Abstract
In Arabidopsis (Arabidopsis thaliana), the ATP-dependent chromatin remodeler PICKLE (PKL) determines expression of genes associated with developmental identity. PKL promotes the epigenetic mark trimethylation of histone H3 lysine 27 (H3K27me3) that facilitates repression of tissue-specific genes in plants. It has previously been proposed that PKL acts indirectly to promote H3K27me3 by promoting expression of the POLYCOMB REPRESSIVE COMPLEX2 complex that generates H3K27me3. We undertook expression and chromatin immunoprecipitation analyses to further characterize the contribution of PKL to gene expression and developmental identity. Our expression data support a critical and specific role for PKL in expression of H3K27me3-enriched loci but do not support a role for PKL in expression of POLYCOMB REPRESSIVE COMPLEX2. Moreover, our chromatin immunoprecipitation data reveal that PKL protein is present at the promoter region of multiple H3K27me3-enriched loci, indicating that PKL directly acts on these loci. In particular, we find that PKL is present at LEAFY COTYLEDON1 and LEAFY COTYLEDON2 during germination, which is when PKL acts to repress these master regulators of embryonic identity. Surprisingly, we also find that PKL is present at the promoters of actively transcribed genes that are ubiquitously expressed such as ACTIN7 and POLYUBIQUITIN10 that do not exhibit PKL-dependent expression. Taken together, our data contravene the previous model of PKL action and instead support a direct role for PKL in determining levels of H3K27me3 at repressed loci. Our data also raise the possibility that PKL facilitates a common chromatin remodeling process that is not restricted to H3K27me3-enriched regions.
Proper regulation of genes that exhibit altered expression during development is dependent on the coordinated action of a variety of chromatin remodeling factors (Clapier and Cairns, 2009; Ho and Crabtree, 2010). One class of remodeler that plays a critical role in controlling expression of genes associated with developmental identity and illustrates the combinatorial nature of chromatin-mediated transcriptional regulation belongs to the CHD family of ATP-dependent chromatin remodelers.
CHD3 and CHD4 proteins are interchangeable components of the Mi-2/NuRD histone deacetylase complex in animals (Hall and Georgel, 2007; Ramírez and Hagman, 2009). Mi-2/NuRD complexes are modular in nature and contain several other interchangeable subunits including a methyl-CpG-binding domain protein (MBD2 or MBD3) and a histone deacetylase (HDAC1 or HDAC2). The Mi-2/NuRD complex is the most abundant histone deacetylase complex in mammalian cells and has been shown to be necessary for repression of a wide variety of developmentally regulated genes in mammals and other animals (Ahringer, 2000; Wolffe et al., 2000). CHD4 is also found in a complex with the histone acetyltransferase p300, however, and in that context promotes expression of CD4 during T-cell development in mice (Williams et al., 2004). Similarly, CHD3 functions as a coactivator for human c-Myb (Saether et al., 2007). Thus CHD3 and CHD4 proteins can participate in multiple remodeling pathways and can either repress or activate gene expression depending on the other factors they associate with.
Initial characterization of the CHD3/4-related gene PICKLE (PKL) in Arabidopsis (Arabidopsis thaliana) indicated that it also plays a significant role in transcriptional repression of developmental identity genes (Eshed et al., 1999; Ogas et al., 1999). In particular, pkl seedlings fail to repress seed-specific genes (Dean Rider et al., 2003; Zhang et al., 2008; Aichinger et al., 2009). As a result, pkl primary roots can express numerous embryonic differentiation characteristics and undergo spontaneous somatic embryogenesis (Henderson et al., 2004). pkl primary roots expressing these traits adopt a green tuberous phenotype and are referred to as pickle roots (Ogas et al., 1997). Microarray analysis of gene expression reveals that derepression of seed-specific genes first occurs during germination in pkl seedlings (Dean Rider et al., 2003; Zhang et al., 2008). Furthermore, use of a conditional PKL construct generated by fusing PKL to the glucocorticoid receptor reveals that PKL acts specifically during germination to repress expression of seed-specific traits (Li et al., 2005).
PKL contributes to other developmental processes in addition to repression of embryonic traits. PKL plays a role in repression of ectopic stipules and meristems in leaf tissue (Hay et al., 2002) and represses meristematic genes in carpel tissue (Eshed et al., 1999). Loss of PKL results in increased responsivity to the plant growth regulator cytokinin with regards to both gene expression and callus formation (Furuta et al., 2011). PKL also is necessary for proper root development and has been found to play two somewhat opposing roles in this context: PKL is a negative regulator of auxin-mediated lateral root initiation (Fukaki et al., 2006) and yet also promotes root growth and expression of root meristem marker genes (Aichinger et al., 2011).
Comparative genomic analyses led to the discovery that in contrast to animal CHD3/4 proteins, PKL promotes trimethylation of Lys 27 of histone H3 (H3K27me3) rather than histone deacetylation (Zhang et al., 2008). In both plants and animals, H3K27me3-mediated gene repression plays a critical role in various developmental processes (Simon and Kingston, 2009; Zheng and Chen, 2011). The POLYCOMB REPRESSIVE COMPLEX2 (PRC2) catalyzes trimethylation of H3K27 (Cao et al., 2002; Kuzmichev et al., 2002; Müller et al., 2002; Schmitges et al., 2011), and characterization of mutants lacking components of PRC2 has contributed greatly to our understanding of the contribution of H3K27me3 to repression of developmental regulators in Arabidopsis. Arabidopsis PRC2 mutants with substantially reduced levels of H3K27me3 exhibit profound developmental defects and extensive derepression of embryonic traits (Chanvivattana et al., 2004; Schubert et al., 2006; Bouyer et al., 2011). Characterization of PRC2 mutants similarly reveals an important role for H3K27me3 in repressing expression of floral activators (Goodrich et al., 1997; Kinoshita et al., 2001; Yoshida et al., 2001; Chanvivattana et al., 2004; Schönrock et al., 2006; Bouyer et al., 2011; Zheng and Chen, 2011) and in imprinting and endosperm development (Chaudhury et al., 1997; Grossniklaus et al., 1998; Hsieh et al., 2011). Intriguingly, however, H3K27me3 is dispensable for development of the embryo (Bouyer et al., 2011). In total, about 4,400 genes are enriched for H3K27me3 in 14-d-old Arabidopsis plants (Zhang et al., 2007; Bouyer et al., 2011). Tissue-specific genes are significantly overrepresented among these 4,400 genes, suggesting that H3K27me3 plays a general role in restricting expression of developmentally regulated genes (Zhang et al., 2007). Importantly, loss of H3K27me3 does not result in global derepression of H3K27me3-enriched loci, suggesting that other processes in addition to H3K27me3 act to restrict expression of H3K27me3-enriched genes (Bouyer et al., 2011).
The discovery that pkl plants exhibit a 2-fold or greater reduction of H3K27me3 at H3K27me3-enriched genes revealed that PKL acts in some fashion to promote this repressive histone modification (Zhang et al., 2008). Furthermore, H3K27me3 levels are reduced at LEAFY COTYLEDON (LEC) genes during germination of pkl seedlings, suggesting that it is the reduction of H3K27me3 that leads to derepression of these genes (Zhang et al., 2008). These data indicate that PKL promotes deposition of H3K27me3, but they do not reveal if PKL acts directly on H3K27me3-enriched loci. Although much of the chromatin machinery associated with H3K27me3 is conserved in animals and plants, there is much that remains to be elucidated regarding H3K27me3 in plants (Hennig and Derkacheva, 2009; Zheng and Chen, 2011). In particular, it is unknown how genes are targeted for H3K27me3 enrichment and how the domains of H3K27me3 enrichment are restricted to specific regions. The presence of the ATP-dependent remodeler PKL at H3K27me3-enriched genes would raise the possibility that PKL plays a role in one or more of these and other processes associated with H3K27me3 homeostasis.
Published analyses by Aichinger and colleagues, however, support an indirect mode of action for PKL (Aichinger et al., 2009). Transcript levels of several subunits of PRC2 are decreased in 5-d-old roots that lack PKL and PICKLE RELATED2 (PKR2), a closely related CHD protein. The authors also used chromatin immunoprecipitation (ChIP) to determine that PKL is present at genes that code for subunits of the PRC2 complex that promotes deposition of H3K27me3 plants. Taken together, their data are consistent with the hypothesis that PKL functions redundantly with PKR2 and together they influence H3K27me3 deposition indirectly by promoting the expression of the PRC2 complex that deposits H3K27me3 (Aichinger et al., 2009). These studies, however, examined seedlings after germination and their biological material included plants that expressed the developmentally aberrant pickle root phenotype. As a result, these data do not address the role of PKL during the critical developmental window of germination when PKL is known to act to repress expression of seed-specific genes (Li et al., 2005). In addition, inclusion of the pickle roots in the sample confounds transcript analysis due to the altered developmental identity and correspondingly altered transcriptome of this tissue (Dean Rider et al., 2003; Rider et al., 2004).
We have used microarray analysis and ChIP to further examine the contribution of PKL to H3K27me3, gene expression, and developmental transcription programs. Microarray analysis of 14-d-old seedlings reveals that although PKL continues to play a role in expression of H3K27me3-enriched genes after germination, it is dispensable for repression of seed-specific genes. Importantly, our expression analyses also reveal that transcript levels of genes that code for PRC2 components are not altered in pkl or pkl pkr2 plants. ChIP with epitope-tagged PKL demonstrates that PKL is present in the promoter region of H3K27me3-enriched loci. In particular, we find that PKL is present at LEC genes during germination, consistent with prior data revealing that PKL acts during this stage of development to repress these key regulators of embryogenesis. Surprisingly, our ChIP analyses also reveal that PKL is present at the promoters of ubiquitously expressed genes. These genes, however, do not exhibit PKL-dependent expression and further do not exhibit alterations in the level of several epigenetic marks. In contrast, all H3K27me3-enriched genes assayed exhibit reduced levels of H3K27me3 in the absence of PKL. Our data thus indicate that PKL acts directly upon H3K27me3-enriched genes to affect levels of H3K27me3 and gene expression at these loci and also raise the possibility that the chromatin remodeling role played by PKL may not be unique to H3K27me3-enriched genes.
RESULTS
PKL Contributes to Transcriptional Regulation of Developmental Pathways Differently in 14-d-old Plants versus Germinating Seedlings
The discovery that loss of PKL preferentially affected expression of genes that are enriched for H3K27me3 was based on comparison of genomic data sets from disparate developmental samples: Microarray analysis was used to identify PKL-dependent genes during germination (Zhang et al., 2008) or in roots (Aichinger et al., 2009) whereas the list of H3K27me3-enriched genes used for comparison was obtained from studies using 14-d-old seedlings (Zhang et al., 2007). It has been shown that the spectrum of genes enriched for H3K27me3 and other epigenetic modifications differs in different developmental samples (Charron et al., 2009; Deal and Henikoff, 2010; Lafos et al., 2011). Furthermore, phenotypic analysis of conditional PKL-GR plants strongly suggests that loss of PKL has distinct effects on the transcriptome during germination and after germination (Li et al., 2005).
We therefore undertook a new microarray analysis of 14-d-old wild-type and pkl seedlings grown on synthetic media so that we could examine the intersection between genes that exhibit PKL-dependent expression and genes subject to various epigenetic modifications in developmentally equivalent samples when available. pkl seedlings with pickle roots (approximately 2% of seedlings) were specifically excluded from the sample to eliminate the confounding issue that results from derepression of the embryo transcriptome in pickle roots (Dean Rider et al., 2003). Affymetrix GeneChip ATH1 arrays were used for the analysis, and the microarray experimental design consisted of two treatments, wild-type plants and pkl plants, and three biological replicates. We identified differentially expressed genes with the criterion that the positive false discovery rate (pFDR) was less than 0.05 (Storey, 2003).
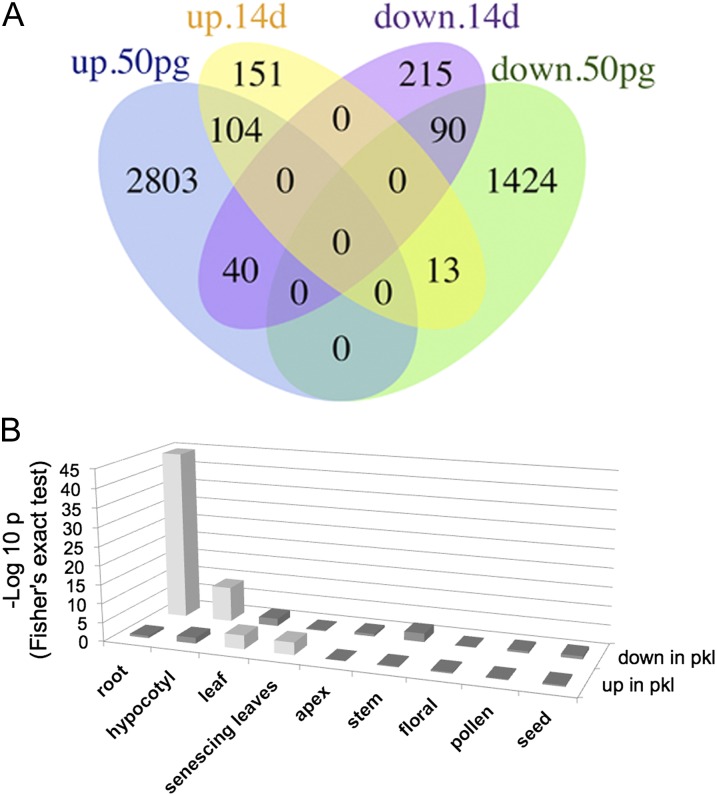
Overall, loss of PKL has a less dramatic effect on the transcriptome of 14-d-old plants than in 50% germinated seedlings (Fig. 1A). In 14-d-old plants, we identified 282 features for which the corresponding transcript is significantly up-regulated in pkl plants and 365 features for which the corresponding transcript is significantly down-regulated in pkl plants (Supplemental Table S1). In contrast, 2,917 features are identified for which the corresponding transcript is significantly up-regulated in 50% germinated pkl seedlings, and 1,557 features are identified for which the corresponding transcript is significantly down-regulated in 50% germinated pkl seedlings (Supplemental Table S2). Although expression of fewer genes is affected in 14-d-old pkl plants, many new PKL-dependent genes are identified in our analysis. Sixty-one percent of the genes that exhibit increased transcript levels in 14-d-old pkl plants do not exhibit increased transcript levels in germinating pkl seedlings and 74% of the genes that exhibit decreased transcript levels in 14-d-old pkl plants do not exhibit decreased transcript levels in germinating pkl seedlings.
Figure 1.
PKL plays distinct roles in expression of transcription programs in 14-d-old pkl plants versus germinating pkl seedlings. A, Venn diagram indicating the number of loci for which the corresponding transcript is expressed at significantly different levels (up or down) in pkl seedlings versus wild-type seedlings at 50% germination and at 14 d. 14d, 14-d-old seedlings; 50pg, 50% germination. For a complete list of PKL-dependent genes identified by our analysis at 50% germination, please see Supplemental Table S2. B, Fisher’s exact test (see “Materials and Methods”) was used to examine the intersection of genes preferentially expressed in a specific tissue (x axis) and genes that exhibit PKL-dependent expression (y axis). Down in pkl refers to genes that exhibit reduced transcript levels in pkl plants relative to wild-type plants whereas up in pkl refers to genes that exhibit increased transcript levels in pkl plants relative to wild-type plants. The log10 of P associated with each intersection as determined by Fisher’s exact test is represented on the z axis. A white bar denotes more genes observed in common between the two sets than expected (at P < 0.001).
Previous analysis of the transcriptome of germinating seedlings revealed that PKL is necessary to repress expression of seed-associated transcripts and that loss of PKL has little if any effect on expression of transcriptional programs associated with other organs or stages of development (Zhang et al., 2008). We repeated this analysis and examined the intersection of genes that exhibit PKL-dependent expression in 14-d-old plants with genes that are preferentially expressed in root, hypocotyl, leaf, senescing leaf, apex, stem, floral, stamen/pollen, and seed as determined by Shannon entropy (Zhang et al., 2008; Supplemental Table S3). This analysis revealed that only leaf-specific genes are modestly overrepresented among genes that exhibit elevated transcript levels in 14-d-old pkl plants (Fig. 1B). Thus although microarray analysis reveals that PKL plays a critical role in repression of the seed-specific transcriptional program during germination, loss of PKL has no detectable effect on repression of this program at the level of the transcriptome at a later stage in development.
In contrast, this analysis did reveal that genes that exhibit root-specific expression and to a lesser extent those that exhibit hypocotyl-specific expression are overrepresented among genes that exhibit decreased transcript levels in pkl plants. Twenty-two percent (54/244) of root-specific genes exhibit decreased transcript levels in pkl plants although only 1.6% (4/244) are expected to do so by chance. Although pkl primary roots can develop aberrantly and become pickle roots, the plants used for microarray analysis were screened to remove plants expressing the pickle root phenotype (“Materials and Methods”). As a result, the pickle root phenotype is not a contributing factor to the observed decrease in expression of root-specific genes in pkl plants.
Given that root elongation is reduced in pkl plants (Aichinger et al., 2011), we examined whether pkl plants exhibit a decrease in root mass relative to shoot mass, which would then provide a simple explanation for the observation of reduced transcript levels of root-specific genes in pkl plants. We measured total root mass and total shoot mass in 14-d-old wild-type and pkl plants grown on synthetic media (Table I). We observed that the ratio of root mass to shoot mass in pkl plants was greater than the ratio of root mass to shoot mass in wild-type plants, indicating that a relative reduction in root tissue in pkl plants was not the cause of the observed decrease in transcript levels of root-specific genes in pkl plants. These data are likely a reflection of the role of PKL in promoting shoot development (Henderson et al., 2004) as well as the role of PKL in repression of lateral root formation (Fukaki et al., 2006).
Table I. The ratio of root to shoot is greater in pkl plants than in wild-type plants.
Fresh weight of root and shoot tissue was determined for 14-d-old wild-type and pkl seedlings grown in synthetic media.
| Genotype | Tissue | Average Weight of 10 Seedlings ± sd | Ratio of Root Weight to Shoot Weight ± sd |
|---|---|---|---|
| mg | |||
| WT | Root | 15.7 ± 4.3 | 0.22 ± 0.07 |
| Shoot | 71.8 ± 13 | ||
| pkl | Root | 19.8 ± 1.6 | 0.33 ± 0.02a |
| Shoot | 59.5 ± 3.0 |
Denotes statistically significant difference (P < 0.05).
Genes That Exhibit PKL-Dependent Expression Are Enriched for Targets of H3K27me3 and Depleted for Targets of H3K4me3
Taken together, the preceding analyses of our microarray data reveal that PKL affects expression of a set of genes in 14-d-old plants that are distinct from those affected in 50% germinated seedlings. We therefore determined whether H3K27me3-enriched genes are overrepresented in this set of PKL-dependent genes as was observed previously for PKL-dependent genes from other developmental samples (Zhang et al., 2008; Aichinger et al., 2009). In particular, we focused on genes identified as H3K27me3 enriched from developmentally equivalent samples (14-d-old plants grown on synthetic media; Zhang et al., 2007; Bouyer et al., 2011).
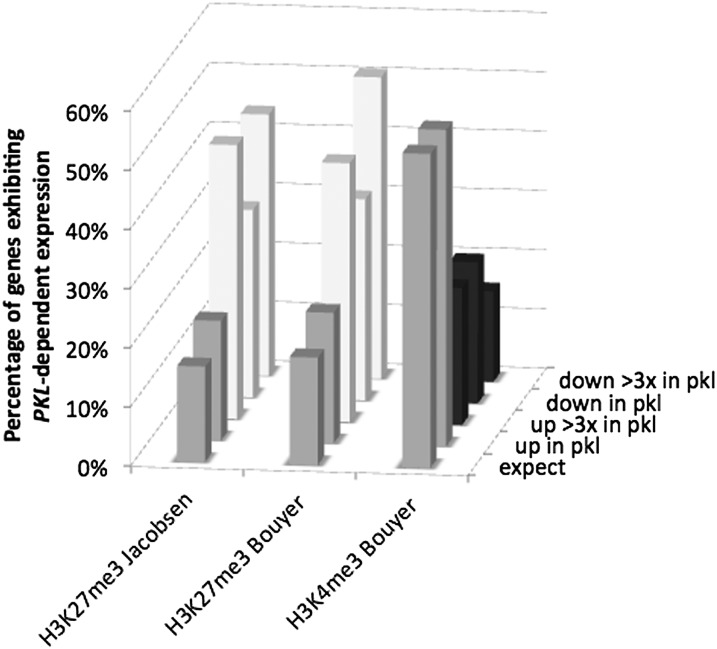
When comparing equivalent developmental samples, we found that H3K27me3 targets are greatly enriched among genes that exhibit PKL-dependent expression (Fig. 2). For genes that exhibit a greater than 3-fold increased transcript levels in pkl plants, more than 40% of genes are also enriched for H3K27me3 as determined by the two independent studies included in our analysis (Zhang et al., 2007; Bouyer et al., 2011). We also observed a significant overlap with genes that exhibit decreased transcript levels in pkl plants: More than 30% of genes that exhibit significantly decreased transcript levels in pkl plants are enriched for H3K27me3. The extent of overlap between H3K27me3-enriched genes and genes that exhibit decreased transcript levels in pkl plants is increased by imposing a threshold requirement: For genes that exhibit a 3-fold or more decrease in transcript levels in pkl plants, the overlap is 44% with loci identified in one study (Zhang et al., 2007) and 50% for the other (Bouyer et al., 2011).
Figure 2.
Genes that exhibit altered expression in the absence of PKL are frequently targets of H3K27me3 and not targets of H3K4me3. Fisher’s exact test (see “Materials and Methods”) was used to examine the intersection of genes linked to an epigenetic pathway (x axis) and genes that exhibit altered expression in response to pkl according to various selection criteria (y axis). The percent of genes that exhibit altered expression in pkl plants that fall within each intersection is represented on the z axis. The expect category on the y axis indicates the percentage of PKL-dependent genes expected to be found in the intersection of the compared sets of genes. A gray bar denotes either the expected number of genes or the number of genes in common between two sets that is not significantly different from the expected number, a white bar denotes more genes observed in common between two sets than expected (at P < 1 × 10−3), and a dark bar denotes fewer genes observed than expected (at P < 1 × 10−3).
Previous studies have suggested that PKL may act by promoting transcription rather than repressing transcription (Aichinger et al., 2009, 2011). H3K4me3-enriched loci are strongly enriched for actively transcribed genes (Shilatifard, 2006; Li et al., 2007), and these loci were also identified in the analysis of 14-d-old plants by Bouyer and colleagues (Bouyer et al., 2011). We therefore examined whether H3K4me3-enriched genes are overrepresented in PKL-dependent genes to test the hypothesis that loss of PKL preferentially affects expression of actively transcribed genes enriched for this mark. Instead, we observed that H3K4me3-enriched loci are substantially underrepresented among genes that exhibit PKL-dependent expression (Fig. 2). Although more than 50% of PKL-dependent genes are expected to be targets of H3K4me3, only 23% of genes for which transcript levels are 3-fold or higher in pkl plants are targets of H3K4me3. Similarly, 23% of genes that exhibit decreased transcript levels in pkl plants are targets of H3K4me3 and only 15% of genes for which transcript levels are decreased more than 3-fold in pkl plants are targets of H3K4me3.
Given that H3K4me3-enriched genes are depleted for H3K27me3 targets (Bouyer et al., 2011) and PKL-dependent genes are enriched for H3K27me3 targets (Fig. 2), we asked if exclusion of H3K27me3-enriched loci from the comparison of PKL-dependent and H3K4me3 targets might alter the observed relationship. We found that exclusion of H3K27me3-enriched loci did not prevent underrepresentation of H3K4me3 targets among genes that exhibit decreased transcript levels in pkl plants: 32% of genes that exhibit decreased transcript levels in pkl plants were enriched for H3K4me3 whereas 62% were expected (P = 1.22E-20 by Fisher’s exact test). These data thus suggest that rather than PKL preferentially promoting expression of H3K4me3-enriched genes, the presence of H3K4me3 at a gene helps to protect against reduced transcript levels in the absence of PKL.
We also examined the intersection of PKL-dependent genes in 14-d-old plants with other epigenomic data sets, including DNA methylation and histone acetylation (Supplemental Fig. S1). This analysis failed to uncover any overlap of PKL-dependent genes with another epigenetic pathway comparable to that observed for H3K27me3-enriched genes. Further, as observed previously (Zhang et al., 2008), this analysis was consistent with the hypothesis that PKL does not function in the plant equivalent of a Mi-2/NuRD complex.
Expression of the PRC2 Machinery That Deposits H3K27me3 Is Not Reduced in pkl Plants
Importantly, one class of genes that was not identified in our microarray analysis as being differentially expressed is the set of genes that code for members of the PRC2 complex. Previous published data suggested that PKL acts to repress expression of H3K27me3-enriched genes not by promoting H3K27me3 at these loci but instead by promoting expression of members of the PRC2 complex that deposits H3K27me3 (Aichinger et al., 2009). In particular, the authors observed that the transcript levels of EMBRYONIC FLOWER2 (EMF2), CURLY LEAF (CLF), and SWINGER (SWN) were reduced in the roots of pkl plants. A confounding factor in this analysis, however, is that the root samples collected from pkl plants included pickle root tissue, which is developmentally distinct from normal root tissue and in which the embryo transcriptome is strongly derepressed (Dean Rider et al., 2003; Rider et al., 2004). Thus altered expression of a gene in these samples could be an indirect consequence of altered developmental identity rather than a direct effect of loss of PKL.
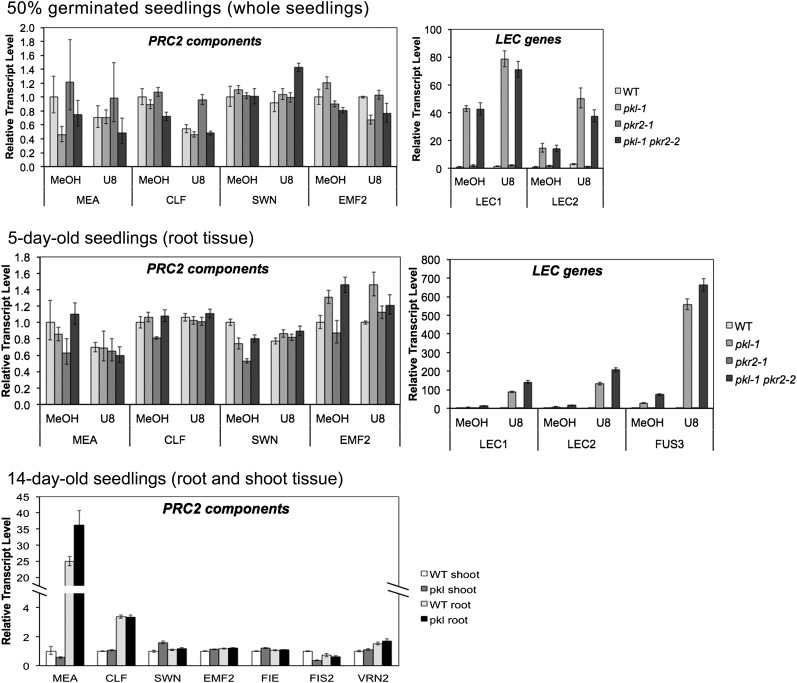
We used quantitative reverse transcription (qRT)-PCR to confirm our microarray analysis and examined the expression of PRC2 components at three developmental stages: in 50% germinated seedlings (seeds were imbibed and then collected when the radicle had emerged from 50% of the seeds [Zhang et al., 2008]), in roots from 5-d-old plants, and in roots and shoots from 14-d-old plants. We have previously shown that PKL acts during germination to repress expression of the embryo-specific developmental program (Li et al., 2005). In addition, both gene expression and H3K27me3 levels have previously been shown to be PKL dependent in 50% germinated seedlings and in 14-d-old plants (Dean Rider et al., 2003; Zhang et al., 2008), whereas 5-d-old roots correspond to the 5-d-old roots used previously to examine expression of PRC2 components (Aichinger et al., 2009). In our analysis, however, visible pickle roots were specifically excluded from roots collected from 14-d-old plants. We determined relative transcript levels in wild-type and pkl plants at all three developmental stages. In addition, we also included pkr2 and pkl pkr2 plants and grew plants in the absence and presence of 10−8 m uniconazole-P for the 50% germinated seedlings and 5-d-old plants so that the assay conditions were more favorable for derepression of embryonic traits.
In every case, we found that expression of genes coding for PRC2 machinery is largely unaffected by loss of PKL or by conditions that favored derepression of embryonic genes such as loss of both PKL and PKR2 and/or addition of uniconazole-P (Fig. 3). In contrast, we consistently observed that the transcript level of LEC genes is strongly elevated under those conditions that favor derepression of embryonic genes. Further, our data reveal that pkl plants do not exhibit decreased transcript levels of PRC2 components in situations in which the level of H3K27me3 at H3K27me3-enriched loci is reduced (Zhang et al., 2008). These data thus indicate that reduced expression of PRC2 is not the cause of reduced levels of H3K27me3 and elevated expression of embryo-specific genes observed in pkl plants in the developmental samples assayed.
Figure 3.
Expression of PRC2 machinery is largely insensitive to loss of PKL and PKR2. qRT-PCR was used to determine the relative transcript level of the indicated genes in 50% germinated seedlings, roots from 5-d-old plants, or roots and shoots from 14-d-old seedlings in the indicated genetic background. Because pickle roots are not readily visible until 7 d, pickle roots are consistently excluded for analysis in 14-d-old seedlings but not necessarily in 50% germinated or 5-d-old seedlings. Fifty percent germinated seedlings and 5-d-old plants were grown in the presence of 10−8 m uniconazole-P (U8) to favor expression of the pickle root phenotype or in 0.01% methanol (MeOH) as a mock treatment. LEC genes (LEC1, LEC2, and FUS3) were included as positive controls for PKL-dependent expression in 50% germinated seedlings and 5-d-old plants. 18S rRNA was used as a standardization control, and expression levels are normalized to the indicated wild-type tissue in each graph. Error bars represent sd calculated from three technical replicates.
PKL Associates with Genes Enriched for H3K27me3
Previous data strongly suggested a role for PKL in repression of genes by promoting H3K27me3 at those loci (Zhang et al., 2008). Previous ChIP analysis of PKL used a polyclonal antibody to PKL and did not detect association of PKL with H3K27me3-enriched loci (Aichinger et al., 2009). We generated an epitope-tagged version of PKL for use in ChIP to reexamine the possibility that PKL associated with loci that are enriched for H3K27me3. Six copies of the c-Myc epitope were fused to the full-length PKL open reading frame to generate a PKL-c-Myc translational fusion. This construct was placed under the control of PKL regulatory sequences (Li et al., 2005) and introduced into pkl-1 plants where it rescued all mutant phenotypes associated with loss of PKL (data not shown).
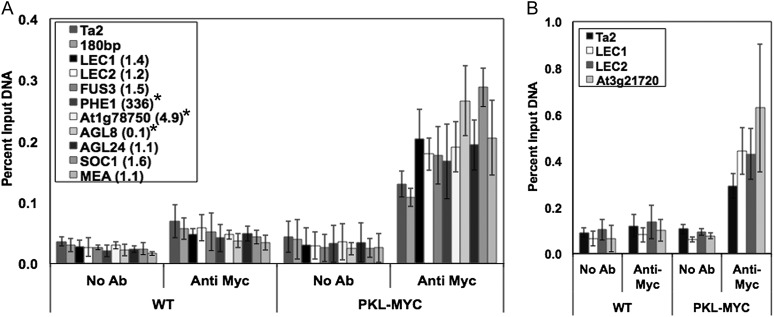
Use of PKL-c-Myc plants for ChIP reveals that PKL is present at the promoters of H3K27me3-enriched loci and that the amount of PKL detected at these loci is elevated relative to two heterochromatic loci: the Ta2 retrotransposon and the 180-bp centromeric repeats. Previous analyses revealed that levels of H3K27me3 are reduced at H3K27me3-enriched loci in pkl plants whether the locus in question exhibits PKL-dependent expression (Zhang et al., 2008). We therefore selected H3K27me3-enriched loci for analysis that exhibit both PKL-dependent and PKL-independent expression (Fig. 4A). Five-day-old seedlings were used for ChIP to take advantage of high levels of PKL protein at this stage of development (Li et al., 2005). For the sake of consistency, we restricted our region of analysis to within 500 bp of the predicted transcription start site. Importantly, our ChIP data revealed the presence of PKL at both LEC1 and LEC2. LEC1 and LEC2 are two master regulators of embryogenesis that we have previously shown are strongly derepressed in germinating pkl seeds and in pickle roots (Ogas et al., 1999; Dean Rider et al., 2003). In addition, we observed that PKL is present at the third LEC gene, FUSCA3 (FUS3), as well as at PHERES1 (PHE1), which is strongly derepressed in pkl plants after germination. Examination of another H3K27me3-enriched locus, At1g78750, that exhibits 5-fold elevation of transcript level in the absence of PKL in 5-d-old plants revealed association of PKL that is comparable to that of the LEC genes. We also examined the floral regulators AGAMOUS-LIKE 8 (AGL8) and AGL24, which exhibit a 10-fold reduction and unchanged transcript levels, respectively, in 5-d-old pkl plants, and found robust association of PKL with both of these H3K27me3-enriched loci. Finally, we also selected two additional H3K27me3-enriched loci for analysis that do not exhibit altered transcript levels in pkl plants: the floral regulator SUPPRESSOR OF OVEREXPRESSION OF CO1 (SOC1) and the E(Z) histone methyltransferase MEDEA (MEA). PKL was present at both of these loci as well. In summary, our ChIP analyses reveal that PKL associates with the promoters of all nine H3K27me3-enriched loci we examined regardless of whether expression of the corresponding gene is increased, decreased, or unchanged in pkl plants.
Figure 4.
PKL associates with H3K27me3-enriched loci. ChIP was used to examine association of PKL in 5-d-old plants (A) and in 75% germinated seedlings (B). ChIP was carried out using no antibody (No Ab) or antic-Myc (Anti Myc) using cross-linked DNA from wild-type (WT) and PKL-c-Myc (PKL-MYC) plants and the indicated loci were examined. The y axis denotes percent of input DNA brought down for a given immunoprecipitation. All data are the average of four biological replicates. The number in parentheses next to the locus represents the ratio of expression in pkl versus wild-type plants (where known) as determined by qRT-PCR. Bars denote sd. Asterisks denote a significant change in transcript level as determined by t test (P < 0.01).
We have previously shown that PKL acts during germination to prevent expression of the pickle root phenotype and to repress LEC1 (Li et al., 2005). To determine if PKL acts directly during this time to repress the LEC genes, we used ChIP to examine association of PKL with LEC1 and LEC2 in germinating PKL-c-Myc seeds (Fig. 4B). Seedlings were collected at 75% germination for ChIP analysis due to increased reproducibility of ChIP at this stage as opposed to 50% germination (data not shown). Our analysis revealed that PKL is present at both loci, indicating that PKL acts directly on both loci during this critical developmental window. In addition, we also examined At3g21720, which is a seed-specific gene coding for a predicted isocitrate lyase that exhibits PKL-dependent deposition of H3K27me3 during germination (Zhang et al., 2008). Our ChIP data indicate that PKL is present at this locus as well during germination.
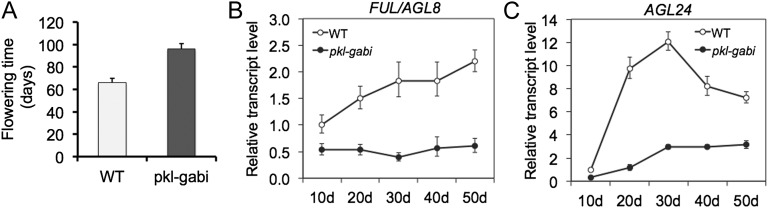
We were intrigued by the discovery that PKL was present at the promoters of AGL8 (FUL) and AGL24, which are H3K27me3-enriched genes and yet exhibit reduced transcript levels in 14-d-old pkl plants according to our microarray analysis (Supplemental Table S1). AGL8 and AGL24 are positive floral regulators, and reduced expression of either is capable of generating a late-flowering phenotype (Gu et al., 1998; Michaels et al., 2003), which is a phenotypic hallmark of pkl plants (Ogas et al., 1997; Henderson et al., 2004). Although our qRT-PCR analysis revealed that the transcript level of AGL24 is not PKL dependent in 5-d-old plants (Fig. 4A), qRT-PCR analysis of 14-d-old plants grown under identical conditions to those used for microarray analysis confirmed that that the transcript level of both genes is reduced in pkl plants; AGL8 is reduced 6-fold whereas AGL24 is reduced 3.3-fold.
To further examine the contribution of PKL to expression of these floral regulators, we used qRT-PCR to follow transcript levels of both genes in wild-type and pkl plants grown under 8-h d to prolong time to flowering. Under these conditions, flowering was delayed more than 45% in pkl plants relative to wild-type plants (Fig. 5A). Our qRT-PCR analysis confirmed that transcript levels of both of these positive floral regulators are reduced in pkl plants (Fig. 5, B and C). In particular, we observed that after 50 d of development, transcript levels of AGL8 largely fail to increase in pkl plants and the transcript level of AGL24 is reduced at least 2-fold. These qRT-PCR data thus confirm that expression of both floral regulators is reduced in pkl plants and further suggest that the late-flowering phenotype of pkl plants may in part be due to reduced expression of AGL8 and/or AGL24. In addition, these expression data further support our observation that PKL associates with the promoters of H3K27me3-enriched loci that exhibit reduced expression in pkl plants.
Figure 5.
FUL/AGL8 and AGL24 exhibit reduced transcript levels in pkl plants. Flowering time (A) and relative transcript levels of FUL (B) and AGL24 (C) were determined for pkl and wild-type plants grown in 8-h d. qRT-PCR was used to determine the relative transcript level of both genes in wild-type and pkl plants. 18S rRNA was used as a standardization control, and expression levels in each graph are normalized to expression of the gene of interest in wild-type plants at day 10. The x axis on sections B and C denotes days after sowing. Error bars = sd of the mean of three technical replicates. Analysis of a second biological replicate was consistent with these results (data not shown).
PKL Associates with Actively Transcribed Genes
Previous ChIP analysis of PKL by Aichinger and colleagues detected association of PKL with the PRC2 components EMF2 and SWN rather than H3K27me3-enriched loci (Aichinger et al., 2009). Although our transcript analysis reveals that expression of both of these genes is not PKL dependent in the absence of pickle root tissue (Fig. 3), this analysis does not contravene their ChIP data. Our ability to detect the presence of PKL at H3K27me3-enriched genes, however, suggested that further ChIP analyses using our PKL-c-Myc plants could reveal additional interactions that were missed by the previous analyses.
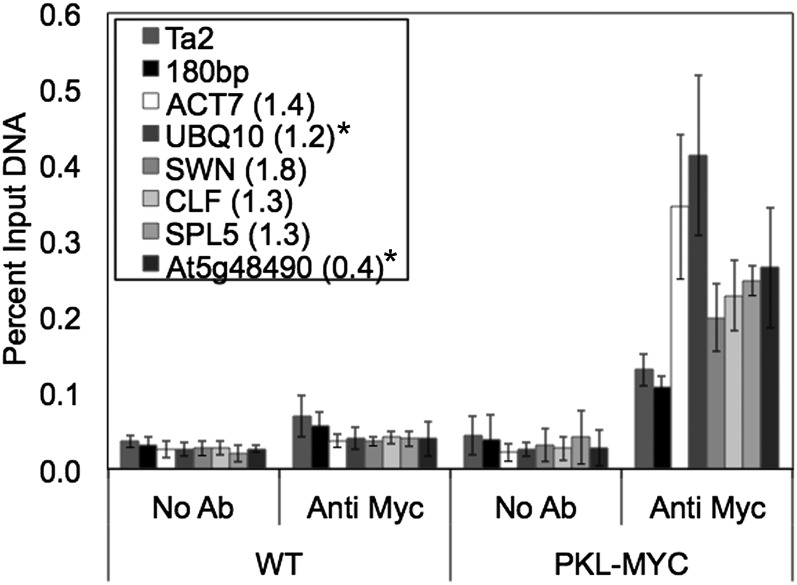
We undertook a ChIP analysis of PRC2 components and other actively transcribed genes. We included genes that exhibit PKL-independent expression as well as genes that exhibit PKL-dependent expression and excluded genes that are enriched for H3K27me3 (Fig. 6). Surprisingly, we found that PKL is present at the promoter of every one of the actively transcribed loci we assayed. In particular, we included ACT7 and UBQ10 in our analysis to serve as controls for the specificity of association of PKL with actively transcribed genes. An analogous control was not included in previous ChIP analyses (Aichinger et al., 2009). We found that PKL is strongly enriched at the promoter of both constitutively expressed genes relative to heterochromatic loci despite the fact that expression of both genes is not PKL dependent. Our ChIP analysis also confirmed that PKL associates with the promoters of the PRC2 components CLF, which exhibits a modest but significant 1.8-fold increase in transcript level in 5-d-old pkl plants, and SWN, which does not exhibit PKL-dependent expression in 5-d-old plants (Fig. 3). Our ChIP analysis also included SPL5, which does not exhibit significantly altered transcript levels, and At5g48490, which exhibits a significant 2-fold reduction in transcript level in 5-d-old pkl plants. We found that PKL robustly associates with both of these loci as well. Viewed in total, our ChIP data reveal that the presence of PKL at the promoter of an actively transcribed gene is not sufficient to confer PKL-dependent expression upon that gene and further suggest that PKL associates ubiquitously with actively transcribed genes.
Figure 6.
PKL associates with actively transcribed loci. ChIP was used to examine recruitment of PKL in 5-d-old plants. ChIP was carried out using no antibody (No Ab) or antic-Myc (Anti Myc) using cross-linked DNA from wild-type (WT) and PKL-c-Myc (PKL-MYC) plants and the indicated loci were examined. The y axis denotes percent of input DNA brought down for a given immunoprecipitation. All data are the average of four biological replicates. The number in parentheses next to the locus represents the ratio of expression in pkl versus wild-type plants (where known) as determined by qRT-PCR. Bars denote sd. Asterisks denote a significant change in transcript level as determined by t test (P < 0.01).
The unexpected finding that PKL associates with both H3K27me3-enriched genes and actively transcribed genes that are not enriched for H3K27me3 prompted us to consider the possibility that some or all of our data are an artifact of the PKL-c-Myc transgenic lines or the c-Myc antisera. We therefore repeated our ChIP analysis using PKL-FLAG plants to determine if our data were reproducible. PKL-FLAG plants were generated in an analogous fashion to PKL-c-Myc plants, and the PKL-FLAG construct was expressed using endogenous PKL regulatory sequences and rescued all pkl-associated phenotypes (data not shown). ChIP using anti-FLAG sera in PKL-FLAG lines replicated the observation that PKL preferentially associates with both H3K27me3-enriched loci (MEA and LEC1) and actively transcribed genes (ACT7 and UBQ10) relative to heterochromatin (Ta2 and CENP; Supplemental Fig. S2).
The H3K27me3 Modification Is Specifically and Consistently Reduced in pkl Plants
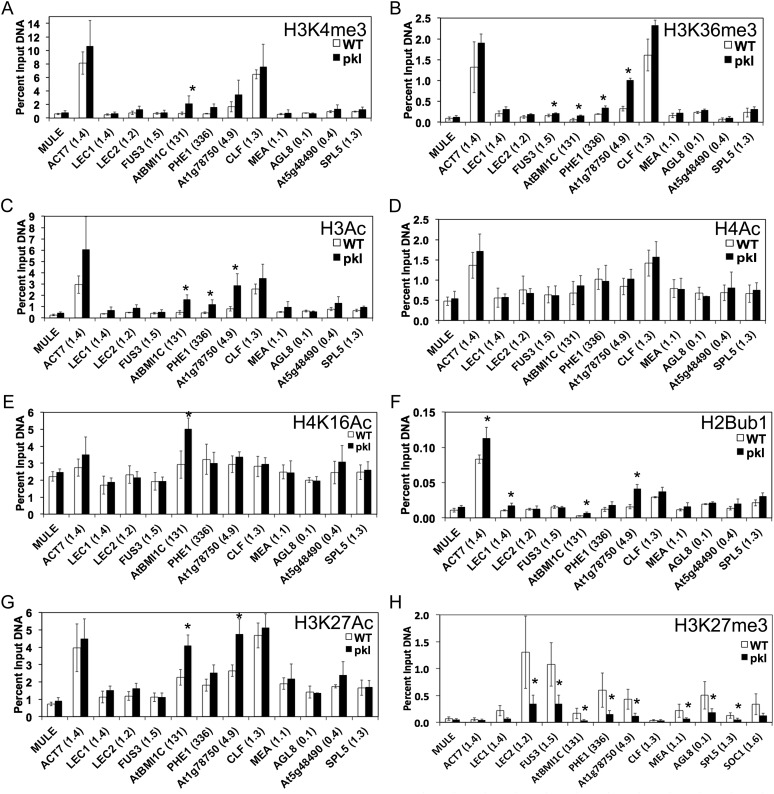
The observation that PKL is present at H3K27me3-enriched genes that exhibit decreased transcript levels (Fig. 4) suggested that PKL might play a role in activation of transcription at these loci as suggested by previous analyses (Aichinger et al., 2009, 2011). The additional discovery that PKL associates with actively transcribed genes (Fig. 6) raised the possibility that such a positive role for PKL in transcription might be more widespread. Given that the ability of PKL to act as a repressor is likely to be conferred in part by promoting H3K27me3 (Zhang et al., 2008; Aichinger et al., 2009), we tested the hypothesis that PKL also promotes an epigenetic modification associated with gene activation. We used ChIP to examine a large number of marks associated with actively transcribed genes: H3K4me3, H3K36me3, H3ac (K9, 14), H4K16ac, H4ac (K5, 8, 12, 16), H3K27ac, and H2Bub1. We also examined H3K27me3 as a positive control for a PKL-dependent epigenetic modification. Our analysis included genes that exhibited PKL-dependent expression and genes that exhibit PKL-independent expression as well as genes were either enriched for H3K27me3 or not.
Our analysis revealed that of all the marks we examined, only the steady-state level of H3K27me3 was consistently affected by loss of PKL (Fig. 7). Every H3K27me3-enriched locus examined exhibits reduced levels of H3K27me3 in pkl plants, whether the locus exhibits PKL-dependent expression (Fig. 7H). In contrast, none of the active marks decreased in pkl plants at any of the loci examined, revealing that PKL is not necessary for promoting these active marks whether in the presence or absence of the H3K27me3 modification. We did observe an increase in active marks (Fig. 7, A–C, F, and G) at the H3K27me3-enriched loci AtBMI1c, PHE1, and the F-box protein-encoding locus At1g78750. All three of these genes exhibit at least 5-fold increased transcript levels in pkl plants, however, suggesting that the observed increase in active marks at these loci is an indirect effect of increased expression of these loci. The PHE1 locus, which is strongly derepressed in pkl plants, was previously found to similarly exhibit increased acetylation in pkl plants (Zhang et al., 2008).
Figure 7.
Deposition of H3K27me3 is consistently reduced in pkl plants. ChIP was used to assay deposition of eight different histone modifications in 5-d-old wild-type and pkl plants. ChIP was carried out using antibodies to H3K4me3 (section A), H3K36me3 (section B), H3Ac (section C), H4Ac (section D), H4K16Ac (section E), H2Bub1 (section F), H3K27Ac (section G), and H3K27me3 (section H) using cross-linked DNA from wild-type (WT) and pkl-gabi (pkl) plants and the indicated loci were examined. Data are normalized for histone loading using a ChIP carried out using antibodies to H3. The y axis denotes percent of input DNA brought down for a given immunoprecipitation. All data are the average of four biological replicates. The number in parentheses next to the locus represents the ratio of expression in pkl versus wild-type plants (where known) as determined by qRT-PCR. Bars denote sd. Asterisks denote a significant change in a mark as determined by t test (P < 0.05).
DISCUSSION
PKL Acts Directly upon H3K27me3-Enriched Genes
H3K27me3 is a repressive epigenetic mark that plays a critical role in restricting expression of tissue-specific genes in Arabidopsis (Chanvivattana et al., 2004; Schubert et al., 2006; Bouyer et al., 2011; Lafos et al., 2011; Zheng and Chen, 2011). We have previously shown that PKL also represses expression of tissue-specific genes in Arabidopsis and that it promotes H3K27me3 (Zhang et al., 2008). Here we demonstrate that PKL protein is present at H3K27me3-enriched loci, indicating that PKL directly acts upon these loci to promote wild-type levels of H3K27me3.
Previous published work had indicated that PKL acts indirectly to promote H3K27me3 by instead promoting expression of the PRC2 machinery that deposits H3K27me3 (Aichinger et al., 2009). Specifically, Aichinger and colleagues found that transcript levels of the PRC2 components EMF2, CLF, and SWN were reduced in the roots of pkl pkr2 plants although not pkl plants (Aichinger et al., 2009). These studies, however, used biological material that included pickle root tissue, which is developmentally aberrant tissue that simultaneously expresses root and embryo transcriptional programs and is not equivalent to wild-type tissue (Dean Rider et al., 2003; Rider et al., 2004). We therefore tested the hypotheses that the observed alteration in transcript levels of PRC2 components was an experimental artifact that depended on the inclusion of pickle root tissue. We examined three developmental samples for PKL-dependent expression of PRC2 components: 50% germinated seedlings, 5-d-old roots, and 14-d-old seedlings. The pickle root phenotype is not visibly expressed in 50% germinated seedlings and is easily visually excluded from 14-d-old seedlings. Importantly, in 50% germinated seedlings and in 14-d-old seedlings the levels of H3K27me3 are reduced at H3K27me3-enriched genes in the absence of PKL (Zhang et al., 2008), revealing that PKL does act in these developmental samples to promote H3K27me3. Further, it has previously been demonstrated that PKL specifically acts during germination and not after germination to repress expression of embryonic traits (Li et al., 2005). In every developmental sample we examined, we found that expression of PRC2 components is not decreased in pkl plants or in pkl pkr2 plants, demonstrating that the decreased levels of H3K27me3 observed in pkl plants is not due to decreased expression of PRC2 components in pkl plants (Fig. 3).
Aichinger and colleagues also examined PKL localization by ChIP using a polyclonal antibody to PKL (Aichinger et al., 2009). They observed that PKL associated with EMF2 and CLF, which code for components of the PRC2 histone methyltransferase complex that deposits H3K27me3. They also found no detectable association of PKL with H3K27me3-enriched loci. These ChIP data were consistent with their hypothesis that PKL acts indirectly to promote H3K27me3 by promoting expression of the PRC2 machinery. It is worth noting, however, that a caveat of their ChIP data are that the control line for their analyses are pkl plants, which exhibit substantial developmental differences from wild-type plants. These developmental differences may lead to differences in signal intensity between wild-type and pkl plants that are indirect effects rather than direct effects that reflect the presence or absence of PKL. We reexamined PKL localization with ChIP using epitope-tagged versions of PKL that were expressed under endogenous regulatory sequences and that rescued the phenotype of pkl plants. As a result, our control lines were wild-type plants that were developmentally indistinguishable from our transgenic lines carrying an epitope-tagged version of PKL. Also, both of the epitopes we used, c-Myc and FLAG, are well-characterized epitopes that are used extensively in ChIP analysis due to the low background and robust signal associated with using these tags.
Our ChIP analyses revealed that PKL associates with both H3K27me3-enriched genes and actively transcribed loci that are not enriched for H3K27me3. Although the scope of targets is unexpected, it has the welcome attribute of being consistent with the data previously obtained by Aichinger et al. (2009). In their ChIP analysis, Aichinger and colleagues did not include a ubiquitously expressed gene as a specificity control (Aichinger et al., 2009). We included two such controls, ACT7 and UBQ10, and observed that PKL strongly associates with the promoter of both genes as well as with the PRC2 components CLF and SWN (Fig. 6). In fact, we observe that PKL preferentially associates with the promoter of all of the actively transcribed genes we examined relative to heterochromatic loci and that this association is not coupled to whether expression of the locus is PKL dependent. Thus our data suggest that the previously observed association of PKL with EMF2 and CLF reflects the tendency of PKL to associate with actively transcribed genes and that this association is not predictive of PKL-dependent expression.
We further found that PKL associated with the promoter region of nine out of nine H3K27me3-enriched genes we examined, regardless of how expression of the gene was affected by loss of PKL (Fig. 4A). Although Aichinger and colleagues reported that their ChIP analysis failed to uncover association of PKL with H3K2me3-enriched genes, this observation is a negative result (Aichinger et al., 2009). ChIP using our epitope-tagged versions of PKL, in contrast, reveals that the association of PKL with each of the nine H3K27me3-enriched loci examined is significantly above background for every locus. Taken in total, our expression and ChIP data thus strongly refute the model that PKL acts indirectly through transcriptional activation of PRC2 components and are instead consistent with the model that PKL acts directly upon H3K27me3-enriched loci to promote wild-type levels of H3K27me3.
PKL Directly Contributes to Repression of LEC Genes during Germination
Our data provide strong support for our model that PKL directly represses seed-specific genes during germination. A developmental hallmark of pkl plants is the inability to repress embryonic traits and the expression of the pickle root phenotype (Ogas et al., 1997). Pickle roots simultaneously express differentiation characteristics of roots and embryos (Rider et al., 2004). PKL protein is required during germination to repress expression of embryo-associated traits (Li et al., 2005). Furthermore, embryo-specific genes are derepressed during germination of pkl seeds and H3K27me3 levels are reduced at embryo-specific loci during germination of pkl seeds including the embryo master regulators LEC1 and LEC2 (Dean Rider et al., 2003; Zhang et al., 2008). Our new data demonstrate that PKL is present at LEC1 and LEC2 during germination (Fig. 4B), consistent with the model that PKL represses expression of the embryo-specific transcriptional program by directly promoting H3K27me3 at these loci during this developmental period.
Although PKL is necessary to repress expression of the seed transcriptional program during germination, microarray analysis of 14-d-old plants reveals that PKL is dispensable for repression of this program after germination. Seed-specific genes are not overrepresented among genes that exhibit increased transcript levels in 14-d-old pkl plants (Fig. 1B). These results are consistent with previous analyses of conditional PKL-GR plants, in which activation of PKL-GR during germination suppressed the pickle root phenotype but not the shoot phenotype as opposed to activation after germination, which suppressed the shoot phenotype but not the pickle root phenotype (Li et al., 2005). Thus loss of PKL generates a window of opportunity during germination throughout which the seed transcriptional program has the potential to become reestablished and generate the pickle root phenotype. After germination, however, other mechanisms are sufficient to maintain repression of seed-specific genes even in the absence of PKL.
The Spectrum of Loci That PKL Associates with Suggests That PKL May Play a Common Role in Chromatin Homeostasis
Our expression and epigenetic ChIP data reveal that loss of PKL preferentially and specifically affects H3K27me3-enriched genes. The level of H3K27me3 is reduced in 5-d-old pkl plants at nine of nine H3K27me3-enriched loci that we examined (Fig. 7H). These data are consistent with previous data demonstrating that the level of H3K27me3 is reduced in pkl plants at 50% germination and at 14 d after sowing (Zhang et al., 2008). In this study, seven other epigenetic modifications were concurrently examined along with H3K27me3 in pkl plants (Fig. 7). These data reveal that H3K27me3 is the only mark that is reduced at every locus examined for which the mark of interest is elevated above background. H3K27me3 is also the only epigenetic modification examined for which an alteration in the level of the mark occurs in the absence of altered expression of the corresponding gene in pkl plants. Furthermore, our microarray analysis reveals that H3K27me3-enriched genes are significantly overrepresented among genes that exhibit significantly increased or decreased transcript levels in 14-d-old pkl plants (Fig. 2). In contrast, H3K4me3-enriched genes are less likely to exhibit PKL-dependent expression (Fig. 2), suggesting that the presence of H3K4me3 renders a gene less susceptible to loss of PKL rather than revealing a critical role for PKL in promoting expression associated with H3K4me3 enrichment.
Our PKL ChIP analyses also reveal, however, that PKL may act generally across the genome rather than specifically in regions enriched for H3K27me3. In this regard, it is important to note that several of the loci we examined are strongly predicted to exhibit uniform expression states and therefore consistent chromatin architecture throughout the plant: ACT7 and UBQ10 are ubiquitously expressed and likely enriched for H3K4me3 throughout the plant whereas LEC1, LEC2, and PHE1 are uniformly repressed and likely enriched for H3K27me3 throughout the plant. Consequently, the association of PKL with these loci is strongly predicted to reflect the ability of PKL to associate with the epigenetic state typically associated with that locus rather than reflect the ability of PKL to associate with an alternate epigenetic state as a result of heterogeneity in the developmental sample. Furthermore, although the association of PKL with the constitutively silenced heterochromatic loci Ta2 and the centromeric repeats is modest, it is still above background in both PKL-c-Myc and PKL-FLAG lines (Supplemental Fig. S2).
Thus our ChIP data suggest that PKL is distributed throughout the Arabidopsis genome and associates with a diverse range of epigenetic chromatin states. Although at first glance the ability of PKL to associate with such a varied spectrum of sites is surprising, there are precedents and functional analyses to support the biological reality of the observed association of PKL with actively transcribed genes and heterochromatic regions. In Drosophila, the related CHD3 remodeler dMi-2 plays an important role in repression of gene expression (Kehle et al., 1998; Murawsky et al., 2001) and yet is distributed throughout actively transcribed regions of polytene chromosomes (Murawska et al., 2008). Loss of the chromatin factor MOM1 reveals a contribution of PKL to transcriptional gene silencing at endogenous chromosomal loci such as transcriptional silent information that are embedded in heterochromatic centromeric regions (Caikovski et al., 2008). Further, the related CHD4 remodeler has been reported to generally associate with chromatin in mammalian cells (Reynolds et al., 2012).
In total, analyses by our lab and others provide robust support for a role for PKL in H3K27me3-enriched regions and raise the possibility of a role for PKL elsewhere in the genome. One possibility is that the observed association of PKL with these other genomic regions is not functionally relevant; PKL associates with these regions but does not act. A contrasting possibility is that PKL does act in these regions but the role has yet to be uncovered or that another remodeling activity compensates for the loss of PKL. Genetic studies in yeast (Saccharomyces cerevisiae) with the related chromatin remodeler CHD1 provide an example of this type of functional redundancy in fungi (Krogan et al., 2002). If PKL does act throughout the genome, the question arises of why loss of PKL specifically affects H3K27me3-enriched regions. One possible explanation is that PKL has a unique activity in this type of chromatin because it is a subunit of a specific complex that is only targeted to H3K27me3-enriched regions. An alternative option is that PKL acts in a common fashion throughout the genome and H3K27me3-enriched regions are uniquely dependent on this activity.
This last model has recently been demonstrated to be relevant with regards to the contribution of Mi-2/NuRD to H3K27me3 deposition in embryonic stem cells from mice (Reynolds et al., 2012). In these cells, Mi-2/NuRD is associated with promoters that are enriched for H3K4me3 as well as promoters that are enriched for both H3K4me3 and H3K27me3 (referred to as bivalent chromatin domains). Mi-2/NuRD promotes deacetylation of H3K27 in bivalent chromatin domains. In the absence of Mi-2/NuRD, H3K27 remains acetylated in bivalent chromatin domains, PRC2 machinery fails to be recruited, and the level of H3K27me3 at the locus is reduced. Importantly, treatment with Trichostatin A, a histone deacetylase inhibitor, does not affect association of Mi-2/NuRD with the region but does reduce association of the PRC2 component Suz12. Together, these data indicate that the histone deacetylase activity of Mi-2/NuRD is necessary to enable the PRC2 complex to promote trimethylation of H3K27 at certain regions. Thus in an analogous fashion to the mode of action proposed above for PKL, Mi-2/NuRD is commonly acting on different epigenetic states of chromatin to promote a remodeling activity (histone deacetylation) that is uniquely required to promote H3K27me3 at some loci.
The discovery that PKL interacts with H3K27me3-enriched loci lends itself to a model in which PKL participates in a multisubunit complex analogous to the histone deacetylase Mi-2/NuRD complex but that instead promotes trimethylation of H3K27. Efforts to date, however, have failed to uncover a direct biochemical connection between PKL and the E(z) proteins that methylate H3K27. The spectrum of loci with which PKL associates raises the interesting possibility that PKL contributes to a yet to be determined remodeling activity that enables specific properties associated with H3K27me3-enriched regions (e.g. trimethylation of H3K27 and regulation of tissue-specific transcription). Regardless of the specific mechanism, our work reveals that the CHD3 remodeler PKL directly contributes to H3K27me3-associated processes in plants. Combining functional studies that exploit the unique role played by PKL at H3K27me3-enriched loci along with biochemical characterization of the remodeling activity of PKL is likely to greatly illuminate one of the basic processes by which transcriptional programs are expressed in a tissue-specific fashion in plants.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Wild type, pkl-1 (Ogas et al., 1997), and pkl-gabi (GABI-Kat line GK_273E06; Kleinboelting et al., 2012) are in the Columbia background. pkl-1 alleles were used for studies unless noted. pkl-gabi is an insertional allele of PKL that does not express detectable PKL protein by western analysis and is phenotypically indistinguishable from pkl-1 (H. Zhang, unpublished data). Seeds used in these studies were obtained from wild-type, mutant, and/or transgenic plants grown in parallel in an AR75 incubator (Percival Scientific) under 24 h of illumination. Seeds were allowed to dry at least a month on the plant prior to collection. No other treatment was applied (i.e. stratification) prior to use of the seeds. For expression and ChIP studies described here not involving determination of flowering time, plants were incubated on synthetic media (Ogas et al., 1997) and grown in a CU36L5 incubator (Percival Scientific) under 24 h of illumination. For studies involving flowering, plants were grown in an AR75 incubator under 8 h of illumination. For all analyses during germination (microarray, qRT-PCR, and ChIP), seeds were sown at a density of 100 mg of seeds (approximately 3,000 seeds) per 150-mm diameter petri dish. Fifty percent of germinated seedlings correspond to seeds that were imbibed and then collected when the radicle had emerged from 50% of the seeds (Zhang et al., 2008), and 75% germinated seedlings correspond to seeds in which the radicle had emerged from 75% of the seeds. Whole plants were collected 5 or 14 d postimbibition for both ChIP and expression analysis. pkl plants exhibiting the pickle root phenotype were excluded from 14-d samples unless otherwise noted. For studies involving weight of root and shoot tissue, plants were grown as described above for ChIP studies. Shoots were manually separated from the roots at the base of the hypocotyl of 5-d-old plants. Shoots or roots from 10 plants were pooled and then weighed on a Denver Instrument Company Scale (TR-64). Four biological replicates were included from wild-type and pkl plants for this analysis.
Plasmid Construction
A complete description of the construction of all recombinant DNA molecules generated for this study can be found in the Supplemental Materials and Methods S1.
RNA Isolation and Analysis
Total RNA was isolated using the RNAqueous kit from Ambion (Ambion, catalog no. AM1912). All subsequent experimental manipulations for microarray analysis were carried out as per the manufacturer’s instructions as described previously (Dean Rider et al., 2003), with the modifications that Affymetrix ATH1 Gene Chips (Arabidopsis Genome Array, catalog no. 900385) were used, and hybridization data were analyzed with the Affymetrix Microarray Suite version 5.0 software. Quantitative PCR was performed on StepOnePlus real-time PCR system (Applied Biosystems), as described by the manufacturer. 18S rRNA was used as a normalization control for the relative quantification of transcript levels using the comparative CT method as described by ABI User Bulletin number 2. All oligonucleotide primer sequences and primer concentrations used can be found in Supplemental Table S4.
Statistical Analysis and Archiving of Array Data
Microarray data were analyzed using the Bioconductor software. Specifically the cell intensity signal from each array was normalized using the GCRMA package, a moderated t test was performed using the limma package, and the q value for each probe set is calculated to control for the pFDR (Storey, 2002). Differentially expressed genes for both 14-d-old seedlings (Supplemental Table S1) and germinating seedlings (Supplemental Table S2) were identified with the criterion that the pFDR was less than 0.05 (Storey, 2003). Genes whose transcript levels are identified as statistically changed in the pkl mutant, together with corresponding fold-change values, can be found in Supplemental Table S1. Only array features that represent single genes are used for later comparisons with other published array data. Unless otherwise noted, Fisher’s exact test is performed for all the intersectional analyses, using the gmodels package from Bioconductor.
ChIP Analysis
Plant tissue harvested and processed based on a modified version of a previously published protocol (Wierzbicki et al., 2008). Please see Supplemental Materials and Methods S1 for a detailed version of the protocol. All oligonucleotide primer sequences and primer concentrations used for quantitative PCR for ChIP can be found in Supplemental Table S5.
The complete array data set has been deposited at the Gene Expression Omnibus of the National Center for Biotechnology Information, accession number GSE31639.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Analysis of the intersection of PKL-dependent genes in 14-d-old plants with other epigenomic data sets is consistent with the hypothesis that PKL does not function in the plant equivalent of a Mi-2/NuRD complex.
Supplemental Figure S2. ChIP using PKL-FLAG plants reproduce data obtained from PKL-c-Myc plants.
Supplemental Table S1. Table of genes that exhibit significantly different expression in 14-d-old pkl plants (pFDR, q < 0.05).
Supplemental Table S2. Table of genes that exhibit significantly different expression in 50% germinated pkl plants (pFDR, q < 0.05).
Supplemental Table S3. Table of genes that exhibit tissue-specific expression.
Supplemental Table S4. List of primers used for qRT-PCR.
Supplemental Table S5. List of primers used for real-time PCR for ChIP.
Supplemental Materials and Methods S1. Experimental details for plasmid construction and ChIP.
Supplementary Material
Acknowledgments
We thank Scott Briggs and Paul South for generously sharing reagents and technical assistance and Yinglin Bai for generating reagents. We thank Craig Peterson, Scott Briggs, and Ann Kirchmaier for thoughtful discussions. We thank Simon Turner for constructive comments during preparation of the manuscript.
Glossary
- ChIP
chromatin immunoprecipitation
- pFDR
positive false discovery rate
- qRT
quantitative reverse transcription
References
- Ahringer J. (2000) NuRD and SIN3 histone deacetylase complexes in development. Trends Genet 16: 351–356 [DOI] [PubMed] [Google Scholar]
- Aichinger E, Villar CB, Di Mambro R, Sabatini S, Köhler C. (2011) The CHD3 chromatin remodeler PICKLE and polycomb group proteins antagonistically regulate meristem activity in the Arabidopsis root. Plant Cell 23: 1047–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aichinger E, Villar CB, Farrona S, Reyes JC, Hennig L, Köhler C. (2009) CHD3 proteins and polycomb group proteins antagonistically determine cell identity in Arabidopsis. PLoS Genet 5: e1000605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouyer D, Roudier F, Heese M, Andersen ED, Gey D, Nowack MK, Goodrich J, Renou JP, Grini PE, Colot V, et al. (2011) Polycomb repressive complex 2 controls the embryo-to-seedling phase transition. PLoS Genet 7: e1002014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caikovski M, Yokthongwattana C, Habu Y, Nishimura T, Mathieu O, Paszkowski J. (2008) Divergent evolution of CHD3 proteins resulted in MOM1 refining epigenetic control in vascular plants. PLoS Genet 4: e1000165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. (2002) Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 298: 1039–1043 [DOI] [PubMed] [Google Scholar]
- Chanvivattana Y, Bishopp A, Schubert D, Stock C, Moon YH, Sung ZR, Goodrich J. (2004) Interaction of Polycomb-group proteins controlling flowering in Arabidopsis. Development 131: 5263–5276 [DOI] [PubMed] [Google Scholar]
- Charron JB, He H, Elling AA, Deng XW. (2009) Dynamic landscapes of four histone modifications during deetiolation in Arabidopsis. Plant Cell 21: 3732–3748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhury AM, Ming L, Miller C, Craig S, Dennis ES, Peacock WJ. (1997) Fertilization-independent seed development in Arabidopsis thaliana. Proc Natl Acad Sci USA 94: 4223–4228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapier CR, Cairns BR. (2009) The biology of chromatin remodeling complexes. Annu Rev Biochem 78: 273–304 [DOI] [PubMed] [Google Scholar]
- Deal RB, Henikoff S. (2010) A simple method for gene expression and chromatin profiling of individual cell types within a tissue. Dev Cell 18: 1030–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean Rider S, Jr, Henderson JT, Jerome RE, Edenberg HJ, Romero-Severson J, Ogas J. (2003) Coordinate repression of regulators of embryonic identity by PICKLE during germination in Arabidopsis. Plant J 35: 33–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshed Y, Baum SF, Bowman JL. (1999) Distinct mechanisms promote polarity establishment in carpels of Arabidopsis. Cell 99: 199–209 [DOI] [PubMed] [Google Scholar]
- Fukaki H, Taniguchi N, Tasaka M. (2006) PICKLE is required for SOLITARY-ROOT/IAA14-mediated repression of ARF7 and ARF19 activity during Arabidopsis lateral root initiation. Plant J 48: 380–389 [DOI] [PubMed] [Google Scholar]
- Furuta K, Kubo M, Sano K, Demura T, Fukuda H, Liu YG, Shibata D, Kakimoto T. (2011) The CKH2/PKL chromatin remodeling factor negatively regulates cytokinin responses in Arabidopsis calli. Plant Cell Physiol 52: 618–628 [DOI] [PubMed] [Google Scholar]
- Goodrich J, Puangsomlee P, Martin M, Long D, Meyerowitz EM, Coupland G. (1997) A Polycomb-group gene regulates homeotic gene expression in Arabidopsis. Nature 386: 44–51 [DOI] [PubMed] [Google Scholar]
- Grossniklaus U, Vielle-Calzada JP, Hoeppner MA, Gagliano WB. (1998) Maternal control of embryogenesis by MEDEA, a polycomb group gene in Arabidopsis. Science 280: 446–450 [DOI] [PubMed] [Google Scholar]
- Gu Q, Ferrándiz C, Yanofsky MF, Martienssen R. (1998) The FRUITFULL MADS-box gene mediates cell differentiation during Arabidopsis fruit development. Development 125: 1509–1517 [DOI] [PubMed] [Google Scholar]
- Hall JA, Georgel PT. (2007) CHD proteins: a diverse family with strong ties. Biochem Cell Biol 85: 463–476 [DOI] [PubMed] [Google Scholar]
- Hay A, Kaur H, Phillips A, Hedden P, Hake S, Tsiantis M. (2002) The gibberellin pathway mediates KNOTTED1-type homeobox function in plants with different body plans. Curr Biol 12: 1557–1565 [DOI] [PubMed] [Google Scholar]
- Henderson JT, Li HC, Rider SD, Mordhorst AP, Romero-Severson J, Cheng JC, Robey J, Sung ZR, de Vries SC, Ogas J. (2004) PICKLE acts throughout the plant to repress expression of embryonic traits and may play a role in gibberellin-dependent responses. Plant Physiol 134: 995–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennig L, Derkacheva M. (2009) Diversity of Polycomb group complexes in plants: same rules, different players? Trends Genet 25: 414–423 [DOI] [PubMed] [Google Scholar]
- Ho L, Crabtree GR. (2010) Chromatin remodelling during development. Nature 463: 474–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh TF, Shin J, Uzawa R, Silva P, Cohen S, Bauer MJ, Hashimoto M, Kirkbride RC, Harada JJ, Zilberman D, et al. (2011) Regulation of imprinted gene expression in Arabidopsis endosperm. Proc Natl Acad Sci USA 108: 1755–1762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehle J, Beuchle D, Treuheit S, Christen B, Kennison JA, Bienz M, Müller J. (1998) dMi-2, a hunchback-interacting protein that functions in polycomb repression. Science 282: 1897–1900 [DOI] [PubMed] [Google Scholar]
- Kinoshita T, Harada JJ, Goldberg RB, Fischer RL. (2001) Polycomb repression of flowering during early plant development. Proc Natl Acad Sci USA 98: 14156–14161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinboelting N, Huep G, Kloetgen A, Viehoever P, Weisshaar B. (2012) GABI-Kat SimpleSearch: new features of the Arabidopsis thaliana T-DNA mutant database. Nucleic Acids Res (Database issue) 40: D1211–D1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NJ, Kim M, Ahn SH, Zhong G, Kobor MS, Cagney G, Emili A, Shilatifard A, Buratowski S, Greenblatt JF. (2002) RNA polymerase II elongation factors of Saccharomyces cerevisiae: a targeted proteomics approach. Mol Cell Biol 22: 6979–6992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D. (2002) Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev 16: 2893–2905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafos M, Kroll P, Hohenstatt ML, Thorpe FL, Clarenz O, Schubert D. (2011) Dynamic regulation of H3K27 trimethylation during Arabidopsis differentiation. PLoS Genet 7: e1002040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Carey M, Workman JL. (2007) The role of chromatin during transcription. Cell 128: 707–719 [DOI] [PubMed] [Google Scholar]
- Li HC, Chuang K, Henderson JT, Rider SD, Jr, Bai Y, Zhang H, Fountain M, Gerber J, Ogas J. (2005) PICKLE acts during germination to repress expression of embryonic traits. Plant J 44: 1010–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels SD, Ditta G, Gustafson-Brown C, Pelaz S, Yanofsky M, Amasino RM. (2003) AGL24 acts as a promoter of flowering in Arabidopsis and is positively regulated by vernalization. Plant J 33: 867–874 [DOI] [PubMed] [Google Scholar]
- Müller J, Hart CM, Francis NJ, Vargas ML, Sengupta A, Wild B, Miller EL, O’Connor MB, Kingston RE, Simon JA. (2002) Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell 111: 197–208 [DOI] [PubMed] [Google Scholar]
- Murawska M, Kunert N, van Vugt J, Längst G, Kremmer E, Logie C, Brehm A. (2008) dCHD3, a novel ATP-dependent chromatin remodeler associated with sites of active transcription. Mol Cell Biol 28: 2745–2757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murawsky CM, Brehm A, Badenhorst P, Lowe N, Becker PB, Travers AA. (2001) Tramtrack69 interacts with the dMi-2 subunit of the Drosophila NuRD chromatin remodelling complex. EMBO Rep 2: 1089–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogas J, Cheng JC, Sung ZR, Somerville C. (1997) Cellular differentiation regulated by gibberellin in the Arabidopsis thaliana pickle mutant. Science 277: 91–94 [DOI] [PubMed] [Google Scholar]
- Ogas J, Kaufmann S, Henderson J, Somerville C. (1999) PICKLE is a CHD3 chromatin-remodeling factor that regulates the transition from embryonic to vegetative development in Arabidopsis. Proc Natl Acad Sci USA 96: 13839–13844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez J, Hagman J. (2009) The Mi-2/NuRD complex: a critical epigenetic regulator of hematopoietic development, differentiation and cancer. Epigenetics 4: 532–536 [DOI] [PubMed] [Google Scholar]
- Reynolds N, Salmon-Divon M, Dvinge H, Hynes-Allen A, Balasooriya G, Leaford D, Behrens A, Bertone P, Hendrich B. (2012) NuRD-mediated deacetylation of H3K27 facilitates recruitment of Polycomb Repressive Complex 2 to direct gene repression. EMBO J 31: 593–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rider SD, Jr, Hemm MR, Hostetler HA, Li HC, Chapple C, Ogas J. (2004) Metabolic profiling of the Arabidopsis pkl mutant reveals selective derepression of embryonic traits. Planta 219: 489–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saether T, Berge T, Ledsaak M, Matre V, Alm-Kristiansen AH, Dahle O, Aubry F, Gabrielsen OS. (2007) The chromatin remodeling factor Mi-2alpha acts as a novel co-activator for human c-Myb. J Biol Chem 282: 13994–14005 [DOI] [PubMed] [Google Scholar]
- Schmitges FW, Prusty AB, Faty M, Stützer A, Lingaraju GM, Aiwazian J, Sack R, Hess D, Li L, Zhou S, et al. (2011) Histone methylation by PRC2 is inhibited by active chromatin marks. Mol Cell 42: 330–341 [DOI] [PubMed] [Google Scholar]
- Schönrock N, Bouveret R, Leroy O, Borghi L, Köhler C, Gruissem W, Hennig L. (2006) Polycomb-group proteins repress the floral activator AGL19 in the FLC-independent vernalization pathway. Genes Dev 20: 1667–1678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert D, Primavesi L, Bishopp A, Roberts G, Doonan J, Jenuwein T, Goodrich J. (2006) Silencing by plant Polycomb-group genes requires dispersed trimethylation of histone H3 at lysine 27. EMBO J 25: 4638–4649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilatifard A. (2006) Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Annu Rev Biochem 75: 243–269 [DOI] [PubMed] [Google Scholar]
- Simon JA, Kingston RE. (2009) Mechanisms of polycomb gene silencing: knowns and unknowns. Nat Rev Mol Cell Biol 10: 697–708 [DOI] [PubMed] [Google Scholar]
- Storey JD. (2002) A direct approach to false discovery rates. J R Stat Soc Series B Stat Methodol 64: 479–498 [Google Scholar]
- Storey JD. (2003) The positive false discovery rate: a Bayesian interpretation and the q-value. Ann Stat 31: 2013–2035 [Google Scholar]
- Wierzbicki AT, Haag JR, Pikaard CS. (2008) Noncoding transcription by RNA polymerase Pol IVb/Pol V mediates transcriptional silencing of overlapping and adjacent genes. Cell 135: 635–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams CJ, Naito T, Arco PG, Seavitt JR, Cashman SM, De Souza B, Qi X, Keables P, Von Andrian UH, Georgopoulos K. (2004) The chromatin remodeler Mi-2beta is required for CD4 expression and T cell development. Immunity 20: 719–733 [DOI] [PubMed] [Google Scholar]
- Wolffe AP, Urnov FD, Guschin D. (2000) Co-repressor complexes and remodelling chromatin for repression. Biochem Soc Trans 28: 379–386 [PubMed] [Google Scholar]
- Yoshida N, Yanai Y, Chen L, Kato Y, Hiratsuka J, Miwa T, Sung ZR, Takahashi S. (2001) EMBRYONIC FLOWER2, a novel polycomb group protein homolog, mediates shoot development and flowering in Arabidopsis. Plant Cell 13: 2471–2481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Rider SD, Jr, Henderson JT, Fountain M, Chuang K, Kandachar V, Simons A, Edenberg HJ, Romero-Severson J, Muir WM, et al. (2008) The CHD3 remodeler PICKLE promotes trimethylation of histone H3 lysine 27. J Biol Chem 283: 22637–22648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Clarenz O, Cokus S, Bernatavichute YV, Pellegrini M, Goodrich J, Jacobsen SE. (2007) Whole-genome analysis of histone H3 lysine 27 trimethylation in Arabidopsis. PLoS Biol 5: e129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng B, Chen X. (2011) Dynamics of histone H3 lysine 27 trimethylation in plant development. Curr Opin Plant Biol 14: 123–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.