Abstract
Reactive oxygen species (ROS) play an important role as triggers of gene expression during biotic and abiotic stresses, among which is low oxygen (O2). Previous studies have shown that ROS regulation under low O2 is driven by a RHO-like GTPase that allows tight control of hydrogen peroxide (H2O2) production. H2O2 is thought to regulate the expression of heat shock proteins, in a mechanism that is common to both O2 deprivation and to heat stress. In this work, we used publicly available Arabidopsis (Arabidopsis thaliana) microarray datasets related to ROS and O2 deprivation to define transcriptome convergence pattern. Our results show that although Arabidopsis response to anoxic and hypoxic treatments share a common core of genes related to the anaerobic metabolism, they differ in terms of ROS-related gene response. We propose that H2O2 production under O2 deprivation is a trait present in a very early phase of anoxia, and that ROS are needed for the regulation of a set of genes belonging to the heat shock protein and ROS-mediated groups. This mechanism, likely not regulated via the N-end rule pathway for O2 sensing, is probably mediated by a NADPH oxidase and it is involved in plant tolerance to the stress.
Oxygen (O2) deprivation in plants occurs frequently, due either to natural flooding events in flood-prone areas (Bailey-Serres et al., 2010) or to the slow diffusion of O2 in bulky organs (Geigenberger et al., 2000; van Dongen et al., 2003). Energy production is tightly regulated in the absence of O2. The respiratory metabolism switches from aerobic to anaerobic, to sustain ATP generation and guarantee survival (for review, see Geigenberger, 2003; Sachs and Vartapetian, 2007; Colmer and Voesenek, 2009). The way that plant morphology and physiology allows plants to withstand O2 deprivation has been a subject of study for many years (for review, see Voesenek et al., 2006).
Recently, considerable progress has been made toward the comprehension of the molecular mechanisms governing these traits and which are responsible for plant sensitivity/tolerance to low-O2 stress (Xu et al., 2006; Hattori et al., 2009; Lee et al., 2009; Licausi, 2011). Moreover, very recently a direct homeostatic low-O2 sensor has been identified in plants (Gibbs et al., 2011; Licausi et al., 2011). Ethylene-responsive factors (ERFs) of group VII, among which HYPOXIA RESPONSIVE1 (HRE1) and HRE2 and RELATED TO AP2 12 (RAP2.12), have been shown to be substrate of the N-rule pathway, where the N-terminal Met-Cys motif is subjected to targeted ubiquitin-dependent protein degradation under normoxic condition, possibly through the oxidation of the tertiary destabilizing residue Cys (Gibbs et al., 2011; Licausi et al., 2011). The stabilization of the N-terminal motif under low O2 leads to increased plant survival, through the control of the expression of hypoxic core genes (Gibbs et al., 2011). Whether Cys (in)stability depends directly on O2 or cellular changes associated to its availability, such as cytosolic pH or reactive oxygen species (ROS) balance, is still unclear (Gibbs et al., 2011).
ROS production has been suggested to be a component of low-O2 signaling. Baxter-Burrell et al. (2002) proved that the activation of a RHO-like small G protein of plant (ROP) under low O2 induces hydrogen peroxide (H2O2) accumulation via an NADPH oxidase mechanism that was shown to be required for ALCOHOL DEHYDROGENASE (ADH) expression and activity, thus tolerance. The ROP family modulates signaling cascades associated with a variety of mechanism in plants and eukaryotic kingdoms in general (for review, see Yang and Fu, 2007). It seems that tolerance to O2 deprivation also requires ROP activation and deactivation and that their activity be controlled by negative feedback regulation that in turn requires a ROP GTPase activating protein, also regulated by H2O2 (Baxter-Burrell et al., 2002). The mechanism that drives the ROP rheostat activation under low O2 is currently not known, but it should involve a novel mechanism because RAC1, the counterpart of ROP, regulates NADPH oxidase through binding to the p47phox regulatory subunit that is absent in plants (Gu et al., 2004).
H2O2 production is observed under both anoxia and heat stress (Banti et al., 2010), suggesting that it can be involved in the induction of heat shock transcription factors (HSFs) and heat shock proteins (HSPs) found to be induced under these stresses (Banti et al., 2010). Indeed, among the families of conserved H2O2-responsive proteins across kingdoms, DNAJ-type HSPs and small HSP proteins were identified (Vandenbroucke et al., 2008). Moreover, a cross-kingdoms comparison of transcriptome regulation under low O2 indicates a general increase of HSP transcripts (Mustroph et al., 2010). HSFs have been proposed to be specific H2O2 sensors in plants (Miller and Mittler, 2006) and Arabidopsis (Arabidopsis thaliana) seedlings that overexpress HsfA2 are markedly more tolerant to anoxia (Banti et al., 2010).
Overall, these results indicate that the response to anoxia and to heat often overlaps, and it seems that ROS production may occur upstream of the signaling pathway required for tolerance. HSFs have not been proposed as directly regulated by the N-end rule pathway, suggesting ROS as possible actors in a side mechanism. However, ROS production due to general oxidative stress generated by low-O2-dependent secondary alterations cannot be excluded.
To verify whether a subset of hypoxic/anoxic transcripts are in fact regulated by ROS, we examined some Arabidopsis Affymetrix genome arrays related to both O2 deprivation and ROS-producing experiments that are available in the literature. Our results demonstrate that a certain overlap exists between genes induced by ROS and genes induced by anoxia. Here we show that genes induced by both ROS and anoxia include some encoding HSPs and ROS-related transcription factors (TFs) and we propose that the expression of those genes under anoxia is regulated by a ROS-dependent way that requires the activation of an NADPH oxidase involved in tolerance.
RESULTS AND DISCUSSION
The Low-O2 Core Transcriptome in Arabidopsis Plants
Transcript profiling datasets related to O2 deprivation (Table I) and ROS-related experiments (Table II) were gathered and analyzed. From cluster analysis of the datasets, it became evident that shoots and roots respond differently to ROS (Supplemental Fig. S1) and low-O2 conditions (Supplemental Fig. S2), as previously observed (Ellis et al., 1999; Mustroph et al., 2009; Lee et al., 2011), and that the seedlings’ response was more similar to the shoot one. Therefore roots and seedlings/shoots datasets were analyzed separately.
Table I. Overview of the O2 deprivation Affymetrix Arabidopsis microarrays studies used for the analysis.
A 6h1, 6 h of anoxia treatment without or with 90 mm Suc (s) added before the treatment (Loreti et al., 2005); A 6h2, 6 h of anoxia treatment (Banti et al., 2010); A 12h, 12 h of anoxia treatment, total and polysomal (p) RNA (Branco-Price et al., 2005); H 4h, 4 h of hypoxia treatment at 1% O2 (Licausi et al., 2010); S 7/24h, 7 and 24 h of submergence treatment (Lee et al., 2011); Hyp, 0.5, 2 and 48 h at 1%, 4%, and 8% O2 (van Dongen et al., 2009); GS, growth stage (Boyes et al., 2001).
| Sample Description | GS | Growth | Treatment | Accession No. | Arrays | Replicates | Reference |
| Colglabra seedlings | 1.0 | Petri dishes, MS liquid medium | 6 h anoxia ± 90 mm Suc, dark conditions | GSE2133 | 8 | 2 | A 6h1 |
| GSE3704 | A 6h1s | ||||||
| Col-0 seedlings | 1.0 | Petri dishes, MS liquid medium (90 mm Suc) | 6 h anoxia, dark conditions | GSE16222 | 4 | 2 | A 6h2 |
| Landsberg erecta seedlings | 1.02 | Vertical plates, MS solid medium (1% Suc) | 12 h O2-free argon atmosphere, dim light | GSE2218 | 4 | 1 | A 12h |
| A 12hp | |||||||
| Col-0 seedlings | 1.02 | Petri dishes, MS liquid medium (1% Suc) | 4 h at 1% O2, dark conditions | GSE17099 | 4 | 2 | H 4h |
| Col-0 roots and shoots | 1.09 | Pot, soil | 7 to 24 h submersion, dark conditions | GSE24077 | 16 | 2 | S 7h |
| S 24h | |||||||
| Col2 roots | 1.02 | Vertical plates, MS solid medium (1% Suc) | 0.5, 2 h and 48 h at 1%, 4%, and 8% O2, dark conditions | GSE11558 | 28 | 2 | Hyp |
Table II. Overview of the ROS-related Affymetrix Arabidopsis microarrays studies used for the analysis.
flu 0.5/1/2h, flu mutant shift from dark to light (op den Camp et al., 2003); AOX1a-AS, AOX1a-AS mutant (Umbach et al., 2005); MV 1/6/12h, MV 1, 6, 12 h treatment (Dr. Kirch, NASCArray repository); O3, 1 h of O3 fumigation at 200 nL L−1 (Dr. Shirras, NASCArray repository); H2O2, 1 h of H2O2 treatment at 20 mm (Dr. Mittler, NASCArray repository); GS, growth stage (Boyes et al., 2001).
| Sample Description | GS | Treatment | ROS | Accession No. | Arrays | Replicates | Reference |
| flu mutant in Landsberg erecta background leaves | 1.02 | Dark to light 0.5, 1 h, 2 h | 1O2 plast | GSE10876 | 6 | 1 | flu 0.5/1/2h |
| AOX1a-AS mutant in Col background leaves | 1.09 | – | O2− mit | GSE2406 | 4 | 2 | AOX1a-AS |
| Col-0 root and shoot | 1.04 | 10 μm MV 0.5, 1, 3, 6, 12, 24 h | O2− mit/chl | NASCARRAYS-143 | 44 | 2 | MV 1/6/12h |
| Seedlings | 1.02 | 1 h O3 200 nL L−1 | O2−H2O2 | NASCARRAYS-26 | 6 | 3 | O3 |
| Seedlings | 1.0 | 20 mmH2O2 1 h | H2O2 | NASCARRAYS-338 | 6 | 3 | H2O2 |
The microarray datasets related to O2 deprivation were investigated first. The majority of the experiments were performed using the Columbia (Col) ecotype at growth stage 1.0 (Boyes et al., 2001), under dark conditions. Treatment in the dark should eliminate the possibility that O2 be produced by photosynthesis (Mustroph et al., 2006; Colmer and Pedersen, 2008).
A core of genes up-regulated under anoxia and hypoxia was defined for both seedlings and shoots, by querying the whole microarray datasets at the arbitrary threshold of log2 ≥ 1 (P < 0.05). For experiments containing several time points (i.e. Lee et al., 2011), we considered the regulation to be significant when at least one of the time points analyzed was significantly up-regulated.
The regulation of anaerobic core genes was also investigated in 35S::HRE1 and 35S::HRE2 Arabidopsis transgenic plants under hypoxia. The HRE1 gene is an ERF acting as positive regulator of a set of anaerobic genes under hypoxia (Licausi et al., 2010). Overexpression of HRE1 results in an increased anoxia tolerance (Licausi et al., 2010). Moreover, we investigated whether low-O2 up-regulated transcripts were ectopically accumulated in the N-end rule mutants prt6 and ate1/2 and thus directly regulated by the N-end rule pathway (Gibbs et al., 2011; Licausi et al., 2011). These mutants lack some of the recognition steps required for proteasomal degradation under normoxia, thus they constitutively express hypoxic core genes related to the anaerobic metabolism such as ADH1, PYRUVATE DECARBOXYLASE1 (PDC1), and SUC SYNTHASE4 (SUS4; Gibbs et al., 2011).
Six genes were found that constitute a core of transcripts significantly up-regulated under all the anoxic experiments but not under hypoxia (Fig. 1A). These genes encoded mostly HSPs (Fig. 1B), most of which are regulated by heat stress and that have been indicated to be HsfA2 targets (Nishizawa et al., 2006). HSPs were previously indicated to be part of the genes in which expression in dependence of low O2 is conserved among different kingdoms (Mustroph et al., 2010). Their expression has been observed since very short anoxia treatment (e.g. 2 h in seedlings; Mustroph et al., 2010), indicating a fast mechanism of response. Moreover, HSP response is suggested to be H2O2 specific (Vandenbroucke et al., 2008). Interestingly, none of these genes was up-regulated in the HRE mutants subjected to hypoxia (Fig. 1B), neither were ectopically accumulated in N-end rule mutants prt6 and ate1/2 (Gibbs et al., 2011). Thus, the expression of these HSPs is likely not directly regulated by this mechanism. A single mitochondrial HSP (HSP23.5-M, At5g51440) is accumulated under normoxia in the ate1/2 mutant only (Gibbs et al., 2011). Secondary signaling molecules to monitor O2 availability might relay on calcium flux, energy charge, and ROS balance. These three parameters seem to be interrelated, thus suggesting the presence of downstream events that could converge (Bailey-Serres and Chang, 2005). Emerging evidence suggests that ROS-mediated activation of plasma membrane calcium (Ca2+) channels is involved in plant signal transduction related to both biotic and abiotic stress and development events (for review, see Lecourieux et al., 2006). Previous results demonstrated the involvement of NADPH-oxidase-dependent H2O2 production under O2 deprivation response (Baxter-Burrell et al., 2002). Moreover, transient changes in Ca2+ during low O2 was found to be involved in ADH regulation (Sedbrook et al., 1996; Subbaiah et al., 1994a, 1994b). A connection between NADPH-oxidase-dependent ROS production and Ca2+ oscillation has been suggested in Arabidopsis, where Ca2+ likely binds to the EF hands of the N-terminal region of NADPH oxidase and, together with phosphorylation, promotes ROS production (Ogasawara et al., 2008; Takeda et al., 2008; Suzuki et al., 2011). Whether and how this mechanism could in part overlap with N-end rule pathway is still unknown.
Figure 1.
Genes regulated by anoxia, hypoxia, and O2 deprivation in the seedlings/shoots of Arabidopsis microarray datasets. The core genes were obtained selecting significantly regulated genes (−1 > log2 >1, P < 0.05) in all the experiments and in at least one time point in the experiments with several time points. A, Venn diagram showing significantly regulated genes under anoxia, hypoxia, and the overlap between anoxia and hypoxia. B, Heat map showing the expression profile of anoxia, O2 deprivation, and hypoxia-regulated genes. Value represents log2 and color indicates significant change (P < 0.05). A 6h1, 6 h of anoxia treatment without or with 90 mm Suc (s) added before the treatment (Loreti et al., 2005); A 6h2, 6 h of anoxia treatment (Banti et al., 2010); A 12h, 12 h of anoxia treatment, total and polysomal (p) RNA (Branco-Price et al., 2005); H 4h, 4 h of hypoxia treatment at 1% O2 (Licausi et al., 2010); S 7/24h, 7 and 24 h of submergence treatment (Lee et al., 2011); HRE1/2, HRE1 and HRE2 overexpressing plants under 4 h of hypoxia treatment at 1% O2 (Licausi et al., 2010); prt6 and ate1/2, N-end rule mutants under air compared with the wild type (Gibbs et al., 2011); M, multiple hit; fp, family protein; P, protein.
Five transcripts were found to be consistently up-regulated by hypoxia. These same genes were also significantly up-regulated in some anoxic microarrays but not necessarily in all (Fig. 1B). This was not true the other way around. Although we found genes that were consistently up-regulated in anoxia, they were not significantly up-regulated in any of the hypoxic microarray experiments. This suggests that the response to anoxia contains all elements of the response to hypoxia but that anoxia also contains additional elements (Fig. 1B), as was previously proposed by Licausi (2011). Among the set of five hypoxic genes, we found 1-AMINOCYCLOPROPANE-1-CARBOXYLATE OXIDASE1 (At2g19590), a gene that plays a key role in ethylene biosynthesis and requires O2 for its catalytic activity. Ethylene increases within minutes in plants subjected to soil waterlogging or complete submergence due to entrapment in submerged organs and reduced catabolism (Kawase, 1972, 1978; Könings and Jackson, 1979; Geisler-Lee et al., 2010).
A specific group of eight genes that are up-regulated after both anoxia and hypoxia were also identified, they include PDC1 (At4g33070), ADH1 (At1g77120), and SUS4 (At3g43190; Fig. 1B). Suc synthase catalyzes the conversion of Suc to UDP-Glc and Fru that will be used in glycolysis (Ricard et al., 1998). ADH1 and PDC1 are involved in catalyzing reactions involved in ethanol fermentation, which occurs in response to O2 deprivation to ensure the regeneration of NAD+ for glycolysis with concomitant production of ATP, in the absence of mitochondrial respiration (Kürsteiner et al., 2003). These genes are regulated by low O2, independently of the degree of O2 deprivation, and they encode enzymes required for the anaerobic respiration. Hypoxia-responsive genes (i.e. ADH1 and SUS4) induction is augmented in HRE1 overexpressor under anoxia (Licausi et al., 2010) and in 35S::Δ13RAP2.12 plants in air, where the first 13 amino acid residues were deleted, resulting in loss of recognition for degradation under normoxia (Licausi et al., 2011). HRE1 and RAP2.12 are members of the group VII of the ERFs Arabidopsis family, composed by five members (Nakano et al., 2006). They share homology to the rice (Oryza sativa) ERF SUBMERGENCEs and SNORKELs, found to be necessary for the activation of the quiescence strategy in lowland rice (Xu et al., 2006) and escape strategy in deepwater rice (Hattori et al., 2009), respectively. Arabidopsis ERFs of group VII are activated under low O2 to support gene expression for acclimation strategy (Gibbs et al., 2011; Licausi et al., 2011). Through the presence of the initiating motif MetCys at the N-terminal site, these ERFs are degraded through ubiquitination under normoxia, by the N-end-rule mediated removal of Met and oxidation of the Cys residue (Gibbs et al., 2011; Licausi et al., 2011). Under low O2, they are stabilized and activated and move to the nucleus to trigger anaerobic gene expression (Licausi et al., 2011). As previously described, anaerobic genes are also activated ectopically in N-end-rule mutants prt6 and ate1/2 (Fig. 1B; Gibbs et al., 2011).
Two other genes were also part of the group up-regulated by both anoxia and hypoxia, namely the LATERAL ORGAN BOUNDARIES DOMAIN CONTAINING PROTEIN41 (LBD41; At3g02550), and a UNIVERSAL STRESS PROTEIN (USP; At3g03270). LBD41 is member of the LBD gene family that encodes plant-specific TFs (Husbands et al., 2007). LBD genes are suggested to be involved in developmental processes, including leaf polarity establishment (Lin et al., 2003; Xu et al., 2003) and lateral root formation (Inukai et al., 2005; Liu et al., 2005; Okushima et al., 2007). Recent results also suggest some LBDs be involved in auxin signaling (Lin et al., 2003; Inukai et al., 2005; Taramino et al., 2007; Mangeon et al., 2011). The USP protein family has been identified and studied in bacteria, but its biochemical role is not yet fully understood. These proteins are believed to play a function in response to a plethora of stresses, including carbon starvation, O2 deprivation, nitrate, phosphate, and sulfate starvation, oxidative stress, and heat shock (Nachin et al., 2008). In plants, they have been classified but not deeply studied (Kerk et al., 2003). Indeed, an USP was found to be induced in rice under submergence and to be positively regulated by ethylene, suggesting a role in adaptation to this stress (Sauter et al., 2002).
We have identified a set of genes that are down-regulated by anoxia but not hypoxia (Fig. 1, A and B). Many of them belong to the cell wall functional class, suggesting a mechanism of general repression of growth under severe O2 deprivation, whereas others belong to the protein posttranslational modification functional class. Intriguing was the behavior of the AUXIN-REGULATED GENE INVOLVED IN ORGAN SIZE (At3g59900), which, while down-regulated under anoxia, was up-regulated in some hypoxic experiments (Fig. 1B). This gene is induced by auxin and controls lateral organ size (Hu et al., 2003).
The available root datasets were screened for significantly regulated genes (Supplemental Fig. S3A). The root material from the submergence experiment of Lee et al. (2011) was defined as anoxic since the measurement of the O2 partial pressure demonstrated its absence. The specific group of genes that are up-regulated after both anoxia and hypoxia were defined to be those significantly regulated in at least five conditions out of the nine analyzed by van Dongen et al. (2009), and one condition out of the two analyzed by Lee et al. (2011); anoxic the genes significantly regulated in both the conditions analyzed by Lee et al. (2011) and none or one from van Dongen et al. (2009), and hypoxic the genes regulated in at least five conditions out of the nine analyzed by van Dongen et al. (2009) and never regulated under the conditions of Lee et al. (2011). Also for root experiments a core of genes up-regulated under both anoxia and hypoxia was defined. Among these genes, six were in common with the up-regulated genes of the seedlings/shoots group and some of them encode for the enzymes required for the anaerobic respiratory pathway (e.g. ADH1 and SUS4).
In roots, like in seedlings/shoots, some genes were found to be up-regulated more specifically under anoxic conditions (Supplemental Fig. S3A). None of them was in common with the corresponding category of the seedlings/shoots group. In particular, the group of HSPs found in seedlings/shoots was not present in the genes up-regulated under anoxia in roots. Checking each single root array, it was evident that none of them showed a positive regulation of these HSP genes (Supplemental Table S1). However, previous results showed the induction of HSPs under low O2 in roots (Mustroph et al., 2010) with an enrichment in the phloem companion cells (Mustroph et al., 2009).
No genes were found to be significantly down-regulated in roots under hypoxia, after our analysis of data (five conditions out of nine, log2 ≥ 1, P < 0.05; Supplemental Fig. S3A), in line with the seedlings/shoots core genes dataset (Fig. 1B). However, genes were down-regulated in single experiments as previously reported (van Dongen et al., 2009) but none of them was present in all the time sets following our criteria. van Dongen et al. (2009) reported a low tendency of genes to be down-regulated, suggesting a preferential reprogramming toward adaptation rather than inhibition. Down-regulation of gene expression under low O2 could be a side effect of low-O2 treatment that does not depend on a direct and conserved modulation of gene expression, as it happens instead for up-regulated genes (van Dongen et al., 2009).
Questioning the organ-specific response of gene regulation, only six genes were found to be in common between roots and seedlings/shoots (Supplemental Fig. S3B; Supplemental Table S1). Ten genes regulated by O2 deprivation conditions in seedlings/shoots were also found to be regulated in some of the root experiments (Supplemental Fig. S3B; Supplemental Table S1). The contrary was observed for seven genes. Thus, a number of genes both up- and down-regulated were organ related. These genes are interesting because they could define an organ-specific response to O2 limitation. Indeed, transcriptome adjustment has been suggested to be organ specific (Ellis et al., 1999; Mustroph et al., 2009; Lee et al., 2011). Because of the absence of a direct ROS signature in the genes commonly regulated in roots, we did not report data about subsequent analysis.
Convergence in the Gene Transcripts Regulated by ROS and O2 Deprivation
Some authors have proposed the involvement of ROS-driven signaling under O2 deprivation (Banti et al., 2010) to be mediated by NADPH oxidase (Baxter-Burrell et al., 2002). This idea might seem counterintuitive because O2 is required for ROS production. However, O2 deprivation at the cellular level occurs later than deficiency in the environment, because internal O2 concentration depends also on the resistance to O2 diffusion through tissue (Gupta et al., 2009). Cytochrome c oxidase catalyzes the reduction of O2 to water with high affinity for O2 [Km(O2) approximately 0.1 μm; Gupta et al., 2009] along the mitochondrial electron transport chain (Cooper, 2002), whereas the oxidative burst that produces ROS via the cell-membrane-located NADPH oxidase occurs in the apoplast (Torres and Dangl, 2005). It is tempting to speculate that this oxidative burst occurs prior to the use of O2 by cytochrome c oxidase inside the cell. Recent findings report the involvement of mitochondria in oxidative burst after anoxia where ROS production is likely to occur at the mitochondrial electron transport chain (Chang et al., 2012).
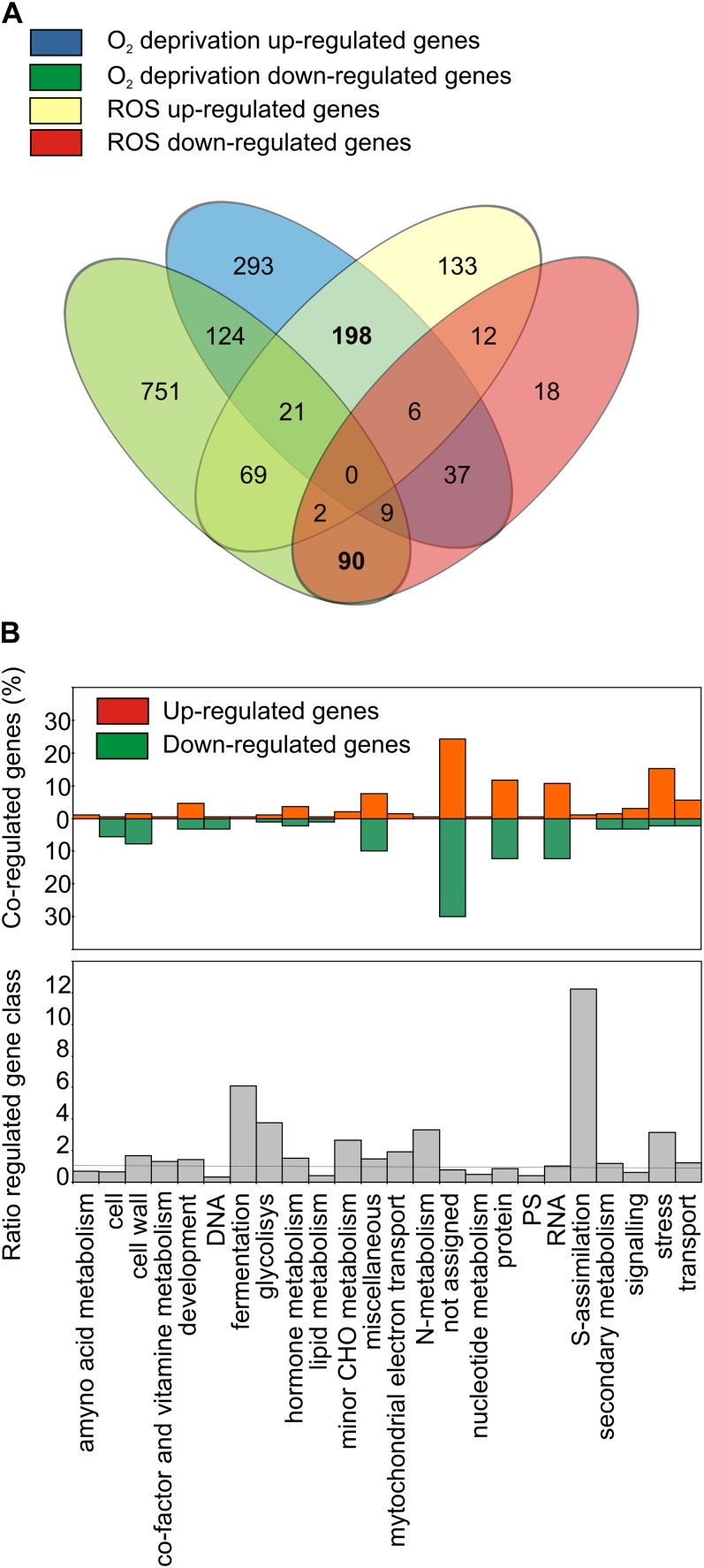
Should ROS be part of the mechanism(s) behind gene regulation under hypoxia or anoxia, we would expect to find a convergence in gene regulation when comparing ROS and low-O2-related datasets. The ROS-related datasets used for our analysis, all of which were performed at growth stage 1.0 (Boyes et al., 2001), are described in Table II. A very detailed analysis on the specificity of ROS signaling was previously published by Gadjev et al. (2006). We instead filtered both the ROS and the O2 deprivation (anoxia and hypoxia) datasets related to seedlings/shoots, and selected genes that were regulated (log2 ≥ 1 and log2 ≤ −1, P < 0.05) in at least one experiment. The genes identified with these criteria were 1,762 (data not shown). We searched this group of genes to identify those found both in O2 deprivation and ROS signaling, identifying 431 genes (Fig. 2A). One hundred and ninety seven of them were commonly up-regulated (Supplemental Fig. S4) and 90 commonly down-regulated (Supplemental Fig. S5). Among the functional classes that were coregulated, S-assimilation, fermentation, N-metabolism, and stress were overrepresented and some of them consisted of up-regulated genes only (Fig. 2B). However, only the stress cluster contained a large number of genes (Fig. 2B). A large number of co-down-regulated genes belonging to the overrepresented cell wall cluster was also found (Fig. 2B).
Figure 2.
Genes coregulated in O2 deprivation and ROS-related seedlings/shoots microarray experiments. A, Venn diagram showing the overlap of up- and down-regulated genes under O2 deprivation and/or ROS-related experiments, considering at least one significantly regulated experiment for each condition. In bold, up and down coregulated transcripts. B, Percentage of up- and down-regulated functional gene categories under O2 deprivation and ROS-related microarray experiments, and ratio of coregulated class percentage divided by the relative percentage of gene classes represented in the microarray. The line indicates ratio = 1, all the classes above this value are overrepresented in the O2 deprivation and ROS-related coregulated gene classes.
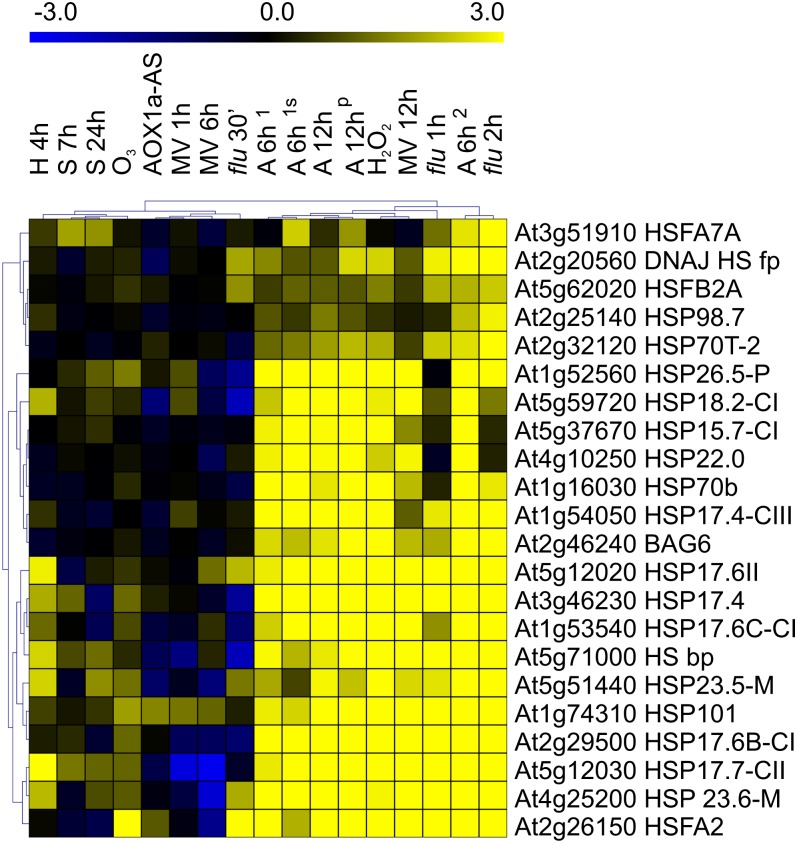
Among the 197 genes up-regulated in both O2 deprivation and ROS datasets, the hierarchical average linkage clustering identified a functional group composed of 22 genes related to heat stress (Fig. 3), including several HSPs but also heat shock TFs (i.e. HsfA2, HsfA7A, and HsfB2A). This group included genes induced both under anoxia and some of the ROS-related experiments. Hsfs are encoded by a large gene family and act by binding to a highly conserved heat shock element in the promoter region of many target genes related to defense (Mittler and Zilinskas, 1992; Storozhenko et al., 1998; Davletova et al., 2005a). Several reports suggest the existence of a direct link between heat shock and oxidative stresses, as they show that Hsfs are activated during various environmental stimuli (Li et al., 2005; Nishizawa et al., 2006; Banti et al., 2008, 2010). Hsfs were also proposed to be ROS sensors in plants (Miller and Mittler, 2006), as suggested for Drosophila and mammals (Zhong et al., 1998; Ahn and Thiele, 2003). Hierarchical average linkage clustering associated the heat-related genes to H2O2 and O2−-producing experiments as well as to the anoxic (but not hypoxic) ones (Fig. 3), a result that is consistent with the seedlings/shoots core gene dataset for hypoxia, showing no significantly up-regulated HSP genes (Fig. 1B).
Figure 3.
Hierarchical average linkage clustering of heat-stress-related genes regulated in seedlings/shoots in both O2 deprivation and ROS-related experiments. H 4h, 4 h of hypoxia treatment at 1% O2 (Licausi et al., 2010); S 7/24h, 7 and 24 h of submergence treatment (Lee et al., 2011); O3, 1 h of O3 fumigation at 200 nL L−1 (Dr. Shirras, NASCArray repository); AOX1a-AS, AOX1a-AS mutant (Umbach et al., 2005); MV 1/6/12h, MV treatment (Dr. Kirch, NASCArray repository); flu 0.5 (30’) /1/2h, flu mutant shift from dark to light (op den Camp et al., 2003); A 6h1, 6 h of anoxia treatment without or with 90 mm Suc (s) added before the treatment (Loreti et al., 2005); A 12h, 12 h of anoxia treatment, total and polysomal (p) RNA (Branco-Price et al., 2005); H2O2, 1 h of H2O2 treatment at 20 mm (Dr. Mittler, NASCArray repository); A 6h2, 6 h of anoxia treatment (Banti et al., 2010); bp, binding protein; fp, family protein.
The expression of HSP gene orthologs was investigated in the available microarrays of rice plants tolerant and sensitive to low O2 (Kottapalli et al., 2007; Mustroph et al., 2010). Indeed, some HSP orthologous genes showed an up-regulation in the tolerant varieties only (Supplemental Table S2). In particular, Os03g14180 likely orthologous to HSP25.3-P of Arabidopsis, was induced in tolerant rice under submergence and was also present in the anoxia up-regulated genes category defined in Figure 1B. Indeed, conserved induction of HSPs under low O2 has been found in several plant species analyzed before (Mustroph et al., 2010). The activation of the HSP way could be a successful strategy under low O2. This is demonstrated by the anoxia tolerance of the HsfA2 overexpressor plants that, although not showing up-regulation of classical anaerobic genes, shows increased transcription and translation of HSPs (Banti et al., 2010). Up-regulation of HSP genes in rice-tolerant plants strengthens this hypothesis. However, it appears that this way is not fully executed in Arabidopsis under anoxia (Banti et al., 2010). This might be related to the energy need of the de novo protein synthesis, that likely is dampened under the energy shortage occurring under anoxia. In this context, some of the HSPs found to be up-regulated under anoxia (Fig. 1B) are part of the small HSP class that likely act in an ATP-independent way (Sun et al., 2002).
The overlap between heat stress and anoxia was already described by Banti et al. (2010), who proposed H2O2 as the signaling element linking anoxia and heat stress. HSPs are induced by anoxic stress in several species, suggesting a conservation of this mechanism across different kingdoms (Vandenbroucke et al., 2008; Mustroph et al., 2010). Anoxia, differently from hypoxia, could harbor a more complex mechanism of response/adaptation rather than a simple switch to fermentative metabolism, as suggested by Licausi (2011) and we suggested that induction of heat-related genes is mostly related to anoxia (Fig. 3).
Suc feeding drastically enhances both the induction of HSPs and anoxia tolerance in Arabidopsis seedlings (Loreti et al., 2005). Suc triggers higher HSP expression, especially when Suc is added to the growth medium some days prior to the anoxic treatment (Supplemental Fig. S6). Transcript convergence in processes involving sugar, ROS, and scavengers suggests that sugar modulation of gene expression is likely related to oxidative stress control (Couée et al., 2006). Suc feeding enhances transcription of HSPs (Loreti et al., 2005) that, acting as chaperones, may prevent aggregation, denaturation, misfolding, and degradation of proteins important for plant survival. Indeed, exogenous sugar strongly enhances low-O2 tolerance in a variety of plant species (Vartapetian and Andreeva, 1986; Perata et al., 1992; Germain et al., 1997; Loreti et al., 2005).
AtRbohD Helps Mediate ROS Signaling in Seedlings/Shoots under Anoxia
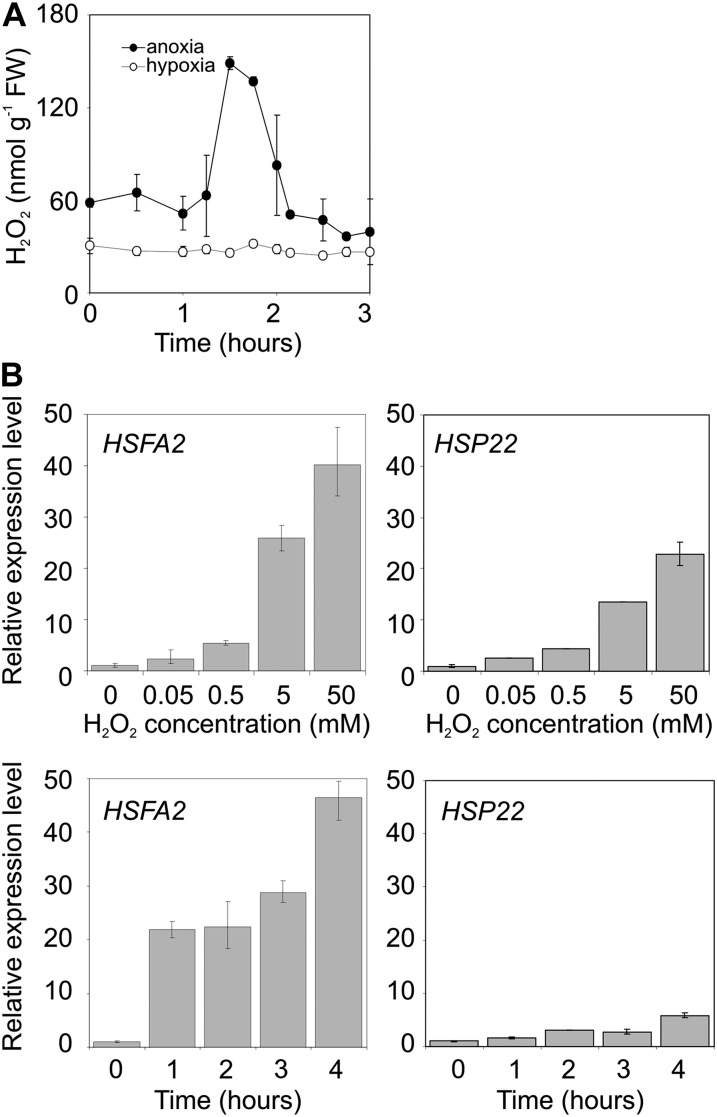
Anoxia triggers an oxidative burst in which H2O2 plays a role (Baxter-Burrell et al., 2002; Banti et al., 2010). Transient, early H2O2 accumulation was observed in Arabidopsis seedlings under anoxia but not hypoxia (Fig. 4A). HsfA2 and HSP22 were up-regulated by H2O2 (Fig. 4B) and the induction of these genes under anoxia but not hypoxia is suggestive of a direct role of the H2O2 produced under anoxia as a trigger for the early expression of these genes (Fig. 3).
Figure 4.
ROS-related experiments under anoxia. A, H2O2 production under anoxia and hypoxia. B, Regulation of HsfA2 and HSP22 gene expression at different exogenous H2O2 concentration and in a time course up to 4 h (H2O2 5 mm).
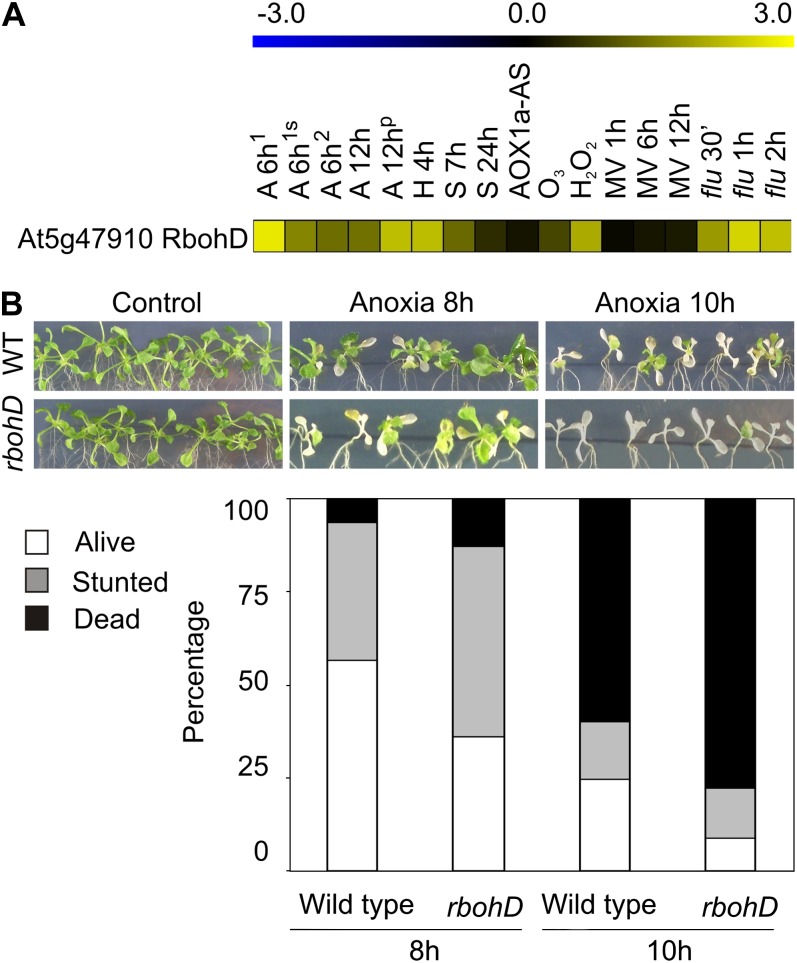
ROS generated by NADPH oxidase regulate plant development as well as biotic and abiotic stress responses (Torres and Dangl, 2005). The NADPH oxidase gene family is composed of 10 Rboh genes (Torres et al., 1998; Dangl and Jones, 2001) encoding plasma membrane proteins that display a cytosolic extension with two Ca2+-binding EF hands, likely responsible for their regulation by Ca2+ (Keller et al., 1998). NADPH oxidase rapidly reduces apoplastic O2 to O2− as a primary product that then may be further converted to H2O2 (Neill et al., 2002). ROS generated via NADPH oxidase might represent the link in common stress signaling (Miller et al., 2009). Previous data suggest that different Rboh genes are active in different cellular contexts. While AtrbohC has a function in root-hair development (Foreman et al., 2003), AtrbohD and AtrbohF are required in response to plant pathogens (Torres et al., 2002) and abscisic acid signaling (Kwak et al., 2003). The survey of Rboh gene expression in the microarray datasets, allowed us to identify RbohD as significantly up-regulated in some of the low-O2 experiments (Fig. 5A). RbohD is also positively regulated in some ROS-related microarrays (Fig. 5A). Interestingly, the rbohD mutant, previously shown to display reduced O2− production and H2O2 accumulation during defense responses (Torres et al., 2002), displayed reduced survival under anoxia (Fig. 5B). We also screened the list of genes commonly up-regulated in some ROS and low-O2 experiments for TF specifically related to ROS signaling (Gadjev et al., 2006). Hierarchical clustering linkage of these genes showed association of gene induction with anaerobic experiments (Fig. 6A). A survey of expression of some of these gene under anoxia, revealed a lower induction of HsfA2 and ZAT12 in the insertional rbohD mutant when compared with the wild type (Fig. 6B), suggesting a role for this NADPH oxidase in inducing these genes under anoxia, and their partial requirement for survival. HsfA2 has previously been shown to be fast up-regulated after 1 h under anoxia (Banti et al., 2010). The regulation of some other genes like ZAT6 and ADH1 was instead mostly not affected by the lack of functional RBOHD. ZAT12 has been found to be potentially involved in ROS because its expression is up-regulated under several stresses including heat, high light, and cold (Rizhsky et al., 2004; Davletova et al., 2005a; Miller et al., 2008; Doherty et al., 2009). Moreover, knockout and overexpressor ZAT12 plants were found to be altered in response to different stresses (Rizhsky et al., 2004; Davletova et al., 2005b).
Figure 5.
NADPH oxidase role under anoxia. A, Regulation of rbohD isoform in the O2 deprivation microarray experiments. B, Arabidopsis wild-type plants and rbohD mutant survival after 8 and 10 h of anoxia; stunted plants were those showing bleaching symptoms on some of the leaves but not all. Percentage of alive plants was significantly different, paired Student’s t test (P < 0.05, n = 6). A 6h1, 6 h of anoxia treatment without or with 90 mm Suc (s) added before the treatment (Loreti et al., 2005); A 6h2, 6 h of anoxia treatment (Banti et al., 2010); A 12h, 12 h of anoxia treatment, total and polysomal (p) RNA (Branco-Price et al., 2005); H 4h, 4 h of hypoxia treatment at 1% O2 (Licausi et al., 2010); S 7/24h, 7 and 24 h of submergence treatment (Lee et al., 2011); flu 0.5 (30’)/1/2h, flu mutant shift from dark to light (op den Camp et al., 2003); AOX1a-AS, AOX1a-AS mutant (Umbach et al., 2005); MV 1/6/12h, MV 1, 6, 12 h treatment (Dr. Kirch, NASCArray repository); O3, 1 h of O3 fumigation at 200 nL L−1 (Dr. Shirras, NASCArray repository); H2O2, 1 h of H2O2 treatment at 20 mm (Dr. Mittler, NASCArray repository).
Figure 6.
ROS-related TF regulation under anoxia. A, Regulation of ROS-related TF in the O2 deprivation microarray experiments. B, Regulation of HsfA2, ZAT12, ZAT6, and ADH1 gene in wild-type and rbohD− plants under anoxia in a time course up to 8 h. A 6h1, 6 h of anoxia treatment without or with 90 mm Suc (s) added before the treatment (Loreti et al., 2005); A 6h2, 6 h of anoxia treatment (Banti et al., 2010); A 12h, 12 h of anoxia treatment, total and polysomal (p) RNA (Branco-Price et al., 2005); H 4h, 4 h of hypoxia treatment at 1% O2 (Licausi et al., 2010); S 7/24h, 7 and 24 h of submergence treatment (Lee et al., 2011); flu 0.5 (30’)/1/2h, flu mutant shift from dark to light (op den Camp et al., 2003); AOX1a-AS, AOX1a-AS mutant (Umbach et al., 2005); MV 1/6/12h, MV 1, 6, 12 h treatment (Dr. Kirch, NASCArray repository); O3, 1 h of O3 fumigation at 200 nL L−1 (Dr. Shirras, NASCArray repository); H2O2, 1 h of H2O2 treatment at 20 mm (Dr. Mittler, NASCArray repository).
Recently, mitochondria-associated ROS production has been found to be involved in Arabidopsis response to O2 deprivation (Chang et al., 2012). O2 deprivation and also reoxygenation promote the rapid and transient activation of mitogen-activated protein kinases 3, 4, and 6 that seem to be dependent on mitochondrial ROS. The overexpression of mitogen-activated protein kinase 6 leads to increased survival to anoxia, but the classic transcripts related to the anaerobic metabolism were not significantly modulated, suggesting a promotion in plant survival probably related to a pathway different from that involving the activity of proteins encoded by the anaerobic core genes (Chang et al., 2012). The link between mitochondrial and membrane-derived ROS production under anoxia is currently unknown. However, a link between functional mitochondria and HSP synthesis has been demonstrated. Mitochondrial dysfunction down-regulates HSP production during mild heat shock in Arabidopsis cell (Rikhvanov et al., 2007). Moreover, in mammals’ pulmonary arteries hypoxia-driven mitochondrial ROS production has been suggested to trigger NADPH oxidase activity, suggesting a mechanism by which mitochondria and cytosol both contribute to the increase in ROS production during low O2 (Rathore et al., 2008). Future studies will elucidate whether this is the case in plants.
CONCLUSION
The balance between cellular ROS production and ROS scavenging rate enables a rapid and dynamic change in tissue and subcellular ROS levels (Mittler et al., 2011). These are necessary features if a signaling molecule is to be efficient. ROS were recently shown to be required in response to diverse abiotic stimuli for rapid cell-to-cell communication over long distances (Miller et al., 2009). Data describing the existence of ROS production under O2 deprivation (Baxter-Burrell et al., 2002; Chang et al., 2012), together with the overlap with the molecular response to heat (Banti et al., 2010), suggest a role for ROS in plant adaptation to low O2. In this work, we identified a group of genes regulated by anoxia but not hypoxia, most of which are heat related. These genes are not part of the group of anaerobic core genes that are regulated by ERFs under the control of the N-end pathway for O2 sensing recently described (Gibbs et al., 2011; Licausi et al., 2011). Furthermore, a set of genes that respond to anoxia overlaps with a group of ROS-regulated genes and we observed an early, transient peak of H2O2 production under anoxia but not hypoxia. We propose that RbohD is partially involved in the signaling cascade responsible for the induction of ROS-related TF under anoxia, a mechanism that allows Arabidopsis to survive longer under O2 absence.
MATERIALS AND METHODS
Microarray Analysis
Publicly available Arabidopsis (Arabidopsis thaliana) microarray databases were screened for ROS and O2 deprivation related experiments (Tables I and II). CEL files were downloaded from the Gene Expression Omnibus repository (http://www.ncbi.nlm.nih.gov/gds) or the European Arabidopsis Stock Centre’s International Affymetrix Service (http://affymetrix.arabidopsis.info/narrays/experimentbrowse.pl). A total of six experiments related to O2 deprivation (6 h anoxia ± Suc feeding, 12 h anoxia, 0.5, 2 and 48 h of 1%, 4%, and 8% of O2, 4 h of 1% O2, and 7 and 24 h of submergence) and another five related to ROS-generating systems (MV treatment, O3 fumigation, H2O2 treatment, AOX1a-AS, and flu mutants) were selected for the analysis. Three of the microarrays had no biological replicates whereas the others included a minimum of two biological replicates. All the experiments were used for all analysis. For specific information on each experiment please refer to the original datasets. Information on the condition of the MV, O3, and H2O2 treatment, performed by the groups of Dr. Kirch, Dr. Shirras, and Dr. Mittler, respectively, can be found in the NASCArrays repository at the respective reference number (Table II).
Data Processing
Raw intensity CEL files were imported in Robin interface for microarray (Lohse et al., 2010) and the quality of the original 133 arrays was checked. Probe level model residual pseudo images showing potential artifacts were excluded from the analysis. Raw normalized expression values of ROS and O2 deprivation arrays, generated from treated samples only, were analyzed using hierarchical clustering based on Pearson correlation. Two experimental groups with a similar global response in gene expression were identified in roots and seedlings/shoots, in both ROS and O2 deprivation conditions (Supplemental Figs. S1 and S2). The two organ-specific clusters were treated separately in the subsequent analysis. For the MV treatment on shoot (NASCARRAY-143), cluster dendrogram revealed three different main clusters of genes (data not shown). The experiments related to 1, 6, and 12 h of treatment were then selected for the seedlings/shoots analysis as representative of the three clusters. Both seedlings/shoots and roots groups data were normalized and the signal intensities were estimated using the Affimetrix Microarray Analysis Suite 5.0. Average expression values and their adjusted P values were calculated using the Benjamini-Hochberg adjustment method (Reiner et al., 2003). For the experiment of Branco-Price et al. (2005), polysomal RNA fold-change in expression values was calculated on the basis of the row CEL data obtained by the GSE2218 experiment. Calls resulting significantly regulated in at least one experimental condition (P < 0.05) were filtered, resulting in 1,762 and 68 significantly differentially expressed genes, in seedlings/shoots and roots, respectively (data not shown).
The core of O2 deprivation, anoxic, and hypoxic related genes was arbitrarily defined to be log2 ≥ 1 (P < 0.05) in the overall seedlings/shoots and roots experiments (Fig. 1; Supplemental Fig. S3).
The relative abundance of functional gene classes was calculated considering for each class the percentage of coregulated genes divided by the percentage of gene in the microarray (Fig. 2B).
The averaged log-normalized values of the selected probesets were hierarchically clustered using the average linkage on the Euclidean distance. The clustering analysis and heatmaps were obtained using The Institute for Genomic Research Multiple Experiment Viewer 3.1 (Saeed et al., 2003).
Plant Material and Treatment Conditions
Arabidopsis, Col-0 ecotype, was used for the experiments. The homozygous line of the transposon-tagged insertional mutant rbohD (N9555) was obtained from the European Arabidopsis Stock Centre, where it was donated by Jonathan Jones (John Innes Centre). Seeds were sterilized 10 min with diluted bleach (1.7% sodium hypochlorite), rinsed, and washed several times in sterile water. For the detection of H2O2 and the H2O2 treatments, the experiments were performed with 4-d-old dark-grown seedlings. Seeds were sown in liquid Murashige and Skoog (MS) one-half-strength medium supplemented with 1% Suc. Seeds were stratified for 72 h in the dark at 4°C and then transferred to 23°C in the dark with shaking.
When fed to seedlings, H2O2 was dissolved in MS medium of 4-d-old seedlings, added to the final concentration of 0.05, 0.5, 5, and 50 mm. For the time-course experiments, 5 mm H2O2 was used (Fig. 4B). Plates were incubated at 23°C in the dark with shaking for 2 h.
Anoxic and hypoxic treatments were carried out in the dark. An enclosed anaerobic workstation (anaerobic system model 1025; Forma Scientific) was used to provide an O2-free environment for seedlings’ incubation. This chamber uses palladium catalyst wafers and desiccant wafers to maintain strict anaerobiosis to less than 10 μg mL−1 O2 (according to the manufacturer’s specifications). High-purity N2 was used to initially purge the chamber, and the working anaerobic gas mixture was N2:H2 with a ratio of 90:10. Hypoxic treatments were carried out using a glovebox flushed with 1% O2. The anoxic and hypoxic treatments lasted up to 3 h, collecting samples every 15 min (Fig. 4A).
Agar plates were used to evaluate plants’ tolerance to anoxia and for gene expression analysis (Figs. 5B and 6B). To obtain 7-d-old plants in vertical plates, seeds were germinated on MS one-half-strength medium containing agar (0.9%) and Suc (1%). Seeds were stratified for 72 h in the dark at 4°C and then transferred at 23°C and 12 h light photoperiod, photosynthetically active radiation approximately 100 μmol m−2 s−2.
Anoxic treatments were performed by transferring the plates in the anaerobic work station and kept in the dark. The plates were then transferred for the postanoxic recovery to monitor the plant survival (23°C and 12 h light photoperiod, photosynthetically active radiation approximately 100 μmol m−2 s−2).
H2O2 Quantification
H2O2 production was measured using the Amplex red H2O2/peroxidase assay kit (Molecular Probes) following the manufacturer’s instructions. Plant material extraction was performed using 50 mg of frozen tissue ground in 200 μL of 20 mm sodium phosphate buffer (pH 6.5), according to the protocol developed by the Schachtman Laboratory (Shin and Schachtman, 2004). After centrifugation at 10,000 rpm for 10 min (4°C), 50 μL of supernatant were used for Amplex red assay.
Molecular Analysis
Total RNA was extracted using a RNAqueous kit (Applied Biosystems), according to the manufacturer’s instructions, and subjected to DNase treatment using TURBO DNA-free kit (Ambion). Five micrograms of RNA were reverse transcribed using the high-capacity cDNA archive kit (Applied Biosystems). Transcript abundance was analyzed by real-time reverse transcription PCR, using TaqMan probes (Applied Biosystems/Ambion) and primers specific for each gene (Supplemental Table S3), using an ABI Prism 7000 sequence detection system (Applied Biosystems). PCR reactions were carried out using 50 ng of cDNA and TaqMan universal PCR master mix (Applied Biosystems/Ambion) or qPCR MasterMix Plus for SYBR green I (Eurogentec), following the manufacturer’s protocol. The relative expression level of each gene was quantified with the comparative threshold cycle method, as described in the ABI PRISM 7700 Sequence Detection System (User Bulletin No. 2; Applied Biosystems), using Ubiquitin10 (At4g05320) as an internal reference. PCR reactions for each of the three biological replicates were performed in duplicate.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Hierarchical clustering based on Pearson correlation of the raw normalized expression values of ROS-related arrays, generated from treated samples only.
Supplemental Figure S2. Hierarchical clustering based on Pearson correlation of the raw normalized expression values of O2 deprivation arrays, generated from treated samples only.
Supplemental Figure S3. Genes regulated by anoxia, hypoxia, and O2 deprivation in Arabidopsis microarray datasets of roots.
Supplemental Figure S4. Hierarchical average linkage clustering of transcripts up-regulated in both the ROS-generating and O2 deprivation experiments.
Supplemental Figure S5. Hierarchical average linkage clustering of transcripts down-regulated in both the ROS-generating and O2 deprivation experiments.
Supplemental Figure S6. Hierarchical average linkage clustering of seedlings/shoots heat-stress-related genes regulated in 6 h anoxic experiments with different Suc treatment.
Supplemental Table S1. List of genes significantly up-regulated both in roots and seedlings/shoots experiments under low O2.
Supplemental Table S2. Regulation of heat stress orthologous genes in tolerant/sensitive rice varieties under submergence.
Supplemental Table S3. List of primers used in this work.
Supplementary Material
Acknowledgments
We would like to acknowledge Dr. Christine X-Chen Chang (Carnegie Institution of Washington, Stanford); Dr. Francesco Licausi and Dr. Federico Giorgi (Max Planck Institute of Potsdam-Golm) for helpful discussion; and Dr. Angelika Mustroph (Bayreuth University) for critically reading the manuscript.
Glossary
- ROS
reactive oxygen species
- O2
oxygen
- H2O2
hydrogen peroxide
- HSF
heat shock transcription factor
- HSP
heat shock protein
- ERF
ethylene-responsive factor
- Col
Columbia
- MS
Murashige and Skoog
References
- Ahn S-G, Thiele DJ. (2003) Redox regulation of mammalian heat shock factor 1 is essential for Hsp gene activation and protection from stress. Genes Dev 17: 516–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey-Serres J, Chang R. (2005) Sensing and signalling in response to oxygen deprivation in plants and other organisms. Ann Bot (Lond) 96: 507–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey-Serres J, Fukao T, Ronald P, Ismail A, Heuer S, Mackill D. (2010) Submergence tolerant rice: SUB1’s journey from landrace to modern cultivar. Rice 3: 138–147 [Google Scholar]
- Banti V, Loreti E, Novi G, Santaniello A, Alpi A, Perata P. (2008) Heat acclimation and cross-tolerance against anoxia in Arabidopsis. Plant Cell Environ 31: 1029–1037 [DOI] [PubMed] [Google Scholar]
- Banti V, Mafessoni F, Loreti E, Alpi A, Perata P. (2010) The heat-inducible transcription factor HsfA2 enhances anoxia tolerance in Arabidopsis. Plant Physiol 152: 1471–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter-Burrell A, Yang Z, Springer PS, Bailey-Serres J. (2002) RopGAP4-dependent Rop GTPase rheostat control of Arabidopsis oxygen deprivation tolerance. Science 296: 2026–2028 [DOI] [PubMed] [Google Scholar]
- Boyes DC, Zayed AM, Ascenzi R, McCaskill AJ, Hoffman NE, Davis KR, Görlach J. (2001) Growth stage-based phenotypic analysis of Arabidopsis: a model for high throughput functional genomics in plants. Plant Cell 13: 1499–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branco-Price C, Kawaguchi R, Ferreira RB, Bailey-Serres J. (2005) Genome-wide analysis of transcript abundance and translation in Arabidopsis seedlings subjected to oxygen deprivation. Ann Bot (Lond) 96: 647–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang R, Jang CJH, Branco-Price C, Nghiem P, Bailey-Serres J. (2012) Transient MPK6 activation in response to oxygen deprivation and reoxygenation is mediated by mitochondria and aids seedling survival in Arabidopsis. Plant Mol Biol 78: 109–122 [DOI] [PubMed] [Google Scholar]
- Colmer TD, Pedersen O. (2008) Oxygen dynamics in submerged rice (Oryza sativa). New Phytol 178: 326–334 [DOI] [PubMed] [Google Scholar]
- Colmer TD, Voesenek LACJ. (2009) Flooding tolerance: suites of plant traits in variable environments. Funct Plant Biol 36: 665–681 [DOI] [PubMed] [Google Scholar]
- Cooper CE. (2002) Nitric oxide and cytochrome oxidase: substrate, inhibitor or effector? Trends Biochem Sci 27: 33–39 [DOI] [PubMed] [Google Scholar]
- Couée I, Sulmon C, Gouesbet G, El Amrani A. (2006) Involvement of soluble sugars in reactive oxygen species balance and responses to oxidative stress in plants. J Exp Bot 57: 449–459 [DOI] [PubMed] [Google Scholar]
- Dangl JL, Jones JDG. (2001) Plant pathogens and integrated defence responses to infection. Nature 411: 826–833 [DOI] [PubMed] [Google Scholar]
- Davletova S, Rizhsky L, Liang H, Shengqiang Z, Oliver DJ, Coutu J, Shulaev V, Schlauch K, Mittler R. (2005a) Cytosolic ascorbate peroxidase 1 is a central component of the reactive oxygen gene network of Arabidopsis. Plant Cell 17: 268–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davletova S, Schlauch K, Coutu J, Mittler R. (2005b) The zinc-finger protein Zat12 plays a central role in reactive oxygen and abiotic stress signaling in Arabidopsis. Plant Physiol 139: 847–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty CJ, Van Buskirk HA, Myers SJ, Thomashow MF. (2009) Roles for Arabidopsis CAMTA transcription factors in cold-regulated gene expression and freezing tolerance. Plant Cell 21: 972–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis MH, Dennis ES, Peacock WJ. (1999) Arabidopsis roots and shoots have different mechanisms for hypoxic stress tolerance. Plant Physiol 119: 57–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foreman J, Demidchik V, Bothwell JH, Mylona P, Miedema H, Torres MA, Linstead P, Costa S, Brownlee C, Jones JD, et al. (2003) Reactive oxygen species produced by NADPH oxidase regulate plant cell growth. Nature 422: 442–446 [DOI] [PubMed] [Google Scholar]
- Gadjev I, Vanderauwera S, Gechev TS, Laloi C, Minkov IN, Shulaev V, Apel K, Inzé D, Mittler R, Van Breusegem F. (2006) Transcriptomic footprints disclose specificity of reactive oxygen species signaling in Arabidopsis. Plant Physiol 141: 436–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geigenberger P. (2003) Response of plant metabolism to too little oxygen. Curr Opin Plant Biol 6: 247–256 [DOI] [PubMed] [Google Scholar]
- Geigenberger P, Fernie AR, Gibon Y, Christ M, Stitt M. (2000) Metabolic activity decreases as an adaptive response to low internal oxygen in growing potato tubers. Biol Chem 381: 723–740 [DOI] [PubMed] [Google Scholar]
- Geisler-Lee J, Caldwell C, Gallie DR. (2010) Expression of the ethylene biosynthetic machinery in maize roots is regulated in response to hypoxia. J Exp Bot 61: 857–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germain V, Ricard B, Raymond P, Saglio PH. (1997) The role of sugars, hexokinase, and sucrose synthase in the determination of hypoxically induced tolerance to anoxia in tomato roots. Plant Physiol 114: 167–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs DJ, Lee SC, Isa NM, Gramuglia S, Fukao T, Bassel GW, Correia CS, Corbineau F, Theodoulou FL, Bailey-Serres J, et al. (2011) Homeostatic response to hypoxia is regulated by the N-end rule pathway in plants. Nature 479: 415–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y, Wang Z, Yang Z. (2004) ROP/RAC GTPase: an old new master regulator for plant signaling. Curr Opin Plant Biol 7: 527–536 [DOI] [PubMed] [Google Scholar]
- Gupta KJ, Zabalza A, van Dongen JT. (2009) Regulation of respiration when the oxygen availability changes. Physiol Plant 137: 383–391 [DOI] [PubMed] [Google Scholar]
- Hattori Y, Nagai K, Furukawa S, Song XJ, Kawano R, Sakakibara H, Wu J, Matsumoto T, Yoshimura A, Kitano H, et al. (2009) The ethylene response factors SNORKEL1 and SNORKEL2 allow rice to adapt to deep water. Nature 460: 1026–1030 [DOI] [PubMed] [Google Scholar]
- Hu Y, Xie Q, Chua N-H. (2003) The Arabidopsis auxin-inducible gene ARGOS controls lateral organ size. Plant Cell 15: 1951–1961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husbands A, Bell EM, Shuai B, Smith HM, Springer PS. (2007) LATERAL ORGAN BOUNDARIES defines a new family of DNA-binding transcription factors and can interact with specific bHLH proteins. Nucleic Acids Res 35: 6663–6671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inukai Y, Sakamoto T, Ueguchi-Tanaka M, Shibata Y, Gomi K, Umemura I, Hasegawa Y, Ashikari M, Kitano H, Matsuoka M. (2005) Crown rootless1, which is essential for crown root formation in rice, is a target of an AUXIN RESPONSE FACTOR in auxin signaling. Plant Cell 17: 1387–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawase M. (1972) Effect of flooding on ethylene concentration in horticultural plants. J Am Soc Hortic Sci 97: 584–588 [Google Scholar]
- Kawase M. (1978) Anaerobic elevation of ethylene concentration in waterlogged plants. Am J Bot 65: 736–740 [Google Scholar]
- Keller T, Damude HG, Werner D, Doerner P, Dixon RA, Lamb C. (1998) A plant homolog of the neutrophil NADPH oxidase gp91phox subunit gene encodes a plasma membrane protein with Ca2+ binding motifs. Plant Cell 10: 255–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerk D, Bulgrien J, Smith DW, Gribskov M. (2003) Arabidopsis proteins containing similarity to the universal stress protein domain of bacteria. Plant Physiol 131: 1209–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Könings H, Jackson MB. (1979) A relationship between rates of ethylene production by roots and the promoting or inhibiting effects of exogenous ethylene and water on root elongation. Z Pflanzenphysiologie 92: 385–397 [Google Scholar]
- Kottapalli KR, Satoh K, Rakwal R, Shibato J, Doi K, Nagata T, Kikuchi S. (2007) Combining in silico mapping and arraying: an approach to identifying common candidate genes for submergence tolerance and resistance to bacterial leaf blight in rice. Mol Cells 24: 394–408 [PubMed] [Google Scholar]
- Kürsteiner O, Dupuis I, Kuhlemeier C. (2003) The pyruvate decarboxylase1 gene of Arabidopsis is required during anoxia but not other environmental stresses. Plant Physiol 132: 968–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak JM, Mori IC, Pei ZM, Leonhardt N, Torres MA, Dangl JL, Bloom RE, Bodde S, Jones JD, Schroeder JI. (2003) NADPH oxidase AtrbohD and AtrbohF genes function in ROS-dependent ABA signaling in Arabidopsis. EMBO J 22: 2623–2633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecourieux D, Ranjeva R, Pugin A. (2006) Calcium in plant defence-signalling pathways. New Phytol 171: 249–269 [DOI] [PubMed] [Google Scholar]
- Lee K-W, Chen P-W, Lu C-A, Chen S, Ho T-HD, Yu S-M. (2009) Coordinated responses to oxygen and sugar deficiency allow rice seedlings to tolerate flooding. Sci Signal 2: ra61. [DOI] [PubMed] [Google Scholar]
- Lee SC, Mustroph A, Sasidharan R, Vashisht D, Pedersen O, Oosumi T, Voesenek LACJ, Bailey-Serres J. (2011) Molecular characterization of the submergence response of the Arabidopsis thaliana ecotype Columbia. New Phytol 190: 457–471 [DOI] [PubMed] [Google Scholar]
- Li C, Chen Q, Gao X, Qi B, Chen N, Xu S, Chen J, Wang X. (2005) AtHsfA2 modulates expression of stress responsive genes and enhances tolerance to heat and oxidative stress in Arabidopsis. Sci China C Life Sci 48: 540–550 [DOI] [PubMed] [Google Scholar]
- Licausi F. (2011) Regulation of the molecular response to oxygen limitations in plants. New Phytol 190: 550–555 [DOI] [PubMed] [Google Scholar]
- Licausi F, Kosmacz M, Weits DA, Giuntoli B, Giorgi FM, Voesenek LA, Perata P, van Dongen JT. (2011) Oxygen sensing in plants is mediated by an N-end rule pathway for protein destabilization. Nature 479: 419–422 [DOI] [PubMed] [Google Scholar]
- Licausi F, van Dongen JT, Giuntoli B, Novi G, Santaniello A, Geigenberger P, Perata P. (2010) HRE1 and HRE2, two hypoxia-inducible ethylene response factors, affect anaerobic responses in Arabidopsis thaliana. Plant J 62: 302–315 [DOI] [PubMed] [Google Scholar]
- Lin WC, Shuai B, Springer PS. (2003) The Arabidopsis LATERAL ORGAN BOUNDARIES-domain gene ASYMMETRIC LEAVES2 functions in the repression of KNOX gene expression and in adaxial-abaxial patterning. Plant Cell 15: 2241–2252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Wang S, Yu X, Yu J, He X, Zhang S, Shou H, Wu P. (2005) ARL1, a LOB-domain protein required for adventitious root formation in rice. Plant J 43: 47–56 [DOI] [PubMed] [Google Scholar]
- Lohse M, Nunes-Nesi A, Krüger P, Nagel A, Hannemann J, Giorgi FM, Childs L, Osorio S, Walther D, Selbig J, et al. (2010) Robin: an intuitive wizard application for R-based expression microarray quality assessment and analysis. Plant Physiol 153: 642–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loreti E, Poggi A, Novi G, Alpi A, Perata P. (2005) A genome-wide analysis of the effects of sucrose on gene expression in Arabidopsis seedlings under anoxia. Plant Physiol 137: 1130–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangeon A, Bell EM, Lin WC, Jablonska B, Springer PS. (2011) Misregulation of the LOB domain gene DDA1 suggests possible functions in auxin signalling and photomorphogenesis. J Exp Bot 62: 221–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller G, Mittler R. (2006) Could heat shock transcription factors function as hydrogen peroxide sensors in plants? Ann Bot (Lond) 98: 279–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller G, Schlauch K, Tam R, Cortes D, Torres MA, Shulaev V, Dangl JL, Mittler R. (2009) The plant NADPH oxidase RBOHD mediates rapid systemic signaling in response to diverse stimuli. Sci Signal 2: ra45. [DOI] [PubMed] [Google Scholar]
- Miller G, Shulaev V, Mittler R. (2008) Reactive oxygen signaling and abiotic stress. Physiol Plant 133: 481–489 [DOI] [PubMed] [Google Scholar]
- Mittler R, Vanderauwera S, Suzuki N, Miller G, Tognetti VB, Vandepoele K, Gollery M, Shulaev V, Van Breusegem F. (2011) ROS signaling: the new wave? Trends Plant Sci 16: 300–309 [DOI] [PubMed] [Google Scholar]
- Mittler R, Zilinskas BA. (1992) Molecular cloning and characterization of a gene encoding pea cytosolic ascorbate peroxidase. J Biol Chem 267: 21802–21807 [PubMed] [Google Scholar]
- Mustroph A, Boamfa EI, Laarhoven LJJ, Harren FJM, Pörs Y, Grimm B. (2006) Organ specific analysis of the anaerobic primary metabolism in rice and wheat seedlings II: light exposure reduces needs for fermentation and extends survival during anaerobiosis. Planta 225: 139–152 [DOI] [PubMed] [Google Scholar]
- Mustroph A, Lee SC, Oosumi T, Zanetti ME, Yang H, Ma K, Yaghoubi-Masihi A, Fukao T, Bailey-Serres J. (2010) Cross-kingdom comparison of transcriptomic adjustments to low-oxygen stress highlights conserved and plant-specific responses. Plant Physiol 152: 1484–1500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustroph A, Zanetti ME, Jang CJ, Holtan HE, Repetti PP, Galbraith DW, Girke T, Bailey-Serres J. (2009) Profiling translatomes of discrete cell populations resolves altered cellular priorities during hypoxia in Arabidopsis. Proc Natl Acad Sci USA 106: 18843–18848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachin L, Brive L, Persson K-C, Svensson P, Nyström T. (2008) Heterodimer formation within universal stress protein classes revealed by an in silico and experimental approach. J Mol Biol 380: 340–350 [DOI] [PubMed] [Google Scholar]
- Nakano T, Suzuki K, Fujimura T, Shinshi H. (2006) Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol 140: 411–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neill S, Desikan R, Hancock J. (2002) Hydrogen peroxide signalling. Curr Opin Plant Biol 5: 388–395 [DOI] [PubMed] [Google Scholar]
- Nishizawa A, Yabuta Y, Yoshida E, Maruta T, Yoshimura K, Shigeoka S. (2006) Arabidopsis heat shock transcription factor A2 as a key regulator in response to several types of environmental stress. Plant J 48: 535–547 [DOI] [PubMed] [Google Scholar]
- Ogasawara Y, Kaya H, Hiraoka G, Yumoto F, Kimura S, Kadota Y, Hishinuma H, Senzaki E, Yamagoe S, Nagata K, et al. (2008) Synergistic activation of the Arabidopsis NADPH oxidase AtrbohD by Ca2+ and phosphorylation. J Biol Chem 283: 8885–8892 [DOI] [PubMed] [Google Scholar]
- Okushima Y, Fukaki H, Onoda M, Theologis A, Tasaka M. (2007) ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis. Plant Cell 19: 118–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- op den Camp RGL, Przybyla D, Ochsenbein C, Laloi C, Kim C, Danon A, Wagner D, Hideg É, Göbel C, Feussner I, et al. (2003) Rapid induction of distinct stress responses after the release of singlet oxygen in Arabidopsis. Plant Cell 15: 2320–2332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perata P, Pozuetaromero J, Akazawa T, Yamaguchi J. (1992) Effect of anoxia on starch breakdown in rice and wheat seeds. Planta 188: 611–618 [DOI] [PubMed] [Google Scholar]
- Rathore R, Zheng YM, Niu CF, Liu QH, Korde A, Ho YS, Wang YX. (2008) Hypoxia activates NADPH oxidase to increase [ROS]i and [Ca2+]i through the mitochondrial ROS-PKCepsilon signaling axis in pulmonary artery smooth muscle cells. Free Radic Biol Med 45: 1223–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiner A, Yekutieli D, Benjamini Y. (2003) Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinformatics 19: 368–375 [DOI] [PubMed] [Google Scholar]
- Ricard B, Toai TV, Chourey P, Saglio P. (1998) Evidence for the critical role of sucrose synthase for anoxic tolerance of maize roots using a double mutant. Plant Physiol 116: 1323–1331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rikhvanov EG, Gamburg KZ, Varakina NN, Rusaleva TM, Fedoseeva IV, Tauson EL, Stupnikova IV, Stepanov AV, Borovskii GB, Voinikov VK. (2007) Nuclear-mitochondrial cross-talk during heat shock in Arabidopsis cell culture. Plant J 52: 763–778 [DOI] [PubMed] [Google Scholar]
- Rizhsky L, Davletova S, Liang H, Mittler R. (2004) The zinc finger protein Zat12 is required for cytosolic ascorbate peroxidase 1 expression during oxidative stress in Arabidopsis. J Biol Chem 279: 11736–11743 [DOI] [PubMed] [Google Scholar]
- Sachs MM, Vartapetian BB. (2007) Plant anaerobic stress. I. Metabolic adaptation to oxygen deficiency. Plant Stress 1: 123–135 [Google Scholar]
- Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, et al. (2003) TM4: a free, open-source system for microarray data management and analysis. Biotechniques 34: 374–378 [DOI] [PubMed] [Google Scholar]
- Sauter M, Rzewuski G, Marwedel T, Lorbiecke R. (2002) The novel ethylene-regulated gene OsUsp1 from rice encodes a member of a plant protein family related to prokaryotic universal stress proteins. J Exp Bot 53: 2325–2331 [DOI] [PubMed] [Google Scholar]
- Sedbrook JC, Kronebusch PJ, Borisy GG, Trewavas AJ, Masson PH. (1996) Transgenic AEQUORIN reveals organ-specific cytosolic Ca2+ responses to anoxia and Arabidopsis thaliana seedlings. Plant Physiol 111: 243–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin R, Schachtman DP. (2004) Hydrogen peroxide mediates plant root cell response to nutrient deprivation. Proc Natl Acad Sci USA 101: 8827–8832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storozhenko S, De Pauw P, Van Montagu M, Inzé D, Kushnir S. (1998) The heat-shock element is a functional component of the Arabidopsis APX1 gene promoter. Plant Physiol 118: 1005–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subbaiah CC, Bush DS, Sachs MM. (1994a) Elevation of cytosolic calcium precedes anoxic gene expression in maize suspension-cultured cells. Plant Cell 6: 1747–1762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subbaiah CC, Zhang J, Sachs MM. (1994b) Involvement of intracellular calcium in anaerobic gene expression and survival of maize seedlings. Plant Physiol 105: 369–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W, Van Montagu M, Verbruggen N. (2002) Small heat shock proteins and stress tolerance in plants. Biochim Biophys Acta 1577: 1–9 [DOI] [PubMed] [Google Scholar]
- Suzuki N, Miller G, Morales J, Shulaev V, Torres MA, Mittler R. (2011) Respiratory burst oxidases: the engines of ROS signaling. Curr Opin Plant Biol 14: 1–9 [DOI] [PubMed] [Google Scholar]
- Takeda S, Gapper C, Kaya H, Bell E, Kuchitsu K, Dolan L. (2008) Local positive feedback regulation determines cell shape in root hair cells. Science 319: 1241–1244 [DOI] [PubMed] [Google Scholar]
- Taramino G, Sauer M, Stauffer JL, Jr, Multani D, Niu X, Sakai H, Hochholdinger F. (2007) The maize (Zea mays L.) RTCS gene encodes a LOB domain protein that is a key regulator of embryonic seminal and post-embryonic shoot-borne root initiation. Plant J 50: 649–659 [DOI] [PubMed] [Google Scholar]
- Torres MA, Dangl JL. (2005) Functions of the respiratory burst oxidase in biotic interactions, abiotic stress and development. Curr Opin Plant Biol 8: 397–403 [DOI] [PubMed] [Google Scholar]
- Torres MA, Dangl JL, Jones JD. (2002) Arabidopsis gp91phox homologues AtrbohD and AtrbohF are required for accumulation of reactive oxygen intermediates in the plant defense response. Proc Natl Acad Sci USA 99: 523–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres MA, Onouchi H, Hamada S, Machida C, Hammond-Kosack KE, Jones JDG. (1998) Six Arabidopsis thaliana homologues of the human respiratory burst oxidase (gp91phox). Plant J 14: 365–370 [DOI] [PubMed] [Google Scholar]
- Umbach AL, Fiorani F, Siedow JN. (2005) Characterization of transformed Arabidopsis with altered alternative oxidase levels and analysis of effects on reactive oxygen species in tissue. Plant Physiol 139: 1806–1820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dongen JT, Fröhlich A, Ramírez-Aguilar SJ, Schauer N, Fernie AR, Erban A, Kopka J, Clark J, Langer A, Geigenberger P. (2009) Transcript and metabolite profiling of the adaptive response to mild decreases in oxygen concentration in the roots of Arabidopsis plants. Ann Bot (Lond) 103: 269–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dongen JT, Schurr U, Pfister M, Geigenberger P. (2003) Phloem metabolism and function have to cope with low internal oxygen. Plant Physiol 131: 1529–1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenbroucke K, Robbens S, Vandepoele K, Inzé D, Van de Peer Y, Van Breusegem F. (2008) Hydrogen peroxide-induced gene expression across kingdoms: a comparative analysis. Mol Biol Evol 25: 507–516 [DOI] [PubMed] [Google Scholar]
- Vartapetian BB, Andreeva IN. (1986) Mitochondrial ultrastructure of three hygrophytes species at anoxia and in anoxic glucose-supplemented medium. J Exp Bot 37: 685–692 [Google Scholar]
- Voesenek LACJ, Colmer TD, Pierik R, Millenaar FF, Peeters AJM. (2006) How plants cope with complete submergence. New Phytol 170: 213–226 [DOI] [PubMed] [Google Scholar]
- Xu K, Xu X, Fukao T, Canlas P, Maghirang-Rodriguez R, Heuer S, Ismail AM, Bailey-Serres J, Ronald PC, Mackill DJ. (2006) Sub1A is an ethylene-response-factor-like gene that confers submergence tolerance to rice. Nature 442: 705–708 [DOI] [PubMed] [Google Scholar]
- Xu L, Xu Y, Dong A, Sun Y, Pi L, Xu Y, Huang H. (2003) Novel as1 and as2 defects in leaf adaxial-abaxial polarity reveal the requirement for ASYMMETRIC LEAVES1 and 2 and ERECTA functions in specifying leaf adaxial identity. Development 130: 4097–4107 [DOI] [PubMed] [Google Scholar]
- Yang Z, Fu Y. (2007) ROP/RAC GTPase signaling. Curr Opin Plant Biol 10: 490–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong M, Orosz A, Wu C. (1998) Direct sensing of heat and oxidation by Drosophila heat shock transcription factor. Mol Cell 2: 101–108 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.