Abstract
RAC/ROP proteins (ρ-related GTPases of plants) are plant-specific small G proteins that function as molecular switches within elementary signal transduction pathways, including the regulation of reactive oxygen species (ROS) generation during early microbial infection via the activation of NADPH oxidase homologs of plants termed RBOH (for respiratory burst oxidase homolog). We investigated the role of Medicago truncatula Jemalong A17 small GTPase MtROP9, orthologous to Medicago sativa Rac1, via an RNA interference silencing approach. Composite M. truncatula plants (MtROP9i) whose roots have been transformed by Agrobacterium rhizogenes carrying the RNA interference vector were generated and infected with the symbiotic arbuscular mycorrhiza fungus Glomus intraradices and the rhizobial bacterium Sinorhizobium meliloti as well as with the pathogenic oomycete Aphanomyces euteiches. MtROP9i transgenic lines showed a clear growth-reduced phenotype and revealed neither ROS generation nor MtROP9 and MtRBOH gene expression after microbial infection. Coincidently, antioxidative compounds were not induced in infected MtROP9i roots, as documented by differential proteomics (two-dimensional differential gel electrophoresis). Furthermore, MtROP9 knockdown clearly promoted mycorrhizal and A. euteiches early hyphal root colonization, while rhizobial infection was clearly impaired. Infected MtROP9i roots showed, in part, extremely swollen noninfected root hairs and reduced numbers of deformed nodules. S. meliloti nodulation factor treatments of MtROP9i led to deformed root hairs showing progressed swelling of its upper regions or even of the entire root hair and spontaneous constrictions but reduced branching effects occurring only at swollen root hairs. These results suggest a key role of Rac1 GTPase MtROP9 in ROS-mediated early infection signaling.
RAC/ROP (ρ-related GTPases of plants) belong to a plant-specific subfamily of conserved ρ-type small GTPases that function as important molecular switches within elementary signal transduction pathways by cycling between active GTP-bound and inactive GDP-bound forms (Schiene et al., 2000; Yang and Fu, 2007; Liu et al., 2010). They mediate a wide range of molecular stimuli to downstream signaling compounds that provoke specific cellular responses. Based on these functional properties, which are well understood in mammalian and yeast cells, small GTPases are grouped into several subfamilies (Bourne et al., 1990, 1991; Schiene et al., 2000). In plants, they are involved in diverse developmental processes, such as polarized cell growth, cell morphogenesis, pollen tube and root hair development, as well as hormone signaling (Carol et al., 2005; Cole et al., 2005; Nibau et al., 2006; Yang and Fu, 2007; Liu et al., 2009).
Besides these functions in plant cellular development, there is increasing evidence that small GTP-binding proteins of plants are key regulatory elements for reactive oxygen species (ROS) generation by plasma membrane-associated NADPH oxidases termed RBOHs (for respiratory burst oxidase homologs) in plants. These oxygen intermediates (superoxide, hydroxyl radicals, and mainly hydrogen peroxide [H2O2]) were shown to provide direct antimicrobial activity via the inhibition of spore germination or of penetration processes of plant tissues (Lamb and Dixon, 1997). In addition, ROS contribute to cell wall strengthening via cross-linkage of cell wall polymers and lignin and represent elementary signal molecules for the activation of plant defense responses (Peng and Kuc, 1992; Ralph et al., 2004). Thus, a relevant role during early defense responses is assigned to RAC/ROP-like GTPases (Agrawal et al., 2003). The GTPase OsRac1 from rice (Oryza sativa) was found to be a positive regulator in disease resistance via the activation of RBOH-mediated ROS signaling through direct binding of Rac1 to the catalytic subunits of the protein’s N-terminal extension, which is specific for plant RBOH proteins (Kawasaki et al., 2006; Wong et al., 2007; Nakashima et al., 2008). Inversely, a dominant-negative form of Rac1 causes reduction of ROS levels, and dominant-negative OsRac1 was monitored to decrease ROS production and tobacco mosaic virus resistance in tobacco (Nicotiana tabacum) plants (Kawasaki et al., 1999; Moeder et al., 2005).
This paper explores the question of whether RAC/ROP-like GTPases also affect early symbiotic associations in plants, especially the symbiosis of rhizobial bacteria and of arbuscular mycorrhiza (AM) fungi with legume species (Markmann and Parniske, 2009; Colditz and Braun, 2010). Although both mutualistic interactions are distinct in nature, their formation depends on overlapping signaling pathways (Harrison, 1999; Markmann and Parniske, 2009). Recently, ROPs of the model legume Medicago truncatula were characterized with special focus on expression profiles during rhizobial infection, indicating an active role of some MtROP members (Liu et al., 2010). In addition, Lohar et al. (2007) reported a transient down-regulation of MtRBOH2 and MtRBOH3 during early treatment with Nod factors (NFs) that is responsible for root hair tip swelling, but the authors did not investigate MtROP expression patterns.
We previously reported the isolation and cloning of MsRac1 from Medicago sativa (Schiene et al., 2000). When expressed as an antisense construct in transgenic tobacco, the plants failed to develop necrotic lesions upon elicitor infiltration, suggesting an involvement in ROS signaling (Schiene et al., 2000). Heterologously expressed MsRac1 then was shown to interact with GTP (Brecht et al., 2004). In this study, we have utilized the previously investigated MsRac1 sequence from M. sativa for RNA interference (RNAi)-mediated gene silencing in the model legume M. truncatula. We identified a M. truncatula sequence ortholog annotated as MtROP9 (TC173331; Dana-Farber Cancer Institute M. truncatula Gene Index [MtGI]; Quackenbush et al., 2001). For RNAi gene knockdown, a gene-specific region of MtROP9 was selected. Roots of composite M. truncatula plants transformed by Agrobacterium rhizogenes carrying the RNAi vector (MtROP9i) exhibited drastically reduced MtROP9 expression. Oxidative burst assays as well as the expression of MtROP9-related genes in transgenic roots infected with symbiotic and pathogenic microorganisms allowed a characterization of the role of the Rac1-type MtROP9 in initial infection-related ROS signaling. Proteomic profiling supplements the expression studies and the physiological analysis. Perhaps most importantly, our RNAi approach indicates a fundamental role of MtROP9 in the establishment of efficient rhizobial infection, since infection was found to be impaired in the MtROP9i transgenic roots.
RESULTS
RNAi-Mediated Silencing of MtROP9 in M. truncatula
To investigate the role of the small GTPase Rac1 during early colonization of M. truncatula roots with symbiotic and pathogenic microbes, we silenced Rac1 orthologous MtROP9 gene expression using an RNAi approach to obtain composite M. truncatula (genotype Jemalong A17) plants with transgenic roots termed MtROP9i. Isolation and characterization of the Rac1 clone from M. sativa were described previously, and expression of an MsRac1 antisense construct in tobacco failed to develop necrotic lesions upon elicitor infiltration (Schiene et al., 2000). The M. truncatula sequence chosen for RNAi-mediated gene knockdown is 100% identical to the MsRac1 sequence and is annotated as MtROP9 (TC173331) according to the Dana-Farber Cancer Institute MtGI (Quackenbush et al., 2001). The sequence was selected for gene knockdown because it exhibits little sequence conservation with other ROP members from M. truncatula. BLAST search in MtGI revealed 82% sequence identity to MtROP3 (TC177831) as the closest relative, followed by MtROP8 (TC186969) and MtROP1 (TC178105), with 75% sequence identity each. Moreover, expression analyses revealed that MtROP3 was not affected by MtROP9 gene knockdown in MtROP9i transgenic roots. These results suggest that no cross-silencing of other ROP genes could occur. The RNAi-inducing construct cloned into the pK7GWIWG2(II)::DsRED binary vector contained the gene-specific MtROP9 sequence encoding parts of the putative effector (G2) and GTPase (G3) domains (Schiene et al., 2000; Limpens et al., 2005). In addition to the nondestructive identification of roots that contain the transgene of interest, this vector also allows the detection of chimeric roots (Limpens et al., 2004). Via A. rhizogenes-mediated root transformation, 12 populations of MtROP9i were constructed independently, each representing at least 100 stably growing composite plants with transgenic roots. Since the root phenotype was found to vary considerably, MtROP9i roots comprising on average 60% and more of transformed (DsRED-positive) roots were considered for all analyses (at least 50 single roots per individual population). For validation of the effective knockdown in MtROP9i transgenic roots, MtROP9 transcript abundance was determined by reverse transcription (RT)-quantitative PCR (Fig. 1). The relative expression of MtROP9 was drastically reduced in these roots after microbial infection when compared with control (Mtvector) roots.
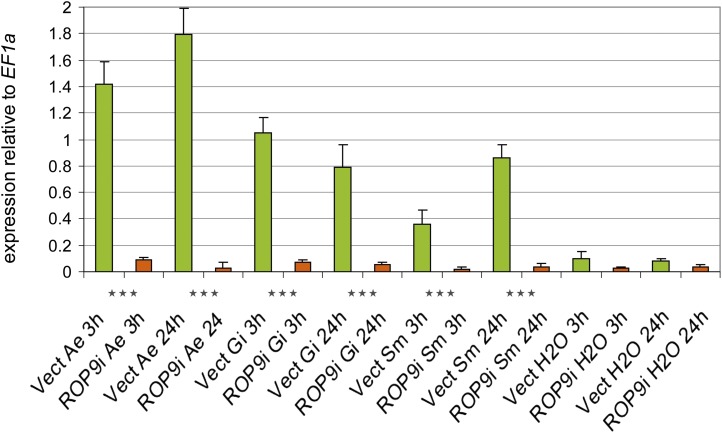
Figure 1.
Relative expression of MtROP9 in MtROP9i and Mtvector transgenic roots after A. euteiches (Ae), G. intraradices (Gi), and S. meliloti (Sm) infection at 3 and 24 hpi (RT-quantitative real-time PCR). The data shown represent averages and se of five biological replicates. Expression rates for MtROP9i are presented as orange bars and those for Mtvector as green bars. Data are standardized for the abundance of the EF1α transcripts. Asterisks indicate levels of significance by Student’s t test on all values evaluated for MtROP9i, Mtvector, and Mtwt: *** P < 0.001. [See online article for color version of this figure.]
Phenotypic Evaluation of M. truncatula MtROP9i Composite Plants with Transgenic Roots
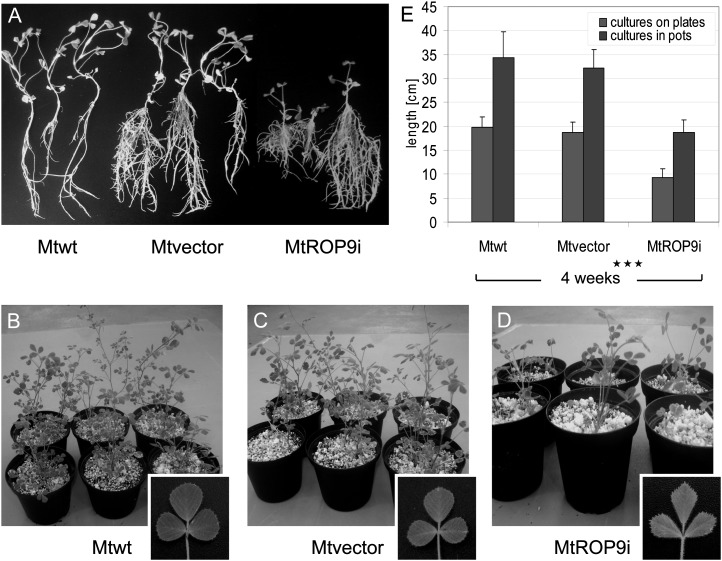
Since Rac/Rop-type small GTPases participate in diverse developmental processes, such as polarized cell growth, pollen tube growth, and root hair development (Chen et al., 2010; Liu et al., 2010), the influence of MtROP9 knockdown on the phenotype was investigated for the transformed MtROP9i and Mtvector composite plants and compared with nontransformed M. truncatula Jemalong A17 wild-type plants (Mtwt) grown under similar cultivation conditions. The initiation time point of transgenic root formation (from 12 d after transformation) as well as the number of adventitious roots were found to be similar for MtROP9i and Mtvector (data not shown). By contrast, composite plants with roots transformed by A. rhizogenes carrying the RNAi vector (MtROP9i) were clearly stunted as compared with the vector control plants, which showed similar growth to the nontransformed wild-type plants, whether grown on plates (Fig. 2A) or in pots (Fig. 2, B–D). Four weeks after cultivation, MtROP9i composite plants showed almost 50% reduction in length (Fig. 2E). By contrast, the general root architecture and anatomy of transgenic roots did not differ between MtROP9i and Mtvector.
Figure 2.
Phenotypic characterization of M. truncatula ROP9-silenced MtROP9i composite plants after 4 weeks. A, Comparisons of Mtwt roots (left), Mtvector roots (middle), and MtROP9i roots (right) grown on plates. B to D, Corresponding plants grown in pots together with detail images of leaves of wild-type and root-transformed composite plants. E, Lengths of Mtwt, Mtvector, and MtROP9i composite plants after 4 weeks of germination. Asterisks indicate levels of significance by Student’s t test on all values evaluated for MtROP9i, Mtvector, and Mtwt: *** P < 0.001.
Important for the analysis of rhizobial infections, MtROP9 knockdown also had no obvious developmental effect on the phenotype of root hairs of MtROP9i transgenic roots, which appeared to grow similar to those from the controls for all populations analyzed (data not shown). Interestingly, MtROP9 knockdown revealed pleiotropic effects even on the epicotylic phenotype of MtROP9i plants, most notably for the leaf shapes: leaves of the MtROP9i composite plants were drawn out but reduced in width, exhibiting a trapezoid-like shape, as compared with leaves of the Mtvector composite plants (Fig. 2, C and D).
MtROP9i Transgenic Roots Show Clear Reductions of ROS Accumulation
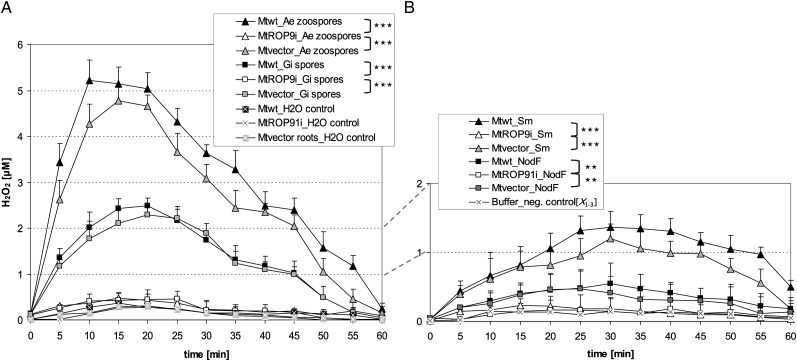
In rice, OsRac1 was found to be involved in ROS production during the early defense response via the activation of membrane-associated RBOH (Wong et al., 2007). Therefore, we examined whether the suppression of MtROP9 alters ROS levels in MtROP9i roots. ROS accumulation was measured in root segments of MtROP9i, Mtvector, and Mtwt after inoculation by a luminol-based chemiluminescence assay to determine the production of H2O2 during a period of 1 h after the application of two spore inocula: (1) 500 μL of lake water containing 250,000 Aphanomyces euteiches vital zoospores, as described previously (Trapphoff et al., 2009); and (2) 500 μL of water inoculum containing 10,000 viable Glomus intraradices spores. These measurements revealed clearly reduced ROS production in the MtROP9i transgenic roots as compared with the Mtwt and Mtvector root segments (Fig. 3A), probably indicating reduced RBOH activity. Induction of an oxidative burst was comparable for Mtwt and Mtvector, with high ROS levels after application of the A. euteiches inoculum (maxima at 10 min [Mtwt] and 15 min [Mtvector]) and a moderate level for the G. intraradices spore inoculum, with a delayed maximum at 20 min. To prove whether germinating hyphae that emerge from G. intraradices spores increase the potential of inducing an oxidative burst in the root segments, we applied a spore inoculum that had been incubated for 2 d at 25°C in the dark. After incubation, germination of spores was tested microscopically. Neither in Mtvector nor in MtROP9i root segments could considerable changes in the H2O2 accumulation be observed (data not shown). By contrast, MtROP9i root segments revealed only basal oxidative burst levels after inoculation with all spore fractions. ROS levels increased marginally to those of water controls, indicating residual RBOH activity in MtROP9i. Interestingly, treatments with Sinorhizobium meliloti bacterial culture induced a small but measurable oxidative burst in the Mtwt and Mtvector root segments, which was significantly higher as compared with ROS accumulation after S. meliloti NF treatment (Fig. 3B). The values for NF treatment were only very slightly increased in comparison with buffer control treatments. By contrast, both treatments failed to induce an oxidative burst in MtROP9i root segments.
Figure 3.
Oxidative burst ROS accumulation in Mtwt, Mtvector, and MtROP9i root fragments after different treatments. A, Root fragments were treated with A. euteiches (Ae) zoospores, G. intraradices (Gi) spores, or water representing the negative control. B, Root fragments were treated with S. meliloti (Sm) bacterial suspension, S. meliloti NF, or dilution buffer as a negative control (X1-3 = means of Mtwt, Mtvector, and MtROP9i measurements). Production of H2O2 was determined utilizing the luminal-based chemiluminesence assay. Asterisks indicate levels of significance by Student’s t test on the five most significant peak values evaluated for Mtwt, MtROP9i, and Mtvector as considered in clamps: *** P < 0.001, ** P < 0.01.
MtROP9i Transgenic Roots Reveal Suppressed Relative Expression of ROP9, RBOH, and Cat Genes
To validate the targeted knockdown of MtROP9 in the MtROP9i transgenic roots, we analyzed the relative expression of MtROP9 and that of selected genes involved in early infection signaling by RT-PCR performed on RNA isolated from five individual MtROP9i and Mtvector transformation populations. Relative expression analyses were carried out with A. euteiches-, G. intraradices-, and S. meliloti-infected roots at 3 and 24 h post inoculation (hpi); the data shown represent averages and se of five biological replicates (Fig. 4). In addition, representative bands of electrophoretically separated amplification products of RT-PCR are shown in Supplemental Figure S1.
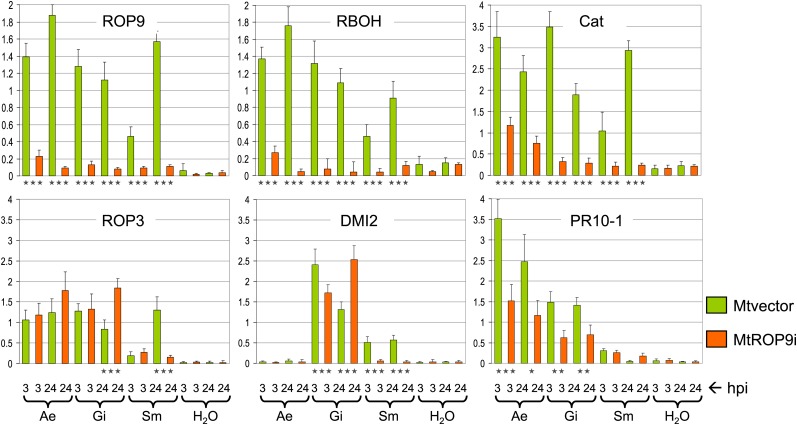
Figure 4.
Expression of MtROP9 and other related genes in MtROP9i and Mtvector transgenic roots after A. euteiches (Ae), G. intraradices (Gi), and S. meliloti (Sm) infection at 3 and 24 hpi. For quantitative evaluation of relative in-gel intensities of the amplified PCR products, intensities were calculated relative to the values for the EF1α reference gene (first two bars of each graph; scaled as 1). The data shown represent averages and se of five biological replicates. Expression rates for MtROP9i are presented as orange bars and those for Mtvector as green bars. Asterisks indicate levels of significance by Student’s t test on all expression values evaluated for Mtvector and MtROP9i: *** P < 0.001, ** P < 0.01, * P < 0.05. [See online article for color version of this figure.]
The relative expression analyses revealed a clear knockdown of MtROP9 expression in the gene-silenced MtROP9i roots as compared with Mtvector roots at both time points during pathogenic A. euteiches and symbiotic G. intraradices and S. meliloti infection, indicating effective MtROP9 knockdown. Interestingly, MtROP9 expression was induced in Mtvector roots not only after pathogenic infection but also during symbiotic mycorrhizal and rhizobial infection (Fig. 4). For expression analysis of MtRBOH involved in the generation of ROS, the MtRBOH3 gene (TC188820/94834) according to Lohar et al. (2007) was chosen. The relative transcript abundance levels were considerably decreased in the MtROP9i transgenic roots with respect to the Mtvector roots during all microbial interactions analyzed, which is coincident with a clearly suppressed oxidative burst in the MtROP9i roots (Fig. 3). Interestingly, the expression pattern for MtRBOH3 in Mtvector roots was similar to that of MtROP9, indicating synergistic induction profiles (Fig. 4). In accordance with the expression patterns for MtROP9 in the Mtvector roots, MtRBOH3 expression was found to be reduced during very early S. meliloti infection (3 hpi) as compared with the later 24-hpi time point. This transient decrease of MtRBOH3 expression during initial treatment with NF was reported previously (Lohar et al., 2007). Coincident with the clearly reduced relative expression of both MtROP9 and MtRBOH3 in the MtROP9i transgenic roots, the antioxidative response also was reduced in terms of reduced catalase MtCat (TC194418) expression in MtROP9i, apart from a slight increase after A. euteiches inoculation. By contrast, its expression was considerably increased in Mtvector roots during all three infections (especially at the 3-hpi time point), indicating elevated ROS levels even during early symbiotic infection due to ROP9-mediated RBOH activation, as also measured for G. intraradices spore inoculation (Fig. 3). As reported previously, PR proteins of class 10 (PR-10) represent marker proteins for established A. euteiches infection in M. truncatula (Colditz et al., 2004, 2005; Schenkluhn et al., 2010). Relative expression of MtPR10-1 is stronger in Mtvector than in MtROP9i roots after A. euteiches infection (Fig. 4). For MtDMI2, a member of symbiosis-specific early signaling, increased relative expression was found specifically during mycorrhizal and rhizobial early infection in Mtvector roots and also during mycorrhiza infection in the MtROP9i transgenic roots. By contrast, MtDMI2 expression was not induced in MtROP9i roots after infection with S. meliloti.
To prove whether MtROP9 knockdown also reveals a suppressive effect on the expression levels of other ROPs from M. truncatula, the expression of the closest relative of MtROP9, MtROP3 (TC177831), was analyzed. These analyses revealed no reduced MtROP3 expression levels in MtROP9i roots when compared with Mtvector roots. Thus, MtROP9 knockdown does not affect MtROP3 expression.
For the validation of relative gene expression data, statistical analyses were performed. Principal component analysis revealed clustering of expression scores according to the different infection treatments (Supplemental Fig. S2A). Interestingly, vectoral alignment of the expression scores showed unidirectional orientation for the genes MtROP9, MtRBOH3, and MtCat, suggesting the coexpression of genes. For mycorrhizal and A. euteiches infections, MtPR10-1 revealed additional coexpression, as did MtDMI2 for S. meliloti infection (Supplemental Fig. S2, B–D).
MtROP9 Expression Is Required for Efficient Rhizobial Symbiosis and Affects NF Signaling
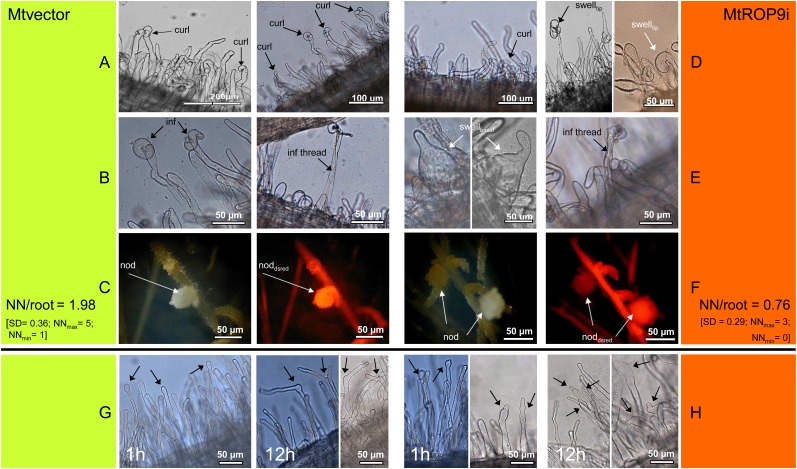
To investigate the effects of MtROP9 knockdown on the establishment of rhizobial symbiosis, infection with S. meliloti was documented for MtROP9i roots deriving from eight different transformation populations (50 roots per population; n = 400) where ROS accumulation as well as MtROP9 and MTRBOH3 gene expression was found clearly suppressed and compared with nodulated Mtvector roots (n = 400). The results obtained indicate a clear decrease in the number of infection events, such as root hair tip curling and branching in MtROP9i relative to Mtvector roots (Fig. 5, A and D). Moreover, bacterial infection was clearly delayed: first symptoms were visible between 4 and 7 d post inoculation (dpi) at MtROP9i roots but at 2 dpi in Mtvector roots (data not shown). At the same time, an abnormal root hair development, such as deformed root hairs revealing extreme basal or tip swelling, was observed on average in 23% of MtROP9i transgenic roots (Fig. 5, D, right, and E, left) where efficient bacterial infection was impaired: in no case of abnormal root hair development was infection progression, as indicated by the formation of infection threads, observed. Infection threads were only identified in nondeformed MtROP9i root hairs, but at clearly lower rates relative to Mtvector roots (Fig. 5E, right). By contrast, infected curled or branched root hairs of the Mtvector roots resemble the normally occurring wild-type competent root hairs exposed to rhizobia or NFs (Fig. 5B). Consistent with the reduction of infection events, MtROP9i roots developed fewer nodules than Mtvector roots did (0.76 nodules per transgenic root for MtROP9i, 1.98 nodules for Mtvector). In addition, the transgenic nodules in the MtROP9i roots also showed deformations such as rippled surfaces (Fig. 5F) in about 25% of the nodules formed.
Figure 5.
Microscopic characterization of S. meliloti infection (A–F) and of root hairs treated with S. meliloti NF for 1 h (left panels) and 12 h (right panels; G and H) in MtROP9i and Mtvector transgenic roots. A, Rhizobia attach to root hairs (curl, curling). B, Early infection (inf, transcellular infection; inf thread, formation of infection thread). C, Early nodule (nod) formation (noddsred, DsRED-positive nodule). D, Rhizobia attach to root hairs (swelltip, abnormal swelling of tips). E, Left, interrupted infection (swellbasal, abnormal basal swelling of root hairs); right, successful infection. F, Early nodule formation. Text at C and F is as follows: number of nodules (NN) per single root at 21 dpi; NNmax, maximum number of nodules found in one root; NNmin, minimum number of nodules found in one root. G, Arrows indicate root hair tip swelling, branching, and reinitiation of polar growth. H, Arrows indicate progressed (tip) swelling, spontaneous constriction, and branching of root hairs. [See online article for color version of this figure.]
To investigate whether S. meliloti signaling also is affected by MtROP9 knockdown, we applied S. meliloti NF to the transgenic roots. NF treatment was documented for MtROP9i and Mtvector root hairs deriving from four different transformation populations each (n = 200). Mtvector roots treated with NF showed typical morphological alterations of root hairs, such as root hair tip swelling (at 1 h after treatments) as well as branching and reinitiation of polar growth (at 12 h after treatments; Fig. 5G). Root hairs from MtROP9i exhibited an asynchronous phenotype with different deformations: a progressed swelling of not just the root hair tips but also at least its upper regions or even the entire root hair (immediately after NF application), accompanied by spontaneous constrictions of swollen root hairs (at 12 h after treatments). Branching effects were observed only at swollen root hairs at this stage, but in clearly reduced abundance as compared with the Mtvector controls. In general, root hair branching in MtROP9i was considerably less frequent as compared with Mtvector, suggesting impaired (re)initiation of polar growth.
Suppression of MtROP9 Expression Stimulates Mycorrhizal Colonization
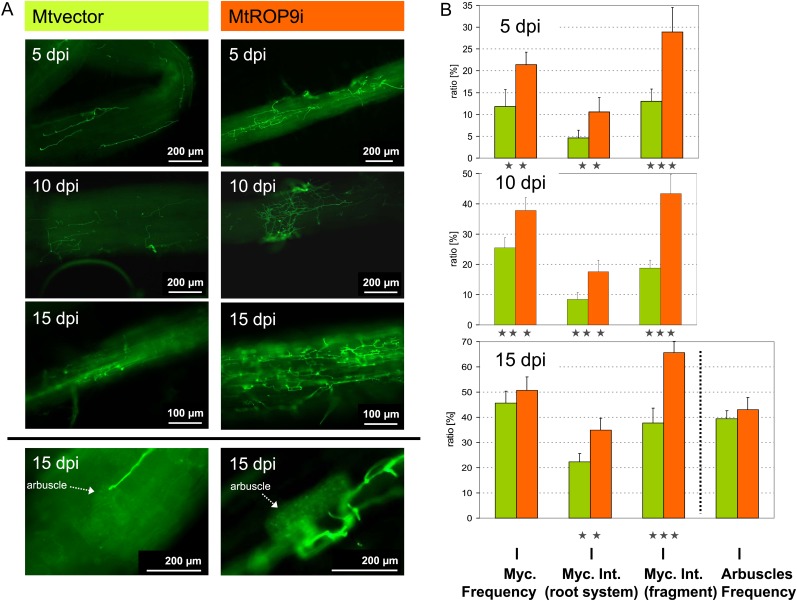
To analyze the effects of MtROP9 knockdown also on the AM symbiosis, infection with G. intraradices was documented for roots of MtROP9i composite plants deriving from six different transformation populations (50 roots per population; n = 300). Roots were considered as colonized when microscopical analysis revealed hyphae penetration through the epidermal cell layers. Contrary to the rhizobial interaction, colonization with G. intraradices was progressed in MtROP9i relative to Mtvector roots (n = 300; Fig. 6). Already at 5 dpi, the frequency of mycorrhization reached values of 20%. During the course of infection, especially the intensity of mycorrhizal colonization in infected root fragments was clearly increased in MtROP9i transgenic roots, with intensity maxima of up to 70% at 15 dpi compared with around 40% in Mtvector roots. Thus, initial colonization as well as progressed colonization with G. intraradices hyphae in already infected root fragments in MtROP9i are stimulated. At later infection stages (10 and 15 dpi), mycorrhization frequency and intensity in the root system of Mtvector roots converge with the values measured for the ROP9-silenced roots. Interestingly, the arbuscle abundance in mycorrhized root fragments was found to be similar for both MtROP9i and Mtvector (around 40% at 15 dpi).
Figure 6.
Fluorescence microscopic characterization of G. intraradices infection in MtROP9i and Mtvector transgenic roots at 5, 10, and 15 dpi. A, Mycorrhizal hyphal structures are WGA-Alexa Fluor 488 stained. B, Ratios of mycorrhization (Myc.) frequency (and arbuscle frequency at 15 dpi), mycorrhization intensity (Int.) in the whole root system, and the intensity in infected root fragments. Asterisks indicate levels of significance by Student’s t test on all values evaluated for Mtvector and MtROP9i: *** P < 0.001, ** P < 0.01. [See online article for color version of this figure.]
Suppression of MtROP9 Expression Stimulates A. euteiches Pathogenic Colonization
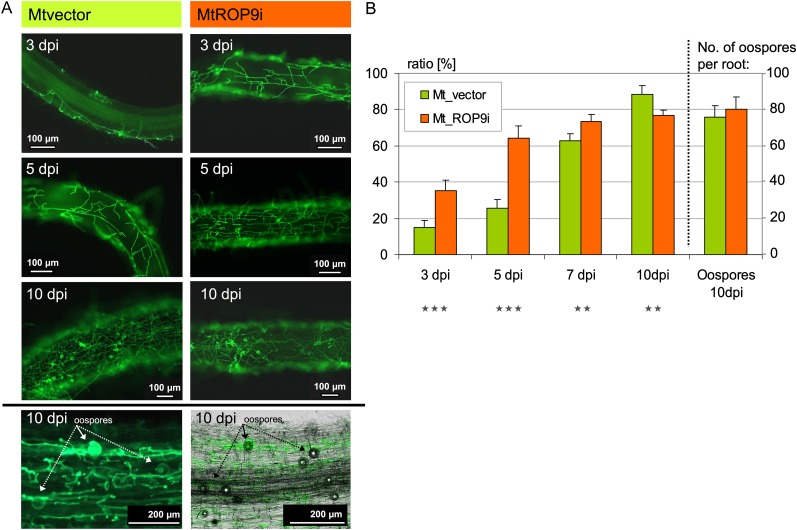
As ROS activity and signaling is an important part of the initial cellular defense responses, inoculation of Mtvector roots with A. euteiches zoospores induced a pronounced oxidative burst (Fig. 3). Thus, we investigated the effect of suppressed MtROP9 gene expression on colonization with this oomycete root pathogen in terms of average colonization per root, since all roots analyzed revealed A. euteiches infection (Fig. 7). The analyses performed on eight different transformation populations (50 roots per population; n = 400) indicate a clearly progressed A. euteiches colonization during early stages of infection in MtROP9i transgenic roots as compared with Mtvector roots (n = 400), which was limited to 5 dpi. At this infection time point, already more than 60% of MtROP9i root area was colonized with A. euteiches hyphae compared with only 25% of the Mtvector roots. Only 48 h later (at 7 dpi), A. euteiches colonization in Mtvector roots was clearly progressed, reaching nearly the infection levels of MtROP9i roots. These results indicate a loss of defense barriers between 5 and 7 dpi in Mtvector roots. By contrast, high A. euteiches colonization in MtROP9i transgenic roots occurred during the first 3 d of infection. At 10 dpi, A. euteiches colonization in Mtvector roots was even slightly increased as compared with the MtROP9i roots. In addition, the appearance of oospores was documented for both MtROP9i and Mtvector roots at this time point in comparable abundances (between 70 and 80 oospores per root).
Figure 7.
Fluorescence microscopic characterization of A. euteiches infection in MtROP9i and Mtvector transgenic roots at 3, 5, and 10 dpi. A, A. euteiches hyphal structures are WGA-Alexa Fluor 488 stained. B, Ratios of average A. euteiches colonization per infected root at 3, 5, 7, and 10 dpi and the number of oospores at 10 dpi. Asterisks indicate levels of significance by Student’s t test on all values evaluated for Mtvector and MtROP9i: *** P < 0.001, ** P < 0.01. [See online article for color version of this figure.]
Proteins of the ROS-Detoxifying System Are Not Induced in MtROP9i Transgenic Roots after Microbial Infection
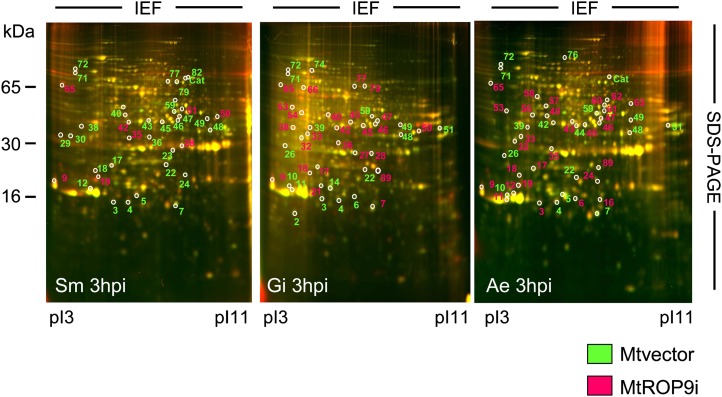
To prove whether altered symbiotic and pathogenic infection establishment in MtROP9i transgenic roots results in clear changes of infection-related protein patterns, a proteomic approach was carried out in order to monitor the protein profiles of MtROP9i and Mtvector root cells infected with S. meliloti, G. intraradices, and A. euteiches. For all infections analyzed, two-dimensional differential gel electrophoresis (2D-DIGE) was applied as reported previously for Mtwt roots (Schenkluhn et al., 2010). For two time points of early microbial infection, 3 and 24 hpi, differential protein induction in MtROP9i and Mtvector root cells was monitored by labeling each protein extract with distinct fluorescent dyes and combining the samples prior to electrophoretic separation. After fluorescence imaging of gels, proteins with increased relative intensity in the MtROP9i samples were detectable as red spots and proteins of increased relative intensity in the Mtvector samples were detectable as green spots. Representative gels for the 3-hpi time point of all three microbial infections are shown in Figure 8, and proteins of different induction are labeled with numbers according to the annotation of proteins as described previously for infected Mtwt roots (Schenkluhn et al., 2010). Protein spots were only considered as differentially induced if they exhibited at least 1.5-fold changes in spot intensity in all biological and technical samples prepared (evaluation according to Schenkluhn et al. [2010]). These proteins are displayed in Table I (beginning with the proteins of highest relative in-gel intensities for each group, followed by those with steadily decreasing intensities).
Figure 8.
Root proteome maps from 2D-DIGE analyses for MtROP9i and Mtvector root tissue at 3 h post infection with S. meliloti (Sm), G. intraradices (Gm), and A. euteiches (Ae). Differentially induced proteins are numbered as annotated for Mtwt infections according to Schenkluhn et al. (2010). Proteins induced in the Mtvector roots appear fluorescent green in the gel image and are labeled green; proteins induced in the MtROP9i roots appear fluorescent red in the gel image and are labeled red. IEF, Isoelectric focusing. [See online article for color version of this figure.]
Table I. Protein induction in Mtvector and MtROP9i transgenic roots after microbial infections.
Proteins are sorted beginning with the proteins of highest relative in-gel intensities, followed by those with steadily decreasing intensities. Protein induction is compared with that of infected Mtwt roots as investigated previously (Schenkluhn et al., 2010).
| Spot No.a | TC No.b | Best Matching Gene Productc | Molecular Weightd | pId | Regulation in Mtwt Rootse | Protein Regulationf |
||
| Regulation in Mtvector/MtROP9i | 3 hpi | 24 hpi | ||||||
| Protein induction in Mtvector transgenic roots | ||||||||
| 10 | 132039 | Class 10 -1 PR protein | 16.8 | 4.3 | Ae | Mtvector (Ae, Gi) | 3.24 (0.3) Ae | 3.82 (0.4) |
| 2.33 (0.2) Gi | 1.63 (0.1) | |||||||
| 71 | 129282 | Peroxidase precursor | 63.5 | 4.1 | Ae | Mtvector (Ae, Sm, Gi) | 3.48 (0.2) Ae | 2.5 (0.3) |
| 2.42 (0.4) Sm | 2.26 (0.2) | |||||||
| 2.37 (0.3) Gi | 2.13 (0.2) | |||||||
| 72 | 129282 | Peroxidase precursor | 64.0 | 4.1 | Ae | Mtvector (Ae, Sm, Gi) | 3.37 (0.4) Ae | 2.23 (0.3) |
| 1.82 (0.2) Sm | 2.46 (0.2) | |||||||
| 2.14 (0.2) Gi | 2.31 (0.3) | |||||||
| 22 | 127667 | Peroxidase | 26.5 | 6.8 | Sm, Sm + Ae | Mtvector (Sm, Gi, Ae) | 2.82 (0.2) Sm | 2.50 (0.3) |
| 2.44 (0.2) Gi | 2.41 (0.1) | |||||||
| 1.77 (0.4) Ae | 2.04 (0.3) | |||||||
| 48 | 132014 | Proteasome subunit β type-1 | 32.0 | 7.9 | Gi, Gi + Ae | Mtvector (Sm, Gi, Ae) | 2.75 (0.3) Sm | 2.47 (0.4) |
| 2.32 (0.2) Gi | 2.36 (0.3) | |||||||
| 2.19 (0.2) Ae | 1.67 (0.3) | |||||||
| 26 | 130855 | Proteasome subunit α type-5 | 28.4 | 4.4 | Ae | Mtvector (Ae, Gi) | 2.24 (0.3) Ae | 3.42 (0.2) |
| 2.25 (0.3) Gi | 2.33 (0.4) | |||||||
| 39 | 116053 | Proteasome subunit α type | 31.5 | 5.0 | Ae | Mtvector (Ae) | 2.4 (0.3) Ae | 2.67 (0.2) |
| 4 | 130380 | Superoxide dismutase (copper-zinc) | 16.0 | 5.4 | Ae, Sm + Ae | Mtvector (Ae, Sm, Gi) | 2.27 (0.4) Ae | 2.34 (0.2) |
| 1.80 (0.2) Sm | 1.76 (0.3) | |||||||
| 1.54 (0.2) Gi | 1.50 (0.1) | |||||||
| 49 | 119714 | Guanine nucleotide-binding protein (β) | 36.0 | 7.0 | Ae | Mtvector (Ae, Sm, Gi) | 2.41 (0.2) Ae | 2.38 (0.2) |
| 1.62 (0.3) Sm | 1.56 (0.2) | |||||||
| 1.49 (0.1) Gi | 1.57 (0.3) | |||||||
| 51 | 127667 | Peroxidase | 39.0 | 10.4 | Ae, Gi + Ae | Mtvector (Ae, Gi) | 2.48 (0.3) Ae | 2.12 (0.2) |
| 2.06 (0.5) Gi | 1.96 (0.3) | |||||||
| 5 | 116948 | Putative ripening-related protein | 17.5 | 5.6 | Ae, Con, Sm + Ae | Mtvector (Sm, Ae) | 2.33 (0.1) Sm | 1.84 (0.3) |
| 1.56 (0.3) Ae | 1.67 (0.1) | |||||||
| 59 | 116827 | Glyceraldehyde 3-phosphate dehydrogenase | 45.4 | 7.3 | Ae | Mtvector (Sm, Gi, Ae) | 2.45 (0.2) Sm | 1.55 (0.4) |
| 1.72 (0.2) Gi | 1.86 (0.3) | |||||||
| 30 | 114090 | Ran-binding protein | 32.0 | 4.4 | Sm | Mtvector (Sm) | 1.54 (0.3) Sm | 2.37 (0.3) |
| 2 | 128617 | Profilin-1 | 14.8 | 4.5 | Ae, Sm + Ae | Mtvector (Gi) | 1.75 (0.2) Gi | 1.77 (0.3) |
| 14 | 136600 | Putative ripening-related protein | 18.0 | 5.4 | Ae, Gi | Mtvector (Gi) | 1.65 (0.3) Gi | 1.54 (0.1) |
| 23 | 118388 | Fru-bisP aldolase | 29.5 | 7.4 | Ae | Mtvector (Sm) | 1.59 (0.3) Sm | 1.81 (0.1) |
| 29 | 139326 | Translational elongation factor 1, subunit B | 32.5 | 4.3 | Sm | Mtvector (Sm) | 1.64 (0.2) Sm | 1.45 (0.4) |
| 36 | 124173 | Short-chain dehydrogenase/reductase | 30.0 | 6.6 | Ae | Mtvector (Sm) | 1.52 (0.3) Sm | 1.51 (0.2) |
| 44 | 139960 | Stromal ascorbate peroxidase | 37.5 | 6.1 | Sm, Sm + Ae | Mtvector (Ae) | 1.69 (0.3) Ae | 1.48 (0.2) |
| 74 | 118164 | Cationic peroxidase 1 (precursor) | 68.0 | 4.8 | Sm, Sm + Ae | Mtvector (Gi) | 1.67 (0.3) Gi | 1.55 (0.3) |
| 76 | 136030 | Stress-induced protein ST1-like protein | 67.8 | 5.9 | Ae | Mtvector (Ae) | 1.50 (0.1) Ae | 1.58 (0.1) |
| 82 | 140211 | β-Glucosidase G1 | 62.0 | 7.4 | Sm, Gi + Ae | Mtvector (Sm) | 1.63 (0.2) Sm | 1.20 (0.4) |
| Protein induction in MtROP9i transgenic roots | ||||||||
| 9 | 134736 | Calmodulin 2 | 17.4 | 4.1 | Gi | MtROP9i (Gi, Sm, Ae) | 3.23 (0.4) Gi | 2.49 (0.3) |
| 1.96 (0.4) Sm | 2.23 (0.1) | |||||||
| 2.21 (0.3) Ae | 2.04 (0.2) | |||||||
| 27 | 119667 | Triosephosphate isomerase | 30.0 | 6.2 | Gi | MtROP9i (Gi) | 2.47 (0.1) Gi | 2.50 (0.3) |
| 35 | 115931 | Vegetative Gal-binding lectin | 30.0 | 5.6 | Ae | MtROP9i (Ae, Sm) | 2.44 (0.2) Ae | 2.36 (0.1) |
| 1.72 (0.3) Sm | 1.56 (0.1) | |||||||
| 33 | 132148 | 14-3-3-like protein | 33.0 | 4.8 | Gi, Sm, Gi + Ae, Sm + Ae | MtROP9i (Gi, Ae) | 2.40 (0.1) Gi | 2.46 (0.1) |
| 1.70 (0.2) Ae | 1.58 (0.2) | |||||||
| 42 | 117071 | Auxin-induced protein | 37.5 | 5.3 | Gi, Sm, Gi + Ae, Sm + Ae | MtROP9i (Gi, Ae, Sm) | 2.23 (0.1) Gi | 2.41 (0.1) |
| 1.53 (0.3) Ae | 2.04 (0.2) | |||||||
| 1.46 (0.2) Sm | 0.23 (0.1) | |||||||
| 89 | 121437 | Osmotin/thaumatin-like protein | 27.0 | 7.1 | Ae | MtROP9i (Gi, Ae) | 2.24 (0.3) Gi | 2.47 (0.1) |
| 2.25 (0.3) Ae | 2.36 (0.4) | |||||||
| 54 | 126275 | Putative adenosine kinase 2 (fragment) | 42.5 | 4.9 | Gi, Sm | MtROP9i (Gi) | 2.50 (0.3) Gi | 2.34 (0.1) |
| 16 | 125444 | Putative ripening-related protein | 16.5 | 7.3 | Ae | MtROP9i (Ae) | 2.48 (0.2) Ae | 2.50 (0.2) |
| 56 | 136010 | Putative narbonin | 42.0 | 5.2 | Ae, Gi + Ae, Gi | MtROP9i (Ae) | 2.41 (0.3) Ae | 1.67 (0.4) |
| 57 | 122880 | Transaldolase | 48.0 | 5.4 | Ae | MtROP9i (Ae) | 2.43 (0.3) Ae | 1.60 (0.1) |
| 62 | 116147 | Pyruvate dehydrogenase E1 component α | 45.0 | 7.8 | Ae | MtROP9i (Ae) | 2.31 (0.1) Ae | 1.53 (0.2) |
| 65 | 113679 | Putative ankyrin-repeat protein | 33.0 | 4.8 | Sm, Gi, Sm + Ae | MtROP9i (Sm, Gi, Ae) | 2.45 (0.2) Sm | 1.55 (0.4) |
| 1.78 (0.1) Gi | 1.88 (0.2) | |||||||
| 1.59 (0.1) Ae | 1.70 (0.5) | |||||||
| 21 | 114909 | ATP synthase subunit d, mitochondrial | 21.0 | 5.3 | Gi, Sm | MtROP9i (Gi) | 1.75 (0.2) Gi | 2.35 (0.2) |
| 28 | 131709 | Acid phosphatase | 30.5 | 6.8 | Sm | MtROP9i (Gi, Sm) | 1.62 (0.2) Gi | 2.46 (0.3) |
| 1.69 (0.1) Sm | 2.27 (0.1) | |||||||
| 58 | 139129 | Ribulose bisphosphate carboxylase/oxygenase activase large protein isoform | 53.5 | 5.4 | Ae, Sm, Sm + Ae | MtROP9i (Ae) | 1.67 (0.1) Ae | 2.03 (0.2) |
| 60 | 121355 | Formate dehydrogenase, mitochondrial | 44.0 | 7.6 | Ae, Gi + Ae | MtROP9i (Ae) | 1.49 (0.3) Ae | 2.16 (0.1) |
| 32 | 132148 | 14-3-3-like protein | 33.0 | 4.7 | Gi, Sm, Gi + Ae, Sm + Ae | MtROP9i (Gi, Ae) | 1.77 (0.1) Gi | 1.84 (0.2) |
| 1.59 (0.2) Ae | 0.57 (0.2) | |||||||
| 50 | 124940 | Glycoside hydrolase, family 17 | 34.0 | 8.3 | Ae | MtROP9i (Sm, Ae) | 1.59 (0.2) Sm | 1.58 (0.2) |
| 1.53 (0.2) Ae | 1.65 (0.3) | |||||||
| 61 | 117244 | Fru-bisP aldolase | 43.5 | 7.3 | Ae | MtROP9i (Ae) | 1.79 (0.1) Ae | 1.53 (0.2) |
| 66 | 120756 | Phosphoglycerate kinase | 55.0 | 5.3 | Gi, Gi + Ae | MtROP9i (Gi) | 1.75 (0.1) Gi | 1.50 (0.3) |
| 53 | 116888 | F2E2.4 | 45.5 | 4.4 | Sm, Sm + Ae | MtROP9i (Gi, Ae) | 1.76 (0.2) Gi | 0.85 (0.2) |
| 63 | 118388 | Fru-bisP aldolase | 43.0 | 8.1 | Ae, Sm + Ae | MtROP9i (Ae) | 0.49 (0.1) Ae | 1.56 (0.2) |
| Protein induction in both MtROP9i and Mtvector transgenic roots | ||||||||
| 11 | 117333 | Disease resistance response protein Pi49 | 17.0 | 4.4 | Ae | MtROP9i (Ae) | 2.84 (0.3) Ae | 2.76 (0.7) |
| Mtvector (Gi) | 1.83 (0.2) Gi | 1.92 (0.2) | ||||||
| 12 | 106314 | Abscisic acid-responsive protein ABR17 | 16.9 | 5.5 | Ae | MtROP9i (Ae) | 2.80 (0.2) Ae | 2.93 (0.1) |
| Mtvector (Sm) | 1.64 (0.3) Sm | 1.81 (0.2) | ||||||
| 40 | 136734 | Isoflavone reductase | 40.0 | 5.3 | Ae | MtROP9i (Ae) | 2.56 (0.1) Ae | 2.30 (0.2) |
| Mtvector (Sm) | 2.61 (0.2) Sm | 1.67 (0.1) | ||||||
| 46 | 126900 | Isoflavone reductase-like NAD(P)H-dependent oxidoreductase | 37.5 | 7.2 | Ae, Gi, Gi + Ae | MtROP9i (Ae, Gi) | 2.32 (0.2) Ae | 2.5 (0.3) |
| 2.46 (0.1) Gi | 2.31 (0.1) | |||||||
| Mtvector (Sm) | 1.73 (0.4) Sm | 2.64 (0.3) | ||||||
| 77 | 121513 | Mitochondrial processing peptidase | 65.0 | 6.6 | Gi, Gi + Ae | MtROP9i (Gi) | 2.42 (0.2) Gi | 2.76 (0.3) |
| Mtvector (Sm) | 2.20 (0.3) Sm | 2.06 (0.2) | ||||||
| 17 | 140281 | Chalcone-flavonone isomerase 1 | 27.0 | 5.4 | Ae, Gi, Gi+ Ae | MtROP9i (Gi) Mtvector (Sm) | 2.21 (0.2) Gi | 2.16 (0.3) |
| 1.63 (0.3) Sm | 2.46 (0.6) | |||||||
| 18 | 117480 | Trypsin protein inhibitor 3 | 24.0 | 4.8 | Ae, Gi + Ae | MtROP9i (Ae, Gi) | 2.42 (0.2) Ae | 2.40 (0.3) |
| 2.47 (0.2) Gi | 2.06 (0.2) | |||||||
| Mtvector (Sm) | 1.84 (0.2) Sm | 0.61 (0.4) | ||||||
| 43 | 118248 | NAD(P)H-dependent 6′-deoxychalcone synthase | 40.5 | 6.3 | Gi, Ae, Gi + Ae | MtROP9i (Gi, Ae) | 2.12 (0.2) Gi | 2.24 (0.3) |
| 2.06 (0.1) Ae | 2.32 (0.2) | |||||||
| Mtvector (Sm) | 1.73 (0.2) Sm | 2.14 (0.2) | ||||||
| 45 | 113451 | Chalcone reductase | 38.0 | 6.3 | Ae, Gi | MtROP9i (Gi, Ae) | 2.26 (0.1) Gi | 2.20 (0.2) |
| 2.00 (0.3) Ae | 2.23 (0.2) | |||||||
| Mtvector (Sm) | 1.66 (0.3) Sm | 1.54 (0.1) | ||||||
| 24 | 121437 | Osmotin/thaumatin-like protein | 20.0 | 7.2 | Ae | MtROP9i (Ae) | 2.06 (0.3) Ae | 2.32 (0.2) |
| Mtvector (Sm) | 2.12 (0.2) Sm | 0.31 (0.1) | ||||||
| 7 | 128250 | Nucleoside diphosphate kinase | 15.8 | 7.3 | Sm | Mtvector (Sm, Ae) | 2.45 (0.2) Sm | 2.5 (0.3) |
| 1.72 (0.2) Ae | 0.58 (0.2) | |||||||
| MtROP9i (Gi) | 1.61 (0.2) Gi | 1.76 (0.3) | ||||||
| 47 | 130358 | Malate dehydrogenase | 38.5 | 7.4 | Gi | MtROP9i (Gi, Ae) | 2.10 (0.2) Gi | 1.64 (0.2) |
| 2.06 (0.3) Ae | 0.52 (0.4) | |||||||
| Mtvector (Sm) | 1.78 (0.2) Sm | 2.01 (0.2) | ||||||
| 38 | 114090 | Ran-binding protein | 36.0 | 4.3 | Gi | MtROP9i (Gi) Mtvector (Sm) | 2.07 (0.3) Gi | 1.63 (0.2) |
| 1.54 (0.2) Sm | 1.71 (0.3) | |||||||
| 6 | 126924 | Actin-depolymerizing factor 2 | 15.9 | 5.8 | Ae, Sm + Ae | MtROP9i (Ae) Mtvector (Gi) | 1.70 (0.1) Ae | 1.65 (0.2) |
| 2.14 (0.3) Gi | 1.52 (0.1) | |||||||
| 19 | 122640 | Kunitz proteinase inhibitor ST1-like | 20.5 | 5.5 | Ae | MtROP9i (Ae, Sm) | 1.56 (0.3) Ae | 1.72 (0.2) |
| 0.54 (0.3) Sm | 1.50 (0.3) | |||||||
| 03 | 114386 | Major latex protein homolog | 16.0 | 5.2 | Ae, Sm, Con, Gi | Mtvector (Sm, Gi) MtROP9i (Ae) | 2.06 (0.1) Sm | 1.53 (0.3) |
| 1.58 (0.3) Gi | 0.32 (0.2) | |||||||
| 79 | 135749 | Δ-1-Pyrroline-5-carboxylate dehydrogenase 12A1, mitochondrial | 64.5 | 6.9 | Sm, Gi | Mtvector (Sm) | 1.59 (0.2) Sm | 1.71 (0.3) |
| MtROP9i (Gi) | 1.49 (0.2) Gi | 0.21 (0.1) | ||||||
Protein number. bBest matching tentative consensus (TC) sequence identifier in the MtGI. cProtein encoded by the best matching gene. dCalculated from the position on the two-dimensional electrophoresis gel. ePredominant protein induction in Mtwt roots following microbial infection as investigated previously (Schenkluhn et al., 2010). Ae, A. euteiches induced; Gi, G. intraradices induced; Sm, S. meliloti induced; Gi + Ae and Sm + Ae, induced in mycorrhized or nodulated roots secondarily infected with A. euteiches; Con, induced in control roots. fProtein regulation in the Mtvector and MtROP9i transgenic roots at 3 and 24 hpi. Specific induction is indicated with the abbreviations of the microbes as described above. Abundances represent relative in-gel abundances (±sd): greater than 2.5-fold, abundance is highly increased; 2- to 2.5-fold, increased abundance; 1.5- to 2-fold, abundance is slightly increased.
Evaluation of different protein patterns revealed that proteins involved in initial cytosolic antioxidative defense and ROS detoxification (several peroxidases [spots 71, 72, 22, 51, 44, and 76], superoxide dismutase [spot 4], and proteasome subunits [spots 48, 26 and 39]) were not induced in MtROP9i roots during all microbial infections analyzed (Table I). Although MtROP9i exhibited increased infection in comparison with Mtvector roots, antioxidative defense and ROS detoxification proteins were found exclusively in the Mtvector control. By contrast, other typically defense-related proteins that are not involved in early antioxidative reactions were found predominantly induced in MtROP9i roots (Fig. 8), which is consistent with progressed A. euteiches infections in these roots (PR10-like proteins [spots 11 and 12], PR5-like proteins [spots 89 and 24], and proteinase inhibitors [spot 18 and 19]). Enzymes of secondary phenylpropanoid and phytoalexin metabolism (spots 40, 46, 17, 43, and 45) were also predominantly induced in MtROP9i roots after A. euteiches and G. intraradices infection. Since both infections are promoted in MtROP9i roots, noninduction of enzymes of the initial antioxidative defense response likely results in the induction of infection-related proteins of a second defense barrier. In addition, the protein induction pattern of MtROP9i roots reflects progressed mycorrhizal infection in terms of increased induction of early mycorrhizal signaling factors as described for Mtwt roots (Schenkluhn et al., 2010): calmodulin 2 (spot 9), proteins involved in energy-regulative processes (spots 27, 21, 54, 66, 77, 7, and 47), proteins of auxin regulation (42 and 65), 14-3-3-like proteins (spots 33 and 32), a Ran-binding protein (spot 38), and the already mentioned enzymes of secondary phytoalexin metabolism.
DISCUSSION
Plant homologs of membrane-associated NADPH oxidases play a pivotal role in early pathogen defense. Their activity is triggered by RAC/ROP GTPases, which mediate RBOH-induced ROS production via their interaction with the RBOH N-terminal extension domain that contains two Ca2+-binding EF-hand motifs (Kawasaki et al., 1999; Schiene et al., 2000; Wong et al., 2007; Nakashima et al., 2008). This prompted us to investigate whether Rac GTPase regulates RBOH activity and ROS production in the model legume M. truncatula not only during microbial pathogenic infection but also during the prominent legume-specific rhizobial symbiosis as well as the widespread mycorrhizal symbiosis. In this study, we have demonstrated that the small GTPase ROP9 from M. truncatula represents an important signaling protein involved in the regulation of RBOH-mediated ROS production, which is required for the establishment of not only early pathogenic but also symbiotic microbial interactions. Since MtROP9i transgenic roots exhibited a wide range of phenotypic traits, it cannot be excluded that alterations in biotic root infections may be also related to broader metabolic changes caused by gene knockdown that not only affect the ROP-RBOH interaction. However, the growth and morphology of MtROP9i roots and root hairs were generally not affected during noninfected stages. Alterations were not observed prior to the infection of transgenic roots with the microbial partners.
Most notably, reduction in MtROP9 expression levels resulted in abnormal root hair development in MtROP9i transgenic roots during inoculation with rhizobial bacteria and caused impairment of infection. The obvious symptom for the cessation of further infection progress in MtROP9i roots is an extreme swelling of root hairs, which is different from the characteristic transiently occurring local swelling at the root hair tips in rhizobial infection and NF treatment, involving the cessation but then the reinitiation of polar root hair growth (Esseling et al., 2003). Since RBOH activity is drastically reduced in MtROP9i roots and ROS production after microbial stimulus is persistently diminished, we postulate that the local swelling of root hair tips as observed in Mtvector roots is expanded to the entire root hair in MtROP9i and that the failure to regain RBOH activity results in nonreinitiation of polar root hair growth or branching. Coincidently, MtROP9i root hairs treated with S. meliloti NF showed progressed swelling combined with spontaneous constrictions. Reinitiation of polar root hair growth was impaired in MtROP9i root hairs, leading to considerably less frequent branching effects. This is consistent with findings by Lohar et al. (2007), who reported that a transient decrease in ROS accumulation caused by the down-regulation of MtRBOH3 (and MtRBOH2) expression leads to root hair tip swelling at 1 h after NF treatment in M. truncatula. Most importantly, root hairs failed to reinitiate polar growth and branching in response to NF if ROS accumulation was persistently inhibited. Taken together, these findings provide further support for a key role of ROS accumulation during initial legume-rhizobia symbiosis. Furthermore, S. meliloti mutants defective in the biosynthesis of extracellular polysaccharides are also defective in the infection of their host plant, yet even so, they are able to induce root hair curling (Leigh at al. 1985). These mutants induce high levels of ROS (Niehaus et al., 1993). Obviously, only a balanced ROS level can promote S. meliloti infection.
Based on our results, we propose the following model for early NF signaling in MtROP9i. When ROS production is persistently diminished, as in MtROP9i, NF signaling is unhampered, resulting in progressed swelling of the root hair tips and almost the entire root hairs. NF signaling does not proceed in a coordinated manner, since the resumption of ROS accumulation fails; therefore, also branching effects and reinitiation of polar growth are clearly disabled. Instead, spontaneous constrictions occur at swollen root hairs, indicating residual RBOH activity in the ROP9-silenced roots at the sites of constriction. Alternatively, spontaneous branching of swollen root hairs was observed that might represent compensatory effects for considerably increased levels of turgor pressure.
By contrast, persistently inhibited ROS elevation in MtROP9i transgenic roots seems to have a promotive effect on the initial colonization of fungal (mycorrhizal) or fungus-like (oomycete) invaders, regardless of their symbiotic or pathogenic character. Thus, ROS signaling is considered to represent an effective host defense barrier preventing early colonization by microbes. For the progressive A. euteiches hyphal colonization, a delay in rapid colonization of approximately 5 d can be estimated in Mtvector roots when compared with MtROP9i. This is consistent with recent studies on the establishment of partial resistance in M. truncatula cultivars toward A. euteiches, which is mediated by initial induction of a high level of antioxidant enzymes accompanied by increased concentrations of soluble phenolics and lignin (Djébali et al., 2011). For AM infection, the infection intensity in mycorrhized MtROP9i root fragments was clearly increased for the first 15 dpi. Since mycorrhizal spores were capable of inducing a moderate oxidative burst in the root segments, evidence is given that ROS signaling is also involved in response to symbiotic mycorrhiza interaction. As a consequence of reduced ROS levels, antioxidative cellular components were not induced during early infection, as reported by noninduction of MtCat gene expression and enzymes of the antioxidative response via differential proteomics. The loss of this first defense barrier likely seems to promote the induction of compounds of further defense lines, including PR proteins and enzymes of antimicrobial phytoalexin synthesis along with infection-mediated phytohormone regulation. Thus, A. euteiches infection did not considerably increase in MtROP9i roots after 5 dpi but did in Mtvector roots, even exhibiting higher infection levels at 10 dpi. In contrast to the general prediction that Rac-related ROPs induce defense-related pathways and their compounds, these findings may indicate that ROP9 does not directly activate defense responses via the induction of multiple pathways (Kawasaki et al., 2006). Moreover, we assume that ROS signaling and cellular defense responses are not directly linked together, so that Rac1 is more a manifold signaling compound during early microbial associations than a strictly defense-related element.
CONCLUSION
In accordance with earlier investigations (Park et al., 2004; Wong et al., 2007; Nakashima et al., 2008), our results indicate that MtROP9, a small ρ-type GTPase, is involved in NADPH-mediated ROS production. First, ROS levels were clearly reduced in MtROP9i roots after microbial stimuli as compared with those in control roots. Second, the expression patterns of MtROB9 and MtRBOH3 were found to be similar after microbial infection in control roots, indicating gene coexpression, and both were clearly suppressed in MtROP9i. Furthermore, noninduction of the antioxidative MtCat gene and enzymes involved in the antioxidative response likely is a result of RBOH suppression in MtROP9i transgenic roots. Silencing of MtROP9i in M. truncatula exhibited considerable changes in early microbial infection patterns not only during pathogenic but also symbiotic interactions. Most notably, rhizobial symbiosis was found to be impaired in MtROP9-silenced roots, indicating the essential role of small GTPases during infection signaling in plants.
MATERIALS AND METHODS
RNAi Vector Construction
The binary vector pK7GWIWG2(II)::DsRED (kindly provided by E. Limpens; Limpens et al., 2005) containing the gene for the red fluorescent marker DsRED1 was used to produce transgenic MtROP9i roots. The vector was modified by the insertion of two sequence cassettes (in the sense-antisense direction) encoding parts of the putative effector (G2) and GTPase (G3) domains of the MsRac1 ortholog MtROP9 (TC173331; GenBank accession no. AF498359). Gateway technology (Invitrogen Life Technologies) was used to construct binary vectors for gene knockdown by RNAi. Therefore, gene-specific oligonucleotides (ROP9attb1_for, 5′-attB1-GTGTTACTGTTGGTGATG-3′; ROP9attb2_rev, 5′-attB2-ACGCCTTCACGTTCTCC-3′) with attached attB adapters were deduced from the Medicago sativa MsRac1 sequence (GenBank accession no. AJ251210; Schiene et al., 2000). These oligonucleotides were used for the amplification of a 461-bp fragment from Medicago truncatula cDNA. The PCR products were purified using the QIAquick PCR purification kit (Qiagen) and cloned into the pDONR221 donor vector (Invitrogen). A second cloning step transferred the inserts into the Gateway-compatible binary vectors mentioned above, which were transformed into Agrobacterium rhizogenes strain ArquaI (Quandt et al., 1993) using standard methods. This construct was sequenced to confirm the presence of the MtROP9 sequence fragments and the sense-antisense orientation of the cloned fragments in the T-DNA. For controls, nonmodified binary vectors were transformed into A. rhizogenes ArquaI.
Generation of Transgenic M. truncatula Roots
The generation of transgenic M. truncatula roots was conducted after the protocol of Boisson-Dernier et al. (2001) using M. truncatula (Jemalong A17) wild-type plantlets as described before (Colditz et al., 2007). Composite M. truncatula plants whose roots had been transformed by A. rhizogenes were cultured stably on M medium (Bécard and Fortin, 1988) containing 25 mg L−1 kanamycin for selection and decreasing concentrations of 350 to 0 mg L−1 ticarcillin disodium/clavulanate potassium (Duchefa) for the elimination of A. rhizogenes bacterial growth. Twelve individual populations of composite plants with MtROP9i and Mtvector transgenic roots were generated independently via A. rhizogenes transformation, containing at least 100 plants each. Populations were cultured either on M medium in growth chambers or in pots containing a 1:2 (v/v) mixture of sterile expanded clay and vermiculite in the greenhouse (22°C, 65% humidity, 16-h photoperiod at 220 μE m−2 s−1).
Nontransgenic Plant Material, Microbial Infections, and Infection Evaluation
M. truncatula (Jemalong A17) seedlings were grown as described previously (Colditz et al., 2004, 2005). Inoculation of MtROP9i, Mtvector, and Mtwt plant roots with Aphanomyces euteiches (ATCC 201684) was carried out as described before (Colditz et al., 2007). Each transgenic root population was infected with 500 μL of lake water containing 250,000 A. euteiches vital zoospores. Inoculation with Glomus intraradices was performed with MtROP9i and Mtvector potted composite plants using a commercially available inoculum (Granular AMF inoculum; BIORIZE), and granulate was mixed 1:10 with the clay:vermiculite mixture that was used to pot the MtROP9i and Mtvector composite plants. Infection with Sinorhizobium meliloti wild-type strain Rm2011 was performed as described previously (Schenkluhn et al., 2010). S. meliloti NFs were isolated from a NF-overproducing strain of S. meliloti strain 1021 p(EK327) as described by Baier et al. (1999). Purified NF (10−7 m) was prepared from a 10−3 m stock (diluted in CH3CN) in PS buffer (7 g of Na2PO4, 5 g of NaCl, and 3 g of KH2PO4 per liter, pH 7) and applied to the entire roots without touching them. PS buffer with CH3CN (in the same volume as the NF stock) was used for mock treatment. NF treatment was documented for MtROP9i and Mtvector root hairs deriving from four different transformation populations each.
For microscopic analysis of microbial infections, a Zeiss Axioscope fluorescence microscope (Carl Zeiss) was used. For visualization of mycorrhizal and A. euteiches hyphal structures, roots were stained with 0.2 μg mL−1 WGA-Alexa Fluor 488 (Invitrogen) according to Liu et al. (2003). Evaluation of G. intraradices infection was performed with the program Mycocalc (Trouvelot et al., 1986). Evaluation of A. euteiches infection was carried out as described previously (Colditz et al., 2004, 2005).
Measurement of ROS Oxidative Burst
Oxidative burst measurements were performed using a luminol-based chemiluminescence assay to determine the production of H2O2 during a period of 1 h after application of microbial fractions to root fragments of MtROP9i, Mtvector, and Mtwt as described previously (Trapphoff et al., 2009). For the measurements, 20 root pieces of 0.5 cm length were collected from MtROP9i, Mtvector, and Mtwt each and transferred to 200 μL of tap water in illuminometer cuvettes. They were kept for 4 h in the dark at room temperature. Afterward, the tap water was removed and root fragments were subjected to different experimental treatments: (1) 500 μL of lake water containing 250,000 A. euteiches vital zoospores as described previously (Trapphoff et al., 2009); (2) 500 μL of water inoculum containing 10,000 viable G. intraradices spores (Premier Tech); (3) 500 μL of S. meliloti bacterial culture (optical density at 600 nm of 1) in PS buffer; (4) 500 μL of S. meliloti NF (10−7 m) in PS buffer; and, as controls, (5) an equal volume of lake water (control for 1); (6) deionized water (control for 2); and (7) PS buffer (controls for 3 and 4). In addition, we applied a G. intraradices spore inoculum that had been incubated for 2 d at 25°C in the dark. After incubation, germination of spores was tested microscopically. After the addition of 1.2 mm luminol (Sigma-Aldrich) and 20 μL of horseradish peroxidase (0.1 mg mL−1), luminescence was determined with a Sirius luminometer (Berthold Technologies) in 5-min intervals during the 1-h treatment. For each treatment, three independent measurements were carried out per transformed MtROP9i and Mtvector population as well as for Mtwt roots. The evaluation of oxidative burst as displayed in Figure 2 is based on measurements for five different populations.
RT-PCR
Bulked transgenic roots (five to 10 individual roots) from microbe-inoculated and mock-inoculated MtROP9i and Mtvector populations were collected at 3 and 24 hpi. Samples from five independently generated populations of transgenic roots were collected separately, each representing one individual biological replicate. Total RNA was extracted from each root sample following a standard procedure. RNA samples (1 μg) were digested with DNase I (amplification grade; Invitrogen), and cDNA was synthesized using SuperScript II reverse transcriptase (Invitrogen). Semiquantitative RT-PCR was performed with 2 μL of cDNA as a template using 1.25 units of Dream Taq DNA Polymerase (Fermentas) according to the manufacturer’s protocol. Gene-specific oligonucleotides used during these PCRs are listed in Table II. Primers were designed using the Primer3 Web interface (Rozen and Skaletsky, 2000) according to the following criteria: product size range of 200 to 250 bp; primer size minimum of 21 bp and maximum of 24 bp; primer minimal melting temperature of 58°C and maximal melting temperature of 61°C. For each primer set, final PCR conditions were selected as follows: 2 min at 95°C (no cycling); 20 s at 95°C; 30 s at 60°C; and 60 s at 72°C; 16 additional cycles. PCR products were separated on a 1% agarose gel and visualized on a Biometradoc Transilluminizer (Biometra). A single product was detected in each case. For quantitative evaluation of relative in-gel intensities of the amplified PCR products, the BioDocAnalyze version 2.1 image software (Biometra) was used. The intensities were calculated relative to the values for the Elongation Factor1α (EF1α) reference gene of the corresponding gel, which were scaled as 1. Relative expression values for selected genes as given in Figure 3 represent averages and se of five biological replicates. For validation of effective MtROP9 knockdown, MtROP9 transcript abundance was additionally measured by RT-quantitative real-time PCR. Reactions were performed using the TaqMan Real Time PCR system (Life Technologies/Invitrogen). Therefore, the Platinum Quantitative PCR Super Mastermix (Life Technologies/Invitrogen) was used for PCR. Data were standardized for EF1α transcript abundance in five biological replicates, and average values were used to calculate relative gene expression levels using the 2-ΔCT value with ΔCT = CTgene − CTMtEf1α.
Table II.
Oligonucleotide primers used in RT-PCR
| Gene Name | TC No. | Left Primer | Right Primer | Expected Size of Amplicon |
| bp | ||||
| MtROP9 | 173331 | 5′-TGGTTGTTAATGGAAGTATTGTGAAT-3′ | 5′-TCTATGCAGAACTGCTTATCATC-3′ | 202 |
| MtROP3 | 177831 | 5′-GGCGCCAATCACCACAGCACA-3′ | 5′-GGAACAGGCTTTTTGCCCCTTTC-3′ | 200 |
| MtCat | 194418 | 5′-CTTGGCCTGAGGACATTATACCCC-3′ | 5′-GTTGTGGTGAGCACACTTGGGAGC-3′ | 200 |
| MtRBOH3 | 188820 (94834) | 5′-AGAGGGATTCAAGGAGGTTCG-3′ | 5′-TCCATGTAATAAGAGAGTCTCGA-3′ | 244 |
| MtPR10-1 | 176506 | 5′-TCCAAAGTATTGAGTTCACTGAAG-3′ | 5′-CTTTCAATTCCTCCTCACTTAGAT-3′ | 221 |
| MtDMI2 | 172456 | 5′-TAAGCAGAAACAATCTTGGTGAC-3′ | 5′-CTGCTCTGACAGTGCCATTTA-3′ | 245 |
| MtEF1α | 178258 | 5′-AAGCGTGTGATCGAGAGATTC-3′ | 5′-TGTCCATCCTTAGAGATACCAG-3′ | 245 |
2D-DIGE
Protein extraction, 2D-DIGE sample preparation, and CyDye labeling (GE Healthcare) as well as two-dimensional gel electrophoresis, image acquisition, and analysis were performed as described before (Schenkluhn et al., 2010). For protein extraction, 0.5 g of noninfected and infected root tissue from five individual MtROP9i and Mtvector populations was applied separately, resulting in five two-dimensional gels for each tissue analyzed. Gel evaluation was performed using the DeCyder version 5.01 software (GE Healthcare) as described previously (Schenkluhn et al., 2010).
Statistical Analysis
Analyses were performed using the R statistical software package (version 2.14.0; http://cran.r-project.org/bin/windows/base/). Scores from all investigations carried out for Mtwt, MtROP9i, and Mtvector were analyzed for significance using Student’s t test. Therefore, a general linear hypothesis testing method according to Hothorn et al. (2008) was used to provide multiplicity-adjusted P values for the Mtvector-MtROP9i comparisons. Additionally, to test the significance of the relative gene expression analysis, a principal component analysis was performed for loge-transformed scaled data. The scores for the first two principal components are presented in a biplot. ANOVA was performed separately on the scores of the first two principal components to test for significant knockdown, time points, infection effects, and their interactions according to Läuter et al. (1996). A multivariate linear model was used to estimate the means for each gene-silenced/nonsilenced-hour-infection level combination. These parameters were estimated in a linear mixed-effects model framework using the R software package (Bates et al., 2011).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number AF498359 for MtROP9.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Expression of MtROP9 and other related genes in MtROP9i and Mtvector transgenic roots after A. euteiches (Ae), G. intraradices (Gi), and S. meliloti (Sm) infection at 3 and 24 hpi.
Supplemental Figure S2. Principal component analysis of the relative expression of MtROP9 and other related genes in MtROP9i and Mtvector transgenic roots for all single infections (A) and separately after A. euteiches (Ae; B), G. intraradices (Gi; C), and S. meliloti (Sm; D) infection at 3 and 24 hpi.
Supplementary Material
Acknowledgments
We thank E. Limpens (Wageningen University) for kindly providing us the binary core vector. Furthermore, we thank K. Krause and C. Peterhänsel (Institute for Botany, Leibniz University) for assistance with the RT-quantitative real-time PCR analysis. We are thankful to D. Gerhard (Institute for Biostatistics, Leibniz University) for assistance with the statistical evaluation and to A. Wiebe (Department for Proteome and Metabolome Research, Bielefeld University) for help with the oxidative burst assays. We thank H. Küster and H.-P. Braun (Institute for Plant Genetics, Leibniz University) for proofreading the manuscript and for fruitful discussion.
Glossary
- ROS
reactive oxygen species
- H2O2
hydrogen peroxide
- AM
arbuscular mycorrhiza
- NF
Nod factor
- RNAi
RNA interference
- MtGI
M. truncatula Gene Index
- RT
reverse transcription
- Mtwt
M. truncatula A17 wild-type plants
- hpi
h post inoculation
- dpi
d post inoculation
- 2D-DIGE
two-dimensional differential gel electrophoresis
References
- Agrawal GK, Iwahashi H, Rakwal R. (2003) Small GTPase ‘Rop’: molecular switch for plant defense responses. FEBS Lett 546: 173–180 [DOI] [PubMed] [Google Scholar]
- Baier R, Schiene K, Kohring B, Flaschel E, Niehaus K. (1999) Alfalfa and tobacco cells react differently to chitin oligosaccharides and Sinorhizobium meliloti nodulation factors. Planta 210: 157–164 [DOI] [PubMed] [Google Scholar]
- Bates D, Maechler M, Bolker B. (2011) lme4: linear mixed-effects models using S4 classes. R package version 0.999375-42. http://CRAN.R-project.org/package=lme4 (December 5, 2011)
- Bécard G, Fortin JA. (1988) Early events of vesicular-arbuscular mycorrhiza formation on Ri T-DNA transformed roots. New Phytol 108: 211–218 [DOI] [PubMed] [Google Scholar]
- Boisson-Dernier A, Chabaud M, Rosenberg C, Barker D. (2001) Agrobacterium rhizogenes-transformed roots of Medicago truncatula for the study of nitrogen-fixing and endomycorrhizal symbiotic associations. Mol Plant Microbe Interact 14: 695–700 [DOI] [PubMed] [Google Scholar]
- Bourne HR, Sanders DA, McCormick F. (1990) The GTPase superfamily: a conserved switch for diverse cell functions. Nature 348: 125–132 [DOI] [PubMed] [Google Scholar]
- Bourne HR, Sanders DA, McCormick F. (1991) The GTPase superfamily: conserved structure and molecular mechanism. Nature 349: 117–127 [DOI] [PubMed] [Google Scholar]
- Brecht M, Sewald K, Schiene K, Keen G, Fricke M, Sauer M, Niehaus K. (2004) The use of surface plasmon resonance (SPR) and fluorescence resonance energy transfer (FRET) to monitor the interaction of the plant G-proteins Ms-Rac1 and Ms-Rac4 with GTP. J Biotechnol 112: 151–164 [DOI] [PubMed] [Google Scholar]
- Carol RJ, Takeda S, Linstead P, Durrant MC, Kakesova H, Derbyshire P, Drea S, Zarsky V, Dolan L. (2005) A RhoGDP dissociation inhibitor spatially regulates growth in root hair cells. Nature 438: 1013–1016 [DOI] [PubMed] [Google Scholar]
- Chen L, Shiotani K, Togashi T, Miki D, Aoyama M, Wong HL, Kawasaki T, Shimamoto K. (2010) Analysis of the Rac/Rop small GTPase family in rice: expression, subcellular localization and role in disease resistance. Plant Cell Physiol 51: 585–595 [DOI] [PubMed] [Google Scholar]
- Colditz F, Braun H-P. (2010) Medicago truncatula proteomics. J Proteomics 73: 1974–1985 [DOI] [PubMed] [Google Scholar]
- Colditz F, Braun H-P, Jacquet C, Niehaus K, Krajinski F. (2005) Proteomic profiling unravels insights into the molecular background underlying increased Aphanomyces euteiches-tolerance of Medicago truncatula. Plant Mol Biol 59: 387–406 [DOI] [PubMed] [Google Scholar]
- Colditz F, Niehaus K, Krajinski F. (2007) Silencing of PR-10-like proteins in Medicago truncatula results in an antagonistic induction of other PR proteins and in an increased tolerance upon infection with the oomycete Aphanomyces euteiches. Planta 226: 57–71 [DOI] [PubMed] [Google Scholar]
- Colditz F, Nyamsuren O, Niehaus K, Eubel H, Braun H-P, Krajinski F. (2004) Proteomic approach: identification of Medicago truncatula proteins induced in roots after infection with the pathogenic oomycete Aphanomyces euteiches. Plant Mol Biol 55: 109–120 [DOI] [PubMed] [Google Scholar]
- Cole RA, Synek L, Zarsky V, Fowler JE. (2005) SEC8, a subunit of the putative Arabidopsis exocyst complex, facilitates pollen germination and competitive pollen tube growth. Plant Physiol 138: 2005–2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djébali N, Mhadhbi H, Lafitte C, Dumas B, Esquerré-Tugayé MT, Aouani ME, Jacquet C. (2011) Hydrogen peroxide scavenging mechanisms are components of Medicago truncatula partial resistance to Aphanomyces euteiches. Eur J Plant Pathol 131: 559–571 [Google Scholar]
- Esseling JJ, Lhuissier FG, Emons AMC. (2003) Nod factor-induced root hair curling: continuous polar growth towards the point of nod factor application. Plant Physiol 132: 1982–1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison MJ. (1999) Molecular and cellular aspects of the arbuscular mycorrhizal symbiosis. Annu Rev Plant Physiol Plant Mol Biol 50: 361–389 [DOI] [PubMed] [Google Scholar]
- Hothorn T, Bretz F, Westfall P. (2008) Simultaneous inference in general parametric models. Biom J 50: 346–363 [DOI] [PubMed] [Google Scholar]
- Kawasaki T, Henmi K, Ono E, Hatakeyama S, Iwano M, Satoh H, Shimamoto K. (1999) The small GTP-binding protein rac is a regulator of cell death in plants. Proc Natl Acad Sci USA 96: 10922–10926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki T, Koita H, Nakatsubo T, Hasegawa K, Wakabayashi K, Takahashi H, Umemura K, Umezawa T, Shimamoto K. (2006) Cinnamoyl-CoA reductase, a key enzyme in lignin biosynthesis, is an effector of small GTPase Rac in defense signaling in rice. Proc Natl Acad Sci USA 103: 230–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb C, Dixon RA. (1997) The oxidative burst in plant disease resistance. Annu Rev Plant Physiol Plant Mol Biol 48: 251–275 [DOI] [PubMed] [Google Scholar]
- Läuter J, Glimm E, Kropf S. (1996) New multivariate tests for data with an inherent structure. Biom J 38: 5–23 [Google Scholar]
- Leigh JA, Signer ER, Walker GC. (1985) Exopolysaccharide-deficient mutants of Rhizobium meliloti that form ineffective nodules. Proc Natl Acad Sci USA 82: 6231–6235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limpens E, Mirabella R, Fedorova E, Franken C, Franssen H, Bisseling T, Geurts R. (2005) Formation of organelle-like N2-fixing symbiosomes in legume root nodules is controlled by DMI2. Proc Natl Acad Sci USA 102: 10375–10380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limpens E, Ramos J, Franken C, Raz V, Compaan B, Franssen H, Bisseling T, Geurts R. (2004) RNA interference in Agrobacterium rhizogenes-transformed roots of Arabidopsis and Medicago truncatula. J Exp Bot 55: 983–992 [DOI] [PubMed] [Google Scholar]
- Liu JY, Blaylock LA, Endre G, Cho J, Town CD, VandenBosch KA, Harrison MJ. (2003) Transcript profiling coupled with spatial expression analyses reveals genes involved in distinct developmental stages of an arbuscular mycorrhizal symbiosis. Plant Cell 15: 2106–2123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Li RL, Zhang L, Wang QL, Niehaus K, Baluska F, Samaj J, Lin JX. (2009) Lipid microdomain polarization is required for NADPH oxidase-dependent ROS signaling in Picea meyeri pollen tube tip growth. Plant J 60: 303–313 [DOI] [PubMed] [Google Scholar]
- Liu W, Chen A-M, Luo L, Sun J, Cao L-P, Yu G-Q, Zhu J-B, Wang Y-Z. (2010) Characterization and expression analysis of Medicago truncatula ROP GTPase family during the early stage of symbiosis. J Integr Plant Biol 52: 639–652 [DOI] [PubMed] [Google Scholar]
- Lohar DP, Haridas S, Gantt JS, VandenBosch KA. (2007) A transient decrease in reactive oxygen species in roots leads to root hair deformation in the legume-rhizobia symbiosis. New Phytol 173: 39–49 [DOI] [PubMed] [Google Scholar]
- Markmann K, Parniske M. (2009) Evolution of root endosymbiosis with bacteria: how novel are nodules? Trends Plant Sci 14: 77–86 [DOI] [PubMed] [Google Scholar]
- Moeder W, Yoshioka K, Klessig DF. (2005) Involvement of the small GTPase Rac in the defense responses of tobacco to pathogens. Mol Plant Microbe Interact 18: 116–124 [DOI] [PubMed] [Google Scholar]
- Nakashima A, Chen L, Thao NP, Fujiwara M, Wong HL, Kuwano M, Umemura K, Shirasu K, Kawasaki T, Shimamoto K. (2008) RACK1 functions in rice innate immunity by interacting with the Rac1 immune complex. Plant Cell 20: 2265–2279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nibau C, Wu HM, Cheung AY. (2006) RAC/ROP GTPases: ‘hubs’ for signal integration and diversification in plants. Trends Plant Sci 11: 309–315 [DOI] [PubMed] [Google Scholar]
- Niehaus K, Kapp D, Pühler A. (1993) Plant defence and delayed infection of alfalfa pseudonodules induced by an exopolysaccharide (EPS I)-deficient Rhizobium meliloti mutant. Planta 190: 415–425 [Google Scholar]
- Park J, Gu Y, Lee Y, Yang Z, Lee Y. (2004) Phosphatidic acid induces leaf cell death in Arabidopsis by activating the Rho-related small G protein GTPase-mediated pathway of reactive oxygen species generation. Plant Physiol 134: 129–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng M, Kuc J. (1992) Peroxidase-generated hydrogen peroxide as a source of antifungal activity in vitro and on tobacco leaf disks. Phytopathology 82: 696–699 [Google Scholar]
- Quackenbush J, Cho J, Lee D, Liang F, Holt I, Karamycheva S, Parvizi B, Pertea G, Sultana R, White J. (2001) The TIGR Gene Indices: analysis of gene transcript sequences in highly sampled eukaryotic species. Nucleic Acids Res 29: 159–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quandt HJ, Pühler A, Broer I. (1993) Transgenic root nodules of Vicia hirsutea: a fast and efficient system for the study of gene expression in indeterminate-type nodules. Mol Plant Microbe Interact 6: 699–706 [Google Scholar]
- Ralph J, Lundquist K, Brunow G, Lu F, Kim H, Schatz PF, Marita JM, Hatfield RD, Ralph SA, Christensen JH, et al. (2004) Lignins: natural polymers from oxidative coupling of 4-hydroxyphenylpropanoids. Phytochem Rev 3: 29–60 [Google Scholar]
- Rozen S, Skaletsky H. (2000) Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol 132: 365–386 [DOI] [PubMed] [Google Scholar]
- Schenkluhn L, Hohnjec N, Niehaus K, Schmitz U, Colditz F. (2010) Differential gel electrophoresis (DIGE) to quantitatively monitor early symbiosis- and pathogenesis-induced changes of the Medicago truncatula root proteome. J Proteomics 73: 753–768 [DOI] [PubMed] [Google Scholar]
- Schiene K, Pühler A, Niehaus K. (2000) Transgenic tobacco plants that express an antisense construct derived from a Medicago sativa cDNA encoding a Rac-related small GTP-binding protein fail to develop necrotic lesions upon elicitor infiltration. Mol Gen Genet 263: 761–770 [DOI] [PubMed] [Google Scholar]
- Trapphoff T, Beutner C, Niehaus K, Colditz F. (2009) Induction of distinct defense-associated protein patterns in Aphanomyces euteiches (Oomycota)-elicited and -inoculated Medicago truncatula cell-suspension cultures: a proteome and phosphoproteome approach. Mol Plant Microbe Interact 22: 421–436 [DOI] [PubMed] [Google Scholar]
- Trouvelot A, Kough JL, Gianinazzi-Pearson V. (1986) Mesure du taux de mycorhization VA d’un système radiculaire: recherche de signall d’estimation ayant une signification fonctionnelle. In V Gianinazzi-Pearson, S Gianinazzi, eds, Physiological and Genetical Aspects of Mycorrhizae. INRA Press, Paris, pp 217–221 [Google Scholar]
- Wong HL, Pinontoan R, Hayashi K, Tabata R, Yaeno T, Hasegawa K, Kojima C, Yoshioka H, Iba K, Kawasaki T.et al (2007) Regulation of rice NADPH oxidase by binding of Rac GTPase to its N-terminal extension. Plant Cell 19: 4022–4034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Fu Y. (2007) ROP/RAC GTPase signaling. Curr Opin Plant Biol 10: 490–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.