Abstract
Abscisic acid is a stress-related phytohormone that has roles in dehydration and rehydration. In Arabidopsis (Arabidopsis thaliana), two genes that inactivate abscisic acid, CYP707A1 and CYP707A3, are rapidly up-regulated upon rehydration. The factors that regulate CYP707A1/3 are not well characterized. We expressed a bZIP protein, VIP1, as a green fluorescent protein fusion protein in Arabidopsis and found that the nuclear localization of VIP1 was enhanced within 10 min after rehydration. A yeast one-hybrid assay revealed that the amino-terminal region of VIP1 has transcriptional activation potential. In a transient reporter assay using Arabidopsis protoplasts, VIP1 enhanced the promoter activities of CYP707A1/3. In gel shift and chromatin immunoprecipitation analyses, VIP1 directly bound to DNA fragments of the CYP707A1/3 promoters. Transgenic plants expressing VIP1-green fluorescent protein were found to overexpress CYP707A1/3 mRNAs. The time course of nuclear-cytoplasmic shuttling of VIP1 was consistent with the time courses of the expression of CYP707A1/3. These results suggest that VIP1 functions as a regulator of osmosensory signaling in Arabidopsis.
Water availability is a key factor determining the growth, productivity, and distribution of plants. Plants have adaptive responses to water availability. Two of the adaptive responses of plants to water availability are the biosynthesis and inactivation of abscisic acid (ABA), which modulates stress tolerance in plants. In Arabidopsis (Arabidopsis thaliana), NCED3, which encodes the rate-limiting enzyme in ABA biosynthesis, is rapidly up-regulated by dehydration and down-regulated by rehydration (Iuchi et al., 2001; Tan et al., 2003; Kushiro et al., 2004; Umezawa et al., 2006). On the other hand, CYP707A1 and CYP707A3, which encode key enzymes in ABA inactivation, are rapidly induced by rehydration (Kushiro et al., 2004; Umezawa et al., 2006; Okamoto et al., 2009). Because the mRNA amounts of NCED3, CYP707A1, and CYP707A3 correlate well with the amount of ABA (Kushiro et al., 2004; Umezawa et al., 2006), the transcriptional regulation of these genes is thought to be crucial for controlling the amount of ABA in response to water availability.
These genes are thought to act downstream in the osmosensory signaling pathway (Tran et al., 2007; Wohlbach et al., 2008). In baker’s yeast (Saccharomyces cerevisiae), the osmosensing signal is transmitted by two independent osmosensory proteins, Sho1 and Sln1 (for review, see O’Rourke et al., 2002). Sho1 is a transmembrane scaffold protein, whereas Sln1 is a transmembrane His kinase. Arabidopsis does not have a Sho1 homolog but it does have some His kinases, one of which, ATHK1, was shown to complement the function of Sln1 in yeast cells (Urao et al., 1999). In Arabidopsis, ATHK1-null plants exhibited decreased expression of ABA-responsive genes and decreased drought tolerance, whereas ATHK1-overexpressing plants exhibited increased expression of ABA-responsive genes and increased drought tolerance (Tran et al., 2007; Wohlbach et al., 2008). However, there is no clear evidence that ATHK1 is a plant osmosensor or that ATHK1 regulates NCED3, CYP707A1, and CYP707A3. In yeast, the HOG mitogen-activated protein kinase (MAPK) cascade acts downstream of Sho1 and Sln1 (for review, see O’Rourke et al., 2002), but in plants, it is unclear whether a MAPK pathway is involved in osmosensory signaling.
VIP1, an Arabidopsis bZIP transcription factor, is involved in the process of Agrobacterium tumefaciens infection in Arabidopsis (Tzfira et al., 2001, 2002; Li et al., 2005) and regulates the expression of Agrobacterium-responsive genes in Arabidopsis (Djamei et al., 2007; Pitzschke et al., 2009). During an investigation of the function of VIP1 in pathogen responses, we discovered that the nuclear localization of VIP1 is rapidly enhanced in response to increased turgor pressure. This led us to investigate whether VIP1 interacts in some way with CYP707A1 and CYP707A3 because of their induction by rehydration. Here, we show that VIP1 directly binds to promoter fragments of CYP707A1 and CYP707A3 and activates their transcription. Our results suggest that VIP1 is a novel upstream component of Arabidopsis osmosensory signaling.
RESULTS
The Nuclear Localization of VIP1 Is Enhanced by Increased Turgor
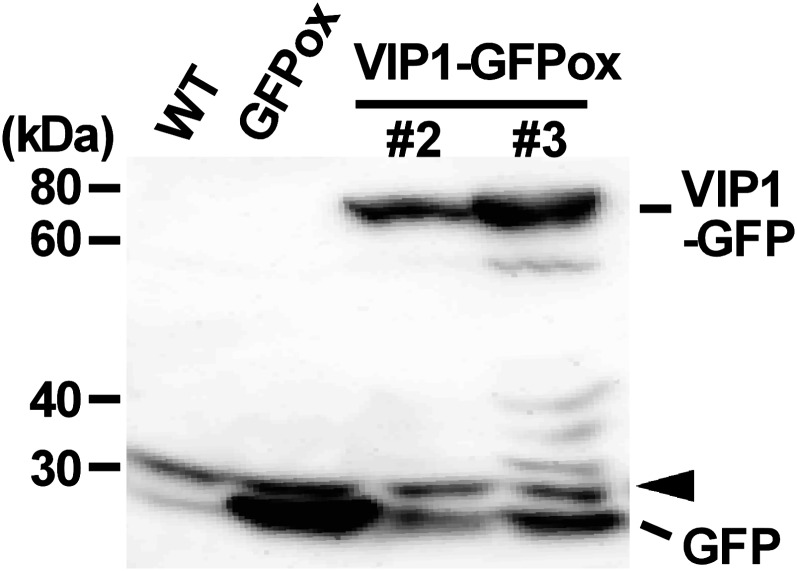
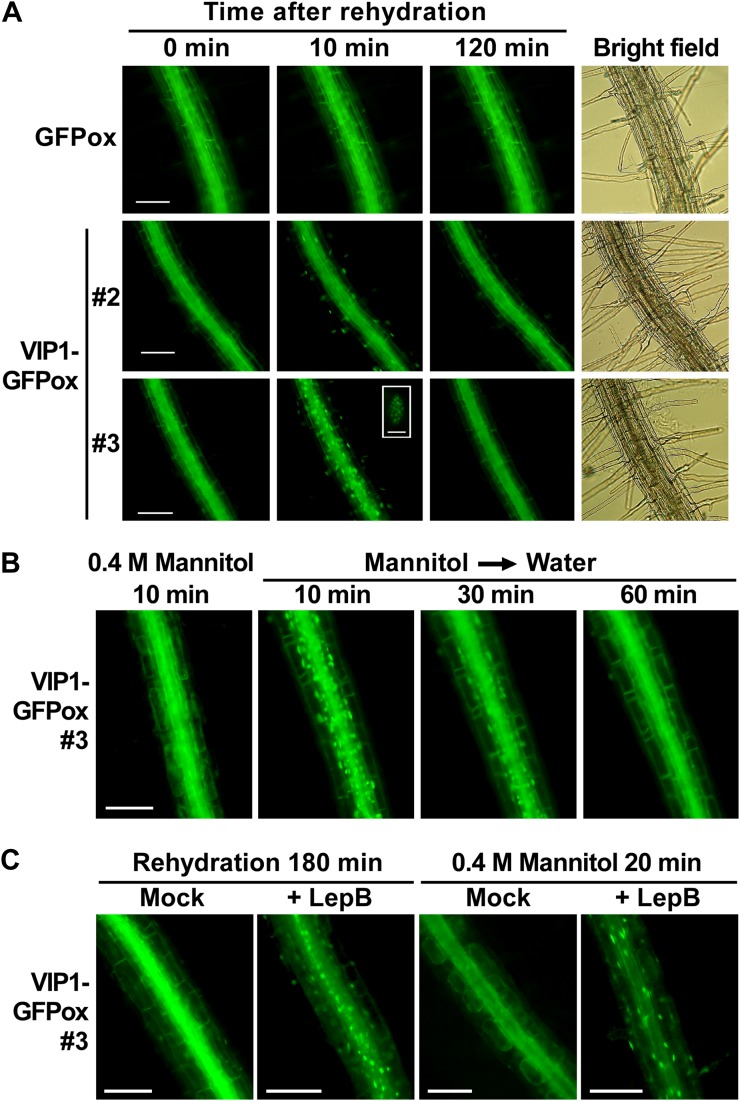
During our investigation of the pathogen-related role of VIP1 in Arabidopsis, we generated transgenic plants expressing VIP1-GFP (VIP1-GFPox). Expression of VIP1-GFP was confirmed by immunoblot analysis, and two lines (VIP1-GFPox #2 and #3) that had different expression levels of VIP1-GFP (Fig. 1) were chosen for further analysis. In the course of our GFP observations, we noticed that VIP1-GFP signals in the nucleus were rapidly (within 10 min) enhanced after VIP1-GFPox plants were submerged in water. When the submergence was prolonged, VIP1-GFP signals in the nucleus of root cells became speckled and then weakened (Fig. 2A). This phenomenon was also observed in hypocotyls and leaf cells, including epidermal, mesophyll, and guard cells (Supplemental Fig. S1). It took longer for VIP1-GFP signals in the nucleus to be attenuated in VIP1-GFPox #3 than in VIP1-GFPox #2 (data not shown). The rapid signal intensification of VIP1-GFP in the nucleus was inhibited in the presence of 0.4 m mannitol (Fig. 2B) but not by the phytohormones ABA (100 μm) and GA3 (0.1 mg mL−1; Supplemental Fig. S2). These results suggest that the nuclear localization of VIP1-GFP is rapidly increased upon an increase of turgor pressure and is slowly decreased after cells are acclimated to the change in turgor pressure. VIP1 has a functional nuclear localization signal (NLS) at the beginning of its bZIP domain (Tzfira et al., 2002; Li et al., 2005) and a putative nuclear export signal (NES) in its C-terminal region (their positions are shown in Fig. 3A). To examine whether active nuclear export is involved in regulating the subcellular localization of VIP1, we examined the effects of leptomycin B on VIP1 localization. In active nuclear export, exportin binds to a cargo protein in the nucleus and transports it to the cytoplasm. The process can be inhibited by leptomycin B. In the presence of leptomycin B, signals of VIP1-GFP were retained in the nucleus even 180 min after plants were submerged in water (Fig. 2C, left panels). In addition, leptomycin B enhanced VIP1-GFP signals in the nucleus even in the presence of 0.4 m mannitol (Fig. 2C, right panels). These results suggest that VIP1 is actively exported from the nucleus when turgor pressure is steady and that the active nuclear export of VIP1 is inhibited when turgor pressure is increased.
Figure 1.
Immunoblot of VIP1-GFP. Plants were transformed with a construct containing CaMV 35S promoter-VIP1-GFP (VIP1-GFPox #2 and #3), and 10-d-old seedlings were subjected to immunoblot analysis using anti-GFP antibody. Wild-type plants (WT) and transgenic plants expressing GFP alone (GFPox) were used as controls. The arrowhead designates a nonspecific band, which served as a loading control.
Figure 2.
Turgor-dependent translocation of VIP1-GFP. A root is shown in each image. A, Roots of GFP- or VIP1-GFP-overexpressing plants (GFPox or VIP1-GFPox, respectively) were observed by epifluorescence microscopy at the indicated time points after being submerged in 20 mm Tris-HCl, pH 6.8 (rehydration). Speckled VIP1-GFP signals in the nucleus in VIP1-GFPox #3 at 30 min after rehydration are shown as a high-magnification image (inset). Bars = 100 μm for low-magnification images and 5 μm for the high-magnification image. B, Roots of VIP1-GFPox #3 were observed 10 min after being submerged in 20 mm Tris-HCl, pH 6.8, with 0.4 m mannitol (0.4 m Mannitol 10 min). Plants were then transferred to 20 mm Tris-HCl, pH 6.8, without mannitol (Mannitol → Water) and observed at the indicated time points. Bar = 100 μm. C, Roots of VIP1-GFPox #3 were observed 180 min after being submerged in 20 mm Tris-HCl, pH 6.8 (rehydration), containing 0 (Mock) or 2 μg mL−1 leptomycin B (+ LepB), or 20 min after being submerged in 0.4 m mannitol, 20 mm Tris-HCl, pH 6.8, containing 0 or 2 μg mL−1 leptomycin B. Bars = 100 μm.
Figure 3.
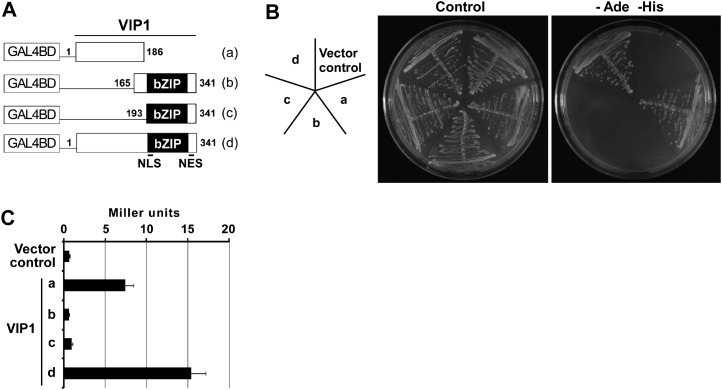
Transcriptional activation function of VIP1 in yeast cells. A, Constructs used for the Y1H assay. Numbers indicate amino acid positions in full-length VIP1. Lowercase letters (a–d) correspond to those in B and C. B, A yeast reporter strain, AH109, was transformed with the constructs shown in A. Transformed cells were grown on synthetic dextrose medium with (Control) or without adenine and His (−Ade −His). For each construct, three individual colonies were tested. A representative result is shown. The image at left indicates the position of each cell line. C, β-Galactosidase assay was performed using three individual colonies for each construct. Values are means ± se.
VIP1 Interacts with CYP707A1/A3 Promoters
The transcriptional activation function of VIP1 was evaluated with a yeast one-hybrid (Y1H) assay. Full-length or truncated VIP1 was fused to GAL4 DNA-binding domain (GAL4BD; Fig. 3A), and the fusion protein was expressed in a yeast reporter strain, AH109, that has GAL4BD-binding sites upstream of the reporter genes ADE2, HIS3, and lacZ. When the N-terminal region of VIP1 was present, transformed yeast cells were able to grow on medium lacking adenine and His (Fig. 3B) and showed higher β-galactosidase activities (Fig. 3C), suggesting that the N-terminal region of VIP1 functions as a transcriptional activation domain in yeast.
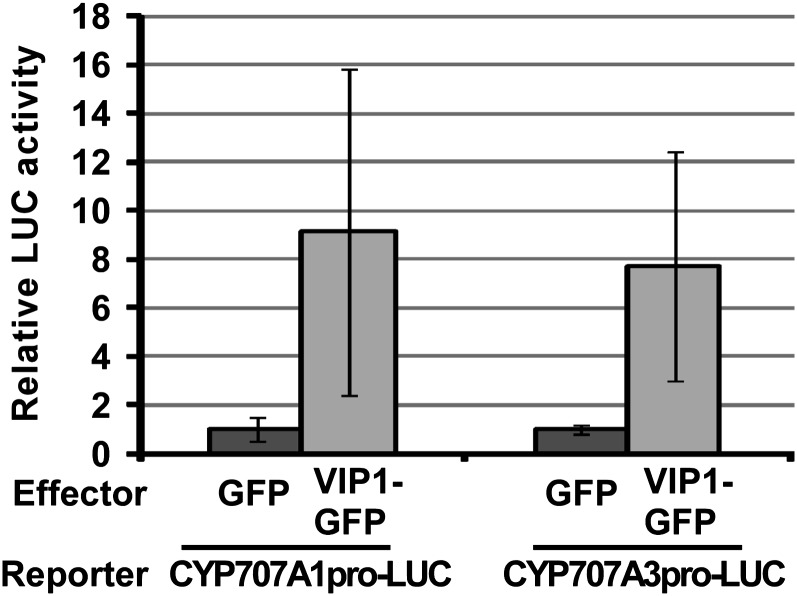
Because the nuclear localization of VIP1 was rapidly enhanced by increased turgor pressure, we examined the relationship between VIP1 and CYP707A1/3, which was reported to be up-regulated upon rehydration or under high-humidity conditions (Kushiro et al., 2004; Umezawa et al., 2006; Okamoto et al., 2009). First, we used VIP1-GFP as an effector and CYP707A1/3 promoter-luciferase as a reporter to examine the effects of VIP1 on the expression of CYP707A1/3. Reporter activities were enhanced only when VIP1 was present in the effector plasmid (Fig. 4), suggesting that VIP1 can activate the promoter activities of both CYP707A1 and CYP707A3.
Figure 4.
A transient reporter assay for CYP707A1/3 promoter-VIP1 interactions in plant cells. A reporter construct containing CYP707A1/3 promoter-luciferase (CYP707A1/3pro-LUC) was introduced into Arabidopsis mesophyll protoplasts with an effector construct to express GFP or VIP1-GFP. Reporter luciferase (firefly luciferase) activities were standardized by activities of coexpressed Metridium luciferase, and relative reporter activities were calculated. Experiments were repeated three or four times for each reporter-effector combination. Values are means ± se.
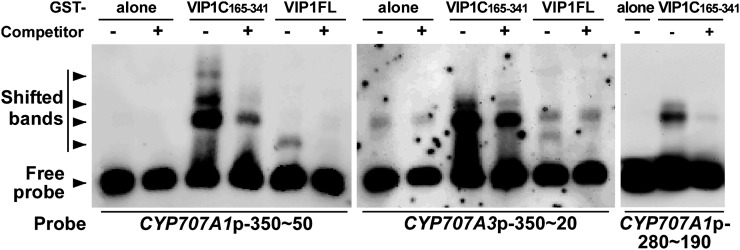
Next, we carried out gel-shift assays using full-length VIP1 or the C-terminal bZIP domain (amino acids 165–341) of VIP1 to examine whether VIP1 directly binds to the CYP707A1 and CYP707A3 promoters in vitro. For the probes, we first used approximately 350 bp of the CYP707A1 and CYP707A3 promoters just upstream of the start codons. When the CYP707A1 approximately 350-bp promoter was used as a probe (Fig. 5, left panel), shifted bands appeared when the VIP1 C terminus was present in the reaction solution (lane 3) and when full-length VIP1 was present in the reaction solution (lane 5). Furthermore, these signals were weakened in the presence of competitor DNA (lanes 4 and 6, respectively). Similar results were obtained when the CYP707A3 approximately 350-bp promoter was used as a probe (Fig. 5, middle panel). An alignment of the approximately 350-bp promoters shows several shared regions (Supplemental Fig. S1), which could explain how VIP1 could bind to both promoters. A smaller fragment (approximately 90 bp, −280 to −190 bp upstream of the start codon) of the CYP707A1 promoter without the G-box, which is known to bind many bZIP proteins, was also shifted in the presence of VIP1 (Fig. 5, right panel), suggesting that this region contains a VIP1-binding sequence.
Figure 5.
Gel-shift assays for CYP707A1/3 promoter-VIP1 interactions in vitro. Full-length VIP1 (VIP1FL) or its C-terminal 165 to 341 amino acids (VIP1C165-341) was expressed in E. coli as a GST fusion protein, purified, and used for the analyses. GST alone was used as a control. Digoxigenin-labeled DNA fragments of an approximately 350-bp region of the CYP707A1 promoter (CYP707A1p-350∼50), an approximately 350-bp region of the CYP707A3 promoter (CYP707A3p-350∼20), and a smaller region of the CYP707A1 promoter (CYP707A1p-280∼190) were used as probes. Competitor + or – indicates the presence or absence, respectively, of nonlabeled DNA fragments of CYP707A1/3 promoter in the reaction solution.
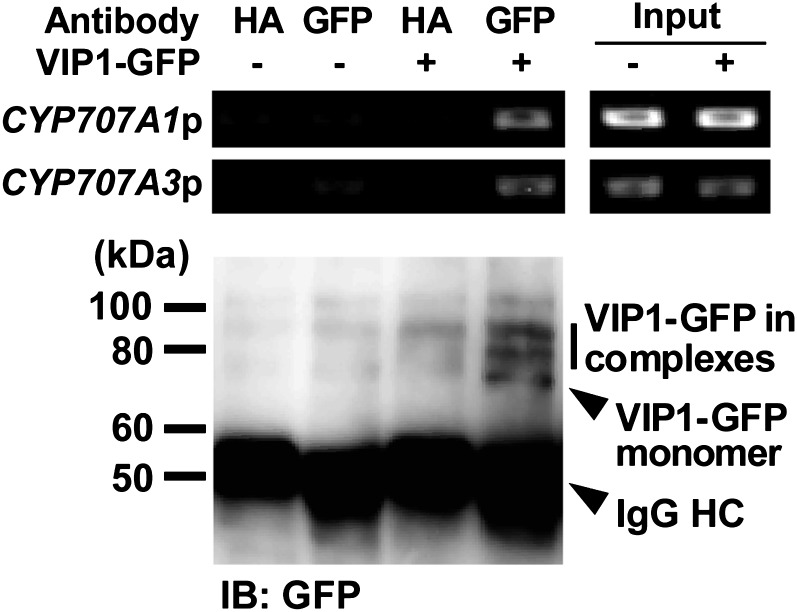
Next, a chromatin immunoprecipitation (ChIP) analysis was performed to examine whether VIP1 interacts with the CYP707A1/3 promoters in vivo. In the ChIP analysis, VIP1-GFPox plants were rehydrated and VIP1-GFP was immunoprecipitated using an anti-GFP antibody after VIP1-GFP was cross-linked with DNA. CYP707A1/3 promoter fragments were detected only when VIP1-GFP was precipitated (Fig. 6), suggesting that VIP1 binds the CYP707A1/3 promoters in vivo.
Figure 6.
ChIP analysis of interactions between VIP1-CYP707A1/3 promoters in vivo. GFPox (VIP1-GFP −) or VIP1-GFPox (VIP1-GFP +) plants were rehydrated in 20 mm Tris-HCl, pH 6.8, for 10 min and used for the experiment. After making cross-links between VIP1-GFP and DNA, chromatin was isolated, sonicated, and used as an input for the subsequent PCR. VIP1-GFP was immunoprecipitated from the sonicated sample using a rabbit anti-GFP antibody. A rabbit anti-hemagglutinin (HA) antibody was used as a negative control. DNA was eluted from immunocomplexes subjected to the PCR analysis of CYP707A1/3 promoter fragments (CYP707A1p and CYP707A3p). Immunocomplexes were then disrupted by incubating them at 100°C in SDS sample buffer, and VIP1-GFP was analyzed by immunoblotting using the anti-GFP antibody (IB: GFP). In the combination of the anti-GFP antibody and VIP1-GFP + (lane 4), two bands were detected above the band of monomeric VIP1-GFP, suggesting that some VIP1-GFP-containing complexes were fixed by the cross-linking reaction. IgG HC, IgG H chain.
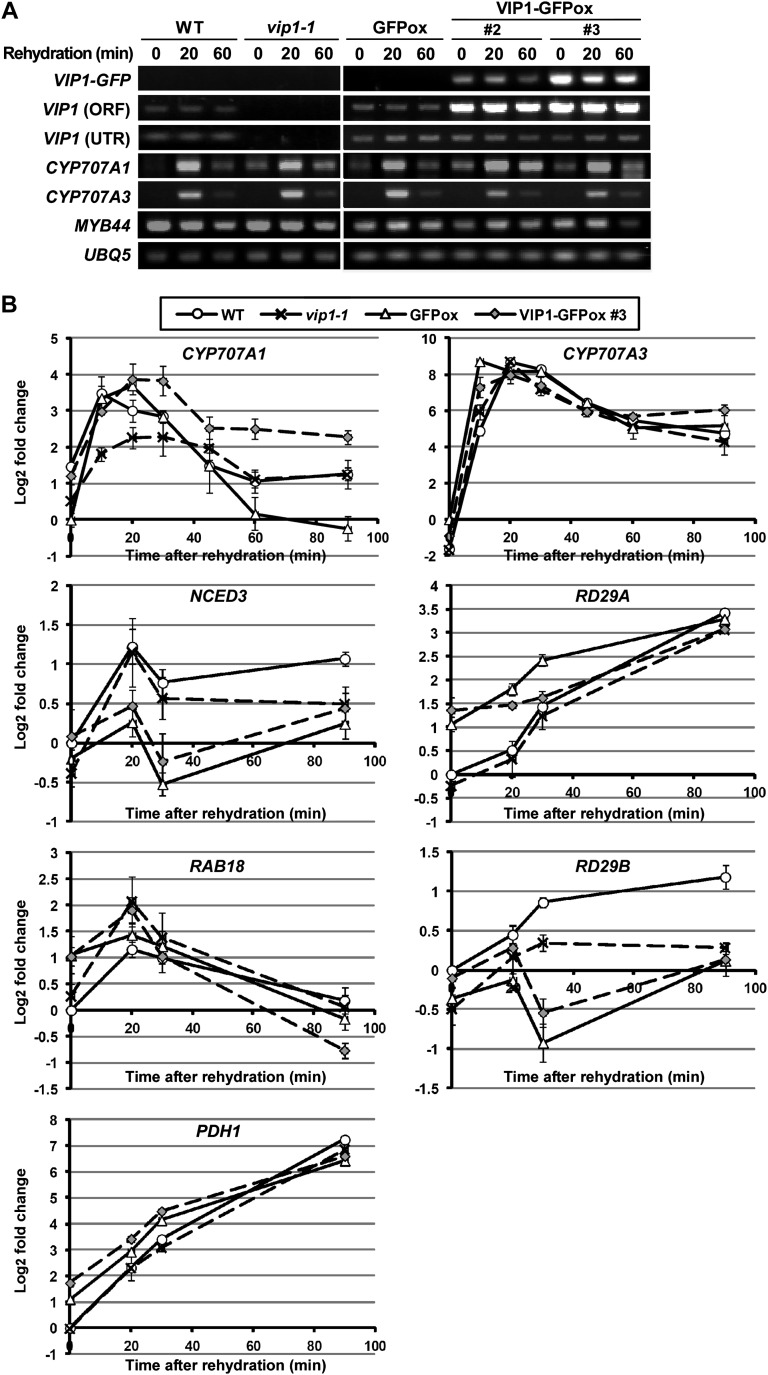
Finally, we performed reverse transcription (RT)-PCR analyses using a VIP1-deficient mutant, vip1-1 (Li et al., 2005), and VIP1-GFPox to evaluate the effects of VIP1 on the transcription of CYP707A1/3 in vivo. Lack of VIP1 transcripts in vip1-1 and overexpression of VIP1 in VIP1-GFPox were confirmed by semiquantitative (semi-q) RT-PCR (Fig. 7A, rows 1–3). In all the genotypes studied, the expression of CYP707A1 and CYP707A3 transcripts was transiently increased by rehydration and then decreased, in agreement with previous reports (Kushiro et al., 2004; Umezawa et al., 2006; Okamoto et al., 2009). The expression levels of CYP707A1/3 were comparable among the five genotypes in semi-qRT-PCR (Fig. 7A, rows 4 and 5). MYB44 was analyzed as a previously reported VIP1 target gene (Pitzschke et al., 2009), but there was little difference in its expression level among the transgenic lines studied (Fig. 7A, row 6). The expression levels of CYP707A1/3 were further analyzed by quantitative (q) RT-PCR. The expression levels of CYP707A3 under rehydration were comparable among all the genotypes studied. In contrast, the expression level of CYP707A1 was lowest in vip1-1 at 10 to 30 min after rehydration, during which the rehydration-dependent induction of CYP707A1 was maximized in all the genotypes. In addition, the expression level of CYP707A1 was highest in VIP1-GFPox #3 at 20 to 90 min after rehydration (Fig. 7B, top panels). These results suggest that VIP1 positively regulates the expression of CYP707A1 under rehydration. The expression levels of dehydration-responsive genes (NCED3 and RD29A), ABA-responsive genes (RAB18 and RD29B), and another rehydration-responsive gene (PDH1; Umezawa et al., 2006) under rehydration did not seem to correlate with the expression level of VIP1 (Fig. 7B), suggesting that VIP1 does not regulate the transcription of these genes under rehydration.
Figure 7.
RT-PCR analyses of gene expression. A, Semi-qRT-PCR analysis of gene expression in vip1-1 and VIP1-GFPox plants. Two-week-old seedlings of each genotype were submerged in 20 mm Tris-HCl (rehydration) for 0, 20, and 60 min and sampled for RNA extraction and cDNA synthesis. For VIP1 (ORF), a primer pair specific to the ORF of VIP1 was used. For VIP1 (UTR), a reverse primer specific to the 3′ untranslated region of VIP1 was used. UBQ5 was used as an internal control gene. B, Real-time qRT-PCR analysis of the expression of dehydration-, rehydration-, and ABA-responsive genes. Two-week-old seedlings of each genotype were submerged in 20 mm Tris-HCl (rehydration) for 0, 10, 20, 30, 45, 60, and 90 min and sampled for RNA extraction and cDNA synthesis. Relative expression levels were calculated by the comparative cycle threshold method using UBQ5 as an internal control gene and are shown as log2 fold changes. Experiments were performed in triplicate. Values are means ± se. WT, Wild type.
Stage and Tissue Specificity of VIP1 Expression
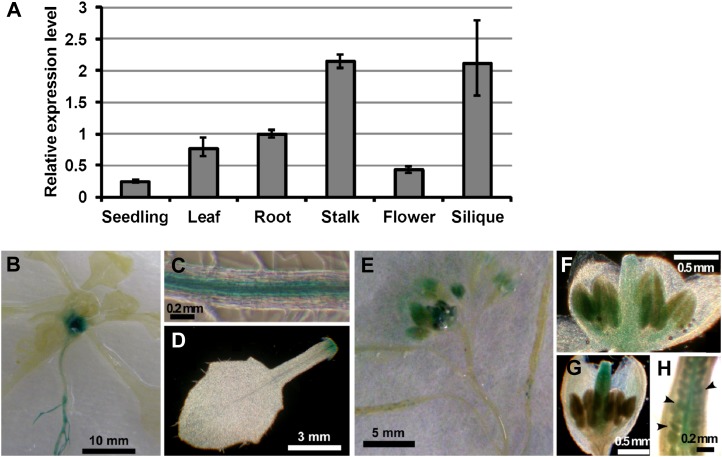
We examined the expression levels of VIP1 in various tissues by qRT-PCR. VIP1 mRNA was undetectable in mature seeds but detectable in all the other tissues studied, being highest in flower stalks and lowest in seedlings (Fig. 8A). To further characterize the stage and tissue specificity of VIP1 expression, we generated transgenic plants carrying the VIP1 promoter (an approximately 2,000-bp fragment upstream from the start codon of VIP1)-GUS and histochemically stained them for GUS activity. During the vegetative stage, GUS activity was detected in vascular tissues in hypocotyls and roots (Fig. 8, B and C) and around leaf joints (Fig. 8, B and D). In flower organs, GUS staining was observed in anthers (Fig. 8F) and pistils (Fig. 8G) according to the developmental stages of flowers. GUS activity was also found in young embryos (Fig. 8, E and H).
Figure 8.
Stage and tissue specificity of VIP1 expression. A, qRT-PCR analysis of VIP1 transcripts in various tissues. Relative expression levels were calculated by the comparative cycle threshold method using UBQ5 as an internal control gene and a root sample as a reference sample. Experiments were performed in triplicate. Values are means ± se. B to H, Histochemical GUS staining of transgenic Arabidopsis carrying VIP1 promoter-GUS. B, A 20-d-old plant. C, Mature leaf. D, Root. E, Flowers and siliques. F and G, Developing flowers. H, Pistil. Arrowheads indicate developing embryos.
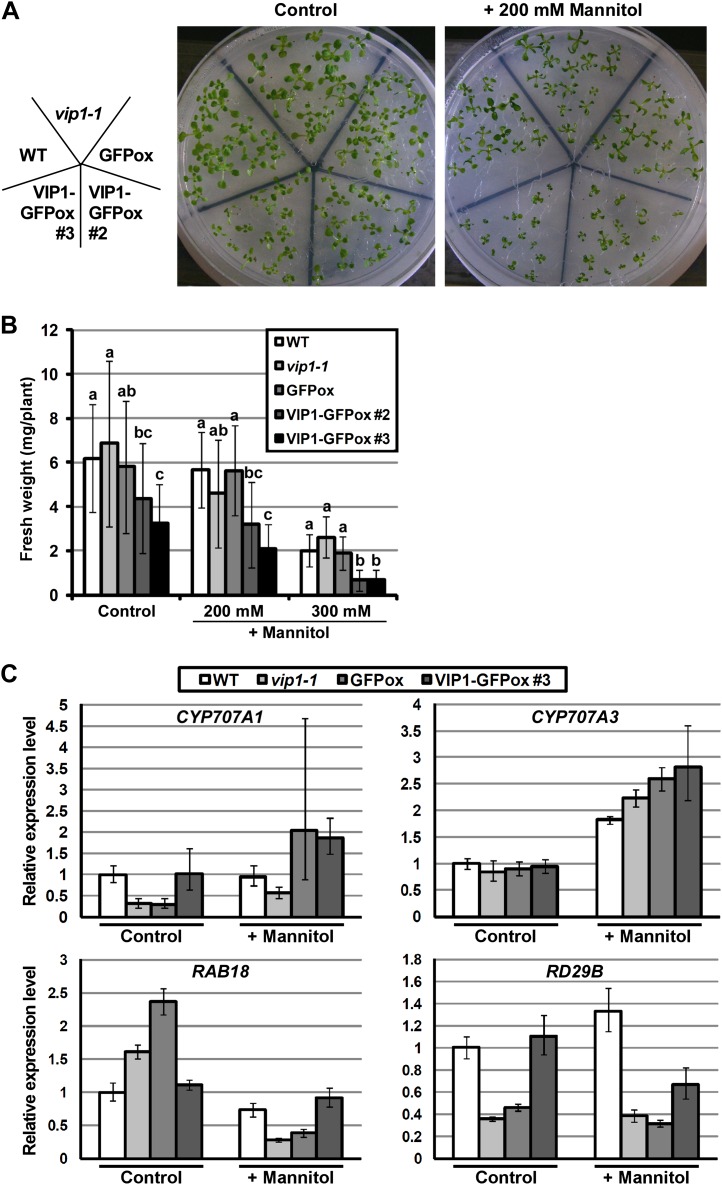
Overexpression of VIP1 Enhances Mannitol-Induced Growth Retardation
To investigate whether VIP1 is involved in osmotic stress responses, the growth of vip1-1, VIP1-GFPox #2, and VIP1-GFPox #3 was compared with the growth of wild-type and GFPox plants in the presence of 200 or 300 mm mannitol or 125 mm NaCl. Even under control conditions, the growth of VIP1-GFPox #3 was significantly retarded. In the presence of mannitol, the growth of VIP1-GFPox #2 and #3 was more highly retarded than in wild-type and GFPox plants (Fig. 9, A and B; Supplemental Fig. S4), suggesting that VIP1-GFPox plants are more sensitive to mannitol stress. On the other hand, no clear difference was observed in their NaCl responses (Supplemental Fig. S5). Under all the conditions, the germination rates of VIP1-GFPox #2 and #3 were comparable to the germination rate of GFPox (data not shown), suggesting that the growth retardation of VIP1-GFPox #2 and #3 was due to decreased water absorption at the postgermination stage. In plant cells immediately after the plants were picked up from the medium, VIP1-GFP signals were visible in the cytoplasm but not in the nucleus (data not shown). Using soil-grown plants, phenotypic analysis under dehydration and rehydration was also conducted, but there was no clear difference in wilting phenotypes during dehydration and recovery after rehydration among the five genotypes studied (data not shown).
Figure 9.
Responses of VIP1-GFPox to mannitol stress. A, Images of 2-week-old plants grown on medium with 0 (Control) and 200 mm mannitol. B, Fresh weights of 2-week-old plants grown on medium in the presence of 0 (Control), 200, and 300 mm mannitol. Values are means ± se (n = 11–16). Means marked with the same letter are not significantly different (P < 0.05) based on an ANOVA test. C, Expression levels of CYP707A1/3 and ABA-responsive genes under mannitol stress. Plants were grown for 3 weeks on medium with 0 (Control) and 200 mm mannitol (+ Mannitol) and used for RNA isolation and cDNA synthesis. Relative expression levels were calculated by the comparative cycle threshold method using UBQ5 as an internal control gene and a sample of the wild type (WT) grown under the control condition as a reference sample. Experiments were performed in triplicate. Values are means ± se.
To examine whether CYP707A1/3 expression and/or altered ABA responses are involved in the mannitol-sensitive phenotype of VIP1-GFPox, the expression of CYP707A1/3 and ABA-responsive genes (RAB18 and RD29B) was compared. The expression levels of RAB18 and RD29B reflect both the endogenous level of ABA and the sensitivity to ABA (Umezawa et al., 2006). None of these genes, however, showed expression patterns specific to VIP1-GFPox (Fig. 9C). Expression levels of dehydration-responsive genes (NCED3 and RD29A) and another rehydration-responsive gene (PDH1) were also compared, but their expression patterns were not specific to VIP1-GFPox (Supplemental Fig. S6). In our qRT-PCR system, NCED3 and RD29A as well as RAB18 and RD29B were up-regulated by ABA treatment (Supplemental Fig. S7). These results suggest that ABA is not involved in the mannitol-sensitive phenotype of VIP1-GFPox.
DISCUSSION
Factors Involved in the Regulation of VIP1 Localization
Our results show that turgor pressure induces intracellular translocation of VIP1 (Fig. 2). VIP1 is classified as a plant group I bZIP protein (Jakoby et al., 2002). Two other group I bZIP proteins, tobacco (Nicotiana tabacum) RSG and rice (Oryza sativa) RF2a, are also reported to be localized to both the nucleus and cytoplasm (Igarashi et al., 2001; Dai et al., 2004; Ishida et al., 2004, 2008), whereas other bZIP proteins, such as AREB1/2 (group A) and HY5 (group H), are limited to the nucleus (Chattopadhyay et al., 1998; Yoshida et al., 2010). Nuclear-cytoplasmic shuttling is probably a mechanism for regulating the functions of these proteins. VIP1 has a functional NLS (Tzfira et al., 2002; Li et al., 2005), and our results suggest that a putative NES in the C-terminal region of VIP1 (Fig. 3A) is functional (Fig. 2C). Thus, the NLS/NES-based transport systems seem to work on VIP1. Osmosensing-dependent posttranslational modifications and/or changes in conformation or interactions with other proteins are likely to be involved in the regulation of VIP1 localization. ATHK1 has been proposed to act as a plant osmosensor (Urao et al., 1999; Tran et al., 2007; Wohlbach et al., 2008); thus, it would be interesting to know whether VIP1 is under the control of ATHK1.
The Ser residue at position 79 (Ser-79) of VIP1 is phosphorylated by a MAPK, MPK3, which promotes the nuclear import of VIP1 (Djamei et al., 2007). We have not examined whether Ser-79 is phosphorylated by increased turgor pressure, but we have confirmed that MAPKs are deactivated by submergence treatment, at least under our experimental conditions (data not shown), suggesting that the turgor-induced enhancement of the nuclear localization of VIP1 is independent of MAPK activation.
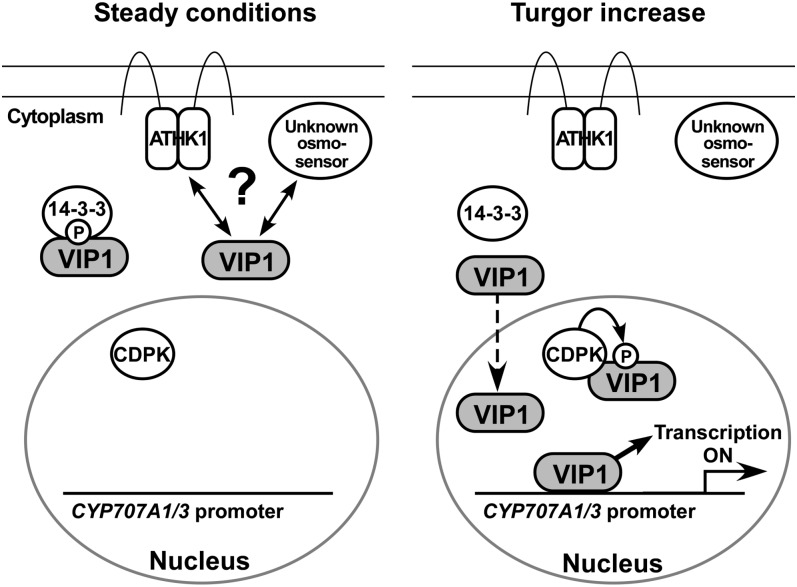
When Ser-114 of tobacco RSG is phosphorylated, the 14-3-3 protein binds to RSG (Igarashi et al., 2001; Ishida et al., 2004), causing RSG to be retained in the cytoplasm. Ser-114 of RSG is phosphorylated by a Ca2+-dependent protein kinase in the nucleus (Ishida et al., 2008). RSG has not been reported to be involved in osmosensory signaling, but Ser-114 and its neighboring residues in RSG are conserved in VIP1, which raises the possibility that the localization of VIP1 is controlled by the 14-3-3 protein and Ca2+-dependent protein kinase in the osmosensory signaling pathway. Factors potentially involved in regulating the localization of VIP1 in Arabidopsis osmosensory signaling are illustrated in Figure 10.
Figure 10.
Diagram of a proposed model of VIP1-mediated osmosensory signaling. Under steady conditions, VIP1 localizes in the cytoplasm, probably by interacting with the 14-3-3 protein, ATHK1, and/or unknown osmosensors. When turgor pressure is increased (Turgor increase), the nuclear localization of VIP1 is rapidly enhanced and VIP1 binds to the CYP707A1/3 promoters and activates their transcription. VIP1 may be phosphorylated by Ca2+-dependent protein kinase (CDPK) in the nucleus, which promotes its interaction with the 14-3-3 protein.
VIP1 as a Transcriptional Activator
We have shown that VIP1 binds to the CYP707A1/3 promoters and activates their promoter activities (Figs. 4–6). It is notable that the time course of VIP1 translocation during rehydration (Fig. 2) was consistent with that of the mRNA expression of CYP707A1/3 (Fig. 7), supporting the idea that VIP1 is a key regulator of CYP707A1/3 transcription in osmosensory signaling. In the RT-PCR analyses, the expression levels of CYP707A3 seemed unrelated to overexpression or deficiency of VIP1 at 10 to 30 min after rehydration (Fig. 7B). In addition to VIP1, Arabidopsis has 12 other group I bZIP proteins (Jakoby et al., 2002) whose subcellular localization and transcriptional activation functions may be similar to those of VIP1. Thus, the expression of CYP707A1/3 may fluctuate or become saturated shortly (10–30 min) after rehydration. It should be noted that the expression levels of CYP707A1 at 20 to 90 min after rehydration were highest in VIP1-GFPox #3 (Fig. 7B), which also had the highest expression of VIP1 (Fig. 7A); thus, a greater amount of VIP1 stayed in the nucleus after VIP1 began to be exported from the nucleus (data not shown).
Some DNA sequences have been reported to be bound by VIP1 or other group I bZIP proteins (Torres-Schumann et al., 1996; Yin et al., 1997; Ringli and Keller, 1998; Fukazawa et al., 2000; Pitzschke et al., 2009). However, none of these sequences are shared between the CYP707A1/3 promoter regions used for our gel-shift assays (Supplemental Fig. S3); thus, these regions must contain a novel VIP1-binding element. On the other hand, it is not known which element is responsible for the turgor-induced enhancement of the activities of the CYP707A1/3 promoters. Identification of the novel VIP1-binding element and the turgor-response elements in the CYP707A1/3 promoters will help to better understand plant osmosensory signaling.
Physiological Roles of VIP1
CYP707A3 is reported to be expressed in vascular tissues, whereas CYP707A1 is expressed mainly in stomata (Okamoto et al., 2009). The tissue specificity of CYP707A3 expression is consistent with our GUS analysis, in which VIP1 was predominantly expressed in vascular tissues and flower organs (Fig. 8B). Although the GUS analysis failed to detect VIP1 expression in stomata (data not shown), VIP1 might be expressed in stomata because loss of function and overexpression of VIP1 affected the expression of CYP707A1 (Fig. 7B). Tomato (Solanum lycopersicum) VSF-1 and rice RF2a and RF2b are also specifically expressed in vascular tissue, where they control vascular-specific gene expression and thus vascular development (Torres-Schumann et al., 1996; Dai et al., 2003, 2004). The growth of VIP1-GFPox and vip1-1 was similar to the growth of wild-type plants (Fig. 9A), which suggests that VIP1 does not strongly affect vascular development, but this does not rule out the possibility that VIP1 regulates the expression of some vascular genes.
Highly regulated expression patterns in flower organs have also been detected in many other Arabidopsis bZIP proteins, such as ABI5 (Brocard et al., 2002), ABF2 (Fujita et al., 2005), ABF3 and ABF4 (Kang et al., 2002), bZIP10 and bZIP25 (Lara et al., 2003), bZIP53 (Alonso et al., 2009), and bZIP60 (Iwata et al., 2008). Some of them act redundantly or synergistically in a common signal transduction pathway, but others participate in different pathways. Nevertheless, the bZIP proteins cooperatively regulate flower and seed development. VIP1 may participate in such processes to regulate CYP707A1/3 or ABA metabolism, which is critical for seed development and embryogenesis.
The growth rates of VIP1-GFPox #2 and #3 were retarded and further decreased in the presence of mannitol (Fig. 9, A and B), suggesting that VIP1 regulates the water potential of the cell. In the presence or absence of mannitol, VIP1-GFP was undetectable in the nucleus unless the plants were rehydrated (data not shown). This raises the possibility that VIP1 functions as a signaling molecule not only in the nucleus but also in the cytoplasm, although the amount of VIP1 in the nucleus may still have been higher in VIP1-GFPox #2 and #3 than in the wild type or GFPox. In previous reports, ectopic expression of bZIP53 or constitutive active forms of ABF2, both of which activate the transcription of ABA-responsive genes, retarded plant growth (Fujita et al., 2005; Furihata et al., 2006; Alonso et al., 2009). We concluded that neither the ABA level nor the sensitivity to ABA is involved in the mannitol-sensitive phenotype of VIP1-GFPox, but this does not rule out the possibility that VIP1 regulates the expression of other ABA-responsive genes.
Although the rehydration-induced nuclear retention of VIP1 and the interactions between VIP1 and CYP707A1/3 promoters suggest that VIP1 regulates osmosensory signaling, the vip1-1 single mutation did not cause a significant difference in either mannitol response (Fig. 9, A and B) or dehydration/rehydration responses (data not shown). On the other hand, vip1-1 showed a lower rate of Agrobacterium infection (Li et al., 2005) and a higher tolerance to low inputs of sulfur (Wu et al., 2010). Thus, VIP1 may have specific roles in Agrobacterium responses and low-sulfur responses but act redundantly with other group I bZIP proteins in osmosensory signaling. Further analysis will be required to elucidate the specificity and redundancy among group I bZIP proteins.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) ecotype Columbia-0 was used throughout the experiments. Seeds of vip1-1 (SALK_001014C; Li et al., 2005) were obtained from the Arabidopsis Biological Resource Center (http://www.arabidopsis.org). T-DNA insertion was confirmed by genomic PCR analysis as described previously (Li et al., 2005). Seeds were surface sterilized and sown on 0.8% agar containing 0.5× Murashige and Skoog (MS) salts (Wako), 1% (w/v) Suc, and 0.5 g L−1 MES, pH 5.8, with 0, 200, or 300 mm mannitol or 125 mm NaCl, chilled at 4°C in the dark for 48 h (stratified), and germinated at 22°C. Plants were grown at 22°C under 16-h-light/8-h-dark conditions (light intensity of 120 μmol m−2 s−1). For phenotypic analysis under dehydration and rehydration, plants were grown on soil for 3 weeks with regular watering of 0.1× MS solution and another 2 weeks without watering. MS solution was supplied again, and subsequent phenotypes were observed. Arabidopsis transformation was performed by the Agrobacterium tumefaciens-mediated floral dip method (Clough and Bent, 1998). T2 generation plants were used for analyses.
Localization Studies
To generate transgenic plants overexpressing GFP or VIP1-GFP, the open reading frame (ORF) of GFP was cloned into pBI121, replacing the GUS gene. The ORF of VIP1 (AT1G43700) was amplified by RT-PCR and inserted between the cauliflower mosaic virus (CaMV) 35S promoter and the GFP of the plasmid above. The resultant plasmids were used for Arabidopsis transformation. For GFP localization studies, 10-d-old transgenic plants expressing GFP or VIP1-GFP were submerged into 20 mm Tris-HCl, pH 6.8, with or without 0.4 m mannitol, 100 μm ABA (Wako), or 0.1 mg mL−1 GA3. Nuclear export was inhibited by leptomycin B at a concentration of 2 μg mL−1. Tissues were observed at the time points indicated in the figures using an epifluorescence microscope (BX51; Olympus). Images were processed with Canvas X software (ACD Systems).
Immunoblotting
Immunoblot analysis to confirm the VIP1-GFP expression was performed as described previously (Tsugama et al., 2011). Briefly, 10-d-old Arabidopsis seedlings showing GFP fluorescence were put in the lysis solution (0.1 m EDTA, 0.12 m Tris-HCl, 4% [w/v] SDS, 10% [v/v] β-mercaptoethanol, and 5% [v/v] glycerol, pH 7.4) and boiled for 5 to 10 min to be lysed. The supernatant was used as a protein sample. Proteins were separated on an 11% SDS-PAGE gel, transferred to a polyvinylidene difluoride membrane, and reacted with rabbit anti-GFP antibody (Medical & Biological Laboratories Co., Ltd. [MBL], Japan) as the primary antibody and horseradish peroxidase-conjugated anti-rabbit IgG (MBL) as the secondary antibody. Signals were detected using SuperSignal West Pico Chemiluminescent Substrate (Thermo Fisher Scientific) and LAS-1000 plus image analyzer (Fuji Film). Images were processed with Canvas X software (ACD Systems).
Y1H Assay
A Y1H assay was performed as described previously (Yamaji et al., 2009). Briefly, the full-length ORF of VIP1 or its truncated version (corresponding to amino acids 1–186, 165–341, or 193–341) was cloned in frame behind the GAL4BD, and this construct was introduced into a yeast reporter strain, AH109. Reporter gene activation was checked by a β-galactosidase assay (Yeast Protocols Handbook; Clontech) and by seeing cell growth on the synthetic dextrose medium without adenine and His.
Transient Reporter Assay
To create the reporter constructs, promoter regions of CYP707A1 and CYP707A3 (approximately 900 bp upstream of the start codon) were amplified by PCR using the following primer pairs: 5′-GGTCTAGAAGCTACTACAGACACGTAAACAGA-3′ (a restriction recognition sequence is underlined) and 5′-CTCGGATCCACAGTGGGAGTAGCTGAACAACGA-3′ for the CYP707A1 promoter, and 5′-GGTCTAGATGACCACACGTGGGTTTTCCCA-3′ and 5′-CTCGGATCCTGGGAATTCGAAGTATGTTTTGTGAGT-3′ for the CYP707A3 promoter. PCR products were digested by XbaI and BamHI and cloned into the NheI-BglII site of pGL3-Basic (Promega). To create effector constructs, the DNA fragment containing the CaMV 35S promoter-GFP-Nos terminator was cloned into pBluescript SK−, and then the coding sequence of VIP1 was inserted between the CaMV 35S promoter and GFP. To create a control construct, the CaMV 35S promoter fragment was cloned into pMetLuc2-Reporter (Clontech). The reporter, effector, and control constructs were cointroduced into Arabidopsis mesophyll protoplasts as described previously (Yoo et al., 2007). Luciferase activities were measured 12 h after protoplast transfection according to the manufacturer’s instructions, and firefly luciferase activities were standardized by Metridium luciferase activities.
Gel-Shift Assay
The coding sequence of full-length VIP1 or amino acids 165 to 341 of VIP1 was cloned into pGEX-6P-3 or pGEX-5X-1, respectively, in frame to the coding sequence of glutathione S-transferase (GST). These constructs were introduced into the Escherichia coli strain BL21 (DE3). To induce GST fusion proteins, cells were cultured at 37°C in Luria-Bertani medium until optical density at 600 nm reached 0.5 and incubated at 28°C for 4 h after the addition of isopropylthio-β-galactoside to a final concentration of 0.1 mm. The cells were then harvested by centrifugation and sonicated in 1× Tris-buffered saline (TBS). GST fusion proteins were bound to Glutathione Sepharose 4 Fast Flow (GE Healthcare), washed four times by an excess amount of 1× TBS (150 mm NaCl, 20 mm Tris-HCl, pH 7.5), and eluted with reduced glutathione, according to the manufacturer’s instructions (GE Healthcare). The presence of GST fusion proteins in the solution was confirmed by immunoblot analysis using a horseradish peroxidase-conjugated anti-GST antibody (Thermo Fisher Scientific), and 2 μL of the solution was used for the reaction. Promoter fragments of CYP707A1 (280 or 350 bp upstream of the start codon) or CYP707A3 (350 bp upstream of the start codon) were amplified by PCR using KOD FX (TOYOBO) with or without PCR DIG Labeling Mix (Roche). Primer sequences are provided in Supplemental Figure S1. PCR products were purified by agarose gel electrophoresis and used as probes after being diluted 50 times with distilled water or as competitors without dilution. A reaction mixture (20 μL) for a gel-shift assay consisted of 4% (v/v) glycerol, 60 mm KCl, 10 mm Tris-HCl, pH 7.5, 50 ng μL−1 poly(deoxyinosinic-deoxycytidylic) acid, 2 μL of protein solution, 1 μL of probe solution, and, in some cases, 2 μL of competitor solution. Reaction mixtures were incubated at room temperature for 20 min and electrophoresed on a 1% agarose gel in 0.25× Tris-borate/EDTA running buffer. DNA was transferred from the gel to a Hybond-N+ nylon membrane (GE Healthcare). The membrane was blocked by 2.5% (w/v) ECL Blocking Agent (GE Healthcare), reacted with alkaline phosphatase-conjugated anti-digoxigenin antibody (Roche) in Tween-TBS (0.1% [v/v] Tween 20 in TBS), washed three times in Tween-TBS, and equilibrated in detection buffer (50 mm NaCl, 50 mm Tris-HCl, pH 9.5). Signals were detected using a chemiluminescent substrate, CDP-Star (Roche), according to the manufacturer’s instructions.
ChIP
ChIP analysis was performed essentially as described previously (Gendrel et al., 2002). Briefly, 15 to 25 seedlings (150–200 mg in total) of GFPox (negative control) or VIP1-GFPox were grown for 2 weeks, submerged in 10 mL of distilled water for 10 min at room temperature, incubated in 10 mL of 1% (w/v) formaldehyde for 10 min to cross-link protein and DNA, washed three times with distilled water, incubated in 180 mm Gly for 5 min to stop the cross-linking reaction, and ground to a fine powder in liquid nitrogen. The powder was suspended in 2 mL of the cell lysis solution (50 mm Tris-HCl, pH 7.5, 1 mm EDTA, and 0.5% [v/v] Triton X-100). The solution was filtered by one layer of Miracloth (Calbiochem) and centrifuged for 3 min at 5,000g at 4°C. Precipitates were resuspended in 2 mL of the cell lysis solution, and the solution was centrifuged for 3 min at 5,000g at 4°C. Precipitates, which contain purified chromatin, were resuspended in 110 μL of the cell lysis solution with Complete Mini (Roche) and sonicated on ice to obtain 0.5- to 1.5-kb fragments of DNA. The solution was centrifuged for 5 min at 15,000g at 4°C to remove the remaining cellular contaminants. The supernatant was divided into two equal (50-μL) portions. Five microliters of the remaining supernatant was diluted 10 times with distilled water and used as a DNA input for subsequent PCR. A rabbit anti-GFP antibody (MBL; 0.5 μL) was added to one of the 50-μL portions, and as a negative control, a rabbit anti-hemagglutinin antibody (MBL; 0.5 μL) was added to the other 50-μL portion. The solutions were incubated for 1 h at room temperature. Then, 20 μL of Protein G Sepharose 4 Fast Flow (GE Healthcare; 50% slurry) was added and the solution was further incubated for 45 min at room temperature with gentle shaking. The Sepharose was washed twice with wash buffer 1 (20 mm Tris-HCl, pH 7.5, 1 mm EDTA, 500 mm NaCl, and 1% [v/v] Triton X-100), twice with wash buffer 2 (20 mm Tris-HCl, pH 7.5, 1 mm EDTA, 500 mm LiCl, and 1% [v/v] Triton X-100), and twice with 1× TBS. Fifty microliters of 300 mm NaCl was then added, and the Sepharose was incubated for 4 h at 65°C to remove the cross-links between protein and DNA. The supernatant was transferred to a new tube and directly used as a template for PCR using KOD FX Neo (TOYOBO). DNA fragments of the CYP707A1/3 promoters were amplified using the following primer pairs: 5′-CTATCTTCGTCTCTCTCAAATG-3′ and 5′-ACAGTGGGAGTAGCTGAACAACGA-3′ for the CYP707A1 promoter, and 5′-TGCCCCTTTCCTTGACTTAATCC-3′ and 5′-TGGGAATTCGAAGTATGTTTTGTGAGT-3′ for the CYP707A3 promoter. After the removal of the cross-links, the remaining Sepharose was incubated at 100°C for 5 min in 30 μL of 2× SDS sample buffer (0.125 m Tris-HCl, pH 6.8, 10% [v/v] β-mercaptoethanol, 4% [w/v] SDS, 10% [w/v] Suc, and 0.01% [w/v] bromphenol blue). VIP1-GFP in the supernatant was analyzed by immunoblotting using the anti-GFP antibody (MBL).
RT-PCR
To analyze gene expression under rehydration, 2-week-old seedlings of each genotype were submerged in 20 mm Tris-HCl, pH 6.8, and sampled at the time points indicated in Figure 7. To analyze stage- and tissue-specific expression of VIP1, 10-d-old seedlings and roots, leaves, inflorescences, flower stalks, and siliques of mature plants were sampled. To analyze the expression of the osmoregulated genes or ABA-responsive genes under mannitol stress, plants of each genotype were grown for 3 weeks on medium with 0 or 200 mm mannitol and sampled. To analyze transcript responses of NCED3, RD29A, RAB18, and RD29B, 14-d-old wild-type seedlings were incubated for 90 min at room temperature in 20 mm Tris-HCl, pH 6.8, with 0 or 100 μm ABA and sampled. Total RNA was extracted as described previously (Chomczynski and Sacchi, 1987), and cDNA was synthesized from 2.5 μg of the total RNA with PrimeScript Reverse Transcriptase (Takara Bio) using an oligo(dT) primer. The reaction mixtures were diluted 20 times with distilled water and used as a template for PCR. The primer pairs used are given in Supplemental Table S1. Semi-qRT-PCR was performed using GoTaq Green Master Mix (Promega). The PCR cycle for semi-qRT-PCR was as follows: 98°C for 2 min, 25 to 35 cycles of 98°C for 10 s, 60°C for 30 s, and 72°C for 30 s, then 72°C for 7 min and 4°C until analysis. qRT-PCR was performed using GoTaq qPCR Master Mix (Promega) and the StepOne Real-Time PCR System (Applied Biosystems).
Histochemical GUS Staining
To generate the transgenic plants, the promoter region of VIP1 was amplified by genomic PCR as described previously (Tsugama et al., 2011) using the following primer pair: 5′-GGAAGGTGGGATGGTTCCTATG-3′ and 5′-GTGGATCCTGATTTTTTTTTTTCCCGGCGGCGAGG-3′. The PCR product was digested by HindIII and BamHI, resulting in a DNA fragment around 1,750 bp long. This fragment was cloned into the HindIII-BamHI site in pBI121, replacing the CaMV 35S promoter region. The resultant plasmid was transformed into Arabidopsis. Histochemical GUS staining was performed as described previously (Tsugama et al., 2009) with slight modifications. Briefly, tissues were soaked in ice-cold 90% (v/v) acetone, rinsed with 100 mm NaPi, pH 7.0, vacuum infiltrated for 30 min with staining solution (100 mm NaPi, pH 7.0, 1 mm K4[Fe(CN)6], 1 mm K3[Fe(CN)6], 0.1% [v/v] Triton X-100, 10% [v/v] methanol, and 1.5 mm 5-bromo-4-chloro-3-indolyl-β-glucuronic acid), and incubated in the dark at 37°C until staining became visible. After staining, the tissues were washed in 70% ethanol to remove the chlorophyll. To obtain high-magnification images, tissues were cleared by mounting them in a mixture of chloral hydrate:water:glycerol (8:2:1, v/v).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Turgor-dependent translocation of VIP1-GFP in a hypocotyl and a leaf.
Supplemental Figure S2. Turgor-dependent translocation of VIP1-GFP is not affected by ABA or GA3.
Supplemental Figure S3. Alignment of DNA sequences of the CYP707A1 promoter and the CYP707A3 promoter.
Supplemental Figure S4. Relative growth of VIP1-GFPox plants under mannitol stress conditions.
Supplemental Figure S5. Responses of VIP1-GFPox plants to NaCl stress.
Supplemental Figure S6. Expression levels of dehydration-responsive genes, NCED3 and RD29A, and a rehydration-responsive gene, PDH1, under mannitol stress.
Supplemental Figure S7. Induction of NCED3, RD29A, RAB18, and RD29B by ABA.
Supplemental Table S1. Primer pairs used for RT-PCR analyses.
Supplementary Material
Acknowledgments
We are grateful to the Arabidopsis Biological Resource Center for providing the Arabidopsis mutant seeds. We thank Mr. Yuichi Matsuo for assistance in fluorescence microscopy.
Glossary
- ABA
abscisic acid
- MAPK
mitogen-activated protein kinase
- NLS
nuclear localization signal
- NES
nuclear export signal
- Y1H
yeast one-hybrid
- ChIP
chromatin immunoprecipitation
- RT
reverse transcription
- Semi-q
semiquantitative
- Semi-qRT
semiquantitative reverse transcription
- q
quantitative
- qRT
quantitative reverse transcription
- MS
Murashige and Skoog
- ORF
open reading frame
- CaMV
cauliflower mosaic virus
- GAL4BD
GAL4 DNA-binding domain
- GST
glutathione S-transferase
- TBS
Tris-buffered saline
References
- Alonso R, Oñate-Sánchez L, Weltmeier F, Ehlert A, Diaz I, Dietrich K, Vicente-Carbajosa J, Dröge-Laser W. (2009) A pivotal role of the basic leucine zipper transcription factor bZIP53 in the regulation of Arabidopsis seed maturation gene expression based on heterodimerization and protein complex formation. Plant Cell 21: 1747–1761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brocard IM, Lynch TJ, Finkelstein RR. (2002) Regulation and role of the Arabidopsis abscisic acid-insensitive 5 gene in abscisic acid, sugar, and stress response. Plant Physiol 129: 1533–1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay S, Ang LH, Puente P, Deng XW, Wei N. (1998) Arabidopsis bZIP protein HY5 directly interacts with light-responsive promoters in mediating light control of gene expression. Plant Cell 10: 673–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. (1987) Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem 162: 156–159 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Dai S, Petruccelli S, Ordiz MI, Zhang Z, Chen S, Beachy RN. (2003) Functional analysis of RF2a, a rice transcription factor. J Biol Chem 278: 36396–36402 [DOI] [PubMed] [Google Scholar]
- Dai S, Zhang Z, Chen S, Beachy RN. (2004) RF2b, a rice bZIP transcription activator, interacts with RF2a and is involved in symptom development of rice tungro disease. Proc Natl Acad Sci USA 101: 687–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djamei A, Pitzschke A, Nakagami H, Rajh I, Hirt H. (2007) Trojan horse strategy in Agrobacterium transformation: abusing MAPK defense signaling. Science 318: 453–456 [DOI] [PubMed] [Google Scholar]
- Fujita Y, Fujita M, Satoh R, Maruyama K, Parvez MM, Seki M, Hiratsu K, Ohme-Takagi M, Shinozaki K, Yamaguchi-Shinozaki K. (2005) AREB1 is a transcription activator of novel ABRE-dependent ABA signaling that enhances drought stress tolerance in Arabidopsis. Plant Cell 17: 3470–3488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukazawa J, Sakai T, Ishida S, Yamaguchi I, Kamiya Y, Takahashi Y. (2000) Repression of shoot growth, a bZIP transcriptional activator, regulates cell elongation by controlling the level of gibberellins. Plant Cell 12: 901–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furihata T, Maruyama K, Fujita Y, Umezawa T, Yoshida R, Shinozaki K, Yamaguchi-Shinozaki K. (2006) Abscisic acid-dependent multisite phosphorylation regulates the activity of a transcription activator AREB1. Proc Natl Acad Sci USA 103: 1988–1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendrel AV, Lippman Z, Yordan C, Colot V, Martienssen RA. (2002) Dependence of heterochromatic histone H3 methylation patterns on the Arabidopsis gene DDM1. Science 297: 1871–1873 [DOI] [PubMed] [Google Scholar]
- Igarashi D, Ishida S, Fukazawa J, Takahashi Y. (2001) 14-3-3 proteins regulate intracellular localization of the bZIP transcriptional activator RSG. Plant Cell 13: 2483–2497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida S, Fukazawa J, Yuasa T, Takahashi Y. (2004) Involvement of 14-3-3 signaling protein binding in the functional regulation of the transcriptional activator REPRESSION OF SHOOT GROWTH by gibberellins. Plant Cell 16: 2641–2651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida S, Yuasa T, Nakata M, Takahashi Y. (2008) A tobacco calcium-dependent protein kinase, CDPK1, regulates the transcription factor REPRESSION OF SHOOT GROWTH in response to gibberellins. Plant Cell 20: 3273–3288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iuchi S, Kobayashi M, Taji T, Naramoto M, Seki M, Kato T, Tabata S, Kakubari Y, Yamaguchi-Shinozaki K, Shinozaki K. (2001) Regulation of drought tolerance by gene manipulation of 9-cis-epoxycarotenoid dioxygenase, a key enzyme in abscisic acid biosynthesis in Arabidopsis. Plant J 27: 325–333 [DOI] [PubMed] [Google Scholar]
- Iwata Y, Fedoroff NV, Koizumi N. (2008) Arabidopsis bZIP60 is a proteolysis-activated transcription factor involved in the endoplasmic reticulum stress response. Plant Cell 20: 3107–3121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakoby M, Weisshaar B, Dröge-Laser W, Vicente-Carbajosa J, Tiedemann J, Kroj T, Parcy F. bZIP Research Group(2002) bZIP transcription factors in Arabidopsis. Trends Plant Sci 7: 106–111 [DOI] [PubMed] [Google Scholar]
- Kang JY, Choi HI, Im MY, Kim SY. (2002) Arabidopsis basic leucine zipper proteins that mediate stress-responsive abscisic acid signaling. Plant Cell 14: 343–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushiro T, Okamoto M, Nakabayashi K, Yamagishi K, Kitamura S, Asami T, Hirai N, Koshiba T, Kamiya Y, Nambara E. (2004) The Arabidopsis cytochrome P450 CYP707A encodes ABA 8′-hydroxylases: key enzymes in ABA catabolism. EMBO J 23: 1647–1656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara P, Oñate-Sánchez L, Abraham Z, Ferrándiz C, Díaz I, Carbonero P, Vicente-Carbajosa J. (2003) Synergistic activation of seed storage protein gene expression in Arabidopsis by ABI3 and two bZIPs related to OPAQUE2. J Biol Chem 278: 21003–21011 [DOI] [PubMed] [Google Scholar]
- Li J, Krichevsky A, Vaidya M, Tzfira T, Citovsky V. (2005) Uncoupling of the functions of the Arabidopsis VIP1 protein in transient and stable plant genetic transformation by Agrobacterium. Proc Natl Acad Sci USA 102: 5733–5738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto M, Tanaka Y, Abrams SR, Kamiya Y, Seki M, Nambara E. (2009) High humidity induces abscisic acid 8′-hydroxylase in stomata and vasculature to regulate local and systemic abscisic acid responses in Arabidopsis. Plant Physiol 149: 825–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Rourke SM, Herskowitz I, O’Shea EK. (2002) Yeast go the whole HOG for the hyperosmotic response. Trends Genet 18: 405–412 [DOI] [PubMed] [Google Scholar]
- Pitzschke A, Djamei A, Teige M, Hirt H. (2009) VIP1 response elements mediate mitogen-activated protein kinase 3-induced stress gene expression. Proc Natl Acad Sci USA 106: 18414–18419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringli C, Keller B. (1998) Specific interaction of the tomato bZIP transcription factor VSF-1 with a non-palindromic DNA sequence that controls vascular gene expression. Plant Mol Biol 37: 977–988 [DOI] [PubMed] [Google Scholar]
- Tan B-C, Joseph LM, Deng W-T, Liu L, Li Q-B, Cline K, McCarty DR. (2003) Molecular characterization of the Arabidopsis 9-cis epoxycarotenoid dioxygenase gene family. Plant J 35: 44–56 [DOI] [PubMed] [Google Scholar]
- Torres-Schumann S, Ringli C, Heierli D, Amrhein N, Keller B. (1996) In vitro binding of the tomato bZIP transcriptional activator VSF-1 to a regulatory element that controls xylem-specific gene expression. Plant J 9: 283–296 [DOI] [PubMed] [Google Scholar]
- Tran L-SP, Urao T, Qin F, Maruyama K, Kakimoto T, Shinozaki K, Yamaguchi-Shinozaki K. (2007) Functional analysis of AHK1/ATHK1 and cytokinin receptor histidine kinases in response to abscisic acid, drought, and salt stress in Arabidopsis. Proc Natl Acad Sci USA 104: 20623–20628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsugama D, Liu S, Takano T. (2009) Stage- and tissue-specific expression of rice OsIsu1 gene encoding a scaffold protein for mitochondrial iron-sulfur-cluster biogenesis. Biotechnol Lett 31: 1305–1310 [DOI] [PubMed] [Google Scholar]
- Tsugama D, Liu S, Takano T. (2011) A rapid chemical method for lysing Arabidopsis cells for protein analysis. Plant Methods 7: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzfira T, Vaidya M, Citovsky V. (2001) VIP1, an Arabidopsis protein that interacts with Agrobacterium VirE2, is involved in VirE2 nuclear import and Agrobacterium infectivity. EMBO J 20: 3596–3607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzfira T, Vaidya M, Citovsky V. (2002) Increasing plant susceptibility to Agrobacterium infection by overexpression of the Arabidopsis nuclear protein VIP1. Proc Natl Acad Sci USA 99: 10435–10440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umezawa T, Okamoto M, Kushiro T, Nambara E, Oono Y, Seki M, Kobayashi M, Koshiba T, Kamiya Y, Shinozaki K. (2006) CYP707A3, a major ABA 8′-hydroxylase involved in dehydration and rehydration response in Arabidopsis thaliana. Plant J 46: 171–182 [DOI] [PubMed] [Google Scholar]
- Urao T, Yakubov B, Satoh R, Yamaguchi-Shinozaki K, Seki M, Hirayama T, Shinozaki K. (1999) A transmembrane hybrid-type histidine kinase in Arabidopsis functions as an osmosensor. Plant Cell 11: 1743–1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlbach DJ, Quirino BF, Sussman MR. (2008) Analysis of the Arabidopsis histidine kinase ATHK1 reveals a connection between vegetative osmotic stress sensing and seed maturation. Plant Cell 20: 1101–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Zhao Q, Gao L, Yu XM, Fang P, Oliver DJ, Xiang CB. (2010) Isolation and characterization of low-sulphur-tolerant mutants of Arabidopsis. J Exp Bot 61: 3407–3422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaji N, Huang CF, Nagao S, Yano M, Sato Y, Nagamura Y, Ma JF. (2009) A zinc finger transcription factor ART1 regulates multiple genes implicated in aluminum tolerance in rice. Plant Cell 21: 3339–3349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Zhu Q, Dai S, Lamb C, Beachy RN. (1997) RF2a, a bZIP transcriptional activator of the phloem-specific rice tungro bacilliform virus promoter, functions in vascular development. EMBO J 16: 5247–5259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S-D, Cho Y-H, Sheen J. (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Yoshida T, Fujita Y, Sayama H, Kidokoro S, Maruyama K, Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K. (2010) AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. Plant J 61: 672–685 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.