Abstract
Nod Factor Receptor5 (NFR5) is an atypical receptor-like kinase, having no activation loop in the protein kinase domain. It forms a heterodimer with NFR1 and is required for the early plant responses to Rhizobium infection. A Rho-like small GTPase from Lotus japonicus was identified as an NFR5-interacting protein. The amino acid sequence of this Rho-like GTPase is closest to the Arabidopsis (Arabidopsis thaliana) ROP6 and Medicago truncatula ROP6 and was designated as LjROP6. The interaction between Rop6 and NFR5 occurred both in vitro and in planta. No interaction between Rop6 and NFR1 was observed. Green fluorescent protein-tagged ROP6 was localized at the plasma membrane and cytoplasm. The interaction between ROP6 and NFR5 appeared to take place at the plasma membrane. The expression of the ROP6 gene could be detected in vascular tissues of Lotus roots. After inoculation with Mesorhizobium loti, elevated levels of ROP6 expression were found in the root hairs, root tips, vascular bundles of roots, nodule primordia, and young nodules. In transgenic hairy roots expressing ROP6 RNA interference constructs, Rhizobium entry into the root hairs did not appear to be affected, but infection thread growth through the root cortex were severely inhibited, resulting in the development of fewer nodules per plant. These data demonstrate a role of ROP6 as a positive regulator of infection thread formation and nodulation in L. japonicus.
Small GTPases of the Rho family play critical roles in regulating cell motility, cell division, and directional growth (Etienne-Manneville and Hall, 2002). Rho GTPases have been grouped into the Rho, Rac, and Cdc24 subfamilies in animals and fungi, but they form a distinct subfamily in plants (Boureux et al., 2007). The plant Rho subfamily has been referred to as ROP, for Rho of plants, or RAC, because they are closely related to nonplant Rac GTPases (Yang, 2002). Rho GTPases from different organisms execute their biochemical functions in a similar manner, acting as molecular switches in signaling pathways (Etienne-Manneville and Hall, 2002; Kost, 2008). Rho GTPases associated with effector proteins in the GTP-bound state can stimulate downstream signaling, whereas those in the GDP-bound conformation are inactive. Rho GTPases are regulated by distinct groups of regulatory proteins, including GTPase-activating proteins (RhoGAPs), guanine nucleotide exchange factors (RhoGEFs), and guanine nucleotide dissociation inhibitors (RhoGDIs; Etienne-Manneville and Hall, 2002; Kost, 2008). ROP proteins in plants have been implicated in controlling cellular processes such as polarized cell growth, cell morphogenesis, hormone signaling, defense, and responses to oxygen deprivation (Yang, 2002; Agrawal et al., 2003; Yang and Fu, 2007). It has been demonstrated that ROP controls the tip growth of pollen tubes and root hairs by regulating actin organization (Molendijk et al., 2001; Fu et al., 2002; Jones et al., 2002; Gu et al., 2005). Members of the RhoGAP, RhoGEF, and RhoGDI families and several downstream effectors have also been characterized in plants (Borg et al., 1999; Kost, 2008; Yalovsky et al., 2008).
Analysis of Lotus EST sequences has revealed the expression of a ROP GTPase in root nodules (Yuksel and Memon, 2008). In Medicago truncatula, the increase in gene expression of MtROP3, MtROP5, and MtROP6 coincides with the induction of the Rhizobium-induced marker genes NOD FACTOR PERCEPTION (NFP), Rip1, and ENOD11. The GUS reporter driven by a ROP promoter has been shown to be induced by rhizobial inoculation (Liu et al., 2010). These observations suggest a role of ROP GTPases in the establishment of symbiotic nitrogen fixation in legumes (Yuksel and Memon, 2009).
The development of root nodules begins with the recognition of rhizobial Nod factors (NFs), which are capable of inducing a series of specific responses, including root hair deformation, cytoplasmic alkalinization, plasma membrane depolarization, and calcium influx and spiking (Kurkdjian, 1995; Ehrhardt et al., 1996; Niwa et al., 2001; Limpens and Bisseling, 2003; Gleason et al., 2006; Miwa et al., 2006). Rhizobial cells enter into the legume root through the curled root hairs and continue to penetrate the root cortex through a newly formed tubular structure known as infection threads (ITs). At the nodule primordium, rhizobia are released from the ITs into the cytoplasm of nodule cells (Schauser et al., 1999; Esseling et al., 2003). The mechanisms underlying the induction of root hair curling and IT formation after the perception of bacterial NFs are largely unknown.
Recent genetic and molecular biology studies have resulted in the identification of a set of host genes essential for NF detection and signal transduction in Lotus japonicus, M. truncatula, and soybean (Glycine max). Recognition of rhizobial NFs is mediated by the LysM-containing receptor-like kinases Nod Factor Receptor1 (NFR1) and NFR5 in L. japonicus (Madsen et al., 2003; Radutoiu et al., 2003, 2007; Bek et al., 2010) and LysM domain-containing receptor-like kinase3 (LYK3) and NFP in M. truncatula (Amor et al., 2003; Arrighi et al., 2006; Smit et al., 2007). Soybean, a tetraploid legume, has duplicated copies of NFR: GmNFR1α, GmNFR1β and GmNFR5α, GmNFR5β. GmNFR1α is capable of fulfilling all similar functions of LjNFR1, whereas GmNFR1β cannot detect NFs at low Bradyrhizobium titers (Indrasumunar et al., 2010). Mutants of GmNFR5α and GmNFR5β can be functionally complemented by wild-type GmNFR5α and GmNFR5β, suggesting that both genes are functional (Indrasumunar et al., 2011).
The extracellular domain of NFR5 contains three LysM domains that are believed to bind specific NFs (Madsen et al., 2003; Bek et al., 2010). Lotus nfr5 mutants are nonnodulating and unresponsive to inoculation with Mesorhizobium loti or to signal molecules of purified bacterial NFs (Radutoiu et al., 2003). In M. truncatula, LYK3 (an ortholog of NFR1) interacts with and phosphorylates PUB1, an E3 ubiquitin ligase (Mbengue et al., 2010). How NFR5 regulates root hair curling, rhizobial infection, and nodulation in L. japonicus is unclear. In this study, we identify ROP6 GTPase as an interactor of NFR5 and provide evidence for a pivotal role of ROP6 as a positive regulator of nodule organogenesis in L. japonicus.
RESULTS
Isolation of ROP6 as an Interactor of NFR5
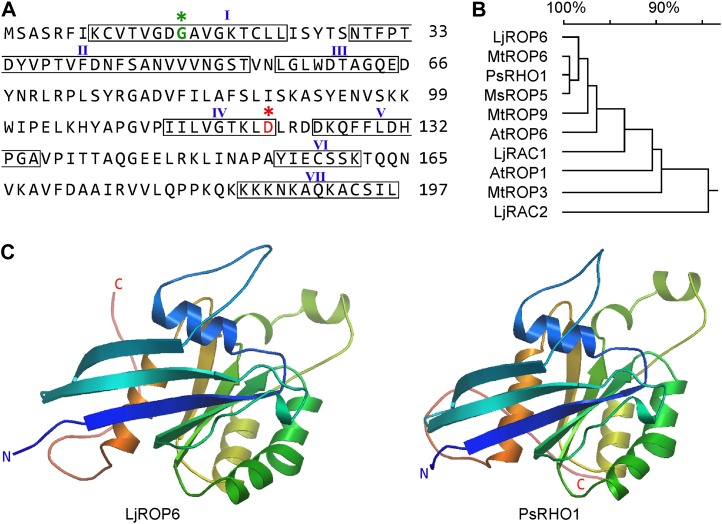
NFR5 is an atypical protein kinase that forms a heteromeric receptor complex with NFR1, which has protein kinase activity (Madsen et al., 2011). We used the atypical kinase domain (NFR5-PK) as bait to screen a Lotus cDNA library constructed in the prey vector pGADT7-Rec (Zhu et al., 2008). Among the 26 positive colonies identified, 12 were found to derive from a cDNA sequence in the L. japonicus database (Fig. 1A). Amino acid sequence analysis revealed that this protein is most closely related to ROP6 of M. truncatula and Arabidopsis (Arabidopsis thaliana; Fig. 1B; Supplemental Fig. S1). This clone is hereafter designated as L. japonicus ROP6 (GenBank accession no. JF260911).
Figure 1.
ROP6 GTPase in L. japonicus. A, Deduced amino acid sequence of ROP6. Boxed are seven conserved domains: GTPase domains (I and III), GDP/GTP-binding domains (IV and VI), effector domain (II), RHO insert region (V), and membrane localization domain (VII) or hypervariable region. B, Phylogenetic tree of ROP6 and its homologs (with GenBank accession numbers in parentheses) from Lotus japonicus (LjRAC1, Z73961; LjRAC2, Z73962), Medicago truncatula (MtROP3, AF498357.1; MtROP6, AF498358.1; MtROP9, AF498359.1), Pisum sativum (PsRHO1, L19093.1), Medicago sativa (MsROP5, AJ966571.1), and Arabidopsis thaliana (AtROP1, NM114989.3; AtROP6, AF031427.1). The phylogenetic tree was generated using DNAMAN software. C, Conservation of protein three-dimensional structures of LjROP6 and PsRHO1. The three-dimensional models were produced using modeler at the PS2 server (http://ps2.life.nctu.edu.tw/). N and C termini are indicated by N and C, respectively. [See online article for color version of this figure.]
The ROP6 cDNA contained a 594-bp open reading frame encoding a peptide of 197 amino acids with a molecular mass of 25 kD. Like other ROP GTPases, ROP6 contains seven functional domains: the GTPase domains (I and III), the GDP/GTP-binding domains (IV and VI), the effector domain (II), and the variable insert region (V) and C-terminal tail (VII). LjROP6 belongs to the type I ROP GTPases, which contain a canonical CaaL box that is prenylated primarily by genranylgeranyl transferase I (Sorek et al., 2007). The C-terminal hypervariable domains of ROP GTPases are determinants of their subcellular localizations. The predicted three-dimensional model of LjROP6 was highly conserved as compared with that of Pisum sativum RHO1 (Fig. 1C). Phylogenetic tree analysis revealed LjROP6 to be closely related to M. truncatula ROP6, P. sativum RHO1, Medicago sativa ROP5, and Arabidopsis ROP6 (Fig. 1B).
Interaction between NFR5 and ROP6 in Vivo and in Vitro
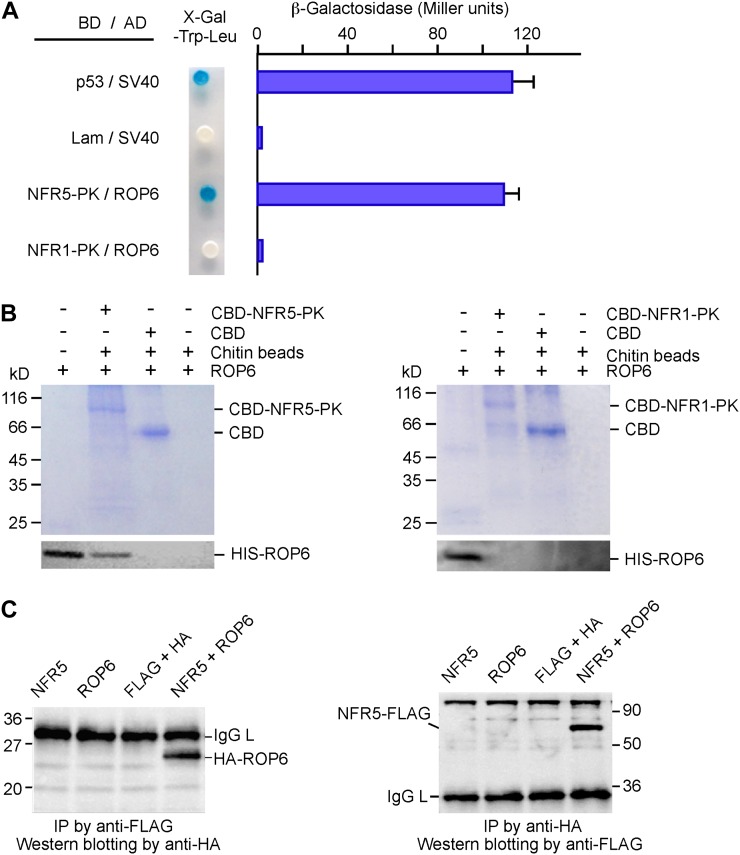
The strength of the ROP6/NFR5-PK interaction was almost as strong as that of the positive control between p53 and SV40, as evaluated by measuring the β-galactosidase activity (Fig. 2A). To assess the specificity of the ROP6/NFR5-PK interaction, we used NFR1-PK to replace NFR5-PK and did not observe any interaction, suggesting that ROP6 specifically recognizes NFR5-PK. We then verified the ROP6/NFR5-PK interaction in an in vitro pull-down assay. Chitin-binding domain (CBD)-tagged NFR5-PK and His-tagged ROP6 were expressed in Escherichia coli and purified using the corresponding affinity tags. NFR5-PK immobilized on chitin beads was incubated with purified soluble ROP6. After washing with buffer, proteins retained on the beads were eluted and resolved by SDS-PAGE. The presence of ROP6 on the beads was detected by immunoblotting with anti-ROP6 antibody. We also used NFR1-PK to substitute for NFR5-PK, and the results show that ROP6 could not be pulled down by NFR1-PK (Fig. 2B).
Figure 2.
Interaction between ROP6 and NFR5. A, Yeast two-hybrid assays. ROP6 was expressed as a fusion protein with the activation domain (AD in pGADT7); NFR1-PK and NFR5-PK were expressed as fusion proteins with the Gal4 DNA-binding domain (BD in pGBKT7). Yeast cells harboring the plasmids were grown on SD/-Leu-Trp medium containing X-gal (left). The strength of the interaction was evaluated by β-galactosidase activity (right). The combinations p53/SV40 and Lam/SV40 served as positive and negative controls, respectively. NFR1-PK was used to replace NFR5-PK for testing interaction specificity. B, In vitro protein pull-down assay. Purified soluble ROP6 was mixed with CBD-tagged NFR5-PK (left), which was immobilized to chitin beads. After washing, proteins pulled down by chitin beads were separated on SDS-PAGE gels. The gels were stained with Coomassie blue (top) or used for western blotting using anti-ROP6 antibodies (bottom). As a control for interaction specificity, CBD-tagged NFR1-PK was used to replace CBD-tagged NFR5-PK in the same assay (right). The positions of CBD and CBD-tagged NFR-PK are indicated. C, Coimmunoprecipitation of proteins expressed in plants. FLAG-tagged NFR5 and HA-tagged ROP6 were expressed in leaves of tobacco via Agrobacterium-mediated transient transformation. A leaf extract or a mixture of two extracts was incubated with anti-FLAG antibody, followed by reaction with protein G beads. Immunoprecipitates (IP) were separated by SDS-PAGE and immunoblotted with anti-HA antibody (left). The same experiment was performed again except that the order of antibody uses was reversed. Anti-HA antibody was used to precipitate the immunocomplex, and anti-FLAG antibody served as the primary antibody in western-blot analysis (right). Molecular masses of marker proteins in kD and the positions of ROP6, NFR5, and IgG light chain (IgG L) are indicated. [See online article for color version of this figure.]
ROP6 was initially identified using the kinase domain of NFR5 (NFR5-PK). The full-length NFR5 contains a transmembrane domain and was difficult to express in E. coli. We changed affinity tags and expressed the full-length proteins of NFR5 and ROP6 in tobacco (Nicotiana benthamiana) leaves. FLAG-tagged NFR5 and hemagglutinin (HA)-tagged ROP6 were coexpressed in leaf cells under the control of the 35S promoter. The presence of the recombinant proteins in plant cells was confirmed on immunoblots using antibodies against the tags (Supplemental Fig. S2), and the interaction was examined by coimmunoprecipitation. Leaf extracts were reacted with immobilized anti-FLAG antibody and anti-HA antibody, respectively. The immunoprecipitated products were resolved by SDS-PAGE, and the presence of the interacting partner was detected by immunoblot analysis using an antibody different from the one used for immunoprecipitation. As shown in Figure 2C, ROP6 could be detected in the coimmunoprecipitation products only when NFR5, but not the empty vector control, was coexpressed in tobacco leaves. We also switched the order of antibodies used in coimmunoprecipitation and western-blot analysis. As shown in Figure 2C, NFR5 could be pulled down by ROP6 but not in the mock controls. These results clearly indicate that the interaction between NFR5 and ROP6 occurs in plant cells.
Colocalization of NFR5 and ROP6 at the Plasma Membrane
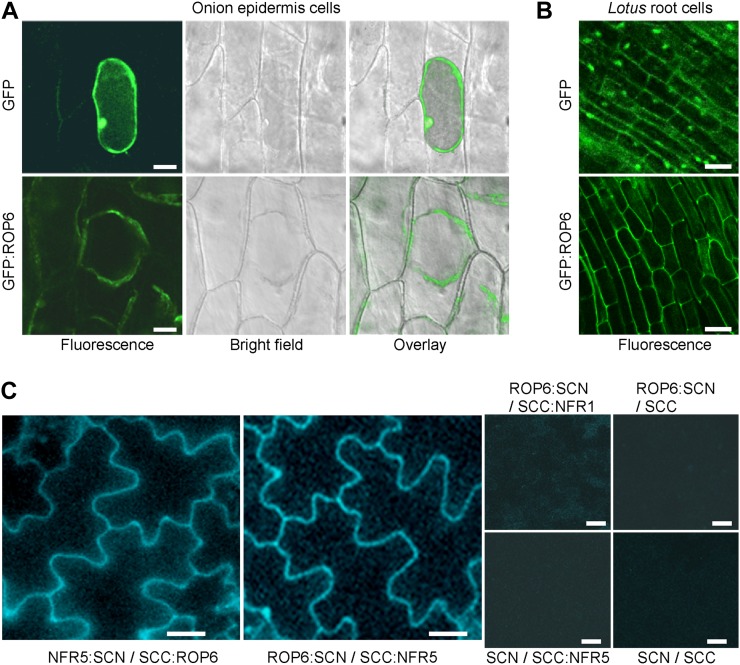
Previous studies have shown that GFP-tagged ROP proteins are associated with the plasma membrane and display enhanced membrane association at growth sites in pollen tubes, root hairs, and leaf epidermal cells (Ivanchenko et al., 2000; Molendijk et al., 2001; Fu et al., 2002; Jones et al., 2002; Lavy and Yalovsky, 2006; Sorek et al., 2007). Different ROPs have been localized to the plasma membrane, perinuclear organelle, and cytoplasm in plants (Bischoff et al., 2000; Ivanchenko et al., 2000; Jones et al., 2002). The activation of ROP GTPase is often accompanied by translocation from the cytoplasm to the plasma membrane, where it interacts with downstream effectors. Recently, NFR1 and NFR5 have been localized to the plasma membrane, where the two proteins form a heteromeric protein complex (Madsen et al., 2011). We expressed GFP-tagged ROP6 under the control of the cauliflower mosaic virus 35S promoter in onion (Allium cepa) epidermal cells through particle bombardment (Fig. 3A). GFP alone was present in the cytoplasm and nuclei, whereas GFP-tagged ROP6 was detected in the cytoplasm and plasma membrane and was excluded from the nucleus. In order to ensure a clear identification of the cell wall from the cytoplasm, we treated onion epidermal cells with 4% NaCl for 5 min to induce plasmolysis right before microscopic observation. The fluorescence signal was localized to the cytoplasm and plasma membrane (Fig. 3A). Similar localization results were also observed in Lotus hairy roots expressing GFP-tagged ROP6 (Fig. 3B). We conclude that ROP6 is present in the cytoplasm and plasma membrane.
Figure 3.
Subcellular localization of ROP6 and its interaction with NFR5 in planta. A, Localization of GFP-tagged ROP6 in onion cells. Plasmids expressing GFP alone or GFP:ROP6 were delivered to the onion epidermal cells via particle bombardment. Onion epidermal cells expressing GFP:ROP6 were treated with 4% NaCl for 5 min to induce plasmolysis before imaging. Bars = 50 μm. B, Localization of GFP-tagged ROP6 in L. japonicus hairy roots. GFP alone served as a control. Bars = 20 μm. C, Interaction of ROP6 and NFR5 in planta. Cyan fluorescent protein (CFP) was split into N and C termini, which were fused to NFR5 and ROP6, respectively. The two constructs (NFR5:SCN and SCC:ROP6) were used to cotransfect tobacco leaves via Agrobacterium-mediated transient expression. The two fusion proteins were swapped with their CFP tags, and the resulting constructs (ROP6:SCN and SCC:NFR5) were coexpressed. Constructs SCC:NFR1 and CFP tags (SCN and SCC) were used to make combinations for coexpression and served as negative controls. Bars = 50 µm.
We performed bimolecular fluorescence complementation to assess the interaction between NFR5 and ROP6 in tobacco leaves. ROP6 and NFR5/NFR1 were fused to the split cyan fluorescent protein N terminus (SCN) and C terminus (SCC; Waadt et al., 2008), respectively. No fluorescence could be detected when the fusion proteins were expressed alone or coexpressed with an empty vector (Fig. 3C). When SCC:NFR5 with ROP6:SCN were coexpressed in the leaf cells, strong cyan fluorescent signals were detected at the plasma membrane. No fluorescence signal was observed when NFR1 was used to replace NFR5, suggesting that ROP6 interacts with NFR5 but not with NFR1 at the plasma membrane of living plant cells.
GTPase Activity of ROP6
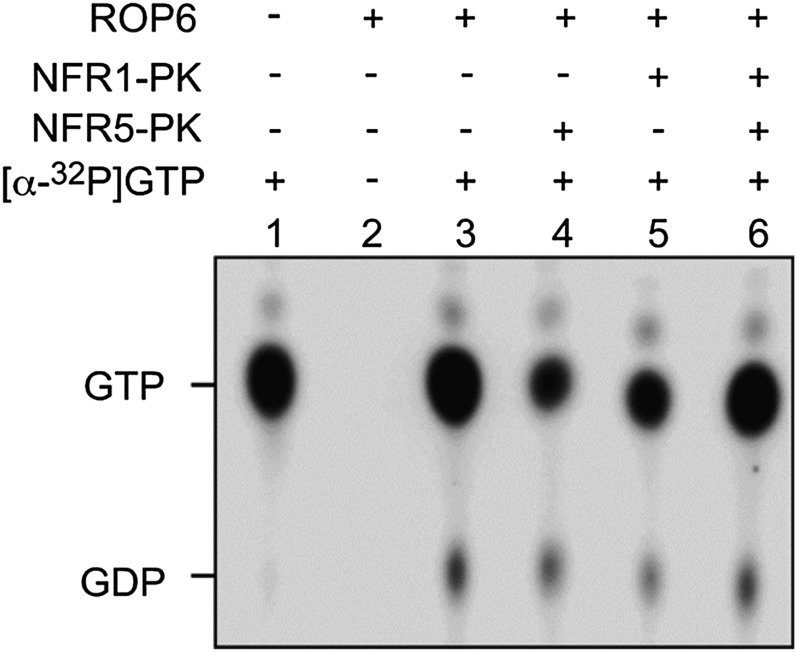
To test if the intrinsic GTPase activity of ROP6 was affected by its interaction partner, we measured the activity in the absence and presence of NFR5 and NFR1. Purified ROP6 was incubated with [α-32P]GTP, and the reaction product [α-32P]GDP was detected by autoradiography on thin-layer chromatography plates. ROP6 was able to hydrolyze GTP to GDP (Fig. 4, lane 3). When NFR1, NFR5, and a combination of both were added to the GTPase assay mix, the amount of GDP product was not changed (Fig. 4, lanes 4–6), suggesting that interaction with NFR5 had no effect on ROP6 GTPase activity.
Figure 4.
ROP6 GTPase activity assay. His-tagged ROP6, NFR1-PK, and NFR5-PK proteins were expressed in E. coli and purified using nickel beads. The GTPase activity of ROP6 protein was assayed in the presence of [α-32P]GTP. NFR1-PK and NFR5-PK were added to the reaction mix as potential effector proteins. The reaction products were separated by thin-layer chromatography on polyethyleneimine cellulose plates. The positions of GTP and GDP are indicated.
Expression of ROP6 in Roots and Nodules
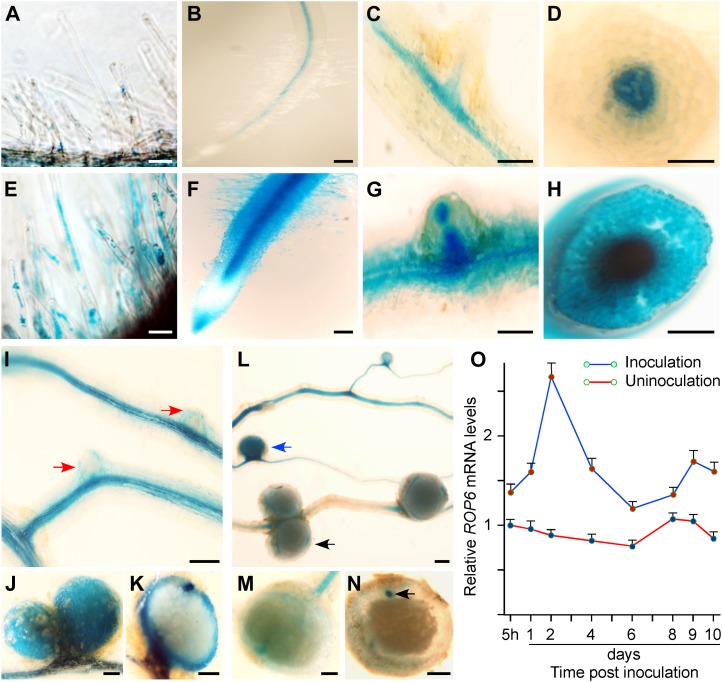
We took the quantitative real-time PCR approach to assess the gene expression pattern of ROP6 during the early stages of symbiosis. The expression level of ROP6 was elevated in roots 5 h after inoculation with M. loti, peaked 2 d after inoculation, and remained constant up to 10 d after inoculation, as compared with that in uninoculated control roots (Fig. 5O). To investigate the spatial pattern of ROP6 expression at the cellular level, the ROP6 gene promoter was used to drive the expression of the GUS reporter in transgenic hairy roots. In the control hairy roots, GUS activity was detected at a very low level in root hairs (Fig. 5A) and was expressed constitutively in the vascular bundles of roots and lateral roots (Fig. 5, B–D). After inoculation with M. loti, GUS reporter activity was significantly enhanced in root vascular bundles, and it was further expanded to the root hairs, root tips, and lateral root primordia (Fig. 5, E–H). The activity was concentrated in the cortex of developing nodules (Fig. 5, I–K) and was down-regulated drastically in mature and senescent nodules (Fig. 5, L–N). These observations were consistent with the results of real-time reverse transcription (RT)-PCR (Fig. 5O) and indicate that ROP6 may be involved in the process of rhizobial infection in L. japonicus.
Figure 5.
Up-regulation of ROP6 gene expression in roots after rhizobial infection and in developing nodules. Construct ROP6Pro:GUS contained a 1.4-kb ROP6 gene promoter fused to the GUS coding region. Transgenic hairy roots expressing ROP6Pro:GUS were stained for GUS reporter activity. A to D, In the absence of Rhizobium, ROP6Pro:GUS was expressed very weakly in root hairs (A) and moderately in root vascular tissues (B and D) and lateral root vascular tissues (C). E to H, After Rhizobium inoculation, ROP6Pro:GUS was highly expressed in most parts of the hairy roots, including root hairs (E and F), epidermal cells, root tips, and vascular tissues (F and G), the apical region of lateral root primordia (G), and root vascular tissues (H). I to N, GUS staining was intense in the cortex of nodule primordia (I) and developing nodules (J and K) and became very weak in mature nodules (L–N). Bars = 100 µm. O, Real-time PCR analysis. Total RNA was extracted from L. japonicus roots at different time intervals without inoculation or with M. loti inoculation. Real-time PCR was performed using ROP6-specific primers on RNA samples from three biological replicates. The expression levels were compared with the expression value of the uninoculated root of the first time point. Average values of relative gene expression levels are presented.
Decreases in Infection Thread Formation and Nodulation by ROP6 RNA Interference
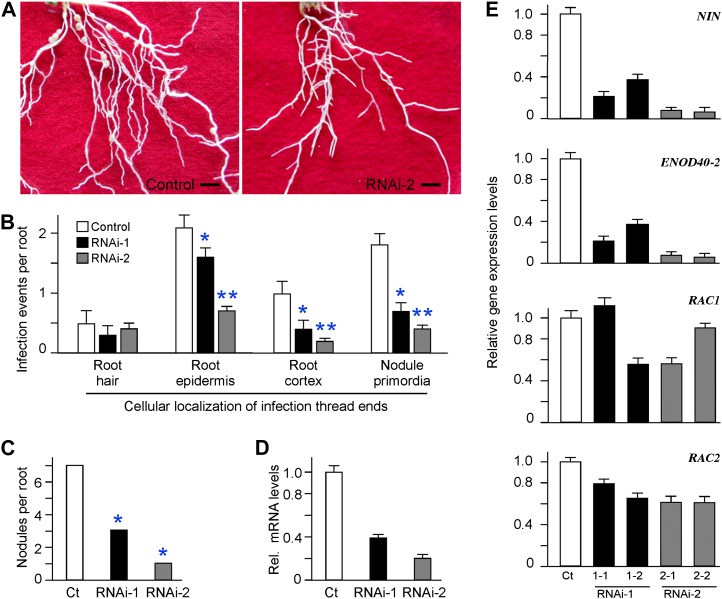
To investigate the effect of down-regulation of ROP6 mRNA on the early nodulation process, we generated transgenic hairy roots expressing ROP6 RNA interference (RNAi). Transgenic hairy roots expressing the empty vector served as a control. Two ROP6-specific RNAi constructs were made, with ROP6 RNAi-1 targeting the 188-bp 5′ untranslated region (UTR) and RNAi-2 targeting the 209-bp 3′ UTR of ROP6 mRNA (Supplemental Fig. S3). The two UTR sequences were chosen because they were specific to ROP6 and were not found in its two closely related homologs, RAC1 and RAC2 in L. japonicus. Real-time PCR analysis confirmed that the mRNA levels of RAC1 and RAC2 were not altered in transgenic ROP6 RNAi hairy roots as compared with that in the control hairy roots (Fig. 6E). In contrast, ROP6 transcripts showed a reduction of 60% in ROP6 RNAi-1 and 80% in RNAi-2 as compared with that in the control hairy roots (Fig. 6D).
Figure 6.
Effects of ROP6 RNAi expression on rhizobial infection and nodulation in L. japonicus. A, Images representing the hairy root systems expressing the empty vector (control) or ROP6 RNAi-2. Photographs were taken 3 weeks after inoculation with M. loti MAFF303099, and plants were grown in the absence of nitrogen fertilizer. Bars = 5 mm. B, Frequencies of infection events in transgenic hairy roots expressing empty vector p1302G (control) or ROP6 RNAi-1 and RNAi-2. Transgenic hairy roots were inoculated with M. loti strain MAFF303099 expressing the lacZ β-galactosidase. Nine days post rhizobial inoculation, the roots were stained with X-gal and the cellular distribution of infection threads was recorded with a microscope. Ten independent transgenic hairy root systems for each construct were generated. Four roots between 4 and 6 cm in length were chosen to represent each root system. Asterisks indicate statistical differences from the value of the control at * P < 0.01 and ** P < 0.001. C, Nodule numbers of transgenic hairy roots expressing the empty vector (control [Ct]) or ROP6 RNAi-1 and RNAi-2. Nodulation phenotypes were examined 3 weeks after inoculation with M. loti MAFF303099. D, Knockdown of ROP6 transcripts in transgenic hairy roots expressing ROP6 RNAi-1 and RNAi-2. The expression levels of ROP6 mRNA were estimated by real-time RT-PCR. The ROP6 expression values for RNAi-1 and RNAi-2 roots were compared with that of the control hairy roots expressing empty vector p1302G (control [Ct]), and relative mRNA levels are shown. E, Real-time PCR analysis of the expression of nodulation marker genes and ROP6 homologs in transgenic hairy roots expressing ROP6 RNAi. Total RNA was isolated from two representative root systems for each RNAi construct and used for real-time PCR amplification. Note that the expression of NIN and ENOD40-2, serving as the marker genes for rhizobial infection and early nodulation, was down-regulated by ROP6 RNAi expression. The expression of RAC1 and RAC2, two ROP6 homolog genes in L. japonicus, was not affected by ROP6 RNAi expression. [See online article for color version of this figure.]
As root growth defects may influence nodule formation, we examined the growth phenotype of ROP6 RNAi roots under nonnodulating conditions, and we did not observe any significant change as compared with the control hairy roots (data not shown). The ROP6 RNAi hairy roots were inoculated with a lacZ-labeled strain of M. loti, which provided a marker for identification of ITs after staining with 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside (X-gal) solution (Kang et al., 2011; Chen et al., 2012). Nine days after Rhizobium inoculation, the cellular locations and the numbers of ITs in the RNAi hairy roots were recorded. We divided ITs into four groups on the basis of the cellular locations of the IT tips: ITs emerging from curled root hair tips; ITs passing from the elongated root hairs to the root epidermis; ITs penetrating through the root cortex; and ITs reaching to the nodule primordia (Supplemental Fig. S4). As shown in Figure 6B, the numbers of ITs emerging from the curled root hairs were not changed significantly in RNAi roots, suggesting that down-regulation of ROP6 gene expression does not affect rhizobial entry into the root hair. The numbers of the rest of the three IT types were significantly reduced in ROP6 RNAi hairy roots as compared with those in the control. The total numbers of all three IT types together were 5.0 ITs per hairy root in the control, 2.6 in ROP6 RNAi-1, and 1.3 in RNAi-2 (Fig. 6B). The ROP6 RNAi effects on the last two types of ITs were very profound, demonstrating that the reduced ROP6 gene expression has deleterious effects on the growth of ITs through the root cortex and the connection of ITs with the nodule primordia.
We also recorded the number of nodules formed on the hairy roots 3 weeks after inoculation with M. loti. The average number of nodules per hairy root from two experiments was 6.3 in the control, 1.6 in ROP6 RNAi-1, and 0.7 in RNAi-2 (Fig. 6C; Table I). We noticed that all of the control hairy roots produced nodules, whereas about 30% of the ROP6 RNAi hairy roots did not develop any nodule at all. These results suggest that down-regulation of ROP6 gene expression by RNAi significantly impairs nodulation by inhibiting infection thread formation and nodule initiation. Thus, ROP6 may act as a positive regulator in the process of symbiosis in L. japonicus.
Table I. Nodulation phenotype of L. japonicus hairy roots expressing ROP6 RNAi.
Transgenic hairy roots of L. japonicus expressing empty vector p1302G (control) or ROP6 RNAi constructs (RNAi-1 and RNAi-2) were inoculated with M. loti. Two independent experiments (1 and 2) were performed. Mean numbers of nodules per root system formed 3 weeks post rhizobial inoculation with sd are shown. The number (n) of scored plants is indicated in parentheses.
| Experiment | Control | RNAi-1 | RNAi-2 | P |
|---|---|---|---|---|
| 1 | 6.8 ± 2.9 (n = 29) | 2.3 ± 2.0 (n = 25) | 0.9 ± 1.1 (n = 28) | <0.01 × 10−4 |
| 2 | 5.8 ± 1.5 (n = 20) | 0.8 ± 1.2 (n = 22) | 0.5 ± 0.8 (n = 18) | <0.01 × 10−3 |
Down-Regulation of Early Nodulation Genes in ROP6 RNAi Roots
The NIN gene is required for IT formation and nodule primordial initiation in L. japonicus (Schauser et al., 1999). A mutation in NIN blocks the entry of symbiotic bacteria into root hair cells completely, thus preventing the formation of nodule primordia. ENOD40-2 is a critical gene for nodule initiation and subsequent organogenesis, but it is not required for early infection by rhizobia (Kumagai et al., 2006). Both NIN and ENOD40-2 transcripts in L. japonicus accumulate rapidly in response to rhizobial inoculation. We selected representative hairy roots expressing ROP6 RNAi-1 and RNAi-2 and analyzed the expression levels of the two nodulation-related marker genes by real-time PCR. Both NIN and ENOD40-2 transcripts decreased markedly in ROP6 RNAi hairy roots as compared with those in the control (Fig. 6E). These results suggest that ROP6 is required for the NF-induced expression of early nodulin genes. Because NFR5 is essential for the perception of rhizobial NF signals by the host cells (Radutoiu et al., 2003), ROP6, as an interacting protein of NFR5, may thus act as a novel regulator of the NF signal recognition process, through which this unique Rho GTPase becomes involved in rhizobial infection and nodulation in L. japonicus.
DISCUSSION
ROP6 as a Possible Missing Link in NF-Mediated Cytoskeleton Reorganization
Both NFR1 and NFR5 are LysM-containing receptor-like kinases that act together as a heteromeric receptor complex that binds with rhizobial NF molecules in L. japonicus (Radutoiu et al., 2003, 2007). NFR5 is an atypical protein kinase that lacks the activation loop in the kinase domain and may not possess kinase activity at all (Madsen et al., 2003). The kinase activity of NFR1 is known to be essential for activating downstream symbiotic signaling pathways (Kouchi et al., 2010). NFR1 is capable of phosphorylating NFR5 in vitro and may function in a similar way in planta to transmit the NF signal to NFR5 upon binding with rhizobial NF molecules (Madsen et al., 2011). How NFR5 signal is relayed to downstream targets is not clear. One of the early observable downstream cellular events regulated by NFs is root hair deformation, which required cytoskeleton reorganization. It is unknown how NF signals are transmitted to initiate and regulate cytoskeleton reorganization. Animal receptor kinases are known to bind to Ras GTPases upon stimulation by hormones. Because plants do not have Ras orthologs, they may use ROP GTPases in a similar manner. There has been so far no direct evidence of the involvement of a Rho GTPase in receptor-mediated symbiotic signal transduction pathways in legumes. In this study, ROP6 GTPase was identified as a specific interaction partner of NFR5 in vitro and in planta (Figs. 2 and 3). ROP6 is present in the cytoplasm and plasma membrane (Fig. 3, A and B), the latter of which is a likely subcellular localization for interaction with the NFR1/NFR5 receptor complex. We speculate that the interaction between ROP6 and NFR5 may be involved in the NF-mediated cytoskeleton reorganization during the early stages of legume-Rhizobium symbiosis establishment.
A Role of ROP6 in Establishing a Symbiotic Relationship
Small GTPases act as switch molecules, turning on and off a great number of biological processes, including the protein trafficking pathway (Takeuchi et al., 2002), high-salinity tolerance (Bolte et al., 2000), and disease resistance (Ono et al., 2001; Agrawal et al., 2003). It is reasonable to assume that some of the small GTPases may play roles to regulate nodulation in legumes. Common bean (Phaseolus vulgaris) RabA2, a small GTPase of the RabA2 subfamily, is isolated as a differentially expressed sequence in root hairs infected with Rhizobium etli. Its protein is associated with vesicles that move along the cell. Expression of RabA2 RNAi results in severe impairment of root hair development, block of nodulation, and lack of induction of early nodulation (ENOD) genes, such as ERN1, ENOD40, and Hap5 (Blanco et al., 2009). Other small GTPases, including a ROP and two ARFs, have also been shown to be expressed in a nodule-specific pattern and implicated in the vacuolar trafficking pathway during symbiosis establishment (Pimpl et al., 2003; Yuksel and Memon, 2008, 2009).
Signal transduction after rhizobial attachment to the legume root hairs is a very complex process that involves specific molecular and cellular responses in different layers of the root, ultimately leading to successful rhizobial infection, nodule organogenesis, and nitrogen fixation. That the expression level of ROP6 was significantly increased after inoculation with M. loti (Fig. 5) indicates that its expression is stimulated by rhizobial infection. ROP6 was highly expressed in the tips of primary and lateral roots after rhizobial infection, which is consistent with the proposed role of ROP6 in regulating the tip growth. In pollen tubes, ROP accumulates at the plasma membrane of the growing tips (Li et al., 1999; Klahre and Kost, 2006).
To uncover the involvement of ROP6 in the symbiosis signal transduction pathway, we generated ROP6 RNAi hairy roots that had 60% to 80% reduction in ROP6 mRNA as compared with that in the control. As a result, nodulation in 30% of transgenic ROP6 RNAi hairy roots was eradicated. The drastic reduction of ITs by ROP6 RNAi indicates that ROP6 may play a pivotal role in the epidermis for the morphological changes to occur that lead to rhizobial infection. Consistent with this assumption is the reduction of early nodulin gene expression upon rhizobial infection in ROP6 RNAi hairy roots. ROP6 also appears to be necessary for molecular responses in both epidermal and cortical cells. We propose that the function of ROP6 in the symbiotic signal pathway is placed soon after the detection of rhizobia by the legume root and upstream of the signal connecting changes in the epidermis with responses in the cortical cells. These results demonstrate a role of ROP6 as a positive regulator in NFR5-mediated symbiotic signaling transduction pathways.
Requirement of ROP6 for NF-Induced ENOD Expression
The perception of NFs occurs primarily in the epidermal cells, where initial induced responses in the plant lead to ENOD induction in the zone of actively growing root hairs. ENOD expression is regulated by several transcription factors (Smit et al., 2005; Andriankaja et al., 2007; Middleton et al., 2007). Our data show that ROP6 is also required, probably indirectly, for ENOD expression, because the induction of NIN and ENOD40-2 was drastically reduced by rhizobial infection in ROP6 RNAi hairy roots (Fig. 6E). Therefore, it appears that ROP6 is required at an early stage after NF perception and prior to early signaling events and root hair deformation. This temporal pattern is consistent with our identification of ROP6 from a cDNA library constructed using mRNA from roots at the early stages of rhizobial infection (Zhu et al., 2008). It is also consistent with the induction pattern of ROP6 transcripts by rhizobial inoculation (Fig. 5). We propose that ROP6 is involved in the control of early infection events, exerts its function at a stage upstream of ENOD gene induction, and may serve as an early molecular marker for the establishment of a symbiotic relationship with compatible rhizobia in L. japonicus.
Potential Downstream Targets of ROP6
Typically, a mammalian Ras GTPase relays an external signal through a mitogen-activated protein kinase (MAPK) pathway. Plants do not have Ras orthologs and may employ Rho GTPases in a similar manner. We propose that ROP6 GTPase may relay the NF signal from NFR5 to a MAPK cascade. Because NFR5 has no kinase activity, the signal relay from NFR5 to ROP6 may not involve phosphorylation. Because the GTPase activity of ROP6 is not affected by NFR5, the signal relay from NFR5 to ROP6 may not involve the regulation of ROP6 GTPase activity. The molecular mechanism of the signal relay from NFR5 to ROP6 remains a subject of future research.
SymRK is a member of a large family of Leu-rich repeat receptor-like kinases and is required for legume roots to perceive NF signals and for root nodule development. SymRK interacts with SymRK-Interacting Protein2 (SIP2), a typical MAPK kinase in L. japonicus (Chen et al., 2012). Signaling from Rho and Rho-associated protein kinase to MAPK pathways is known to be a key step in regulating cell migration and proliferation in Glioblastoma multiforme (Zohrabian et al., 2009). The Ste20-related kinase and Rac-type GTPase via the JNK-like MAPK signaling pathway participate in the heavy metal stress response in Caenorhabditis elegans (Fujiki et al., 2010). OsRac1, a ROP6 homolog from rice (Oryza sativa), forms a complex with MAPK6, RAR1, SGT1, Hsp90, and RACK1, thereby transmitting signals of innate immunity to downstream components (Lieberherr et al., 2005; Thao et al., 2007; Nakashima et al., 2008; Fujiwara et al., 2009). It would be of interest to explore how ROP GTPases relay the NF signal to the SIP2 MAPK signaling cascade in L. japonicus.
In this report, we have described the identification of ROP6 as an interacting partner of NFR5. Despite the high sequence similarity, NFR1 cannot replace NFR5 in this interaction. Our results demonstrate a pivotal role of ROP6 in IT growth through the root cortex and in nodule development in L. japonicus. Because the interaction with NFR5 did not change ROP6 GTPase activity, some other GTPase-regulating proteins and downstream target(s) may be involved in the initiation of signals from NFs to cytoskeleton reorganization during symbiosis establishment. The identification of these protein components would facilitate elucidation of the mechanisms underlying rhizobial infection and nodule organogenesis. This work has identified a new player in the NF signaling arena and opened a new avenue for future research in the fields of ROP signaling and biological nitrogen fixation.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Seeds of Lotus japonicus MG-20 were surface sterilized in 75% ethanol for 1 min, followed by 15 min in 5% sodium hypochlorite and washing six times with sterile water. The seeds were placed on sterile water-soaked filter paper in petri dishes for germination for 48 h at 22°C in the dark. Seedlings were planted in pots on sterile sands supplemented with nitrogen-free Fahraeus medium (Fahraeus, 1957) and were grown in a growth chamber maintained at 22°C with a 16-h-light/8-h-dark cycle. Five-day-old seedlings were inoculated with approximately 107 colony-forming units mL−1 Mesorhizobium loti MAFF303099. Roots collected at different time intervals after rhizobial inoculation were immediately frozen in liquid nitrogen. Wild-type tobacco (Nicotiana benthamiana) plants were grown in a growth chamber at 22°C and 70% relative humidity under a 16-h-light/8-h-dark photoperiod for about 1 to 1.5 months before infiltration with Agrobacterium tumefaciens. After infiltration, plants were maintained under the same growth conditions.
Library Screening for Interaction Clones
The NFR5 cDNA fragment (GenBank accession no. AJ575255.1) encoding the kinase domain (amino acid residues 272–595) was amplified by PCR using primers 5′-GGGAATTCATGCGCAGAAAGAAGGCT-3′ and 5′-GGGGTCGACTTAACGTGCAGTAATGG-3′. The PCR product was fused in frame with the GAL4 DNA-binding domain in pGBKT7. Screening of interaction clones was carried out according to the manufacturer’s instructions (Clontech). A total of 1 × 107 transformants were selected for growth on synthetic dextrose (SD)/-Leu-Trp-His-Ade medium. Positive clones were verified by retransformation of the rescued plasmids into yeast cells containing the bait plasmid (pGBKT7-NFR5-PK). Plasmid pGBKT7-Lam (Clontech) was used as a negative control. Colonies growing on SD/-Leu-Trp-His-Ade medium were transferred to selective medium containing X-gal (80 μg mL−1) or were lifted on filters as described (Ma et al., 2007).
β-Galactosidase Assay
Yeast cells grown in liquid selection medium were pelleted and washed twice with Z-buffer (60 mm Na2HPO4, 40 mm NaH2PO4, 10 mm KCl, and 1.0 mm MgSO4, pH 7.0). The cells were resuspended in 100 μL of Z-buffer and permeabilized by three freeze-thaw cycles in liquid nitrogen and a 37°C water bath. Cell extracts were added to 0.7 mL of Z-buffer containing 50 mm β-mercaptoethanol and 160 μL of O-nitrophenyl β-d-galactopyranoside (4 mg mL−1 in Z-buffer). After incubation at 30°C for 30 min, the reaction was terminated by the addition of 0.4 mL of 1.0 m Na2CO3. The reaction mixture was centrifuged for 10 min at 14,000g to remove cell debris. β-Galactosidase activity was measured at an optical density (OD) at 420 nm.
Expression and Purification of Fusion Proteins
Full-length ROP6 cDNA was amplified by PCR and inserted in frame at the NdeI/EcoRI site of pET28a vector (Novagen), generating pET-ROP6. The coding regions corresponding to the kinase domain of NFR1 (amino acids 325–623) and NFR5 (amino acids 272–595) were cloned into pTYB12 (New England Biolabs), yielding pTYB-NFR1-PK and pTYB-NFR5-PK. For protein expression, Escherichia coli BL21-Codon Plus (DE3)-RIL (Stratagene) harboring plasmids were induced with 0.1 mm isopropylthio-β-galactoside for 5 h at 20°C. His-tagged proteins were purified using nickel-agarose beads (Qiagen) under native conditions and eluted with an imidazole solution (137 mm NaCl, 2.7 mm KCl, 10 mm Na2HPO4, 2 mm KH2PO4, and 200 mm imidazole, pH 8.0). CBD-tagged proteins were purified using chitin beads (New England Biolabs). Purified proteins were desalted by dialysis in phosphate-buffered saline buffer and concentrated with polyethylene glycol-8000 powder.
In Vitro Protein-Protein Interaction
To assay in vitro protein-protein interaction, His-tagged ROP6 was purified using nickel-agarose beads and eluted as a soluble protein. CBD-tagged NFR5-PK was absorbed on chitin beads and incubated with 20 μg of soluble ROP6 protein in 1 mL of interaction buffer (20 mm Tris-HCl, 100 mm KCl, 2 mm MgCl2, and 5% glycerol, pH 8.0) for 1 h on ice with gentle shaking. The chitin beads were washed three times with 1.0 mL of TEG buffer (20 mm Tris-HCl, 1 mm EDTA, and 5% glycerol, pH 8.0). Retained proteins were eluted by boiling for 5 min in 1× SDS sample buffer (2% SDS, 29.1 mm L−1 Tris, pH 6.8, 10% glycerol, and 0.01% bromphenol blue) and analyzed by 12% SDS-PAGE. Proteins on the gel were visualized by staining with Coomassie Brilliant Blue R250 or were transferred to nitrocellulose membrane for detection using anti-His antibody. To assay the interaction between NFR1 and ROP6, CBD-tagged NFR1-PK was used to replace NFR5-PK.
Infiltration of Tobacco Leaves with Agrobacterium
Cells of Agrobacterium strain EHA105 containing plasmids were selected on Luria-Bertani (LB) medium containing appropriate antibiotics at 28°C. A single colony was inoculated to 5 mL of LB medium, and the culture was grown at 28°C in a shaker for 48 h. The cells were transferred to fresh LB medium containing 10 mm MES (pH 5.6) and 40 μm acetosyringone (1:100 ratio, v/v). After 16 h of growth at 28°C or when the culture reached an OD600 of 3.0, the bacterial cells were spun down gently at 3,200g for 10 min. The pellet was resuspended in 10 mm MgCl2 at a final OD600 of 1.5. For Agrobacterium strain p19, a final OD600 of 1.0 was used instead. Acetosyringone at a final concentration of 200 μm was added to the Agrobacterium solution, which was kept at room temperature for at least 3 h without shaking. For coinfiltration, an equal volume of different Agrobacterium strains carrying plasmids was mixed together prior to infiltration. Infiltration of leaves with Agrobacterium cells was conducted by slowly depressing the plunger of a 1-mL disposable syringe to the surface of fully expanded leaves.
Preparation of Polyclonal Antibodies against ROP6
His-tagged recombinant ROP6 protein was expressed in E. coli and purified on nickel beads. The protein was separated by SDS-PAGE. Gel slices containing ROP6 protein were used to inject New Zealand rabbits. Titers of the antibody were assayed using ELISA. The anti-ROP6 antibody was used to detect ROP6 protein on western blots.
Protein Extraction and Western-Blot Analysis
Infiltrated leaves of tobacco plants were harvested and ground in liquid nitrogen. Leaf powders were resuspended in extraction buffer on ice. Leaf extract was centrifuged at 16,000g at 4°C for 30 min, and the supernatant was used for western-blot analysis. The denaturing buffer used for protein extraction contained 50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 0.1% Nonidet P-40, 4 m urea, and 1 mm phenylmethylsulfonyl fluoride. The native extraction buffer contained 50 mm Tris-MES, pH 8.0, 0.5 m Suc, 1 mm MgCl2, 10 mm EDTA, 5 mm dithiothreitol, and protease inhibitor cocktail Complete Mini tablets (Roche; http://www.roche.com/). Proteins of leaf extracts were separated by 12% PAGE and electroblotted to a nitrocellulose membrane at 25 V for 40 min. The membrane was blocked with Tris-buffered saline plus Tween 20 containing 5% skim milk powder for 2 h at room temperature. After incubation with primary antibody and then with secondary antibody, the membrane was transferred for protein detection using a Thermo SuperSignal West Pico kit (Thermo Scientific). The sources and dilutions of antibodies were as follows: anti-HA antibody (sc-7392AC [Santa Cruz] at 1:1,000), anti-FLAG antibody (F3165 [Sigma] at 1:1,000), and goat anti-mouse horseradish peroxidase-conjugated antibody (00001-1 [Proteintech] at 1:2,500).
Immunoprecipitation
Three days after infiltration, leaves of tobacco plants expressing HA-tagged ROP6 and FLAG-tagged NFR5 were harvested and ground in liquid nitrogen separately in native extraction buffer. The two leaf extracts were mixed together in a volume ratio of 1:1. Corresponding antibodies were added to the cell lysates (10 μg mL−1), and MG132 was added at a final concentration of 50 μm to prevent protein degradation. The mixtures were kept at 4°C with gentle shaking for 3 h to overnight. The immunocomplex was immobilized by adding 20 μL mL−1 protein G-agarose beads (Millipore) and shaking at 4°C for another 3 h. The beads were pelleted by centrifugation at 14,000g for 5 s and washed three times with cold phosphate-buffered saline. Proteins were solubilized with SDS sample buffer and detected by western-blot analysis.
Bimolecular Fluorescence Complementation Analysis
The full-length NFR5 coding region was amplified by PCR and cloned into the SpeI/XhoΙ site of pSCYCE(R) to obtain the NFR5:SCC fusion. Full-length ROP6 was cloned into the BamHI/SmaΙ site of pSCYNE to obtain the ROP6:SCN fusion. The constructs were transferred into Agrobacterium strain GV3101 by electroporation and used for transient protein expression in tobacco. Different Agrobacterium strains were mixed to a final OD600 of 0.5 at a 1:1 ratio. The mixture was kept at room temperature for 2 to 4 h and infiltrated into the top leaves of 6-week-old tobacco plants. Cyan fluorescence was observed 40 h after infiltration using a confocal microscope with a cyan fluorescent protein filter set of excitation at 405 nm and emission at 477 nm.
In Vitro GTPase Activity Assay
GTPase activity was assayed in a 25-μL reaction mixture containing 50 mm Tris-HCl (pH 8.0), 5 mm MgCl2, 0.1 mm dithiothreitol, 13 μm cold GTP, 13 nm [α-32P]GTP (1 μCi, 3,000 Ci mmol−1), and 0.1 μg of purified proteins. The reaction was carried out at 37°C for 60 min. Aliquots (0.5 μL) of the reaction products were applied to polyethyleneimine cellulose plates. GTP was separated from GDP by thin-layer chromatography in a solution containing 1.6 m LiCl and 1 m HAc. Radioactive signal was scanned with a Fujifilm FLA-5100 phophorimager.
Gene Expression Analysis
The expression profile of the ROP6 gene was analyzed by quantitative real-time PCR using RNA isolated from L. japonicus roots after inoculation with M. loti MAFF303099. Roots were collected at different time intervals. Total RNA was isolated using TRIZOL reagent (Invitrogen) and treated with DNase I (Promega), followed by extraction with phenol:chloroform. First-strand cDNA was synthesized from 0.5 ng of total RNA using oligo(dT) primer. Reactions of quantitative real-time PCR were performed using the one-step SYBR PrimeScript RT-PCR kit II (Takara) on a Roche LightCycler apparatus according to the manufacturer’s instructions. ROP6 transcripts were amplified using forward primer 5′-TACCAGCAACACCTTCCCCACCG-3′ and reverse primer 5′-TCCGGCAGTGTCCCATAAACCCAG-3′. The Polyubiquitin (AW720576) transcript served as an internal control and was amplified using forward primer 5′-TTCACCTTGTGCTCCGTCTTC-3′ and reverse primer 5′-AACAACAGCACACACAGACAATC-3′. The thermal cycle was set as follows: 95°C for 10 s, and 40 cycles of 95°C for 5 s and 60°C for 30 s. Three independent biological replicates were performed for each condition tested.
Subcellular Localization of ROP6 in Onion Epidermal Cells
The full-length ROP6 coding sequence was cloned into the NcoI/SpeI site of pCAMBIA1302 vector (CAMBIA). The plasmid was used for transient expression in onion (Allium cepa) epidermal cells by particle bombardment using the Biolistic PDS-1000/He Particle Delivery System (Bio-Rad). After incubation for 12 to 24 h at 23°C in the dark, the epidermal cell layers were examined using a confocal laser-scanning microscope with a filter set of excitation at 488 nm and emission at 550 nm.
GUS Staining
For ROP6Pro:GUS reporter analysis, a 1.3-kb genomic DNA upstream of the ROP6 coding region (−1,379 to −86 bp from ATG) was amplified by PCR using primers 5′-GCGTCGACGAAAGTGTATGCTGGACACG-3′ and 5′-CCGGAATTCCAGATCTTCAACCTGAGGAC-3′. The promoter was cloned into the SalI/EcoRΙ site of the p1391Z vector. Transgenic hairy roots of L. japonicus expressing ROP6Pro:GUS were incubated with 5-bromo-4-chloro-3-indolyl-β-D-glucuronide (X-Gluc) solution (50 mm sodium phosphate buffer, pH 7.2, 0.05% Triton X-100, 2 mm potassium ferrocyanide, 2 mm potassium ferricyanide, and 2 mm X-Gluc [Sigma]). GUS activity of hairy roots without rhizobial inoculation served as a control.
Construction of the ROP6 RNAi Plasmid
For the construction of ROP6 RNAi-1, a 188-bp fragment of the 5′ UTR of ROP6 was amplified by RT-PCR using primers 5′-GCGTCGTCGTCATTGATTC-3′ and 5′-CCCTTTTACTCTTCTTCTC-3′. For ROP6 RNAi-2 construction, a 209-bp fragment of the 3′ UTR was amplified using 5′-TTCGCTGCGTCTGAACATTTC-3′ and 5′-GAAATCACAATAATTCCA-3′. The forward primers were attached to a SmaI or PstI site and the reverse primers to a BamHI or XbaI site. PCR products were digested with SmaI-BamHI or with PstI-XbaI and ligated into pCAMBIA1301-35S-int-T7. The resulting construct contained both the sense and antisense ROP6 cDNA fragments in a tandem arrangement. The sense and antisense sequences were separated by an intron sequence from the Arabidopsis (Arabidopsis thaliana) Actin11 gene. The RNAi construct was placed behind the cauliflower mosaic virus 35S promoter. The constructs were transferred into Agrobacterium rhizogenes LBA1334 by electroporation. Plants containing transformed hairy roots were transferred to pots filled with vermiculite and sand (1:1) and inoculated with M. loti MAFF303099. The plants were grown in a chamber in a 16-h/8-h day/night cycle at 22°C for 5 to 7 d.
Hairy Root Transformation and Identification
Hairy root transformation of L. japonicus MG-20 using A. rhizogenes strain LBA1334 was performed essentially as described previously (Kang et al., 2011; Chen et al., 2012). Seedlings maintained in a growth chamber were cut at the base of the hypocotyls and placed in the A. rhizogenes suspension in a petri dish for several minutes. The seedlings with hypocotyls were transferred onto Murashige and Skoog (Sigma) plates with 1.5% Suc and cocultivated for 5 d in a growth chamber. The plants were transferred onto fresh Murashige and Skoog medium containing 100 µg mL−1 cefotaxime and grown for another 10 d until hairy roots developed from the section of hypocotyls. For the selection of transformed hairy roots, a short tip (2–3 mm) of each hairy root was excised for GUS staining [100 mm sodium phosphate buffer, pH 7.0, 0.1% Triton X-100, 0.1% N-laurylsarcosine, 10 mm Na2EDTA, 1 mm K3Fe(CN)6, 1 mm K4Fe(CN)6, and 0.5 mg mL−1 X-Gluc] at 37°C overnight in the dark. Each hairy root was labeled properly. The whole hairy root was discarded if the tip was GUS negative. A hairy root was allowed to develop into a root system if its tip was GUS positive.
Rhizobial Infection Assay
For the rhizobial infection assay, transgenic hairy roots were inoculated with M. loti strain MAFF303099 constitutively expressing the lacZ reporter gene and grown in a pot containing sand:vermiculite (1:1, v/v). Nine days after inoculation, hairy roots were stained for β-galactosidase activity as described elsewhere (Tansengco et al., 2003). Infection threads were observed using an Olympus BX51 microscope under bright-field illumination.
Sequence data from this article can be found in GenBank under accession number JF260911.2.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Conserved domains of LjROP6 protein and its homologs.
Supplemental Figure S2. Detection of recombinant ROP6 and NFR5 proteins in transfected leaves of tobacco.
Supplemental Figure S3. DNA sequences of ROP6 RNAi constructs.
Supplemental Figure S4. Classification of infection threads in transgenic hairy roots.
Supplementary Material
Acknowledgments
We thank Dr. Guojiang Wu for the M. loti MAFF303099 strain and Dr. Da Luo for pCAMBIA1301-35S-int-T7. We also thank Yao Hang for technical advice on transient expression and microscopy.
Glossary
- NF
Nod factor
- IT
infection thread
- HA
hemagglutinin
- RNAi
RNA interference
- UTR
untranslated region
- ENOD
early nodulation
- MAPK
mitogen-activated protein kinase
- SD
synthetic dextrose
- OD
optical density
- LB
Luria-Bertani
- X-Gluc
5-bromo-4-chloro-3-indolyl-β-glucuronic acid
- RT
reverse transcription
References
- Agrawal GK, Iwahashi H, Rakwal R. (2003) Small GTPase ‘Rop’: molecular switch for plant defense responses. FEBS Lett 546: 173–180 [DOI] [PubMed] [Google Scholar]
- Amor BB, Shaw SL, Oldroyd GE, Maillet F, Penmetsa RV, Cook D, Long SR, Dénarié J, Gough C. (2003) The NFP locus of Medicago truncatula controls an early step of Nod factor signal transduction upstream of a rapid calcium flux and root hair deformation. Plant J 34: 495–506 [DOI] [PubMed] [Google Scholar]
- Andriankaja A, Boisson-Dernier A, Frances L, Sauviac L, Jauneau A, Barker DG, de Carvalho-Niebel F. (2007) AP2-ERF transcription factors mediate Nod factor dependent Mt ENOD11 activation in root hairs via a novel cis-regulatory motif. Plant Cell 19: 2866–2885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrighi JF, Barre A, Ben Amor B, Bersoult A, Soriano LC, Mirabella R, de Carvalho-Niebel F, Journet EP, Ghérardi M, Huguet T, et al. (2006) The Medicago truncatula lysine motif-receptor-like kinase gene family includes NFP and new nodule-expressed genes. Plant Physiol 142: 265–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bek AS, Sauer J, Thygesen MB, Duus JØ, Petersen BO, Thirup S, James E, Jensen KJ, Stougaard J, Radutoiu S. (2010) Improved characterization of nod factors and genetically based variation in LysM receptor domains identify amino acids expendable for nod factor recognition in Lotus spp. Mol Plant Microbe Interact 23: 58–66 [DOI] [PubMed] [Google Scholar]
- Bischoff F, Vahlkamp L, Molendijk A, Palme K. (2000) Localization of AtROP4 and AtROP6 and interaction with the guanine nucleotide dissociation inhibitor AtRhoGDI1 from Arabidopsis. Plant Mol Biol 42: 515–530 [DOI] [PubMed] [Google Scholar]
- Blanco FA, Meschini EP, Zanetti ME, Aguilar OM. (2009) A small GTPase of the Rab family is required for root hair formation and preinfection stages of the common bean-Rhizobium symbiotic association. Plant Cell 21: 2797–2810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolte S, Schiene K, Dietz KJ. (2000) Characterization of a small GTP-binding protein of the rab 5 family in Mesembryanthemum crystallinum with increased level of expression during early salt stress. Plant Mol Biol 42: 923–936 [DOI] [PubMed] [Google Scholar]
- Borg S, Pødenphant L, Jensen TJ, Poulsen C. (1999) Plant cell growth and differentiation may involve GAP regulation of Rac activity. FEBS Lett 453: 341–345 [DOI] [PubMed] [Google Scholar]
- Boureux A, Vignal E, Faure S, Fort P. (2007) Evolution of the Rho family of ras-like GTPases in eukaryotes. Mol Biol Evol 24: 203–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Zhu H, Ke D, Cai K, Wang T, Gou H, Hong Z, Zhang Z. (2012) A MAP kinase kinase interacts with SymRK and regulates nodule organogenesis in Lotus japonicus. Plant Cell 24: 823–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrhardt DW, Wais R, Long SR. (1996) Calcium spiking in plant root hairs responding to Rhizobium nodulation signals. Cell 85: 673–681 [DOI] [PubMed] [Google Scholar]
- Esseling JJ, Lhuissier FG, Emons AM. (2003) Nod factor-induced root hair curling: continuous polar growth towards the point of nod factor application. Plant Physiol 132: 1982–1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etienne-Manneville S, Hall A. (2002) Rho GTPases in cell biology. Nature 420: 629–635 [DOI] [PubMed] [Google Scholar]
- Fahraeus G. (1957) The infection of clover root hairs by nodule bacteria studied by a simple glass slide technique. J Gen Microbiol 16: 374–381 [DOI] [PubMed] [Google Scholar]
- Fu Y, Li H, Yang Z. (2002) The ROP2 GTPase controls the formation of cortical fine F-actin and the early phase of directional cell expansion during Arabidopsis organogenesis. Plant Cell 14: 777–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiki K, Mizuno T, Hisamoto N, Matsumoto K. (2010) The Caenorhabditis elegans Ste20-related kinase and Rac-type small GTPase regulate the c-Jun N-terminal kinase signaling pathway mediating the stress response. Mol Cell Biol 30: 995–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara M, Hamada S, Hiratsuka M, Fukao Y, Kawasaki T, Shimamoto K. (2009) Proteome analysis of detergent-resistant membranes (DRMs) associated with OsRac1-mediated innate immunity in rice. Plant Cell Physiol 50: 1191–1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleason C, Chaudhuri S, Yang T, Muñoz A, Poovaiah BW, Oldroyd GE. (2006) Nodulation independent of rhizobia induced by a calcium-activated kinase lacking autoinhibition. Nature 441: 1149–1152 [DOI] [PubMed] [Google Scholar]
- Gu Y, Fu Y, Dowd P, Li S, Vernoud V, Gilroy S, Yang Z. (2005) A Rho family GTPase controls actin dynamics and tip growth via two counteracting downstream pathways in pollen tubes. J Cell Biol 169: 127–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indrasumunar A, Kereszt A, Searle I, Miyagi M, Li D, Nguyen CD, Men A, Carroll BJ, Gresshoff PM. (2010) Inactivation of duplicated nod factor receptor 5 (NFR5) genes in recessive loss-of-function non-nodulation mutants of allotetraploid soybean (Glycine max L. Merr.). Plant Cell Physiol 51: 201–214 [DOI] [PubMed] [Google Scholar]
- Indrasumunar A, Searle I, Lin MH, Kereszt A, Men A, Carroll BJ, Gresshoff PM. (2011) Nodulation factor receptor kinase 1α controls nodule organ number in soybean (Glycine max L. Merr). Plant J 65: 39–50 [DOI] [PubMed] [Google Scholar]
- Ivanchenko M, Vejlupkova Z, Quatrano RS, Fowler JE. (2000) Maize ROP7 GTPase contains a unique, CaaX box-independent plasma membrane targeting signal. Plant J 24: 79–90 [DOI] [PubMed] [Google Scholar]
- Jones MA, Shen JJ, Fu Y, Li H, Yang Z, Grierson CS. (2002) The Arabidopsis Rop2 GTPase is a positive regulator of both root hair initiation and tip growth. Plant Cell 14: 763–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H, Zhu H, Chu X, Yang Z, Yuan S, Yu D, Wang C, Hong Z, Zhang Z. (2011) A novel interaction between CCaMK and a protein containing the Scythe_N ubiquitin-like domain in Lotus japonicus. Plant Physiol 155: 1312–1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klahre U, Kost B. (2006) Tobacco RhoGTPase ACTIVATING PROTEIN1 spatially restricts signaling of RAC/Rop to the apex of pollen tubes. Plant Cell 18: 3033–3046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kost B. (2008) Spatial control of Rho (Rac-Rop) signaling in tip-growing plant cells. Trends Cell Biol 18: 119–127 [DOI] [PubMed] [Google Scholar]
- Kouchi H, Imaizumi-Anraku H, Hayashi M, Hakoyama T, Nakagawa T, Umehara Y, Suganuma N, Kawaguchi M. (2010) How many peas in a pod? Legume genes responsible for mutualistic symbioses underground. Plant Cell Physiol 51: 1381–1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai H, Kinoshita E, Ridge RW, Kouchi H. (2006) RNAi knock-down of ENOD40s leads to significant suppression of nodule formation in Lotus japonicus. Plant Cell Physiol 47: 1102–1111 [DOI] [PubMed] [Google Scholar]
- Kurkdjian AC. (1995) Role of the differentiation of root epidermal cells in Nod factor (from Rhizobium meliloti)-induced root-hair depolarization of Medicago sativa. Plant Physiol 107: 783–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavy M, Yalovsky S. (2006) Association of Arabidopsis type-II ROPs with the plasma membrane requires a conserved C-terminal sequence motif and a proximal polybasic domain. Plant J 46: 934–947 [DOI] [PubMed] [Google Scholar]
- Li H, Lin Y, Heath RM, Zhu MX, Yang Z. (1999) Control of pollen tube tip growth by a Rop GTPase-dependent pathway that leads to tip-localized calcium influx. Plant Cell 11: 1731–1742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberherr D, Thao NP, Nakashima A, Umemura K, Kawasaki T, Shimamoto K. (2005) A sphingolipid elicitor-inducible mitogen-activated protein kinase is regulated by the small GTPase OsRac1 and heterotrimeric G-protein in rice. Plant Physiol 138: 1644–1652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limpens E, Bisseling T. (2003) Signaling in symbiosis. Curr Opin Plant Biol 6: 343–350 [DOI] [PubMed] [Google Scholar]
- Liu W, Chen AM, Luo L, Sun J, Cao LP, Yu GQ, Zhu JB, Wang YZ. (2010) Characterization and expression analysis of Medicago truncatula ROP GTPase family during the early stage of symbiosis. J Integr Plant Biol 52: 639–652 [DOI] [PubMed] [Google Scholar]
- Ma L, Hong Z, Zhang Z. (2007) Perinuclear and nuclear envelope localizations of Arabidopsis Ran proteins. Plant Cell Rep 26: 1373–1382 [DOI] [PubMed] [Google Scholar]
- Madsen EB, Antolín-Llovera M, Grossmann C, Ye J, Vieweg S, Broghammer A, Krusell L, Radutoiu S, Jensen ON, Stougaard J, et al. (2011) Autophosphorylation is essential for the in vivo function of the Lotus japonicus Nod factor receptor 1 and receptor-mediated signalling in cooperation with Nod factor receptor 5. Plant J 65: 404–417 [DOI] [PubMed] [Google Scholar]
- Madsen EB, Madsen LH, Radutoiu S, Olbryt M, Rakwalska M, Szczyglowski K, Sato S, Kaneko T, Tabata S, Sandal N, et al. (2003) A receptor kinase gene of the LysM type is involved in legume perception of rhizobial signals. Nature 425: 637–640 [DOI] [PubMed] [Google Scholar]
- Mbengue M, Camut S, de Carvalho-Niebel F, Deslandes L, Froidure S, Klaus-Heisen D, Moreau S, Rivas S, Timmers T, Hervé C, et al. (2010) The Medicago truncatula E3 ubiquitin ligase PUB1 interacts with the LYK3 symbiotic receptor and negatively regulates infection and nodulation. Plant Cell 22: 3474–3488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middleton PH, Jakab J, Penmetsa RV, Starker CG, Doll J, Kaló P, Prabhu R, Marsh JF, Mitra RM, Kereszt A, et al. (2007) An ERF transcription factor in Medicago truncatula that is essential for Nod factor signal transduction. Plant Cell 19: 1221–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miwa H, Sun J, Oldroyd GE, Downie JA. (2006) Analysis of Nod-factor-induced calcium signaling in root hairs of symbiotically defective mutants of Lotus japonicus. Mol Plant Microbe Interact 19: 914–923 [DOI] [PubMed] [Google Scholar]
- Molendijk AJ, Bischoff F, Rajendrakumar CS, Friml J, Braun M, Gilroy S, Palme K. (2001) Arabidopsis thaliana Rop GTPases are localized to tips of root hairs and control polar growth. EMBO J 20: 2779–2788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima A, Chen L, Thao NP, Fujiwara M, Wong HL, Kuwano M, Umemura K, Shirasu K, Kawasaki T, Shimamoto K. (2008) RACK1 functions in rice innate immunity by interacting with the Rac1 immune complex. Plant Cell 20: 2265–2279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa S, Kawaguchi M, Imazumi-Anraku H, Chechetka SA, Ishizaka M, Ikuta A, Kouchi H. (2001) Responses of a model legume Lotus japonicus to lipochitin oligosaccharide nodulation factors purified from Mesorhizobium loti JRL501. Mol Plant Microbe Interact 14: 848–856 [DOI] [PubMed] [Google Scholar]
- Ono E, Wong HL, Kawasaki T, Hasegawa M, Kodama O, Shimamoto K. (2001) Essential role of the small GTPase Rac in disease resistance of rice. Proc Natl Acad Sci USA 98: 759–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimpl P, Hanton SL, Taylor JP, Pinto-daSilva LL, Denecke J. (2003) The GTPase ARF1p controls the sequence-specific vacuolar sorting route to the lytic vacuole. Plant Cell 15: 1242–1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radutoiu S, Madsen LH, Madsen EB, Felle HH, Umehara Y, Grønlund M, Sato S, Nakamura Y, Tabata S, Sandal N, et al. (2003) Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature 425: 585–592 [DOI] [PubMed] [Google Scholar]
- Radutoiu S, Madsen LH, Madsen EB, Jurkiewicz A, Fukai E, Quistgaard EM, Albrektsen AS, James EK, Thirup S, Stougaard J. (2007) LysM domains mediate lipochitin-oligosaccharide recognition and Nfr genes extend the symbiotic host range. EMBO J 26: 3923–3935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauser L, Roussis A, Stiller J, Stougaard J. (1999) A plant regulator controlling development of symbiotic root nodules. Nature 402: 191–195 [DOI] [PubMed] [Google Scholar]
- Smit P, Limpens E, Geurts R, Fedorova E, Dolgikh E, Gough C, Bisseling T. (2007) Medicago LYK3, an entry receptor in rhizobial nodulation factor signaling. Plant Physiol 145: 183–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit P, Raedts J, Portyanko V, Debellé F, Gough C, Bisseling T, Geurts R. (2005) NSP1 of the GRAS protein family is essential for rhizobial Nod factor-induced transcription. Science 308: 1789–1791 [DOI] [PubMed] [Google Scholar]
- Sorek N, Poraty L, Sternberg H, Bar E, Lewinsohn E, Yalovsky S. (2007) Activation status-coupled transient S acylation determines membrane partitioning of a plant Rho-related GTPase. Mol Cell Biol 27: 2144–2154 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Takeuchi M, Ueda T, Yahara N, Nakano A. (2002) Arf1 GTPase plays roles in the protein traffic between the endoplasmic reticulum and the Golgi apparatus in tobacco and Arabidopsis cultured cells. Plant J 31: 499–515 [DOI] [PubMed] [Google Scholar]
- Tansengco ML, Hayashi M, Kawaguchi M, Imaizumi-Anraku H, Murooka Y. (2003) crinkle, a novel symbiotic mutant that affects the infection thread growth and alters the root hair, trichome, and seed development in Lotus japonicus. Plant Physiol 131: 1054–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thao NP, Chen L, Nakashima A, Hara S, Umemura K, Takahashi A, Shirasu K, Kawasaki T, Shimamoto K. (2007) RAR1 and HSP90 form a complex with Rac/Rop GTPase and function in innate-immune responses in rice. Plant Cell 19: 4035–4045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waadt R, Schmidt LK, Lohse M, Hashimoto K, Bock R, Kudla J. (2008) Multicolor bimolecular fluorescence complementation reveals simultaneous formation of alternative CBL/CIPK complexes in planta. Plant J 56: 505–516 [DOI] [PubMed] [Google Scholar]
- Yalovsky S, Bloch D, Sorek N, Kost B. (2008) Regulation of membrane trafficking, cytoskeleton dynamics, and cell polarity by ROP/RAC GTPases. Plant Physiol 147: 1527–1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. (2002) Small GTPases: versatile signaling switches in plants. Plant Cell (Suppl) 14: S375–S388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Fu Y. (2007) ROP/RAC GTPase signaling. Curr Opin Plant Biol 10: 490–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuksel B, Memon AR. (2008) Comparative phylogenetic analysis of small GTP-binding genes of model legume plants and assessment of their roles in root nodules. J Exp Bot 59: 3831–3844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuksel B, Memon AR. (2009) Legume small GTPases and their role in the establishment of symbiotic associations with Rhizobium spp. Plant Signal Behav 4: 257–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Chen T, Zhu M, Fang Q, Kang H, Hong Z, Zhang Z. (2008) A novel ARID DNA-binding protein interacts with SymRK and is expressed during early nodule development in Lotus japonicus. Plant Physiol 148: 337–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zohrabian VM, Forzani B, Chau Z, Murali R, Jhanwar-Uniyal M. (2009) Rho/ROCK and MAPK signaling pathways are involved in glioblastoma cell migration and proliferation. Anticancer Res 29: 119–123 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.