Abstract
The syncytium is a unique plant root organ whose differentiation is induced by plant-parasitic cyst nematodes to create a source of nourishment. Syncytium formation involves the redifferentiation and fusion of hundreds of root cells. The underlying regulatory networks that control this unique change of plant cell fate are not understood. Here, we report that a strong down-regulation of Arabidopsis (Arabidopsis thaliana) microRNA396 (miR396) in cells giving rise to the syncytium coincides with the initiation of the syncytial induction/formation phase and that specific miR396 up-regulation in the developed syncytium marks the beginning of the maintenance phase, when no new cells are incorporated into the syncytium. In addition, our results show that miR396 in fact has a role in the transition from one phase to the other. Expression modulations of miR396 and its Growth-Regulating Factor (GRF) target genes resulted in reduced syncytium size and arrested nematode development. Furthermore, genome-wide expression profiling revealed that the miR396-GRF regulatory system can alter the expression of 44% of the more than 7,000 genes reported to change expression in the Arabidopsis syncytium. Thus, miR396 represents a key regulator for the reprogramming of root cells. As such, this regulatory unit represents a powerful molecular target for the parasitic animal to modulate plant cells and force them into novel developmental pathways.
Pathogens alter their hosts’ biology to ensure successful infection. Such modifications range from moderate to extensive, and in the case of plant pathogens, few infections result in more dramatic changes than those of sedentary endoparasitic nematodes, which include the cyst nematodes (Heterodera and Globodera spp.). Cyst nematodes are obligate parasitic roundworms that induce the formation of novel plant cell types that are associated in a unique feeding organ, the syncytium. After root penetration, the infective and migratory second-stage Heterodera cyst nematode juveniles (J2) become sedentary and induce the formation of a syncytium in the vascular cylinder and then mature through three molts into the third-stage (J3), fourth-stage (J4), and adult male or female. This nematode development is enabled by feeding from the syncytium (Sijmons et al., 1994).
Syncytium development can be separated into an induction/formation phase followed by a maintenance phase. Induction/formation involves effector-mediated communication between the nematode and an initial feeding cell leading to cytoplasmic and nuclear changes followed by successive fusions of the cells surrounding the initial feeding cell (Sobczak and Golinowski, 2009). Through continuous cell fusions, syncytium formation and enlargement continue and then stop once the syncytium is mature. During the ensuing maintenance phase, no additional cells are incorporated and syncytial cells have undergone their developmental changes and now are fully engaged in maintaining the syncytium function of feeding the developing nematode (Sobczak and Golinowski, 2009). However, the exact mechanisms of how plant cells are directed to reenter a novel developmental program and how this process is ended are unknown.
Syncytium formation encompasses the reprogramming of differentiated root cells, and these redifferentiations are accompanied and mediated by massive gene expression changes, which have been documented in diverse research approaches using soybean (Glycine max) and the soybean cyst nematode Heterodera glycines (Alkharouf et al., 2006; Ithal et al., 2007; Klink et al., 2009) and probably most extensively in Arabidopsis (Arabidopsis thaliana) infected by the sugar beet cyst nematode Heterodera schachtii (Szakasits et al., 2009). These gene expression changes clearly require powerful mechanisms of concerted regulation, and the existence of major regulatory checkpoints can be hypothesized, but none have been documented to date.
The finding that microRNAs (miRNAs) and other small RNAs are involved in plant developmental and biological processes that are similar to those implicated in syncytium formation and function raised the specter that small RNAs, including miRNAs, could play fundamental roles in the etiology of the syncytium (Hewezi et al., 2008a). miRNAs initially have been shown to be involved in the regulation of a variety of plant developmental processes, including phase transition, hormone synthesis and signaling, pattern formation, and morphogenesis (Chen, 2009). Recent studies indicate that miRNAs and small endogenous RNAs also are involved in biotic stress responses in plants (Katiyar-Agarwal et al., 2006, 2007; Navarro et al., 2006; Fahlgren et al., 2007; Lu et al., 2007; He et al., 2008; Hewezi et al., 2008a; Pandey et al., 2008; Li et al., 2010). Also, consistent with a role of small RNAs in the development of plant diseases, Arabidopsis mutants deficient in small interfering RNA or miRNA biogenesis affected plant susceptibility to bacteria (Navarro et al., 2008) and the sugar beet cyst nematode H. schachtii (Hewezi et al., 2008a). Collectively, these emerging data indicate that small RNA-mediated gene regulation, not surprisingly, is a fundamental mechanism in plant-pathogen interactions.
Despite these advances, little is known about the molecular mechanisms controlling cell differentiation and development in the nematode-induced syncytium. In this context, we previously identified microRNA396 (miR396) as a potentially interesting regulator of target gene expression during nematode parasitism (Hewezi et al., 2008a). The miR396 family, miR396a and miR396b, governs the expression of seven Growth-Regulating Factor (GRF) genes (Jones-Rhoades and Bartel, 2004). The GRF gene family in Arabidopsis is known to act in a functionally redundant fashion to positively control cell proliferation and size in leaves (Kim et al., 2003; Kim and Kende, 2004; Horiguchi et al., 2005; Kim and Lee, 2006). Consistent with the fact that miR396 acts as a negative regulator of GRF gene expression, overexpression of miR396 negatively impacted cell proliferation in leaves and meristem size (Liu et al., 2009; Rodriguez et al., 2010). However, the roles of the miR396/GRF regulatory module in controlling developmental events during plant-pathogen interactions or in roots are completely unknown. In this study, we demonstrate that miR396 is differentially expressed in the syncytium and that this miRNA functions as an important master switch in syncytium formation.
RESULTS
miR396a and miR396b Have Similar Spatial Expression Patterns
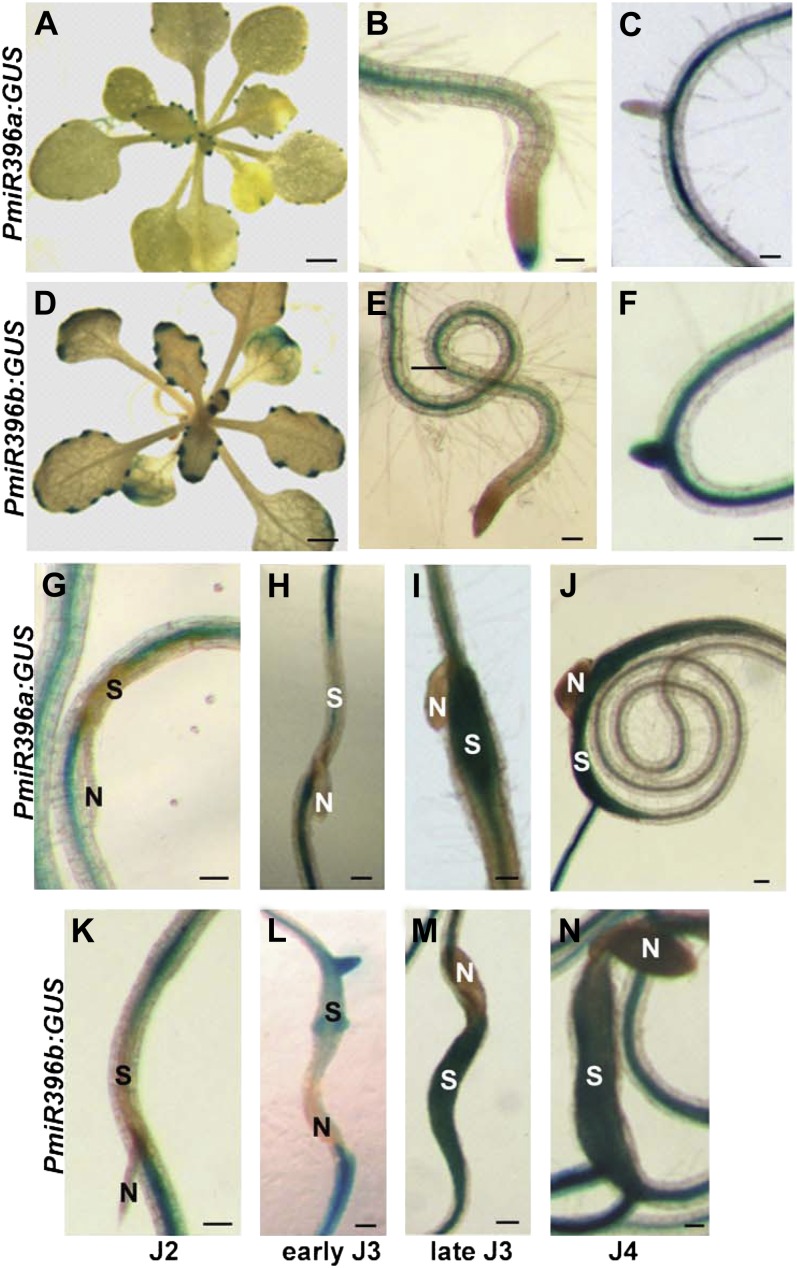
In Arabidopsis, miR396 is encoded by two genes, miR396a (AT2G10606) and miR396b (AT5G35407), and regulates the expression of seven of the nine Arabidopsis growth-regulating transcription factor genes (GRF1–4 and GRF7–9) that share the miR396-binding site (Jones-Rhoades and Bartel, 2004). To examine tissue-specific expression patterns of the two miR396 genes, we generated transgenic plants expressing constructs containing the regions upstream of miR396 precursor sequences fused to the GUS reporter gene (PmiR396a:GUS and PmiR396b:GUS). GUS staining of at least four independent lines for each construct revealed that the miR396a and miR396b promoters have very similar spatial expression patterns, both in leaf and root tissues (Fig. 1, A–F). Despite the fact that miR396a and miR396b have similar spatial expression patterns, GUS staining of PmiR396b:GUS lines was in general much stronger than that of PmiR396a:GUS lines. This was confirmed by real-time reverse transcription (RT)-PCR analysis of miR396 precursors (pri-miR396) in roots of 2-week-old ecotype Columbia-0 (Col-0) plants. We found an mRNA abundance of pri-miR396b about 70-fold higher than that of pri-miR396a.
Figure 1.
Histochemical localization of GUS activity directed by miR396 promoters. A to F, Spatial expression patterns of miR396a and miR396b. A to C, GUS staining of miR396a:GUS plants showing expression in leaf hydathodes (A), vascular root tissues and the root cap (B), and absence of expression in the emerging lateral roots (C). D to F, GUS staining of miR396b:GUS plants showing expression in leaf hydathodes (D), vascular root tissues (E), and emerging lateral roots (F). G to N, Promoter activity of miR396a and miR396b during H. schachtii infection. Time-course experiments compared the expression of PmiR396a:GUS (G–J) and PmiR396b:GUS (K–N) transgenic plants at the second-stage (J2), early and late third-stage (J3), and fourth-stage (J4) juvenile time points. N indicates nematode, and S indicates syncytium. Bars = 2 mm in A and D and 100 µm in B, C, and E to N.
miR396 Expression Changes Delineate the Syncytium Induction/Formation Phase
We previously observed marked RNA abundance changes for miR396 following root infection by H. schachtii (Hewezi et al., 2008a); thus, it was of great interest to identify the locations of these altered miR396 expressions. We explored this question by analyzing the promoter activities of miR396a and miR396b at different time points after H. schachtii infection using our transgenic Arabidopsis GUS lines. Most remarkably, the activities of the promoters of both miR396a and miR396b were strongly down-regulated in developing syncytia at early time points of H. schachtii infection (i.e. the parasitic J2 and early J3 stages; Fig. 1, G, H, K, and L). Maybe more interestingly, after this initial early phase, the promoters of both miR396a and miR396b became very active in the syncytia of late J3 and J4 nematodes (Fig. 1, I, J, M, and N), which is the developmental stage when syncytia reach their maximum size. Thus, miR396 expression changes delineate the syncytium induction/formation phase, whereby a down-regulation marks the beginning of syncytium induction/formation and a subsequent strong up-regulation coincides with the transition to the maintenance phase.
GRF1 and GRF3 Are miR396 Targets during H. schachtii Infection
Obviously, the immediate question became whether any GRF target genes were coexpressed in the syncytium, which would make them subject to miR396 regulation and could potentially suggest a role for GRFs in syncytium induction/formation. To determine which GRF genes could be targeted by miR396 in roots, we measured mRNA steady-state levels in root tissues of 10-d-old seedlings of all nine GRF genes by quantitative real-time RT-PCR. GRF1, GRF2, and GRF3 showed by far the highest root expression levels (Supplemental Fig. S1). This observation implies that miR396 has the potential to be active in posttranscriptional gene regulation also in Arabidopsis roots, which has not been documented to date. In order to determine if any GRFs are subject to posttranscriptional regulation in the syncytium, we generated transgenic plants expressing promoter/GUS fusion constructs of the seven GRF genes (GRF1–4 and GRF7–9) that are targeted by miR396. The promoter activities of these plants were assayed at different time points after H. schachtii infection. In these assays, the promoters of GRF4, GRF7, and GRF8 were completely inactive in syncytia during all time points (Fig. 2, D–F). The promoter of GRF2 showed very weak syncytial GUS activity at late J3 and J4 stages, with no GUS activity detectable in early syncytia during the J2 and early J3 infective stages (Fig. 2B). Similarly, only weak and very infrequent syncytial GUS activity could be detected during the J3 stage for GRF9 (Fig. 2G). However, the GRF1 and GRF3 promoters showed very strong GUS activity in syncytia at all stages, with only the GRF3 promoter becoming inactive at the J4 stage (Fig. 2, A and C). The fact that only the GRF1 and GRF3 promoters show strong GUS activity in the syncytium coinciding with those of the miR396 promoters, particularly a strong expression when the negative miR396 regulators are remarkably decreased, indicates that GRF1 and GRF3 could be targets of miR396 during nematode infection. On the other hand, GRF2, GRF4, GRF7, GRF8, and GRF9 clearly do not work in concert with GRF1 and GRF3 during the early induction/formation period of the syncytium.
Figure 2.
Promoter activity of seven GRF genes during H. schachtii infection. Time-course experiments compared the expression of PGRF1:GUS (A), PGRF2:rGRF2-GUS (B), PGRF3:GUS (C), PGRF4:GUS (D), PGRF7:GUS (E), PGRF8:GUS (F), and PGRF9:GUS (G) transgenic plants at the sedentary second-stage (J2), early and late third-stage (J3), and fourth-stage (J4) juvenile time points. N indicates nematode, and S indicates syncytium. Bars = 100 µm.
GRF1 and GRF3 Are Posttranscriptionally Regulated by miR396 during Nematode Infection
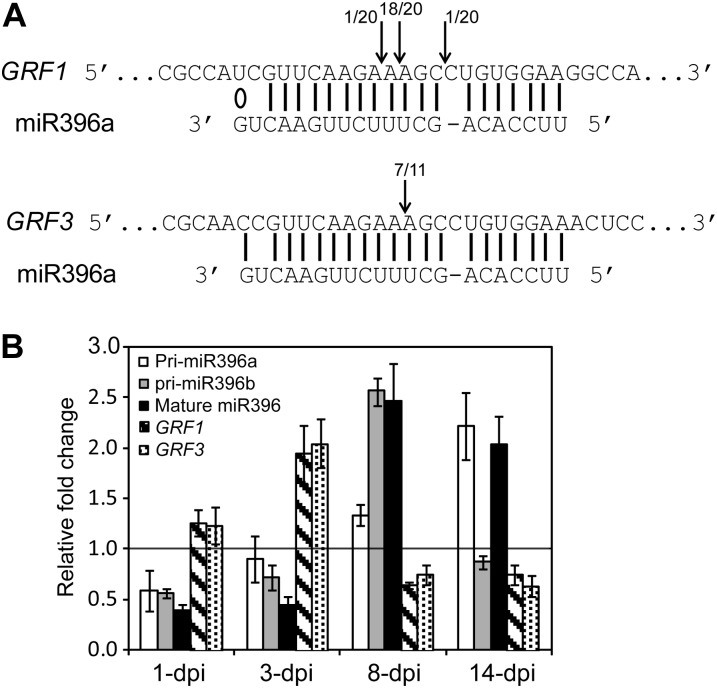
The coexpression of miR396 with its GRF1 and GRF3 target genes in the syncytium suggests a posttranscriptional GRF expression regulation following nematode infection. To confirm that GRF1 and GRF3 genes are cleaved by miR396 in root tissues, total RNA was isolated from wild-type (Col-0) plants at 8 d post H. schachtii infection and used for the 5′ RNA ligase-mediated (RLM)-RACE assay. Sequence analysis of cloned RLM-RACE products demonstrated that cleavage of GRF1 and GRF3 transcripts occurs predominantly between nucleotides 10 and 11 of the miR396 pairing region (Fig. 3A), confirming previous results obtained by Jones-Rhoades and Bartel (2004). These data provide direct evidence that miR396 mediates GRF1/3 mRNA cleavage in roots and during H. schachtii infection. To examine whether the posttranscriptional regulation of GRF1 and GRF3 by miR396 impacts the steady-state mRNA levels of GRF1 and GRF3, we quantified the abundances of miR396 precursors (pri-miR396a and pri-miR396b) and mature miRNAs (miR396) along with GRF1 and GRF3 mRNA levels in response to H. schachtii infection over time using quantitative (q)PCR. Ten-day-old wild-type Arabidopsis seedlings were inoculated with H. schachtii, and root tissues were collected from inoculated and noninoculated control plants at 1, 3, 8, and 14 d post inoculation (dpi) for RNA isolation and cDNA synthesis. Data from three independent experiments revealed that the accumulation of pri-miR396a, pri-miR396b, and mature miR396 was down-regulated in H. schachtii-inoculated roots at the 1- and 3-dpi time points when compared with noninoculated roots (Fig. 3B), confirming the down-regulation of the miR396a/b promoters in developing syncytia (Fig. 1, G and K). Consistent with a posttranscriptional regulation of GRF1 and GRF3, this down-regulation was accompanied by elevated mRNA abundance for both GRF1 and GRF3 (Fig. 3B), most probably as a result of decreased cleavage of GRF1 and GRF3 mRNA by miR396. In contrast, at 8 and 14 dpi, miRNA precursors and mature miR396 were elevated more than 2-fold in infected roots (Fig. 3B). Again consistent with a posttranscriptional regulation of GRF1 and GRF3, this miR396 increase was correlated with low transcript abundance of GRF1 and GRF3 (Fig. 3B). In other words, despite the fact that the nematode strongly induced the activity of the GRF1 and GRF3 promoters in the syncytium (Fig. 2, A and C), GRF1 and GRF3 steady-state mRNA levels decreased at the time when syncytial expression of miR396 increased (Fig. 3B).
Figure 3.
Posttranscriptional regulation of GRF1 and GRF3 by miR396 in response to H. schachtii infection. A, Mapping of the cleavage sites of GRF1 and GRF3 using 5′ RLM-RACE. Arrows indicate the positions of cleavage sites in the sequenced 5′ RACE clones. The number of clones corresponding to each cleavage site is indicated. Perfect base pairing between miR396 and GRF1 and GRF3 mRNA is shown by vertical lines, and G:U wobble pairing is indicated by a circle. B, miR396 mediates the down-regulation of GRF1 and GRF3 expression in response to H. schachtii infection. The expression level of pri-miR396a, pri-miR396b, mature miR396, GRF1, and GRF3 was measured by qPCR in wild-type (Col-0) root tissues. Infected and noninfected tissues were collected at 1, 3, 8, and 14 dpi. Down-regulation of miR396 at 1 and 3 dpi was associated with up-regulation of both GRF1 and GRF3. In contrast, up-regulation of miR396 at 8 and 14 dpi was associated with low transcript accumulation of GRF1 and GRF3. U6 small nuclear RNA was used as an internal control to normalize the expression levels of miR396, whereas Actin8 was used to normalize the expression levels of GRF1 and GRF3. The relative fold-change values represent changes of the expression levels in infected tissues relative to noninfected controls. Data are averages of three biologically independent experiments ± se.
Overexpression of miR396 and Altered GRF Expression Modulate Nematode Susceptibility
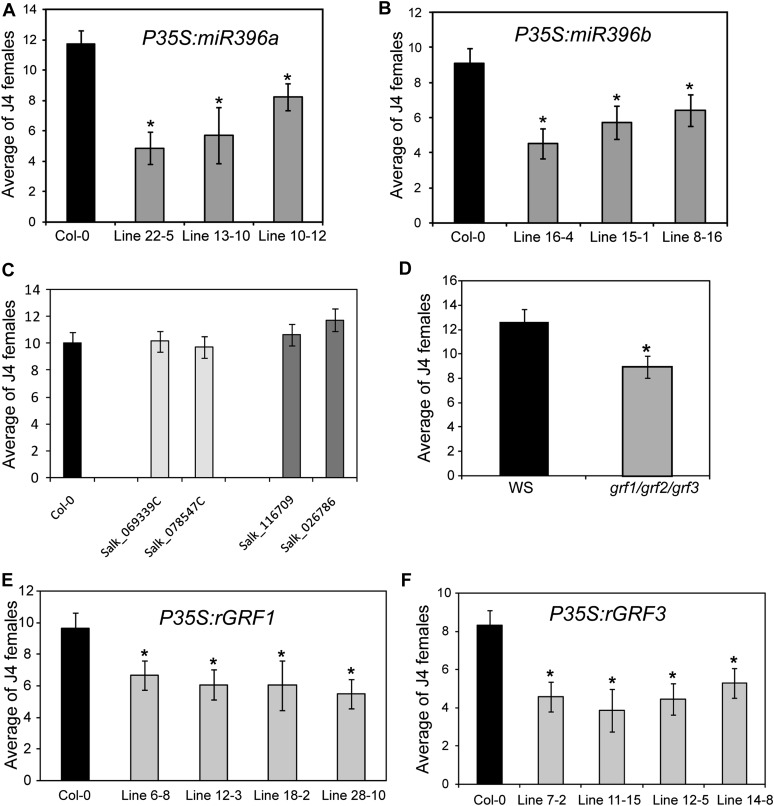
Our findings that miR396 expression is significantly down-regulated at the onset of the syncytium induction/formation phase, which results in elevated syncytial expression of GRF1 and GRF3, and that there is a coordinated posttranscriptional repression of GRF1 and GRF3 by miR396 at the onset of the maintenance phase imply a possible function of this regulatory module in syncytium formation and the transition to syncytium maintenance. If the coordinated regulation of miR396 and GRF1 and GRF3 is critical for correct cell fate specification and differentiation in the developing syncytium, as has been shown in leaves and meristems, it is reasonable to hypothesize that any changes in this equilibrium will result in reduced cyst nematode parasitic success. As a first test of this hypothesis, we expressed the primary miRNA sequences of both miR396a and miR396b in Arabidopsis under the control of the 35S promoter. Independent homozygous T3 lines expressing between 2- and 5-fold higher miRNA levels relative to the wild type were identified (Supplemental Fig. S2, A and B). We first determined whether miR396 overexpression resulted in the expected decreased mRNA abundance of GRF target genes by using qPCR to quantify the mRNA levels of the GRF gene family in the roots of transgenic miR396b overexpression plants (line 16-4). As expected, GRF mRNA abundances were reduced as a consequence of this manipulation (Supplemental Fig. S3). Subsequently, 10-d-old plants were inoculated with H. schachtii J2 nematodes, and the number of adult females was counted 3 weeks after inoculation for both the transgenic lines and the wild-type control and used to quantify plant susceptibility. A remarkable effect of miR396 overexpression on nematode susceptibility was observed. All transgenic lines overexpressing miR396a (Fig. 4A) or miR396b (Fig. 4B) were dramatically less susceptible than the wild-type control, as shown by the statistically significant reduction in the number of females per root system.
Figure 4.
Nematode susceptibility assays of miR396 overexpression lines and GRF mutants. A and B, Nematode susceptibility assays of miR396 overexpression lines. Transgenic plants overexpressing miR396a (A) or miR396b (B) exhibited reduced susceptibility to H. schachtii. Homozygous T3 lines overexpressing miR396a (lines 22-5, 13-10, and 10-12) or miR396b (lines 16-4, 15-1, and 8-16) were planted on modified Knop’s medium, and 10-d-old seedlings were inoculated with approximately 200 surface-sterilized J2 H. schachtii nematodes. Three weeks after inoculation, the number of J4 female nematodes per root system was determined. Data are presented as means ± se (n = 20). Mean values significantly different from the wild type (Col-0) were determined by unadjusted paired t tests (P < 0.05) and are indicated by asterisks. Identical results were obtained from at least two independent experiments. The expression levels of miR396a and miR396b in the transgenic lines used in this analysis are provided in Supplemental Figure S2. C, Nematode susceptibility is not significantly altered in the grf1 or grf3 single mutant. The mutant alleles of grf1 (Salk_069339C and Salk_078547C) and gfr3 (Salk_116709 and Salk_026786) along with wild-type Col-0 plants were planted on modified Knop’s medium and assayed for nematode susceptibility. No statistically significant differences between these mutant lines and the wild type were observed. Data are presented as means ± se (n = 20). Similar results were obtained from at least three independent experiments. D, The grf1/grf2/grf3 triple mutant exhibited reduced susceptibility to H. schachtii. Seeds of the grf1/grf2/grf3 triple mutant and the wild type (Ws) were planted on modified Knop’s medium and assayed for nematode susceptibility. Data are presented as means ± se (n = 20), and the statistically significant difference between the grf1/grf2/grf3 mutant and the wild type (Ws) is denoted by the asterisk, as determined by unadjusted paired t tests (P < 0.05). Identical results were obtained from two independent experiments. E and F, Transgenic plants overexpressing rGRF1 (E) or rGRF3 (F) revealed reduced susceptibility to H. schachtii. Four independent homozygous T3 lines for each construct were assayed for nematode susceptibility. All lines showed significantly reduced susceptibility compared with wild-type plants. Data are presented as means ± se (n = 20). Mean values significantly different from the wild type (Col-0) were determined by unadjusted paired t tests (P < 0.05) and are indicated by asterisks. Identical results were obtained from at least two independent experiments. The expression levels of the GRF1 and GRF3 transgenes of all the transgenic lines used in this analysis are provided in Supplemental Figure S2.
It appeared most logical that this reduction of susceptibility in miR396 overexpression lines is mediated through a resultant down-regulation of GRFs, particularly GRF1 and GRF3. Therefore, we hypothesized that mutants of GRF1 and GRF3 (Supplemental Fig. S4) will phenocopy the decreased nematode susceptibility of miR396 overexpression lines. To test this hypothesis, we subjected grf1 and grf3 single mutants as well as the grf1/grf2/grf3 triple mutant of Kim et al. (2003) to the susceptibility assays described above. Single knockdown mutants of GRF1 and GRF3 exhibited small or no effects on nematode susceptibility (Fig. 4C), confirming the previously reported results (Kim et al., 2003) that GRF gene family members are functionally redundant. However, the grf1/grf2/grf3 triple knockout mutant (Kim et al., 2003) showed a statistically significant decrease in susceptibility to H. schachtii relative to the wild-type control (Fig. 4D), thereby phenocopying the miR396 overexpression lines.
Our expression data described above suggest that GRF1 and GRF3 regulation by miR396 is important for syncytium formation. Therefore, interfering with the posttranscriptional regulation of GRF1 and GRF3 should negatively influence syncytium formation and, thus, lead to reduced susceptibility. In order to test this hypothesis, we generated plants harboring miR396-resistant noncleavable variants of these two genes (P35S:rGRF1 and P35S:rGRF3; Supplemental Figs. S2, C and D, and S5). We tested homozygous T3 lines for each of these constructs in nematode susceptibility assays, and all tested lines exhibited significantly reduced susceptibility relative to wild-type plants (Fig. 4, E and F). These results strengthen our hypothesis and again firmly connect GRF transcription factors, particularly GRF1 and GRF3, to determining the outcome of the cyst nematode-Arabidopsis interaction.
Coordinated miR396-GRF1/3 Expression Is Required for Normal Root Development
Having identified that miR396 and GRF1 and GRF3 are highly expressed also in uninfected roots, it was of interest to determine if a perturbation of the miR396-GRF equilibrium also alters root development, as has been documented for shoot tissues (Rodriguez et al., 2010). We found that overexpression of miR396 or the GRF1 and GRF3 variants described above resulted in reproducible root length reductions (Fig. 5; Supplemental Fig. S6). These data show that the miR396/GRF regulatory module functions to maintain the expression equilibrium critical also for normal root development. Given the fact that GRF1 and GRF3 are the most abundantly expressed gene family members in roots, their roles appear most prominent in this developmental pathway.
Figure 5.
Overexpression of miR396 and the target genes GRF1 and GRF3 negatively impacts root development. A and B, Transgenic plants overexpressing miR396a (line 22-5; A) or miR396b (line 15-1; B) develop shorter roots than the wild type (Col-0). C, Transgenic plants overexpressing rGRF1 (line 18-2) or rGRF3 (line 11-15) develop shorter roots than the wild type (Col-0). Homozygous T3 plants were planted on modified Knop’s medium along with the wild type, and 10-d-old seedlings were photographed. Root length measurements are provided in Supplemental Figure S6.
miR396 and Its Target Genes GRF1 and GRF3 Control Syncytium Size and Nematode Development
In addition to merely determining the number of females that mature on the different Arabidopsis genotypes, it is of particular interest to elucidate when and how altered susceptibility phenotypes are established. Also, it was of interest to determine if the reduced root size that we observed for the transgenic lines was responsible for lower female counts. For this purpose, we determined the number of penetrating J2 nematodes at 4 dpi in the transgenic plants overexpressing miR396b or the resistant versions of GRF1 or GRF3 as well as in wild-type Arabidopsis. As shown in Figure 6A, the number of penetrating nematodes in the transgenic lines was not different from that of the wild type, indicating that the mode of action responsible for the reduced nematode susceptibility in these transgenic lines is post penetration and, thus, independent of root length/size.
Figure 6.
Overexpression of miR396, GRF1, or GRF3 negatively impacts syncytium size and nematode development. A, Reduced nematode susceptibility in transgenic plants overexpressing miR396, rGRF1, or rGRF3 is independent of root length. Three homozygous T3 lines overexpressing miR396b (line 16-4), rGRF1 (line 12-3), or rGRF3 (line 12-5) and showing significant decreases in root length were inoculated with J2 H. schachtii nematodes, and at 4 dpi the total number of penetrated nematodes was counted. Data obtained from two independent experiments showed no statistically significant differences between the transgenic lines and the wild-type control. Data are presented as means ± se (n = 20). B, Transgenic plants overexpressing miR396, rGRF1, or rGRF3 developed smaller syncytia than the wild type. Seedlings of the above-indicated lines along with the wild type (Col-0) were inoculated with J2 H. schachtii nematodes, and 2 weeks post inoculation at least 20 single nematode syncytia were randomly selected and measured. Data are presented as means ± se. Asterisks indicate statistically significance differences from wild-type plants at P < 0.05. C and D, Overexpression of miR396, rGRF1, or rGRF3 negatively impacts nematode development. Seeds of the above-indicated lines along with the wild type (Col-0) were planted, and seedlings were inoculated with J2 H. schachtii nematodes. After inoculation, the number of parasitic J2/J3 (C) and J4 (D) females was counted in the same plants. Data are presented as means ± se (n = 20). Asterisks indicate statistically significance differences from wild-type plants at P < 0.05.
Second, we measured syncytium size and quantified different nematode developmental stages at different assessment times. Two weeks post inoculation, we measured the size of fully formed syncytia in the transgenic plants mentioned above as well as in wild-type Arabidopsis. Interestingly, the syncytia formed in the transgenic lines were significantly smaller than those in the wild-type control (Fig. 6B). The average reduction in syncytium size was up to 33% in miR396 overexpression plants and 19% and 14% in the transgenic plants expressing rGRF1 and rGRF3, respectively. These results indicate that the mode of action responsible for the reduced susceptibility in the transgenic lines overexpressing miR396 or the target genes rGRF1 and rGRF3 is manifested during the formation phase of the syncytium (i.e. at early stages of parasitism).
To investigate whether the activity of miR396 and its target genes GRF1 and GRF3 have an effect on nematode development, we counted the number of parasitic J2/J3 nematodes at 7 dpi in the transgenic lines overexpressing miR396 or the target genes rGRF1 and rGRF3. The number of developing (i.e. already swollen) J2 and J3 nematodes was significantly reduced in these transgenic plants relative to the wild-type control (Fig. 6C), and the reduction ranged between 42% for miR396-overexpressing plants and 20% and 39% for the transgenic plants expressing rGRF1 and rGRF3, respectively. These reductions in nematode numbers were also evident when the number of J4 nematodes was counted at 21 dpi in the same plants (Fig. 6D). In fact, the reduction percentages of these nematode stages were not significantly changed from the 7-dpi assessment. These data indicate that the reduced susceptibility of these transgenic lines is associated with early arrested nematode development before the J2/J3 stages, which again points to a mode of action during the early stages of parasitism when the syncytium is being formed.
Identification of Genes Regulated by GRF1 and GRF3 Using Microarray Analysis
Because both GRF1 and GRF3 function as transcription factors, identifying the genes regulated by these transcription factors will elucidate the pathways in which GRF1 and GRF3 function. To this end, we used Arabidopsis Affymetrix ATH1 GeneChips to compare the mRNA profiles of root tissues of the grf1/grf2/grf3 triple mutant and transgenic plants expressing rGRF1 (line 18-2) or rGRF3 (line 12-5) with those of the corresponding wild type (Col-0 or Wassilewskija [Ws]). We identified 3,944, 2,293, and 2,410 genes as differentially expressed in the grf1/grf2/grf3 triple mutant and rGRF1 and rGRF3 plants, respectively, at a false discovery rate of less than 5% and P < 0.05 (Fig. 7A; Supplemental Tables S1–S3). After eliminating duplicates among these three cohorts of differentially expressed genes, a total of 6,385 Arabidopsis genes changed expression as a result of our manipulations of GRFs.
Figure 7.
Functional classification of the differentially expressed genes identified in P35S:rGRF1, P35S:rGRF3, and grf1/grf2/grf3 mutants. A, Venn diagram showing overlaps between differentially expressed genes in P35S:rGRF1, P35S:rGRF3, and grf1/grf2/grf3 mutants when compared with the wild type. The total number of differentially expressed genes in each set is shown in parentheses. Gene identities are listed in Supplemental Tables S1 to S3. B and C, Gene Ontology categorization of the molecular functions (B) or the biological processes (C) of the genes regulated by GRF1 or GRF3. Gene identities used for this categorization are listed in Supplemental Tables S2 and S3. D, Venn diagrams showing overlaps between differentially expressed genes in the syncytium and those identified in P35S:rGRF1, P35S:rGRF3, and grf1/grf2/grf3 mutants. The total number of differentially expressed genes in each set is shown in parentheses. Gene identities are listed in Supplemental Table S4.
GRF1 and GRF3 Regulate Genes with Similar Functions
In addition to apparently targeting identical genes, careful examination of the putative function/annotation of the GRF1- and GRF3-regulated genes revealed that both transcription factors target genes with similar function or different members belonging to the same gene family. When classifying GRF-regulated genes into different groups by molecular function using the Gene Ontology categorization from The Arabidopsis Information Resource (http://www.Arabidopsis.org), we discovered a high proportion of genes associated with other enzyme activity, binding activity, hydrolase activity, transferase activity, and transcription factor activity (Fig. 7B) for both GRF1 and GRF3. When these genes were grouped by associated biological processes, the most abundant groups corresponded to metabolism and other cellular processes, while protein metabolism, response to stress, response to abiotic or biotic stimuli, and developmental processes also represented significant groups (Fig. 7C). When scrutinizing the GRF-regulated genes for gene family membership, different members coding for NAC (for NAM, ATAF1,2, CUC2) domain-containing proteins, AP2 domain-containing transcription factors, auxin-responsive proteins, MYB transcription factors, zinc finger family proteins, cytochrome P450 family proteins, glycosyl hydrolase family proteins, invertases, oxidoreductases, pectin esterase family proteins, peroxidases, nodulin family proteins, UDP-glucosyl transferase family proteins, ubiquitination-related proteins, Cys proteinases, and expansins were found to be similarly regulated by GRF1 and GRF3 (Supplemental Tables S2 and S3). These data provide strong evidence for a functional overlap between GRF1 and GRF3 in the regulation of gene expression. Furthermore, these data provide valuable insight into the molecular functions of GRF1 and GRF3 as transcriptional regulators.
A Key Role for GRF1 and GRF3 in Regulating Syncytial Gene Expression
Considering the fact that GRF1 and GRF3 change expression in the H. schachtii syncytium as a result of miR396 expression dynamics, one can postulate that a certain proportion of the differentially expressed genes identified in our microarray experiments should change expression actually in the syncytium and, in fact, be involved in syncytium induction/formation. Identifying such genes has been attempted in several research approaches, and the study of Szakasits et al. (2009) represents the most comprehensive such analyses in Arabidopsis infected with H. schachtii. Therefore, we next compared the GRF-regulated genes identified by us with the 7,225 genes differentially expressed in Arabidopsis syncytia reported by Szakasits et al. (2009). We found for rGRF1 1,131 genes (49.32%, χ2 = 346.13, P = 2.95E-77) and for rGRF3 1,165 genes (48.34%, χ2 = 325.27, P = 1.03E-72) as overlapping with the 7,225 syncytium-regulated genes (Fig. 7D). After eliminating duplicates between these two pools of regulated genes, a unique set of 1,965 genes remained, and these genes represent 27.2% (χ2 = 605.47, P = 1.08E-133) of the total number of syncytium-regulated genes (Fig. 7D). Furthermore, 2,073 genes overlapped between syncytium-regulated genes and those found to be differentially regulated in the grf1/grf2/grf3 triple mutant (Fig. 7D), which means that 28.7% (χ2 = 916.26, P = 2.87E-201) of the total number of syncytium-regulated genes change expression in the triple mutant. The 1,965 unique syncytial genes identified in rGRF1 and rGRF3 overexpression lines along with the 2,073 syncytial genes identified in the triple mutant make up a unigene set of 3,160 syncytial genes that change expression as a result of altered GRF expression (Fig. 7D; Supplemental Table S4). This number represents an astonishing 44% (χ2 = 1,234.13, P = 2.33E-270) of all syncytial genes reported by Szakasits et al. (2009). In other words, the modulations of GRFs performed by us account for almost half of the reported syncytial gene expression changes in Arabidopsis. Thus, GRFs play important roles in syncytium induction/formation. Considering that the GRFs in question change expression in the syncytium as a function of miR396, as we have shown above, this regulatory module represents a key regulator of syncytial gene expression changes.
DISCUSSION
The formation of functional syncytia must require a tightly fine-tuned coordination of multiple developmental and cellular processes to achieve the redifferentiation and fusion of hundreds of root cells into a functional new organ. The mechanisms and underlying regulatory networks that control the integration of these processes remain poorly understood. In this paper, we report on the biological role of miR396 in syncytium formation and function. In response to H. schachtii, both miR396a and miR396b genes are transcriptionally regulated in the syncytium. Similarly, we determined the expression characteristics of the seven GRF genes that are targeted by miR396 and established that, of these, only GRF1 and GRF3 can be considered major contributors to the outcome of the plant-cyst nematode interaction. Interestingly, miR396 and its target genes GRF1 and GRF3 showed opposite steady-state mRNA abundance patterns at the time of early-developing syncytia during the parasitic J2 and early J3 stages, when miR396 was down-regulated and GRF1 and GRF3 showed up-regulation. At later stages, we established that up-regulation of miR396 at 8 and 14 dpi causes a posttranscriptional down-regulation of GRF1 and GRF3 (Fig. 3B). miR396, therefore, has a stage-specific function in the spatial activation/restriction of GRF1 and GRF3 expression in the syncytium. The fact that miR396 up-regulation and GRF modulations lead to smaller syncytia and reduced susceptibilities shows that the coordinated regulation of miR396 and GRF1 and GRF3 is required for correct cell fate specification and differentiation in the developing syncytium. Our data suggest that maintenance of the homeostasis of miR396 and the target genes at specific threshold levels is critical for syncytium development. This suggestion is supported by our finding that down-regulation of GRFs through overexpression of miR396a/b, or overexpression of miR396-resistant versions of GRF1/GRF3, resulted in reduced nematode susceptibility.
The implication of miRNA/target gene regulatory mechanisms in the control of plant growth and differentiation has been explored for plant organs (Gutierrez et al., 2009; Wu et al., 2009; Marin et al., 2010). Here, we show that the miR396/GRF1 and GRF3 regulatory module is involved in the control of syncytium size and development. We established that transgenic plants overexpressing miR396b, rGRF1, or rGRF3 developed smaller feeding sites than those of the wild type. Our experiments determining penetration rates and assessing nematode development document that the reduced nematode susceptibility is manifested post penetration and is independent of root size. Furthermore, we document that a certain proportion of penetrated nematodes fail to develop and that this arrested development occurs very early on (i.e. when the reprogramming of root cells into syncytial cells is initiated). Together, these observations strongly suggest that the reduced susceptibilities of plants with altered miR396-GRF expression are due to an inability to develop mature and fully functional syncytia.
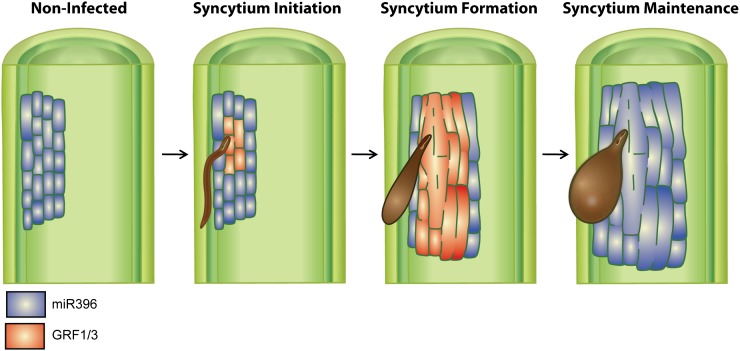
We propose that during the early stage of syncytium development, inactivation of miR396 activity in the syncytium increases GRF1 and GRF3 mRNA abundance to a threshold that enables these transcription factors to regulate gene expression reprogramming events that direct the differentiation and formation of the nematode feeding site (Fig. 8). Once the syncytium is established, the cyst nematode infection-mediated miR396 repression is removed. The resultant miR396 induction in the feeding site posttranscriptionally reduces the mRNA abundance of GRF1 and GRF3, thereby ending the induction/formation phase of the syncytium and leading syncytial cells to enter the maintenance phase (Fig. 8). In other words, the transcriptional regulation of miR396 controls a phase transition from a low-miR396/high-GRF1/3 expression during syncytium initiation to a high-miR396/low-GRF1/3 expression state leading the cells to enter the maintenance phase after the differentiation events have been completed. The opposite expression patterns of miR396 during the syncytium initiation/formation and maintenance stages are similar to those of Arabidopsis miR156 and miR172 during the juvenile-to-adult phase transition, where miR156 is expressed at high levels during shoot development and then decreases with time, while miR172 has an inverse expression pattern (Aukerman and Sakai, 2003; Wu and Poethig, 2006; Jung et al., 2007).
Figure 8.
The miR396-GRF1/3 regulatory module likely controls the phase transition from syncytium initiation/formation to maintenance stages. Under noninfected conditions, miR396 is highly expressed in vascular root tissues (highlighted in blue). Upon H. schachtii infection, miR396 is strongly down-regulated during syncytium initiation/formation phases, and this down-regulation increases GRF1 and GRF3 expression (highlighted in red) to a specific threshold that allows these transcription factors to regulate gene expression reprogramming events that could be required for the redifferentiation of mature root cells to form a very specialized and novel syncytial cell type. Once syncytium formation is completed, miR396 expression increases to high levels in the feeding site, which posttranscriptionally silences the expression of GRF1 and GRF3. Down-regulation of GRF1 and GRF3 effectively ends the induction/formation phase of the syncytium and conditions syncytial cells to enter the maintenance phase.
The Roles of GRF1 and GRF3 in Mediating Gene Expression in the Syncytium
Despite ongoing efforts to identify the biological processes regulated by GRFs during plant development, only a very limited number of target genes have been identified and characterized to date (Kim and Kende, 2004); thus, our microarray study addresses an important area. Functional classification of the differentially expressed genes identified in P35S:rGRF1 and P35S:rGRF3 plants revealed that genes coding for transcription factors or proteins with binding activity represent 43.3% and 40%, respectively (Fig. 7B), which documents a continuous amplification of the GRF response by targeting regulatory genes. Therefore, the enrichment of transcription factors belonging to zinc finger, Myb, WRKY, bHLH, AP2 domain-containing, CCAAT-binding, or NAC domain transcription factor families among the GRF1- or GRF3-regulated genes represents a powerful mechanism to trigger a massive signaling response to GRF1 or GRF3 expression. As a case in point, syncytium formation has to be associated with a modulation of host defense responses (Gheysen and Fenoll, 2002; Davis et al., 2004; Williamson and Kumar, 2006), and we found a number of genes involved in different aspects of plant defenses among the genes regulated by GRF1 or GRF3. Similarly, plant hormones, including auxin, have been implicated in syncytium development (Grunewald et al., 2009), and GRF1 or GRF3 appears to regulate a set of genes involved in hormone biosynthesis or the signaling pathways of auxin, brassinosteroids, cytokinins, ethylene, gibberellins, and jasmonates. Furthermore, cell wall modifications are obvious mechanisms of syncytium formation, and a high proportion of genes with cell wall-related functions also are enriched among the GRF-regulated target genes. In other words, GRF1 and GRF3 likely impact a wide spectrum of physiological processes associated with syncytium formation. This assessment becomes even more concrete when considering our finding that almost half of the GRF1- and GRF3-regulated genes were previously identified as changing expression in the syncytium (Szakasits et al., 2009). Collectively, these data provide the mechanistic basis for GRF1 and GRF3 to directly influence a variety of signaling and developmental pathways required to govern the redifferentiation of nematode-parasitized root cells into a functional new organ.
While it is remarkable to consider that a large percentage of the genes identified in our microarray experiments also change expression in the syncytium, as we would have surmised from the syncytium-localized GRF expression characteristics uncovered in this paper, the truly interesting finding is made when performing this analysis in the opposite direction. Not only are more than 49% of the GRF-regulated genes implicated in syncytium events, but more importantly, the expression of 44% of the 7,225 genes reported by Szakasits et al. (2009) to change expression in the Arabidopsis syncytium is altered by our GRF manipulations and, thus, by miR396. Consequently, almost half of the known syncytial gene expression events in Arabidopsis can be modulated by the miR396/GRF module as a single regulatory unit. No other known mechanism is able to exert the same powerful control over syncytial gene expression events.
When observing the complexity of cyst nematode-plant interactions, particularly the concerted regulation of syncytium formation, it always is fascinating to wonder how such complex events can be regulated by the parasite. How can the nematode possibly change so many aspects of cell biology at once? Here, we have shown that the exploitation of miR396 as a key regulator is one mechanism employed by the nematode. In other words, miR396 provides the nematode with a single molecular target to wield power over a substantial proportion of syncytium developmental events.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) wild-type Col-0 was used in all experiments except for the grf1/grf2/grf3 triple knockout mutant, which is in the Ws background (Kim et al., 2003). Plants were grown in long days (16 h of light/8 h of dark) at 23°C.
Plasmid Construction and Generation of Transgenic Arabidopsis Plants
For the miR396a and miR396b overexpression constructs, 205- and 189-bp genomic sequences containing the precursors of miR396a and miR396b, respectively, were amplified from Arabidopsis genomic DNA using gene-specific primers designed to create the BamHI and SacI restriction sites in the forward and reverse primers, respectively. For overexpression of the miR396-resistant forms of GRF1 and GRF3, we introduced mutations in the miR396-binding sites by three sequential PCR cycles using oligonucleotides containing mismatches in the miR396-binding site. The first round of PCR was performed to amplify the 5′ region containing the mutated miR396-binding site using GRF1-XbaI_F1 and GRF1_R1 primers for GRF1 or GRF3-XbaI_F1 and GRF3_R1 for GRF3. The second round of PCR was performed to amplify the 3′ region containing the mutated miR396-binding site using GRF1_F2 and GRF1-SacI_R2 primers for GRF1 or GRF3_F2 and GRF3-SalI_R2 for GRF3. The purified PCR products from the first and second reactions were used as a template to amplify the full length of GRF1 or GRF3 containing the mutated miR396-binding site using GRF1-XbaI_F1 and GRF1-SacI_R2 for GRF1 or GRF3-XbaI_F1 and GRF3-SalI_R2 for GRF3. PCR amplification was performed using the Expand High Fidelity plus PCR System (Roche) according to the manufacturer’s instructions. The PCR products were digested, gel purified, ligated into the binary vector pBI121, and verified by sequencing.
For the miR396a and miR396b promoter constructs, 2.245- and 2.307-kb fragments upstream of the precursor sequence of miR396a and miR396b, respectively, were amplified from Arabidopsis genomic DNA using PmiR396a-XbaI_F/PmiR396a-BamHI_R and PmiR396b-XbaI_F/PmiR396b-BamHI_R. For the promoter constructs of GRF1, GRF3, GRF4, GRF7, GRF8, and GRF9, 2.0-, 1,960-, 2,403-, 2,181-, 1,993-, and 1,471-kb fragments, respectively, upstream of the start codon were amplified from Arabidopsis genomic DNA using the primers listed in Supplemental Table S5. The purified PCR products were digested, gel purified, cloned into the corresponding restriction sites of binary vector pBI101, and confirmed by sequencing. Agrobacterium tumefaciens strain C58 was transformed with the binary plasmids by the freeze-thaw method and used to transform Arabidopsis wild-type Col-0 as described previously by Clough and Bent (1998). Transformed T1 plants were screened on Murashige and Skoog medium containing 50 mg L–1 kanamycin, and transgenic plants were identified. Homozygous T3 seeds were harvested from T2 lines after segregation analysis on Murashige and Skoog medium containing kanamycin. The primer sequences used for plasmid construction are listed in Supplemental Table S5.
Identification of T-DNA Mutants of GRF1 and GRF3
Two independent T-DNA insertional alleles of GRF1 (Salk_069339C and Salk_078547C) or GRF3 (Salk_026786 and Salk_116709) in the Col-0 background were obtained from the Salk T-DNA insertional mutant collection (Alonso et al., 2003). For details, see Supplemental Figure S4.
Histochemical Analysis of GUS Activities
The histochemical staining of GUS enzyme activity was performed according to Jefferson et al. (1987). Tissue samples were viewed using a Zeiss SV-11 microscope, and the images were captured using a Zeiss AxioCam MRc5 digital camera and then processed using Zeiss Axiovision software (release 4.8).
Nematode Infection Assay
Arabidopsis seeds were surface sterilized and planted in a random-block design on 12-well Falcon tissue culture plates (BD Biosciences) containing modified Knop’s medium (Sijmons et al., 1991) solidified with 0.8% Daishin agar (Research Products International Corp.). Plants were grown at 24°C under 16-h-light/8-h-dark conditions. Ten-day-old seedlings were inoculated with approximately 200 surface-sterilized J2 Heterodera schachtii nematodes per plant, as described previously by Baum et al. (2000). The inoculated plants were maintained under the same conditions described above for an additional 3 weeks before counting the early adult females. Mean values significantly different from that of the wild type were determined in a modified t test using the statistical software package SAS (P < 0.05).
Nematode Penetration Assay
The penetration rate of H. schachtii second-stage juveniles (J2) was performed in homozygous T3 lines overexpressing miR396b (line 16-4), rGRF1 (line 12-3), or rGRF3 (line 12-5) as well as the wild type (Col-0). The four lines were planted in a random-block design on modified Knop’s medium on 12-well culture plates as described above. Ten days after planting, each plant was inoculated with approximately 200 surface-sterilized J2 H. schachtii nematodes. Four days post inoculation, the number of penetrating nematodes in each root system was counted as described previously by Hewezi et al. (2008b). Each line was replicated 12 times, and two independent experiments were carried out. Average numbers of penetrating J2 nematodes were calculated, and statistically significant differences between the transgenic plants and the wild type were determined in a modified t test using the statistical software package SAS.
Nematode Development Assay
The development of H. schachtii parasitic-stage juveniles at 5 d post infection was compared with that of early adult females at 21 d post infection. The transgenic plants overexpressing miR396b (line 16-4), the resistant versions of GRF1 (line 12-3) and GRF3 (line 12-5), as well as wild-type Col-0 were planted in a random-block design on modified Knop’s medium on 12-well culture plates as described above. At 10 d, each plant was inoculated with 200 surface-sterilized J2 H. schachtii nematodes, and the plates were placed back in the incubator for an additional 5 d. Then, each well was examined for developing parasitic-stage juveniles under bright-field illumination using a Zeiss Axiovert 100 microscope. The number of parasitic-stage juveniles was counted for each well of each line, and the plates were placed back in the incubator for an additional 16 d. A 5-d time point was chosen so that a distinction could be made between those nematodes that were actively feeding (swollen) and those that were not feeding. At 21 d post infection, the plates were once again examined, this time with a Zeiss Stemi 2000 dissecting microscope, and the number of early adult females that developed on each line was counted. Each plant line was replicated 20 times, and two independent experiments were conducted. Average numbers of developing nematodes were calculated for each time point, and statistically significant differences were determined in a modified t test using the statistical software package SAS.
Syncytial Measurements
Arabidopsis seeds were planted on modified Knop’s medium, and 10-d-old seedlings were inoculated with approximately 200 surface-sterilized J2 H. schachtii nematodes. For each line, at least 20 single female syncytia were randomly selected, photographed, and measured as described previously by Hewezi et al. (2008b).
Root Length Measurements
Arabidopsis plants were grown vertically on modified Knop’s medium for 10 d, and then the root length of at least 30 plants per line was measured as the distance between the crown and the tip of the main root in three independent experiments. Statistically significant differences between lines were determined by unadjusted paired t tests (P < 0.01).
RNA Isolation and qPCR
Total RNA was extracted from frozen ground root tissues using the TRIzol reagent (Invitrogen) following the manufacturer’s instructions. DNase treatment of total RNA was carried out using DNase I (Invitrogen). The treated total RNA (5 μg) was polyadenylated and reverse transcribed using the Mir-X miRNA First-Strand Synthesis Kit (Clontech) following the manufacturer’s instructions. The synthesized cDNAs then were diluted to a concentration equivalent to 10 ng total RNA µL−1 and used as a template in real-time RT-PCR to quantify both miRNA and GRF expression levels using the two-step RT-PCR kit (Bio-Rad) according to the manufacturer’s protocol. Mature miRNA quantification was performed using a universal reverse primer (mRQ; supplied with the Mir-X miRNA First-Strand Synthesis Kit), complementary to the poly(T) adapter along with miRNA-specific oligonucleotides as forward primers, which were extended by two A residues on the 3′ end to ensure the binding of the primer to the poly(T) region of the mature miRNA cDNA and to avoid its hybridization on the miRNA precursor cDNA, as recently described by Gutierrez et al. (2009). Quantification of the primary transcripts of miR396a and miR396b was performed using forward primers specific to miRNA precursors and the universal mRQ reverse primer. The U6 small nuclear RNA was used as an internal control to normalize the expression levels of miRNA. The PCRs were run in an iCycler (Bio-Rad) using the following program: 50°C for 10 min, 95°C for 5 min, and 40 cycles of 95°C for 30 s and 60°C for 30 s. Following PCR amplification, the reactions were subjected to a temperature ramp to create the dissociation curve, determined as changes in fluorescence measurements as a function of temperature, by which the nonspecific products can be detected. The dissociation program was 95°C for 1 min, 55°C for 10 s, followed by a slow ramp from 55°C to 95°C. Note that the annealing temperatures used to amplify the primary transcripts of miR396a and miR396b are between 6°C and 7°C higher than the actual temperatures to ensure specific annealing to the templates. The specificity of the amplification products was verified by analyzing the melting curves and by agarose gel electrophoresis. No amplification was obtained with the control reactions or with primers designed to bind to the reverse complement strand of primary miRNA transcripts.
For quantification of the expression levels of GRF1 to GRF9, gene-specific primers were designed to discriminate between different gene family members. To measure the levels of transgene mRNA of GRF1 and GRF3 in the P35S:rGRF1 and P35S:rGRF3 transgenic plants, we designed primers corresponding to the miRNA-binding site to distinguish between the endogenous and transgene transcripts. Primer sequences containing the wild-type miR396-binding site were used to quantify the endogenous transcripts, whereas primer sequences containing the mutated miR396-binding site in which 10 mismatches were introduced were used to quantify the expression levels of the transgenes. Arabidopsis Actin8, as a constitutively expressed gene, was used as an internal control to normalize gene expression levels. In all cases, at least three independent experiments, each with four technical replicates of each reaction, were performed. Quantification of the relative changes in gene expression was performed using the 2-ΔΔCT method (Livak and Schmittgen, 2001). The primer sequences used in qPCR analysis are listed in Supplemental Table S5.
Mapping the Cleavage Sites of GRF1 and GRF3
To map the cleavage sites of GRF1 and GRF3, a modified method for RLM 5′-RACE was carried out using the FirstChoice RLM-RACE kit (Ambion) following the manufacturer’s instructions. Total RNA was isolated from root tissues of wild-type (Col-0) plants at 8 d post H. schachtii infection and ligated to 5′-RACE RNA adaptor without calf intestine alkaline phosphatase treatment. The ligated RNA was subsequently used for cDNA synthesis using GRF1 and GRF3 gene-specific outer primers (Supplemental Table S5). The initial PCR amplification was performed using the 5′-RACE outer primer provided with the kit and GRF1 and GRF3 gene-specific outer primer. The nested PCR amplification was performed using the 5′-RACE inner primer included in the kit and GRF1 and GRF3 gene-specific inner primer (Supplemental Table S5). The PCR products were gel purified, cloned into pGEM-T Easy vector (Promega), and sequenced.
Microarray Analysis
Wild-type Arabidopsis (ecotypes Col-0 and Ws), the triple mutant grf1/grf2/grf3, and transgenic plants overexpressing rGRF1 (line 18-2) or rGRF3 (line 12-5) were grown in vertical culture dishes on modified Knop’s medium for 2 weeks, and then root tissues were collected for RNA extraction using the method described by Verwoerd et al. (1989). Affymetrix Arabidopsis GeneChips (ATH1) were used to compare gene expression in the wild type with that in the triple mutant and the rGRF1 or rGRF3 plants. Probe preparation was performed as described in the GeneChip 3′ IVT Express Kit (Affymetrix; part no. 901229) technical manual. Hybridization and washes were performed in the GeneChip facility at Iowa State University as described by Affymetrix. Data were normalized using the robust multiarray average method (Irizarry et al., 2003). The first of two experiments used a completely randomized design with three independent biological replications for each of the plant types, Col-0, grf1/grf2/grf3, rGRF1, and rGRF3. A linear model analysis of the normalized data was conducted for each gene using the empirical Bayes method implemented in the Bioconductor R package limma (Smyth, 2004, 2005). Each linear model included effects for the four plant types. As part of each linear model analysis, tests for differential expression between Col-0 and rGRF1 and between Col-0 and rGRF3 were conducted. The P values from each of these tests were converted to q values using the method of Nettleton et al. (2006) and used to control the false discovery rate at the 0.05 level as described by Storey and Tibshirani (2003). The second experiment used a randomized complete-block design with three independent biological replications of Ws and the triple mutant grf1/grf2/grf3. A linear model analysis using fixed effects for blocks and plant types was conducted for each gene using limma to identify genes differentially expressed between Ws and the triple mutant. The resulting P values were converted to q values to control the false discovery rate at the 0.05 level as described above.
Testing for the Significance of Gene List Overlaps
χ2 tests of independence were used to evaluate the significance of overlaps in gene lists. For example, consider a comparison of the 2,293 genes identified as differentially expressed in rGRF1-overexpressing plants and the 7,225 syncytium-regulated genes identified by Szakasits et al. (2009). The overlap between these two gene lists was 1,131. Given that the ATH1 gene chips have 22,500 probe sets, the expected overlap is only 22,500 × (2,293/22,500) × (7,225/22,500) = 736.3 under the assumption of independence among genes and independence between membership on one gene list and membership on the other. The discrepancy between the observed and expected overlap is highly significant (χ2 = 346.1, P < 0.0001). Analogous calculations indicate the statistical significance of the overlaps reported in “Results.”
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: miR396a (AT2G10606), miR396b (AT5G35407), GRF1 (At2g22840), GRF2 (At4g37740), GRF3 (At2g36400), GRF4 (At3g52910), GRF5 (At3g13960), GRF6 (At2g06200), GRF7 (At5g53660), GRF8 (At4g24150), GRF9 (At2g45480), and Actin8 (AT1G49240).
The microarray data reported in this study were deposited at the National Center for Biotechnology Information Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE31593 and at the Plant Expression Database (http://www.plexdb.org//modules/PD_general/publications.php#citeplex) under accession number AT109.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Expression profiles of GRF gene family members in Arabidopsis roots.
Supplemental Figure S2. qPCR quantification of transgene expression levels in transgenic Arabidopsis lines described in this study.
Supplemental Figure S3. Overexpression of miR396 reduces GRF gene expression.
Supplemental Figure S4. Characterization of Arabidopsis grf1 and grf3 mutants.
Supplemental Figure S5. Schematic representation of miR396-resistant versions of GRF1 and GRF3 transcripts.
Supplemental Figure S6. Overexpression of miR396 and the target genes GRF1 and GRF3 reduces root length.
Supplemental Table S1. List of differentially expressed genes identified in the grf1/grf2/grf3 triple mutant.
Supplemental Table S2. List of differentially expressed genes identified in P35S:rGRF1 plants.
Supplemental Table S3. List of differentially expressed genes identified in P35S:rGRF3 plants.
Supplemental Table S4. List of 3,160 genes differentially expressed in the syncytium that are directly or indirectly regulated by GRFs.
Supplemental Table S5. Primer sequences used in this study.
Supplementary Material
Acknowledgments
We thank Javier Palatnik for the GRF2:rGRF2-GUS line, Jeong Hoe Kim for the grf1/grf2/grf3 triple mutant, Jiqing Peng for microarray hybridization, Peter Howe, Diane May, and Brad Hurst for help with generation of the transgenic plants, and Megan Clements and Jason Noon for technical assistance.
Glossary
- miRNA
microRNA
- Col-0
ecotype Columbia-0
- RT
reverse transcription
- RLM
RNA ligase-mediated
- qPCR
quantitative PCR
- dpi
d post inoculation
- Ws
Wassilewskija
- J2
second-stage juvenile
- J3
third-stage juvenile
- J4
fourth-stage juvenile
References
- Alkharouf NW, Klink VP, Chouikha IB, Beard HS, MacDonald MH, Meyer S, Knap HT, Khan R, Matthews BF. (2006) Timecourse microarray analyses reveal global changes in gene expression of susceptible Glycine max (soybean) roots during infection by Heterodera glycines (soybean cyst nematode). Planta 224: 838–852 [DOI] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Aukerman MJ, Sakai H. (2003) Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes. Plant Cell 15: 2730–2741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum TJ, Wubben MJE, II, Hardyy KA, Su H, Rodermel SR. (2000) A screen for Arabidopsis thaliana mutants with altered susceptibility to Heterodera schachtii. J Nematol 32: 166–173 [PMC free article] [PubMed] [Google Scholar]
- Chen X. (2009) Small RNAs and their roles in plant development. Annu Rev Cell Dev Biol 25: 21–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Davis EL, Hussey RS, Baum TJ. (2004) Getting to the roots of parasitism by nematodes. Trends Parasitol 20: 134–141 [DOI] [PubMed] [Google Scholar]
- Fahlgren N, Howell MD, Kasschau KD, Chapman EJ, Sullivan CM, Cumbie JS, Givan SA, Law TF, Grant SR, Dangl JL, Carrington JC. (2007) High-throughput sequencing of Arabidopsis microRNAs: evidence for frequent birth and death of MIRNA genes. PLoS ONE 2: e219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gheysen G, Fenoll C. (2002) Gene expression in nematode feeding sites. Annu Rev Phytopathol 40: 191–219 [DOI] [PubMed] [Google Scholar]
- Grunewald W, van Noorden G, Van Isterdael G, Beeckman T, Gheysen G, Mathesius U. (2009) Manipulation of auxin transport in plant roots during Rhizobium symbiosis and nematode parasitism. Plant Cell 21: 2553–2562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez L, Bussell JD, Pacurar DI, Schwambach J, Pacurar M, Bellini C. (2009) Phenotypic plasticity of adventitious rooting in Arabidopsis is controlled by complex regulation of AUXIN RESPONSE FACTOR transcripts and microRNA abundance. Plant Cell 21: 3119–3132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He XF, Fang YY, Feng L, Guo HS. (2008) Characterization of conserved and novel microRNAs and their targets, including a TuMV-induced TIR-NBS-LRR class R gene-derived novel miRNA in Brassica. FEBS Lett 582: 2445–2452 [DOI] [PubMed] [Google Scholar]
- Hewezi T, Howe P, Maier TR, Baum TJ. (2008a) Arabidopsis small RNAs and their targets during cyst nematode parasitism. Mol Plant Microbe Interact 21: 1622–1634 [DOI] [PubMed] [Google Scholar]
- Hewezi T, Howe P, Maier TR, Hussey RS, Mitchum MG, Davis EL, Baum TJ. (2008b) Cellulose binding protein from the parasitic nematode Heterodera schachtii interacts with Arabidopsis pectin methylesterase: cooperative cell wall modification during parasitism. Plant Cell 20: 3080–3093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiguchi G, Kim GT, Tsukaya H. (2005) The transcription factor AtGRF5 and the transcription coactivator AN3 regulate cell proliferation in leaf primordia of Arabidopsis thaliana. Plant J 43: 68–78 [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. (2003) Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4: 249–264 [DOI] [PubMed] [Google Scholar]
- Ithal N, Recknor J, Nettleton D, Maier T, Baum TJ, Mitchum MG. (2007) Developmental transcript profiling of cyst nematode feeding cells in soybean roots. Mol Plant Microbe Interact 20: 510–525 [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. (1987) GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones-Rhoades MW, Bartel DP. (2004) Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol Cell 14: 787–799 [DOI] [PubMed] [Google Scholar]
- Jung JH, Seo YH, Seo PJ, Reyes JL, Yun J, Chua NH, Park CM. (2007) The GIGANTEA-regulated microRNA172 mediates photoperiodic flowering independent of CONSTANS in Arabidopsis. Plant Cell 19: 2736–2748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katiyar-Agarwal S, Gao S, Vivian-Smith A, Jin HL. (2007) A novel class of bacteria-induced small RNAs in Arabidopsis. Genes Dev 21: 3123–3134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katiyar-Agarwal S, Morgan R, Dahlbeck D, Borsani O, Villegas A, Jr, Zhu JK, Staskawicz BJ, Jin HL. (2006) A pathogen-inducible endogenous siRNA in plant immunity. Proc Natl Acad Sci USA 103: 18002–18007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Choi D, Kende H. (2003) The AtGRF family of putative transcription factors is involved in leaf and cotyledon growth in Arabidopsis. Plant J 36: 94–104 [DOI] [PubMed] [Google Scholar]
- Kim JH, Kende H. (2004) A transcriptional coactivator, AtGIF1, is involved in regulating leaf growth and morphology in Arabidopsis. Proc Natl Acad Sci USA 101: 13374–13379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Lee BH. (2006) GROWTH-REGULATING FACTOR4 of Arabidopsis thaliana is required for development of leaves, cotyledons, and shoot apical meristem. J Plant Biol 49: 463–468 [Google Scholar]
- Klink VP, Hosseini P, Matsye P, Alkharouf NW, Matthews BF. (2009) A gene expression analysis of syncytia laser microdissected from the roots of the Glycine max (soybean) genotype PI 548402 (Peking) undergoing a resistant reaction after infection by Heterodera glycines (soybean cyst nematode). Plant Mol Biol 71: 525–567 [DOI] [PubMed] [Google Scholar]
- Li Y, Zhang Q, Zhang J, Wu L, Qi Y, Zhou J-M. (2010) Identification of microRNAs involved in pathogen-associated molecular pattern-triggered plant innate immunity. Plant Physiol 152: 2222–2231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Song Y, Chen Z, Yu D. (2009) Ectopic expression of miR396 suppresses GRF target gene expression and alters leaf growth in Arabidopsis. Physiol Plant 136: 223–236 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Δ Δ C(T)) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Lu S, Sun Y-H, Amerson H, Chiang VL. (2007) MicroRNAs in loblolly pine (Pinus taeda L.) and their association with fusiform rust gall development. Plant J 51: 1077–1098 [DOI] [PubMed] [Google Scholar]
- Marin E, Jouannet V, Herz A, Lokerse AS, Weijers D, Vaucheret H, Nussaume L, Crespi MD, Maizel A. (2010) miR390, Arabidopsis TAS3 tasiRNAs, and their AUXIN RESPONSE FACTOR targets define an autoregulatory network quantitatively regulating lateral root growth. Plant Cell 22: 1104–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro L, Dunoyer P, Jay F, Arnold B, Dharmasiri N, Estelle M, Voinnet O, Jones JDG. (2006) A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science 312: 436–439 [DOI] [PubMed] [Google Scholar]
- Navarro L, Jay F, Nomura K, He SY, Voinnet O. (2008) Suppression of the microRNA pathway by bacterial effector proteins. Science 321: 964–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nettleton D, Hwang JTG, Caldo RA, Wise RP. (2006) Estimating the number of true null hypotheses from a histogram of p-values. J Agric Biol Environ Stat 11: 337–356 [Google Scholar]
- Pandey SP, Shahi P, Gase K, Baldwin IT. (2008) Herbivory-induced changes in the small-RNA transcriptome and phytohormone signaling in Nicotiana attenuata. Proc Natl Acad Sci USA 105: 4559–4564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez RE, Mecchia MA, Debernardi JM, Schommer C, Weigel D, Palatnik JF. (2010) Control of cell proliferation in Arabidopsis thaliana by microRNA miR396. Development 137: 103–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sijmons PC, Atkinson HJ, Wyss U. (1994) Parasitic strategies of root nematodes and associated host cell responses. Annu Rev Phytopathol 32: 235–259 [Google Scholar]
- Sijmons PC, Grundler FMW, Von Mende N, Burrows PR, Wyss U. (1991) Arabidopsis thaliana as a new model host for plant parasitic nematodes. Plant J 1: 245–254 [Google Scholar]
- Smyth GK (2004) Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3: Article 3 [DOI] [PubMed] [Google Scholar]
- Smyth GK (2005) limma: Linear Models for Microarray Data. In R Gentleman, V Carey, S Dudoit, R Irizarry, W Huber, eds, Bioinformatics and Computational Biology Solutions Using R and Bioconductor. Springer, New York, pp 397–420 [Google Scholar]
- Sobczak M, Golinowski W. (2009) Structure of cyst nematode feeding sites. Berg RH, Taylor CG, , Cell Biology of Plant Nematode Parasitism. Plant Cell Monographs; Springer, Berlin, pp 153–187 [Google Scholar]
- Storey JD, Tibshirani R. (2003) Statistical significance for genomewide studies. Proc Natl Acad Sci USA 100: 9440–9445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szakasits D, Heinen P, Wieczorek K, Hofmann J, Wagner F, Kreil DP, Sykacek P, Grundler FM, Bohlmann H. (2009) The transcriptome of syncytia induced by the cyst nematode Heterodera schachtii in Arabidopsis roots. Plant J 57: 771–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verwoerd TC, Dekker BMM, Hoekema A. (1989) A small-scale procedure for the rapid isolation of plant RNAs. Nucleic Acids Res 17: 2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson VM, Kumar A. (2006) Nematode resistance in plants: the battle underground. Trends Genet 22: 396–403 [DOI] [PubMed] [Google Scholar]
- Wu G, Park MY, Conway SR, Wang JW, Weigel D, Poethig RS. (2009) The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 138: 750–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Poethig RS. (2006) Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3. Development 133: 3539–3547 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.