Abstract
We used a comparative genomics approach to investigate the evolution of a complex nucleotide-binding (NB)-leucine-rich repeat (LRR) gene cluster found in soybean (Glycine max) and common bean (Phaseolus vulgaris) that is associated with several disease resistance (R) genes of known function, including Rpg1b (for Resistance to Pseudomonas glycinea1b), an R gene effective against specific races of bacterial blight. Analysis of domains revealed that the amino-terminal coiled-coil (CC) domain, central nucleotide-binding domain (NB-ARC [for APAF1, Resistance genes, and CED4]), and carboxyl-terminal LRR domain have undergone distinct evolutionary paths. Sequence exchanges within the NB-ARC domain were rare. In contrast, interparalogue exchanges involving the CC and LRR domains were common, consistent with both of these regions coevolving with pathogens. Residues under positive selection were overrepresented within the predicted solvent-exposed face of the LRR domain, although several also were detected within the CC and NB-ARC domains. Superimposition of these latter residues onto predicted tertiary structures revealed that the majority are located on the surface, suggestive of a role in interactions with other domains or proteins. Following polyploidy in the Glycine lineage, NB-LRR genes have been preferentially lost from one of the duplicated chromosomes (homeologues found in soybean), and there has been partitioning of NB-LRR clades between the two homeologues. The single orthologous region in common bean contains approximately the same number of paralogues as found in the two soybean homeologues combined. We conclude that while polyploidization in Glycine has not driven a stable increase in family size for NB-LRR genes, it has generated two recombinationally isolated clusters, one of which appears to be in the process of decay.
The nucleotide-binding (NB)-leucine-rich repeat (LRR) family of plant disease resistance genes (R) is known for both its size and its rapid evolution (Meyers et al., 2003; Leister, 2004). The products of these genes mediate the detection of pathogen virulence proteins via both direct and indirect mechanisms (DeYoung and Innes, 2006; Jones and Dangl, 2006; Eitas and Dangl, 2010). NB-LRR genes can be subdivided into two classes based on their N-terminal domains, the coiled-coil (CC) class and the TIR class (for Toll, Interleukin1 receptor, and Resistance genes). The NB domains from both CC and TIR class genes share a strong similarity to the NB domains of the mammalian (for APOPTOTIC PROTEASE ACTIVATING FACTOR1) protein and the Caenorhabditis elegans CED4 (for CELL DEATH ABNORMALITY4) protein (van der Biezen and Jones, 1998a). Because of this sequence similarity, this type of NB domain is often referred to as an NB-ARC domain (for APAF1, Resistance genes, and CED4).
Current models predict that the NB-ARC domain functions as a molecular switch, with the ATP-bound form representing the “on” state (activating defenses) and the ADP-bound form the “off” state (Takken et al., 2006). Crystal structures of the APAF1 and CED4 proteins revealed that NB-ARC can be further subdivided into a subdomain containing the NB pocket and two additional subdomains referred to as ARC1 and ARC2, which appear to interact with the NB subdomain to regulate nucleotide exchange (Albrecht and Takken, 2006). This supposition is supported by the identification of autoactivating mutations in numerous plant NB-LRR genes that map to the ARC1 and ARC2 domains and that are predicted to increase the rate of nucleotide exchange by disrupting interactions between the NB and ARC subdomains (Takken et al., 2006; van Ooijen et al., 2007).
Intramolecular interactions between NB-LRR functional domains have been demonstrated (Moffett et al., 2002; Leister et al., 2005), and progress has been made toward delimiting the regions involved (Rairdan et al., 2008). However, the precise nature and role of these interdomain contacts remain incompletely understood. The structure of the CC domain from the barley (Hordeum vulgare) MLA10 (for POLYMORPHIC BARLEY MILDEW A10) R protein has recently been solved, and the domain was shown to form a homodimer, with dimerization being functionally important (Maekawa et al., 2011).
It is generally assumed that the rapid evolution of R genes is driven by an evolutionary arms race between pathogens and their hosts, in which changes in the repertoire of pathogen virulence proteins select for the creation of new R gene specificities. Defining the molecular mechanisms underpinning this arms race is central to our understanding of the evolution of disease resistance and to the development of crop plants with durable resistance. Recombination, positive selection, and local duplications/deletions have all been shown to have important roles in R gene evolution (for review, see Bent and Mackey, 2007).
Another mechanism impacting R gene evolution is whole genome duplication (WGD). WGD events should enable the evolution of new traits by relaxing selective pressures on gene duplicates, freeing them to evolve new functions and/or expression patterns (Lynch and Katju, 2004; Adams and Wendel, 2005). Genome duplication thus might be expected to cause an increase in R gene number and diversity. However, analyses of the Arabidopsis (Arabidopsis thaliana) genome, which is believed to have undergone at least two WGD events (Bowers et al., 2003), indicate that R genes were preferentially lost following polyploidy (Cannon et al., 2004; Nobuta et al., 2005), leading to the retention of almost no duplicated R genes following WGD. This enhanced loss of R genes suggests that there may be a fitness cost associated with R genes following duplication. In cases where genome duplication is the result of allopolyploidy (i.e. combining genomes from two different species or subspecies), fitness costs could stem in part from autoimmune-type responses in which R genes from one genome are activated in the genomic context of the other genome (Bomblies and Weigel, 2007).
We have been evaluating the impact of WGD on the evolution of a complex R gene cluster in soybean (Glycine max; Innes et al., 2008). The genome of the ancestor of extant Glycine species, including soybean, underwent a WGD, likely as a consequence of allopolyploidy (Gill et al., 2009), and therefore is an excellent species with which to investigate the consequences of this type of event. Analysis of silent site substitution rates (Ks) between gene duplicates has been used to estimate a homeologue divergence time of approximately 13 million years (Schlueter et al., 2004; Egan and Doyle, 2010; Schmutz et al., 2010), which thus provides a maximum age for this WGD event (Gill et al., 2009).
Prior to the release of the complete soybean genome sequence (Schmutz et al., 2010), we sequenced an approximately 1-Mb region of soybean cv Williams 82 centered on the Rpg1b (for Resistance to Pseudomonas glycinea1b), disease resistance gene on chromosome 13 (Innes et al., 2008). This region contains numerous NB-LRR-type genes, one of which encodes Rpg1b, a gene effective against certain races of bacterial blight (Pseudomonas syringae pv glycinea; Ashfield et al., 2004). Also mapping to the vicinity are other R genes of known function, including those effective against bacterial, viral, and oomycete pathogens (Diers et al., 1992; Yu et al., 1994; Ashfield et al., 1998; Sandhu et al., 2005).
To allow us to assess the impact of polyploidy on the Rpg1b region, we also sequenced the homeologous (duplicated by polyploidy) segment on chromosome 15 (Innes et al., 2008). This allowed us to compare the evolution of this R gene-rich region in the two duplicated segments generated by polyploidy. In addition, to gain information on the likely ancestral state of this region prior to polyploidy, we sequenced the single orthologous (separated by speciation) region from common bean (Phaseolus vulgaris; Innes et al., 2008), which diverged from Glycine approximately 19 million years ago (mya; Lavin et al., 2005) and has not undergone the less than 13-mya WGD event. In common bean, this region is referred to as the Co-2 locus and contains R genes effective against diverse pathogens, including the fungus Colletotrichum lindemuthianum (Geffroy et al., 1998) and P. syringae pv phaseolicola expressing AvrRpm1 (Chen et al., 2010). Finally, to allow us to assess more recent changes within the Glycine homeologues, we sequenced the orthologous regions in a second soybean accession (PI96983) and also in a perennial Glycine species, Glycine tomentella (Innes et al., 2008). The soybean and G. tomentella lineages are thought to have diverged about 6 mya (Egan and Doyle, 2010). The phylogenetic relationships among these taxa are shown in Figure 1A.
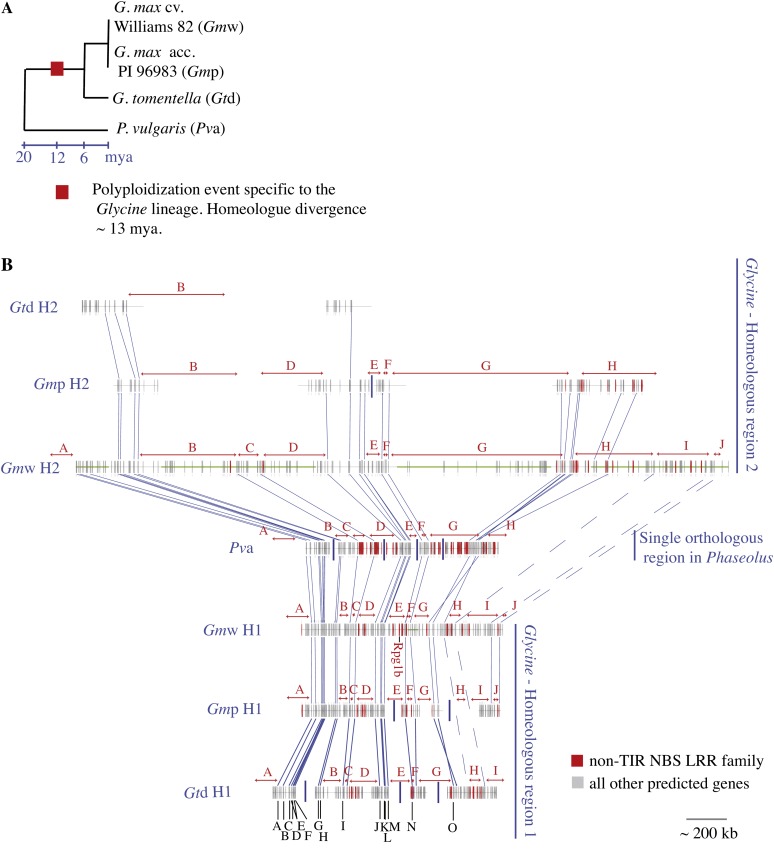
Figure 1.
Distribution of CC-NB-LRR genes across the homeologous and orthologous sequences corresponding to the Rpg1b region in soybean. A, Species tree of the legume species included in this study. B, Alignment of predicted genes across Rpg1b homologous regions. Gray horizontal lines represent the available sequence, with gaps indicated by vertical blue lines. The sequence derived from the soybean whole genome sequencing project is indicated with a horizontal green line. Vertical red rectangles positioned on the horizontal lines represent predicted CC-NB-LRR genes, and vertical gray rectangles represent all other genes. Putative low-copy orthologous/homeologous genes are linked by blue lines, and where these relationships have been confirmed phylogenetically (Innes et al., 2008), a black letter is assigned to the gene set. Orthologous/homeologous intervals containing CC-NB-LRR genes in at least one of the plants sequenced are indicated by red letters over red double-ended arrows. Gtd, G. tomentella; Gmp, soybean accession PI96983; Gmw, soybean ′Williams82′; Pva, common bean Andean accession. H1, Glycine homeologue 1; H2, Glycine homeologue 2. This figure is adapted from Innes et al. (2008).
Comparison of the Glycine homeologues revealed a remarkably high retention of non-NB-LRR gene duplicates in this region following polyploidy (approximately 77%), most of which are still expressed (Innes et al., 2008). This was interesting given that the target region in Glycine homeologue 2 now resides in a pericentromeric region rich in retrotransposons and displaying suppressed recombination (Innes et al., 2008; Wawrzynski et al., 2008a).
In contrast, although NB-LRRs have been retained in both the duplicated regions, we found evidence for partitioning of ancestral NB-LRR lineages (lineages defined by phylogenetic analysis of the NB-ARC region; van der Biezen and Jones, 1998a) between the two homeologues in Glycine. Also apparent was a relatively recent species/homeologue-specific expansion of individual lineages (Innes et al., 2008).
Although we reported an initial analysis of the above genomic regions (Innes et al., 2008), we now have expanded our analysis by delving further into the evolutionary history of the NB-LRR genes in this region of the Glycine genome. In particular, we now ask the following questions. (1) Following polyploidy, are NB-LRRs generally lost to bring copy number back to the diploid state? (2) What do phylogenetic analyses and physical arrangements reveal about the duplication history of NB-LRR sequences in Glycine and Phaseolus? (3) How frequent is recombination among loci, and which parts of the NB-LRRs are more prone to recombination? (4) Do NB-LRR genes on separate homeologues recombine? (5) Does partitioning of NB-LRR lineages between homeologues in Glycine reduce concerted evolution of the family and therefore facilitate the generation/retention of greater diversity as compared with that in a diploid ancestor? (6) Is there evidence for diversifying selection acting on specific codons of NB-LRR genes, and if so, can combining this information with modeling of R protein tertiary structure provide insights into NB-LRR protein function?
RESULTS
Different Fates of the NB-LRR Family in the Two Homeologous Regions Derived from the Ancestral Rpg1b Region following Polyploidization
To better understand the evolutionary processes that have driven the divergence of the CC-NB-LRR genes (henceforth referred to simply as NB-LRR open reading frames [ORFs]) found in the Rpg1b region in soybean and its relatives, we first defined 10 physical intervals within the approximately 1-Mb target region centered on the soybean Rpg1b gene and the corresponding orthologous and homeologous regions that contained NB-LRR ORFs (regions A–J in Fig. 1B). Each interval is defined by flanking low-copy gene(s) conserved in most/all of the homologous regions being compared.
Although NB-LRR ORFs are present in both soybean homeologous regions H1 and H2, these ORFs display only limited conservation of synteny, based on alignments of flanking low-copy genes (Fig. 1B; Table I). This lack of colinearity suggests that the NB-LRR genes have undergone many deletions and/or duplications following the divergence of the homeologous segments in the presumed allopolyploid (Gill et al., 2009; Fig. 1B; Table I). Of the nine intervals represented in both soybean cv Williams 82 (Gmw) H1 and Gmw H2, seven contain NB-LRR ORFs in H1 versus five intervals in H2. However, only four intervals contain NB-LRR ORFs in both Gmw H1 and Gmw H2. If duplications are at least partially responsible for this pattern, there must have also been translocations to account for the presence of homeologue-specific NB-LRR ORF locations.
Table I. Distribution of CC-NB-LRR genes, and ancestral NB-ARC clades, across the approximately 1-Mb Rpg1b study region in soybean and the corresponding homologous regions.
“Yes” indicates that the physical interval in that homolog contains at least one CC-NB-LRR ORF, and “no” indicates the absence of CC-NB-LRR ORFs. com complete sequence coverage of interval; none no sequence coverage of interval; part partial sequence coverage of interval. Lowercase italic letters indicate which ancestral NB-ARC clades are represented within that physical interval.
| Homologous Region |
NB-LRR Gene-Containing Physical Intervala |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| A | B | C | D | E | F | G | H | I | J | Totalsb | |
| Soybean ′Williams82′ H1 CNLsc | Yespar | Nocom | Nocom | Yescom | Yescom | Yescom | Yescom | Yescom | Yescom | Yescom | |
| Cladesd | c | – | – | i | c, f | f | g | b, c, e | c, h | c | b, c, e, f, g, h, i |
| Soybean ′PI96983′ H1 CNLsc | Yespar | Nocom | Nocom | Yescom | Yespar | Yescom | Yespar | Gapnone | Gappar | Yescom | |
| Cladesd | c | – | – | i | f | f | – | – | – | c | c, f, i |
| G. tomentella H1 CNLsc | Nopar | Nocom | Nocom | Yescom | Yespar | Yescom | Yespar | Yescom | Gappar | Gapnone | |
| Cladesd | – | – | – | i | f | f | g | c, e | – | – | c, e, f, i, g |
| Bean (Andean) CNLsc | Gapnone | Nocom | Yescom | Yespar | Gappar | Nocom | Yespar | Yespar | Gapnone | Gapnone | |
| Cladesd | – | – | i | i | – | – | c, d | d | – | – | c, d, i |
| Soybean ′PI96983′ H2 CNLsc | Gapnone | Gappar | Gapnone | Gappar | Gappar | Nocom | Gappar | Yespar | Gapnone | Gapnone | |
| Cladesd | – | – | – | – | – | – | d | e | – | – | d, e |
| Soybean ′Williams82′ H2 CNLsc | Gapnone | Yescom | Nocom | Yescom | Nocom | Nocom | Yescom | Yescom | Yescom | Nocom | |
| Cladesd | – | – | – | i | – | – | – | e | a, e | – | a, e, i |
We have previously shown that, contrary to our expectations, most of the low-copy, non-NB-LRR genes in this region appear to be expressed in both soybean homeologues, despite the fact that the target region in H2 is found in the pericentromeric region of chromosome 15 (Innes et al., 2008). To assess the expression pattern of the NB-LRR family in the pericentromeric H2 region, we used the soybean EST data set to estimate what proportion of the NB-LRR genes in Gmw H1 and Gmw H2 are expressed (Table II; Supplemental Table S1). Unlike the low-copy genes (Innes et al., 2008), very few of the predicted NB-LRR ORFs in Gmw H2 are expressed, with only three out of the 18 predicted ORFs having matching ESTs (17%). This contrasts with 12 out of 15 ORFs (80%) having EST support in Gmw H1, which is not in a pericentromeric region.
Table II. Abundance of intact and expressed CC-NB-LRR genes in soybean (cv Williams 82) homeologous regions 1 and 2 and in the orthologous region in bean.
|
NB-LRR-Containing Intervala |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| A | B | C | D | E | F | G | H | I | J | Totalsb | |
| Gmw H1 | |||||||||||
| Tc | 1 | 0 | 0 | 3 | 3 | 1 | 1 | 3 | 2 | 1 | 15 |
| Id | 1 | 0 | 0 | 2 | 3 | 1 | 1 | 2 | 2 | 1 | 13 (87%) |
| ESTe | 1 | 0 | 0 | 2 | 2 | 1 | 1 | 2 | 2 | 1 | 12 (80%) |
| I/ESTf | 1 | 0 | 0 | 2 | 3 | 1 | 1 | 2 | 2 | 1 | 13 (87%) |
| Bean Andean accession | |||||||||||
| Tc | Gap | 0 | 5 | 7g,h | Gap | 0 | 14g | 4g,h | Gap | Gap | 30 |
| Id | Gap | 0 | 1 | 3 | Gap | 0 | 7 | 1 | Gap | Gap | 12 (40%) |
| ESTe | Gap | 0 | 0 | 0 | Gap | 0 | 0 | 0 | Gap | Gap | 0 |
| I/ESTf | Gap | 0 | 1 | 3 | Gap | 0 | 7 | 1 | Gap | Gap | 12 (40%) |
| Gmw H2 | |||||||||||
| Tc | Gap | 1h | 0 | 1 | 0 | 0 | 2 | 9 | 5 | 0 | 18 |
| Id | Gap | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 2 | 0 | 4 (22%) |
| ESTe | Gap | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 1 | 0 | 3 (17%) |
| I/ESTf | Gap | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 2 | 0 | 5 (28%) |
The 10 physical intervals (labeled A–J) as defined in Figure 1B.
Total for the entire homologous region.
All NB-LRR ORFs (intact NB-LRR genes and pseudogenes) predicted by the fgenesh gene-prediction software, not counting partial NB-LRRs at the end of contigs.
Intact NB-LRR genes (defined as containing a single exon encoding CC, NB-ARC, and LRR domains with no obvious deletions of the P loop, kin2, GLPL, RNBS-D, or MHD motifs).
Corresponding EST available in database (matches greater than 98% nucleotide identity over a minimum of 100 nucleotides in length, with the supported NB-LRR paralogue being the top BLAST hit in the soybean whole genome predicted proteome).
We further investigated the fate of the NB-LRR genes in Gmw H2 by partitioning the family into ORFs that appear to encode intact NB-LRR genes versus those encoding disrupted and/or truncated genes (Table II). Consistent with the EST analysis, even though numerous NB-LRR ORFs are distributed across the target region in Gmw H2, few appear to correspond to intact, full-length genes (Table II; Supplemental Table S1). In fact, of the 18 predicted NB-LRR ORFs within Gmw H2, only four appear to encode full-length genes with no obvious deletions of key domains or motifs (22% of the total). This compares with 13 out of the 15 NB-LRR ORFs in Gmw H1 appearing to be full length and intact (87%). There is substantial, but not complete, overlap between the sets of paralogues that appear to be intact and those having EST support (Supplemental Table S1).
Interestingly, of the 30 predicted NB-LRR ORFs found in the single orthologous region in common bean (henceforth referred to as bean), only 12 (40%) appear to be intact (Table II). In reality, the actual number of intact NB-LRRs in bean is probably somewhat larger, because gaps in the bacterial artificial chromosome (BAC) contig for this species may harbor additional paralogues (two NB-LRR genes at the end of BAC contigs were not included in these totals, as we could not determine whether they were intact). Given that the total number of intact NB-LRR genes in both Gmw H1 and Gmw H2 combined is 17, it is apparent that the polyploidization event in the Glycine lineage does not correlate with a large, stable increase in the number of intact NB-LRR genes derived from the ancestral Rpg1b region over that seen in bean. As described later, this observation was investigated further by assessing the relative importance of polyploidy, versus rates of local duplications and deletions, in the evolution of the NB-LRR family in Glycine and Phaseolus.
Sequence Exchanges between Paralogues Are Common within the CC and LRR Domains and Less Common in the NB-ARC
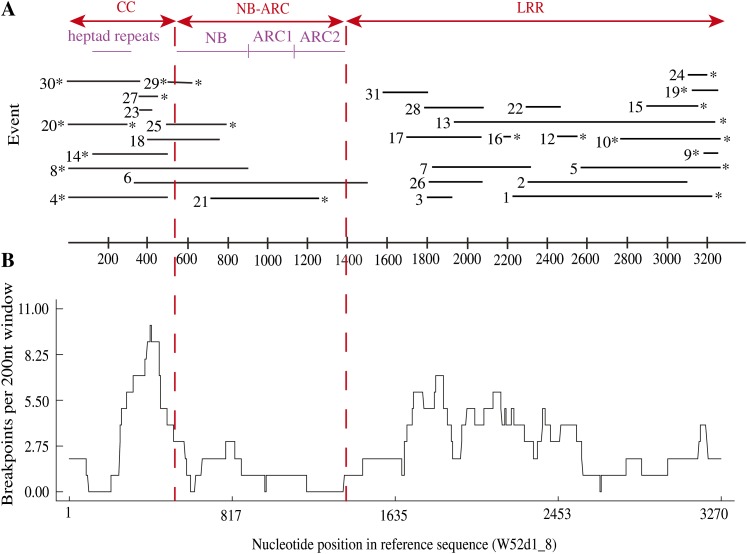
NB-LRR gene clusters are thought to undergo frequent unequal crossing-over events, leading to expansion and contraction of copy number within a cluster and likely giving rise to recombinant NB-LRR genes with novel specificities for pathogen recognition (Collins et al., 1999; McDowell and Simon, 2008). In addition, gene conversion events among NB-LRR genes may also give rise to new specificities by shuffling subregions of NB-LRR sequence (Dodds et al., 2001; Mondragon-Palomino and Gaut, 2005). Both unequal crossing over and gene conversion create chimeric sequences that cannot be meaningfully analyzed in a tree-building framework, such as traditional phylogenetic analysis (Huson and Bryant, 2006). To assess the frequency of recombination among NB-LRR genes in our data set, we aligned them using a combination of automated and manual alignment tools (see “Materials and Methods”) and then detected likely recombination break points using several methods implemented in Recombination Detection Program version 3.15 (Martin et al., 2005b). As expected, these analyses uncovered multiple NB-LRR genes with strong evidence of recombination. Plotting the positions of the recombination break points relative to the conserved domains of NB-LRR genes revealed a strong bias against recombination within the NB-ARC domain (Fig. 2), with only three of 38 (8%) defined break points falling within the NB-ARC, although this stretch represents about 25% of the total sequence length. In contrast, the LRR region, which is generally considered a zone of high recombination, contains 68% of the break points while only representing approximately 58% of the gene sequence. Interestingly, recombination events are also overrepresented in the CC domain, which contains 24% of the break points while only representing 17% of the gene. However, no defined break points were detected in the first 300 nucleotides of the CC, suggesting that recombination is poorly tolerated in this region.
Figure 2.
Sequence exchanges between the NB-LRR paralogues are common in the CC and LRR domains while being comparatively rare within the NB-ARC. A, Positions of the 30 events detected by RDP analyses are shown. Each horizontal line represents a distinct event. One event (no. 6) resulted in the transfer of the complete NB-ARC domain between paralogues. The events are drawn relative to the W52d1_8 sequence, and the position in this sequence is shown on the x axis. Asterisks indicate the ends of sequence exchanges in which the actual break point position could not be determined by RDP. B, Frequency (y axis) of break points among all sequences plotted against nucleotide position in the alignment (x axis). Events are supported by at least two of the four methods utilized within RDP version 3.15 at P < 0.001.
The paucity of recombination break points within the NB-ARC domain raised the question of whether reduced DNA polymorphism in this domain might be limiting our ability to detect such events. Therefore, we determined the total DNA polymorphism levels in the CC, NB-ARC, and LRR domains separately. Despite higher amino acid sequence conservation in the NB-ARC domain (Supplemental Table S2), compared with the CC and LRR domains, the level of nucleotide polymorphism per site was only slightly lower in the NB-ARC domain. Overall, we identified 539 polymorphic sites in the NB-ARC domain compared with only 279 in the CC domain, indicating that our failure to detect recombination break points within the NB-ARC domain was not due to a lack of polymorphism. We thus conclude that the lack of recombination break points within the NB-ARC domain is real and likely reflects functional constraints.
The length of sequence-exchange events detected was highly variable, ranging from 71 nucleotides (event 23) to 1,166 nucleotides (event 6) for events in which both break points could be defined (Supplemental Table S3). It should be noted that in many cases, the positions of the break points could not be accurately determined (asterisks in Fig. 2A), so some exchange events may be even longer. Interestingly, most of the events are confined predominantly to the CC or LRR domains, with several possibly extending into the flanking 5′ and 3′ regions. Only one event extends completely across the NB-ARC (Fig. 2A, event 6). Also striking is the absence of events that extend from the LRRs into the NB-ARC domain. Overall, these observations suggest that events leading to chimeric NB-ARC domains are poorly tolerated.
For a subset of the recombination events shown in Figure 2, the program RDP was able to identify both of the likely parent sequences and their genomic locations were known (Table III). In all such cases, these sequences were within the same physical region (Table III). Events involving sequence transfer within Glycine (i.e. soybean or G. tomentella) H1, Glycine H2, and the single orthologous region in bean were detected (nine, one, and two events, respectively), but no unambiguous exchanges between the Glycine homeologous regions were identified (Table III; Supplemental Table S3). So while a similar number of intact NB-LRR paralogues have been maintained in the soybean and bean accessions studied, in soybean the paralogues are distributed between two recombinationally isolated clusters that are free to evolve independently.
Table III. Sequence exchanges detected between NB-LRR ORFs.
| Description | No. |
|---|---|
| Total number of recombination events detected | 208 |
| Total number of independent events | 31 |
| Events in which a parental sequence is unknown | 19 |
| Events exclusively represented in Glycine H1 | 13 |
| Events exclusively represented in Glycine H2 | 8 |
| Events represented exclusively in Phaseolus | 8 |
| Events represented in both Glycine H1 and H2 | 0 |
| Events involving exchanges within Glycine H1 | 9 |
| Events involving exchanges within Glycine H2 | 1 |
| Events involving exchanges within Phaseolus | 2 |
| Events involving exchanges between Glycine H1 and H2 | 0 |
| Events involving exchanges within the same ancestral clade | 2 |
| Events involving exchanges between different ancestral clades | 8 |
| Events involving only the CC domain | 6 |
| Events involving only the NB-ARC domain | 1 |
| Events involving only the LRR domain | 18 |
| Events involving both CC and NB-ARC | 4 |
| Events involving both NB-ARC and LRRs | 0 |
| Events involving CC, NB-ARC, and LRRs | 1 |
A Subset of NB-LRR Paralogues Display No Evidence of Recombination or Recent Duplications
Using the stringent criteria used to detect recombinant paralogues described here, most but not all paralogues were found to be recombinant in at least one of their three domains. For example, in Gmw H1, only two out of the 12 NB-LRR genes included in the analyses contain no detectable recombination events. Both of the nonrecombinant genes (Gmw21f22_7 and Gmw221b6_15) are intact, expressed, and contain NB-ARC domains that are relatively isolated in the NB-ARC phylogenetic tree (Fig. 3; described below). Their sequence divergence likely accounts for their lack of recombination with other genes in this region.
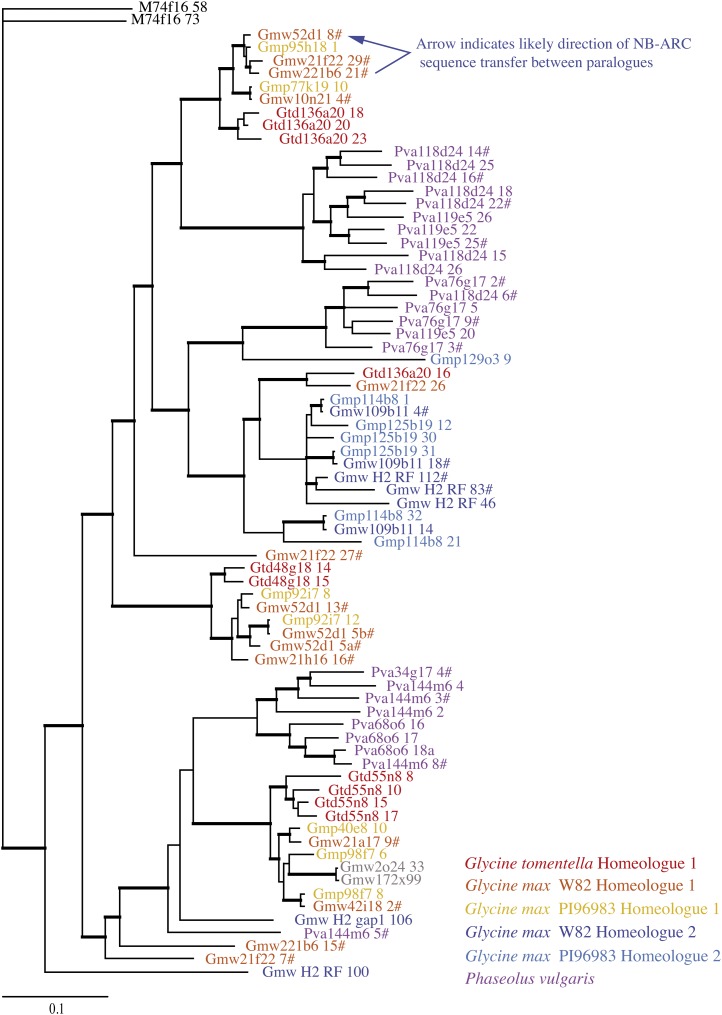
Figure 3.
Bayesian phylogeny of NB-ARC domains derived from the Rpg1b region. Thick branches indicate posterior probabilities of 0.95 or greater. Sequences derived from soybean Williams 82 WGS scaffolds (7× draft assembly) are preceded by “GmwRF” or “GmwGap1.” The remaining sequences are BAC derived and are labeled with the BAC name and gene number corresponding to those described by Innes et al. (2008). # indicates a soybean or bean NB-ARC sequence from an intact NB-LRR paralogue (i.e. not a pseudogene). Note that sequences from two soybean accessions are included (with prefixes Gmw and Gmp), and putatively allelic pairs should not be confused with local duplications. The sequences shown in gray are derived from Gmw H1 but are outside the 1-Mb target region.
Phylogenetic Analysis Reveals Homeologue-Specific Expansions of NB-ARC Clades
To avoid the errors introduced by chimeric sequences, we focused our phylogenetic analyses on the NB-ARC region, which displayed the lowest levels of sequence exchange. The recombination test was repeated on this region alone, and any recombined NB-LRR sequences, or sequences missing most or all of this region, were excluded, leaving 72 of 93 sequences. These 72 sequences were then subjected to Bayesian analysis to construct a phylogenetic tree (Fig. 3).
Prominent features of the resulting tree include the identification of several NB-ARC lineages that predate the Glycine/Phaseolus split, together with terminal clades consisting exclusively of groups of relatively similar paralogues from individual species or homeologous regions. This pattern of recent, species-specific duplications is particularly striking in bean, where all but one of the paralogues are found in three well-supported clades containing sequences only from this species. Consistent with rampant species-specific local duplications, we observed only a single orthologous gene pair, even when comparing paralogues from soybean and G. tomentella, while multiple groups of likely co-orthologous genes are present (Fig. 3). Taken together, these observations suggest that a rapid turnover (i.e. local duplications and deletions) of paralogues within each species has resulted in groups of co-orthologous genes while still preserving a limited number of ancestral NB-ARC lineages over a substantial evolutionary time period (i.e. prior to the Phaseolus/Glycine split).
Also striking is the apparent partitioning of ancestral NB-ARC lineages between the two soybean homeologous regions. As discussed in the section below, this partitioning of lineages between the homeologous regions points to deletions subsequent to the polyploidization event (or, more accurately, subsequent to the separation of the parental lineages, as recent evidence indicates this was likely an allopolyploid event; Gill et al., 2009), eliminating the majority of NB-LRR gene duplicates generated by the WGD.
Gene and Species Tree Reconciliation Reveals a Partitioning of Ancestral NB-ARC Lineages between the Glycine Homeologous Regions
To more accurately define how many loci were present in the common ancestor of Glycine and Phaseolus, we reexamined the NB-ARC phylogeny to look for putatively orthologous groups of sequences. The number of these groups indicates how many loci (at a minimum) would have been present prior to the divergence of Glycine and Phaseolus, thereby providing an indication of the diversity that existed prior to this speciation event, which occurred approximately 19 mya (Lavin et al., 2005). We used the program GeneTree (Page, 1998) to reconcile the species and gene trees while minimizing the gene duplications and losses required to account for the observed data.
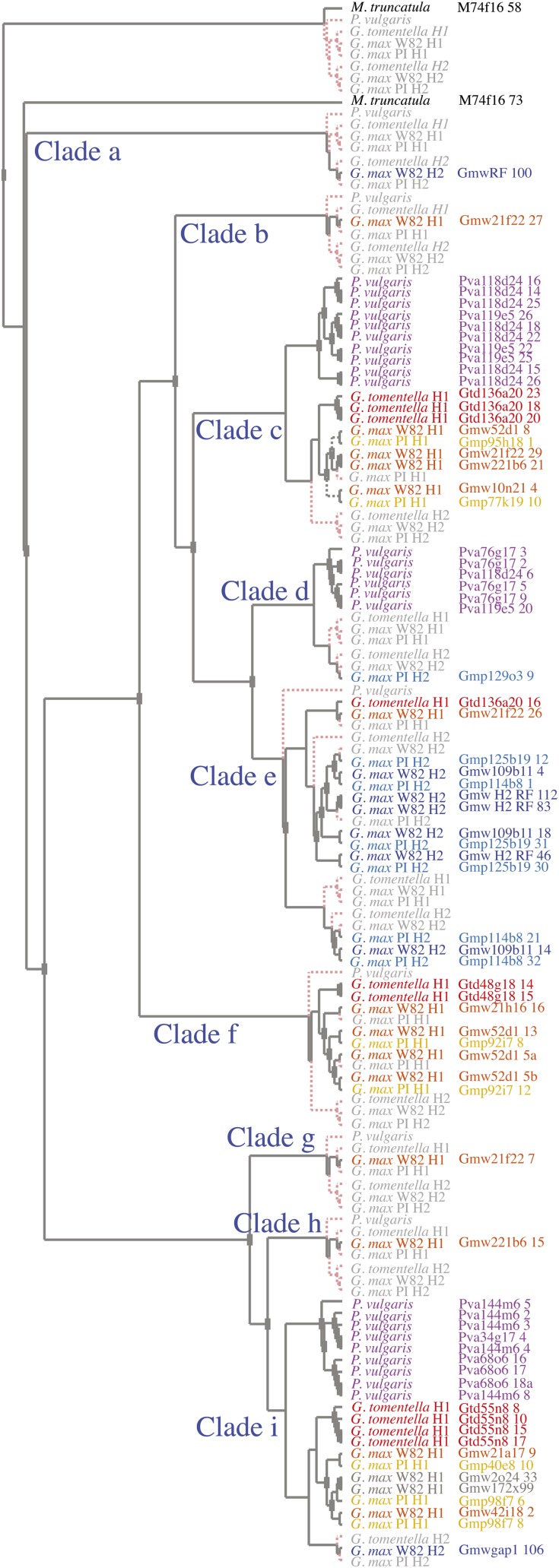
Using the consensus Bayesian tree (Fig. 3) as input for GeneTree, an estimate of 10 NB-ARC lineages in this genomic location in the most recent common ancestor of Phaseolus and Glycine was obtained (Supplemental Fig. S1). However, when weakly supported nodes were resolved in favor of fewer duplications and losses (the optimized tree), only nine loci were predicted in the most recent common ancestor of these taxa (Fig. 4).
Figure 4.
Reconciled gene and species trees based on the optimized NB-ARC phylogeny. Squares at the nodes represent hypothesized duplication events within the evolutionary history of the sampled genes. Dashed lines and gray text represent lineages that theoretically exist(ed) as a result of hypothesized duplication events but are otherwise missing due to being unsampled or through gene loss. Extant genes and taxa are color coded as in Figure 3. Clades a to i represent nine ancestral NB-ARC lineages predicted to have persisted since before the Glycine/Phaseolus split. H1 and H2 refer to homeologues 1 and 2, respectively. The right-hand column provides gene names labeled with BAC name and gene number as described by Innes et al. (2008).
Interestingly, only four of the nine ancestral NB-ARC lineages have been retained in soybean H2, whereas seven have been retained in H1. Only two clades are represented in both homeologues (Fig. 4, clades e and i). The partitioning of clades between the homeologues is even more striking when only intact paralogues are considered. Neither the single Gmw H1 paralogue (Gmw21f22_26) found in clade e nor the single Gmw H2 paralogue found in clade i (GmwGap1_106) is intact or has EST support, so it appears that no clade has functional representatives in both homeologues (Supplemental Table S1). Further examination of the data reveals that all four of the intact paralogues in Gmw H2 belong to a single clade (Table I, clade e), whereas Gmw H1 contains intact paralogues from six different clades (Table I, clades b, c, and f–i; Supplemental Table S1). Thus, while the ancestral clades have been partitioned between the two homeologous regions, all nine clades have been retained in soybean, seven of them represented by at least one intact paralogue in the accessions studied here.
In contrast, all of the paralogues from the single orthologous region in bean are found within three ancestral clades (Fig. 4; Table I). However, caution must be used when interpreting this observation, as our sequence coverage in bean is incomplete and additional clades may be represented by paralogues present in contig gaps.
As noted earlier, although sequence exchanges within the NB-ARC domain are rare, events within the CC and LRR domains are more common. We find that sequence exchanges are not restricted to transfers between paralogues that share the same NB-ARC ancestral lineage. Although two events were detected involving parental paralogues sharing the same class of NB-ARC, eight events occurred between paralogues containing NB-ARC domains belonging to different lineages (Table III; Supplemental Table S3). So while several distinct lineages of NB-ARC domains have been maintained since before Glycine and Phaseolus diverged (this was likely facilitated by the rarity of interparalogue recombination involving this domain), sequence exchanges within the CC and LRR domains are still occurring between paralogues containing different NB-ARC lineages. As a result, gene trees based on just the NB-ARC domain do not necessarily reflect the relatedness of the CC and LRR domains from the same genes.
Duplications and Deletions of NB-LRR Genes in the Rpg1b Region Are Ongoing
The GeneTree analysis also allowed us to estimate the number of duplications and deletions that have occurred in the different orthologous and homeologous segments derived from the ancestral Rpg1b region. Using the optimized tree, eight duplications of NB-LRR loci are suggested to have occurred in the Phaseolus + Glycine lineage after its divergence from the Medicago lineage, but before these two genera diverged, creating the nine labeled clades shown in Figure 4. In contrast, at least 53 duplications (including both local duplications and those resulting from polyploidy) are suggested to have occurred in total in the Phaseolus and Glycine lineages after their divergence (Fig. 4). However, the number of loci and therefore duplications that occurred prior to the Phaseolus + Glycine divergence from Medicago is probably underestimated, due to losses that may occur without leaving any obvious trace. Clearly, duplications and deletions of NB-LRR loci have been ongoing throughout the history of these legumes.
In all, the GeneTree analysis indicates that at least 25 duplications occurred in the soybean lineage subsequent to the split from Phaseolus. Of these, 10 duplications were the result of the Glycine-specific polyploidization event, whereas 15 were “local” duplications (i.e. they occurred within one of the homeologous regions examined here or in the ancestral sequence after divergence from Phaseolus but prior to polyploidization; Fig. 4). It is worth noting that at least one of these apparent local duplications likely reflects the conversion of a preexisting paralogue rather than a WGD, as this event (Gmw52d1_8; Fig. 2A, event 6) spans the entire NB-ARC region. If only intact paralogues are considered, there is no evidence that any of the nine ancestral lineages have been retained in both soybean homeologous regions following the polyploidization event (although the possibility cannot be excluded based on the analysis of only two accessions). So although polyploidization has been responsible for approximately 40% of the NB-LRR duplication events that have occurred in the soybean lineage, subsequent loss of lineages has prevented the WGD from driving a sustained increase in family size.
Local duplications that have occurred subsequent to polyploidization are observed in both the soybean homeologues and have occurred at a similar rate in the two locations. This is an interesting observation, as the target region in H2 is found in a pericentromeric location known to exhibit suppressed recombination (Innes et al., 2008; Schmutz et al., 2010). Recombination would be required for unequal exchange, one mechanism proposed to generate local duplications in NB-LRR clusters (Leister, 2004). Considering the soybean sequences alone, six duplications among H2 sequences were found (all in clade e) versus eight duplications among H1 sequences (three each in clades c and f and two in clade i; Fig. 4). However, as noted earlier, in contrast to the situation observed in soybean H1, few of the paralogues generated by the local duplications in H2 have remained expressed or intact (Table II).
Interestingly, local duplication rates associated with specific NB-ARC lineages in Phaseolus and Glycine appear to have differentiated following speciation. Bean NB-LRR genes within the target region have duplicated at least 22 times since divergence (the actual number of duplications may be higher due to unobserved losses and unsampled paralogues in contig gaps) while also having been lost at least six times. Excluding duplications caused by polyploidy, soybean NB-LRRs have duplicated only 15 times. When specific NB-ARC lineages are considered, the difference is even more striking (Fig. 4, clades c and i).
In conclusion, although polyploidization accounts for a significant proportion of the NB-LRR duplications that have occurred during the evolution of the soybean lineage, subsequent deletion of most the duplicated NB-LRR lineages has ensured that it has not driven a large, sustained increase in family size over that seen in bean. Instead, the genome duplication in Glycine has partitioned the ancestral NB-ARC families between two genomic locations that appear to be recombinationally isolated from one another.
Positive Selection Is Acting on Predicted Solvent-Exposed Residues in the CC, NB-ARC, and LRR Domains
From the above analyses, it is clear that a rapid turnover of NB-LRR paralogues, primarily as a consequence of local duplications and deletions, is one factor driving evolution of the NB-LRR gene family in the vicinity of Rpg1b. To investigate further the evolutionary forces driving changes in the family, we attempted to identify what role positive selection has played.
We first investigated the NB-ARC region utilizing the large set of aligned nonrecombinant sequences used to generate the Bayesian phylogeny shown in Figure 3. Sites under either negative or positive selection were identified using three different methods implemented by the DataMonkey server (Fig. 5; see “Materials and Methods”; Single Likelihood Ancestor Counting [SLAC] P < 0.05, Fixed Effects Likelihood [FEL] P < 0.05, Random Effects Likelihood [REL] Bayes Factor [BF] > 50). These analyses identified 12 codons under positive selection and 92 codons under negative selection as determined by the FEL method (Fig. 5B; Supplemental Table S4). Using the more stringent criteria of requiring support by all three methods, five and 71 codons were still found to be under positive and negative selection, respectively, within the NB-ARC domain (Fig. 5C).
Figure 5.
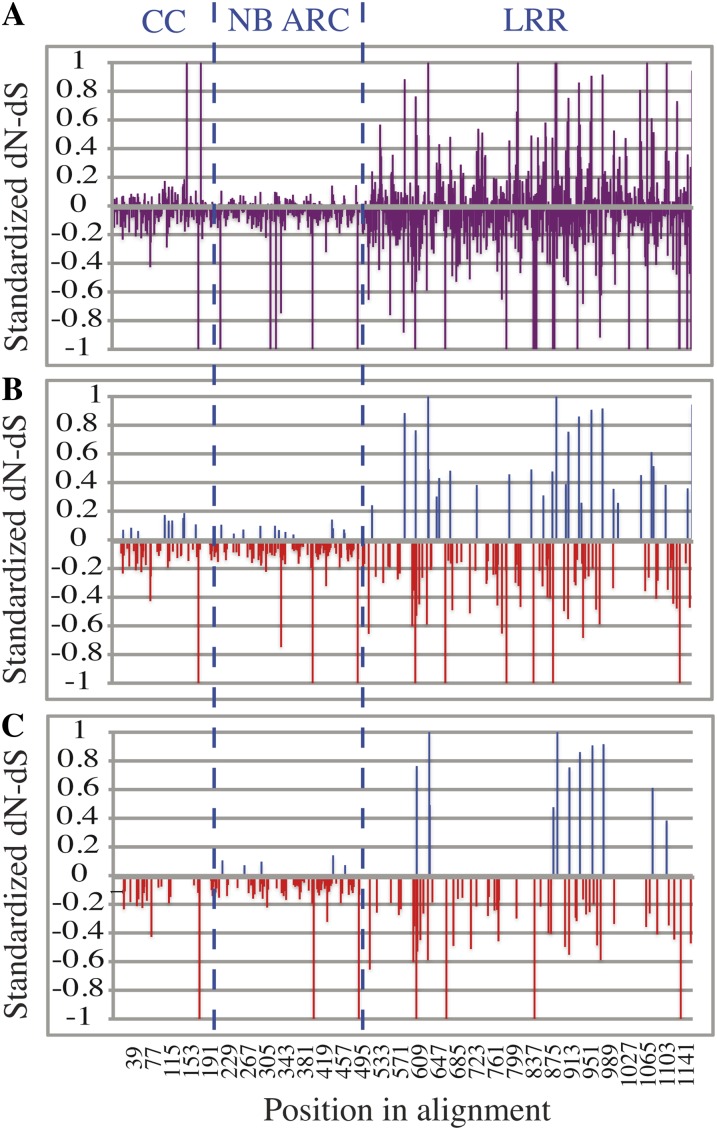
Distinct patterns of selection detected in the CC, NB-ARC, and LRR domains. Graphs represent standardized nonsynonymous (dN) minus synonymous (dS) substitution rates (dN − dS) values calculated using the FEL method (Pond and Frost, 2005b) plotted against the position of each codon in the alignment. The y axis scale is limited from −1 to 1, although some standardized dN-dS values extend beyond this range. A, Standardized dN-dS values for all codons. B, Only those codons where nonneutral selection is supported by FEL (P < 0.05). C, Only those codons where nonneutral selection is supported by FEL (P < 0.05), SLAC (P < 0.05), and REL (BF > 50).
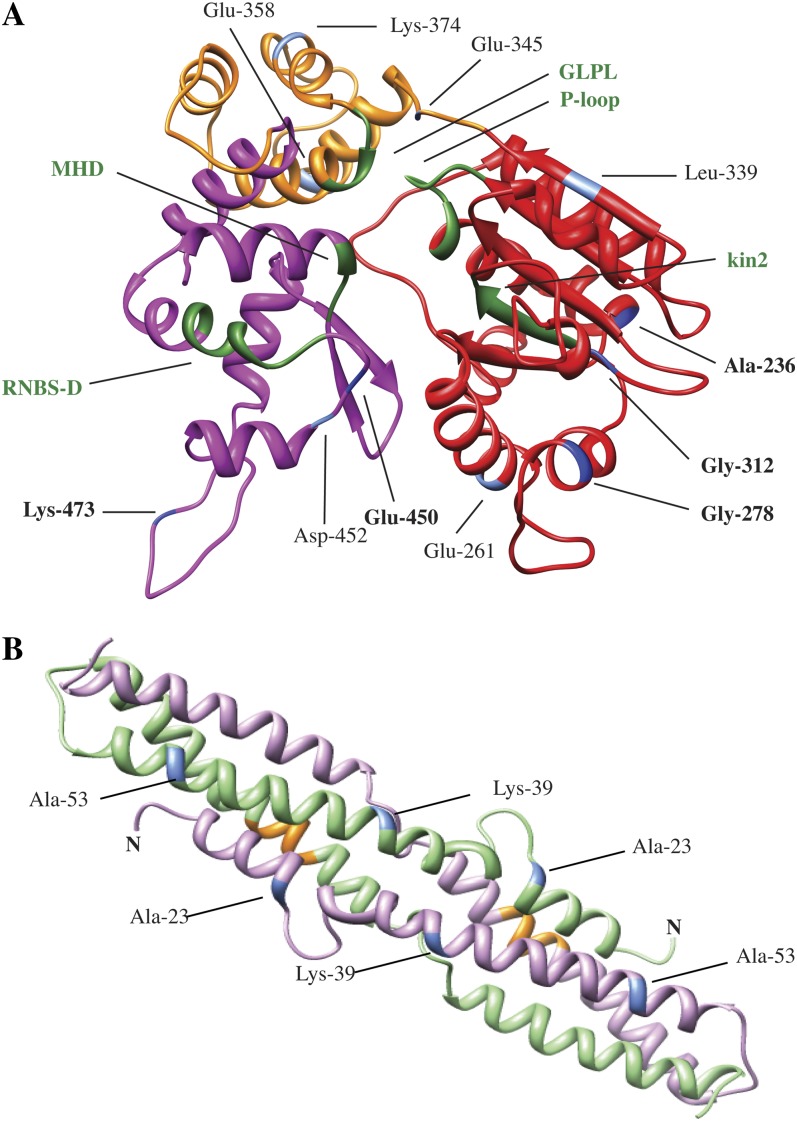
Not surprisingly, the five codons experiencing positive selection (as defined by the more stringent criteria; Fig. 5C) are located outside the several previously defined conserved motifs (P loop, kin2, GLPL, RNBS-D, MHD; Meyers et al., 1999) characteristic of the NB-ARC domain (Fig. 6). To gain insights into the possible functional significance of the codons under positive selection, the tertiary structure of the NB-ARC domain from a representative paralogue (Gmw52d1-8; encodes Rpg1b) was modeled. Interestingly, analysis of the predicted structure indicates that the majority of the sites under positive selection are located on the surface of the folded protein, consistent with a role in interactions with the CC or LRR domains or with other proteins (Fig. 7A; Supplemental Movie S1).
Figure 6.
Sites under positive (diversifying) selection are overrepresented in the predicted solvent-exposed surface of the LRRs. Residues under selection are indicated in the consensus sequences corresponding to the alignments used in the selection analysis. Sites under overall positive selection are shown in blue, and those under negative selection are shown in red. Where selection is supported by FEL (P < 0.05), the colored residue is shown in normal type; where the prediction is also supported by SLAC (P < 0.05) and REL (BF > 50), the residue is shown in boldface type. A specific amino acid is shown in the consensus sequence when it is present in at least 51% of the sequences represented at that location in the alignment (gaps excluded). x indicates all other sites in the alignment. A, CC domain. Underlined residues are predicted to form CCs (P > 95% and P > 89% for the first and second underlined regions, respectively). The predicted positions of amino acids within each CC heptad repeat are indicated with lowercase letters, with expected hydrophobic positions printed in bold. B, NB-ARC domain. Boxed residues indicate previously defined conserved motifs (from the N-terminal end: P loop, kin2, GLPL, RNBS-D, MHD). C, LRR domain. Individual LRRs are shown on separate lines. The predicted solvent-exposed face is boxed. D, Consensus sequence for intracellular LRRs. The region predicted to form part of a solvent-exposed face is underlined.
Figure 7.
Comparative modeling of the tertiary structures of the Rpg1b CC and NB-ARC domains suggests that the majority of residues under positive selection are located on the surface. A, Predicted tertiary structure of the Rpg1b NB-ARC domain modeled after the APAF-1 NB-ARC domain (PDB code 1Z6T). The NB, ARC1, and ARC2 subdomains are shown in red, yellow, and purple, respectively. Previously defined conserved motifs are shown in green. B, Predicted tertiary structure of the Rpg1b CC domain homodimer modeled after the MLA10 CC domain (PDB code 3QFL). The conserved EDVID motif is shown in orange. Residues with statistical support for positive selection in the NB-ARC and CC structures are indicated in blue in both panels (dark blue and boldface labels when supported by FEL, SLAC, and REL; light blue and regular labels when supported by at least FEL). Residue numbers correspond to positions in the full-length Rpg1b protein (Williams 82 allele).
To identify codons under selection within the CC and LRR domains, we partitioned the master, full-length NB-LRR ORF alignment containing the entire set of NB-LRR sequences from this study into three regions corresponding to the CC, NB-ARC, and LRR domains. The parts of the alignment corresponding to the CC and LRR domains were then screened for recombination events individually using RDP 3.44, and recombinant sequences were removed. The resulting alignments were then subjected to SLAC, FEL, and REL analyses to detect sites under selection. Numerous sites in both the CC and LRR domains were found to be under selection, although the overall trend was for the strength of selection, both positive and negative, to be stronger within the LRRs (Fig. 5A).
To investigate the functional significance of the sites under selection within the CC domain, the COILs program was used to screen the region for the presence of CCs. The software predicted two intervals encoding CCs when a consensus sequence from the N-terminal alignment was used as the input (Supplemental Fig. S2). As might be expected, several of the predicted locations for conserved hydrophobic residues within the CC heptad repeats are predicted to be under purifying (negative) selection (Fig. 6A). Such purifying selection likely reflects a requirement of the CC domain to fold into a CC structure, similar to that recently described for the CC domain of MLA10, which forms a homodimer (Maekawa et al., 2011). We thus used the MLA10 CC domain as a template to create a structural model for the Rpg1b CC domain (Fig. 7B; Supplemental Movie S2). Using this structure, we next asked whether the sites under positive selection within the CC domain preferentially localized to the surface. Intriguingly, all sites under positive selection (blue in Fig. 7B and Supplemental Movie S2) appeared to be on the surface and all map to the same face, opposite to the face occupied by the conserved EDVID motif (orange in Fig. 7B and Supplemental Movie S2). This motif has been implicated in mediating interaction between the CC and NB-ARC domains (Rairdan et al., 2008; Maekawa et al., 2011), suggesting that the sites under positive selection may mediate interaction with other proteins rather than with the NB-ARC domain.
The LRR regions of NB-LRR genes have been implicated in recognition specificity, and previous studies have reported a bias for the predicted solvent-exposed residues within these regions to be under positive selection (Bittner-Eddy et al., 2000; Mondragón-Palomino et al., 2002). Therefore, we investigated whether positive selection may also be driving divergence within the solvent-exposed faces of the NB-LRR genes from the Rpg1b region. A consensus sequence was derived from the alignment of nonrecombinant sequences corresponding to the LRR region, and putative repeat units were identified and aligned. As can be seen in Figure 6C, a high percentage of the residues under positive selection within the LRR domain are found within the predicted solvent-exposed face (“x” residues printed in blue within the xxLxLxx consensus).
DISCUSSION
WGD should initially result in a doubling of gene number in a polyploid relative to its diploid progenitor(s). This is certainly true of an autopolyploid; the prediction for an allopolyploid would be the sum of the two progenitors. In this study, we compared the evolution of an NB-LRR gene cluster in two homeologous segments found in Glycine with its evolution in the single orthologous region found in the diploid bean, a stand-in for the ancestor(s) of Glycine. We found that while NB-LRRs have been retained in both Glycine homeologous regions, this does not result in a large stable increase in functional paralogue number over that seen in bean (17 full-length paralogues in Gmw H1 + H2 versus at least 12 paralogues in bean). Instead, the polyploidy event in Glycine most likely has resulted in the partitioning of ancestral NB-ARC lineages between two physically and recombinationally isolated clusters that are now evolving independently.
These observations are consistent with previous whole-genome studies of the NB-LRR family in Arabidopsis, cotton (Gossypium hirsutum), rice (Oryza sativa), and soybean indicating that, at least in these species, polyploid events have not driven a stable increase in gene family size (Cannon et al., 2004; Nobuta et al., 2005; Schmutz et al., 2010; Zhang et al., 2010, 2011). In fact, it appears that few duplicated NB-LRRs in Arabidopsis have been retained in both homeologous (duplicated) segments following the most recent genome doubling (thought to have occurred 20–40 mya; Bowers et al., 2003). Of 153 segmental duplication events found to contain NB-LRR gene(s) in at least one of the duplicated segments, only 22 (14%) of these events have NB-LRR genes retained in both homeologous segments in Arabidopsis, and in only one of these did phylogenetic analyses support the hypothesis that homeologous NB-LRR lineages were retained in both duplicated segments (Nobuta et al., 2005).
Together, these observations suggest that there is no selective advantage to maintaining a doubled number of NB-LRR genes following polyploidization; thus, whole-genome duplications are not a key mechanism determining family size. Furthermore, NB-LRR family size appears to be highly variable within a species, with the estimated gene number in soybean ranging from 501 to 1,801 among 11 accessions analyzed (Zhang et al., 2010). Such large variation in gene content is consistent with our phylogenetic analyses that show many duplication and loss events when comparing species and when comparing the two soybean accessions, Williams 82 and PI96983. Intriguingly, there is a positive correlation between warmer climates and NB-LRR family size in both soybean and rice, suggesting that selection by pathogens may lead to rapid increases in NB-LRR number (Zhang et al., 2010).
The lack of conservation of NB-LRR genes between the Rpg1 region and its homeologous region in soybean stands in striking contrast to the low-copy genes that are interspersed throughout the Rpg1 region. We previously determined that 77% of single-copy genes in the Rpg1 region have been retained in soybean following the 13-mya polyploidy event (Innes et al., 2008). The retention of these low-copy genes in syntenic order between the two soybean homeologues and bean (Fig. 1) indicates that unequal crossing over between NB-LRR clusters in the Rpg1 region has not occurred, as this would have resulted in the deletion or duplication of intervening low-copy genes. Despite this lack of unequal crossing-over events, we detected sequence exchange between NB-LRR genes in separate clusters. We infer from this observation that gene conversion between NB-LRR genes is more readily tolerated than unequal crossing over, presumably due to a need to retain the low-copy genes. It is also notable that many NB-LRR clusters have been entirely lost in homeologue 2 (intervals C, E, and F in Fig. 1), while synteny has been retained, suggesting that NB-LRRs can be lost by mechanisms other than unequal crossing over.
The large intraspecies variation in NB-LRR copy number also implies that in the absence of pathogen pressure, there may be a rapid loss of NB-LRR genes. Several lines of evidence suggest that a large increase in NB-LRR paralogue number following a polyploid event could impose a significant cost on the host plant. For example, in one landmark study, transgenic expression of the Arabidopsis RPM1 (for RESISTANCE TO PSEUDOMONAS SYRINGAE MACULICOLA1) NB-LRR gene in an ecotype that normally does not express this gene resulted in a 9% decrease in seed number (Tian et al., 2003). It should be noted, however, that as Arabidopsis expresses more than 150 NB-LRR genes, this cost cannot be typical of all R genes and perhaps reflects an autoactivation of RPM1 in a novel genetic background. An even more dramatic cost could result from inappropriate activation of NB-LRRs following the combination of genomes by an allopolyploid event, the likely origin of the Glycine genome (Gill et al., 2009). Supporting this hypothesis, Bomblies and Weigel (2007) observed that in 2% of Arabidopsis intraspecific crosses, hybrid necrosis was triggered in the F1 generation. In at least one instance, this phenotype was shown to be dependent on an epistatic interaction between an NB-LRR gene and a second, unlinked gene. One explanation for these observations could be provided by the “guard model,” which proposes that at least some NB-LRRs monitor/guard the plant proteins targeted by pathogen virulence factors (van der Biezen and Jones, 1998b; Dangl and Jones, 2001; Innes, 2004). The NB-LRRs and the proteins they monitor presumably coevolve, and it is possible that when NB-LRRs are introduced into a novel genetic background, some will autoactivate in response to inappropriate alleles of the guarded proteins.
As mentioned above, our data indicate a high rate of “birth and death” of NB-LRR paralogues as the result of local duplications and deletions (“local” being defined in our study as occurring within one of the approximately 1-Mb homeologous or orthologous regions analyzed). For example, eight local duplications were identified in soybean H1 and six in H2. The duplications in soybean H2 are especially interesting, as this region resides in a pericentromeric location that is strongly suppressed for recombination and therefore unlikely to experience unequal exchange events (Innes et al., 2008; Schmutz et al., 2010). Strikingly, in bean, the rate of local duplications appears to be significantly higher than in either of the soybean homeologues, with 22 duplications detected within the available sequence. This increased rate of local duplications presumably reflects an increased rate of unequal crossing over that can lead to an expansion of gene clusters. Consistent with this hypothesis, this NB-LRR cluster in bean is located in a subtelomeric region, which are known to display elevated recombination rates in soybean and maize (Zea mays; Gore et al., 2009; Schmutz et al., 2010).
Our observations are consistent with several whole-genome studies in Arabidopsis that indicate that most NB-LRR paralogues found in this species were generated by local duplications (Richly et al., 2002; Baumgarten et al., 2003; Cannon et al., 2004). For example, Cannon et al. (2004) combined phylogenetic analyses with the identification of segmental duplication events to investigate the relative importance of tandem/local and “segmental” (the result of polyploidy or duplication of large chromosomal segments) duplications in the evolution of 50 large gene families in Arabidopsis (Cannon et al., 2004). In all, 54 local and only six segmental duplication events were identified involving the NB-LRR family. It is thought that the Arabidopsis lineage includes at least two polyploidization events (Bowers et al., 2003), so it is clear that the vast majority of NB-LRRs duplicated by these events have subsequently been lost. From these data, the authors concluded that the rate of local duplications and losses, not polyploidy, has been the key factor in determining NB-LRR family size. Our results echo these findings for a more recent polyploidy event. Notably, our identification of numerous NB-LRR pseudogenes at the tips of the phylogenetic tree suggests that gene loss is ongoing and can occur shortly after duplication. These pseudogenes may provide some selective advantage, however, in functioning as a sequence reservoir for gene conversion events that could give rise to new specificities in nearby functional NB-LRR genes.
A second possible beneficial role for polyploid events in NB-LRR evolution could be in creating recombinationally isolated NB-LRR clusters (Baumgarten et al., 2003; Leister, 2004). Following a polyploid event, the number of NB-LRR genes/clusters will initially be increased (doubled in the case of an autopolyploid event). Subsequent deletions of the duplicated NB-LRR paralogues may then result in a return to the original NB-LRR family size. However, for the minority of clusters surviving in both duplicated segments, partitioning of the remaining paralogues between the homeologues has the potential to distribute genes between an increased number of genomic locations (as compared with the prepolyploid state). Providing that sequence exchanges between unlinked NB-LRR loci are rare, the duplicated loci should be free to evolve independently, potentially permitting the maintenance of greater NB-LRR diversity in the genome. Furthermore, this could provide a mechanism by which R genes encoding novel recognition specificities are separated from closely related paralogues and so can be protected from disruption by the recombination events that are often common within complex R gene loci. Our observation that ancestral NB-ARC lineages from the Rpg1b cluster have been partitioned between the two homeologous segments supports this model.
In support of the above model, we found that while interparalogue sequence exchange has occurred within each of the two Glycine homeologous regions, no convincing interhomeologue events could be detected. Numerous previous studies have identified sequence exchanges between linked paralogues within NB-LRR clusters, and it has been reported that the frequency of events correlates with physical distance (negatively), sequence similarity, gene orientation, and recombination rate (Mondragon-Palomino and Gaut, 2005). It is thought that such recombination may be important for shuffling accumulated sequence diversity within the cluster to facilitate the evolution of new recognition specificities. It is interesting that we observe recombination tracts in soybean H2 paralogues, as the target region in homeologue 2 is located in a pericentromeric region that displays suppressed recombination (Innes et al., 2008; Schmutz et al., 2010). However, this apparent paradox can be explained by the observation that while crossing-over events are suppressed within pericentromeric regions, gene conversion is not (Shi et al., 2010; Talbert and Henikoff, 2010).
Although physical proximity is a key factor modulating rates of sequence exchange between NB-LRR genes, rare recombination events have been detected between paralogues found in different NB-LRR clusters (Kuang et al., 2008) and even between genes located on different chromosomes (Baumgarten et al., 2003; Kuang et al., 2005; Mondragon-Palomino and Gaut, 2005). It has been noted, however, that at least some of these apparent interchromosomal events could reflect recombination between linked genes prior to polyploidy and subsequent losses of parental sequences (Baumgarten et al., 2003; Kuang et al., 2005). Our failure to detect interhomeologue exchanges between NB-LRR paralogues in soybean H1 and H2 could reflect the high density of retrotransposons in, and the associated expansion of, soybean H2, which might be expected to inhibit interhomeologue pairing (Innes et al., 2008; Wawrzynski et al., 2008; Wang and Paterson, 2011).
Significantly, we observed exchanges between paralogues that contain NB-ARC domains belonging to different ancestral lineages. This observation is in contrast to a previous report that, at least in some clusters, exchanges only occur between NB-LRRs that are highly similar in sequence, irrespective of physical proximity, permitting the independent evolution of subfamilies of NB-LRRs within a single cluster (Kuang et al., 2005). Our finding of interlineage exchange among NB-LRR genes highlights the need to exclude recombinant sequences from phylogenetic analyses, as such sequences will make it impossible to construct a tree that reflects ancestral relationships. Indeed, many of the genes included in the tree shown in Figure 3 contain recombination sites outside the NB-ARC domain; thus, trees based on just the CC domain or the LRR domain of this gene set would look quite different.
In addition to a rapid rate of paralogue turnover and gene conversion, we also found evidence that positive selection has helped drive the evolution of the Rpg1b-associated NB-LRR cluster. Strong diversifying selection provides one possible explanation for the retention of paralogue diversity in NB-LRR clusters and has been previously implicated in NB-LRR evolution in plants and the mammalian major histocompatibility complex (MHC) cluster (Hughes and Yeager, 1998a, 1998b; Mondragón-Palomino et al., 2002; Zhang et al., 2011). As has been observed for other NB-LRR proteins (Mondragón-Palomino et al., 2002; Geffroy et al., 2009; Zhang et al., 2011), many of the sites under positive selection in the Rpg1b cluster are located within the predicted solvent-exposed face of the LRR domain. These observations are consistent with the model that the solvent-exposed residues interact with pathogen-derived ligands or other components of the recognition complex. Evidence to support this hypothesis has been provided by structural modeling of the interaction between the flax (Linum usitatissimum) L5/L6 R proteins and the corresponding Avr proteins from flax rust (Dodds et al., 2006; Wang et al., 2007).
Interestingly, we also identified several sites under positive selection in the NB-ARC domain. Although similar observations have been made before (Mondragón-Palomino et al., 2002; Zhang et al., 2011), this was a somewhat surprising finding, as the NB-ARC domain has not been implicated in determining recognition specificity. A possible explanation is provided by the discovery that intramolecular interactions can occur between the NB-ARC, CC, and LRR domains (Moffett et al., 2002; Ade et al., 2007). Furthermore, these interactions appear to be a key factor in maintaining the R protein in an inactive state in the absence of a corresponding pathogen-dependent signal. It seems plausible that as the LRRs evolve to acquire the ability to detect new pathogen signals, the NB-ARC domain must coevolve to maintain the required intramolecular interactions, perhaps explaining the presence of sites under positive selection in both R protein domains. Consistent with this hypothesis, domain swaps between the tomato (Solanum lycopersicum) Mi1.1 (for Meloidogyne incognita resistance1.1) and Mi1.2 genes, which are 91% identical, generated an autoactive allele that triggered cell death (Hwang and Williamson, 2003). A similar finding has also been described for the potato (Solanum tuberosum) Rx (for Resistance to potato virus X) gene (Rairdan and Moffett, 2006). Modeling of the tertiary structure of the Rpg1b NB-ARC domain suggests that the majority of residues under positive selection are located on the surface of the folded protein, where they would be available to interact with other domains (Fig. 7).
We also identified several sites under positive selection within the CC domain. Mapping of these residues onto a predicted tertiary structure of an Rpg1b homodimer revealed that all sites were in the α1 helix facing outward and were on the side opposite to the conserved EDVID motif (Fig. 7B). The EDVID motif is required for interaction between the CC and NB-ARC domains in the potato Rx protein (Rairdan et al., 2008) and participates in the formation of homodimers in the barley MLA10 protein (Maekawa et al., 2011). The face opposite to the EDVID domain would thus not be expected to interact with the NB-ARC domain. We speculate that this surface of the CC domain may be interacting with another host protein such as a homologue(s) of the Arabidopsis RIN4 (for RPM1-INTERACTING PROTEIN4), which has been shown to interact with the CC domain of Rpg1b (Selote and Kachroo, 2010), or with a downstream signaling protein. In support of the latter hypothesis, the α1 helix of MLA10 has been shown to interact with WRKY transcription factors (Shen et al., 2007; Maekawa et al., 2011). If this face of the CC homodimer does indeed interact with other host proteins, our finding that positive selection is occurring on this face implies that such protein-protein interactions are rapidly evolving. The Arabidopsis RIN4 protein is modified by at least three different bacterial effector proteins, and it is these modifications that appear to activate the RPM1 or RPS2 (for RESISTANT TO PSEUDOMONAS SYRINGAE2) NB-LRR proteins (Axtell and Staskawicz, 2003; Chung et al., 2011). It is thus plausible that the evolution of new ways to modify RIN4 by pathogens selects for changes in RIN4-CC interactions.
In summary, we propose a model for the evolution of the Rpg1b cluster in Glycine in which a relatively rapid deletion of duplicated NB-LRR paralogues followed polyploidization, but in a manner that resulted in the partitioning of most of the ancestral lineages between two sister clusters. The H2 cluster became subsumed within a pericentromeric region. Low levels of unequal crossing over in this region removed a key driver of NB-LRR gene births at the same time that transposon insertions led to gene deaths; the associated accumulation of retrotransposons in H2 may have also suppressed interhomeologue sequence exchanges with H1. Subsequently, the two sister clusters evolved independently with local duplications/deletions of paralogues, sequence exchange, and positive selection, all playing roles in the divergence of the NB-LRR family. While the cluster in Glycine H1 has retained much of the diversity found in the ancestral cluster, only a few closely related paralogues have survived in H2. In comparison with Glycine, Phaseolus has likely lost diversity in terms of extant NB-LRR lineages in this region, possibly due to an enhanced rate of gene birth and death resulting from unequal crossing-over events and frequent gene conversion, both of which will lead to the homogenization of NB-LRR sequences in the absence of strong diversifying selection.
MATERIALS AND METHODS
DNA Sequence Sources
Our analyses focused on CC-NB-LRR genes localized within a 1-Mb genomic region around the soybean (Glycine max ) Rpg1b disease resistance gene, located in molecular linkage group F on chromosome 13 in cv Williams 82 and the region homeologous to it (in linkage group E on chromosome 15), produced by polyploidy within the last approximately 13 million years (Innes et al., 2008; Schmutz et al., 2010). These regions were compared with orthologous regions in a second soybean accession (PI96983); in a congener, Glycine tomentella (diploid accession G1403), that shares the less than 13-mya polyploidy event with soybean; in the common bean (Phaseolus vulgaris Andean accession G19833); and in the model legume Medicago truncatula (accession A17 Jemalong). Assembly of BAC contigs, DNA sequencing, and gene identification have been described previously (Innes et al., 2008), and BAC accession numbers are available in the supplementary materials associated with that publication. For purposes of discussing homeologues, we refer to the reference sequence of soybean Williams 82 containing Rpg1b and its corresponding regions in soybean PI96983 and G. tomentella as homeologue 1 (H1) and the duplicated region that arose from the less than 13-mya polyploid event as homeologue 2 (H2). The majority of the low-copy genes used to define the physical intervals shown in Figure 1 have been previously analyzed phylogenetically to confirm the predicted orthologous and homeologous relationships (Innes et al., 2008).
Expression and Gene Fragmentation Analyses
Previously identified soybean fgenesh-predicted ORFs from Gmw H1 and Gmw H2 with significant sequence similarity to CC-NB-LRR genes (Innes et al., 2008) were compared with the soybean EST data set maintained at the National Center for Biotechnology Information, the soybean transcript assembly database at The Institute for Genomic Research (http://plantta.jcvi.org/), and the set of predicted soybean genes (Glyma 1.0) from the soybean genome project (http://www.phytozome.net/soybean; Schmutz et al., 2010) using the BLASTN algorithm (Altschul et al., 1990). ORFs were recorded as EST supported when matches were 98% or greater nucleotide identity over 100 or more nucleotides and the BLAST score was 1e-10 or better. Matches were further verified by querying the soybean whole genome proteome (gene set Glyma1.0) with the ESTs using BLASTX and confirming that the expected NB-LRR paralogues were the top hits. CC-NB-LRR genes were considered “intact” (full length) when the predicted ORF was encoded by a single exon, all three major domains (CC, NB-ARC, and LRR) were represented, and the P-loop, kin2, GLPL, RNBS-D, and MHD motifs (Meyers et al., 1999) within the NB-ARC domain (Albrecht and Takken, 2006) were not obviously deleted.
DNA Sequence Polymorphism Analysis
We estimated polymorphism statistics for each of the CC, NB-ARC, and LRR domains using DNAsp version 5 using default parameters (Librado and Rozas, 2009). We also estimated synonymous and nonsynonymous substitution rates, as well as the ratio of the nonsynonymous changes per nonsynonymous site to the synonymous changes per synonymous site (Ka/Ks ratios) in DNAsp version 5. Most analyses in DNAsp remove columns with gaps and/or missing data. Therefore, we removed sequences from each region that included a large string of gaps or missing data.
Phylogenetic and Recombination Analyses
Multiple sequence alignments at the amino acid level were generated using ClustalX (Larkin et al., 2007) and BLAST (Altschul et al., 1997). Aligned protein sequences were used as a guide to align the corresponding DNA sequences, and alignments were maximized by manual editing. Recombination among loci was assessed using several methods implemented in RDP version 3.15 (Martin et al., 2005b): RDP (Martin and Rybicki, 2000), Geneconv (Padidam et al., 1999), Chimaera (Posada and Crandall, 2001), and Bootscan (Martin et al., 2005a). Default parameter settings were used for each method except as follows: RDP (internal reference sequence), Bootscan (window = 150, step = 20, neighbor-joining trees, 200 replicates, 95% cutoff, Jin and Nei model with a transition:transversion rate ratio (Ti:Tv) = 2, coefficient of variation = 2). The maximum P value for accepting recombination was set at 0.001 (after Bonferroni correction).
After finding a high degree of recombination in the CC and LRR domains, the alignment was trimmed to the NB-ARC domain from the P loop motif to the MHD motif (i.e. from amino acids VGMGG to MHDLL in AY452685). Recombination testing was repeated (as above), and any remaining recombined sequences were also excluded, leaving a subset of 72 out of 93 sequences.
Phylogenetic analyses were performed using MrBayes version 3.1.2 (Ronquist and Huelsenbeck, 2003). Two runs using 10 chains each were analyzed to 5 million generations, sampling every 10,000 generations for each analysis. Trees sampled from the first 4 million generations were discarded as the burn in and in each case were found to have reached stability by examining the likelihood versus generation plots. Convergence between runs using the remaining 200 trees within an analysis was checked by three methods: a visual inspection of mixing of samples from each run in the aforementioned plot; that the potential scale reduction factor was at or close to 1; and that the average sd of split frequencies was 0.01 or less.
We explored the effect of model choice on trees produced by MrBayes using four different models: (1) HKY (Hasegawa, Kishina, and Yano, 1985 model), (2) HKY + G (includes a γ-distributed site rate heterogeneity parameter), (3) GTR (generalized time-reversible) + I (includes an invariant sites parameter) + G, (4) GTR + I + G with codons. Each analysis, except the codon model, was repeated a total of three times, and trees and posterior probabilities of clades were checked for consistency among analyses. Model likelihood scores (the best of three replicates) are given in Supplemental Table S5. Within each model, the three replicates did not differ in qualitative results. Supported clades (P ≥ 0.95) differed in posterior probabilities by between 0 and 0.03, although some poorly supported clades differed by more than this. Differences among models were of similar magnitude, suggesting that model fit is not critical for these data. The codon model analysis was performed only once, because of computing restrictions, but failed to better the likelihood score of the simpler models described above and therefore was not pursued.
Gene Tree/Species Tree Reconciliation
GeneTree version 1.3.0 (Page, 1998) was used to plot the positions of gene duplications and losses in the course of evolutionary history (phylogeny) and within the framework of assumed relationships of the taxa (i.e. the species tree). This program incorporates gene duplications and losses as well as speciation events for large gene families and attempts to reduce the number of events required to explain the observed data (i.e. extant genes present in the genomes of the sampled species). This program also provided an estimate of the number of NB-LRR loci present prior to the divergence of Phaseolus and Glycine approximately 19 mya (Lavin et al., 2005). We used the species tree shown in Figure 1A. For simplicity, soybean accessions were coded as separate species, but this distinction was ignored for the counts of gene duplications within soybean listed in “Results.” The model allowing gene duplications and losses was used.
Selection Analyses
Selection acting on individual codons within the NB-LRR genes was assessed using packages available through the DataMonkey server (http://www.datamonkey.org/; Pond and Frost, 2005a; Pond et al., 2005; Delport et al., 2010). These analyses required alignments of nonrecombinant sequences. Because few of the NB-LRR genes from the Rpg1b region are nonrecombinant across their entire ORFs, the master alignment of full-length ORFs was divided into three segments representing the CC, NB-ARC, and LRR domains that were then analyzed separately.
Selection within the NB-ARC domain was assessed using the set of nonrecombinant NB-ARC sequences used for the phylogenetic analyses described above. Using DataMonkey, we determined that the closely related sequence pairs W10N21_4/P77K19_10 and W52D1_5B/P92I7_20 were noninformative and removed one sequence from each before initiating analyses using the SLAC, FEL, and REL methods (Pond and Frost, 2005b) implemented on the DataMonkey server and performed using the Bayesian tree presented in Figure 3 and the default settings, with DataMonkey selecting the optimum nucleotide substitution model.
To identify sets of nonrecombinant sequences for selection analyses within the CC and LRR domains, the sections of the full-length master alignment (93 sequences) corresponding to these domains were screened for recombinant sequences using RDP version 3.44 and the parameters described earlier (“Phylogenetic and Recombination Analyses”). All recombinant sequences were then removed. In this manner, alignments of nonrecombinant sequences containing 53 and 20 sequences were identified for the CC and LRR domains, respectively. These alignments were then used as input to MrBayes (version 3.1.2) for generating Bayesian trees (nst = 6 rates = invgamma, 1,000,000 generations, samplefreq 100, Nchains = 4 Nruns = 2). These alignments and phylogenetic trees were then used as inputs for selection analyses using the DataMonkey server as described above.
Prediction of CCs
A consensus sequence was generated from the alignment of 53 nonrecombinant sequences corresponding to the CC domain (selected as described above) such that only specific amino acids present in greater than 50% of the sequences represented at a given position in the alignment were retained. This consensus was then used as input for the COILS server (http://www.ch.embnet.org/software/COILS_form.html; Lupas et al., 1991) using the MTIDK matrix (matrix derived from myosins, paramyosins, tropomyosins, intermediate filaments types I to V, desmosomal proteins, and kinesins; for further details, see COILS server documentation). The analysis was repeated with and without weighting (2.5-fold weighting for positions a and d of the heptad repeat) to facilitate the identification of false positives.
Comparative Modeling of the Rpg1b CC and NB-ARC Tertiary Structures
To examine the positions of positively selected sites within the NB-ARC domain in the context of tertiary structure, the Rpg1b Williams 82 allele (referred to as paralogue W52d1_8 elsewhere in this paper; Ashfield et al., 2004) and APAF1 (Uniprot sequence no. O14727) amino acid sequences were aligned using ClustalX (Larkin et al., 2007), and the alignment was trimmed to positions 108 to 450 in APAF1. Secondary structure was predicted for both proteins using the PSIPRED Protein Structure Prediction Server (http://bioinf.cs.ucl.ac.uk/psipred/; Jones, 1999; Bryson et al., 2005) to help validate the alignment. The previously described structure for APAF1 (Protein Data Bank [PDB] code 1Z6T; Riedl et al., 2005) was obtained from the Research Collaboratory for Structural Bioinformatics PDB database (www.pdb.org; Berman et al., 2000). The tertiary structure for Rpg1b was then modeled using the MODELER software (http://www.salilab.org/modeller/; Eswar et al., 2006) as implemented through the UCSF Chimera interface (http://www.cgl.ucsf.edu/chimera/; Pettersen et al., 2004) and using APAF1 as the known structure for comparison. Structures were visualized and manipulated using UCSF Chimera. Modeling of the Rpg1b CC domain was accomplished using a similar approach but using the known structure of the MLA10 CC domain as reference (PDB code 3QFL; Maekawa et al., 2011). The Rpg1b CC monomer structure was first modeled, and the dimer structure was subsequently inferred by applying the symmetry matrix present in the MLA10 PDB file. The Rpg1b and MLA10 CC domains display a similar pattern of secondary structures but only share 14.9% amino acid identity (41% similarity) over the modeled region. While the presence of conserved EDVID and heptad repeat regions facilitated the sequence alignment, alternative structures cannot be excluded due to ambiguities in the alignment. The structure shown was the one most similar to the MLA10 structure.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. GeneTree analyses of the NB-ARC domain: nonoptimized tree.
Supplemental Figure S2. COILS analysis of a consensus sequence derived from the alignment of nonrecombinant sequences corresponding to the CC domain.
Supplemental Table S1. NB-LRR genes characterized in this study.
Supplemental Table S2. Polymorphism statistics for the CC, NB-ARC, and LRR domains.
Supplemental Table S3. Recombination events.
Supplemental Table S4. Selection analysis statistics.
Supplemental Table S5. Bayesian analyses model likelihood scores, tree length (TL) variance, and sd of split frequencies (SD) between runs.
Supplemental Movie S1. Predicted tertiary structure of the Rpg1b NB-ARC domain showing amino acids under positive selection (for further details on the structure shown, see the Fig. 7A legend).
Supplemental Movie S2. Predicted tertiary structure of the Rpg1b CC domain showing amino acids under positive selection (for further details on the structure shown, see the Fig. 7B legend).
Supplementary Material
Acknowledgments
Computer support was provided by the Indiana University Information Technology Services Research Database Complex, the Computational Biology Service Unit from Cornell University (which is partially funded by Microsoft Corporation), and the Oklahoma University Advanced Center for Genome Technology. Molecular graphics images were produced using the UCSF Chimera package from the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by National Institutes of Health grant no. P41 RR001081).
Glossary
- NB
nucleotide-binding
- LRR
leucine-rich repeat
- CC
coiled-coil
- WGD
whole genome duplication
- mya
million years ago
- ORFs
open reading frames
- Gmw
soybean cv Williams 82
- BAC
bacterial artificial chromosome
- SLAC
Single Likelihood Ancestor Counting
- FEL
Fixed Effects Likelihood
- REL
Random Effects Likelihood
- PDB
Protein Data Bank
References
- Adams KL, Wendel JF. (2005) Polyploidy and genome evolution in plants. Curr Opin Plant Biol 8: 135–141 [DOI] [PubMed] [Google Scholar]
- Ade J, DeYoung BJ, Golstein C, Innes RW. (2007) Indirect activation of a plant nucleotide binding site-leucine-rich repeat protein by a bacterial protease. Proc Natl Acad Sci USA 104: 2531–2536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht M, Takken FL. (2006) Update on the domain architectures of NLRs and R proteins. Biochem Biophys Res Commun 339: 459–462 [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. (1990) Basic local alignment search tool. J Mol Biol 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashfield T, Danzer JR, Held D, Clayton K, Keim P, Saghai Maroof MA, Webb PM, Innes RW. (1998) Rpg1, a soybean gene effective against races of bacterial blight, maps to a cluster of previously identified disease resistance genes. Theor Appl Genet 96: 1013–1021 [Google Scholar]
- Ashfield T, Ong LE, Nobuta K, Schneider CM, Innes RW. (2004) Convergent evolution of disease resistance gene specificity in two flowering plant families. Plant Cell 16: 309–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell MJ, Staskawicz BJ. (2003) Initiation of RPS2-specified disease resistance in Arabidopsis is coupled to the AvrRpt2-directed elimination of RIN4. Cell 112: 369–377 [DOI] [PubMed] [Google Scholar]
- Baumgarten A, Cannon S, Spangler R, May G. (2003) Genome-level evolution of resistance genes in Arabidopsis thaliana. Genetics 165: 309–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bent AF, Mackey D. (2007) Elicitors, effectors, and R genes: the new paradigm and a lifetime supply of questions. Annu Rev Phytopathol 45: 399–436 [DOI] [PubMed] [Google Scholar]
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. (2000) The Protein Data Bank. Nucleic Acids Res 28: 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bittner-Eddy PD, Crute IR, Holub EB, Beynon JL. (2000) RPP13 is a simple locus in Arabidopsis thaliana for alleles that specify downy mildew resistance to different avirulence determinants in Peronospora parasitica. Plant J 21: 177–188 [DOI] [PubMed] [Google Scholar]
- Bomblies K, Weigel D. (2007) Hybrid necrosis: autoimmunity as a potential gene-flow barrier in plant species. Nat Rev Genet 8: 382–393 [DOI] [PubMed] [Google Scholar]
- Bowers JE, Chapman BA, Rong J, Paterson AH. (2003) Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 422: 433–438 [DOI] [PubMed] [Google Scholar]
- Bryson K, McGuffin LJ, Marsden RL, Ward JJ, Sodhi JS, Jones DT. (2005) Protein structure prediction servers at University College London. Nucleic Acids Res 33: W36–W38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon SB, Mitra A, Baumgarten A, Young ND, May G. (2004) The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol 4: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen NW, Sévignac M, Thareau V, Magdelenat G, David P, Ashfield T, Innes RW, Geffroy V. (2010) Specific resistances against Pseudomonas syringae effectors AvrB and AvrRpm1 have evolved differently in common bean (Phaseolus vulgaris), soybean (Glycine max), and Arabidopsis thaliana. New Phytol 187: 941–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung EH, da Cunha L, Wu AJ, Gao Z, Cherkis K, Afzal AJ, Mackey D, Dangl JL. (2011) Specific threonine phosphorylation of a host target by two unrelated type III effectors activates a host innate immune receptor in plants. Cell Host Microbe 9: 125–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins N, Drake J, Ayliffe M, Sun Q, Ellis J, Hulbert S, Pryor T. (1999) Molecular characterization of the maize Rp1-D rust resistance haplotype and its mutants. Plant Cell 11: 1365–1376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangl JL, Jones JD. (2001) Plant pathogens and integrated defence responses to infection. Nature 411: 826–833 [DOI] [PubMed] [Google Scholar]
- Delport W, Poon AF, Frost SD, Pond SLK. (2010) DataMonkey 2010: a suite of phylogenetic analysis tools for evolutionary biology. Bioinformatics 26: 2455–2457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeYoung BJ, Innes RW. (2006) Plant NBS-LRR proteins in pathogen sensing and host defense. Nat Immunol 7: 1243–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diers BW, Mansur L, Imsande J, Shoemaker RC. (1992) Mapping Phytophthora resistance loci in soybean with restriction fragment length polymorphism markers. Crop Sci 32: 377–383 [Google Scholar]
- Dodds PN, Lawrence GJ, Catanzariti AM, Teh T, Wang CI, Ayliffe MA, Kobe B, Ellis JG. (2006) Direct protein interaction underlies gene-for-gene specificity and coevolution of the flax resistance genes and flax rust avirulence genes. Proc Natl Acad Sci USA 103: 8888–8893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds PN, Lawrence GJ, Ellis JG. (2001) Contrasting modes of evolution acting on the complex N locus for rust resistance in flax. Plant J 27: 439–453 [DOI] [PubMed] [Google Scholar]
- Egan AN, Doyle J. (2010) A comparison of global, gene-specific, and relaxed clock methods in a comparative genomics framework: dating the polyploid history of soybean (Glycine max). Syst Biol 59: 534–547 [DOI] [PubMed] [Google Scholar]
- Eitas TK, Dangl JL. (2010) NB-LRR proteins: pairs, pieces, perception, partners, and pathways. Curr Opin Plant Biol 13: 472–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eswar N, Webb B, Marti-Renom MA, Madhusudhan MS, Eramian D, Shen MY, Pieper U, Sali A (2006) Comparative protein structure modeling using Modeller. Curr Protoc Bioinformatics Chapter 5: Unit 5.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geffroy V, Creusot F, Falquet J, Sévignac M, Adam-Blondon AF, Bannerot H, Gepts P, Dron M. (1998) A family of LRR sequences in the vicinity of the Co-2 locus for anthracnose resistance in Phaseolus vulgaris and its potential use in marker-assisted selection. Theor Appl Genet 96: 494–502 [DOI] [PubMed] [Google Scholar]
- Geffroy V, Macadré C, David P, Pedrosa-Harand A, Sévignac M, Dauga C, Langin T. (2009) Molecular analysis of a large subtelomeric nucleotide-binding-site-leucine-rich-repeat family in two representative genotypes of the major gene pools of Phaseolus vulgaris. Genetics 181: 405–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill N, Findley S, Walling JG, Hans C, Ma J, Doyle J, Stacey G, Jackson SA. (2009) Molecular and chromosomal evidence for allopolyploidy in soybean. Plant Physiol 151: 1167–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gore MA, Chia JM, Elshire RJ, Sun Q, Ersoz ES, Hurwitz BL, Peiffer JA, McMullen MD, Grills GS, Ross-Ibarra J, et al (2009) A first-generation haplotype map of maize. Science 326: 1115–1117 [DOI] [PubMed] [Google Scholar]
- Hughes AL, Yeager M. (1998a) Natural selection and the evolutionary history of major histocompatibility complex loci. Front Biosci 3: d509–d516 [DOI] [PubMed] [Google Scholar]
- Hughes AL, Yeager M. (1998b) Natural selection at major histocompatibility complex loci of vertebrates. Annu Rev Genet 32: 415–435 [DOI] [PubMed] [Google Scholar]
- Huson DH, Bryant D. (2006) Application of phylogenetic networks in evolutionary studies. Mol Biol Evol 23: 254–267 [DOI] [PubMed] [Google Scholar]
- Hwang CF, Williamson VM. (2003) Leucine-rich repeat-mediated intramolecular interactions in nematode recognition and cell death signaling by the tomato resistance protein Mi. Plant J 34: 585–593 [DOI] [PubMed] [Google Scholar]
- Innes RW. (2004) Guarding the goods: new insights into the central alarm system of plants. Plant Physiol 135: 695–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innes RW, Ameline-Torregrosa C, Ashfield T, Cannon E, Cannon SB, Chacko B, Chen NW, Couloux A, Dalwani A, Denny R, et al. (2008) Differential accumulation of retroelements and diversification of NB-LRR disease resistance genes in duplicated regions following polyploidy in the ancestor of soybean. Plant Physiol 148: 1740–1759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DT. (1999) Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol 292: 195–202 [DOI] [PubMed] [Google Scholar]
- Jones JD, Dangl JL. (2006) The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Kuang H, Caldwell KS, Meyers BC, Michelmore RW. (2008) Frequent sequence exchanges between homologs of RPP8 in Arabidopsis are not necessarily associated with genomic proximity. Plant J 54: 69–80 [DOI] [PubMed] [Google Scholar]
- Kuang H, Wei F, Marano MR, Wirtz U, Wang X, Liu J, Shum WP, Zaborsky J, Tallon LJ, Rensink W, et al. (2005) The R1 resistance gene cluster contains three groups of independently evolving, type I R1 homologues and shows substantial structural variation among haplotypes of Solanum demissum. Plant J 44: 37–51 [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948 [DOI] [PubMed] [Google Scholar]
- Lavin M, Herendeen PS, Wojciechowski MF. (2005) Evolutionary rates analysis of Leguminosae implicates a rapid diversification of lineages during the tertiary. Syst Biol 54: 575–594 [DOI] [PubMed] [Google Scholar]
- Leister D. (2004) Tandem and segmental gene duplication and recombination in the evolution of plant disease resistance gene. Trends Genet 20: 116–122 [DOI] [PubMed] [Google Scholar]
- Leister RT, Dahlbeck D, Day B, Li Y, Chesnokova O, Staskawicz BJ. (2005) Molecular genetic evidence for the role of SGT1 in the intramolecular complementation of Bs2 protein activity in Nicotiana benthamiana. Plant Cell 17: 1268–1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Librado P, Rozas J. (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25: 1451–1452 [DOI] [PubMed] [Google Scholar]
- Lupas A, Van Dyke M, Stock J. (1991) Predicting coiled coils from protein sequences. Science 252: 1162–1164 [DOI] [PubMed] [Google Scholar]
- Lynch M, Katju V. (2004) The altered evolutionary trajectories of gene duplicates. Trends Genet 20: 544–549 [DOI] [PubMed] [Google Scholar]
- Maekawa T, Cheng W, Spiridon LN, Töller A, Lukasik E, Saijo Y, Liu P, Shen QH, Micluta MA, Somssich IE, et al. (2011) Coiled-coil domain-dependent homodimerization of intracellular barley immune receptors defines a minimal functional module for triggering cell death. Cell Host Microbe 9: 187–199 [DOI] [PubMed] [Google Scholar]
- Martin D, Rybicki E. (2000) RDP: detection of recombination amongst aligned sequences. Bioinformatics 16: 562–563 [DOI] [PubMed] [Google Scholar]
- Martin DP, Posada D, Crandall KA, Williamson C. (2005a) A modified bootscan algorithm for automated identification of recombinant sequences and recombination breakpoints. AIDS Res Hum Retroviruses 21: 98–102 [DOI] [PubMed] [Google Scholar]
- Martin DP, Williamson C, Posada D. (2005b) RDP2: recombination detection and analysis from sequence alignments. Bioinformatics 21: 260–262 [DOI] [PubMed] [Google Scholar]
- McDowell JM, Simon SA. (2008) Molecular diversity at the plant-pathogen interface. Dev Comp Immunol 32: 736–744 [DOI] [PubMed] [Google Scholar]
- Meyers BC, Dickerman AW, Michelmore RW, Sivaramakrishnan S, Sobral BW, Young ND. (1999) Plant disease resistance genes encode members of an ancient and diverse protein family within the nucleotide-binding superfamily. Plant J 20: 317–332 [DOI] [PubMed] [Google Scholar]
- Meyers BC, Kozik A, Griego A, Kuang H, Michelmore RW. (2003) Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell 15: 809–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffett P, Farnham G, Peart J, Baulcombe DC. (2002) Interaction between domains of a plant NBS-LRR protein in disease resistance-related cell death. EMBO J 21: 4511–4519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondragon-Palomino M, Gaut BS. (2005) Gene conversion and the evolution of three leucine-rich repeat gene families in Arabidopsis thaliana. Mol Biol Evol 22: 2444–2456 [DOI] [PubMed] [Google Scholar]
- Mondragón-Palomino M, Meyers BC, Michelmore RW, Gaut BS. (2002) Patterns of positive selection in the complete NBS-LRR gene family of Arabidopsis thaliana. Genome Res 12: 1305–1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobuta K, Ashfield T, Kim S, Innes RW. (2005) Diversification of non-TIR class NB-LRR genes in relation to whole-genome duplication events in Arabidopsis. Mol Plant Microbe Interact 18: 103–109 [DOI] [PubMed] [Google Scholar]
- Padidam M, Sawyer S, Fauquet CM. (1999) Possible emergence of new geminiviruses by frequent recombination. Virology 265: 218–225 [DOI] [PubMed] [Google Scholar]
- Page RD. (1998) GeneTree: comparing gene and species phylogenies using reconciled trees. Bioinformatics 14: 819–820 [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. (2004) UCSF Chimera: a visualization system for exploratory research and analysis. J Comput Chem 25: 1605–1612 [DOI] [PubMed] [Google Scholar]
- Pond SL, Frost SD. (2005a) DataMonkey: rapid detection of selective pressure on individual sites of codon alignments. Bioinformatics 21: 2531–2533 [DOI] [PubMed] [Google Scholar]
- Pond SL, Frost SD, Muse SV. (2005) HyPhy: hypothesis testing using phylogenies. Bioinformatics 21: 676–679 [DOI] [PubMed] [Google Scholar]
- Pond SLK, Frost SDW. (2005b) Not so different after all: a comparison of methods for detecting amino acid sites under selection. Mol Biol Evol 22: 1208–1222 [DOI] [PubMed] [Google Scholar]
- Posada D, Crandall KA. (2001) Evaluation of methods for detecting recombination from DNA sequences: computer simulations. Proc Natl Acad Sci USA 98: 13757–13762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rairdan GJ, Collier SM, Sacco MA, Baldwin TT, Boettrich T, Moffett P. (2008) The coiled-coil and nucleotide binding domains of the potato Rx disease resistance protein function in pathogen recognition and signaling. Plant Cell 20: 739–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rairdan GJ, Moffett P. (2006) Distinct domains in the ARC region of the potato resistance protein Rx mediate LRR binding and inhibition of activation. Plant Cell 18: 2082–2093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richly E, Kurth J, Leister D. (2002) Mode of amplification and reorganization of resistance genes during recent Arabidopsis thaliana evolution. Mol Biol Evol 19: 76–84 [DOI] [PubMed] [Google Scholar]
- Riedl SJ, Li W, Chao Y, Schwarzenbacher R, Shi Y. (2005) Structure of the apoptotic protease-activating factor 1 bound to ADP. Nature 434: 926–933 [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574 [DOI] [PubMed] [Google Scholar]
- Sandhu D, Schallock KG, Rivera-Velez N, Lundeen P, Cianzio S, Bhattacharyya MK. (2005) Soybean Phytophthora resistance gene Rps8 maps closely to the Rps3 region. J Hered 96: 536–541 [DOI] [PubMed] [Google Scholar]
- Schlueter JA, Dixon P, Granger C, Grant D, Clark L, Doyle JJ, Shoemaker RC. (2004) Mining EST databases to resolve evolutionary events in major crop species. Genome 47: 868–876 [DOI] [PubMed] [Google Scholar]
- Schmutz J, Cannon SB, Schlueter J, Ma J, Mitros T, Nelson W, Hyten DL, Song Q, Thelen JJ, Cheng J, et al. (2010) Genome sequence of the palaeopolyploid soybean. Nature 463: 178–183 [DOI] [PubMed] [Google Scholar]
- Selote D, Kachroo A. (2010) RPG1-B-derived resistance to AvrB-expressing Pseudomonas syringae requires RIN4-like proteins in soybean. Plant Physiol 153: 1199–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen QH, Saijo Y, Mauch S, Biskup C, Bieri S, Keller B, Seki H, Ulker B, Somssich IE, Schulze-Lefert P. (2007) Nuclear activity of MLA immune receptors links isolate-specific and basal disease-resistance responses. Science 315: 1098–1103 [DOI] [PubMed] [Google Scholar]
- Shi J, Wolf SE, Burke JM, Presting GG, Ross-Ibarra J, Dawe RK. (2010) Widespread gene conversion in centromere cores. PLoS Biol 8: e1000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takken FL, Albrecht M, Tameling WI. (2006) Resistance proteins: molecular switches of plant defence. Curr Opin Plant Biol 9: 383–390 [DOI] [PubMed] [Google Scholar]
- Talbert PB, Henikoff S. (2010) Centromeres convert but don’t cross. PLoS Biol 8: e1000326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian D, Traw MB, Chen JQ, Kreitman M, Bergelson J. (2003) Fitness costs of R-gene-mediated resistance in Arabidopsis thaliana. Nature 423: 74–77 [DOI] [PubMed] [Google Scholar]
- van der Biezen EA, Jones JDG. (1998a) The NB-ARC domain: a novel signalling motif shared by plant resistance gene products and regulators of cell death in animals. Curr Biol 8: R226–R227 [DOI] [PubMed] [Google Scholar]
- van der Biezen EA, Jones JDG. (1998b) Plant disease-resistance proteins and the gene-for-gene concept. Trends Biochem Sci 23: 454–456 [DOI] [PubMed] [Google Scholar]
- van Ooijen G, van den Burg HA, Cornelissen BJC, Takken FLW. (2007) Structure and function of resistance proteins in solanaceous plants. Annu Rev Phytopathol 45: 43–72 [DOI] [PubMed] [Google Scholar]
- Wang CI, Guncar G, Forwood JK, Teh T, Catanzariti AM, Lawrence GJ, Loughlin FE, Mackay JP, Schirra HJ, Anderson PA, et al. (2007) Crystal structures of flax rust avirulence proteins AvrL567-A and -D reveal details of the structural basis for flax disease resistance specificity. Plant Cell 19: 2898–2912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Paterson AH. (2011) Gene conversion in angiosperm genomes with an emphasis on genes duplicated by polyploidization. Genes 2: 1–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wawrzynski A, Ashfield T, Chen NW, Mammadov J, Nguyen A, Podicheti R, Cannon SB, Thareau V, Ameline-Torregrosa C, Cannon E, et al. (2008) Replication of nonautonomous retroelements in soybean appears to be both recent and common. Plant Physiol 148: 1760–1771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu YG, Saghai Maroof MA, Buss GR, Maughan PJ, Tolin SA. (1994) RFLP and microsatellite mapping of a gene for soybean mosaic virus resistance. Phytopathology 84: 60–64 [Google Scholar]
- Zhang M, Wu YH, Lee MK, Liu YH, Rong Y, Santos TS, Wu C, Xie F, Nelson RL, Zhang HB. (2010) Numbers of genes in the NBS and RLK families vary by more than four-fold within a plant species and are regulated by multiple factors. Nucleic Acids Res 38: 6513–6525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Feng Y, Cheng H, Tian D, Yang S, Chen JQ. (2011) Relative evolutionary rates of NBS-encoding genes revealed by soybean segmental duplication. Mol Genet Genomics 285: 79–90 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.