Abstract
Iron (Fe) is an essential mineral nutrient for plants, but often it is not available in sufficient quantities to sustain optimal growth. To gain insights into adaptive processes to low Fe availability at the posttranslational level, we conducted a quantitative analysis of Fe deficiency-induced changes in the phosphoproteome profile of Arabidopsis (Arabidopsis thaliana) roots. Isobaric tags for relative and absolute quantitation-labeled phosphopeptides were analyzed by liquid chromatography-tandem mass spectrometry on an LTQ-Orbitrap with collision-induced dissociation and high-energy collision dissociation capabilities. Using a combination of titanium dioxide and immobilized metal affinity chromatography to enrich phosphopeptides, we extracted 849 uniquely identified phosphopeptides corresponding to 425 proteins and identified several not previously described phosphorylation motifs. A subset of 45 phosphoproteins was defined as being significantly changed in abundance upon Fe deficiency. Kinase motifs in Fe-responsive proteins matched to protein kinase A/calcium calmodulin-dependent kinase II, casein kinase II, and proline-directed kinase, indicating a possible critical function of these kinase classes in Fe homeostasis. To validate our analysis, we conducted site-directed mutagenesis on IAA-CONJUGATE-RESISTANT4 (IAR4), a protein putatively functioning in auxin homeostasis. iar4 mutants showed compromised root hair formation and developed shorter primary roots. Changing serine-296 in IAR4 to alanine resulted in a phenotype intermediate between mutant and wild type, whereas acidic substitution to aspartate to mimic phosphorylation was either lethal or caused an extreme dwarf phenotype, supporting the critical importance of this residue in Fe homeostasis. Our analyses further disclose substantial changes in the abundance of phosphoproteins involved in primary carbohydrate metabolism upon Fe deficiency, complementing the picture derived from previous proteomic and transcriptomic profiling studies.
Iron (Fe) often limits plant growth because of its tendency to form complexes of low solubility. To improve the acquisition of Fe from pools of limited availability, plants have developed a suite of responses that readjust cellular homeostasis, comprising changes in developmental programs, metabolism, and expression of transporters mediating the uptake and distribution of Fe. In Arabidopsis (Arabidopsis thaliana), Fe acquisition is controlled by two basic helix-loop-helix (bHLH) transcription factors, FER-LIKE IRON DEFICIENCY-INDUCED TRANSCRIPTION FACTOR (FIT) and POPEYE (PYE), regulating nonoverlapping subsets of genes with various roles in Fe uptake and metabolism (Colangelo and Guerinot, 2004; Bauer et al., 2007; Long et al., 2010; Schmidt and Buckhout, 2011). Disruption of FIT or PYE function leads to severe growth reduction and chlorosis under Fe-limited conditions, indicating that the function of these genes is critical for regulating Fe homeostasis. FIT forms heterodimers with bHLH38 and bHLH39 and positively regulates a subset of Fe-responsive genes, including two key genes required for Fe acquisition that encode the ferric reductase FERRIC REDUCTION OXIDASE2 and the Fe transporter IRT1 (Eide et al., 1996; Robinson et al., 1999; Vert et al., 2002; Colangelo and Guerinot, 2004; Yuan et al., 2008). PYE is preferentially expressed in the pericycle and aids in maintaining Fe homeostasis by positively regulating a separate cluster of genes. The expression of BRUTUS (BTS), encoding a putative E3 ligase protein that negatively regulates the Fe deficiency responses, is tightly correlated with PYE gene activity. Both proteins interact with the PYE homologs IAA-LEU-RESISTANT3 (ILR3) and bHLH115, suggesting a complex and dynamic regulatory circuit that adapts plants to fluctuating availability of Fe.
The signaling processes that are upstream of or parallel to FIT, PYE, and BTS are largely unknown. All three genes are regulated by the plant’s Fe status, indicating that other components are involved in Fe sensing and signaling. A study of Fe deficiency-induced protein profile changes revealed that many of the differentially expressed proteins were not associated with changes in their corresponding transcripts, indicating that posttranscriptional and posttranslational processes are employed to acclimate the plant to low Fe availability (Lan et al., 2011). Homozygous mutations in the UBIQUITIN CONJUGASE13A (UBC13A) locus caused alterations in Fe deficiency-induced root hair formation and in the expression of several Fe-responsive genes, including early signaling components such as bHLH38/39 (Li and Schmidt, 2010). UBC13 proteins catalyze the formation of noncanonical Lys-63-linked polyubiquitin chains, suggesting that posttranslational modifications of proteins are crucial for the Fe deficiency response (Hofmann and Pickart, 1999; Pickart and Fushman, 2004; Wen et al., 2006).
The possible function of other posttranslational processes in the Fe deficiency response has not been investigated. An estimated one-third of all eukaryotic proteins could be regulated by reversible phosphorylation via kinases/phosphatases, demonstrating the importance of this posttranslational protein modification. Modifications of protein with phosphate can affect protein structure, activity, localization, and stability, thereby regulating crucial processes such as metabolism and development. The reversible phosphorylation of proteins on Ser, Thr, and Tyr residues is the most common and conserved posttranslational modification and is one of the key regulators of the cellular responses to external stimuli. In transcriptional profiling experiments on Fe-deficient roots, several protein kinases were found to be differentially expressed upon Fe deficiency, suggesting that alterations in protein phosphorylation patterns induced by Fe deficiency are involved in the control of Fe homeostasis (Colangelo and Guerinot, 2004; Dinneny et al., 2008; Buckhout et al., 2009; García et al., 2010; Yang et al., 2010). Their exact role and substrates, however, remain enigmatic.
In contrast to the analysis of global changes in transcript abundance during development or in response to external stimuli, the role of posttranslational modification in these processes is understudied. Recent technological advances in mass spectrometry (MS)-based proteomics techniques allow the generation of a system-wide analysis of phosphoproteomics data via improved enrichment and analysis methodologies for phosphorylated proteins. A highly specific, unbiased enrichment of phosphopeptides is crucial for understanding regulatory processes that are mediated by reversible phosphorylation of proteins. Deciphering specific signaling pathways in response to external cues, however, not only requires the identification but also the quantification of phosphorylated peptides. Quantitative phosphoproteomics in plants remain a major challenge, mainly due to the low abundance of phosphorylated proteins and the neutral loss of phosphoric acid during the collision-induced dissociation (CID) process.
Here, we present a large-scale analysis of phosphopeptides that differentially accumulated upon Fe deficiency. It is shown that with a combination of two complementary phosphoprotein enrichment methods and identification/quantification of isobaric tags for relative and absolute quantitation (iTRAQ)-labeled proteins on an Orbitrap with CID/high-energy collision dissociation (HCD) capability, an accurate detection and determination of quantitative changes in the phosphoproteome upon Fe deficiency can be monitored from a relatively small sample input. Using this strategy, we show that Fe deficiency induces substantial, previously unnoted changes in the relative abundance of phosphoproteins. These changes are likely to be associated with functions in auxin homeostasis, RNA metabolism, carbohydrate flow, nitrate assimilation/amino acid synthesis, and transport processes at the plasma membrane.
RESULTS
Identification of Phosphopeptides in Arabidopsis Roots
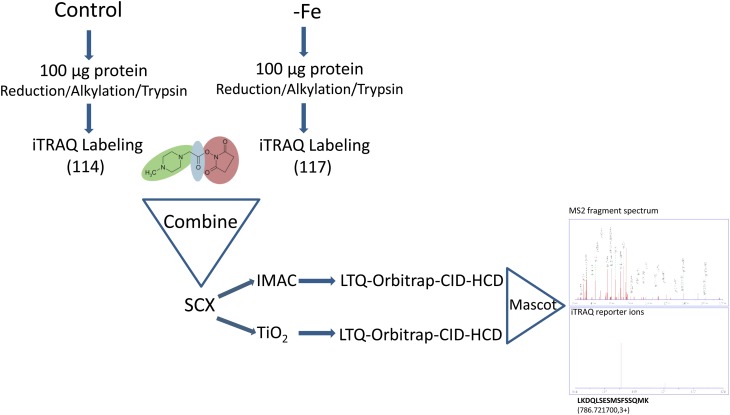
To gain insights into changes of protein phosphorylation patterns upon Fe deficiency, we quantitatively compared phosphopeptides isolated from roots of Fe-sufficient plants and roots from plants that were grown on Fe-depleted medium for 3 d. This time point was chosen because it mirrors an acclimation phase in which a new homeostasis is established (Kosová et al., 2011) and allows for a comparison with previous transcriptional and proteomic profiling studies using the same material and growth conditions (Yang et al., 2010; Lan et al., 2011). The workflow of the analysis is shown in Figure 1. Proteins were digested in solution and labeled with iTRAQ tags. The protein extraction protocol deviated from the manufacturer’s recommendation and was described previously (Lan et al., 2011). Samples were separated by strong cation-exchange (SCX) chromatography, and phosphopeptides were enriched by means of immobilized metal affinity chromatography (IMAC) and titanium dioxide (TiO2) chromatography. The two enrichment methods are partially complementary, yielding a higher coverage of the phosphopeptides in the sample (Bodenmiller et al., 2007). Peptides were sequenced by liquid chromatography-tandem mass spectrometry (MS/MS) on an LTQ-Orbitrap XL coupled to a linear ion trap that collects CID spectra for identification and determination of modification sites and HCD spectra for quantification of the iTRAQ reporter ions. CID in linear ion traps allows an accurate identification of phosphopeptides but is incompatible with iTRAQ labeling. HCD widens the scan range and can detect fragment ions with low mass-to-charge ratio (m/z). The 2.3.02 version of the Mascot software was used to simultaneously identify and quantify proteins. Using this strategy, we identified a total of 654 unique phospho-sites, spanning 849 phosphopeptides on 427 proteins (Supplemental Table S1). The distribution of phosphorylation events was Ser (87.2%), Thr (11.9%), and Tyr (0.9%; Fig. 2, inset). The vast majority of the peptides (98%) were singly phosphorylated.
Figure 1.
Experimental strategy for the enrichment, analysis, and quantification of phosphopeptides. [See online article for color version of this figure.]
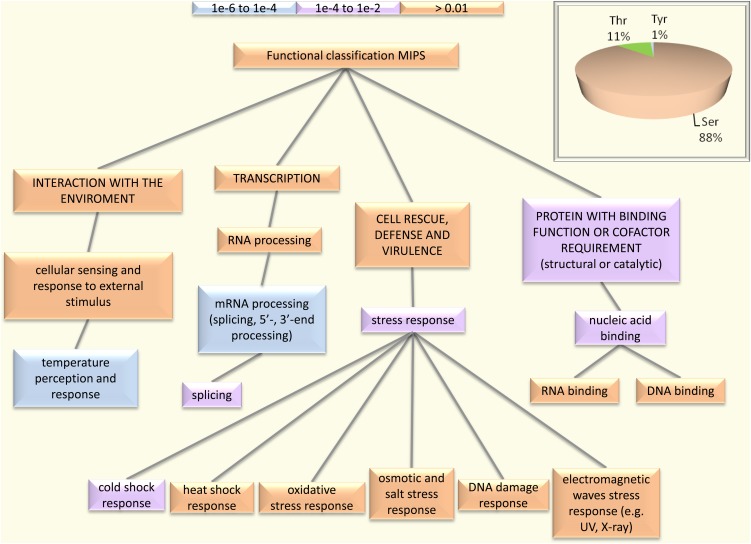
Figure 2.
Functional classification of Arabidopsis root phosphoproteins according to the Munich Information Center for Protein Sequences. The BioMap tool package provided at http://virtualplant.bio.nyu.edu/ was used for the analysis. [See online article for color version of this figure.]
Twenty-one significantly enriched phosphorylation motifs (grouped into 10 classes) were extracted from phosphopeptide data sets and identified using the Motif-X algorithm (Schwartz and Gygi, 2005). Nineteen motifs represent Ser phosphorylation, and two motifs were Thr phosphorylation sites (Table I; Supplemental Fig. S1). Some of these motifs, such sXH and KSXsXXN, to our knowledge, have not been reported previously.
Table I. Phosphorylation motifs identified in Arabidopsis roots.
The phosphorylated residue is indicated in lowercase underlined letters (s for Ser and t for Thr), and X denotes any amino acid. For sequence alignment, see Supplemental Figure S1.
| No. | Motif | Motif Score | No. | Fe Responsive | No. of Fe-Responsive Peptides | Putative Kinasea |
|---|---|---|---|---|---|---|
| I | ||||||
| 1 | E.....sP..... | 21.21 | 21 | Yes | 2 | Pro-directed kinase |
| 2 | .E....sP..... | 19.84 | 16 | No | ||
| 3 | ...... sP..... | 16.00 | 93 | Yes | 12 | |
| II | ||||||
| 4 | ...... sD.E... | 28.07 | 27 | No | CK II | |
| 5 | ...... sEDE... | 21.89 | 7 | No | ||
| 6 | ...... s.E.EE. | 16.73 | 8 | No | ||
| 7 | ...... sDDD... | 15.18 | 7 | Yes | 2 | |
| 8 | ...... s.E.... | 3.95 | 15 | No | ||
| 9 | .....As.E.... | 9.22 | 8 | No | ||
| 10 | ...... s.KE... | 9.65 | 9 | Yes | 1 | |
| 11 | .....Ds.D.... | 8.24 | 7 | No | ||
| III | ||||||
| 12 | ...H.. s.S.... | 10.82 | 8 | Yes | 1 | GSK3-like kinase |
| 13 | ....S. s...... | 4.06 | 26 | Yes | 1 | |
| IV | ||||||
| 14 | ...... s.S.... | 3.34 | 30 | Yes | 2 | PKA |
| V | ||||||
| 15 | ...... s.H.... | 3.56 | 8 | Yes | 2 | Protein kinase C |
| 16 | ...KS. s..N... | 14.43 | 6 | Yes | 1 | |
| VI | ||||||
| 17 | .....Gs...... | 4.51 | 27 | Yes | 2 | Novel CK II |
| VII | ||||||
| 18 | ...... s.....X | 16.00 | 7 | Yes | 1 | Unknown |
| VIII | ||||||
| 19 | ...R.. s...... | 16.00 | 74 | Yes | 5 | PKA/calcium calmodulin-dependent kinase II |
| IX | ||||||
| 20 | ......tD..... | 3.36 | 10 | Yes | 2 | CK II |
| X | ||||||
| 21 | ......tP..... | 3.55 | 10 | Yes | 2 | Pro-directed kinase |
Predicted by NetPhosK 1.0 (http://www.cbs.dtu.dk/services/NetPhosK/). The putative kinase with the highest score was chosen.
Proteins Involved in RNA Processing, Translation, and Carbohydrate Metabolism Form Major Hubs in the Root Phosphoproteome Interaction Network
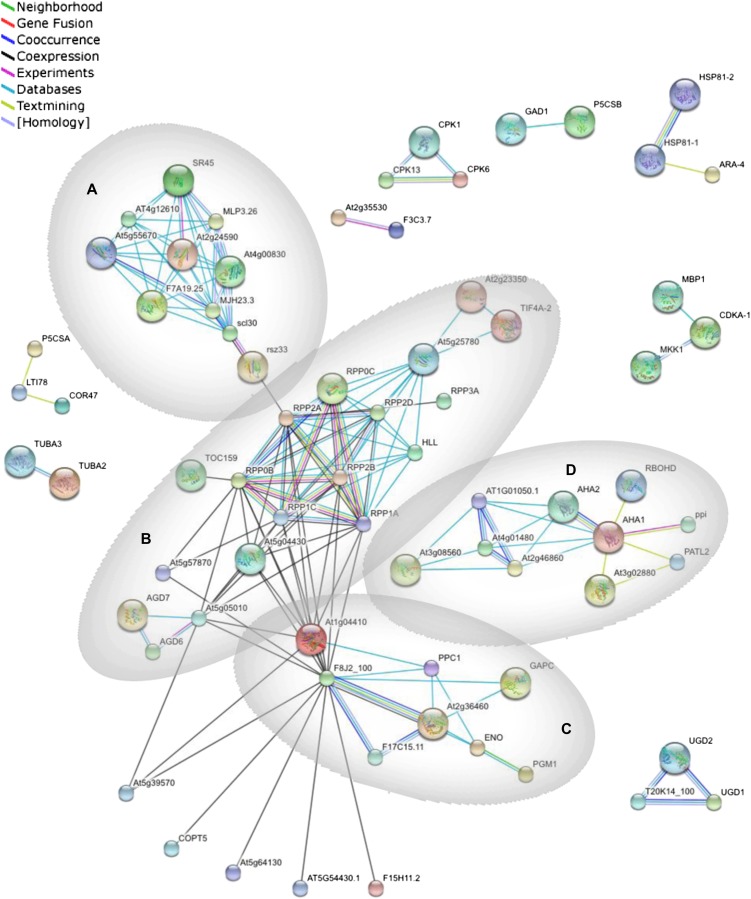
Functional classification of phosphoproteins according to the Munich Information Center for Protein Sequences revealed highest P values for the Gene Ontology categories “temperature perception and response” and “RNA processing” (Fig. 2). To get a better understanding of root-related processes that are regulated by phosphorylation, a protein-protein interaction network was constructed using the STRING system (http://string-db.org; Szklarczyk et al., 2011). This software package functionally links proteins based on predictions, neighborhood analysis, experimental results, and literature mining. Four major clusters, derived from the mapping of interologs from the root phosphoproteome, were associated with mRNA processing, translation, carbohydrate metabolism, and proton transport (Fig. 3, clusters A, B, C, and D, respectively). Cluster A is composed of five proteins with RNA-binding motifs: an RNA polymerase II transcription factor and four Ser-Arg-rich splicing factors that belong to a highly conserved family of splicing regulators involved in constitutive and alternative splicing of eukaryotic mRNA that are responsive to stresses and hormones (Palusa et al., 2007). The second cluster (B) consists of 18 proteins, nine of which are ribosomal proteins, indicating that translation is regulated and/or biased by phosphorylation. Several proteins with evolutionarily conserved phosphorylation sites involved in primary carbohydrate metabolism showed close interactions and constitute the third cluster (C). The remaining cluster (D) contains among other proteins the two major plasma membrane H+-ATPases AHA1 and AHA2, the proton pump interactor 1, and the three putative pyrophosphatases PPA1, PPA3, and PPA5.
Figure 3.
Interaction network of the identified phosphoproteins. Mapping of the network was performed with the STRING system (http://string.embl.de) based on known and predicted interactions. Lines of different colors represent different types of evidence for the associations. Four major clusters associated with mRNA processing (A), translation (B), carbohydrate metabolism (C), and proton transport (D) were derived from the associations of the proteins. Proteins without interactions have been removed from the graph. [See online article for color version of this figure.]
Fe Deficiency Alters Carbohydrate Flow at Different Regulatory Levels
Phosphopeptides that had a 117-114 iTRAQ ratio outside the range of 0.74- to 1.36-fold and that were detected in both biological experiments with similar accumulation patterns were classified as being significantly changed in abundance upon Fe deficiency. The cutoff values were selected by power analysis with a power index of 7.84 (95% confidence; for details, see “Materials and Methods”). From the motif patterns of the Fe-responsive phosphopeptides, it can be concluded that the Pro-directed kinase, casein kinase II (CK II), and protein kinase A (PKA)/calcium calmodulin-dependent kinase II kinase-mediated phosphorylation may play critical roles in the acclimation to Fe deficiency (Table I).
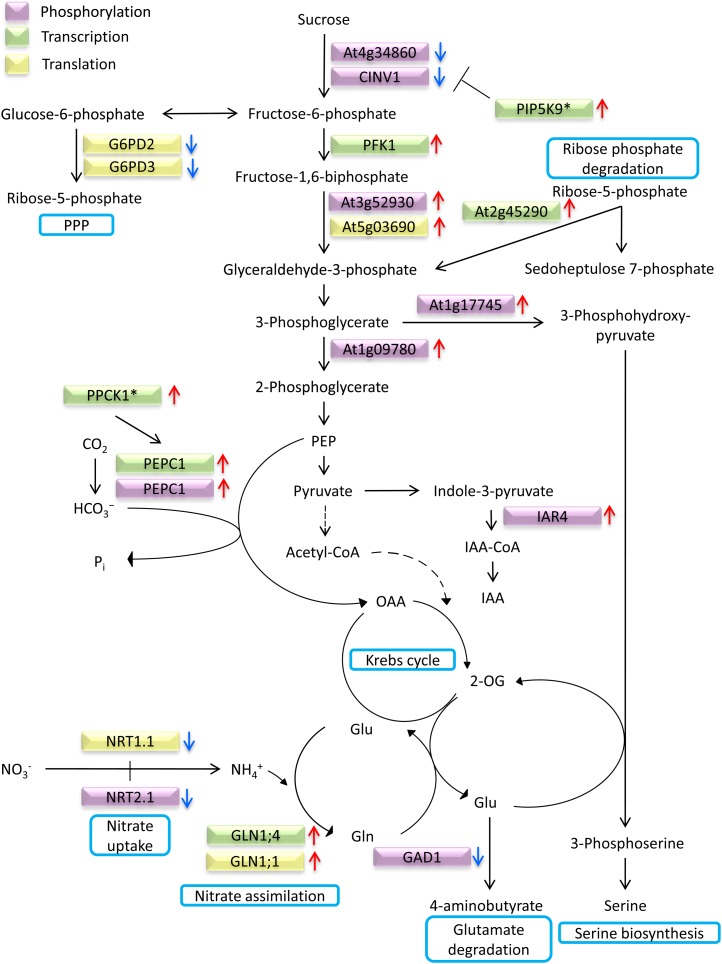
Several phosphoproteins that passed our criteria are related to carbohydrate metabolism (Table II). Figure 4 illustrates alterations in the flow of carbohydrate induced by Fe deficiency under different regulatory levels, integrating data obtained from transcriptional and proteomic profiling studies conducted earlier on plants that were grown under similar conditions (Yang et al., 2010; Lan et al., 2011). Decreased abundance of glucose-6-phosphate dehydrogenase (G6PD2/G6PD3) protein indicates that diversion of the hexose phosphate pool into the nonoxidative pentose phosphate pathway is reduced upon Fe deficiency. Increased abundance of phosphorylated 3-phosphoglycerate dehydrogenase (At1g17745) is indicative of increased Ser biosynthesis. Phosphorylation of phosphoglycerate mutase, catalyzing the conversion of 3-phosphoglycerate into 2-phosphoglycerate, also increased under Fe-deficient conditions. Rib-phosphate degradation may contribute to replenish the triose phosphate pool that is depleted by these two enzymes (Fig. 4). A gene encoding a transketolase (At2g45290) catalyzing the conversion of Rib-5-P and Xyl-5-P to sedoheptolose-7-phosphate and glyceraldehyde-3-phosphate was found to be up-regulated upon Fe deficiency in a previous study (Yang et al., 2010). Phosphoenolpyruvate carboxylase (PEPC) is a ubiquitous cytosolic enzyme in vascular plants that mediates the β-carboxylation of phosphoenolpyruvate, yielding oxaloacetate and inorganic phosphate. PEPC is tightly controlled transcriptionally by allosteric effectors and by reversible phosphorylation at the conserved N-terminal seryl residue. Phosphorylated PEPC1 protein and PEPC1 transcript accumulated under Fe-deficient conditions, indicating multiple regulatory levels of PEPC activity (Yang et al., 2010; this study).
Table II. Quantification of Fe deficiency-regulated phosphopeptides.
Phosphorylated residues are indicated by p preceding the phospho-amino acid. Oxidized Met is labeled by °M.
| Locus | Protein Description | Peptide | 117-114 ± sd |
|---|---|---|---|
| At1g04820.1 | Tortifolia 2, tubulin α-4 chain (TUA4) | TIQFVDWCPpTGFK | 0.61 ± 0.06 |
| At1g08800.1 | Unknown protein | pSVSFNHLPDVGYTELK | 0.59 ± 0.18 |
| At1g08800.1 | Unknown protein | SVpSFNHLPDVGYTELK | 0.59 ± 0.10 |
| At1g09780.1 | Phosphoglycerate mutase | AHGTAVGLPSEDD°MGNpSEVGHNALGAGR | 2.15 ± 0.85 |
| At1g09780.1 | Phosphoglycerate mutase | AHGTAVGLPSEDDMGNpSEVGHNALGAGR | 2.67 ± 1.80 |
| At1g09780.1 | Phosphoglycerate mutase | AHGTAVGLPSEDDMGNpSEVGHNALGAGR | 2.18 ± 0.64 |
| At1g17745.1 | 3-Phosphoglycerate dehydrogenase (PGDH) | FSTVGpSDSDEYNPTLPKPR | 1.36 ± 0.2 |
| At3g52930.1 | Fru-bisP aldolase | LGDGAAEpSLHVK | 1.61 ± 0.31 |
| At4g34860.1 | Plant neutral invertase family protein | APDHADYVIpSPSFGR | 0.66 ± 0.01 |
| At5g17330.1 | Glu decarboxylase 1 (GAD1) | VLSHAVpSESDVpSVHSTFASR | 0.56 ± 0.24 |
| At1g20110.1 | RING/FYVE/PHD zinc finger superfamily protein | SRpSDLGSDLYGK | 1.39 ± 0.42 |
| At1g20110.1 | RING/FYVE/PHD zinc finger superfamily protein | pSRSDLGSDLYGK | 1.41 ± 0.39 |
| At1g20440.1 | Cold-regulated 47 (COR47) | AQIpSEPELAAEHEEVKENK | 1.67 ± 0.33 |
| At1g24180.1 | IAA-conjugate-resistant 4 (IAR4) | YHGHpS°MSDPGSTYR | 1.66 ± 0.006 |
| At1g24180.1 | IAA-conjugate-resistant 4 (IAR4) | YHGHpSMSDPGSTYR | 1.61 ± 0.07 |
| At1g35580.1 | Cytosolic invertase 1 (CINV1) | AVGpSHCSLSEMDDLDLTR | 0.67 ± 0.19 |
| At3g07660.1 | Kinase-related protein | SGPDpSDGRVSPFVSPGVASK | 0.72 ± 0.10 |
| At3g07660.1 | Kinase-related protein | SGPDSDGRVpSPFVSPGVASK | 0.73 ± 0.27 |
| At3g11330.1 | Plant intracellular ras group-related lrr 9 (PIRL9) | TYVADVSEYLGSNpSPRDPYLER | 1.37 ± 0.16 |
| At1g53310.1 | Phosphoenolpyruvate carboxylase 1 (PEPC1) | MApSIDVHLR | 1.41 ± 0.21 |
| At3g12140.1 | Emsy N terminus (ENT)/plant Tudor-like domains-containing protein | HATIQPFDVLPpSPTFSAAR | 0.72 ± 0.002 |
| At1g69040.1 | ACT repeat 4 (ACR4) | SKpSFVNFGLVR | 0.73 ± 0.31 |
| At1g72160.1 | Sec14p-like phosphatidylinositol transfer family protein | S°MIPQNLGpSFKEESSK | 0.71 ± 0.12 |
| At2g18960.1 | AHA1 | GLDIDTAGHHpYTV | 0.74 ± 0.02 |
| At2g46630.1 | Unknown protein | VLpSPYSLPASLLHSER | 0.70 ± 0.007 |
| At3g05090.1 | Lateral root stimulator 1 (LRS1) | TVGSpSNNISVQSSPSHGYTPTIAK | 0.68 ± 0.21 |
| At3g15450.1 | Aluminum-induced protein | RGpSEANWALANSR | 0.56 ± 0.15 |
| At3g15450.1 | Aluminum-induced protein | IDpSEGVLCGASFK | 0.37 ± 0.15 |
| At3g16560.1 | Protein phosphatase 2C family protein | KVPpSSPALSK | 0.60 ± 0.09 |
| At3g48450.1 | RPM1-interacting protein 4 (RIN4) family protein | TAVAGPESIVpSPPRNEEPPK | 3.02 ± 0.14 |
| At3g48890.1 | Membrane steroid-binding protein 2 (MSBP2) | KDVApTDDDDAAKE | 1.60 ± 0.51 |
| At3g55460.1 | SC35-like splicing factor 30 (SCL30) | RpSYpSPGYEGAAAAAPDR | 0.66 ± 0.18 |
| At1g80930.1 | MIF4G domain-containing protein | RKEpTSDDEELAR | 1.45 ± 0.38 |
| At4g17720.1 | RNA-binding (RRM/RBD/RNP motifs) family protein | VHLSEpSPK | 1.58 ± 0.27 |
| At4g35785.1 | RNA-binding (RRM/RBD/RNP motifs) family protein | pTPTPGHYLGLK | 1.96 ± 1.0 |
| At3g18240.1 | Ribosomal protein S24/S35 | HAEpTDDELLEK | 1.47 ± 0.39 |
| At4g22670.1 | HSP70-interacting protein 1 (HIP1) | SFVVEEpSDDD°MDETEEVKPK | 0.70 ± 0.29 |
| At5g53330.1 | Ubiquitin-associated/translation elongation factor EF1B protein | NSpSFQHNTSPSSGIGIR | 1.38 ± 0.15 |
| At4g39150.1 | NAJ heat shock N-terminal domain-containing protein | ENpSLRHEEETGVK | 1.45 ± 0.41 |
| At4g35890.1 | Winged-helix DNA-binding transcription factor family protein | ISGNHGSPTASVAQpSPR | 1.83 ± 0.26 |
| At5g13890.1 | Family of unknown function (DUF716) | TGpSYEALPTNNADSNHIQMK | 0.66 ± 0.35 |
| At5g13890.1 | Family of unknown function (DUF716) | TGpSYEALPTNNADSNHIQMK | 0.62 ± 0.05 |
| At5g17920.1 | Cobalamin-independent Met synthase (ATCIMS) | YGAGIGPGVYDIHpSPR | 0.60 ± 0.04 |
| At5g19770.1 | Tubulin α-3 (TUA3) | TVQFVDWCPpTGFK | 0.56 ± 0.11 |
| At5g40390.1 | Seed imbibition 1-like SIP1 | SDpSGINGVDFTEK | 0.69 ± 0.02 |
| At5g49400.1 | Zinc knuckle (CCHC-type) family protein | TKPSVDDLDGpSDDDDEEERPDATNGK | 2.64 ± 1.94 |
| At5g49400.1 | Zinc knuckle (CCHC-type) family protein | TKPSVDDLDGpSDDDDEEERPDATNGK | 1.82 ± 0.70 |
| At5g61150.1 | Vernalization independence 4 (VIP4) | NDVEQDEHRpSPIEDEEGSEK | 1.47 ± 0.33 |
| At5g64680.1 | Unknown protein | SMFSGFTETPKpSPK | 0.63 ± 0.10 |
| At3g60600.1 | Vesicle-associated protein 27-1 (VAP27) | VTYVAPPRPPpSPVHEGSEEGSpSPR | 0.74 ± 0.10 |
| At4g11740.1 | SAY4 (suppressor of Ara4) | AASGSLAPPNADRpSR | 1.38 ± 0.02 |
| At5g43350.1 | Phosphate transporter 1 (PHT1;1) | SLEELSGEAEVpSHDEK | 0.74 ± 0.15 |
| At5g49890.1 | Chloride channel C (CLC-C) | KIpSGILDDGSVGFR | 0.71 ± 0.006 |
| At1g08090.1 | Nitrate transporter 2.1 (NRT2.1) | EQSFAFSVQpSPIVHTDK | 0.56 ± 0.01 |
Figure 4.
Fe deficiency-induced alterations in carbohydrate metabolism at different regulator levels. Up-pointing arrows indicate up-regulated processes, and arrows pointing downward denote down-regulated processes. See text for details. Asterisks indicate genes that were significantly up-regulated more than 1.5-fold but less than 2-fold. Protein data were taken from Lan et al. (2011); transcript changes from Yang et al. (2010). OAA, Oxaloacetate; 2-OG, 2-oxoglutarate; PEP, phosphoenolpyruvate; PPP, pentose phosphate pathway.
Ser-296 Phosphorylation of IAR4 Is Important for Auxin and Fe Homeostasis
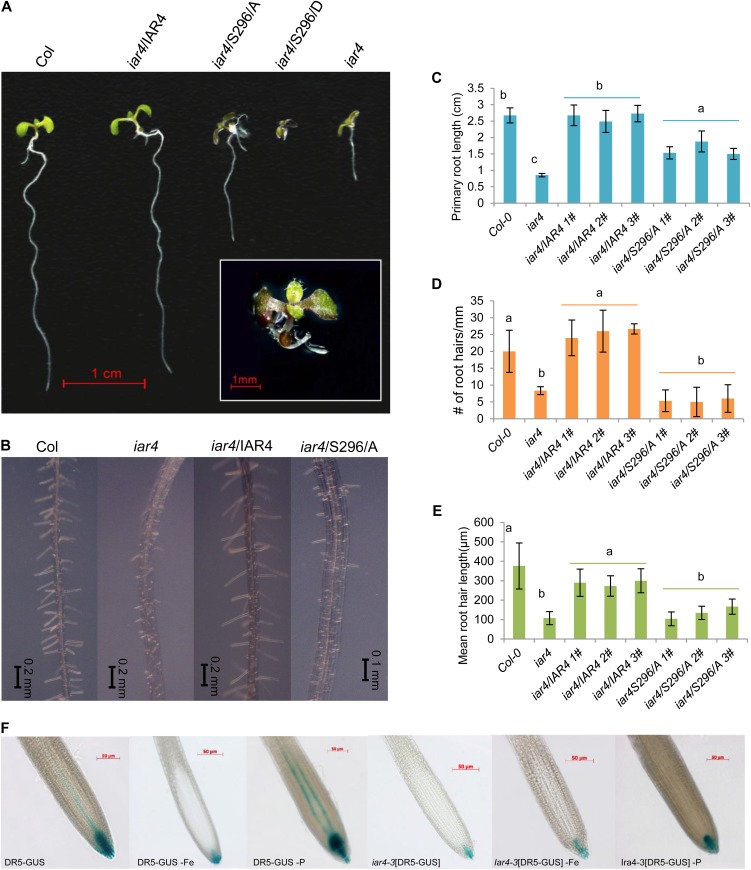
The mitochondrial pyruvate dehydrogenase E1α subunit, IAA-CONJUGATE-RESISTANT4 (IAR4), is a further phosphoprotein with increased abundance upon Fe deficiency. Pyruvate dehydrogenase catalyzes the conversion of pyruvate to acetyl-CoA; the precise function of IAR4, however, is not clear. It has been speculated that IAR4 catalyzes the conversion of indole-3-pyruvate to indole-3-acetic acid (IAA)-CoA, an intermediate of IAA biosynthesis in microbes (LeClere et al., 2004). The accumulation of IAR4 phosphopeptides could thus be related to the synthesis of auxin, the level of which increases under conditions of Fe deficiency in Arabidopsis (Chen et al., 2010). To assess the role of Ser-296 phosphorylation of IAR4, we have changed this residue by site-directed mutagenesis to Ala (S296A) or Asp (S296D) and introduced these constructs into an iar4 knockout line (SALK_091909) harboring a T-DNA insertion located in the first exon. In line with previous reports, both the number and length of root hairs are reduced in iar4 plants when compared with the wild type (Fig. 5, A–E; Quint et al., 2009). In addition, the length of the primary root is severely decreased in the mutant. Complementation lines in which Ser-296 was changed to Ala (iar4/S296A) showed an intermediate phenotype between the wild type and mutant, whereas acidic substitution for Ser-296 to mimic phosphorylation (iar4/S296D) was either lethal (approximately 50% of the seedlings) or had severe consequences on growth, resulting in an extreme dwarf phenotype (Fig. 5). Most of the kanamycin-resistant iar4/429D seedlings were purple colored and formed very short primary roots (Fig. 5A). Complementation of the iar4 mutant with the wild-type gene restored normal growth (Fig. 5A).
Figure 5.
IAR4 is critical in root development, root hair formation, and auxin response. A, Phenotypes of the wild type (Col-0), the iar4 mutant, an iar4/IAR4 complementation line, and substitution lines in which Ser-296 was changed to Ala (iar4/S296A) or Asp (iar4/S296D). Analysis was performed on 6-d-old plants. B, Root hair phenotypes of Col-0, iar4, iar4/IAR4, and iar4/S296A. C to E, Quantification of primary root length (C), root hair number (D), and root hair length (E) of the investigated lines. Different letters indicate significant differences at P < 0.05. F, DR5-GUS expression in Col-0 plants and iar4 mutants (iar4-3[DR5-GUS]) under control, Fe-deficient, and phosphate-deficient conditions. Thirteen-day-old plants were analyzed for GUS expression.
To elucidate whether auxin homeostasis is affected by Fe deficiency, we grew transgenic plants containing the DR5-GUS reporter construct under control and Fe-deficient conditions. Contrary to our expectations, GUS staining was markedly reduced in Fe-deficient seedlings (Fig. 5F). Phosphate-deficient DR5-GUS plants exhibited markedly higher levels of GUS staining compared with plants grown on phosphate-replete medium (Fig. 5F). These results also indicate that the decrease in auxin response upon Fe deficiency is not part of a nonspecific stress response but rather associated with the growth type. Similar to what has been reported previously (Quint et al., 2009), GUS staining of iar4-3[DR5-GUS] plants was markedly lower when compared with the wild type, supporting a critical function of IAR4 in auxin homeostasis. The DR5 expression pattern in the mutant among the different growth types was generally similar to the wild type, but lower staining intensity was observed under all tested conditions (Fig. 5F).
Fe Deficiency Alters the Phosphorylation Status of Plasma Membrane-Bound Transporters
The reduction-based Fe acquisition system of strategy I plants is dependent on an acidic rhizosphere to allow an optimal function of the plasma membrane-bound reductase and to increase the solubility of Fe, which decreases 1,000-fold for each unit of pH increase. The apoplastic pH is dictated by P-type ATPase-mediated proton extrusion and by the anion-uptake pattern. P-type ATPases are regulated posttranslationally in a complex way, including phosphorylation (Palmgren, 2001). We previously showed that in Arabidopsis, AHA2 is strongly up-regulated by Fe deficiency and encodes the isoform involved in rhizosphere acidification, whereas AHA1 plays a housekeeping function (Santi and Schmidt, 2009). Contrary to the anticipated increased abundance of AHA2 phosphopeptides, only AHA1 phosphopeptides accumulated differentially, being less abundant under Fe-deficient conditions.
Anion uptake is decreased upon Fe deficiency, probably to avoid alkalization of the apoplast associated with the proton-coupled uptake of anions. Phosphorylated PHT1;1 and NRT2.1 proteins, catalyzing the transport of phosphate and nitrate, decreased in abundance under Fe-deficient conditions. Consistent with a reduced nitrate uptake, the dual-affinity transporter NRT1.1 was found to be down-regulated upon Fe deficiency at the protein level (Lan et al., 2011). Ammonium assimilation via Gln synthetase is increased at the transcript (GLN1;4) and protein (GLN1;1) levels, possibly due to a decrease in activity of the Fe-containing enzymes nitrate reductase and nitrite reductase. Decreased accumulation of phosphorylated GAD1 protein, likely associated with decreased GAD1 activity and, thus, Glu degradation, prevents the depletion of the Glu pool in Fe-deficient plants. GAD1 was predicted to interact with FAB1B, a homolog of the Fab1 phosphatidylinositol-3-phosphate 5-kinase from yeast.
DISCUSSION
Accuracy and Precision of iTRAQ Quantitation
Since its invention in 2004 (Ross et al., 2004), iTRAQ has become a popular technique for large-scale quantitative proteome analysis. This success is based on its capability of multiplexing up to eight samples in one experimental setup, its unbiased peptide labeling (iTRAQ isobaric tags label Lys side groups, and all free terminal amino groups of the peptides are present in the sample), and its support by several software packages, such as Mascot, Pro Quant, SpectrumMill, I-Tracker, and Libra, among others (Lacerda et al., 2008). The extensive exploration of the iTRAQ technique over the past few years, however, also unearthed some caveats of this technique. In particular, the precision (i.e. reproducibility) and accuracy (i.e. closeness to true values) of quantitative expression data from iTRAQ experiments have been challenged. Generally, precision is affected by random errors and unpredictable fluctuations around the true value (Karp et al., 2010). In our study here, this issue was addressed by the removal of outliers and the use of weighted means in data analysis. Summed intensities were set as normalization to further improve the precision. In addition, detected peptides that showed a contradicting accumulation pattern in the two biological experiments were excluded from further analysis. Accuracy concerns, by contrast, are caused by predictable systematic biases (Karp et al., 2010). In the iTRAQ methodology, concerns regarding accuracy derived from the possibility that coeluted, near-isobaric ions that are isolated and fragmented together with the target ions lead to a distortion in reporter ion intensities (Ow et al., 2009; Karp et al., 2010; Ting et al., 2011; Yang et al., 2011). Several strategies have been employed to decrease or eliminate the accuracy bias. One of these strategies comprises a sample pretreatment by SCX chromatography, which decreases sample complexity and enriches low-abundance peptides (Yang et al., 2011). A second strategy aims at eliminating protein quantification interference by applying triple-stage MS (Ting et al., 2011). Here, a selected fragment from the tandem mass scans used for peptide identification serves as a reporter in a third scan, a procedure that significantly decreases ratio distortion. However, as a drawback of this strategy, protein coverage is decreased (Ting et al., 2011). Other means, such as narrowing the precursor selecting window, were also reported to increase the accuracy of iTRAQ (Ting et al., 2011). In our study, peptide mixtures were cleaned by SCX chromatography after iTRAQ labeling. The phosphopeptides were further enriched by TiO2 and IMAC chromatography, followed by immediate MS/MS analysis. All these procedures lower the sample complexity and enrich low-abundance peptides, leading to higher accuracy of the reporter ion intensity and higher signal-to-noise ratio.
So far, no generally accepted standardization for the significance of protein/peptide abundance change has been implemented in quantitative proteomic analysis using the iTRAQ method. Different fold changes were chosen as cutoffs to denote the significance of a protein/peptide abundance change. In this and a previous study (Lan et al., 2011), we adopted a method described by Cox and Mann (2008) and further confirmed the calculated cutoff values by power analysis. Based on the above considerations, we consider the fold changes used here as conservative and as accurate as possible without introducing work-intensive and thus limiting techniques, such as confirmation by western blotting with specific antibodies or reporter-based strategies such as absolute quantification (Gerber et al., 2003). It should also be noted that, in principle, differences in phosphopeptide accumulation could reflect changes in the total protein abundance rather than in the stoichiometric phosphorylation level per se.
Phosphoproteomics Reveals Novel Aspects in Carbohydrate Metabolism of Fe-Deficient Plants
In support of previous studies at the protein or transcript level, glycolysis appears to be activated upon Fe deficiency (Thimm et al., 2001; Brumbarova et al., 2008; Li et al., 2008, 2009; Rellán-Alvarez et al., 2010). The data from this study support this finding and add several novel aspects regarding changes of carbon flow during Fe deficiency. For example, phosphopeptides associated with invertase (CINV1 and At4g34860) showed decreased abundance, suggesting that the hexose phosphate pool fueling the increased glycolytic flux was derived rather from starch degradation and not from Suc delivered from the leaves. CINV1 was shown to be negatively regulated by phosphatidylinositol monophosphate 5-kinase 9 (PIP5K9), possibly by compromising CINV1 activity through interactions at the C terminus (Lou et al., 2007). Transcript levels of PIP5K9 were found to be increased upon Fe starvation (Yang et al., 2010), indicating that Suc breakdown is down-regulated both by decreased phosphorylation at the N terminus and by binding to PIP5K9 at the C terminus, probably in response to a reduced photosynthetic rate of Fe-deficient plants. Phosphorylation of CINV1 may thus represent a major regulatory hub for the flux of carbohydrate in response to environmental conditions.
A hallmark of Fe deficiency responses of strategy I plants is the increase of CO2 dark fixation via PEPC activity upon Fe deficiency, probably to support anaplerotic replenishment of carboxylic acid cycle intermediates and to balance the intracellular pH that is alkalinized by active proton extrusion via P-type ATPases (Landsberg, 1986; Rabotti et al., 1995; De Nisi and Zocchi, 2000). Unlike in inorganic phosphate-deficient conditions, where PEPC is phosphorylated by PPCK1 and PPCK2 (Chen et al., 2007; Gregory et al., 2009), in Fe-deficient plants, PPCK1 but not PPCK2 was induced, indicating that the increased abundance of phosphorylated PEPC1 upon Fe deficiency is mediated chiefly by PPCK1.
Auxin Metabolism Is Critical for Fe Homeostasis
To validate our findings regarding the importance of the identified phosphorylation sites, we changed the phosphorylation site at Ser-296 of pyruvate dehydrogenase E1α subunit IAR4. iar4 mutants have short roots and are less sensitive to IAA-amino acid conjugates (LeClere et al., 2004). Mutations in the second E1α subunit gene, encoding a protein 81% identical to IAR4, did not lead to the phenotypes of iar4, pointing to a different or additional function of the latter protein. The importance of the mutation is evident from the extreme phenotype of transgenic lines with acidic substitution of Ser-296, which mimics phosphorylation. It is tempting to speculate that this phenotype is derived from a dramatically altered auxin homeostasis, an assumption that is supported by previous studies on the iar4 mutant (LeClere et al., 2004; Quint et al., 2009). Interestingly, another mutant with reduced sensitivity to IAA conjugates, IAA-Leu-Resistant3 (ilr3), showed altered Fe and manganese uptake patterns and reduced expression of three CCC1-like genes that we recently identified as putative vacuolar Fe transporters (Rampey et al., 2006; Gollhofer et al., 2011). ILR3 encodes the bHLH transcription factor bHLH105 and interacts with the transcription factor PYE, indicating close linkage of IAA and Fe metabolism (Long et al., 2010). Although the precise roles of ILR3 and IAR4 in Fe homeostasis remain elusive, our findings underline the importance of metal homeostasis for auxin metabolism reported by Rampey et al. (2006). These results indicate further that the Fe-dependent phosphorylation of IAR4 may be important for IAA metabolism by directing the carbon flux from pyruvate to IAA under Fe-limited conditions.
An unanticipated finding was the reduced GUS staining in Fe-deficient plants. This finding provides a possible explanation for the branched root hair phenotype that is typical of Fe-deficient plants (Müller and Schmidt, 2004). The formation of branched root hairs has been observed in the auxin-resistant mutant axr2 (Wilson et al., 1990; Nagpal et al., 2000) and in mutants defective in the expression of the RING domain ligases RGLG1/RGLG2 causing compromised auxin transport (Yin et al., 2007; Li and Schmidt, 2010). Interestingly, rglg1 rglg2 double mutants produce constitutively branched root hairs independent on the Fe supply, but root hair development was similar to the wild type when the plants were grown on phosphate-depleted medium (Li and Schmidt, 2010). Phosphate deficiency increased GUS staining in DR5-GUS plants, suggesting that the branching of hairs is inhibited by higher levels of or increased responses to auxin. The lower GUS activity of Fe-deficient plants does not correlate well with the reportedly increased auxin levels in Fe-deficient roots (Chen et al., 2010). Expression of the auxin-responsive reporter DR5 does not necessarily reflect auxin concentration, which provides a possible explanation for the seemingly contrasting results.
Differential Phosphorylation of RNA-Binding Proteins Indicates Posttranscriptional Regulation of RNA Metabolism
The abundance of phosphopeptides of four RNA-binding proteins (RBPs) was found to be significantly altered by Fe deficiency, and three RBPs showed increased phosphorylation sites under Fe-deficient conditions (Table II). Overrepresentation of RNA-binding proteins in the phosphoproteomes has been reported earlier, indicating the importance of protein phosphorylation in RNA processing (de la Fuente van Bentem et al., 2006; Sugiyama et al., 2008). RBPs are important in posttranscriptional processes including RNA stability, export, splicing, and translation. RBPs were shown to be involved in adaptation to environmental conditions, but the mechanisms are not well understood (Lorković, 2009). Binding of RBPs to mRNA may inhibit translation and direct mRNAs to storage. Alternatively, binding of mRNAs could increase translation by protecting the respective mRNA from endonucleolytic cleavage. Phosphorylation could be a means to regulate the affinity of RBPs to mRNA. The MIF4G domain-containing protein At1g80930 is predicted to interact with the splicing factor At5g64270 based on experimentally verified interactions of putative homologs in other organisms (STRING), indicating a possible function of At1g80930 in the regulation of splicing. Another potentially important function in RNA metabolism is related to the RING/FYVE/PHD zinc finger superfamily protein At1g20110, which is predicted to interact with the eukaryotic translation initiation factor 5A3 (eIF5A3). Two out of three members of the eIF5A family, eIF5A1 and eIF5A3, were found to be posttranscriptionally up-regulated upon Fe deficiency (Lan et al., 2011). eIF5A proteins are important in the regulation of translation, mRNA turnover, and cell proliferation under stress conditions in eukaryotes and may be a critical factor for the acclimation of plants under nonfavorable growth conditions. At1g20110 phosphoprotein accumulates under Fe-deficient conditions (Table II), pointing to a possible control of mRNA-related processes under Fe deficiency via the phosphorylation of this protein.
CONCLUSION
Despite the rapid development in MS instrumentation for proteomic studies, quantitative phosphoproteomics remains a major challenge. By combining phosphopeptide enrichment by IMAC and titanium affinity chromatography, quantification by iTRAQ labeling and identification/quantification by dual CID and HCD scans, phosphopeptides from a relatively small sample input can be efficiently analyzed with high reproducibility. An unbiased quantitative analysis of phosphopeptides is crucial for completing the picture of the responses to Fe deficiency that has derived from transcriptomics and proteomics approaches. This is in particular evident for the Fe deficiency-induced alterations in carbohydrate metabolism. Previous studies have indicated that glycolytic flux is increased upon Fe deficiency, but proteomic and transcriptomic studies yielded a fragmentary picture. In addition to providing a database for phosphopeptides in Arabidopsis roots, our analysis disclosed a new level of information that helps us understand how plants acclimate to changing levels in available Fe pools.
MATERIALS AND METHODS
Plants and Growth Conditions
Arabidopsis (Arabidopsis thaliana) plants were grown in a growth chamber on an agar-based medium as described by Estelle and Somerville (1987). Seeds of the accession Columbia (Col-0) and the iar4 mutant (SALK_091909) were obtained from the Arabidopsis Biological Resource Center at Ohio State University. Seeds of DR5-GUS and iar4-3[DR5-GUS] plants were kindly provided by T. Guilfoyle and B. Gray. Seeds were surface sterilized by immersing them in 5% (v/v) NaOCl for 5 min and 70% ethanol for 7 min, followed by four rinses in sterile water. Seeds were placed onto petri dishes and kept for 1 d at 4°C in the dark, before the plates were transferred to a growth chamber and grown at 21°C under continuous illumination (50 μmol m−2 s−1; Phillips TL lamps). The medium was composed of (mm): KNO3 (5), MgSO4 (2), Ca (NO3)2 (2), KH2PO4 (2.5); (µm): H3BO3 (70), MnCl2 (14), ZnSO4 (1), CuSO4 (0.5), NaCl (10), Na2MoO4 (0.2), and 40 µM FeEDTA, solidified with 0.3% Phytagel (Sigma-Aldrich). Suc (43 mm) and 4.7 mm MES were included, and the pH was adjusted to 5.5. After 10 d of precultivation, plants were transferred to fresh agar medium either with 40 μm FeEDTA (+Fe plants) or without Fe and with 100 μm 3-(2-pyridyl)-5,6-diphenyl-1,2,4-triazine sulfonate (ferrozine; −Fe plants) to trap residual Fe. Plants were grown for 3 d on Fe-free medium before analysis. Phosphate deficiency was applied by growing plants on phosphate-free medium for 3 d.
Protein Extraction, In-Solution Trypsin Digestion, and iTRAQ Labeling
Total protein from roots of approximately 1,000 13-d-old Col-0 seedlings grown in the presence or absence of Fe for 3 d was extracted as described previously (Lan et al., 2011). Total protein (100 μg) was reduced by adding dithiothreitol (DTT) to a final concentration of 10 mm and incubated for 1 h at room temperature. Subsequently, IAA was added to a final concentration of 40 mm, and the mixture was incubated for 1 h at room temperature in the dark. Then, DTT (10 mm) was added to the mixture to consume any free IAA by incubating the mixture for 1 h at room temperature in the dark. Proteins were then diluted by 50 mm triethylammonium bicarbonate and 1 mm CaCl2 to reduce the urea concentration to less than 0.6 m and digested with 40 μg of modified trypsin (Promega) at 37°C overnight. The resulting peptide solution was acidified with 10% trifluoroacetic acid and desalted on a C18 solid-phase extraction cartridge. Desalted peptides were then labeled with iTRAQ reagents (Applied Biosystems) according to the manufacturer’s instructions. Control samples (proteins extracted from roots of +Fe plants) were labeled with reagent 114; samples from Fe-deficient roots were labeled with reagent 117. Two independent biological experiments with three technical repeats each were performed.
Cation-Exchange Chromatography
The combined iTRAQ-labeled peptides were cleaned by cation-exchange chromatography according to the manufacturer’s instructions. This cleaning step removed residual SDS, CaCl2, DTT, excess iTRAQ reagents, and undigested proteins such as trypsin. After cleanup, the mixtures were divided into two parts and desalted using Sep-Pak C18 reverse-phase cartridges (Waters). Phosphopeptides were enriched by a combination of TiO2 affinity chromatography and IMAC.
TiO2 Affinity Chromatography
Phosphopeptides were enriched from desalted peptides using TiO2 affinity chromatography according to the manual for the Titansphere Phos-TiO kit (GL Sciences) and finally eluted with 5% NH4OH followed by 5% pyrrolidine. Eluted peptides were desalted and lyophilized in a centrifugal speed vacuum concentrator.
IMAC
Phosphopeptide enrichment by IMAC was performed according to the protocol of the PHOS-Select Iron Affinity Gel kit (P9740; Sigma). Briefly, desalted peptides were acidified with 250 mm acetic acid with 30% acetonitrile (final pH < 3.0), loaded to preequilibrated 80 μL of 50% slurry, and incubated for 30 min at room temperature with mixing (end-over-end rotation). Samples were washed five times with washing buffer (250 mm acetic acid with 30% acetonitrile) and once with deionized water. Phosphopeptides were eluted with 400 mm NH4OH. The pH of all samples was adjusted to 3 by adding drops of 20% trifluoroacetic acid followed by desalting and concentration in a centrifugal speed vacuum concentrator.
MS/MS Analysis
Nano-HPLC-MS/MS analysis was performed on a nanoAcquity system (Waters) connected to an LTQ-Orbitrap XL hybrid mass spectrometer (Thermo Electron) equipped with a PicoView nanospray interface (New Objective). Peptide mixtures were loaded onto a 75-μm i.d., 25-cm length C18 BEH column (Waters) packed with 1.7-μm particles with a pore size of 130 Å and were separated using a segmented gradient in 90 min from 5% to 40% solvent B (acetonitrile with 0.1% formic acid) at a flow rate of 300 nL min−1 and a column temperature of 35°C. Solvent A was 0.1% formic acid in water. The LTQ-Orbitrap XL hybrid mass spectrometer was operated in positive ionization mode. The MS survey scan for all experiments was performed in the Fourier transform cell, recording a window between 350 and 1,600 m/z. The resolution was set to 60,000 at m/z 400, and the automatic gain control was set to 500,000 ions. The m/z values triggering MS/MS were put on an exclusion list for 90 s. The minimum MS signal for triggering MS/MS was set to 10,000. In all cases, one microscan was recorded. The CID-HCD acquisition method consisted of three sets of independent MS/MS scans, with a CID and a HCD MS/MS experiment triggered from the same precursor ion. CID was done with a target value of 7,000 in the linear ion trap, collision energy of 35%, Q value of 0.25, and activation time of 30 ms. To improve the fragmentation spectra of the phosphopeptides, “multistage activation” at 97.97, 48.99, and 32.66 Thompson relative to the precursor ion was enabled in CID events. HCD-generated ions were detected in the Orbitrap using a target value of 100,000, collision energy of 45%, and activation time of 30 ms.
Database Search and Quantification
The 2.3.02 version of the Mascot software (Matrix Science) was used to simultaneously identify and quantify proteins. In this version, only unique peptides used for protein quantification can be chosen, which is more precise to quantify proteins. Searches were made against The Arabidopsis Information Resource (TAIR 10; 35,386 sequences, 14,482,855 residues; ftp://ftp.arabidopsis.org/home/tair/Sequences/blast_datasets/TAIR10_blastsets/) concatenated with a decoy database containing the randomized sequences of the original database. Peak list data (mgf) files used for database searches were generated from Xcalibur raw files using a program in the MassMatrix conversion tools (version 1.4; http://www.massmatrix.net/). Intensities of iTRAQ reporter ions were then extracted from each HCD spectrum with a mass accuracy of 100 ppm and merged with the corresponding multistage activation spectrum using the QuantMerge program (Köcher et al., 2009). The protein sequences in the database were searched with trypsin digestion at both ends and two missed cleavages allowed, fixed modifications of carbamidomethylation at Cys, iTRAQ 4plex at Lys and the N terminus, variable modifications of oxidation at Met, and phosphorylation at Ser, Thr, and Tyr. The merged mgf files were searched with 10 ppm of precursor peptide mass tolerance (monoisotopic) and 0.8 D of MS/MS mass tolerance. The search results were passed through additional filters before exporting the data. For protein identification, the filters were set as follows: significance threshold P < 0.05 (with 95% confidence) and ion score or expect cutoff less than 0.05 (with 95% confidence). For protein quantitation, the filters were set as follows: “weighted” was chosen for protein ratio type (http://mascot-pc/mascot/help/quant_config_help.html); minimum precursor charge was set to 2; unique peptides with the highest score were used to quantify peptides. Summed intensities were set as normalization, and outliers were removed automatically. The peptide threshold was set as above homology. These filters resulted in a false discovery rate of less than 1% after decoy database searches were performed.
Statistical Analysis
Phosphopeptides with differential abundance upon Fe deficiency were selected using power analysis as described by deSouza et al. (2007). Phosphopeptides that fit the following criteria were selected for power analysis: first, phosphopeptides were detected in both biological experiments; second, the ratio did not show a contradicting accumulation pattern in the two biological experiments. A power index of 7.84 was used for confidence limits of 95% for type I error (α) and 80% for type II error (β). The deviation from unity, Δ, beyond which differential expression is indicated is given by [2 × S.D.2 × (Zα + Zβ)2/N]0.5, where S.D. is the averaged sd of 260 phosphopeptides, (Zα + Zβ)2 is the power index, and N is the number of sample sets. Thus, for N = 2, the ratios must be <1/(1 + Δ) or >(1 + Δ) = <0.74 or >1.36 to indicate differential expression.
Phosphorylation Motif Analysis Using Motif X
The Motif X algorithm was used (Schwartz and Gygi, 2005) to extract significantly enriched phosphorylation motifs from phosphopeptide data sets whose phosphorylation sites were confidently identified. The width of the peptides is 13 amino acids, with the phosphorylated site centered by retrieving the sequence context from the TAIR 10 database or by filling up the required number of X (where X stands for any amino acid), if necessary. Next, the centered peptides were aligned and used to extract the motif with the probability threshold set to P < 10−5, the occurrence threshold set to 5, and the default International Protein Index Arabidopsis Proteome data set used as the background data set (which has the same motif patterns as used in the protein sequences of the whole-genome Arabidopsis database TAIR 8 in Fasta format (in a shortened version due to upload restrictions to 10 Mb), as described by Reiland et al. (2009).
Complementation of iar4 Mutants with the Wild-Type IAR4 Gene
A 4,192-bp genomic DNA fragment corresponding to the wild-type IAR4 (At1g24180) gene was amplified from Col-0 genomic DNA by PCR with engineered BamHI sites and cloned into the vector pCR2.1TOPO to generate pTOPO-IAR4. The construct was verified by sequencing, cut with BamHI, and subcloned into BamHI-digested pCAMBIA2300 (http://www.cambia.org/daisy/cambia/585) to generate the pCAMBIA-IAR4 binary vector. This vector was introduced into Agrobacterium tumefaciens strain GV3101 and used to transform iar4 plants (SALK_091909) via floral dipping. The following primers were used for genomic DNA cloning: IAR4F, 5′-GTTGGATCCCATACGTGTGGTCATTCTTC-3′; IAR4R, 5′-CAGGGATCCTCTTTCTCTTAACTACTCTC-3′.
Generation of Transgenic Lines Expressing IAR4 Substitution Mutants
IAR4 substitution mutants were generated by changing the Ser at site 296 into Ala or Asp with the QuikChange Lightning Site-Directed Mutagenesis kit (Stratagene; catalog no. 210518) according to the manufacturer’s protocol. PCR was performed using Pfu polymerase (Promega) on the basis of plasmid pTOPO-IAR4, which created the plasmids pTOPO-IAR4S296A and pTOPO-IAR4S296D. These two plasmids were then subcloned into pCAMBIA2300 and transformed into iar4 mutant plants. The primers 5′-CAGATACCACGGTCACGCTATGTCTGACCCAGG-3′ (sense) and 5′-CCTGGGTCAGACATAGCGTGACCGTGGTATCTG-3′ (antisense) were used for the construction of IAR4S296A, and the primers 5′-TTACAGATACCACGGTCACGATATGTCTGACCCAGGAAGC-3′ (sense) and 5′-GCTTCCTGGGTCAGACATATCGTGACCGTGGTATCTGTAA-3′ (antisense) were used for the construction of IAR4S296D.
Root Hair Length and Density Measurements
Root hair length was determined by using a light microscope equipped with a scale in the ocular. One hundred root hairs from five roots were measured for each genotype and growth type. Root hair density was analyzed by counting the root hairs of 10 roots per time point and treatment in serial 2-mm sections starting from the root tip. Statistically significant deviations from the wild type were determined by Student’s t test (P < 0.001).
Histochemical Localization of GUS Activity
Histochemical GUS staining was performed by incubating whole seedlings of DR5-GUS and iar4-3[DR5-GUS] transgenic plants in staining buffer containing 50 mm sodium phosphate (pH 7.2), 0.5 mm potassium ferrocyanide, 0.2% (v/v) Triton X-100, and 1 mm 5-bromo-4-chloro-3-indolyl-d-glucuronide at 37°C for 20 min. The constructs and transgenic plants were described by Ulmasov et al. (1997) and Quint et al. (2009).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Phosphorylation motif analysis.
Supplemental Table S1. All phosphopeptides identified in Arabidopsis roots.
Supplemental Table S2. Phosphopeptides identified in both experiments with consistent response patterns.
Supplementary Material
Acknowledgments
LTQ-Orbitrap data were acquired at the Academia Sinica Common Mass Spectrometry Facilities, located at the Institute of Biological Chemistry, Academia Sinica. Expert technical help was provided by Ya-Yun Liao, Kalen Lin (Schmidt laboratory), Jeng-Yuan Shiau, and Selena Chen (Proteomics Core Laboratory, Institute of Plant and Microbial Biology). We thank Cole Lu (Schmidt laboratory) for figure artwork and Prof. Thomas J. Buckhout (Humboldt University) for critical comments on the manuscript. DR5-GUS and iar4-3[DR5-GUS] plants were kindly provided by T. Guilfoyle (University of Missouri) and B. Gray (University of Minnesota).
Glossary
- Fe
iron
- bHLH
basic helix-loop-helix
- MS
mass spectrometry
- iTRAQ
isobaric tags for relative and absolute quantitation
- CID
collision-induced dissociation
- HCD
high-energy collision dissociation
- IMAC
immobilized metal affinity chromatography
- SCX
strong cation-exchange
- MS/MS
tandem mass spectrometry
- m/z
mass-to-charge ratio
- CK II
casein kinase II
- PKA
protein kinase A
- PEPC
phosphoenolpyruvate carboxylase
- IAA
indole-3-acetic acid
- RBPs
RNA-binding proteins
- Col-0
accession Columbia
- DTT
dithiothreitol
References
- Bauer P, Ling HQ, Guerinot ML. (2007) FIT, the FER-LIKE IRON DEFICIENCY INDUCED TRANSCRIPTION FACTOR in Arabidopsis. Plant Physiol Biochem 45: 260–261 [DOI] [PubMed] [Google Scholar]
- Bodenmiller B, Mueller LN, Mueller M, Domon B, Aebersold R. (2007) Reproducible isolation of distinct, overlapping segments of the phosphoproteome. Nat Methods 4: 231–237 [DOI] [PubMed] [Google Scholar]
- Brumbarova T, Matros A, Mock HP, Bauer P. (2008) A proteomic study showing differential regulation of stress, redox regulation and peroxidase proteins by iron supply and the transcription factor FER. Plant J 54: 321–334 [DOI] [PubMed] [Google Scholar]
- Buckhout TJ, Yang TJ, Schmidt W. (2009) Early iron-deficiency-induced transcriptional changes in Arabidopsis roots as revealed by microarray analyses. BMC Genomics 10: 147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WW, Yang JL, Qin C, Jin CW, Mo JH, Ye T, Zheng SJ. (2010) Nitric oxide acts downstream of auxin to trigger root ferric-chelate reductase activity in response to iron deficiency in Arabidopsis. Plant Physiol 154: 810–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZH, Nimmo GA, Jenkins GI, Nimmo HG. (2007) BHLH32 modulates several biochemical and morphological processes that respond to Pi starvation in Arabidopsis. Biochem J 405: 191–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colangelo EP, Guerinot ML. (2004) The essential basic helix-loop-helix protein FIT1 is required for the iron deficiency response. Plant Cell 16: 3400–3412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J, Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 26: 1367–1372 [DOI] [PubMed] [Google Scholar]
- de la Fuente van Bentem S, Anrather D, Roitinger E, Djamei A, Hufnagl T, Barta A, Csaszar E, Dohnal I, Lecourieux D, Hirt H. (2006) Phosphoproteomics reveals extensive in vivo phosphorylation of Arabidopsis proteins involved in RNA metabolism. Nucleic Acids Res 34: 3267–3278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Nisi PD, Zocchi G. (2000) Phosphoenolpyruvate carboxylase in cucumber (Cucumis sativus L.) roots under iron deficiency: activity and kinetic characterization. J Exp Bot 51: 1903–1909 [DOI] [PubMed] [Google Scholar]
- deSouza N, Cui J, Dura M, McDonald TV, Marks AR. (2007) A function for tyrosine phosphorylation of type 1 inositol 1,4,5-trisphosphate receptor in lymphocyte activation. J Cell Biol 179: 923–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinneny JR, Long TA, Wang JY, Jung JW, Mace D, Pointer S, Barron C, Brady SM, Schiefelbein J, Benfey PN. (2008) Cell identity mediates the response of Arabidopsis roots to abiotic stress. Science 320: 942–945 [DOI] [PubMed] [Google Scholar]
- Eide D, Broderius M, Fett J, Guerinot ML. (1996) A novel iron-regulated metal transporter from plants identified by functional expression in yeast. Proc Natl Acad Sci USA 93: 5624–5628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estelle MA, Somerville C. (1987) Auxin-resistant mutants of Arabidopsis thaliana with an altered morphology. Mol Gen Genet 206: 200–206 [Google Scholar]
- García MJ, Lucena C, Romera FJ, Alcántara E, Pérez-Vicente R. (2010) Ethylene and nitric oxide involvement in the up-regulation of key genes related to iron acquisition and homeostasis in Arabidopsis. J Exp Bot 61: 3885–3899 [DOI] [PubMed] [Google Scholar]
- Gerber SA, Rush J, Stemman O, Kirschner MW, Gygi SP. (2003) Absolute quantification of proteins and phosphoproteins from cell lysates by tandem MS. Proc Natl Acad Sci USA 100: 6940–6945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gollhofer J, Schläwicke C, Jungnick N, Schmidt W, Buckhout TJ. (2011) Members of a small family of nodulin-like genes are regulated under iron deficiency in roots of Arabidopsis thaliana. Plant Physiol Biochem 49: 557–564 [DOI] [PubMed] [Google Scholar]
- Gregory AL, Hurley BA, Tran HT, Valentine AJ, She YM, Knowles VL, Plaxton WC. (2009) In vivo regulatory phosphorylation of the phosphoenolpyruvate carboxylase AtPPC1 in phosphate-starved Arabidopsis thaliana. Biochem J 420: 57–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann RM, Pickart CM. (1999) Noncanonical MMS2-encoded ubiquitin-conjugating enzyme functions in assembly of novel polyubiquitin chains for DNA repair. Cell 96: 645–653 [DOI] [PubMed] [Google Scholar]
- Karp NA, Huber W, Sadowski PG, Charles PD, Hester SV, Lilley KS. (2010) Addressing accuracy and precision issues in iTRAQ quantitation. Mol Cell Proteomics 9: 1885–1897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köcher T, Pichler P, Schutzbier M, Stingl C, Kaul A, Teucher N, Hasenfuss G, Penninger JM, Mechtler K. (2009) High precision quantitative proteomics using iTRAQ on an LTQ Orbitrap: a new mass spectrometric method combining the benefits of all. J Proteome Res 8: 4743–4752 [DOI] [PubMed] [Google Scholar]
- Kosová K, Vítámvás P, Prášil IT, Renaut J. (2011) Plant proteome changes under abiotic stress: contribution of proteomics studies to understanding plant stress response. J Proteomics 74: 1301–1322 [DOI] [PubMed] [Google Scholar]
- Lacerda CM, Xin L, Rogers I, Reardon KF. (2008) Analysis of iTRAQ data using Mascot and Peaks quantification algorithms. Brief Funct Genomics Proteomics 7: 119–126 [DOI] [PubMed] [Google Scholar]
- Lan P, Li W, Wen TN, Shiau JY, Wu YC, Lin W, Schmidt W. (2011) iTRAQ protein profile analysis of Arabidopsis roots reveals new aspects critical for iron homeostasis. Plant Physiol 155: 821–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landsberg EC. (1986) Function of rhizodermal transfer cells in the Fe stress response mechanism of Capsicum annuum L. Plant Physiol 82: 511–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeClere S, Rampey RA, Bartel B. (2004) IAR4, a gene required for auxin conjugate sensitivity in Arabidopsis, encodes a pyruvate dehydrogenase E1α homolog. Plant Physiol 135: 989–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Wong WS, Zhu L, Guo HW, Ecker J, Li N. (2009) Phosphoproteomic analysis of ethylene-regulated protein phosphorylation in etiolated seedlings of Arabidopsis mutant ein2 using two-dimensional separations coupled with a hybrid quadrupole time-of-flight mass spectrometer. Proteomics 9: 1646–1661 [DOI] [PubMed] [Google Scholar]
- Li J, Wu XD, Hao ST, Wang XJ, Ling HQ. (2008) Proteomic response to iron deficiency in tomato root. Proteomics 8: 2299–2311 [DOI] [PubMed] [Google Scholar]
- Li W, Schmidt W. (2010) A lysine-63-linked ubiquitin chain-forming conjugase, UBC13, promotes the developmental responses to iron deficiency in Arabidopsis roots. Plant J 62: 330–343 [DOI] [PubMed] [Google Scholar]
- Long TA, Tsukagoshi H, Busch W, Lahner B, Salt DE, Benfey PN. (2010) The bHLH transcription factor POPEYE regulates response to iron deficiency in Arabidopsis roots. Plant Cell 22: 2219–2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorković ZJ. (2009) Role of plant RNA-binding proteins in development, stress response and genome organization. Trends Plant Sci 14: 229–236 [DOI] [PubMed] [Google Scholar]
- Lou Y, Gou JY, Xue HW. (2007) PIP5K9, an Arabidopsis phosphatidylinositol monophosphate kinase, interacts with a cytosolic invertase to negatively regulate sugar-mediated root growth. Plant Cell 19: 163–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller M, Schmidt W. (2004) Environmentally induced plasticity of root hair development in Arabidopsis. Plant Physiol 134: 409–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagpal P, Walker LM, Young JC, Sonawala A, Timpte C, Estelle M, Reed JW. (2000) AXR2 encodes a member of the Aux/IAA protein family. Plant Physiol 123: 563–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ow SY, Salim M, Noirel J, Evans C, Rehman I, Wright PC. (2009) iTRAQ underestimation in simple and complex mixtures: “the good, the bad and the ugly”. J Proteome Res 8: 5347–5355 [DOI] [PubMed] [Google Scholar]
- Palmgren MG. (2001) Plant plasma membrane H+-ATPases: powerhouses for nutrient uptake. Annu Rev Plant Physiol Plant Mol Biol 52: 817–845 [DOI] [PubMed] [Google Scholar]
- Palusa SG, Ali GS, Reddy AS. (2007) Alternative splicing of pre-mRNAs of Arabidopsis serine/arginine-rich proteins: regulation by hormones and stresses. Plant J 49: 1091–1107 [DOI] [PubMed] [Google Scholar]
- Pickart CM, Fushman D. (2004) Polyubiquitin chains: polymeric protein signals. Curr Opin Chem Biol 8: 610–616 [DOI] [PubMed] [Google Scholar]
- Quint M, Barkawi LS, Fan KT, Cohen JD, Gray WM. (2009) Arabidopsis IAR4 modulates auxin response by regulating auxin homeostasis. Plant Physiol 150: 748–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabotti G, De Nisi P, Zocchi G. (1995) Metabolic implications in the biochemical responses to iron deficiency in cucumber (Cucumis sativus L.) roots. Plant Physiol 107: 1195–1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampey RA, Woodward AW, Hobbs BN, Tierney MP, Lahner B, Salt DE, Bartel B. (2006) An Arabidopsis basic helix-loop-helix leucine zipper protein modulates metal homeostasis and auxin conjugate responsiveness. Genetics 174: 1841–1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiland S, Messerli G, Baerenfaller K, Gerrits B, Endler A, Grossmann J, Gruissem W, Baginsky S. (2009) Large-scale Arabidopsis phosphoproteome profiling reveals novel chloroplast kinase substrates and phosphorylation networks. Plant Physiol 150: 889–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rellán-Alvarez R, Andaluz S, Rodríguez-Celma J, Wohlgemuth G, Zocchi G, Alvarez-Fernández A, Fiehn O, López-Millán AF, Abadía J. (2010) Changes in the proteomic and metabolic profiles of Beta vulgaris root tips in response to iron deficiency and resupply. BMC Plant Biol 10: 120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson NJ, Procter CM, Connolly EL, Guerinot ML. (1999) A ferric-chelate reductase for iron uptake from soils. Nature 397: 694–697 [DOI] [PubMed] [Google Scholar]
- Ross PL, Huang YN, Marchese JN, Williamson B, Parker K, Hattan S, Khainovski N, Pillai S, Dey S, Daniels S, et al. (2004) Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol Cell Proteomics 3: 1154–1169 [DOI] [PubMed] [Google Scholar]
- Santi S, Schmidt W. (2009) Dissecting iron deficiency-induced proton extrusion in Arabidopsis roots. New Phytol 183: 1072–1084 [DOI] [PubMed] [Google Scholar]
- Schmidt W, Buckhout TJ. (2011) A hitchhiker’s guide to the Arabidopsis ferrome. Plant Physiol Biochem 49: 462–470 [DOI] [PubMed] [Google Scholar]
- Schwartz D, Gygi SP. (2005) An iterative statistical approach to the identification of protein phosphorylation motifs from large-scale data sets. Nat Biotechnol 23: 1391–1398 [DOI] [PubMed] [Google Scholar]
- Sugiyama N, Nakagami H, Mochida K, Daudi A, Tomita M, Shirasu K, Ishihama Y. (2008) Large-scale phosphorylation mapping reveals the extent of tyrosine phosphorylation in Arabidopsis. Mol Syst Biol 4: 193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D, Franceschini A, Kuhn M, Simonovic M, Roth A, Minguez P, Doerks T, Stark M, Muller J, Bork P, et al. (2011) The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res 39: D561–D568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimm O, Essigmann B, Kloska S, Altmann T, Buckhout TJ. (2001) Response of Arabidopsis to iron deficiency stress as revealed by microarray analysis. Plant Physiol 127: 1030–1043 [PMC free article] [PubMed] [Google Scholar]
- Ting L, Rad R, Gygi SP, Haas W. (2011) MS3 eliminates ratio distortion in isobaric multiplexed quantitative proteomics. Nat Methods 8: 937–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ. (1997) Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9: 1963–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vert G, Grotz N, Dédaldéchamp F, Gaymard F, Guerinot ML, Briat JF, Curie C. (2002) IRT1, an Arabidopsis transporter essential for iron uptake from the soil and for plant growth. Plant Cell 14: 1223–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen R, Newton L, Li G, Wang H, Xiao W. (2006) Arabidopsis thaliana UBC13: implication of error-free DNA damage tolerance and Lys63-linked polyubiquitylation in plants. Plant Mol Biol 61: 241–253 [DOI] [PubMed] [Google Scholar]
- Wilson AK, Pickett FB, Turner JC, Estelle M. (1990) A dominant mutation in Arabidopsis confers resistance to auxin, ethylene and abscisic acid. Mol Gen Genet 222: 377–383 [DOI] [PubMed] [Google Scholar]
- Yang TJ, Lin WD, Schmidt W. (2010) Transcriptional profiling of the Arabidopsis iron deficiency response reveals conserved transition metal homeostasis networks. Plant Physiol 152: 2130–2141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Qiang X, Owsiany K, Zhang S, Thannhauser TW, Li L. (2011) Evaluation of different multidimensional LC-MS/MS pipelines for isobaric tags for relative and absolute quantitation (iTRAQ)-based proteomic analysis of potato tubers in response to cold storage. J Proteome Res 10: 4647–4660 [DOI] [PubMed] [Google Scholar]
- Yin XJ, Volk S, Ljung K, Mehlmer N, Dolezal K, Ditengou F, Hanano S, Davis SJ, Schmelzer E, Sandberg G, et al. (2007) Ubiquitin lysine 63 chain forming ligases regulate apical dominance in Arabidopsis. Plant Cell 19: 1898–1911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y, Wu H, Wang N, Li J, Zhao W, Du J, Wang D, Ling HQ. (2008) FIT interacts with AtbHLH38 and AtbHLH39 in regulating iron uptake gene expression for iron homeostasis in Arabidopsis. Cell Res 18: 385–397 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.