Over the past decade, plant heat shock factors (Hsfs) have been associated mainly with responses to abiotic stress (von Koskull-Döring et al., 2007), yet, until this report, they have not been linked to primed defense gene activation or pathogen-induced systemic acquired resistance (SAR). Hsfs are a diverse group of proteins that regulate genes encoding heat shock proteins (von Koskull-Döring et al., 2007). They do so by binding to a regulatory DNA motif called the heat stress element (Pelham, 1982). Arabidopsis (Arabidopsis thaliana) has more than 20 Hsfs that have been divided into three classes (A, B, and C) based on the amino acid sequence of their flexible linkers and oligomerization domains (HR-A/B regions; Nover et al., 2001). Although members of class A Hsfs seem to be bona fide transcription coactivators, class B Hsfs do not contain a transcription-promoting, C-terminal activation domain (Nover et al., 2001). However, they still can serve as transcription coactivators (Bharti et al., 2004).
A recent report with Arabidopsis revealed that HsfB1 (also referred to as HSF4 or TBF1) represses the general heat shock response in the absence of excessive heat but supports the development of acquired thermotolerance in times of heat stress (Ikeda et al., 2011). HsfB1 was also reported to be a negative regulator of the jasmonic acid/ethylene-responsive defensin genes PDF1.2a and PDF1.2b and to suppress the innate immune response of Arabidopsis to the necrotrophic fungus Alternaria brassicicola (Kumar et al., 2009). AtGenExpress microarray data suggested also a positive regulatory role for HsfB1 in the plant response to pathogens and microbe-associated molecular patterns (von Koskull-Döring et al., 2007), but this has not been investigated further. Moreover, except for one very recent report (Pajerowska-Mukhtar et al., 2012), HsfB1 and other Hsfs have not been associated with SAR. This type of plant immune response is induced upon localized infection with a necrotizing pathogen and develops systemically throughout the plant (Hammerschmidt, 2009). SAR is associated with the systemic accumulation of the plant hormone salicylic acid, which primes the plant for a more robust and rapid activation of defense genes upon further pathogen attack (Conrath et al., 2006). Here, we show that HsfB1 positively regulates the primed expression of defense-related genes and that it is required for bona fide SAR to bacterial pathogens in Arabidopsis.
To investigate whether HsfB1 could play a role in the development of defense priming and SAR, we first assessed whether benzo(1,2,3)thiadiazole-7-carbothioic acid S-methyl ester (BTH) could induce the expression of the HsfB1 gene in Arabidopsis. BTH is a synthetic mimic of salicylic acid and a potent inducer of priming and SAR in Arabidopsis (Lawton et al., 1996; Beckers et al., 2009).
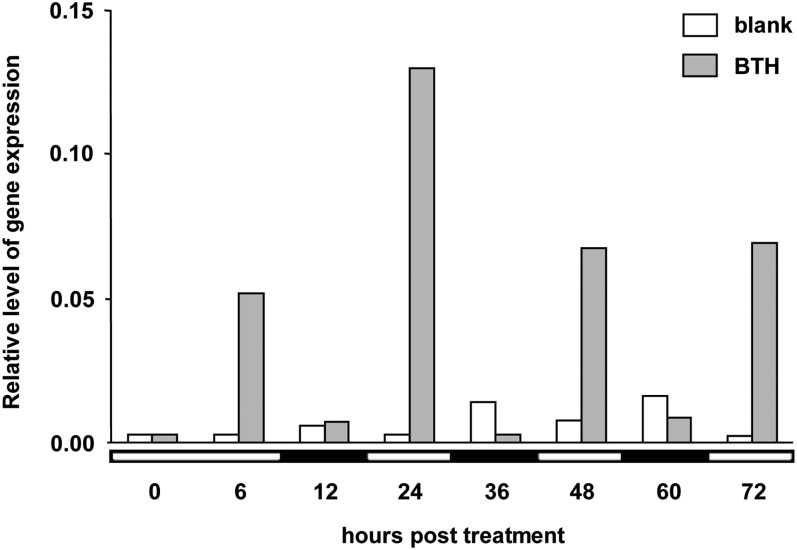
As shown in Figure 1, treatment of Arabidopsis plants with a formulation of BTH caused diurnal expression of the HsfB1 gene, with HsfB1 being activated in the light and dropping to basal expression levels in the dark. When plants were treated with a blank formulation devoid of BTH, little HsfB1 expression was observed in both the light and dark periods (Fig. 1). Together, these results supported our assumption that HsfB1 plays a role in BTH-induced defense priming of Arabidopsis and suggested that HsfB1’s role in BTH-induced defense priming depends on light. The latter conclusion is consistent with earlier findings demonstrating that SAR (Zeier et al., 2004; Griebel and Zeier, 2008), as well as the contribution of methyl salicylate to SAR (Liu et al., 2011), are light dependent.
Figure 1.
Diurnal activation of the HsfB1 gene by BTH. Five-week-old Arabidopsis (accession Columbia-0) plants were grown on soil at 8 h of light/16 h of dark, 20°C, and 60% to 70% relative humidity. Plants were sprayed at 10 am (that is 2 h after start of the light period) with a formulation of BTH (100 µm) or with a blank formulation devoid of BTH (blank). At various times after treatment, RNA was extracted from sprayed leaves and assayed for the accumulation of HsfB1 transcripts by quantitative reverse transcription (qRT)-PCR using gene-specific primers (Supplemental Table S1) as described by Beckers et al. (2009). Relative amounts were normalized to those of ACTIN2. Bars represent means of two replicate measurements (n = 2). White and black bars below the x axis indicate whether the harvest of leaves was done in the light or the dark. The experiment was repeated two times with similar results.
To elucidate whether HsfB1 indeed is required for the primed expression of defense-related genes, and to provide genetic evidence for this, we included the Arabidopsis hsfb1-1 (Ikeda et al., 2011) and hsfb1-3 (Supplemental Fig. S1) mutants in our analyses. We investigated whether hsfB1-1 and hsfB1-3 would or would not display primed expression of the defense-related genes encoding Phe ammonia lyase 1 (PAL1) and WRKY29. The PAL enzyme catalyzes the committed step in the phenylpropanoid pathway, with important roles in the overall plant defense response (Hahlbrock and Scheel, 1989), whereas WRKY29 encodes a transcription coactivator that is crucial for the regulation of plant defense genes (Eulgem and Somssich, 2007). These two loci were previously identified as reliable marker genes for the Arabidopsis stress response, and it was shown that wound stress-induced PAL1 and WRKY29 expression was more robust in primed and subsequently wounded plants than in wounded plants without previous priming (Kohler et al., 2002; Beckers et al., 2009; Jaskiewicz et al., 2011).
Wild-type plants (Columbia-0) and the hsfb1-1 and hsfb1-3 mutants were treated with BTH or the blank formulation for 3 d. Then, leaves were either left untreated or infiltrated with water. Water infiltration into leaves elicits a cell-collapse response and activates the expression of wound-responsive genes (Young et al., 1996; Kohler et al., 2002). Three hours after leaf infiltration, RNA was extracted from leaves and assayed for the accumulation of transcripts encoding PAL1 (Fig. 2A) or WRKY29 (Fig. 2B).
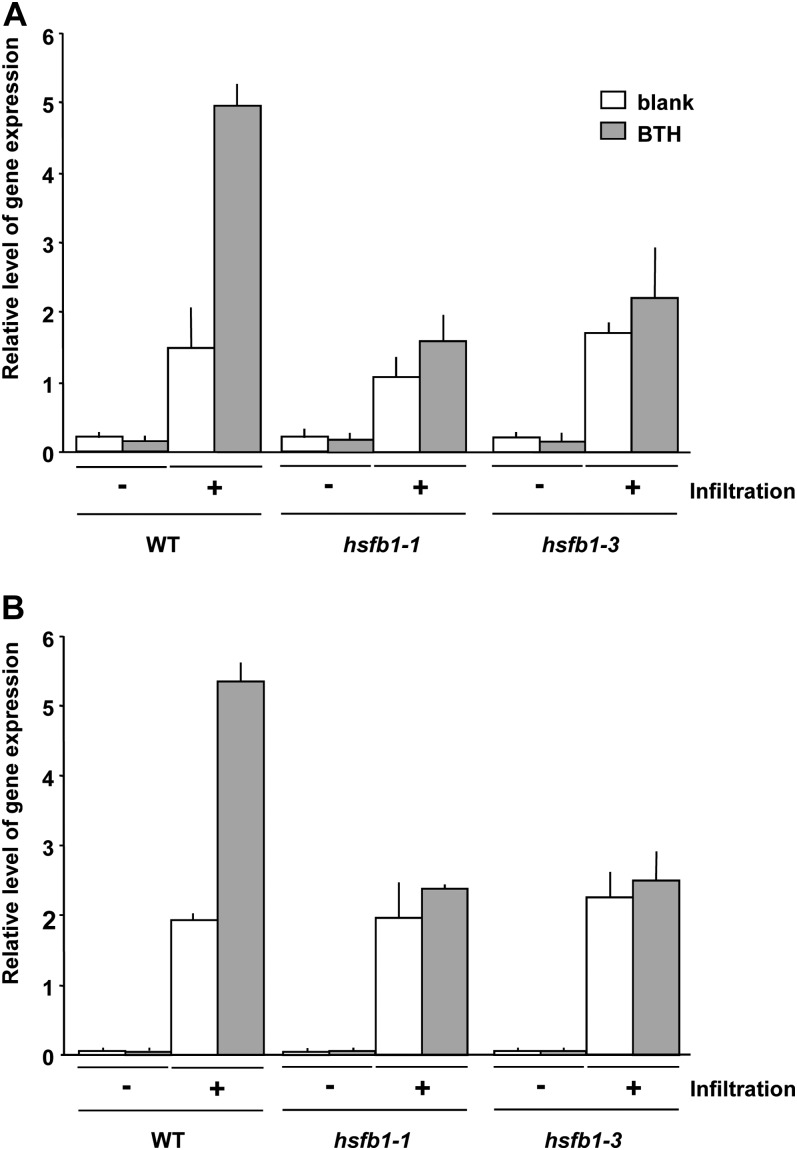
Figure 2.
Absence of primed defense gene activation in hsfb1 mutants. Five-week-old wild-type (WT), hsfb1-1, and hsfb1-3 plants were grown as described in Figure 1 and treated with 100 µm BTH or the blank formulation for 3 d. Then, leaves were either left untreated (−) or infiltrated with water (+). Three hours later, leaves were assayed for the accumulation of PAL1 (A) or WRKY29 (B) transcripts by qRT-PCR. Relative amounts of gene expression were normalized to those of ACTIN2. Error bars indicate sd (n = 3). The experiment was repeated three times with similar results.
In wild-type Arabidopsis, pretreatment with BTH primed the infiltration-activated expression of both PAL1 and WRKY29 almost 3-fold when compared with the response in plants treated with the blank formulation without BTH (Fig. 2). By contrast, pretreatment with BTH did not significantly prime the infiltration-induced PAL1 and WRKY29 expression in leaves of the hsfb1-1 and hsfb1-3 mutants (Fig. 2). However, the two mutants responded to the infiltration stimulus itself to a similar extent as wild-type plants (Fig. 2). Thus, the perception of and responsiveness to water infiltration per se seem not to be affected in hsfb1-1 and hsfb1-3, but HsfB1 is required for priming.
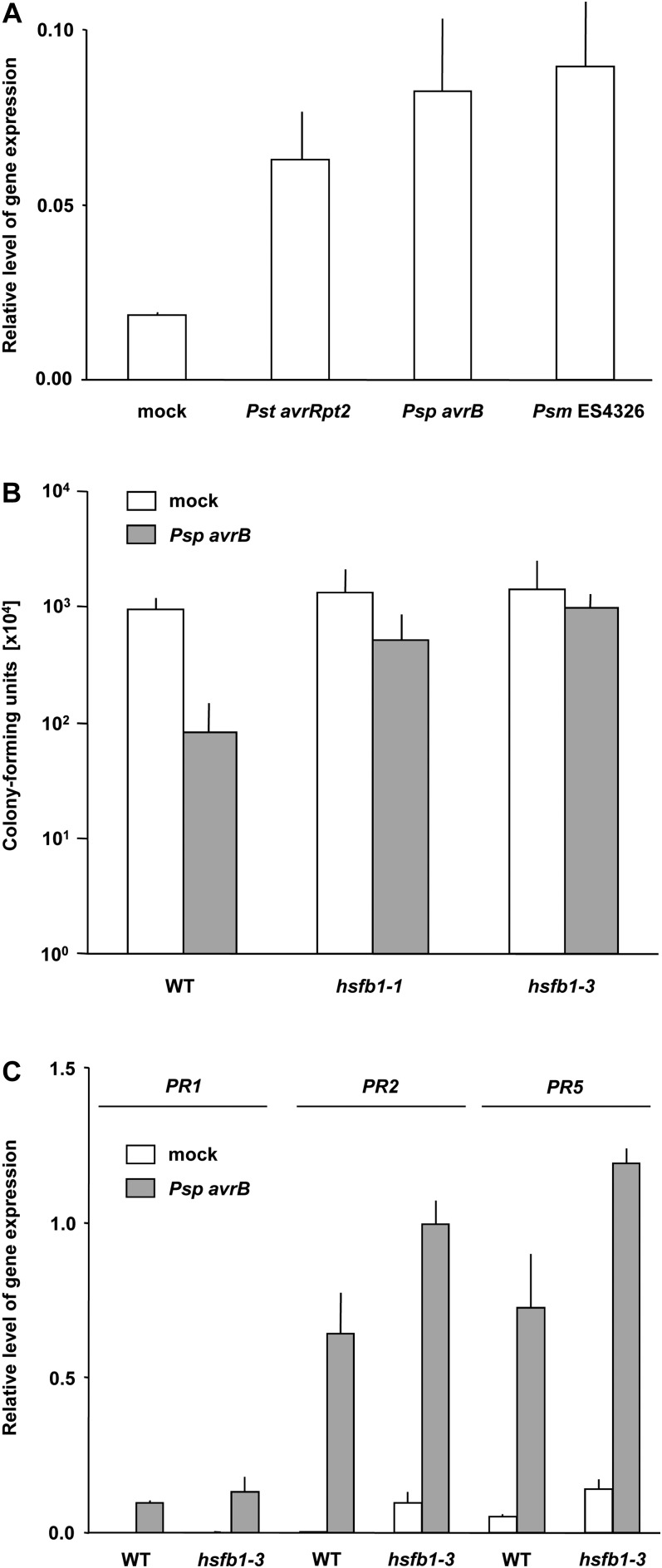
In addition to abiotic stress tolerance, primed defense gene activation has been associated with SAR to pathogen attack (Conrath et al., 2006; Jung et al., 2009; Conrath, 2011). Furthermore, HsfB1 was recently found to be important for salicylic acid-induced disease resistance in Arabidopsis (Pajerowska-Mukhtar et al., 2012). Therefore, we investigated whether HsfB1, in addition to conferring acquired thermotolerance (Ikeda et al., 2011), chemically activated resistance (Pajerowska-Mukhtar et al., 2012), and primed defense gene expression (Fig. 2), might also be pivotal for pathogen-induced, bona fide SAR. Arabidopsis plants were inoculated on three lower leaves with SAR-inducing avirulent Pseudomonas syringae pv tomato (Pst) DC3000 harboring the avirulence gene avrRpt2 (Pst avrRpt2), Pseudomonas syringae pv phaseolicola (Psp) carrying avrB (Psp avrB), or Pseudomonas syringae pv maculicola (Psm) strain ES4326 (Psm ES4326). Pst avrRpt2 and Psp avrB are avirulent to the Columbia-0 accession of Arabidopsis and trigger a hypersensitive response (Whalen et al., 1991; Gopalan et al., 1996), whereas Psm ES4326 is virulent to Columbia-0 plants (Dong et al., 1991). Three days after primary inoculation with any of these three bacterial strains, systemic leaves were harvested and assayed for the accumulation of HsfB1 transcripts. As depicted in Figure 3A, the three SAR-inducing bacterial strains activate the expression of HsfB1 in systemic leaves of locally infected Arabidopsis plants. This finding suggested that HsfB1 indeed could be an important component of bacteria-induced SAR in Arabidopsis.
Figure 3.
Bacteria-induced gene expression and bacterial multiplication in systemic leaves of primary inoculated plants. A, Five-week-old Arabidopsis wild-type plants were pressure infiltrated on four lower leaves with suspensions of Pst avrRpt2 (optical density at 600 nm [OD600] = 0.002), Psm ES4326 (OD600 = 0.002), or Psp avrB (OD600 = 0.002) in 10 mm MgCl2. Control plants were infiltrated with 10 mm MgCl2 in the absence of bacteria (mock). After 3 d, systemic leaves were assayed for the presence of HsfB1 transcripts by qRT-PCR. B, Five-week-old wild-type (WT), hsfb1-1, and hsfb1-3 plants were inoculated on four lower leaves with Psp avrB (OD600 = 0.002). Three days later, systemic leaves were challenge inoculated with Psm ES4326 (OD600 = 0.0002). After another 3 d, Psm ES4326 bacteria were isolated from a homogenized 0.5-cm disc from infected systemic leaves, spread on King’s B agar plates containing 100 µg mL−1 streptomycin as a selection marker, incubated at 28°C for 2 d, and analyzed for the number of developing colonies. C, Five-week-old wild-type and hsfb1-3 plants were infiltrated on four lower leaves with Psp avrB (OD600 = 0.002). Three days later, three systemic leaves were harvested from each plant and assayed for the presence of transcripts for PR1, PR2, and PR5 using gene-specific primers (Supplemental Table S1). In all experiments, plants were grown as described in Figure 1. In A and C, relative amounts were normalized to those of ACTIN2. Error bars indicate sd (n = 3). The experiments were repeated three times with similar results.
To test whether this is the case, wild-type, hsfb1-1, and hsfb1-3 plants were inoculated on three lower leaves with Psp avrB. After 3 d, systemic leaves were challenge inoculated with Psm ES4326. After another 3 d, Psm ES4326-infected systemic leaves were assayed for bacterial multiplication. As shown in Figure 3B, the primary inoculation of wild-type plants with Psp avrB reduced the number of Psm ES4326 bacteria reisolated from systemic leaves about 10-fold. This finding verified the presence of SAR in Psp avrB-inoculated wild-type plants. In contrast, the reduction in bacterial counts reisolated from systemic leaves of the primary inoculated hsfb1-1 and hsfb1-3 mutants was much lower and did not show significant differences from bacterial multiplication in mock-inoculated wild-type or hsfb1 mutant plants (Fig. 3B). These findings demonstrated that HsfB1 is a critical component in Psp avrB-induced SAR to Psm ES4326 in Arabidopsis. Because localized infection with Psp avrB did not significantly affect (PR1) or even enhance (PR2 and PR5) the activation of the SAR marker genes PR1, PR2, and PR5 in systemic leaves of the hsfb1-3 mutant (Fig. 3C), HsfB1 seems to be associated with the priming and not simply the activation of defense genes during SAR. This result also indicates that systemic signaling is not affected in hsfb1-3. Also, as wild-type and hsfB1-3 plants in the absence of previous SAR activation did not show differences in supporting bacterial growth after primary infection with virulent Psm ES4326 or avirulent Psp avrB (Supplemental Fig. S2), the perception of and response to these two bacterial strains per se seem not to be affected in hsfb1-3.
CONCLUSION
It was shown recently that HsfB1 is a crucial component of acquired thermotolerance and salicylic acid-mediated resistance in Arabidopsis. We complemented these findings by demonstrating that HsfB1 plays a pivotal role also in primed defense gene activation and pathogen-induced SAR in this plant. Together, these data disclose HsfB1 as a common key player in acquired tolerance to both biotic and abiotic stresses. These findings also provide new knowledge on the molecular mechanism of defense priming in plants.
The Arabidopsis Information Resource stock names for hsfb1-1 and hsfb1-3 are SALK_104713 and SALK_106223.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Analysis of T-DNA-tagged hsfB1-1 and hsfB1-3 lines.
Supplemental Figure S2. Multiplication of Psm ES4326 and Psp avrB in the wild type and the hsfB1-3 mutant.
Supplemental Table S1. List of gene-specific forward and reverse primers used for real-time qRT-PCR analysis.
Supplementary Material
Glossary
- Hsf
heat shock factor
- SAR
systemic acquired resistance
- BTH
benzo(1,2,3)thiadiazole-7-carbothioic acid S-methyl ester
- Pst
Pseudomonas syringae pv tomato
- Psp
Pseudomonas syringae pv phaseolicola
- Psm
Pseudomonas syringae pv maculicola
- qRT
quantitative reverse transcription
- OD600
optical density at 600 nm
References
- Beckers GJM, Jaskiewicz M, Liu Y, Underwood WR, He SY, Zhang S, Conrath U. (2009) Mitogen-activated protein kinases 3 and 6 are required for full priming of stress responses in Arabidopsis thaliana. Plant Cell 21: 944–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharti K, Von Koskull-Döring P, Bharti S, Kumar P, Tintschl-Körbitzer A, Treuter E, Nover L. (2004) Tomato heat stress transcription factor HsfB1 represents a novel type of general transcription coactivator with a histone-like motif interacting with the plant CREB binding protein ortholog HAC1. Plant Cell 16: 1521–1535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrath U. (2011) Molecular aspects of defence priming. Trends Plant Sci 16: 524–531 [DOI] [PubMed] [Google Scholar]
- Conrath U, Beckers GJ, Flors V, García-Agustín P, Jakab G, Mauch F, Newman MA, Pieterse CM, Poinssot B, Pozo MJ, et al. , Prime-A-Plant Group (2006) Priming: getting ready for battle. Mol Plant Microbe Interact 19: 1062–1071 [DOI] [PubMed] [Google Scholar]
- Dong X, Mindrinos M, Davis KR, Ausubel FM. (1991) Induction of Arabidopsis defense genes by virulent and avirulent Pseudomonas syringae strains and by a cloned avirulence gene. Plant Cell 3: 61–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulgem T, Somssich IE. (2007) Networks of WRKY transcription factors in defense signaling. Curr Opin Plant Biol 10: 366–371 [DOI] [PubMed] [Google Scholar]
- Gopalan S, Bauer DW, Alfano JR, Loniello AO, He SY, Collmer A. (1996) Expression of the Pseudomonas syringae avirulence protein AvrB in plant cells alleviates its dependence on the hypersensitive response and pathogenicity (Hrp) secretion system in eliciting genotype-specific hypersensitive cell death. Plant Cell 8: 1095–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griebel T, Zeier J. (2008) Light regulation and daytime dependency of inducible plant defenses in Arabidopsis: phytochrome signaling controls systemic acquired resistance rather than local defense. Plant Physiol 147: 790–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahlbrock K, Scheel D. (1989) Physiology and molecular biology of phenylpropanoid metabolism. Annu Rev Plant Physiol Plant Mol Biol 40: 347–369 [Google Scholar]
- Hammerschmidt R (2009) Systemic acquired resistance. In LC van Loon, ed, Advances in Botanical Research, Vol 51, Plant Innate Immunity, Ed 1. Academic Press, London, pp 174–222
- Ikeda M, Mitsuda N, Ohme-Takagi M. (2011) Arabidopsis HsfB1 and HsfB2b act as repressors of the expression of heat-inducible Hsfs but positively regulate the acquired thermotolerance. Plant Physiol 157: 1243–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaskiewicz M, Conrath U, Peterhänsel C. (2011) Chromatin modification acts as a memory for systemic acquired resistance in the plant stress response. EMBO Rep 12: 50–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung HW, Tschaplinski TJ, Wang L, Glazebrook J, Greenberg JT. (2009) Priming in systemic plant immunity. Science 324: 89–91 [DOI] [PubMed] [Google Scholar]
- Kohler A, Schwindling S, Conrath U. (2002) Benzothiadiazole-induced priming for potentiated responses to pathogen infection, wounding, and infiltration of water into leaves requires the NPR1/NIM1 gene in Arabidopsis. Plant Physiol 128: 1046–1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M, Busch W, Birke H, Kemmerling B, Nürnberger T, Schöffl F. (2009) Heat shock factors HsfB1 and HsfB2b are involved in the regulation of Pdf1.2 expression and pathogen resistance in Arabidopsis. Mol Plant 2: 152–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawton KA, Friedrich L, Hunt M, Weymann K, Delaney T, Kessmann H, Staub T, Ryals J. (1996) Benzothiadiazole induces disease resistance in Arabidopsis by activation of the systemic acquired resistance signal transduction pathway. Plant J 10: 71–82 [DOI] [PubMed] [Google Scholar]
- Liu P-P, von Dahl CC, Klessig DF. (2011) The extent to which methyl salicylate is required for signaling systemic acquired resistance is dependent on exposure to light after infection. Plant Physiol 157: 2216–2226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nover L, Bharti K, Döring P, Mishra SK, Ganguli A, Scharf KD. (2001) Arabidopsis and the heat stress transcription factor world: how many heat stress transcription factors do we need? Cell Stress Chaperones 6: 177–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajerowska-Mukhtar KM, Wang W, Tada Y, Oka N, Tucker CL, Fonseca JP, Dong X. (2012) The HSF-like transcription factor TBF1 is a major molecular switch for plant growth-to-defense transition. Curr Biol 22: 103–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelham HRB. (1982) A regulatory upstream promoter element in the Drosophila hsp 70 heat-shock gene. Cell 30: 517–528 [DOI] [PubMed] [Google Scholar]
- von Koskull-Döring P, Scharf KD, Nover L. (2007) The diversity of plant heat stress transcription factors. Trends Plant Sci 12: 452–457 [DOI] [PubMed] [Google Scholar]
- Whalen MC, Innes RW, Bent AF, Staskawicz BJ. (1991) Identification of Pseudomonas syringae pathogens of Arabidopsis and a bacterial locus determining avirulence on both Arabidopsis and soybean. Plant Cell 3: 49–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young SA, Wang X, Leach JE. (1996) Changes in the plasma membrane distribution of rice phospholipase D during resistant interactions with Xanthomonas oryzae pv oryzae. Plant Cell 8: 1079–1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeier J, Pink B, Mueller MJ, Berger S. (2004) Light conditions influence specific defence responses in incompatible plant-pathogen interactions: uncoupling systemic resistance from salicylic acid and PR-1 accumulation. Planta 219: 673–683 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.