Chandler). J Exp Bot 53: 1723–1734 [DOI] [PubMed] [Google Scholar]
- Bringhurst RS. (1990) Cytogenetics and evolution in American Fragaria. Hort Sci 106: 679–683 [Google Scholar]
- Bustamante CA, Civello PM, Martínez GA. (2009) Cloning of the promoter region of β-xylosidase (FaXyl1) gene and effect of plant growth regulators on the expression of FaXyl1 in strawberry fruit. Plant Sci 177: 49–56 [Google Scholar]
- Camacho D, de la Fuente A, Mendes P. (2005) The origin of correlations in metabolomics data. Metabolomics 1: 53–63 [Google Scholar]
- Carrasco B, Hancock J, Beaudry R, Retamales J. (2005) Chemical composition and inheritance patterns of aroma in Fragaria × ananassa and Fragaria virginiana progenies. Hort Sci 40: 1649–1650 [Google Scholar]
- Cartharius K, Frech K, Grote K, Klocke B, Haltmeier M, Klingenhoff A, Frisch M, Bayerlein M, Werner T. (2005) MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics 21: 2933–2942 [DOI] [PubMed] [Google Scholar]
- Causse M, Saliba-Colombani V, Lesschaeve I, Buret M. (2001) Genetic analysis of organoleptic quality in fresh market tomato. 2. Mapping QTLs for sensory attributes. Theor Appl Genet 102: 273–283 [Google Scholar]
- Churchill GA, Doerge RW. (1994) Empirical threshold values for quantitative trait mapping. Genetics 138: 963–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darrow G. (1966) The Strawberry: History, Breeding and Physiology. Holt Rinehart & Winston, New York [Google Scholar]
- Doligez A, Audiot E, Baumes R, This P. (2006) QTLs for muscat flavor and monoterpenic odorant content in grapevine (Vitis vinifera L.). Mol Breed 18: 109–125 [Google Scholar]
- Doyle JJ, Doyle JL. (1990) Isolation of plant DNA from fresh tissue. Focus 12: 13–15 [Google Scholar]
- Dudareva N, Pichersky E, Gershenzon J. (2004) Biochemistry of plant volatiles. Plant Physiol 135: 1893–1902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunemann F, Ulrich D, Boudichevskaia A, Grafe C, Weber WE. (2009) QTL mapping of aroma compounds analyzed by headspace solid-phase microextraction gas chromatography in the apple progeny ‘Discovery’ × ‘Prima’. Mol Breed 23: 501–521 [Google Scholar]
- Forney C, Kalt W, Jordan M. (2000) The composition of strawberry aroma is influenced by cultivar, maturity and storage. Hort Sci 35: 1022–1026 [Google Scholar]
- Gar O, Sargent DJ, Tsai C-J, Pleban T, Shalev G, Byrne DH, Zamir D. (2011) An autotetraploid linkage map of rose (Rosa hybrida) validated using the strawberry (Fragaria vesca) genome sequence. PLoS ONE 6: e20463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Given NK, Venis MA, Gierson D. (1988) Hormonal regulation of ripening in the strawberry, a non-climacteric fruit. Planta 174: 402–406 [DOI] [PubMed] [Google Scholar]
- Grattapaglia D, Sederoff R. (1994) Genetic linkage maps of Eucalyptus grandis and Eucalyptus urophylla using a pseudo-testcross: mapping strategy and RAPD markers. Genetics 137: 1121–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann U, Sagasser M, Mehrtens F, Stracke R, Weisshaar B. (2005) Differential combinatorial interactions of cis-acting elements recognized by R2R3-MYB, BZIP, and BHLH factors control light-responsive and tissue-specific activation of phenylpropanoid biosynthesis genes. Plant Mol Biol 57: 155–171 [DOI] [PubMed] [Google Scholar]
- Higo K, Ugawa Y, Iwamoto M, Korenaga T. (1999) Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res 27: 297–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illa E, Eduardo I, Audergon J, Barale F, Dirlewanger E, Li X, Moing A, Lambert P, Le Dantec L, Gao Z, et al. (2011a) Saturating the Prunus (stone fruits) genome with candidate genes for fruit quality. Mol Breed 28: 667–682 [Google Scholar]
- Illa E, Sargent DJ, Lopez Girona E, Bushakra J, Cestaro A, Crowhurst R, Pindo M, Cabrera A, van der Knaap E, Iezzoni A, et al. (2011b) Comparative analysis of rosaceous genomes and the reconstruction of a putative ancestral genome for the family. BMC Evol Biol 11: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata H, Gaston A, Remay A, Thouroude T, Jeauffre J, Kawamura K, Oyant LH, Araki T, Denoyes B, Foucher F. (2012) The TFL1 homologue KSN is a regulator of continuous flowering in rose and strawberry. Plant J 69: 116–125 [DOI] [PubMed] [Google Scholar]
- James C, Wilson F, Hadonou A, Tobutt K. (2003) Isolation and characterization of polymorphic microsatellites in diploid strawberry (Fragaria vesca L.) for mapping, diversity studies and clone identification. Mol Ecol Notes 3: 171–173 [Google Scholar]
- Jetti RR, Yang E, Kurnianta A, Finn C, Qian MC. (2007) Quantification of selected aroma-active compounds in strawberries by headspace solid-phase microextraction gas chromatography and correlation with sensory descriptive analysis. J Food Sci 72: S487–S496 [DOI] [PubMed] [Google Scholar]
- Jia HF, Chai YM, Li CL, Lu D, Luo JJ, Qin L, Shen YY. (2011) Abscisic acid plays an important role in the regulation of strawberry fruit ripening. Plant Physiol 157: 188–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klee HJ. (2010) Improving the flavor of fresh fruits: genomics, biochemistry, and biotechnology. New Phytol 187: 44–56 [DOI] [PubMed] [Google Scholar]
- Knott SA, Haley CS. (1992) Maximum likelihood mapping of quantitative trait loci using full-sib families. Genetics 132: 1211–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Botstein D. (1989) Mapping mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics 121: 185–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen M, Poll L. (1992) Odour thresholds of some important aroma compounds in strawberries. Z Lebensm Unters Forsch 195: 120–123 [Google Scholar]
- Larsen M, Poll L, Olsen C. (1992) Evaluation of the aroma composition of some strawberry (Fragaria × ananassa Duch) cultivars by use of odour threshold values. Z Lebensm Unters Forsch 195: 536–539 [Google Scholar]
- Latrasse A. (1991) Fruits III. In H Maarse, ed, Volatile Compounds in Foods and Beverages. Marcel Dekker, Inc., New York, pp 334–340 [Google Scholar]
- Lerceteau-Köhler E, Guérin G, Laigret F, Denoyes-Rothan B. (2003) Characterization of mixed disomic and polysomic inheritance in the octoploid strawberry (Fragaria × ananassa) using AFLP mapping. Theor Appl Genet 107: 619–628 [DOI] [PubMed] [Google Scholar]
- Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouzé P, Rombauts S. (2002) PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res 30: 325–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loughrin JH, Kasperbauer MJ. (2002) Aroma of fresh strawberries is enhanced by ripening over red versus black mulch. J Agric Food Chem 50: 161–165 [DOI] [PubMed] [Google Scholar]
- Lunkenbein S, Bellido M, Aharoni A, Salentijn EMJ, Kaldenhoff R, Coiner HA, Muñoz-Blanco J, Schwab W. (2006a) Cinnamate metabolism in ripening fruit: characterization of a UDP-glucose:cinnamate glucosyltransferase from strawberry. Plant Physiol 140: 1047–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunkenbein S, Salentijn EMJ, Coiner HA, Boone MJ, Krens FA, Schwab W. (2006b) Up- and down-regulation of Fragaria × ananassa O-methyltransferase: impacts on furanone and phenylpropanoid metabolism. J Exp Bot 57: 2445–2453 [DOI] [PubMed] [Google Scholar]
- Maliepaard C, van Ooijen JW. (1994) QTL mapping in a full-sib family of an outcrossing species. In JW Van Ooijen, J Jansen, eds, Biometrics in Plant Breeding: Applications of Molecular Markers. Proc meeting of the Eucarpia section biometrics in plant breeding, Wageningen, The Netherlands, pp 140–146 [Google Scholar]
- Manning K. (1991) Isolation of nucleic acids from plants by differential solvent precipitation. Anal Biochem 195: 45–50 [DOI] [PubMed] [Google Scholar]
- Manning K. (1994) Changes in gene expression during strawberry fruit ripening and their regulation by auxin. Planta 194: 62–68 [Google Scholar]
- Mathieu S, Cin VD, Fei Z, Li H, Bliss P, Taylor MG, Klee HJ, Tieman DM. (2009) Flavour compounds in tomato fruits: identification of loci and potential pathways affecting volatile composition. J Exp Bot 60: 325–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ménager I, Jost M, Aubert C. (2004) Changes in physicochemical characteristics and volatile constituents of strawberry (Cv. Cigaline) during maturation. J Agric Food Chem 52: 1248–1254 [DOI] [PubMed] [Google Scholar]
- Mitchell W, Jelenkovic G. (1995) Characterizing NAD- and NADP-dependent alcohol dehydrogenase enzymes of strawberries. J Am Soc Hortic Sci 120: 798–801 [Google Scholar]
- Muñoz C, Hoffmann T, Escobar NM, Ludemann F, Botella MA, Valpuesta V, Schwab W. (2010) The strawberry fruit Fra a allergen functions in flavonoid biosynthesis. Mol Plant 3: 113–124 [DOI] [PubMed] [Google Scholar]
- Olbricht K, Grafe C, Weiss K, Ulrich D. (2008) Inheritance of aroma compounds in a model population of Fragaria × ananassa Duch. Plant Breed 127: 87–93 [Google Scholar]
- O’Reilly-Wapstra JM, Freeman JS, Davies NW, Vaillancourt RE, Fitzgerald H, Potts BM. (2011) Quantitative trait loci for foliar terpenes in a global eucalypt species. Tree Genet Genomes 7: 485–498 [Google Scholar]
- Osorio S, Muñoz C, Valpuesta V. (2010) Physiology and biochemistry of fruit flavors. In YH Hui, ed, Handbook of Fruit and Vegetable Flavors. John Wiley & Sons, Inc., New York, pp 25–43 [Google Scholar]
- Pawliszyn J. (1997) Solid Phase Microextraction: Theory and Practice. Wiley VCH, New York [Google Scholar]
- Perkins-Veazie P. (1995) Growth and ripening of strawberry fruit. Hortic Rev (Am Soc Hortic Sci) 17: 267–297 [Google Scholar]
- Pérez A, Olías R, Sanz C. (1996) Furanones in strawberries: evolution during ripening and postharvest shelf life. J Agric Food Chem 44: 3620–3624 [Google Scholar]
- Pérez A, Rios J, Sanz C, Olías J. (1992) Aroma components and free amino acids in strawberry variety Chandler during ripening. J Agric Food Chem 40: 2232–2235 [Google Scholar]
- Pérez A, Sanz A. (2010) Strawberry flavor. In HY Hui, ed, Handbook of Fruit and Vegetable Flavors. Wiley, New York, pp 437–455 [Google Scholar]
- Pérez AG, Olías R, Luaces P, Sanz C. (2002) Biosynthesis of strawberry aroma compounds through amino acid metabolism. J Agric Food Chem 50: 4037–4042 [DOI] [PubMed] [Google Scholar]
- Pflieger S, Lefebvre V, Causse M. (2001) The candidate gene approach in plant genetics: a review. Mol Breed 7: 275–291 [Google Scholar]
- Pyysalo T, Honkanen E, Hirvi T. (1979) Volatiles of wild strawberries, Fragaria vesca L., compared to those of cultivated berries, Fragaria × ananassa cv Senga Sengana. J Agric Food Chem 27: 19–22 [Google Scholar]
- Raab TL, López-Ráez JA, Klein D, Caballero JL, Moyano E, Schwab W, Muñoz-Blanco J. (2006) FaQR, required for the biosynthesis of the strawberry flavor compound 4-hydroxy-2,5-dimethyl-3(2H)-furanone, encodes an enone oxidoreductase. Plant Cell 18: 1023–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raamsdonk LM, Teusink B, Broadhurst D, Zhang N, Hayes A, Walsh MC, Berden JA, Brindle KM, Kell DB, Rowland JJ, et al. (2001) A functional genomics strategy that uses metabolome data to reveal the phenotype of silent mutations. Nat Biotechnol 19: 45–50 [DOI] [PubMed] [Google Scholar]
- Rousseau-Gueutin M, Gaston A, Aïnouche A, Aïnouche ML, Olbricht K, Staudt G, Richard L, Denoyes-Rothan B. (2009) Tracking the evolutionary history of polyploidy in Fragaria L. (strawberry): new insights from phylogenetic analyses of low-copy nuclear genes. Mol Phylogenet Evol 51: 515–530 [DOI] [PubMed] [Google Scholar]
- Rousseau-Gueutin M, Lerceteau-Köhler E, Barrot L, Sargent DJ, Monfort A, Simpson D, Arús P, Guérin G, Denoyes-Rothan B. (2008) Comparative genetic mapping between octoploid and diploid Fragaria species reveals a high level of colinearity between their genomes and the essentially disomic behavior of the cultivated octoploid strawberry. Genetics 179: 2045–2060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowan DD, Hunt MB, Alspach PA, Whitworth CJ, Oraguzie NC. (2009a) Heritability and genetic and phenotypic correlations of apple (Malus × domestica) fruit volatiles in a genetically diverse breeding population. J Agric Food Chem 57: 7944–7952 [DOI] [PubMed] [Google Scholar]
- Rowan DD, Hunt MB, Dimouro A, Alspach PA, Weskett R, Volz RK, Gardiner SE, Chagné D. (2009b) Profiling fruit volatiles in the progeny of a ‘Royal Gala’ × ‘Granny Smith’ apple (Malus × domestica) cross. J Agric Food Chem 57: 7953–7961 [DOI] [PubMed] [Google Scholar]
- Saliba-Colombani V, Causse M, Langlois D, Philouze J, Buret M. (2001) Genetic analysis of organoleptic quality in fresh market tomato. 1. Mapping QTLs for physical and chemical traits. Theor Appl Genet 102: 259–272 [Google Scholar]
- Sargent DJ, Cipriani G, Vilanova S, Gil-Ariza D, Arús P, Simpson DW, Tobutt KR, Monfort A. (2008) The development of a bin mapping population and the selective mapping of 103 markers in the diploid Fragaria reference map. Genome 51: 120–127 [DOI] [PubMed] [Google Scholar]
- Sargent DJ, Fernandez-Fernandez F, Ruiz-Roja JJ, Sutherland BG, Passey A, Whitehouse AB, Simpson DW. (2009) A genetic linkage map of the cultivated strawberry (Fragaria × ananassa) and its comparison to the diploid Fragaria reference map. Mol Breed 24: 293–303 [Google Scholar]
- Sargent DJ, Hadonou AM, Simpson DW. (2003) Development and characterization of polymorphic microsatellite markers from Fragaria viridis, a wild diploid strawberry. Mol Ecol Notes 3: 550–552 [Google Scholar]
- Sargent DJ, Rys A, Nier S, Simpson DW, Tobutt KR. (2007) The development and mapping of functional markers in Fragaria and their transferability and potential for mapping in other genera. Theor Appl Genet 114: 373–384 [DOI] [PubMed] [Google Scholar]
- Schauer N, Semel Y, Roessner U, Gur A, Balbo I, Carrari F, Pleban T, Perez-Melis A, Bruedigam C, Kopka J, et al. (2006) Comprehensive metabolic profiling and phenotyping of interspecific introgression lines for tomato improvement. Nat Biotechnol 24: 447–454 [DOI] [PubMed] [Google Scholar]
- Schieberle P, Hofmann T. (1997) Evaluation of the character impact odorants in fresh strawberry juice by quantitative measurements and sensory studies on model mixtures. J Agric Food Chem 45: 227–232 [Google Scholar]
- Schwab W, Davidovich-Rikanati R, Lewinsohn E. (2008) Biosynthesis of plant-derived flavor compounds. Plant J 54: 712–732 [DOI] [PubMed] [Google Scholar]
- Shapiro S, Wilk M. (1965) An analysis of variance test for normality (complete samples). Biometrika 52: 591–611 [Google Scholar]
- Shulaev V, Sargent DJ, Crowhurst RN, Mockler TC, Folkerts O, Delcher AL, Jaiswal P, Mockaitis K, Liston A, Mane SP, et al. (2011) The genome of woodland strawberry (Fragaria vesca). Nat Genet 43: 109–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, Rastogi S, Dwivedi UN. (2010) Phenylpropanoid metabolism in ripening fruits. Compr Rev Food Sci Food Safety 9: 398–416 [DOI] [PubMed] [Google Scholar]
- Spiller M, Berger RG, Debener T. (2010) Genetic dissection of scent metabolic profiles in diploid rose populations. Theor Appl Genet 120: 1461–1471 [DOI] [PubMed] [Google Scholar]
- Spiller M, Linde M, Hibrand-Saint Oyant L, Tsai C-J, Byrne DH, Smulders MJM, Foucher F, Debener T. (2011) Towards a unified genetic map for diploid roses. Theor Appl Genet 122: 489–500 [DOI] [PubMed] [Google Scholar]
- Stam P. (1993) Construction of integrated genetic linkage maps by means of a new computer package: Join Map. Plant J 3: 739–744 [Google Scholar]
- StatSoft, Inc (2007) Electronic Statistics Textbook. http://www.statsoft.com/textbook/ (January 12, 2011)
- Steuer R, Kurths J, Fiehn O, Weckwerth W. (2003) Observing and interpreting correlations in metabolomic networks. Bioinformatics 19: 1019–1026 [DOI] [PubMed] [Google Scholar]
- Taylor A, Hort J. (2004) Measuring proximal stimuli involved in flavor perception. In AJ Taylor, DD Roberts, eds, Flavor Perception. Blackwell, Oxford, pp 1–38 [Google Scholar]
- Terzaghi WB, Cashmore AR. (1995) Light-regulated transcription. Annu Rev Plant Physiol Plant Mol Biol 46: 445–474 [Google Scholar]
- Tieman DM, Zeigler M, Schmelz EA, Taylor MG, Bliss P, Kirst M, Klee HJ. (2006) Identification of loci affecting flavour volatile emissions in tomato fruits. J Exp Bot 57: 887–896 [DOI] [PubMed] [Google Scholar]
- Tisza V, Kovács L, Balogh A, Heszky L, Kiss E. (2010) Characterization of FaSPT, a SPATULA gene encoding a bHLH transcriptional factor from the non-climacteric strawberry fruit. Plant Physiol Biochem 48: 822–826 [DOI] [PubMed] [Google Scholar]
- Toledo-Ortiz G, Huq E, Quail PH. (2003) The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 15: 1749–1770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich D, Hoberg E, Rapp A, Kecke S. (1997) Analysis of strawberry flavour—discrimination of aroma types by quantification of volatile compounds. Z Lebensm Unters Forsch 205: 218–223 [Google Scholar]
- Ulrich D, Komes D, Olbricht K, Hoberg E. (2007) Diversity of aroma patterns in wild and cultivated Fragaria accessions. Genet Resour Crop Evol 54: 1185–1196 [Google Scholar]
- van Ooijen J. (1992) Accuracy of mapping quantitative trait loci in autogamous species. Theor Appl Genet 84: 803–811 [DOI] [PubMed] [Google Scholar]
- van Ooijen J. (2004) MapQTL 5, Software for the Mapping of Quantitative Trait Loci in Experimental Populations. Kyazma BV, Wageningen, The Netherlands [Google Scholar]
- van Ooijen J. (2006) JoinMap 4, Software for the Calculation of Genetic Linkage Maps in Experimental Populations. Kyazma BV, Wageningen, The Netherlands [Google Scholar]
- Voorrips RE. (2002) MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered 93: 77–78 [DOI] [PubMed] [Google Scholar]
- Weckwerth W, Loureiro ME, Wenzel K, Fiehn O. (2004) Differential metabolic networks unravel the effects of silent plant phenotypes. Proc Natl Acad Sci USA 101: 7809–7814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wein M, Lavid N, Lunkenbein S, Lewinsohn E, Schwab W, Kaldenhoff R. (2002) Isolation, cloning and expression of a multifunctional O-methyltransferase capable of forming 2,5-dimethyl-4-methoxy-3(2H)-furanone, one of the key aroma compounds in strawberry fruits. Plant J 31: 755–765 [DOI] [PubMed] [Google Scholar]
- Weisshaar B, Armstrong GA, Block A, da Costa e Silva O, Hahlbrock K. (1991) Light-inducible and constitutively expressed DNA-binding proteins recognizing a plant promoter element with functional relevance in light responsiveness. EMBO J 10: 1777–1786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita I, Iino K, Nemoto Y, Yoshikawa S. (1977) Studies on flavor development in strawberries. 4. Biosynthesis of volatile alcohol and esters from aldehyde during ripening. J Agric Food Chem 25: 1165–1168 [Google Scholar]
- Yamashita I, Nemoto Y, Yoshikawa S. (1976) Formation of volatile alcohols and esters from aldehydes in strawberries. Phytochemistry 15: 1633–1637 [Google Scholar]
- Zabetakis I, Holden M. (1997) Strawberry flavour: analysis and biosynthesis. J Sci Food Agric 74: 421–434 [Google Scholar]
- Zanor MI, Rambla J-L, Chaïb J, Steppa A, Medina A, Granell A, Fernie AR, Causse M. (2009) Metabolic characterization of loci affecting sensory attributes in tomato allows an assessment of the influence of the levels of primary metabolites and volatile organic contents. J Exp Bot 60: 2139–2154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zini E, Biasioli F, Gasperi F, Mott D, Aprea E, Märk T, Patocchi A, Gessler C, Komjanc M. (2005) QTL mapping of volatile compounds in ripe apples detected by proton transfer reaction-mass spectrometry. Euphytica 145: 269–279 [Google Scholar]
- Zorrilla-Fontanesi Y, Cabeza A, Domínguez P, Medina JJ, Valpuesta V, Denoyes-Rothan B, Sánchez-Sevilla JF, Amaya I. (2011a) Quantitative trait loci and underlying candidate genes controlling agronomical and fruit quality traits in octoploid strawberry (Fragaria × ananassa). Theor Appl Genet 123: 755–778 [DOI] [PubMed] [Google Scholar]
- Zorrilla-Fontanesi Y, Cabeza A, Torres A, Botella M, Valpuesta V, Monfort A, Sánchez-Sevilla J, Amaya I. (2011b) Development and bin mapping of strawberry genic-SSRs in diploid Fragaria and their transferability across the Rosoideae subfamily. Mol Breed 27: 137–156 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.