Abstract
Certain plant receptor-like cytoplasmic kinases were reported to interact with small monomeric G-proteins of the RHO of plant (ROP; also called RAC) family in planta and to be activated by this interaction in vitro. We identified a barley (Hordeum vulgare) partial cDNA of a ROP binding protein kinase (HvRBK1) in yeast (Saccharomyces cerevisiae) two-hybrid screenings with barley HvROP bait proteins. Protein interaction of the constitutively activated (CA) barley HvROPs CA HvRACB and CA HvRAC1 with full-length HvRBK1 was verified in yeast and in planta. Green fluorescent protein-tagged HvRBK1 appears in the cytoplasm and nucleoplasm, but CA HvRACB or CA HvRAC1 can recruit green fluorescent protein-HvRBK1 to the cell periphery. Barley HvRBK1 is an active kinase in vitro, and activity is enhanced by CA HvRACB or GTP-loaded HvRAC1. Hence, HvRBK1 might act downstream of active HvROPs. Transient-induced gene silencing of barley HvRBK1 supported penetration by the parasitic fungus Blumeria graminis f. sp. hordei, suggesting a function of the protein in basal disease resistance. Transient knockdown of HvRBK1 also influenced the stability of cortical microtubules in barley epidermal cells. Hence, HvRBK1 might function in basal resistance to powdery mildew by influencing microtubule organization.
ROP (RHO, RAT SARCOMA HOMOLOG, of plants, also called RAC, RAT SARCOMA-related C3 botulinum toxin substrate) small GTP-binding proteins are signal transduction proteins that are considered as molecular switches for signaling toward cell development, hormone responses, and the cytoskeleton (Nibau et al., 2006). Additionally, ROPs are involved in resistance and susceptibility to plant diseases (Chen et al., 2010a, 2010b; Trusov et al., 2010). ROPs exist in a GTP-bound active form for targeting of downstream effectors and in a GDP-bound inactive form. The C terminus of ROPs is posttranslationally lipid modified for membrane association. Depending on their lipid modification motifs, ROPs are categorized in two phylogenetic subgroups: type I ROP proteins possess a CAAX prenylation motif, whereas those of type II lack a typical CAAX box but are palmitoylated. Activated type I ROPs can be further acylated at a conserved Cys residue, and these lipid modifications influence localization and function of ROPs (Sorek et al., 2007, 2011; Berken and Wittinghofer, 2008). In metazoans, RHO proteins physically interact with a plethora of proteins that regulate many processes in a cell type-specific manner (Bustelo et al., 2007). In plants, several proteins have been identified that bind to and signal upstream or downstream from ROPs (Berken and Wittinghofer, 2008; Yalovsky et al., 2008; Mucha et al., 2011). However, our knowledge of protein interaction partners of ROPs and their physiological relevance is still incomplete. In metazoans, RHO signaling involves upstream and downstream protein kinases (Denhardt, 1996). Similarly, plant ROPs can be activated via receptor-like kinases that activate plant-specific ROP nucleotide exchangers for exchange of ROP-bound GDP for GTP (Zhang and McCormick, 2007; Löcke et al., 2010; Humphries et al., 2011). Active ROP-GTP interacts with downstream factors (also called ROP effectors) to trigger cellular responses. ROP-GTP binds putative downstream protein kinases in yeast (Saccharomyces cerevisiae) and in planta. One type of a protein kinase that interacts with active ROPs is a Cys-rich receptor kinase, named AtNCRK (Molendijk et al., 2008). A second type of Arabidopsis (Arabidopsis thaliana) ROP binding kinases (AtRBKs) is otherwise known as receptor-like cytoplasmic kinase of the VIA subfamily (Arabidopsis AtRLCK VIA) or ROP-interacting receptor-like kinases (Medicago truncatula MtRRKs; Molendijk et al., 2008; Dorjgotov et al., 2009). These RLCKs are activated in vitro by ROPs (Dorjgotov et al., 2009).
Plant receptor-like kinases and downstream kinases are key to plant immunity (Tena et al., 2011). However, the picture of plant kinases involved in pathogen signal transduction or in modulation of the immune response is complex, and no function has yet been described for ROP binding kinases. However, transcripts of Arabidopsis AtRLCK VIAs accumulate in response to stress and hormones and, in the case of AtRBK1, in response to the pathogens Botrytis cinerea and Phytopthora infestans (Jurca et al., 2008; Molendijk et al., 2008).
The barley (Hordeum vulgare) ROP protein HvRACB is required for full susceptibility to powdery mildew disease caused by the biotrophic fungal pathogen Blumeria graminis f. sp. hordei (Schultheiss et al., 2002; Hoefle et al., 2011). Additionally, the barley ROPs HvRACB, HvRAC3, and HvRAC1 have the potential to support susceptibility to fungal penetration when expressed as constitutively activated (CA) mutant variants (Schultheiss et al., 2003; Pathuri et al., 2008). Barley CA HvRAC1, however, also supports callose deposition as well as the oxidative burst and the hypersensitive reaction in those cells of barley, which successfully defend penetration by B. graminis f. sp. hordei (Pathuri et al., 2008). This is similar to the function of rice (Oryza sativa) OsRAC1 in resistance to Magnaporthe grisea (Ono et al., 2001; Chen et al., 2010b). HvRACB further influences polar growth processes in barley leaf epidermis and root hairs (Pathuri et al., 2008; Hoefle et al., 2011). HvRACB regulates polarity of filamentous F-actin in barley epidermal cells during interaction with B. graminis f. sp. hordei and modulates the number of cells to which fungal infection structures, haustoria, get access (Schultheiss et al., 2002; Opalski et al., 2005). Barley HvRIC171 interacts with HvRACB in yeast and in planta (Schultheiss et al., 2008). HvRIC171 is a member of the so-called ROP-interactive CRIB (Cdc42/Rac-interactive binding) motif-containing protein family (RIC). RICs contain the conserved RHO-binding CRIB motif, which is also found in nonplant RHO effectors. Otherwise, RICs are structurally diverse (Wu et al., 2001). Barley HvRIC171 is located at the cell periphery where it further accumulates upon coexpression of CA HvRACB and upon attack from B. graminis f. sp. hordei. This may indicate activation of HvROPs and recruitment of downstream effectors at the site of fungal attack in barley (Schultheiss et al., 2008; Hückelhoven and Panstruga, 2011). Accordingly, HvRIC171 enhances susceptibility to haustorium invasion when overexpressed (Schultheiss et al., 2008). Another barley HvRACB-binding protein is HvMAGAP1, a microtubule (MT)-associated ROP GTPase-activating protein. ROPGAPs inactivate ROPs by the stimulation of GTP hydrolysis (Wu et al., 2000). HvMAGAP1 antagonizes HvRACB in susceptibility to penetration by B. graminis f. sp. hordei and supports polar organization of cortical MTs during fungal attack (Hoefle et al., 2011). Similarly, Arabidopsis AtROPGAP1 and AtROPGAP4 limit susceptibility to the powdery mildew fungus Erysiphe cruciferarum (Hoefle et al., 2011; Huesmann et al., 2011). However, little is known about other ROP-binding proteins of barley that are potentially involved in signaling toward pathophysiological or cell developmental responses.
Here, we describe identification of HvRBK1, a potential ROP effector kinase from barley. CA HvRACB and HvRAC1-GTP enhance kinase activity of HvRBK1 in vitro. CA HvRACB and CA HvRAC1 interact with HvRBK1 in planta. Transient knockdown of HvRBK1 destabilizes MTs and enhances susceptibility to penetration by B. graminis f. sp. hordei. Data suggest a function of barley ROP binding kinase1 in basal resistance to powdery mildew.
RESULTS
Identification and Characterization of Barley HvRBK1
To identify potential ROP binding proteins of barley, we carried out yeast two-hybrid (YTH) screenings of a cDNA library from a pool of near-isogenic barley lines resistant or susceptible to powdery mildew infected with B. graminis f. sp. hordei. We used barley HvRACB, CA HvRACB, and CA HvRAC1 as bait proteins (for more details, see Hoefle et al., 2011). One partial cDNA was repeatedly fished with all three ROP baits out of the cDNA library. A corresponding full-length cDNA was generated by 5′ RACE. We identified a matching genomic contig sequence of 7,372 bp in the draft sequence of the barley genome (contig_6719 at http://webblast.ipk-gatersleben.de/barley/index.php) and generated a corresponding gene model. The gene model identified a corresponding TATA box and was consistent with the full-length character of our cDNA. The gene is on the long arm of barley chromosome 7H and contains seven introns and eight exons (Supplemental Fig. S1A).
The cDNA encodes a protein of 543 amino acids with similarity to ROP binding kinases of Arabidopsis (AtRLCK VIA2, AtRLCK VIA3, AtRLCK VIA-4 [AtRBK1], and AtRLCK VIA6 [AtRBK2]), and M. truncatula (MtRRK1 and MtRRK2). Therefore, we named the predicted protein barley ROP binding kinase1 (HvRBK1; Supplemental Fig. S1B).
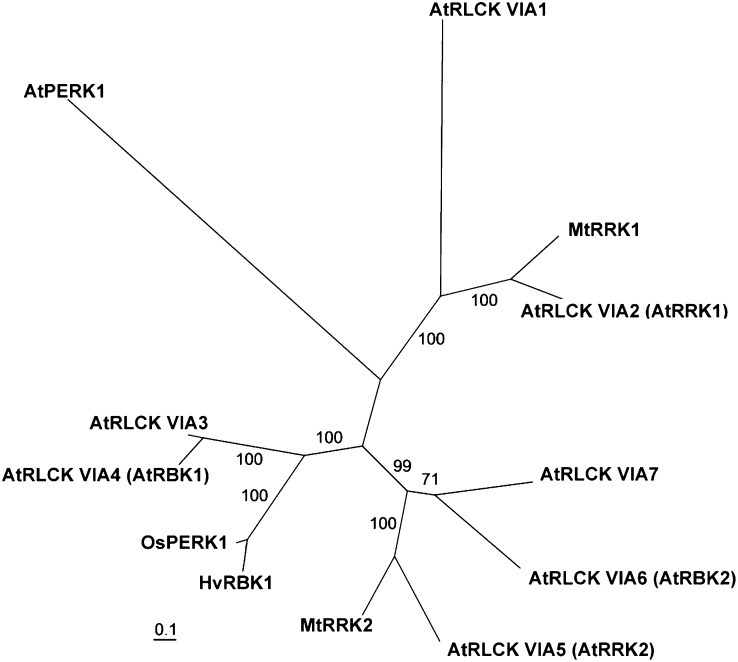
Among similar kinases, a rice protein annotated as OsPERK1 was the most similar protein found in monocots (Supplemental Fig. S1B, Fig. 1). Barley EST libraries and the draft genome contain HvRBK1-similar sequences, hinting at a possible small gene family of barley ROP binding kinases similar to the AtRLCK VIA family in Arabidopsis. Among the Arabidopsis kinases, AtRLCK VIA3 was most similar to the barley HvRBK1 (55% identical between amino acids 149 and 542) followed by AtRLCK VIA4 (synonymous to AtRBK1, 52% identical between amino acids 142 and 542; Supplemental Fig. S1B). Arabidopsis AtRLCK VIA3 (At5g65530) and AtRLCK VIA4 (AtRBK1, AT5G10520) genes also have an HvRBK1-like gene structure with seven introns and eight exons, which differs from that of the other RLCK VIA genes. We carried out a phylogenetic analysis based on protein sequences using protein alignments created by custalw2 (Thompson et al., 1994) and analyzed by RaXML program (Stamatakis et al., 2008). This further supported that HvRBK1 might be the ortholog of AtRLCK VIA3 or of its paralog AtRBK1 (Fig. 1). Bos taurus interleukin-1 receptor-associated kinase4 is the most similar kinase from metazoan species that is represented in GenBank.
Figure 1.
Unrooted phylogenetic tree (best scoring tree after the bootstraps) of predicted RLCKs of the Arabidopsis RLCKVIA family (At5G57670 = AtRLCK VIA1, At2G188900 = AtRLCK VIA2, At5G65530 = AtRLCK VIA3, At5G10520 = AtRLCK VIA4, At5G35960 = AtRLCK VIA5, At3G05140 = AtRLCK VIA6, At5G18910 = AtRLCK VIA7), M. truncatula MtRRKs (MtRRK1, MtRRK2), barley HvRBK1, rice OsPERK1, and the predicted transmembrane receptor-like kinase AtPERK1 (AT3G24550) taken as an outgroup. Numbers indicate support values after bootstraps.
The kinase domain of plant ROP binding kinases is highly conserved in mono- and dicots. It is positioned at the C-terminal part of the protein. By contrast, the N terminus of HvRBK1 is not strongly conserved in rice OsPERK1 and highly dissimilar to that of the related Arabidopsis ROP binding kinases AtRLCK VIA3 or AtRBK1 (Supplemental Fig. S1B). The N terminus of these ROP binding kinases is enriched with Ser residues in all three species. In HvRBK1, however, this is less pronounced when compared with the other three RLCKs from rice and Arabidopsis. The predicted open reading frames of Arabidopsis AtRBK1 and AtRLCK VIA3 code proteins shorter than barley HvRBK1, which possesses an N-terminal extension (Supplemental Fig. S1B).
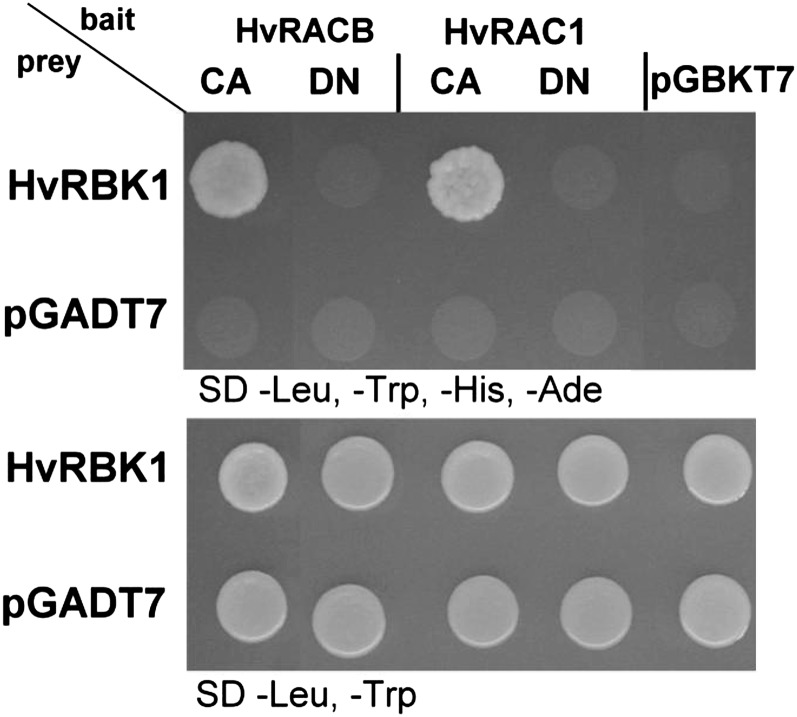
In independent YTH assays, the interaction of the full-length HvRBK1 with barley HvROPs was confirmed and auto-activation was excluded (Fig. 2). Further YTH assays with the partial HvRBK1 from the original screening showed that amino acids 210 to 543 of the barley HvRBK1 C terminus were sufficient for the interaction (not shown). Hence, the kinase domain of HvRBK1 may directly interact with barley ROPs. In all YTH assays, HvRBK1 preferentially interacted with CA HvROPs when compared with corresponding dominant negative (DN) HvROPs (Fig. 2, and see below).
Figure 2.
Targeted YTH assay of HvRBK1 and barley HvROPs in yeast. Targeted YTH assay in yeast strain AH109 cotransformed with HvRBK1 in the pGADT7 prey vector and barley HvROPs in pGBKT7 bait vector. Bait and prey interaction allows growth on selective synthetic dextrose media without −Leu, −Trp, −His, and adenine (−Ade). No growth was observed on selective media when yeast was cotransformed with expression constructs of HvROPs and the empty vector pGADT7 or with HvRBK1 and empty vector pGBKT7. As control for successful cotransformation, yeast cells were dropped on selective synthetic dextrose media −Leu, −Trp (bottom picture).
Together, barley HvRBK1 is similar to Arabidopsis ROP binding AtRLCK VIAs in sequence and in the capacity to bind ROPs in the YTH assay.
Activated Barley HvRACB and HvRAC1 Increase the Activity of HvRBK1 in Vitro
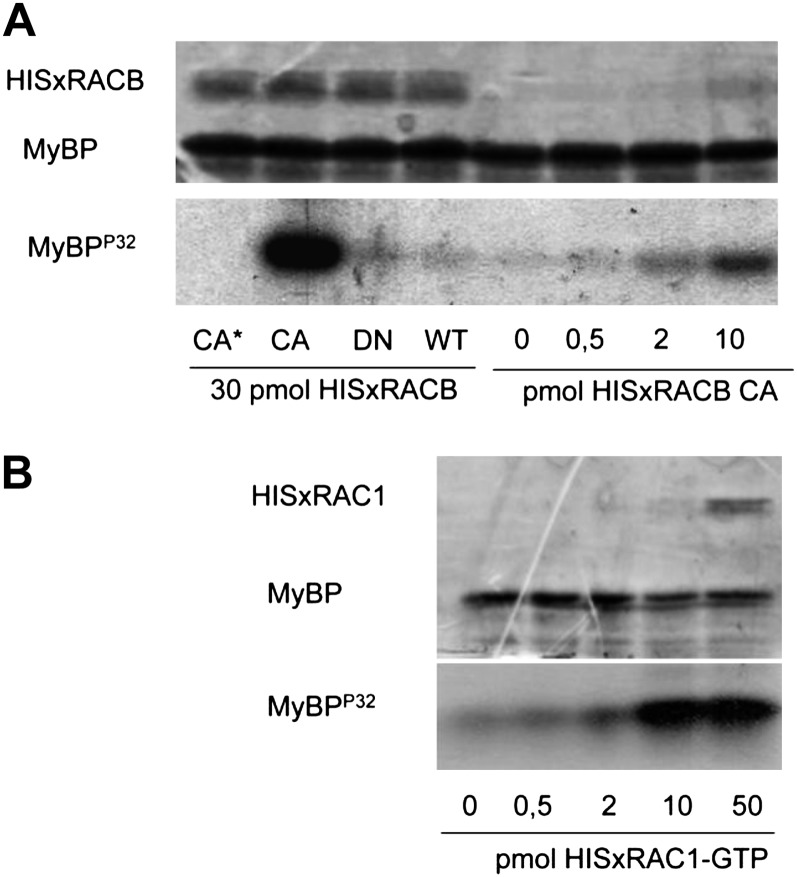
It was previously described that the in vitro activity of Arabidopsis and M. truncatula kinases belonging to the RLCK class VIA is dependent on the presence of active ROPs (Dorjgotov et al., 2009). Therefore, we tested whether the barley RLCK VIA-like kinase HvRBK1 has similar biochemical properties. We investigated the ability of the recombinant HvRBK1 protein to phosphorylate the myelin basic protein in vitro in the presence and absence of HvROP GTPases (HvRACB and HvRAC1, respectively) in various conformations or concentrations (Fig. 3). Activity of HvRBK1 was considerably increased in the presence of the CA HvRACB mutant in a concentration-dependent way. Under the same conditions, the wild-type or DN forms of HvRACB had no significant effect on the kinase activity (Fig. 3A). The effect of GTP-loaded HvRAC1 on the in vitro activity of HvRBK1 was also tested, and a ROP concentration-dependent increase in activity was evident in these assays (Fig. 3B).
Figure 3.
Active HvRACB/HvRAC1 GTPases increase the activity of HvRBK1 in vitro. The in vitro myelin basic protein (MyBP) phosphorylating activity of HvRBK1 is shown in the presence of purified HIS-tagged barley HvRACB (A) and HvRAC1 (B) GTP-binding proteins. A, HISxHvRACB was added to the kinase reaction in CA, DN, and wild-type (WT) forms (30 pmol each). As a negative control (CA*), the same amount of the CA HvRACB GTPase was also added to a reaction mixture not containing the kinase protein (CA*). Moreover, kinase reactions were carried out in the presence of various CA HvRACB GTPase amounts (0–10 pmol). Protein loading is shown by Coomassie Brilliant Blue staining of the proteins in the kinase reaction after their separation in a polyacrylamide gel (top image), whereas kinase activity is demonstrated by the autoradiographic detection of radioactive MyBPP32 in the same gel (bottom image). B, HISxHvRAC1 was added to the kinase reaction at the indicated concentrations after loading the wild-type protein with GTP. The top image shows the result of the CBB staining and the bottom one the autoradiography of the gel used for the separation of proteins in the kinase reactions.
HvROPs Recruit HvRBK1 to the Cell Periphery for Protein-Protein Interaction
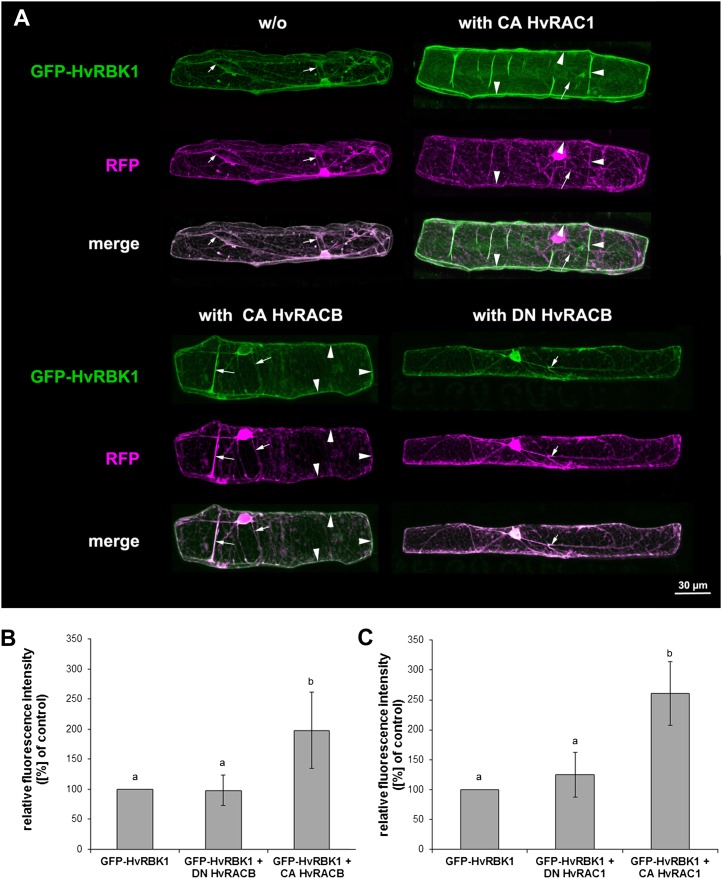
To see where HvRBK1 might interact with barley HvROPs in the cell, we fused the GFP to the N terminus of the protein. Confocal laser scanning microscopy (CLSM) localized GFP-HvRBK1 in the cytoplasm and nucleoplasm when transiently expressed in barley epidermal cells after biolistic transformation (Fig. 4A). The subcellular localization of GFP-HvRBK1 changed upon coexpression of the CA HvRACB or CA HvRAC1 but not upon coexpression of DN HvRACB taken as a control that does not bind HvRBK1 (Figs. 2 and 4A). Both barley CA HvROPs led to the accumulation of GFP-HvRBK1 at the cell periphery. Translocation to the cell periphery was partial when CA HvRACB was coexpressed and complete when CA HvRAC1 was coexpressed (Fig. 4A, Supplemental Fig. S2). Quantitative measurement of fluorescence intensities of GFP-HvRBK1 supported that CA HvRACB and CA HvRAC1 but not DN versions recruit HvRBK1 to the cell periphery (Figs. 4, B and C). Strength of translocation largely reflected the subcellular localization of the respective CA HvROP proteins at the cell periphery as indicated by imaging GFP-CA HvRACB or GFP-CA HvRAC1, respectively (Supplemental Fig. S3; see also Schultheiss et al., 2003).
Figure 4.
Recruitment of GFP-HvRBK1 by HvROPs in barley epidermal cells. A, Subcellular localization of GFP-HvRBK1 in epidermal cells of barley. A, Confocal laser-scanning micrographs of barley epidermal cell expressing GFP-HvRBK1 (green) and RFP (magenta). Soluble RFP was cotransformed as marker for cytoplasmic and nuclear localization. White color in the merged channels demonstrates similar localization of GFP-HvRBK1 and RFP in cytoplasmic strands (arrows). GFP-HvRBK1 was expressed alone (−/−) or together with the unlabeled HvROPs CA HvRACB, CA HvRAC1, or DN HvRACB. Coexpression of GFP-HvRBK1 and CA HvRACB or CA HvRAC1 results in recruitment of GFP-HvRBK1 to the cell periphery/plasma membrane (arrowheads) as demonstrated by color separation and the green cell periphery in the fluorescent merged pictures. DN HvRACB does not alter cytoplasmic localization of GFP-HvRBK1. Pictures are maximum projections of 20 to 30 optical sections at 2-μm increments. B and C, Fluorescence intensity of GFP-HvRBK1 at the cell periphery is greatly modified upon coexpression of different HvROPs. To quantify recruitment of GFP-HvRBK1 by CA HvRACB and CA HvRAC1, both proteins were coexpressed with GFP-HvRBK1 and mCherry as a marker for cytoplasmic and nuclear localization. As negative control, CA HvROP variants were replaced by DN HvRACB or DN HvRAC1. Additionally, the GFP-HvRBK1 was expressed with mCherry and empty vector as control. Mean pixel intensity was measured at the cell periphery and normalized against mCherry pixel intensity in the nucleus. Columns show means of three independent experiments with 95% confidence intervals as error bars. Intensities were significantly different between the CA HvRACB or CA HvRAC1 expressing cells versus the respective controls after ANOVA (Tukey test, P < 0.05, as indicated by different letters in the figure).
The measurement of fluorescence resonance energy transfer (FRET) further supported interaction of HvRBK1 and HvRACB in planta. We generated fusion protein constructs with cyan fluorescent protein (CFP) and yellow fluorescent protein (YFP) for acceptor photobleaching. We coexpressed HvRBK1-YFP with either CFP-CA HvRACB or CFP-DN HvRACB and bleached the acceptor HvRBK1-YFP at the periclinal cell periphery facing the surface of the leaf epidermis. This led to significant fluorescent enhancement of CFP-CA HvRACB but not of CFP-DN HvRACB used as donors (Table I). This corroborated results from the YTH and the in planta recruitment assays. Activated HvRACB may thus recruit HvRBK1 by direct protein interaction at the periphery of barley epidermal cells.
Table I. Efficiency of FRET between HvRBK1-YFP and variants of CFP-HvRACB.
Quantitative analysis of acceptor photobleaching FRET measurements. Data show the means of three independent experiments. Seven to 10 cells were analyzed in each experiment. FRET efficiency was significantly elevated in cells transformed with HvRBK1-YFP and CFP-CA HvRACB compared with cells cotransformed with HvRBK1-YFP and CFP-DN HvRACB (two-tailed Student’s t test; ***, P > 0.001). Errors represent the sds.
Barley HvRBK1 Is Expressed in Pathogen Response
To test whether barley HvRBK1 is expressed in response to B. graminis f. sp. hordei, we extracted total RNA from barley first leaves densely inoculated with conidia of B. graminis f. sp. hordei. Reverse transcription-PCR showed that transcripts of HvRBK1 slightly accumulated from 8 h after inoculation with B. graminis f. sp. hordei onward when compared with noninoculated controls from the same batch of plants (Supplemental Fig. S4). These data were confirmed by checking pathogen-responsive expression of HvRBK1 in publically available transcriptome data at the database PLEXDB (Wise et al., 2007). We identified the Genechip probe set contig14061_at as the one that represents the sequence of HvRBK1. Expression profiles of the microarray dataset BB10, which contains transcription profiling of barley plants differing at MLA1 and MLA6 powdery mildew resistance loci (Meng et al., 2009), revealed a slightly increased expression of HvRBK1 in all investigated plant genotypes inoculated with powdery mildew, from 8 h after inoculation onward. According to that, expression of HvRBK1 increases when barley is challenged by both virulent and avirulent B. graminis f. sp. hordei.
Barley HvRBK1 Influences MT Stability and Fungal Penetration
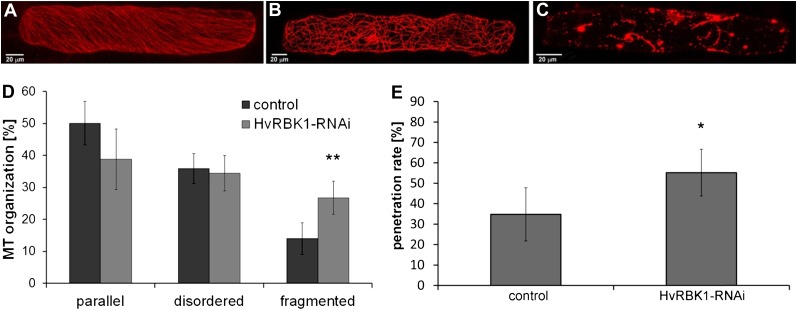
The MT cytoskeleton can be regulated via ROP signaling and has key functions in plant defense against invading pathogens. To test whether HvRBK1 influences the cytoskeleton, the MT arrangement was investigated upon transient-induced gene silencing (TIGS) (Douchkov et al., 2005) of HvRBK1. For this, the TIGS construct pIPKTA30N-HvRBK1 was generated and cotransformed with pGY-1-RFP-HvMAGAP-Cter as a reliable red fluorescent protein (RFP) MT marker in barley (Hoefle et al., 2011). Off-target analysis using the Si-Fi program (http://labtools.ipk-gatersleben.de/; Nowara et al., 2010) indicated that HvRBK1 may be the only target for this TIGS construct in the barley transcriptome. Microscopic evaluation was performed by confocal laser-scanning microscopy 48 h after bombardment. Three types of MT arrangement were distinguished: well-ordered parallel arrangement (Fig. 5A), disordered/randomized arrangement (Fig. 5B), and fragmented MTs (Fig. 5C). In repeated independent experiments, TIGS of HvRBK1 significantly enhanced the frequency of cells with fragmented MTs in comparison to the empty vector control (Fig. 5D). The marker RFP-HvMAGAP1-Cter represents the MT-associating domain of HvMAGAP1 fused to RFP. It was selected as a MT marker because it lacks any ROP interaction domain and thus may not interfere with ROP signaling in the assays (Hoefle et al., 2011). We alternatively used full-length RFP-HvMAGAP1 or DsRED-MBD, a DsRED fusion of the MT-associated protein4 MT-binding domain (Marc et al., 1998). With both alternative MT markers, silencing of HvRBK1 caused similar effects, leading to more cells with fragmented MTs (Supplemental Fig. S5). We then used the same TIGS construct to assess a potential function of HvRBK1 in interaction with B. graminis f. sp. hordei. TIGS of HvRBK1 rendered barley more susceptible to penetration by virulent B. graminis f. sp. hordei in single epidermal cells after biolistic transformation. The penetration success of B. graminis f. sp. hordei increased from 35% in control cells to 55% in cells bombarded with the TIGS construct of HvRBK1 (Fig. 5E).
Figure 5.
Influences of TIGS of HvRBK1 on fungal penetration success and MT organization in epidermal cells of barley. Epidermal cells were transiently transformed with the MT marker RFP-HvMAGAP1 to image typical MT arrays in epidermal cells. A, Parallel arrangement of MTs. B, Disordered/randomized arrangement of MTs. C, Fragmented MTs. D, Frequencies of cells falling into the categories of different MT arrays (A–C). Columns represent means of four independent experiments (50 cells for each plasmid combination were investigated per experiment), with significantly more fragmented MTs after TIGS of HvRBK1. We counted MT arrays after projections of 20 to 25 optical sections at 2-μm increments with the MT marker RFP-HvMAGAP1-Cter. Similar results were obtained with two alternative MT markers (Supplemental Fig. S5). E, TIGS of HvRBK1, when compared with empty TIGS vector control, led to enhanced fungal penetration rate as indicated by more haustoria-containing cells per total cells attacked by B. graminis f. sp. hordei. Columns show the mean of five independent biological experiments. Error bars show sd of the mean (two-sided Student’s t test; *, P < 0.05 and **, P < 0.01). [See online article for color version of this figure.]
DISCUSSION
ROP binding kinases of class RLCK VIA are candidates for downstream effector proteins in plant ROP signaling (Molendijk et al., 2008; Dorjgotov et al., 2009). This hypothesis is supported by our data showing that ROPs bind an RLCK VIA-like barley protein HvRBK1 in yeast and in planta and are capable of increasing its activity in vitro. TIGS of barley HvRBK1 led to enhanced susceptibility to powdery mildew, suggesting a function of this kinase in interaction with the biotrophic fungal leaf pathogen. Data establish a function of the barley ROP binding kinase HvRBK1 in basal resistance to powdery mildew and further support an important function of ROP signaling in plant-microbe interactions.
We identified HvRBK1 in nonbiased YTH screenings using HvRACB or HvRAC1 proteins as baits. YTH assays with full-length HvRBK1 supported that it can interact with HvRAC1 and the powdery mildew susceptibility factor HvRACB (Schultheiss et al., 2002; Hoefle et al., 2011). CA forms of HvRACB and HvRAC1 were able to change subcellular localization of HvRBK1. CA HvRAC1 completely and CA HvRACB partially recruited HvRBK1 to the cell periphery. Differences in the strength of recruitment may be explained by the fact that CA HvRAC1 is more exclusively associated with the cell periphery than CA HvRACB, of which a portion is often visible in the cytoplasm and nucleoplasm when fused to GFP (Schultheiss et al., 2003; Supplemental Fig. S3). Hence, subcellular localization of HvRBK1 seems to mirror that of the coexpressed activated barley ROP. The recruitment experiments support that HvRBK1 is bound by ROPs in planta. FRET experiments further added direct evidence for the in planta interaction of activated HvRACB and HvRBK1 (Table I).
The capability for the phosphorylation of an artificial substrate, myelin basic protein, by purified HvRBK1 confirmed that HvRBK1 is an active kinase. The constitutively active but not the DN mutant or the wild-type form of HvRACB significantly enhanced kinase activity. HvRAC1-GTP also enhanced the kinase activity in vitro in a concentration-dependent way. Similarly, Medicago and Arabidopsis RLCK VIA2 kinases exhibit active ROP-dependent activity in vitro (Dorjgotov et al., 2009). Therefore, the regulation of the activity of RLCK VIA kinases by active ROPs might be a general feature in plants.
Based on the YTH assay and in planta and in vitro experiments, HvRBK1 can be considered as a potential ROP-effector kinase that may be involved in HvRACB/HvRAC1-dependent downstream signaling in barley. The fact that CA HvRACB but not DN HvRACB activated the kinase in vitro and recruited GFP-HvRBK1 to the cell periphery supports the idea that active ROPs signal through HvRBK1 for downstream events. Possibly HvRBK1 resides in the cytoplasm when ROPs are inactive. Activation of ROPs might then recruit HvRBK1 to the cell periphery where downstream substrates of HvRBK1 might be phosphorylated for local regulation. The physiological substrates of plant RLCK VIA kinases are actually unknown. Phosphomimetic mutations in ROPs alter their function and interaction with upstream plant-specific ROP nucleotide exchanger proteins (Fodor-Dunai et al., 2011). However, thus far, there is no evidence that ROPs themselves are substrates of RLCK VIA kinases.
Considering the involvement of the barley ROPs in plant-fungal pathogen interactions (Schultheiss et al., 2002; Pathuri et al., 2008; Hoefle et al., 2011), as well as the elevated expression of the HvRBK1 gene in infected leaves, it was reasonable to suppose a function of HvRBK1 in pathogenesis. It was unexpected, however, that barley HvRBK1 appeared to be required for basal resistance rather than susceptibility to powdery mildew because of strong evidence that barley ROP signaling is involved in susceptibility to penetration by the barley powdery mildew fungus (Schultheiss et al., 2002, 2008; Pathuri et al., 2008; Hoefle et al., 2011). The fact that ROP signaling proteins are involved in both susceptibility and basal resistance in plants would make it an attractive target for a fungal virulence strategy. A function of HvRBK1 in basal resistance could be explained by a negative feedback regulation of the susceptibility factor HvRACB by HvRBK1. Absence of the kinase then would lead to enhanced activation of downstream events, which are independent of HvRBK1. Alternatively, HvRBK1 might be activated by ROPs that function in pathogen defense rather than in susceptibility. HvRBK1-interacting HvRAC1 has a potential role in defense. Expression of CA HvRAC1 enhances callose deposition and oxidative defense at the sites of attack from B. graminis f. sp. hordei. However, this is not sufficient to cause resistance, and the expression of CA HvRAC1 in barley even weakens basal penetration resistance to B. graminis f. sp. hordei. By contrast, the CA HvRAC1 genotypes show enhanced basal penetration resistance to the rice blast fungus Magnaporthe oryza (Pathuri et al., 2008). Barley HvRAC1 is highly similar to rice OsRAC1 (Schultheiss et al., 2003), which is a key factor of plant immunity, including resistance to rice blast, and of pathogen-triggered cell death (Ono et al., 2001; Chen et al., 2010a, 2010b; Trusov et al., 2010). OsRAC1 is involved in immunity triggered by nonspecific pathogen-associated molecular patterns and by race-specific virulence effectors (Chen et al., 2010a; Kawano et al., 2010). Together, plant ROPs and their interactors are pivotal factors in both susceptibility and immunity to diverse fungal pathogens.
MTs reorganize upon attack from B. graminis f. sp. hordei in barley epidermal cells (Baluska et al., 1995; Hoefle et al., 2011), and MT depolymerizing drugs weaken penetration resistance of barley coleoptiles to nonadapted powdery mildew fungi (Kobayashi et al., 1997). A polarized reorganization of the MTs correlates with basal resistance to fungal penetration. This is partially regulated by HvMAGAP1 that supports polarization of MTs under fungal attack (Hoefle et al., 2011). The fact that HvMAGAP1 fulfills a ROP-antagonistic function in association with MTs supports the hypothesis that MTs themselves are targets of ROP signaling (Hoefle et al., 2011; Mucha et al., 2011). Transient silencing of HvRBK1 weakened stability of MTs and supported fungal penetration (Fig. 5). This is consistent with a function of MTs in building penetration barriers against B. graminis f. sp. hordei. In Arabidopsis, ROP-mediated MT organization involves different ROPs that have partially antagonistic functions in the development of epidermal pavement cells (Fu et al., 2005, 2009; Xu et al., 2010). One may therefore speculate that different barley ROP pathways can also have MT destabilizing or organizing functions to which HvRBK1 contributes. Taken together, barley HvROPs, HvMAGAP1, and HvRBK1 may function in MT dynamics in the interaction with invading fungal pathogens. Parasitic B. graminis f. sp. hordei, however, might take advantage of this by manipulating ROP signaling in barley for either suppression of basal penetration defense or for getting support from the host when forming the haustorial complex within an intact barley cell (Hoefle et al., 2011). In such a scenario, HvRBK1 might play a role in penetration defense by supporting MT stability. However, our data do not exclude that a virulent fungus could also make use of HvRBK1, depending on the spatiotemporal pattern of ROP activity during fungal invasion.
MATERIALS AND METHODS
Plant Growth, Pathogens, and Inoculation Conditions
Barley (Hordeum vulgare) plants of the cultivar Golden Promise were grown in a growth chamber at 18°C with 60% relative humidity and a photoperiod of 16 h with 150 μmol m−2 s−1. Blumeria graminis f. sp. hordei, race A6 was maintained on Golden Promise under the above-described conditions.
In transient transformation experiments, detached primary leaf segments of barley were placed on 0.5% (w/v) H2O-agar 7 d after germination and inoculated with >100 conidia mm−2.
Isolation of HvRBK1
The complete coding sequence of HvRBK1 was amplified by 5′-RACE from a barley cDNA pool with the 5′/3′-RACE Kit, 2nd Generation (Roche). From total RNA, 1.5 μg were transcribed into first-strand cDNA via the gene-specific primer SP1 5′-GCGACTCCAAGCGCGATATTG-3′. The isolated single-strand cDNA-containing mix was purified with the High Pure PCR Product Purification Kit (Roche). Purified single-strand cDNA was used for poly(A) tailing of the 3′-end by a terminal transferase and subsequent amplification of the HvRBK1 coding sequence according to the manufacturer’s instructions. The HvRBK1 sequence was amplified on the dA-tailed cDNA with an oligo(dT)-anchor primer and a second gene-specific primer, SP2 5′-CATTTGGGTGGTTTACGTGC-3′. In a second nested PCR with a PCR-anchor primer and a third gene-specific primer, SP3 5′-GAACTGAACCTGTCAGTGGC-3′, the specific HvRBK1 5′-RACE product was obtained and ligated into the pGEM-T-vector (Qiagen) and sequenced.
YTH Screening and Targeted YTH Assay
YTH screening and transformation of yeast (Saccharomyces cerevisiae) was performed according to the yeast protocols handbook and the Matchmaker library construction and screening kits manual (Clontech). The YTH screening for HvRACB and HvRAC1 interaction partners is described in detail in Hoefle et al. (2011). For the targeted YTH assay, HvRBK1 was fused in the YTH vector pGADT7 with the GAL4 activation domain. Transformed yeast cells were selected on synthetic dextrose medium lacking Leu and Trp. Selection of yeast expressing interacting proteins was performed on synthetic dextrose medium lacking Leu, Trp, His, and adenine.
TIGS
Transient transformation of 7-d-old barley leaves of cultivar Golden Promise was performed as described earlier (Douchkov et al., 2005; Eichmann et al., 2010) with the PDS-1000/He System (Bio-Rad Laboratories) plant transformation gun with hepta-adapter. The RNAi construct for TIGS pIPKTA30N-HvRBK1 was produced by blunt end insertion of a 370-bp (bp 636–1,006 of the coding sequence) fragment of HvRBK1 in antisense orientation into the Gateway compatible entry vector pIPKTA38. Then the cDNA fragment was transferred as inverted repeats into the pIPKTA30N destination vector by a Gateway clonase reaction (Douchkov et al., 2005). Seven micrograms per shot of the RNAi construct pIPKTA30N-HvRBK1, the empty vector pIPKTA30N, and 3.5 μg of the reporter plasmid pGY-1-GFP were used in TIGS experiments.
Protein Localization and Protein-Protein Interaction in Planta
For localization studies, leaves of Golden Promise were transiently transformed with GFP-HvRBK1 fusion constructs under control of the 35S promoter via particle bombardment. N-terminal fusion constructs of HvRBK1 with GFP were achieved by insertion of the coding sequence in frame with GFP lacking the stop codon into the expression vector pGY-1 by the BamHI and SalI restriction sites. Soluble RFP, or mCherry in pGY-1, under the control of the P35S promoter was cotransformed as transformation marker. Each shot delivered 1 μg of the fusion construct and 0.5 μg of the transformation marker. In transient coexpression experiments, each shot delivered 1 μg of GFP-HvRBK1 fusion construct together with 1.1 μg of pGY-1-CAHvRACB or 1.1 μg of pGY1-CAHvRAC1. Leaves were inspected 24 h after bombardment by CLSM. Pictures were generated by sequential scanning to avoid channel crosstalk. GFP was excited by a 488-nm laser line and detected at 500 to 550 nm, whereas RFP was excited by a 561-nm laser line and detected at 571 to 610 nm.
Interaction of CFP-HvRACB and HvRBK1-YFP in planta was verified by FRET analysis. A C-terminal fusion of HvRBK1 with YFP was achieved by amplification of HvRBK1 with the primer pair HvKinFW 5′-AACCCGGGATGAAACTAAGGGAGTATTTCC-3′ and HvKinREV 5′-AACCCGGGGACACTGCTCCAACGC-3′ introducing SmaI restriction sites and insertion into the SmaI restriction sites of the pGY-1 vector containing YFP. The construction of CFP-CA HvRACB and CFP-DN HvRACB was described in Hoefle et al. (2011). Leaves of 7-d-old barley plants were cotransformed by particle bombardment with 1.2 μg of pGY-1-CFP-CAHvRACB or pGY-1-CFP-DNHvRACB together with 1.2 μg of pGY-1-HvRBK1-YFP. FRET analysis was performed 24 h after bombardment by the acceptor photobleaching method using the Leica Application Suite, Advanced Fluorescence 1.8.0 software (Leica Microsystems). For description of the FRET analysis, see Hoefle et al. (2011).
For subcellular fluorescence intensity measurements, leaves of barley were transiently transformed via particle bombardment with soluble mCherry (pGY1-mCherry) as cytoplasmic and nucleoplasmic marker, the GFP-HvRBK1 fusion construct, and respective constructs with the CA or DN variants of HvRACB and HvRAC1 under control of the 35S promoter. As control, mCherry and the GFP-HvRBK1 construct were cotransformed. Leaves were analyzed by CLSM 24 h after bombardment. All cells within an experiment were scanned with the same microscope settings. Mean pixel intensities were measured in the red channel (mCherry) in the nucleus and in the green channel (GFP-HvRBK1) at the cell periphery. The values of the peripheral intensities were normalized against the values of the nucleus and relative intensities compared with the control were calculated and plotted.
Analysis of MT Organization
To evaluate MT arrays in cells, leaves of Golden Promise were transiently transformed via particle bombardment with the HvRBK1 RNAi construct pIPKTA30N-HvRBK1 and a GFP expression construct as transformation marker (as described above). To observe the MT dynamics, the cells were also cotransformed with pGY1-RFP-HvMAGAP1 expressing the MT-associating barley HvMAGAP1 as an RFP-fusion protein. Alternatively, cells were cotransformed with the MT marker DsRED-MBD (Marc et al., 1998) or with the RFP fused C-terminal MT-associating part of HvMAGAP1 (Hoefle et al., 2011). The leaves were evaluated 48 h after bombardment by CSLM as described above. Each cell was distributed in one of the three categories of MT organization: parallel, disordered, or fragmented.
Gene Expression Analysis
Total RNA from leaves of 3-week-old barley plants was extracted from frozen plant material using TRIzol reagent (Invitrogen). From each sample, 1 μg of total RNA was reverse transcribed into cDNA using the QuantiTect reverse transcription kit (Qiagen). For semiquantitative two-step reverse transcription-PCR reactions, 1 μg of cDNA from each sample was used. To monitor differences in the initial template amounts, PCR reactions were stopped in the exponential phase. The UBIQUITIN transcript of barley was amplified in parallel as quality and quantity control with specific primers Ubi-5′ 5′-ACCCTCGCCGACTACAACAT-3′ and Ubi-3′ 5′-CAGTAGTGGCGGTCGAAGTG-3′. To amplify the HvRBK1 transcript, the primer pair Kinasefisch 5′-GCCATGAAACTAAGGGAGTAT-3′ and wt26SP3 5′-GAACTGAACCTGTCAGTGGC-3′ was used. As control for normal proliferation of the powdery mildew fungus, transcripts of the β-subunit of MTs were amplified with fungus-specific primers Bgh_beta-tub_F 5′-TCTGCCATTTTCCGCGGTAA-3′ and Bgh_beta-tub_R 5′-CGTTGCTTACTTCCTCTGGA-3′.
Kinase Activity Measurements
The cDNA clones of the barley proteins have been inserted into bacterial expression vectors in order to allow the purification of 6x-His-tagged (6xHIS) proteins from bacterial cultures. The vectors used were pET28a (for HvRBK1), pET28b (for HvRacB) from Novagen (Merck KGaA) and pQE-70 (for HvRac1) from Qiagen. For recombinant protein production, the ArcticExpress (DE3)RIL competent cells were used according to the supplier’s advice (Agilent Technologies). Protein purification was achieved using HisSelect Sepharose (Sigma-Aldrich) as described elsewhere (Dorjgotov et al., 2009, Fodor-Dunai et al., 2011) and the removal of contaminating chaperons derived from the ArcticExpress(DE3)RIL competent cells according to Joseph and Andreotti (2008). For the in vitro kinase assays, the reaction mix was set up as 1 pmol of purified 6xHIS kinase, 0,5-50 pmol of purified 6xHIS GTPase, 20 mm of Tris-HCl, pH = 7.6, 5 mm of MgCl2, 50 mm of NaCl, 1 mm of DTT, 10 μm of ATP, 0.2 MBq [γ-32P] of ATP, and 0.25 μg/μL of myelin basic protein. The reactions were stopped by 5 μL of 5× SDS loading buffer after 60 min at room temperature. Proteins were separated on SDS-polyacrylamide gels that were stained by Coomassie Brilliant Blue, dried, and exposed to x-ray films using standard methods.
The HvRAC1 protein was loaded by GTP after using EDTA for nucleotide removal as described by Rodriguez-Viciana and McCormick (2006).
Sequence data from this article can be found in the GenBank data libraries under the following accession numbers: barley, HvRBK1 (HE611049), HvRACB (AJ344223), HvRACD (AJ439334), HvRAC1 (AJ518933), HvRAC3 (AJ518932), HvROP6 (AJ439333), HvMAGAP1 (AK371854), and HvUBIQUITIN (M60175); Arabidopsis (Arabidopsis thaliana), AtROP6 (At4g35020, NP_829654), AtPERK1 (NP_189098, At3g24550), AtRLCKVIA1 (NP_001078762, At5g57670), AtRLCKVIA2 (AtRRK1, AAT99800, At2g188900), AtRLCKVIA3 (AAO63452, At5g65530), AtRLCKVIA4 (AtRBK1, AED91559, At5g10520), AtRLCKVIA5 (AtRRK2, AED94033, At5g35960), AtRLCKVIA6 (AtRBK2, NP_187165, At3g05140), and AtRLCKVIA7 (AAO64835, At5g18910); rice (Oryza sativa), OsPERK1 (BAD45880) and Medicago truncatula: MtRRK1 (FM886833) and MtRRK2 (FM886834); and Medicago sativa, MsROP6 (CAI84892).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Genomic structure of the HvRBK1 gene and the comparison of the deduced amino acid sequence of HvRBK1 to related proteins.
Supplemental Figure S2. Quantification of GFP-HvRBK1 signal intensity in cells coexpressing different versions of HvROPs.
Supplemental Figure S3. Subcellular localization of GFP-CA HvRACB and GFP-CA HvRAC1.
Supplemental Figure S4. HvRBK1 expression pattern in the interaction of barley with B. graminis f. sp. hordei.
Supplemental Figure S5. Influences of TIGS of HvRBK1 on MT organization in epidermal cells of barley.
Supplementary Material
Acknowledgments
We thank Verena Klingl for outstanding technical assistance. We thank the barley sequencing consortium (GABI BARLEX) for providing data prior to publication.
Glossary
- CA
constitutively activated
- CRIB
Cdc42/Rac-interactive binding
- RIC
ROP-interactive CRIB motif-containing protein
- MT
microtubule
- YTH
yeast two hybrid
- DN
dominant negative
- CLSM
confocal laser scanningmicroscopy
- FRET
fluorescence resonance energy transfer
- CFP
cyan fluorescent protein
- YFP
yellow fluorescent protein
- RFP
red fluorescent protein
- TIGS
transient-induced gene silencing
References
- Baluska F, Bacigálová K, Oud JL, Hauskrecht M, Kubica G. (1995) Rapid reorganization of microtubular cytoskeleton accompanies early changes in nuclear ploidy and chromatin structure in postmitotic cells of barley leaves infected with powdery mildew. Protoplasma 185: 140–151 [Google Scholar]
- Berken A, Wittinghofer A. (2008) Structure and function of Rho-type molecular switches in plants. Plant Physiol Biochem 46: 380–393 [DOI] [PubMed] [Google Scholar]
- Bustelo XR, Sauzeau V, Berenjeno IM. (2007) GTP-binding proteins of the Rho/Rac family: regulation, effectors and functions in vivo. Bioessays 29: 356–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Hamada S, Fujiwara M, Zhu T, Thao NP, Wong HL, Krishna P, Ueda T, Kaku H, Shibuya N, et al. (2010a) The Hop/Sti1-Hsp90 chaperone complex facilitates the maturation and transport of a PAMP receptor in rice innate immunity. Cell Host Microbe 7: 185–196 [DOI] [PubMed] [Google Scholar]
- Chen L, Shiotani K, Togashi T, Miki D, Aoyama M, Wong HL, Kawasaki T, Shimamoto K. (2010b) Analysis of the Rac/Rop small GTPase family in rice: expression, subcellular localization and role in disease resistance. Plant Cell Physiol 51: 585–595 [DOI] [PubMed] [Google Scholar]
- Denhardt DT. (1996) Signal-transducing protein phosphorylation cascades mediated by Ras/Rho proteins in the mammalian cell: the potential for multiplex signalling. Biochem J 318: 729–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorjgotov D, Jurca ME, Fodor-Dunai C, Szucs A, Otvös K, Klement E, Bíró J, Fehér A. (2009) Plant Rho-type (Rop) GTPase-dependent activation of receptor-like cytoplasmic kinases in vitro. FEBS Lett 583: 1175–1182 [DOI] [PubMed] [Google Scholar]
- Douchkov D, Nowara D, Zierold U, Schweizer P. (2005) A high-throughput gene-silencing system for the functional assessment of defense-related genes in barley epidermal cells. Mol Plant Microbe Interact 18: 755–761 [DOI] [PubMed] [Google Scholar]
- Eichmann R, Bischof M, Weis C, Shaw J, Lacomme C, Schweizer P, Duchkov D, Hensel G, Kumlehn J, Hückelhoven R. (2010) BAX INHIBITOR-1 is required for full susceptibility of barley to powdery mildew. Mol Plant Microbe Interact 23: 1217–1227 [DOI] [PubMed] [Google Scholar]
- Fodor-Dunai C, Fricke I, Potocký M, Dorjgotov D, Domoki M, Jurca ME, Otvös K, Zárský V, Berken A, Fehér A. (2011) The phosphomimetic mutation of an evolutionarily conserved serine residue affects the signaling properties of Rho of plants (ROPs). Plant J 66: 669–679 [DOI] [PubMed] [Google Scholar]
- Fu Y, Gu Y, Zheng Z, Wasteneys G, Yang Z. (2005) Arabidopsis interdigitating cell growth requires two antagonistic pathways with opposing action on cell morphogenesis. Cell 120: 687–700 [DOI] [PubMed] [Google Scholar]
- Fu Y, Xu T, Zhu L, Wen M, Yang Z. (2009) A ROP GTPase signaling pathway controls cortical microtubule ordering and cell expansion in Arabidopsis. Curr Biol 19: 1827–1832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoefle C, Huesmann C, Schultheiss H, Börnke F, Hensel G, Kumlehn J, Hückelhoven R. (2011) A barley ROP GTPase ACTIVATING PROTEIN associates with microtubules and regulates entry of the barley powdery mildew fungus into leaf epidermal cells. Plant Cell 23: 2422–2439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hückelhoven R, Panstruga R. (2011) Cell biology of the plant-powdery mildew interaction. Curr Opin Plant Biol 14: 738–746 [DOI] [PubMed] [Google Scholar]
- Huesmann C, Hoefle C, Hückelhoven R. (2011) ROPGAPs of Arabidopsis limit susceptibility to powdery mildew. Plant Signal Behav 6: 1691–1694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphries JA, Vejlupkova Z, Luo A, Meeley RB, Sylvester AW, Fowler JE, Smith LG. (2011) ROP GTPases act with the receptor-like protein PAN1 to polarize asymmetric cell division in maize. Plant Cell 23: 2273–2284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph RE, Andreotti AH. (2008) Bacterial expression and purification of interleukin-2 tyrosine kinase: single step separation of the chaperonin impurity. Protein Expr Purif 60: 194–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurca ME, Bottka S, Fehér A. (2008) Characterization of a family of Arabidopsis receptor-like cytoplasmic kinases (RLCK class VI). Plant Cell Rep 27: 739–748 [DOI] [PubMed] [Google Scholar]
- Kawano Y, Akamatsu A, Hayashi K, Housen Y, Okuda J, Yao A, Nakashima A, Takahashi H, Yoshida H, Wong HL, et al. (2010) Activation of a Rac GTPase by the NLR family disease resistance protein Pit plays a critical role in rice innate immunity. Cell Host Microbe 7: 362–375 [DOI] [PubMed] [Google Scholar]
- Kobayashi Y, Kobayashi I, Funaki Y, Fujimoto S, Takemoto T, Kunoh H. (1997) Dynamic reorganization of microfilaments and microtubules is necessary for the expression of non-host resistance in barley coleoptile cells. Plant J 11: 525–537 [Google Scholar]
- Löcke S, Fricke I, Mucha E, Humpert ML, Berken A. (2010) Interactions in the pollen-specific receptor-like kinases-containing signaling network. Eur J Cell Biol 89: 917–923 [DOI] [PubMed] [Google Scholar]
- Marc J, Granger CL, Brincat J, Fisher DD, Kao Th, McCubbin AG, Cyr RJ. (1998) A GFP-MAP4 reporter gene for visualizing cortical microtubule rearrangements in living epidermal cells. Plant Cell 10: 1927–1940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Y, Moscou MJ, Wise RP. (2009) Blufensin1 negatively impacts basal defense in response to barley powdery mildew. Plant Physiol 149: 271–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molendijk AJ, Ruperti B, Singh MK, Dovzhenko A, Ditengou FA, Milia M, Westphal L, Rosahl S, Soellick TR, Uhrig J, et al. (2008) A cysteine-rich receptor-like kinase NCRK and a pathogen-induced protein kinase RBK1 are Rop GTPase interactors. Plant J 53: 909–923 [DOI] [PubMed] [Google Scholar]
- Mucha E, Fricke I, Schaefer A, Wittinghofer A, Berken A. (2011) Rho proteins of plants: functional cycle and regulation of cytoskeletal dynamics. Eur J Cell Biol 90: 934–943 [DOI] [PubMed] [Google Scholar]
- Nibau C, Wu HM, Cheung AY. (2006) RAC/ROP GTPases: ‘hubs’ for signal integration and diversification in plants. Trends Plant Sci 11: 309–315 [DOI] [PubMed] [Google Scholar]
- Nowara D, Gay A, Lacomme C, Shaw J, Ridout C, Douchkov D, Hensel G, Kumlehn J, Schweizer P. (2010) HIGS: host-induced gene silencing in the obligate biotrophic fungal pathogen Blumeria graminis. Plant Cell 22: 3130–3141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono E, Wong HL, Kawasaki T, Hasegawa M, Kodama O, Shimamoto K. (2001) Essential role of the small GTPase Rac in disease resistance of rice. Proc Natl Acad Sci USA 98: 759–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opalski KS, Schultheiss H, Kogel KH, Hückelhoven R. (2005) The receptor-like MLO protein and the RAC/ROP family G-protein RACB modulate actin reorganization in barley attacked by the biotrophic powdery mildew fungus Blumeria graminis f.sp. hordei. Plant J 41: 291–303 [DOI] [PubMed] [Google Scholar]
- Pathuri IP, Zellerhoff N, Schaffrath U, Hensel G, Kumlehn J, Kogel KH, Eichmann R, Hückelhoven R. (2008) Constitutively activated barley ROPs modulate epidermal cell size, defense reactions and interactions with fungal leaf pathogens. Plant Cell Rep 27: 1877–1887 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Viciana P, McCormick F. (2006) Characterization of interactions between ras family GTPases and their effectors. Methods Enzymol 407: 187–194 [DOI] [PubMed] [Google Scholar]
- Schultheiss H, Dechert C, Kogel KH, Hückelhoven R. (2002) A small GTP-binding host protein is required for entry of powdery mildew fungus into epidermal cells of barley. Plant Physiol 128: 1447–1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultheiss H, Dechert C, Kogel KH, Hückelhoven R. (2003) Functional analysis of barley RAC/ROP G-protein family members in susceptibility to the powdery mildew fungus. Plant J 36: 589–601 [DOI] [PubMed] [Google Scholar]
- Schultheiss H, Preuss J, Pircher T, Eichmann R, Hückelhoven R. (2008) Barley RIC171 interacts with RACB in planta and supports entry of the powdery mildew fungus. Cell Microbiol 10: 1815–1826 [DOI] [PubMed] [Google Scholar]
- Sorek N, Gutman O, Bar E, Abu-Abied M, Feng X, Running MP, Lewinsohn E, Ori N, Sadot E, Henis YI, et al. (2011) Differential effects of prenylation and s-acylation on type I and II ROPS membrane interaction and function. Plant Physiol 155: 706–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorek N, Poraty L, Sternberg H, Bar E, Lewinsohn E, Yalovsky S. (2007) Activation status-coupled transient S acylation determines membrane partitioning of a plant Rho-related GTPase. Mol Cell Biol 27: 2144–2154 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Stamatakis A, Hoover P, Rougemont J. (2008) A rapid bootstrap algorithm for the RAxML Web servers. Syst Biol 57: 758–771 [DOI] [PubMed] [Google Scholar]
- Tena G, Boudsocq M, Sheen J. (2011) Protein kinase signaling networks in plant innate immunity. Curr Opin Plant Biol 14: 519–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trusov Y, Jordá L, Molina A, Botella JR. (2010) G proteins and plant innate immunity. In Yalovsky S, Baluška F, Jones A, eds, Integrated G Proteins Signaling in Plants. Series: Signaling and Communication in Plants, Ed 1. Springer-Verlag, Berlin, Heidelberg, pp 221–250 [Google Scholar]
- Wise RP, Caldo RA, Hong L, Shen L, Cannon EK, Dickerson JA. (2007) BarleyBase/PLEXdb. Methods Mol Biol 406: 347–363 [DOI] [PubMed] [Google Scholar]
- Wu G, Gu Y, Li S, Yang Z. (2001) A genome-wide analysis of Arabidopsis Rop-interactive CRIB motif-containing proteins that act as Rop GTPase targets. Plant Cell 13: 2841–2856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Li H, Yang Z. (2000) Arabidopsis RopGAPs are a novel family of rho GTPase-activating proteins that require the Cdc42/Rac-interactive binding motif for rop-specific GTPase stimulation. Plant Physiol 124: 1625–1636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T, Wen M, Nagawa S, Fu Y, Chen JG, Wu MJ, Perrot-Rechenmann C, Friml J, Jones AM, Yang Z. (2010) Cell surface- and rho GTPase-based auxin signaling controls cellular interdigitation in Arabidopsis. Cell 143: 99–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yalovsky S, Bloch D, Sorek N, Kost B. (2008) Regulation of membrane trafficking, cytoskeleton dynamics, and cell polarity by ROP/RAC GTPases. Plant Physiol 147: 1527–1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, McCormick S. (2007) A distinct mechanism regulating a pollen-specific guanine nucleotide exchange factor for the small GTPase Rop in Arabidopsis thaliana. Proc Natl Acad Sci USA 104: 18830–18835 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.