Plant scientists face the difficult challenge of increasing food production without further degradation of the environment. In order to protect drinking water resources and prevent the proliferation of harmful algal blooms and “dead zones” in coastal marine ecosystems, it is imperative to reduce anthropogenic nutrient inputs (Conley et al., 2009). These challenges are further compounded by the goal of utilizing agriculture to provide replacement fuels such as biodiesel and alcohol for our oil-based economy. Phosphate (Pi) is an essential macronutrient required for plant growth and development (Chen et al., 2008). Plant nutrient acquisition and partitioning depend on the H+ gradients generated by the plasma membrane H+-ATPases (Palmgren, 2001; Fuglsang et al., 2011). In this update, we discuss the potential role that the type I H+-proton-pyrophosphatase (PPase) could play in optimizing Pi use efficiency in plants.
A BROKEN CYCLE
The need to improve phosphorus (P) use efficiency by crops arises from two main issues: long-standing concerns about the effects of fertilizer runoff on aquatic ecosystems, and more recent attention to geological and geopolitical availability of P reserves for the production of fertilizers. These dimensions have been the subject of several recent papers (Elser and Bennett, 2011; Vaccari, 2011, and refs. cited therein), so we only briefly touch on the main issues here. The global P cycle is a long-term process in geologic time whereby Pi mineral deposits are distributed to terrestrial habitats by weathering, cycled through flora and fauna, carried to surface water and marine habitats by runoff, and by geologic processes, including tectonic uplift, P-enriched ocean sediments are deposited on land and the process begins anew (Filippelli, 2002). However, due to our growing demand for food, we are depleting our Pi reserves at a rate that will create a future shortage of Pi on the much smaller time scale relevant to our sustainability.
Globally, less than 50% of the P added to fields as fertilizer and manure leaves the field in the harvested crop (Cordell et al., 2009). Of the remainder, much is tied up by soil chemical processes in forms not available to plants and in both the short and longer term lost from the field during soil erosion and leaching. Thus, Pi is transported downwind and downstream, entering streams, rivers, lakes, and oceans. Because Pi is limiting to production in aquatic ecosystems (Elser et al., 2007) as it is on farms, this loading of Pi (as well as nitrogen) leads to eutrophication (Smith et al., 2006). In lakes and marine ecosystems, eutrophication outcomes include algal blooms that often involve toxic species that impair drinking water quality and result in significant economic losses associated with recreation, real estate values, and remediation and ecosystem restoration (Hoagland et al., 2002; Diaz and Rosenberg, 2008; Dodds et al., 2009).
A more recent sustainability dimension of human Pi use concerns the extraction of Pi from fossil phosphorite deposits: how long can this last? Recent considerations on patterns of overall global Pi use in relation to estimated Pi reserves (as of approximately 2005) suggest that “peak phosphorus” (the date at which the global Pi extraction rate would have its maximum) might occur as soon as approximately 2035 (Cordell et al., 2009). This estimate attracted considerable attention, given its coincidence with major increases in 2007 and 2008 in the price of phosphate rock (more than 500%) and phosphate fertilizers (more than 250%). Also important in driving concerns is the fact that Pi reserves are geographically confined to just a few countries (Morocco, China, the United States, Jordan, and South Africa contained more than 85% of the 2005 reserve estimates), and thus many countries must rely on imports to sustain agricultural productivity. The two sides of the Pi sustainability coin, pollution and scarcity, call attention to the need for new approaches, such as genetic modification of crops toward enhanced Pi use efficiency.
SOIL Pi CHEMISTRY
Soils contain both organic and inorganic P compounds. Sources of soil organic Pi includes floral residues, dead and decaying fauna, and microbial matter (cells and metabolic products). Approximately 1% to 5% of the organic P is found in the phospholipid fraction, 0.2% to 2% as nucleic acids or degradation products, and up to 50% make up the inositol polyphosphate fraction (Anderson, 1967, 1975; Halstead and McKercher, 1975). A significant portion of the soil organic fraction is unidentified. P released to the soil solution from the mineralization of organic matter might be taken up by the microbial population or by growing plants, transferred to the soil inorganic pool, or lost by leaching and runoff. It has generally been presumed that P availability to plants is largely controlled by the inorganic P fraction (Sanchez, 2006).
Inorganic P entering the soil solution, by mineralization or by fertilizer additions, is rapidly converted to forms less available to plants. Sorption and precipitation reactions are involved. The sorption of inorganic P from solution is related to the presence of iron and aluminum oxides and hydrous oxide minerals (Fried and Dean, 1955; Fox and Kamprath, 1971; Cogger and Duxbury, 1984) and CaCO3 (Cole et al., 1953; Griffin and Jurinak, 1973; Holford and Mattingly, 1975). P sorption is limited to relatively low initial P solution concentrations, and precipitation becomes a more important mechanism of P removal from the soil solution at higher P concentrations (Cole et al., 1953). The nature of the reaction products formed when P fertilizer is added to soil depends on the pH of the saturated solution, the coexisting cations, the quantity of P fertilizer added, and the chemical characteristics of the soil (Lindsay, 1979). In acid soils, iron and aluminum will generally precipitate the Pi. In calcareous soils, the acidic fertilizer solution would readily dissolve calcium, and it is expected that most of the added phosphorous fertilizer would precipitate initially as dicalcium phosphate dihydrate and dicalcium phosphate (Lindsay and Stephenson, 1959). These products are only meta-stable and undergo a slow conversion to compounds such as octacalcium phosphate, tricalcium phosphate, or one of the apatites. In calcareous soils, the reactions of added fertilizer Pi are often referred to as reversion, because thermodynamics directs the added Pi toward mineral species similar to those found in mined phosphate (Gowariker et al., 2009) and these forms are typically not available to plants. Soil P transformations are complex and poorly defined for any given soil. P availability is often characterized in general terms as solution Pi, readily available or labile Pi, and nonlabile Pi. The labile fraction might include easily mineralizable organic Pi, low-energy sorbed Pi, and relatively soluble mineral Pi. The nonlabile fraction might include resistant organic Pi, high-energy sorbed Pi, and relatively insoluble P minerals. As plants absorb P from the soil solution, it is fairly rapidly replenished from the labile fraction, which in turn is more slowly replenished by the nonlabile fraction but often at rates insufficient to meet plant demand. P availability to plants is optimal at soil pH levels of 6.5. The capacity of the soil-labile fraction to replenish solution Pi is reduced by the formation of stable iron and aluminum P minerals below pH 6.5 and by calcium minerals above pH 6.5. Agriculture research of the past century has focused on seeking methods of modifying the soil so that P is not limiting crop production (Sanchez, 2006), but much less effort has been directed at modifying the plant for improved P use efficiency.
ENHANCED SALT TOLERANCE IS AN ANTICIPATED PHENOTYPE TRIGGERED BY THE GENETIC UP-REGULATION OF A VACUOLAR H+-PPASE
Prototypical plant H+-PPases are highly conserved, with amino acid sequence identities of 85% or greater (Drozdowicz and Rea, 2001). Plants have two phylogenetically distinct H+-PPases: type I and type II. Type I H+-PPases depend on cytosolic K+ for their activity and are moderately sensitive to inhibition by Ca2+, and type II H+-PPases are K+ insensitive but extremely Ca2+ sensitive. Plant type I H+-PPases were first isolated from vacuoles and were considered to be bona fide vacuolar markers (Rea et al., 1992; Maeshima, 2000). Type II H+-PPases localize and function in the Golgi apparatus, and their total amount in tissues is very low, less than 0.2% of the type I H+-PPase (Segami et al., 2010).
Vacuolar sodium sequestration is a conserved mechanism used by salt-tolerant plant species. Initial studies demonstrated that overexpression of the type I H+-PPase AVP1 (AVP1-OX; cDNA of At1g15690) from the cauliflower mosaic virus 35S promoter in Arabidopsis (Arabidopsis thaliana) resulted in enhanced salt tolerance and drought resistance (Gaxiola et al., 2001). The salt-tolerant phenotype was explained by an increased capacity for Na+ uptake into vacuoles, and drought resistance was attributed to an enhanced vacuolar osmoregulatory capacity (Gaxiola et al., 2001, 2002). It has been subsequently demonstrated that overexpression of AVP1 and other plant type I H+-PPase genes can increase both salt and drought tolerance in diverse systems, including tobacco (Nicotiana tabacum; Gao et al., 2006), cotton (Gossypium hirsutum; Lv et al., 2008, 2009), alfalfa (Medicago sativa; Bao et al., 2008), maize (Zea mays; Li et al., 2008), and creeping bentgrass (Agrostis stolonifera; Li et al., 2010). Interestingly, a study on the variation of salinity tolerance in Arabidopsis ecotypes reported a positive relationship between salt tolerance and AVP1 expression (Jha et al., 2010). Furthermore, an Arabidopsis mutant that solely relies on the activity of the H+-PPase for energizing vacuolar transport remained salt tolerant, confirming the supposition that AVP1 is important for salt tolerance (Gaxiola et al., 2001, 2002, 2007; Krebs et al., 2010).
ENHANCED ROOT AND SHOOT BIOMASS ARE SURPRISING PHENOTYPES TRIGGERED BY THE GENETIC UP-REGULATION OF A VACUOLAR H+-PPASE
Increased root and shoot proliferation are two unforeseen but consistent phenotypes triggered by the overexpression of either the Arabidopsis (AVP1) or the Thellungiella halophila (TsVP) H+-PPase ortholog, under the control of either the cauliflower mosaic virus 35S or the Ubiquitin promoter (Li et al., 2005, 2008; Park et al., 2005; Yang et al., 2007; Lv et al., 2008, 2009). In Arabidopsis, the production of more rosette leaves (three to 16, depending on AVP1 expression level) resulted in significantly greater leaf area (40%–60%) than in wild-type plants (Li et al., 2005; Gonzalez et al., 2010). Further analysis showed that leaf enlargement in AVP1-OX plants results from increased cell numbers (Li et al., 2005; Gonzalez et al., 2010), and the observed 1.6- to 8.4-fold increase in root growth likely does as well (Li et al., 2005). In rice, shoot and root biomass were enhanced 82% and 109%, respectively, when engineered with a 35Sp:AVP1 cassette (Yang et al., 2007). Notably, an elite inbred maize line, with overexpression of TsVP, resulted in 21% higher accumulation of total soluble sugars and enhanced root development. These differences were elevated to approximately 50% after 3 d of osmotic stress treatment (Li et al., 2008), and the 1,000-grain weight from plants subjected to 6 weeks of drought stress was 30% higher than in controls (Li et al., 2008). In cotton overexpressing TsVP, the photosynthesis rate, stomatal conductance, and root development were higher than in controls under nonstress conditions, whereas these parameters were significantly lower than in controls in antisense TsVP plants (Lv et al., 2008). Of note, these TsVP transgenic cotton plants are also salt tolerant (Lv et al., 2008). In an independent experiment, it was shown that 35Sp:AVP1 cotton plants also developed larger root systems, and under dry-land conditions, their fiber yield was at least 20% higher than that of controls (Pasapula et al., 2011). These data further confirm that despite the origin of the H+-PPase and the recipient of the expression chimera, its overexpression triggers similar phenotypes, namely those that are easily explained by a vacuole-localized H+-PPase (salt tolerance) and those that are not (enhanced shoot and root biomass).
HINTS TOWARD H+-PPASE PARTICIPATION IN Pi NUTRITION
The existence of a complicated transcriptional regulation system involved in a plant’s responses to Pi starvation is well documented (Franco-Zorrilla et al., 2004). Interestingly, the overexpression of a transcription factor (TF; OsPTF1) in rice resulted in enhanced plant performance under Pi limitation. It was shown that this TF is expressed in phloem cells of primary and lateral roots and leaves (Yi et al., 2005). Microarray data on this OsPTF1 transgenic rice plant showed a concomitant enhanced expression of a rice H+-PPase gene (Yi et al., 2005). Recent work in maize reported that the overexpression of another TF (ZmPTF1) improved the plant’s performance under Pi-limiting conditions by regulating carbon metabolism and root growth (Li et al., 2011). The authors concluded that the overexpression of ZmPTF1 triggers an enhanced allocation of carbohydrates to the roots, allowing better growth of roots under Pi limitation. Interestingly, the expression of the vacuolar H+-PPase in the above ZmPTF1 lines is constitutively enhanced, and after 15 d of Pi deprivation, its mRNA levels are 1-fold higher than in unmodified controls (Li et al., 2011). Similarly, an induction of AVP1 mRNA in wild-type Arabidopsis plants transferred to limiting Pi conditions has been documented (Yang et al., 2007). Furthermore, western blots of microsomal fractions of the above plants probed with polyclonal antibodies raised against the H+-PPase showed that the abundance of the AVP1 was increased 4-fold by Pi starvation (Yang et al., 2007). A conspicuous result from the same work came from the analysis of an AVP1 promoter-GUS (AVP1p:GUS) reporter. The reporter chimera was induced under Pi starvation, and its expression was restricted to the plant’s vasculature (Yang et al., 2007). Early observations suggested that expression levels of the H+-PPase are precisely controlled at the transcriptional level in response to various environmental conditions or developmental stages (Maeshima, 2000). It has been shown that two TFs, AtVOZ1 and AtVOZ2 (for Arabidopsis vascular plant one zinc finger protein), bind to the cis-acting region of the AVP1 promoter; the expression of AtVOZ1 is restricted to the phloem (Mitsuda et al., 2004). Furthermore, the presence of cis-regulatory Pi response elements in the 1.7-kb promoter region used to generate the AVP1p:GUS reporter (i.e. one PHOSPHATE STARVATION RESPONSE1 element at position –540 and two thymine cytosine-rich elements at positions –79 and –103) is consistent with the responsiveness of the H+-PPase to Pi limitation (Yang et al., 2007).
UP-REGULATION OF THE H+-PPASE ENHANCES ARABIDOPSIS PERFORMANCE UNDER LIMITING Pi CONDITIONS
As described earlier, Arabidopsis plants engineered with the 35Sp:AVP1 cassette (AtAVP1-OX) develop more robust shoot and root systems than wild-type plants grown under nonlimiting nutrient conditions (Li et al., 2005). The characterization of these AtAVP1-OX plants under limiting Pi conditions (i.e. synthetic medium and/or low-Pi soil) revealed a clear advantage in terms of both shoot and root development. A closer analysis showed that AtAVP1-OX roots were longer and developed more lateral roots than controls. Furthermore, root hairs were 2.5-fold larger and 1.5-fold denser than those of controls under Pi-deficient conditions (Yang et al., 2007). This observed vigorous root development displayed by AtAVP1-OX plants under Pi limitation resembles, somewhat, the behavior of white lupin (Lupinus albus), a noted Pi scavenger. As part of a complex response to limiting Pi, white lupin develops proteoid roots (densely clustered lateral roots with abundant root hairs) that significantly enhance both the surface area for Pi uptake and rhizosphere acidification capacity (Yan et al., 2002; Cheng et al., 2011). Of particular note is the fact that AtAVP1-OX plants displayed a stronger root acidification capacity than control plants when challenged with Pi limitation (Yang et al., 2007). Furthermore, this enhanced rhizosphere acidification capacity is sensitive to vanadate, suggesting that the up-regulation of the H+-PPase has a positive effect on the activity of the plasma membrane (PM) H+-ATPase (Yang et al., 2007). A recent report on the proton-pumping activity of the PM H+-ATPase from roots of AtAVP1-OX plants grown under nonlimiting nutrient conditions showed that the initial velocity and steady state of the proton gradients generated by the PM H+-ATPase are significantly greater (approximately 50%–70%; α = 0.05) than in the wild type (Undurraga et al., 2012). Rhizosphere acidification is a central mechanism for plant mineral nutrition, because it contributes to nutrient solubility and the PM proton motive force (PMF; Palmgren, 1998; Marschner, 2002). Enhanced rhizosphere acidification capacity linked to PM H+-ATPase activity has also been demonstrated in white lupin proteoid under Pi deficiency (Yan et al., 2002). White lupin proteoid roots release substantial amounts of carboxylates and concomitantly acidify the rhizosphere. Specifically, it was shown that citrate exudation increased transiently and reached a maximum after 5 h, and this effect was accompanied by a strong acidification of the external medium and alkalinization of the cytosol. Fusicoccin stimulated citrate exudation, whereas vanadate, an inhibitor of the H+-ATPase, reduced citrate exudation. The increase in proton secretion was due to both an increased transcription level of an H+-ATPase gene and activating posttranslational modifications of this proton pump (Tomasi et al., 2009).
H+-PPASE UP-REGULATION FUNCTIONS IN OTHER PLANTS
Further data consistent with the universality of this H+-PPase-mediated response to Pi limitation come from the characterization of tomato (Solanum lycopersicum) and rice plants engineered with a H+-PPase overexpression chimera (Yang et al., 2007; Gaxiola et al., 2011). Transgenic tomatoes overexpressing the E229D gain-of-function intragenic mutant (AVP1D) of the Arabidopsis H+-PPase under the control of the 35S promoter developed larger shoots, roots, and fruit yields than controls when grown under Pi-deficient conditions (Yang et al., 2007). Shoot and root dry weights of plants grown in the presence of 100 mg/L NaH2PO4 were, on average, 20% and 17% higher (P < 0.01) in 35Sp:AVP1D tomatoes than in controls, respectively. Furthermore, under the same low-Pi conditions, fruit dry weight data and Pi content per plant were 82% and 30% higher (P < 0.01) in the AVP1-modified plants compared with unmodified controls, respectively (Yang et al., 2007). These enhanced shoot and fruit yields suggest that, under the conditions tested, the physiological costs incurred by the development of larger root systems did not jeopardize the 35Sp:AVP1D tomato’s capacity to allocate sufficient photosynthates for shoot and fruit development, a concern expressed by others (Lynch, 1995). The enhanced H+-PPase expression displayed by both OsPTF1- and ZmPTF1-overexpressing rice and maize plants, and their improved performance under Pi limitations (Yi et al., 2005; Li et al., 2011), are consistent with the advantages shown by japonica rice (cv Taipei 309) plants engineered with the 35Sp:AVP1D cassette (Yang et al., 2007; Gaxiola et al., 2011). These 35Sp:AVP1D rice lines exhibited sustained shoot growth under Pi-deficient (10 μm) conditions, whereas the controls grew poorly. Here again, these lines developed more robust root systems than controls in both Pi-sufficient and Pi-deficient conditions. The dry plant biomass data showed that the 35Sp:AVP1D rice seedlings grown under limiting Pi conditions developed significantly larger shoots (50%; P < 0.01) and roots (90%; P < 0.01) than unmodified controls (Yang et al., 2007). Interestingly, 35Sp:AVP1D rice seedlings grown under Pi-sufficient conditions accumulated 18% higher Pi content than controls. Furthermore, quantification of root organic acid exudates (citrate and malate) in the 35Sp:AVP1D tomato and rice plants grown in the presence of 20 μm AlPO4 showed an increment of about 60% in the levels of organic acid exudates, whereas the increases in unmodified control plants subjected to the same treatment were not statistically significant (Yang et al., 2007). Similarly, organic acid exudation has been well documented in white lupin as a response to limiting Pi (Watt and Evans, 1999). Further information consistent with an enhanced proton extrusion capacity by the roots of 35Sp:AVP1 Arabidopsis as well as 35Sp:AVP1D rice and tomato plants is the 1-fold increase in root K+ content either under normal or Pi-limiting conditions for all the above compared with unmodified controls (Yang et al., 2007).
THE SUGAR AND PHOSPHATE CONNECTION
The enhanced PM H+-ATPase activity, increased organic acid exudation, and augmented root biomass displayed by 35Sp:AVP1 or 35Sp:AVP1D plants are consistent with an increased sugar supply, as described by Hammond and White (2011) and documented in genetically modified ZmPTF1 maize plants (Li et al., 2011). Of note, an elegant stem-girdling experiment showed that white lupin required phloem sugar transport for the expression of Pi deficiency-induced genes in cluster roots (Liu et al., 2005). The characterization of an Arabidopsis mutant hypersensitive to Pi starvation (hps1) caused by the constitutive overexpression of the Suc transporter gene Suc2 further emphasizes the key role that sugar transport/metabolism plays in plant responses to Pi limitation. These hps1 mutants display constitutive phenotypes that mimic plant responses to Pi starvation and accumulate more Suc than controls in shoots and roots. A loss-of-function suc2 mutant hinders plant responses to Pi deficiency (Lei et al., 2011). Furthermore, microarray analysis indicated that about 73% of the genes that are induced by Pi deficiency in control plants are constitutively induced by the elevated levels of Suc in the hps1 mutants even under Pi-sufficient conditions (Lei et al., 2011). Interestingly, sugar-responsive cis-elements (i.e. AMY, BOX1 and -2 CGACG boxes) are present in the regulatory region of the AVP1 promoter (Mitsuda et al., 2001). Up-regulation of H+-PPase genes has been reported in Suc-starved cells of rice (Wang et al., 2007). Of significant note are the transcriptome data from a comparative analysis of Arabidopsis transgenic lines (AVP1, GRF5, JAW, BRI1, and GA20OX1) that develop enlarged leaves (Gonzalez et al., 2010). In AtAVP1-OX, the constitutive up-regulated expression of genes involved in Suc transport/metabolism and Pi transport (Table I) further emphasizes the relationship between the aforementioned physiological processes (Gonzalez et al., 2010). Some of the more conspicuous are genes directly related to Suc biosynthesis, phloem loading, and Pi transport (Table I). Sucrose Synthase (SUS1) has been implicated in the inorganic pyrophosphate (PPi)-dependent metabolism of Suc during phloem loading (Fig. 1; Martin et al., 1993). Interestingly, a SUS1 promoter:GUS reporter showed the induction of expression in phloem cells under conditions of limiting ATP supply and/or increased demand for translocation of carbohydrates (Martin et al., 1993). SUC1 has been recently shown to be involved in cell-to-cell Suc movement in Arabidopsis (Wippel and Sauer, 2012). Also relevant is the constitutive up-regulation of the PHO1 gene (Table I) that has been shown to be involved in long-distance transfer of Pi from the root to the shoot, likely at xylem loading. It is worth noting that a recent report showed that overexpression of PHO1 alone results in reduced rosette growth, opposite to the phenotype triggered by H+-PPase up-regulation (Stefanovic et al., 2011).
Table I. Genes up-regulated in AtAVP1-OX relevant to carbohydrates and Pi metabolism (modified from Gonzalez et al., 2010).
| Gene | Accession No. | Induction |
|---|---|---|
| SUC1, Suc proton symporter 1 | AT1G71880 | 2.24-fold (P < 0.05) |
| Major facilitator superfamily protein, sugar/hydrogen symporter activity | AT5G18840 | 1.86-fold (P < 0.05) |
| PHO1, involved in Pi root-to-shoot transfer, likely the xylem loading | AT3G23430 | 1.74-fold (P < 0.05) |
| PGDH, phosphoglycerate dehydrogenase localized to chloroplast | AT1G17745 | 1.73-fold (P < 0.05) |
| SUS1, Suc synthase 1, expressed in phloem of leaves and roots | AT5G20830 | 1.66-fold (P < 0.05) |
| Suc phosphate synthase 1 | AT5G11110 | 1.66-fold (P < 0.05) |
| Major facilitator superfamily protein, sugar/hydrogen symporter activity | AT4G04750 | 1.52-fold (P < 0.05) |
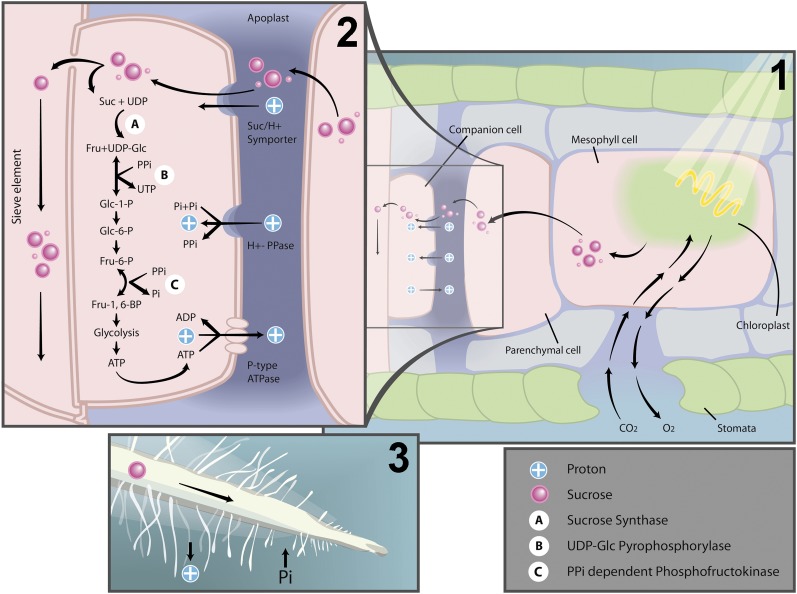
Figure 1.
Model for the function of a PM-localized H+-PPase in Suc partitioning. 1, Suc is synthesized in chloroplast of mesophyll cells and transported to CCs, where it can be hydrolyzed or delivered to the sieve elements. 2, Detail of Suc transport and metabolism in CCs and sieve elements. An adequate supply of ATP for the maintenance of the transmembrane proton gradient generated by the PM H+-ATPase requires PPi-dependent Suc metabolism. The most relevant enzymes and transporters are indicated. 3, Suc travels through sieve elements from source to sink tissues such as roots. Its presence in roots activates the rhizosphere acidification instrumental for Pi uptake.
LOCATION, LOCATION, LOCATION
How can a vacuolar H+-PPase influence sugar and Pi metabolism? Are we missing something? The binding of the phloem-specific AtVOZ1 TF to the cis-acting region of the AVP1 promoter (Mitsuda et al., 2004), together with the conspicuously localized expression of the AVP1p:GUS reporter under control or Pi limitation conditions in Arabidopsis, are consistent with a relevant role for the H+-PPase in Arabidopsis vascular tissue (Yang et al., 2007). A number of studies have shown a dual membrane localization of the type I H+-PPase in plants: (1) immunogold electron microscopy observations confirmed the presence of a PM H+-PPase in cauliflower (Brassica oleracea) inflorescence cells (Ratajczak et al., 1999); (2) double labeling epifluorescence experiments showed that the H+-PPase and the PM H+-ATPase localized in close proximity at the PM of Ricinus communis sieve elements (Langhans et al., 2001); (3) interestingly, H+-PPase immunogold studies with R. communis seedlings showed strong PM staining at phloem tissues of cotyledons and roots, whereas in the mesophyll and cortical cells, the staining was mainly vacuolar (Long et al., 1995); (4) a recent study provided immunohistochemical and immunogold labeling data consistent with a PM localization of the type I H+-PPase in sieve element and companion cell (CC) complexes of Arabidopsis source leaves (Paez-Valencia et al., 2011).
CAN A PM-LOCALIZED H+-PPASE FUNCTION IN PHLOEM CELLS?
An initial model to explain the potential role for a PM-localized H+-PPase in phloem of plants suggested that both H+ pumps (H+-PPase and H+-ATPase) could work together to generate the PMF to maintain high Suc, K+, and amino acid concentrations in the phloem (Langhans et al., 2001). This model of H+-PPases contributing to PMF across the PM in the phloem is an attractive hypothesis to explain the diverse phenotypes observed in various plants overexpressing H+-PPases: more PMF would energize phloem loading and enhance long-distance transport, and more efficient transport would in turn provide more resources for growth and nutrient acquisition and help maintain water balance.
Although attractive, H+-PPases on the PM of phloem cells face a very steep H+ gradient, and their hydrolytic operation is unlikely due to thermodynamic constraints (Davies et al., 1997). However, it has been argued that the reverse reaction, in which the PM PMF is used to synthesize PPi, is thermodynamically feasible (Davies et al., 1997). Therefore, it is highly noteworthy that in vivo data obtained with the H+-PPase from the gram-negative proteobacterium Rhodospirillum rubrum are consistent with the capacity of this enzyme to play two distinct roles depending on location and conditions: it can act as an intracellular H+ pump in the acidocalcisomes (Seufferheld et al., 2004) or as a PPi synthase in the chromatophore membranes during illumination (Baltscheffsky et al., 1966). Furthermore, Rocha Facanha and de Meis (1998) presented in vitro evidence consistent with the reverse function (PPi synthase) of the H+-PPase, utilizing tonoplast fractions of maize coleoptiles and seeds. These authors suggested that, given the appropriate thermodynamic conditions in vivo, the H+-PPase could operate as a system of energy conservation with a role in the maintenance of cytosolic PPi levels (Rocha Facanha and de Meis, 1998).
PROPOSED MODEL OF ACTION FOR A PM H+-PPASE IN PHLOEM ENERGY METABOLISM
Based on the above information regarding the versatility of the H+-PPase, it is tempting to speculate that plant H+-PPases can work as H+ pumps or as PPi synthases. In the phloem, a PM-localized H+-PPase could use the PMF prevailing in the apoplast of CCs to maintain the cytosolic PPi levels required for Suc respiration via SUS1, UDP-Glc pyrophosphorylase, and PPi:Fru-6-P 1-phosphotransferase (Fig. 1). Sonnewald (1992) suggested that the cytosolic concentration of PPi was essential for Suc transport. Lerchl et al. (1995) further tested this hypothesis via the phloem-specific expression of a soluble pyrophosphatase from Escherichia coli (ppa1) in tobacco plants. Characterization of these ppa1 plants revealed that removal of cytosolic PPi from phloem cells triggered the accumulation of Suc in source leaves, chlorophyll loss, and reduced shoot and root growth. Interestingly, the phloem-specific expression of a yeast invertase (Suc2) circumvented the metabolic block of the ppa1 plants, restoring wild-type phenotypes (Lerchl et al., 1995). These data are consistent with a model where Suc phloem loading depends on the levels of cytosolic PPi in CCs (Fig. 1). Suc must be transported actively from mesophyll cells to companion cells via Suc/H+ symporters (Table I) that depend on the proton gradient generated by the plasma membrane H+-ATPase (Fig. 1; Srivastava et al., 2008). In order to have an adequate ATP supply for the maintenance of this transmembrane proton gradient, a percentage of the incoming Suc must be cleaved into Fru and UDP-Glc (Lerchl et al., 1995). Of note, both the PPi:Fru-6-P 1-phosphotransferase and the UDP-Glc pyrophosphorylase enzymes work near equilibrium, so a decrease in the cytosolic concentration of PPi should prevent the reactions leading to glycolysis and therefore compromise the energy production required to maintain the PMF generated by the PM H+-ATPase (Lerchl et al., 1995).
We hypothesize that the up-regulation of AVP1 has different and seemingly contradictory effects in different tissues. In nonphloem cells, type I H+-PPases localize predominantly to and energize the endomembrane system while removing PPi produced as a by-product of metabolism. PPi hydrolysis in source leaf mesophyll could promote Suc synthesis (Sonnewald, 1992; Ferjani et al., 2011). In phloem CCs, however, type I H+-PPases predominantly localize to the PM (Paez-Valencia et al., 2011) and could function as PPi synthases, regulating Suc respiration for the ATP and PMF required for phloem loading (Fig. 1). More efficient Suc transport to sink organs in H+-PPase-overexpressing plants may explain the larger and more energized root systems with higher water and Pi-uptake capacities (Li et al., 2005, 2008; Park et al., 2005; Yang et al., 2007; Lv et al., 2008, 2009).
Some predictions related to the overexpression of the type I H+-PPases that emanate from the proposed model are as follows: (1) engineered plants should have an increased transport of sugars and amino acids from source to sinks (Geigenberger et al., 1996; Gonzalez et al., 2010); (2) the expression of root K+, NO3−, and Pi transporter genes, which has been shown to be coordinated with photosynthesis and the carbon status of the plant, should be up-regulated (Lejay et al., 2008). Plants should have enhanced water (Park et al., 2005), Pi (Yang et al., 2007), and nitrogen (Hermans et al., 2006) use efficiencies.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AT1G71880, AT5G18840, AT3G23430, AT1G17745, AT5G20830, AT5G11110, and AT4G04750.
Acknowledgments
We thank E.G. Baxter (Arizona State University School of Life Sciences Visualization Laboratory) for help with the model figure.
Glossary
- Pi
phosphate
- P
phosphorus
- TF
transcription factor
- PM
plasma membrane
- PPi
inorganic pyrophosphate
- PMF
proton motive force
References
- Anderson G (1967) Nucleic acids, derivatives, and organic phosphorus. In AD McLaren, GH Peterson, eds, Soil Biochemistry, Vol 1. Marcel Dekker, New York, pp 67–90
- Anderson G (1975) Other organic phosphorus compounds. In JE Gieseking, ed, Soil Organic Components, Vol 1. Organic Components. Springer Verlag, New York, Berlin, pp 305–331
- Baltscheffsky H, Von Stedingk LV, Heldt HW, Klingenberg M. (1966) Inorganic pyrophosphate: formation in bacterial photophosphorylation. Science 153: 1120–1122 [DOI] [PubMed] [Google Scholar]
- Bao A-K, Wang S-M, Wu G-Q, Xi J-J, Zhang J-L, Wang C-M. (2008) Overexpression of the Arabidopsis H+-PPase enhanced resistance to salt and drought stress in transgenic alfalfa (Medicago sativa L.). Plant Sci 176: 232–240 [Google Scholar]
- Chen Y-F, Wang Y, Wu W-H. (2008) Membrane transporters for nitrogen, phosphate and potassium uptake in plants. J Integr Plant Biol 50: 835–848 [DOI] [PubMed] [Google Scholar]
- Cheng L, Bucciarelli B, Liu J, Zinn K, Miller S, Patton-Vogt J, Allan D, Shen J, Vance CP. (2011) White lupin cluster root acclimation to phosphorus deficiency and root hair development involve unique glycerophosphodiester phosphodiesterases. Plant Physiol 156: 1131–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogger C, Duxbury JM. (1984) Factors affecting phosphorus losses from cultivated organic soils. J Environ Qual 13: 111–114 [Google Scholar]
- Cole CV, Olsen SR, Scott CO. (1953) The nature of phosphate sorption by calcium carbonate. Soil Sci Soc Am Proc 17: 352–356 [Google Scholar]
- Conley DJ, Paerl HW, Howarth RW, Boesch DF, Seitzinger SP, Havens KE, Lancelot C, Likens GE. (2009) Controlling eutrophication: nitrogen and phosphorus. Science 323: 1014–1015 [DOI] [PubMed] [Google Scholar]
- Cordell D, Drangert JO, White S. (2009) The story of phosphorus: global food security and food for thought. Glob Environ Change 19: 292–305 [Google Scholar]
- Davies JM, Darley CP, Sanders D. (1997) Energetics of the plasma membrane pyrophosphatase. Trends Plant Sci 2: 9–10 [Google Scholar]
- Diaz RJ, Rosenberg R. (2008) Spreading dead zones and consequences for marine ecosystems. Science 321: 926–929 [DOI] [PubMed] [Google Scholar]
- Dodds WK, Bouska WW, Eitzmann JL, Pilger TJ, Pitts KL, Riley AJ, Schloesser JT, Thornbrugh DJ. (2009) Eutrophication of U.S. freshwaters: analysis of potential economic damages. Environ Sci Technol 43: 12–19 [DOI] [PubMed] [Google Scholar]
- Drozdowicz YM, Rea PA. (2001) Vacuolar H(+) pyrophosphatases: from the evolutionary backwaters into the mainstream. Trends Plant Sci 6: 206–211 [DOI] [PubMed] [Google Scholar]
- Elser JJ, Bennett E. (2011) Phosphorus cycle: a broken biogeochemical cycle. Nature 478: 29–31 [DOI] [PubMed] [Google Scholar]
- Elser JJ, Bracken MES, Cleland EE, Gruner DS, Harpole WS, Hillebrand H, Ngai JT, Seabloom EW, Shurin JB, Smith JE. (2007) Global analysis of nitrogen and phosphorus limitation of primary producers in freshwater, marine and terrestrial ecosystems. Ecol Lett 10: 1135–1142 [DOI] [PubMed] [Google Scholar]
- Ferjani A, Segami S, Horiguchi G, Muto Y, Maeshima M, Tsukaya H. (2011) Keep an eye on PPi: the vacuolar-type H+-pyrophosphatase regulates postgerminative development in Arabidopsis. Plant Cell 23: 2895–2908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippelli GM. (2002) The global phosphorus cycle. Rev Mineral Geochem 48: 391–425 [Google Scholar]
- Fox RL, Kamprath EJ. (1971) Adsorption and leaching of P in acid organic soils and high organic matter sand. Soil Sci Soc Am Proc 35: 154–156 [Google Scholar]
- Franco-Zorrilla JM, González E, Bustos R, Linhares F, Leyva A, Paz-Ares J. (2004) The transcriptional control of plant responses to phosphate limitation. J Exp Bot 55: 285–293 [DOI] [PubMed] [Google Scholar]
- Fried M, Dean LA. (1955) Phosphate retention by iron and aluminum in cation exchange systems. Soil Sci Soc Am Proc 19: 142–147 [Google Scholar]
- Fuglsang AT, Paez-Valencia J, Gaxiola RA (2011) Plant proton pumps: regulatory circuits involving H+-ATPase and H+-PPase. In M Geisler, K Venema, eds, Transporters and Pumps in Plant Signaling: Signaling and Communication in Plants, Vol 7. Springer-Verlag, Heidelberg, pp 39–64
- Gao F, Gao Q, Duan XG, Yue GD, Yang AF, Zhang JR. (2006) Cloning of an H+-PPase gene from Thellungiella halophila and its heterologous expression to improve tobacco salt tolerance. J Exp Bot 57: 3259–3270 [DOI] [PubMed] [Google Scholar]
- Gaxiola RA, Edwards M, Elser JJ. (2011) A transgenic approach to enhance phosphorus use efficiency in crops as part of a comprehensive strategy for sustainable agriculture. Chemosphere 84: 840–845 [DOI] [PubMed] [Google Scholar]
- Gaxiola RA, Fink GR, Hirschi KD. (2002) Genetic manipulation of vacuolar proton pumps and transporters. Plant Physiol 129: 967–973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaxiola RA, Li J, Undurraga S, Dang LM, Allen GJ, Alper SL, Fink GR. (2001) Drought- and salt-tolerant plants result from overexpression of the AVP1 H+-pump. Proc Natl Acad Sci USA 98: 11444–11449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaxiola RA, Palmgren MG, Schumacher K. (2007) Plant proton pumps. FEBS Lett 581: 2204–2214 [DOI] [PubMed] [Google Scholar]
- Geigenberger P, Lerchl J, Stitt M, Sonnewald U. (1996) Phloem-specific expression of pyrophosphatase inhibits long-distance transport of carbohydrates and amino acids in tobacco plants. Plant Cell Environ 19: 43–55 [Google Scholar]
- Gonzalez N, De Bodt S, Sulpice R, Jikumaru Y, Chae E, Dhondt S, Van Daele T, De Milde L, Weigel D, Kamiya Y, et al. (2010) Increased leaf size: different means to an end. Plant Physiol 153: 1261–1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowariker V, Krishnamurthy VN, Gowariker S, Dhanorkar M, Paranjape K (2009) The Fertilizer Encyclopedia. John Wily & Sons, Hoboken, NJ
- Griffin RA, Jurinak JJ. (1973) The interaction of phosphate with calcite. Soil Sci Soc Am Proc 37: 847–850 [Google Scholar]
- Halstead RL, McKercher RB (1975) Biochemistry and cycling of phosphorus. In EA Paul, AD McLaren, eds, Soil Biochemistry, Vol 4. Marcel Dekker, New York, pp 31–64
- Hammond JP, White PJ. (2011) Sugar signaling in root responses to low phosphorus availability. Plant Physiol 156: 1033–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermans C, Hammond JP, White PJ, Verbruggen N. (2006) How do plants respond to nutrient shortage by biomass allocation? Trends Plant Sci 11: 610–617 [DOI] [PubMed] [Google Scholar]
- Hoagland P, Anderson DM, Kaoru Y, White AW. (2002) The economic effects of harmful algal blooms in the United States: estimates, assessment issues, and information needs. Estuaries 25: 819–837 [Google Scholar]
- Holford ICR, Mattingly GEG. (1975) Phosphate sorption by Jurassic Oolitic limestones. Geoderma 13: 257–264 [Google Scholar]
- Jha D, Shirley N, Tester M, Roy SJ. (2010) Variation in salinity tolerance and shoot sodium accumulation in Arabidopsis ecotypes linked to differences in the natural expression levels of transporters involved in sodium transport. Plant Cell Environ 33: 793–804 [DOI] [PubMed] [Google Scholar]
- Krebs M, Beyhl D, Görlich E, Al-Rasheid KA, Marten I, Stierhof YD, Hedrich R, Schumacher K. (2010) Arabidopsis V-ATPase activity at the tonoplast is required for efficient nutrient storage but not for sodium accumulation. Proc Natl Acad Sci USA 107: 3251–3256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langhans M, Ratajczak R, Lützelschwab M, Michalke W, Wächter R, Fischer-Schliebs E, Ullrich CI. (2001) Immunolocalization of plasma-membrane H+-ATPase and tonoplast-type pyrophosphatase in the plasma membrane of the sieve element-companion cell complex in the stem of Ricinus communis L. Planta 213: 11–19 [DOI] [PubMed] [Google Scholar]
- Lei M, Liu YC, Zhang B, Zhao Y, Wang XF, Zhou YH, Raghothama KG, Liu D. (2011) Genetic and genomic evidence that sucrose is a global regulator of plant responses to phosphate starvation in Arabidopsis. Plant Physiol 156: 1116–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lejay L, Wirth J, Pervent M, Cross JM-F, Tillard P, Gojon A. (2008) Oxidative pentose phosphate pathway-dependent sugar sensing as a mechanism for regulation of root ion transporters by photosynthesis. Plant Physiol 146: 2036–2053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerchl J, Geigenberger P, Stitt M, Sonnewald U. (1995) Impaired photoassimilate partitioning caused by phloem-specific removal of pyrophosphate can be complemented by a phloem-specific cytosolic yeast-derived invertase in transgenic plants. Plant Cell 7: 259–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Wei A, Song C, Li N, Zhang JR. (2008) Heterologous expression of the TsVP gene improves the drought resistance of maize. Plant Biotechnol J 6: 146–159 [DOI] [PubMed] [Google Scholar]
- Li J, Yang H, Peer WA, Richter G, Blakeslee J, Bandyopadhyay A, Titapiwantakun B, Undurraga S, Khodakovskaya M, Richards EL, et al. (2005) Arabidopsis H+-PPase AVP1 regulates auxin-mediated organ development. Science 310: 121–125 [DOI] [PubMed] [Google Scholar]
- Li Z, Baldwin CM, Hu Q, Liu H, Luo H. (2010) Heterologous expression of Arabidopsis H+-pyrophosphatase enhances salt tolerance in transgenic creeping bentgrass (Agrostis stolonifera L.). Plant Cell Environ 33: 272–289 [DOI] [PubMed] [Google Scholar]
- Li Z, Gao Q, Liu YC, He C, Zhang X, Zhang J. (2011) Overexpression of transcription factor ZmPTF1 improves low phosphate tolerance of maize by regulating carbon metabolism and root growth. Planta 233: 1129–1143 [DOI] [PubMed] [Google Scholar]
- Lindsay WL (1979) Chemical Equilibrium in Soil. Wiley Interscience, New York
- Lindsay WL, Stephenson HF. (1959) Nature of the reactions of monocalcium phosphate monohydrate in soils. II. Dissolution and precipitation reactions involving iron, aluminum, manganese and calcium. Soil Sci Soc Am Proc 26: 446–452 [Google Scholar]
- Liu J, Samac DA, Bucciarelli B, Allan DL, Vance CP. (2005) Signaling of phosphorus deficiency-induced gene expression in white lupin requires sugar and phloem transport. Plant J 41: 257–268 [DOI] [PubMed] [Google Scholar]
- Long AR, Williams LE, Nelson SJ, Hall J. (1995) Localization of membrane pyrophosphatase activity in Ricinus communis seedlings. J Plant Physiol 146: 629–638 [Google Scholar]
- Lv S, Zhang K, Gao Q, Lian L, Song Y, Zhang JR. (2008) Overexpression of an H+-PPase gene from Thellungiella halophila in cotton enhances salt tolerance and improves growth and photosynthetic performance. Plant Cell Physiol 49: 1150–1164 [DOI] [PubMed] [Google Scholar]
- Lv S-L, Lian L-J, Tao P-L, Li Z-X, Zhang K-W, Zhang J-R. (2009) Overexpression of Thellungiella halophila H(+)-PPase (TsVP) in cotton enhances drought stress resistance of plants. Planta 229: 899–910 [DOI] [PubMed] [Google Scholar]
- Lynch J. (1995) Root architecture and plant productivity. Plant Physiol 109: 7–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeshima M. (2000) Vacuolar H(+)-pyrophosphatase. Biochim Biophys Acta 1465: 37–51 [DOI] [PubMed] [Google Scholar]
- Marschner H (2002) Mineral Nutrition in Higher Plants, Ed 2. Academic Press, London
- Martin T, Frommer WB, Salanoubat M, Willmitzer L. (1993) Expression of an Arabidopsis sucrose synthase gene indicates a role in metabolization of sucrose both during phloem loading and in sink organs. Plant J 4: 367–377 [DOI] [PubMed] [Google Scholar]
- Mitsuda N, Enami K, Nakata M, Takeyasu K, Sato MH. (2001) Novel type Arabidopsis thaliana H(+)-PPase is localized to the Golgi apparatus. FEBS Lett 488: 29–33 [DOI] [PubMed] [Google Scholar]
- Mitsuda N, Hisabori T, Takeyasu K, Sato MH. (2004) VOZ: isolation and characterization of novel vascular plant transcription factors with a one-zinc finger from Arabidopsis thaliana. Plant Cell Physiol 45: 845–854 [DOI] [PubMed] [Google Scholar]
- Paez-Valencia J, Patron-Soberano A, Rodriguez-Leviz A, Sanchez-Lares J, Sanchez-Gomez C, Valencia-Mayoral P, Diaz-Rosas G, Gaxiola R. (2011) Plasma membrane localization of the type I H(+)-PPase AVP1 in sieve element-companion cell complexes from Arabidopsis thaliana. Plant Sci 181: 23–30 [DOI] [PubMed] [Google Scholar]
- Palmgren MG (1998) Proton gradients and plant growth: role of the plasma membrane H+-ATPase. Adv Bot Res 28: 1–70
- Palmgren MG. (2001) Plant plasma membrane H+-ATPases: powerhouses for nutrient uptake. Annu Rev Plant Physiol Plant Mol Biol 52: 817–845 [DOI] [PubMed] [Google Scholar]
- Park S, Li J, Pittman JK, Berkowitz GA, Yang H, Undurraga S, Morris J, Hirschi KD, Gaxiola RA. (2005) Up-regulation of a H+-pyrophosphatase (H+-PPase) as a strategy to engineer drought-resistant crop plants. Proc Natl Acad Sci USA 102: 18830–18835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasapula V, Shen G, Kuppu S, Paez-Valencia J, Mendoza M, Hou P, Chen J, Qui X, Zhu L, Zhang X, et al. (2011) Expression of an Arabidopsis vacuolar H+-pyrophosphatase gene (AVP1) in cotton improves drought- and salt tolerance and increases fiber yield in field conditions. Plant Biotechnol J 9: 88–99 [DOI] [PubMed] [Google Scholar]
- Ratajczak R, Hinz G, Robinson DG. (1999) Localization of pyrophosphatase in membranes of cauliflower inflorescence cells. Planta 208: 205–211 [DOI] [PubMed] [Google Scholar]
- Rea PA, Kim Y, Sarafian V, Poole RJ, Davies JM, Sanders D. (1992) Vacuolar H(+)-translocating pyrophosphatases: a new category of ion translocase. Trends Biochem Sci 17: 348–353 [DOI] [PubMed] [Google Scholar]
- Rocha Facanha A, de Meis L. (1998) Reversibility of H+-ATPase and H+-pyrophosphatase in tonoplast vesicles from maize coleoptiles and seeds. Plant Physiol 116: 1487–1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez CA (2006) Phosphorus. In AV Barker, DJ Pilbeam, eds, Handbook of Plant Nutrition. CRC Press/Taylor and Francis, Boca Raton, FL, pp 51–90
- Segami S, Nakanishi Y, Sato MH, Maeshima M. (2010) Quantification, organ-specific accumulation and intracellular localization of type II H(+)-pyrophosphatase in Arabidopsis thaliana. Plant Cell Physiol 51: 1350–1360 [DOI] [PubMed] [Google Scholar]
- Seufferheld M, Lea CR, Vieira M, Oldfield E, Docampo R. (2004) The H(+)-pyrophosphatase of Rhodospirillum rubrum is predominantly located in polyphosphate-rich acidocalcisomes. J Biol Chem 279: 51193–51202 [DOI] [PubMed] [Google Scholar]
- Smith VH, Joye SB, Howarth RW. (2006) Eutrophication of freshwater and marine ecosystems. Limnol Oceanogr 51: 351–355 [Google Scholar]
- Sonnewald U. (1992) Expression of E. coli inorganic pyrophosphatase in transgenic plants alters photoassimilate partitioning. Plant J 2: 571–581 [PubMed] [Google Scholar]
- Srivastava AC, Ganesan S, Ismail IO, Ayre BG. (2008) Functional characterization of the Arabidopsis AtSUC2 sucrose/H+ symporter by tissue-specific complementation reveals an essential role in phloem loading but not in long-distance transport. Plant Physiol 148: 200–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanovic A, Arpat AB, Bligny R, Gout E, Vidoudez C, Bensimon M, Poirier Y. (2011) Over-expression of PHO1 in Arabidopsis leaves reveals its role in mediating phosphate efflux. Plant J 66: 689–699 [DOI] [PubMed] [Google Scholar]
- Tomasi N, Kretzschmar T, Espen L, Weisskopf L, Fuglsang AT, Palmgren MG, Neumann G, Varanini Z, Pinton R, Martinoia E, et al. (2009) Plasma membrane H-ATPase-dependent citrate exudation from cluster roots of phosphate-deficient white lupin. Plant Cell Environ 32: 465–475 [DOI] [PubMed] [Google Scholar]
- Undurraga SF, Santos PM, Paez-Valencia J, Yang H, Hepler PK, Facanha AR, Hirschi KD, Gaxiola RA. (2012) Arabidopsis sodium dependent and independent phenotypes triggered by H+-PPase up-regulation are SOS1 dependent. Plant Sci 183: 96–105 [DOI] [PubMed] [Google Scholar]
- Vaccari DA. (2011) Chemosphere phosphorus cycle issue: introduction. Chemosphere 84: 735–736 [DOI] [PubMed] [Google Scholar]
- Wang HJ, Wan AR, Hsu CM, Lee KW, Yu SM, Jauh GY. (2007) Transcriptomic adaptations in rice suspension cells under sucrose starvation. Plant Mol Biol 63: 441–463 [DOI] [PubMed] [Google Scholar]
- Watt M, Evans JR. (1999) Linking development and determinacy with organic acid efflux from proteoid roots of white lupin grown with low phosphorus and ambient or elevated atmospheric CO2 concentration. Plant Physiol 120: 705–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wippel K, Sauer N. (2012) Arabidopsis SUC1 loads the phloem in suc2 mutants when expressed from the SUC2 promoter. J Exp Bot 63: 669–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan F, Zhu Y, Müller C, Zörb C, Schubert S. (2002) Adaptation of H+-pumping and plasma membrane H+ ATPase activity in proteoid roots of white lupin under phosphate deficiency. Plant Physiol 129: 50–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Knapp J, Koirala P, Rajagopal D, Peer WA, Silbart LK, Murphy A, Gaxiola RA. (2007) Enhanced phosphorus nutrition in monocots and dicots over-expressing a phosphorus-responsive type I H+-pyrophosphatase. Plant Biotechnol J 5: 735–745 [DOI] [PubMed] [Google Scholar]
- Yi K, Wu Z, Zhou J, Du L, Guo L, Wu Y, Wu P. (2005) OsPTF1, a novel transcription factor involved in tolerance to phosphate starvation in rice. Plant Physiol 138: 2087–2096 [DOI] [PMC free article] [PubMed] [Google Scholar]