Abstract
Plant oils are stored in oleosomes or oil bodies, which are surrounded by a monolayer of phospholipids embedded with oleosin proteins that stabilize the structure. Recently, a structural protein, Oleosin3 (OLE3), was shown to exhibit both monoacylglycerol acyltransferase and phospholipase A2 activities. The regulation of these distinct dual activities in a single protein is unclear. Here, we report that a serine/threonine/tyrosine protein kinase phosphorylates oleosin. Using bimolecular fluorescence complementation analysis, we demonstrate that this kinase interacts with OLE3 and that the fluorescence was associated with chloroplasts. Oleosin-green fluorescent protein fusion protein was exclusively associated with the chloroplasts. Phosphorylated OLE3 exhibited reduced monoacylglycerol acyltransferase and increased phospholipase A2 activities. Moreover, phosphatidylcholine and diacylglycerol activated oleosin phosphorylation, whereas lysophosphatidylcholine, oleic acid, and Ca2+ inhibited phosphorylation. In addition, recombinant peanut (Arachis hypogaea) kinase was determined to predominantly phosphorylate serine residues, specifically serine-18 in OLE3. Phosphorylation levels of OLE3 during seed germination were determined to be higher than in developing peanut seeds. These findings provide direct evidence for the in vivo substrate selectivity of the dual-specificity kinase and demonstrate that the bifunctional activities of oleosin are regulated by phosphorylation.
Triacylglycerols (TAGs) are stored in a specialized intracellular organelle-like structure called oleosomes or oil bodies in plants that are surrounded by a monolayer of phospholipids containing embedded proteins that stabilize their structures (Huang, 1992; Napier et al., 1996; Murphy, 2011). A predominant protein in the oleosome is oleosin. The central long hydrophobic core of oleosin has a unique Pro knot (PX5SPX3P) that is conserved across various species (Abell et al., 2004; Huang et al., 2009). Oleosins are postulated to stabilize the oil body by preventing coalescence, particularly during seed desiccation. In addition, these proteins act as binding sites for lipases during the mobilization of stored TAGs during seed germination (Beisson et al., 2001; Murphy, 2011). Major oleosin protein suppression in Arabidopsis (Arabidopsis thaliana; Siloto et al., 2006) and soybean (Glycine max; Schmidt and Herman, 2008) resulted in an aberrant phenotype of embryo cells containing abnormally large oil bodies and delayed germination. Reintroducing recombinant oleosin was observed to reverse the aberrant phenotype. Gene coexpression network analysis of the transcriptome of developing Arabidopsis seeds indicated that the expression profiles of diacylglycerol acyltransferase and oleosin are similar, suggesting the involvement of these genes in oil accumulation (Peng and Weselake, 2011). Recently, we have identified a microsomal membrane-bound monoacylglycerol acyltransferase (MGAT) from immature peanut (Arachis hypogaea) seeds. The MGAT was solubilized from the microsomal membranes using a combination of a chaotropic agent and a zwitterionic detergent, and a functionally active 14S multiprotein complex was isolated and characterized. Oleosin3 (OLE3) was identified as part of the multiprotein complex, which is capable of performing bifunctional activities such as acylating monoacylglycerol (MAG) to diacylglycerol (DAG) and phospholipase A2 (PLA2; Parthibane et al., 2012). The regulation of dual activities in a single protein is unclear.
The reversible phosphorylation of Ser, Thr, and Tyr residues on target proteins plays a key role in the regulation of metabolic activity and modulates the dynamics of plant growth and development. Approximately 20 different mitogen-activated protein kinases have been identified. These kinases are likely to be involved in growth, development, and various stress responses (Zhang and Klessig, 2001). Dual-specificity kinases, such as the Arabidopsis thaliana gene ATN1, Arabidopsis protein kinase1 (APK1), and Arabidopsis dual specificity kinase1(ADK1) from Arabidopsis, phosphorylate both Ser/Thr and Tyr residues, which are involved in the regulation of cellular and metabolic events (Hirayama and Oka, 1992; Ali et al., 1994; Tregear et al., 1996). In soybean, the phytohormones auxin and ethylene synergistically regulate the Ser/Thr/Tyr protein kinases (STYKs) involved in leaf senescence (Xu et al., 2011). A non-mitogen-activated protein kinase cascade dual-specificity kinase or STYK involved in cold, salt stress and seed development was identified in peanut, in which autophosphorylation regulates kinase activity (Rudrabhatla and Rajasekharan, 2002, 2003). We have previously reported the isolation and characterization of an STYK from Arabidopsis (Rudrabhatla et al., 2006; Reddy and Rajasekharan, 2007). However, the substrate of this dual-specificity kinase has not been identified. Most dual-specificity kinases have been identified and biochemically characterized with nonplant proteins, such as histones and myelin basic protein. However, there is only one report of a plant protein substrate for a dual-specificity kinase that phosphorylates the preprotein involved in chloroplast differentiation (Lamberti et al., 2011).
Oleosin was shown to have both MGAT and PLA2 activities (Parthibane et al., 2012), and the regulation of enzyme activities was investigated in this study. Here, we report that OLE3 is phosphorylated by peanut STYK (AhSTYK). We have also gained more insight on the possible role of OLE3 phosphorylation in the regulation of MGAT and PLA2 activities. This regulation could be involved in the biosynthesis and mobilization of TAGs during seed maturation and germination.
RESULTS
Identification of a Kinase That Phosphorylates OLE3
OLE3 is bifunctional, exhibiting both MGAT and PLA2 activities. Notably, any posttranslational modification could modulate these activities. Phosphorylation sites in OLE3 were predicted using the Net phos 2.0 program. Initially, an immuno-pull-down assay was performed with 14S multiprotein complex proteins from immature peanut microsomal membranes, which were precleaned with protein A-agarose and immunoprecipitated with anti-STYK antibodies. The immunoprecipitate was diluted and used as a substrate for AhSTYKs. Immunoblotting with OLE3-specific polyclonal antibodies verified that the immuno-pull-down protein was OLE3.
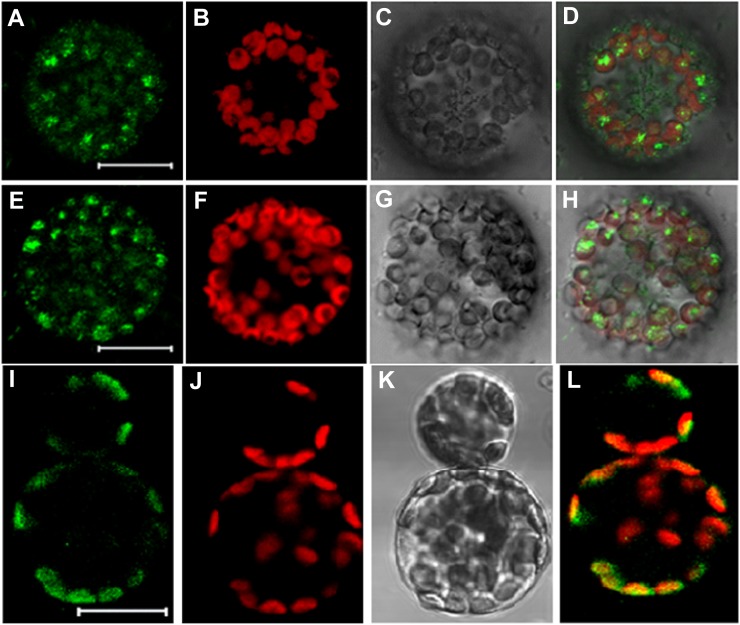
To detect in vivo protein-protein interactions between OLE3 and STYK, a bimolecular fluorescence complementation (BiFC) assay (Citovsky et al., 2006, 2008) was used. The coexpression of Arabidopsis STYKs (AtSTYK) fused to the N terminus of enhanced yellow fluorescent protein (EYFP) and peanut OLE3 fused to the C terminus of EYFP in Arabidopsis protoplasts resulted in an interaction that led to EYFP fluorescence. The pattern of EYFP fluorescence from AtSTYK-nEYFP and AhOLE3-cEYFP interactions resembled the fluorescence generated from the interactions of these proteins (Fig. 1, A–H). Fluorescence was never observed outside chloroplasts in the protein-protein interaction experiments. Analysis of the OLE3 protein sequence using WoLF, PSORT, and ChloroP softwares predicted the presence of a chloroplast-targeting signal sequence, which could explain the EYFP fluorescence observed in the chloroplasts. To assess this prediction, we performed a localization of OLE3, and the cDNA of OLE3 tagged at the C terminus with GFP and the localization of the fusion proteins were analyzed by transient expression in Arabidopsis leaf protoplasts. OLE3 was localized as a discrete structure that was colocalized with the red autofluorescence of chlorophyll. This shows a clear localization of OLE3 to chloroplasts (Fig. 1, I–L). The localization is consistently observed using confocal microscopy. This protein-protein interaction suggested that STYK interacts with OLE3.
Figure 1.
Interaction of OLE3 with STYK and localization of OLE3. A to H, BiFC detection of AtSTYK and AhOLE3 protein-protein interactions in Arabidopsis leaf protoplasts. The leaf protoplast was transformed with pSAT4-nEYFP-N1-AtSTYK and pSAT6-cEYFP-N1-AhOLE3 and transient expression of the YFP fusion proteins and was observed 16 to 20 h after transformation. A and E represent reconstructed YFP fluorescence resulting from AtSTYK and AhOLE3 interactions detected in the green channel. B and F show the autofluorescence of chlorophyll detected in the red channel. C and G show bright-field images. D and H are overlaid images of reconstructed YFP and autofluorescence of chlorophyll. I to L, Subcellular localization of OLE3-GFP in Arabidopsis leaf protoplasts. I shows the GFP fluorescence resulting from AhOLE3-GFP detected in the green channel. J shows the autofluorescence of chlorophyll detected in the red channel. K shows a bright-field image. L is an overlaid image of GFP and autofluorescence of chlorophyll. Bars = 20 μm.
Characterization of OLE3 Phosphorylation by AhSTYK
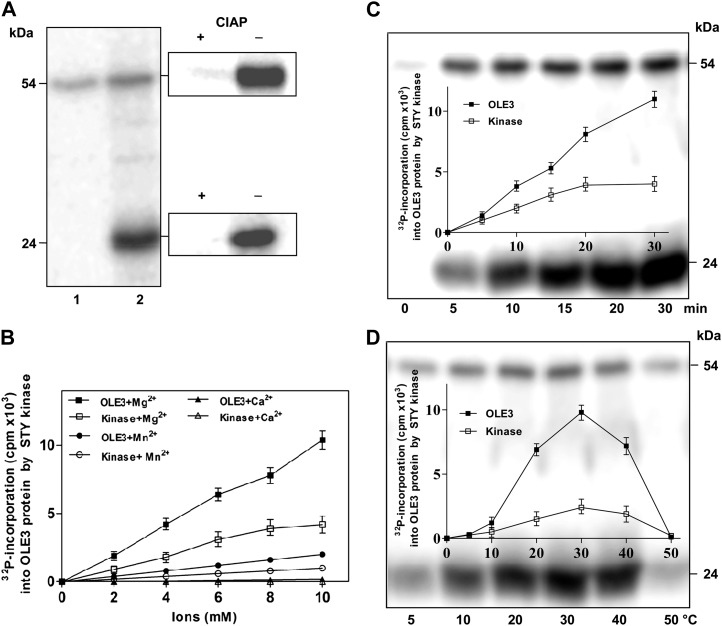
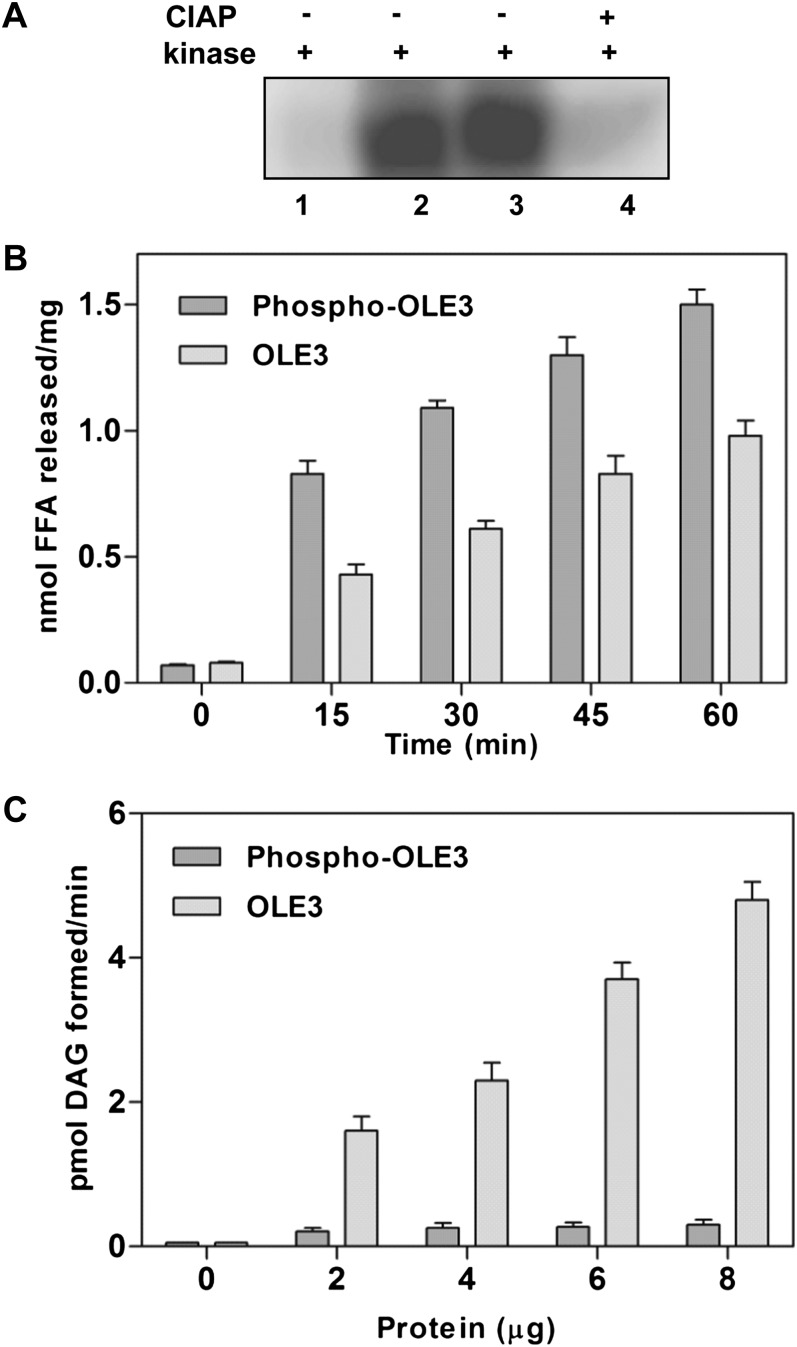
After the expression and purification of recombinant full-length OLE3, the protein was determined to be phosphorylated by recombinant peanut STYK and dephosphorylated by calf intestinal alkaline phosphatase treatment (Fig. 2A). Furthermore, the effects of various metal ions on OLE3 phosphorylation and STYK autophosphorylation were determined. Increasing concentrations of Mg2+ ions were observed to increase both phosphorylations linearly. The amount of phosphate incorporated into oleosin was approximately 2-fold higher (6.4 mol phosphate mol−1 oleosin) than that of STYK in the presence of the Mg2+ ions. However, lower rates of OLE3 phosphorylation were measured in the presence of Ca2+ and Mn2+ ions (Fig. 2B). In addition, the time-dependent phosphorylation of oleosin by AhSTYK was measured, and linear kinetics of phosphorylation was observed. However, AhSTYK autophosphorylation was saturated at all time points studied (Fig. 2C). In the presence of increasing amounts of substrate (OLE3) and a fixed amount of AhSTYK, an increase in the phosphorylation of substrate and autophosphorylation was observed. The optimum temperature of the phosphorylation of oleosin by AhSTYK was 30°C, and phosphorylation was observed even at low temperatures (Fig. 2D). Maximum activity was observed at pH 7.5, and relatively less activity was observed at mild alkaline and acidic pH ranges. These data suggest that the peanut STYK phosphorylates oleosin.
Figure 2.
OLE3 phosphorylation by AhSTYK. A, In vitro phosphorylation of OLE3 by AhSTYK. Lane 1, AhSTYK with vector lysate; lane 2, OLE3 with AhSTYK. Panels at right show calf intestinal alkaline phosphatase (CIAP)-treated AhSTYK and OLE3. When OLE3 was incubated with [γ-32P]ATP, there was no phosphorylation observed. B, Effect of metal ions (Mg2+, Mn2+, and Ca2+) on AhSTYK phosphorylation of OLE3. Values are means ± sd for three independent determinations. C and D, Time-dependent (C) and temperature-dependent (D) AhSTYK phosphorylation of OLE3. Values represent means ± sd of three independent experiments. The kinase assay was performed, and the resulting phospho-proteins were resolved on a 12% (w/v) SDS-polyacrylamide gel. The proteins on the gel were then electroblotted onto a polyvinylidene difluoride membrane and visualized by autoradiography.
The Effect of Lipids on OLE3 Phosphorylation by Peanut STYK
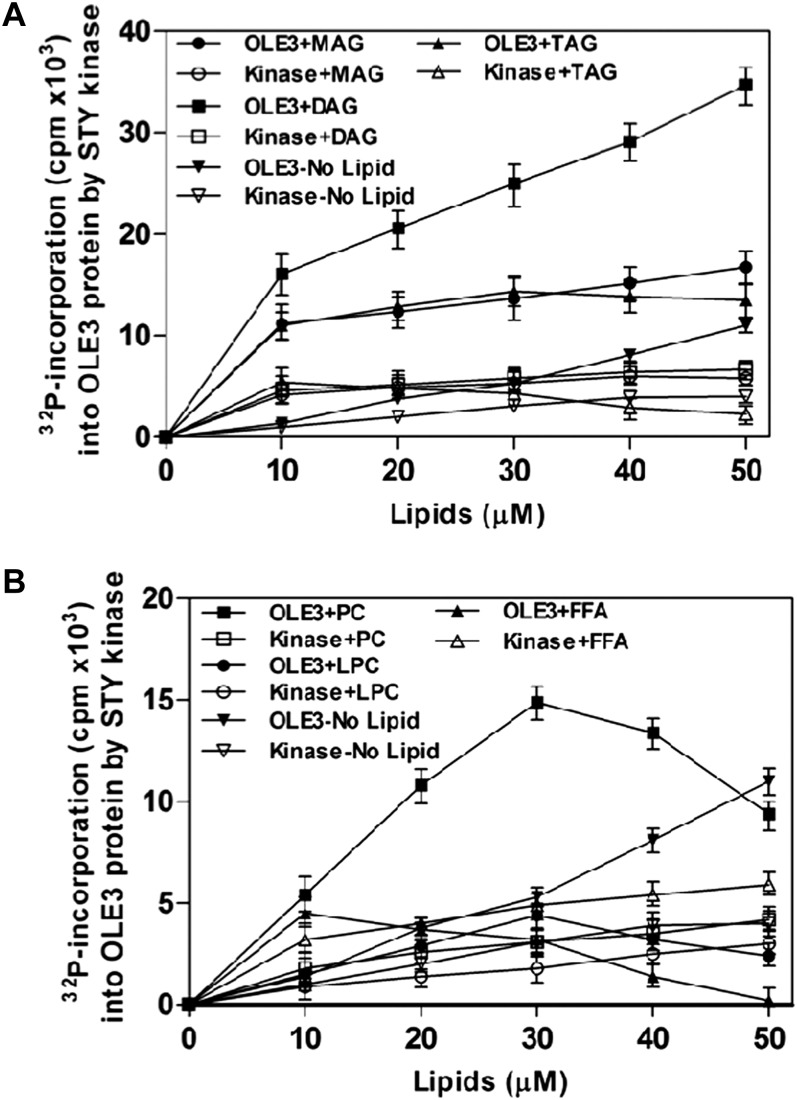
Because OLE3 was shown to metabolize lipids, OLE3 phosphorylation by AhSTYK was characterized in the presence of various lipids. In the presence of MAG, DAG, and TAG, OLE3 phosphorylation was observed to increase compared with that of the 5 mm CHAPS control. Among the neutral lipids, DAG showed a 2.5-fold increase in OLE3 phosphorylation (Fig. 3A). However, there was no change in AhSTYK autophosphorylation in the presence of these lipids. Inhibition of OLE3 phosphorylation (2.2- and 3.2-fold) was observed in the presence of lysophosphatidylcholine (LPC) and oleic acid, respectively. Nonetheless, phosphatidylcholine (PC) activated OLE3 phosphorylation (1.5-fold) and autophosphorylation was not altered by PC or oleic acid. However, LPC was observed to decrease autophosphorylation considerably (Fig. 3B).
Figure 3.
The effect of lipids on the phosphorylation of OLE3 by AhSTYK. A, The effects of MAG, DAG, and TAG on the phosphorylation of OLE3 by AhSTYK. B, The effects of PC, LPC, and free fatty acid (FFA) oleic acid on AhSTYK phosphorylation of OLE3. White symbols represent autophosphorylation in the presence of OLE3, and black symbols represents substrate phosphorylation of OLE3 in the presence of lipid. Values are means ± sd for three independent determinations, and each experiment was performed in duplicate.
Identification of Phospho-Amino Acid Residues of OLE3
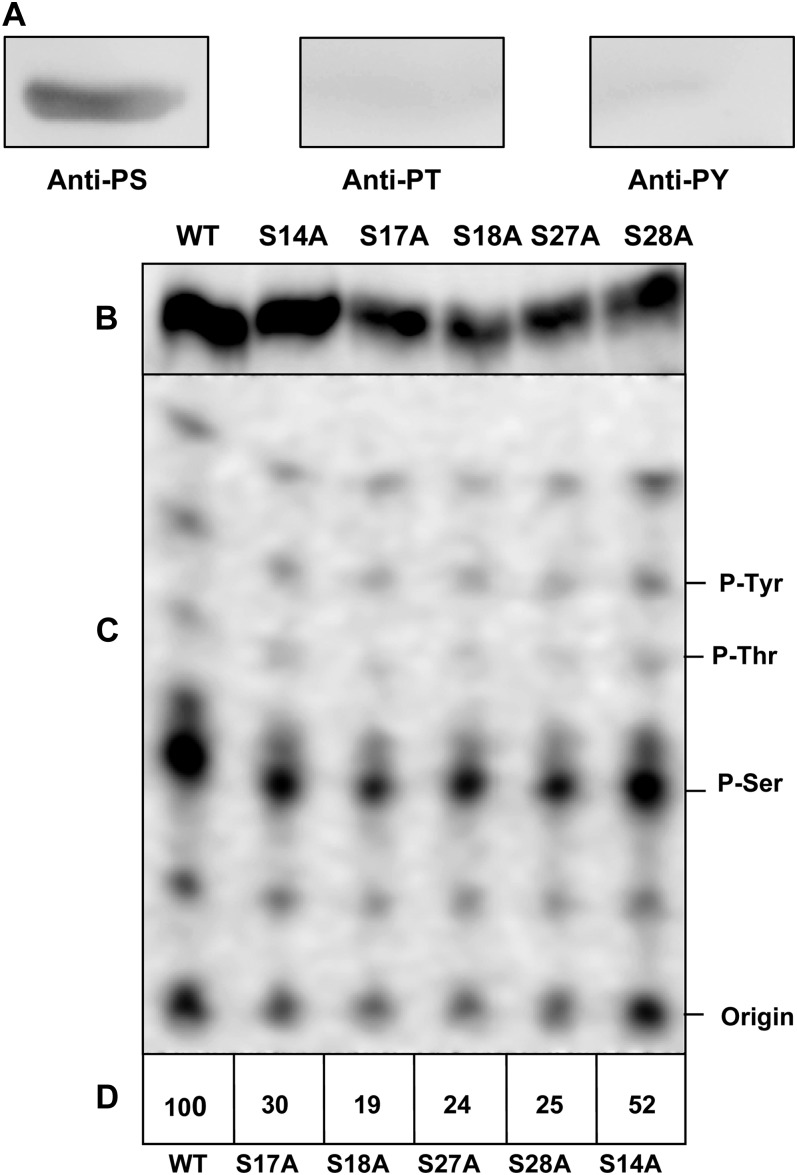
To determine the specificity of OLE3 phosphorylation, phosphorylated OLE3 was subjected to immunoblot analysis with phospho-Ser, phospho-Thr, and phospho-Tyr monoclonal antibodies. Only the phospho-Ser antibody was observed to bind OLE3, suggesting that STYK targets the Ser residues of OLE3 (Fig. 4A). Using the Net phos 2.0 program, we predicted the following Ser residues to be phosphorylated: Ser-14, -17, -18, -27, and -28. These Ser residues were mutated to Ala by site-directed mutagenesis. OLE3 and mutants were then phosphorylated by AhSTYK (Fig. 4B). Repeated phospho-amino acid analysis of the resulting oleosin indicated that predominantly Ser residues were phosphorylated. However, phospho-Thr and phospho-Tyr were not detected (Fig. 4C). Among the mutants, S18A exhibited a significant (81%) decrease in the incorporation of the 32P label in the Ser residues of OLE3 (Fig. 4D). These results suggest that Ser-18 could possibly play a key role in regulating the bifunctional activities of oleosin.
Figure 4.
Site-directed mutagenesis and phospho-amino acid analysis of phospho-OLE3. A, Phospho-OLE3 was electrophoretically transferred onto a nitrocellulose membrane and cross-reacted with anti-phospho-Ser, anti-phospho-Thr, and anti-phospho-Tyr monoclonal antibodies. B, Recombinant OLE3 and mutants (1 µg) were phosphorylated by AhSTYK, and the resulting phosphorylation products were visualized by autoradiography. C, Phosphorylated OLE3 and mutants were hydrolyzed, and the resulting phospho-amino acids were separated by silica-TLC. D, Relative incorporation of the 32P radiolabel into Ser residues of OLE3 and mutants. WT, Wild type.
Regulation of MGAT and PLA2 Activities by Phosphorylation
Microsomal membranes from yeast cells overexpressing OLE3 were incubated with purified recombinant AhSTYK, and phosphorylation was confirmed by 32P incorporation into the target protein (Fig. 5A). After phosphorylation, the time-dependent hydrolysis of PC was measured, and a linear increase in the liberation of fatty acid was observed (Fig. 5B). PLA2 activity with phospho-OLE3 was observed to be significantly higher (1.7-fold) than with OLE3 (unphosphorylated), which served as a control for PLA2 activity. A similar observation was also made with the bacterially expressed recombinant OLE3 (data not shown). In vitro phosphorylated oleosin was also assayed for MGAT activity under standard assay conditions. Phospho-OLE3 exhibited low MGAT activity compared with that of unphosphorylated OLE3 (Fig. 5C). These data suggest that PLA2 and MGAT activities associated with OLE3 are modulated by phosphorylation.
Figure 5.
Phosphorylation regulates the bifunctional activity of OLE3. A, Recombinant OLE3 expressed in S. cerevisiae was phosphorylated by the purified recombinant AhSTYK. Lane 1, vector control with 1.0 µg of AhSTYK; lane 2, OLE3 overexpressed (10 µg of microsomal protein) and phosphorylated by AhSTYK; lane 3, OLE3 overexpressed (20 µg of microsomal protein) and phosphorylated by AhSTYK; lane 4, OLE3 overexpressed and phosphorylated by AhSTYK followed by calf intestinal alkaline phosphatase (CIAP) treatment. Proteins were resolved on a 12% (w/v) SDS-polyacrylamide gel. The proteins were electroblotted onto a polyvinylidene difluoride membrane and visualized by autoradiography. B, The PLA2 assay was performed in a reaction mixture consisting of 100 μm sonicated vesicles of [1,2-palmitoyl-9,10-3H]PC and 20 μg of enzyme in a final volume of 100 μL. Reactions with OLE3 (yeast microsomal membranes without the kinase treatment) and phospho-OLE3 (yeast microsomal membranes with kinase treatment) were performed at 30°C over various time periods. FFA, Free fatty acid. C, MGAT activity was measured by incubating 50 µm MAG and 10 µm [14C]oleoyl-CoA with increasing amounts of OLE3 microsomes with or without kinase treatment under standard conditions. Values are means ± sd of three independent determinations, and each experiment was performed in duplicate.
Possible Role of OLE3 during Seed Maturation and Germination
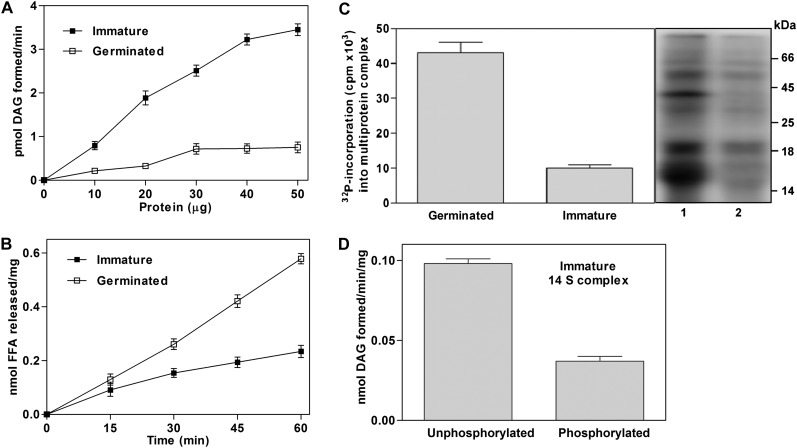
Multiprotein complexes from immature and germinated peanut seed microsomes were purified, and MGAT activity was measured under standard assay conditions. The protein-dependent accumulation of DAG was observed in immature peanut seeds but not in germinated seeds (Fig. 6A). In vitro activity of MGAT from immature multiprotein complexes was measured to be 90 pmol min−1 mg−1, which is 5.6-fold higher than that of germinated seeds. Time-dependent hydrolysis of PC was also observed. In contrast to MGAT activity, the magnitude of hydrolysis was higher (2.3-fold) in germinating seeds than in immature developing seeds (Fig. 6B). Seed slices of immature and germinated seeds were incubated with [32P]orthophosphate followed by the purification of the multiprotein complex. The extent of phosphate incorporation into multiprotein complexes from germinated peanut was 4.3-fold higher than in immature seeds (Fig. 6C). The purified multiprotein complex from immature seeds was phosphorylated with STYK, and the MGAT assay was performed. The activity was 2.6-fold lower than that of the unphosphorylated protein complex (Fig. 6D). These results suggest that oleosin could potentially act as an acyltransferase during seed maturation and hydrolyze phospholipids during seed germination.
Figure 6.
The physiological roles of OLE3 during seed maturation and germination. A, Graphical representation of protein-dependent MGAT activities in purified multiprotein complexes from peanut immature and germinated seeds. Enzyme activity data were derived from at least three independent experiments and are shown as means ± sd. B, Time-dependent hydrolysis of [3H]PC by purified multiprotein complexes from immature and germinated peanut. FFA, Free fatty acid. C, Autoradiography image of 32P incorporation into purified multiprotein complexes from immature and germinated peanut. Lane 1, complex from germinated seeds; lane 2, protein complex from immature seeds. D, MGAT assays of phosphorylated and unphosphorylated purified multiprotein complexes from immature peanut. The values represent at least three independent experiments and are shown as means ± sd.
DISCUSSION
STYK (or dual-specificity kinase) has been characterized, but its plant protein substrate remains to be identified (Rudrabhatla and Rajasekharan, 2002). Many plant dual-specificity kinases have been reported, and their in vivo substrates have not yet been identified (Sessa et al., 1996; Xu et al., 2011). A Ser/Thr protein kinase from Arabidopsis performed in vitro phosphorylation of the Ser and Thr residues within the cleavable presequence of chloroplast-destined precursor proteins. However, mitochondria-destined precursor proteins were not phosphorylated by kinases (Martin et al., 2006). Cytosolic STYKs play a vital role in chloroplast differentiation (Lamberti et al., 2011) and phosphorylate specific substrates. In this study, we demonstrated that OLE3 is phosphorylated by STYK, regulating its bifunctional activities of lipid synthesis and degradation. Chen et al. (1999) previously reported that caleosin, an oil body-associated protein, was phosphorylated at its C terminus by a predicted Tyr kinase.
AhSTYK (Rudrabhatla and Rajasekharan, 2002) and Ca2+-dependent OsCDPK7 (Wan et al., 2007) were induced at low temperatures, and their corresponding mRNA transcripts were expressed abundantly. Arabidopsis oleosin-deficient mutants were sensitive to freezing after imbibition at 4°C, resulting in seed mortality (Shimada et al., 2008), which could be due to the stability of oil bodies at low temperatures. The overexpression of oleosins contributed to the freezing tolerance of seeds (Ellis et al., 1991; Leprince et al., 1998), suggesting a possible relationship between oleosins and freezing tolerance. At low temperatures, a significant amount of AhSTYK phosphorylation of OLE3 was observed.
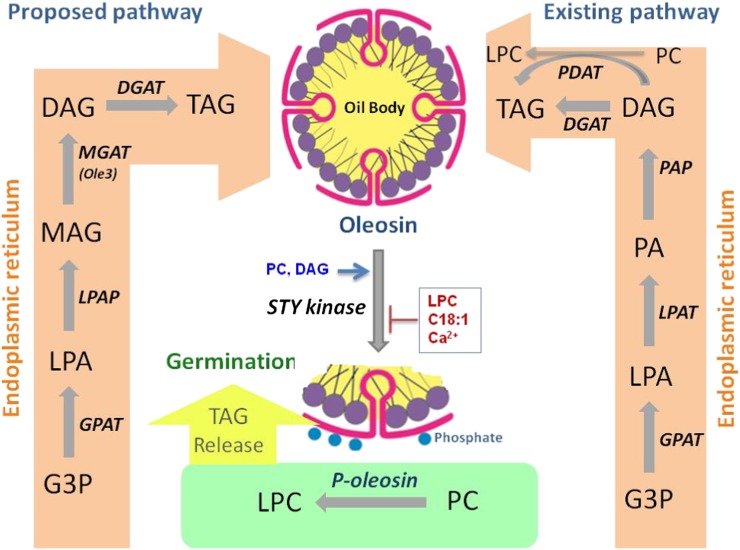
Intracellular free Ca2+ ions evoke rapid changes in effector functions upon various stimuli (Sanders et al., 2002). OLE3 phosphorylation was significantly inhibited by calcium, suggesting that STYK could operate via a calcium-independent mechanism of signal transduction. In soybean, CPK11 autophosphorylation was stimulated by calcium, PC did not have any effect, and substrate phosphorylation was stimulated by phospholipids. In comparison with previous data (Szczegielniak et al., 2005), a similar effect of PC was observed on autophosphorylation, but DAG also increased OLE3 phosphorylation. LPC was shown to increase substrate phosphorylation by AtCPK1 (Binder et al., 1994). However, our results suggest that LPC and oleic acid decreased OLE3 phosphorylation by AhSTYK. Kinase activity increased with PC and DAG but decreased with LPC, oleic acid, and calcium (Fig. 7).
Figure 7.
Schematic representation of the proposed roles of oleosin. The existing pathway occurs in a PA-dependent manner, wherein PA is synthesized by the sequential acylation of glycerol 3-phosphate (G3P) by the substrate-specific acyltransferases. The synthesized PA is dephosphorylated to DAG by PA phosphatase, and the resultant DAG is further acylated by DAG acyltransferase (DGAT) to form TAG. TAG can also be synthesized from DAG in an acyl-CoA-independent manner by an enzyme, phospholipid:diacylglycerol acyltransferase (PDAT). The proposed MAG pathway is involved in the formation of TAG during the maturation of seeds. In this pathway, G3P is acylated to LPA by G3P acyltransferase (GPAT). LPA is dephosphorylated by LPA phosphatase (LPAP) to produce MAG, which is sequentially acylated to TAG by MGAT and DGAT. The regulation of MGAT activity is achieved by phosphorylation catalyzed by STYK, and the phospho-OLE3 exhibits PLA2 activity. The hydrolysis of PC to LPC disturbs the membrane structure, inducing the release of TAG from oil bodies, which is used as the source of energy for germination. PC and DAG increase substrate phosphorylation, whereas LPC, oleic acid, and Ca2+ inhibit phosphorylation.
Substrate phosphorylation at specific Ser residues could modulate various biochemical functions of target proteins. Phosphorylation at the N terminus of a specific Ser residue of plant phosphoenolpyruvate carboxylase by phosphoenolpyruvate carboxylase kinase was shown to regulate its activity and modify allosteric properties of the enzyme (Taybi et al., 2000). The regulation of the bifunctional activities of OLE3 (MGAT and PLA2) could be controlled by STYK phosphorylation of Ser-18.
The regulation of bifunctional activities is controlled by phosphorylation. Upon phosphorylation, perilipin, a scaffold of a lipid droplet-associated protein, interacts with Abhd5 and regulates Abhd5 lipase activity (Granneman et al., 2009). Unlike perilipin, OLE3 directly increases PLA2 activity and eliminates MGAT activity after phosphorylation. During seed germination, oleosins undergo a 4.3-fold increase in phosphorylation in immature multiprotein complexes over those during maturation. This increase could be the result of an increase in PLA2 activity during seed germination and a simultaneous decrease in MGAT activity. In vitro phosphorylation of immature multiprotein complexes resulted in a reduction in MGAT activity. MGAT in plants could be involved in TAG biosynthesis and degradation.
When the sunflower (Helianthus annuus) oleosin protein was expressed ectopically in transgenic Arabidopsis, it caused an accumulation of this protein in the microsomal membrane fraction, and the overexpression of N-terminal-deleted oleosin caused an impaired transfer from the endoplasmic reticulum to the oil body (Beaudoin and Napier, 2000). Some of the seed-specific oleosins in Arabidopsis (At3g01570 and At5g51210) have been shown to be targeted to the chloroplasts, mitochondria, and plasma membrane (Cell eFP Browser; http://bar.utoronto.ca/cell_efp/cgi-bin/cell_efp.cgi?primaryGene). Wahlroos et al. (2003) reported that oleosin-GFP chimeric protein is synthesized in the endoplasmic reticulum and that the protein is associated with oil bodies in tobacco (Nicotiana tabacum) leaf protoplasts. Our results here indicate that OLE3-GFP chimeric protein, when transiently expressed in Arabidopsis leaf protoplasts, is trafficked to chloroplasts. This could be because the ectopic expression of seed-specific oleosin in non-oil-storing tissue may cause the chloroplastic localization. Alternatively, the peanut seed-specific OLE3 is targeted to the lipid bodies in chloroplasts (plastoglobules), which is structurally analogous to seed oil bodies (Martin and Wilson, 1984; Austin et al., 2006) and plastoglobulin (oil body-associated structural protein) functions like oleosin (Huang, 1996; Bréhélin et al., 2007). A similar observation was reported earlier, that oleosin was found largely on the periphery of the plastoglobules in the tapetum cells of Arabidopsis (Wang et al., 1997) and tobacco protoplasts (De Domenico et al., 2011).
The existing pathway for the biosynthesis of TAG occurs by the first acylation of glycerol 3-phosphate by a glycerol 3-phosphate acyltransferase to form lysophosphatidic acid (LPA), which is then acylated to PA by LPA acyltransferase. The synthesized PA is dephosphorylated to DAG by a PA phosphatase. DAG is further acylated to form TAG by DAG acyltransferase. TAG can also be synthesized by phospholipid:diacylglycerol acyltransferase.
In summary, we propose an alternative phosphatidic acid-independent pathway for TAG biosynthesis (Fig. 7). In this pathway, glycerol 3-phosphate is acylated to LPA by an acyl acceptor-specific acyltransferase. LPA is dephosphorylated to MAG by LPA phosphatase (Shekar et al., 2002), which is sequentially acylated to DAG and TAG by MGAT and DAG acyltransferase, respectively. STYK regulates MGAT activity, and phospho-OLE3 hydrolyzes membrane lipids into lysophospholipids, which disturbs the membrane structure and eventually leads to the release of TAG from the oil bodies. Notably, MGAT (OLE3) plays a pivotal role during these physiological processes by altering its biochemical substrates with respect to seed cellular physiology.
MATERIALS AND METHODS
Materials
[1-14C]Monooleoyl-rac-glycerol and [2-palmitoyl-9,10-3H]PC were obtained from American Radiolabeled Chemicals. Lipids and acyl-CoAs were purchased from Avanti Polar Lipids. [γ-32P]ATP was obtained from the Board of Radiation and Isotope Technology, Bhabha Atomic Research Centre, in Mumbai, India. Restriction endonucleases and Pfu polymerase were purchased from New England Biolabs. Oligonucleotides, monoclonal antibodies, phospho-amino acids, calf intestinal alkaline phosphatase, and all other reagents were purchased from Sigma. Silica-thin-layer chromatography (TLC) plates were obtained from Merck. Nickel-nitrilotriacetic acid agarose was purchased from Qiagen. Field-grown immature peanut (Arachis hypogaea) seeds (JL-24) were harvested at 20 to 24 d after flowering. Mature peanut seeds were germinated for 24 h in the dark.
Cloning and Expression of OLE3
A seed-specific cDNA library of peanut was constructed in a λ-ZAP II vector (Stratagene). The open reading frame of OLE3 was PCR amplified and subcloned in a bacterial vector. OLE3 was subcloned into the pRSET C vector and expressed in Escherichia coli. The expressed protein was purified using a nickel-nitrilotriacetic acid agarose matrix. To overexpress OLE3 in Saccharomyces cerevisiae, a pYES2 construct harboring the OLE3 gene was transformed into yeast cells using the lithium acetate method (Schiestl and Gietz, 1989). The resulting transformants were confirmed by colony PCR using OLE3 sequence-specific forward and reverse primers. Successfully transformed yeast was grown to the late log phase in synthetic minimal medium without uracil containing 2% (w/v) Glc. Cells were harvested by centrifugation and inoculated at a concentration of A600 = 0.1 in an induction medium (synthetic minimal medium without uracil containing 2% [w/v] Gal). Protein expression was induced for 24 h and confirmed by immunoblotting using anti-OLE3 antibodies at a dilution of 1:1,000 (v/v). Protein concentrations were determined by the protein-dye binding assay using bovine serum albumin as a standard.
Expression and Purification of His-6-STYK and OLE3
The expression and purification of AhSTYK were described previously (Rudrabhatla and Rajasekharan, 2002). The cDNA encoding OLE3 was amplified using cognate primers and subcloned into the His-tagged expression vector pRSET C at PstI and EcoRI cloning sites (Parthibane et al., 2012). The resulting construct was expressed in E. coli BL-21 cells by induction with 0.5 mm isopropyl-1-thio-β-d-galactopyranoside for 4 h. The recombinant protein was purified by nickel-nitrilotriacetic acid agarose chromatography (Qiagen). Purified fractions containing the eluted protein were analyzed by 12% (w/v) SDS-PAGE followed by Coomassie blue staining.
Plasmid Constructs and Protoplast Transformation
The pSAT vectors for BiFC studies were obtained from the Arabidopsis Biological Resource Center stock center at Ohio State University. The open reading frames of Arabidopsis (Arabidopsis thaliana) STYK and peanut OLE3 were fused in frame and upstream of the N-terminal half of EYFP in pSAT4-nEYFP-N1 and the C-terminal half of EYFP in pSAT6-cEYFP-N1, respectively. AtSTYK was cloned in the NcoI-BamHI cloning sites of pSAT4-nEYFP-N1, and peanut OLE3 was cloned in the NcoI-BamHI cloning sites of pSAT6-cEYFP-N1 vector. BiFC constructs were generated using the following primers: AtSTYK-F, 5′-GGAATTCATGCTAGAAGGAGCAAAGTTCAAC-3′; AtSTYK-R, 5′-CGGATCCCGTCAATCGTCATCGGCTGGCTCAAG-3′; AhOLE3-F, 5′-ATATGAATTCATGGTTATGTCTGATCAAACAAGGAC-3′; AhOLE3-R, 5′-ATATGGATCCTCAATACCCTGGGGTGCCCTC-3′. The AmGPPS.SSU-nEYFP and NtGGPPS1-cEYFP constructs (Orlova et al., 2009) were used as positive controls for interaction. GFP fusion with the AhOLE3 construct was obtained by cloning into XbaI-BamHI sites of the plasmid (326-sGFP). The GFP construct was generated using the following primers: AhOLE3-F, 5′-TCTAGAATGGTTATGTCTGATCAAAC-3′; OLE3-R, 5′-GGATCCATACCCTGGGGTGCCCTC-3′. The accuracy of fusions was confirmed by nucleotide sequencing.
For protoplast preparation (Abdel-Ghany et al., 2005), Arabidopsis plants (ecotype Columbia) were grown on Murashige and Skoog medium for 3 weeks and 2 g of fresh leaf tissue was placed in 15 mL of filter-sterilized enzyme solution (Serva Electrophoresis) containing 1% (w/v) cellulase R-10, 0.25% (w/v) macerozyme R-10, 0.4 m mannitol, 20 mm MES (pH 5.7), and 20 mm KCl incubated for 10 min at 55°C, and then 0.1% (w/v) bovine serum albumin and 10 mm CaCl2 were added. The cut leaf tissues were incubated for 5 h at room temperature. The clear digest was filtered through 100-µm nylon, and the protoplasts were harvested by centrifugation for 2 min at 200 rpm (Eppendorf 5810R; rotor A-4-62), washed twice in 10 mL of ice-cold wash solution (154 mm NaCl, 125 mm CaCl2, 5 mm KCl, 5 mm Glc, and 2 mm MES, pH 5.7), and then incubated with the wash solution for 30 min in ice. The pellet (protoplasts) was suspended in 2 mL of mannitol solution (15 mm MgCl2, 0.4 m mannitol, and 4mm MES, pH 5.7). Plasmid DNA (10–20 µg) was added to 100 µL of protoplast, and 110 µL of polyethylene glycol solution containing 40% (w/v) polyethylene glycol-4000, 0.4 m mannitol, and 0.1 m CaCl2 was added, very gently mixed, and incubated for 30 min at room temperature. The solution was very slowly diluted with 440 µL of wash solution and then pelleted by centrifugation for 3 min at 3,000 rpm. The pellet was finally resuspended in 100 µL of wash solution. Transient expression of the yellow fluorescent protein (YFP) and the GFP fusion proteins was observed 16 to 20 h after transformation. Images were acquired using a Zeiss LSM 510 confocal laser scanning microscope with a 60× oil lens. The YFP was excited with the 514-nm line of the four-line argon laser, and the emission was collected with a 530- to 560-nm band-pass filter. Chlorophyll was excited by the 637-nm red diode laser, and emission greater than 660 nm in wavelength was collected.
Immunoprecipitation and Immune Complex Kinase Assay
The soluble and membrane proteins (200 µg) from immature peanut were precleared with 25 µL of protein A-agarose beads for 1 h. The agarose beads were removed by centrifugation followed by the addition of 2 µg of affinity-purified antibodies (Rudrabhatla and Rajasekharan, 2002), which were incubated with agitation for 6 h at 4°C. After brief centrifugation, the precipitate was washed and phosphorylated, and MGAT activity was measured.
Immunoblotting
The phospho-OLE3 protein was resolved on a 12% (w/v) SDS-polyacrylamide gel and transferred onto a nitrocellulose membrane. The blots were blocked with 0.5% (w/v) gelatin in phosphate-buffered saline. The primary antibody (anti-phospho-Ser, anti-phospho-Thr, or anti-phospho-Tyr monoclonal antibody) was diluted (1:500, v/v), and the blots were subsequently washed in phosphate-buffered saline with 0.05% (v/v) Tween 20 followed by the addition of secondary anti-mouse antibodies (1:2,000, v/v). Proteins were detected by chemiluminescence using luminol as a substrate.
In Vitro Kinase Assay
The autophosphorylation assay was conducted as described previously (Rudrabhatla and Rajasekharan, 2002). The substrate phosphorylation assay mixture consisted of 1 µg of STYK, 2 µg of OLE3, 50 mm Tris-HCl (pH 7.5), 10 mm MgCl2, and 25 µm [γ-32P]ATP (3,000 dpm pmol−1). The assay was incubated for 30 min at 30°C. The autophosphorylation and substrate phosphorylation were stopped by the addition of 2× SDS-PAGE loading buffer. Phosphorylated proteins were resolved on a 12% (w/v) SDS-polyacrylamide gel, and labeled proteins were detected by autoradiography.
Effect of Lipid on Kinase Activity
Kinase assay was performed in a reaction mixture of 1.0 µg of peanut STYK (autophosphorylation), 2.0 µg of peanut OLE3 (substrate phosphorylation), 25 µm [γ-32P]ATP, 10 mm MgCl2, and respective lipids for 30°C at 30 min. The lipids (MAG, DAG, TAG, PC, LPC, and oleic acid) were prepared at a stock concentration of 1.0 mm in 5 mm CHAPS with 50 mm Tris-HCl, pH 7.5. In control kinase assay, the reaction mixture contained all the components except lipid. After the kinase assay, the reaction was stopped by the addition of 2× SDS-PAGE loading dye. The reaction mixtures were resolved by 12% SDS-PAGE and quantified.
MGAT Assay
Enzyme activity was measured using either [1-14C]oleoyl-CoA or [1-14C]MAG (oleoyl) into DAG. The reaction mixture contained 50 mm Tris-HCl, pH 8.0, 1 mm MgCl2, 50 μm monoacylglycerol (5 μL of sonicated vesicles in 5 mm CHAPS), enzyme, and either 10 μm [14C]oleoyl-CoA (55,000 dpm) or 20 μm [1-14C]MAG (110,000 dpm) in a total volume of 100 μL. The reaction was initiated by the addition of enzyme and terminated after 30 min by adding 0.2 mL of acidified water and 0.6 mL of chloroform:methanol (1:2, v/v). Lipids were extracted and separated by TLC and viewed with a phosphorimager (Parthibane et al., 2012).
PLA2 Assay
The reaction mixture contained 1 mm sonicated vesicles of dipalmitoyl-PC and 10 μg of enzyme in assay buffer (0.05 m Tris-HCl, pH 7.5, and 2 mm dithiothreitol) for a total volume of 100 μL. The reaction was incubated for 45 min at 30°C and terminated by extracting the lipids with butanol. Radiometric assay buffer consisted of 100 μm sonicated vesicles of [2-palmitoyl-9,10-3H]PC (1 μCi per reaction) and the enzyme source in a total volume of 100 μL. After the reaction was terminated, lipids were isolated and analyzed by silica-TLC and visualized with a phosphorimager (Parthibane et al., 2012).
Isolation of the 14S Multiprotein Complex from Peanut Seeds
Immature/germinated seeds (20 g) were ground with a pestle in a prechilled mortar with 2.5 g of acid-washed sand and 50 mL of extraction buffer containing 50 mm Tris-HCl (pH 8.0), 1 mm EDTA, 10 mm KCl, 1 mm MgCl2, 1 mm β-mercaptoethanol, 0.1 mm phenylmethylsulfonyl fluoride, and 0.25 m Suc. Microsomal membranes were washed with 2 m urea-containing extraction buffer followed by solubilization with 8 m urea and 10 mm CHAPS. The 14S multiprotein complex was isolated from peanut as described previously (Parthibane et al., 2012).
Radiolabeling of Seed Proteins
The sliced seeds (immature/germinated) were incubated with 50 µCi of [32P]orthophosphate in 50 mm Tris-HCl (pH 8.0 for 1 h) at 30°C. The slices were then transferred to nonradioactive orthophosphate medium for 4 h. The solubilized 14S multiprotein complex was isolated from sliced seeds and run on a 12% (w/v) SDS-polyacrylamide gel, which was imaged by autoradiography. The amount of radioactivity associated with OLE3 was measured by liquid scintillation counting.
Phospho-Amino Acid Analysis
OLE3 mutants were labeled in vitro with [γ-32P]ATP by STYK as described above and electroblotted onto a polyvinylidine difluoride membrane. After autoradiography, radioactive OLE3 was excised and hydrolyzed in 200 µL of 6 m HCl for 2 h at 110°C. The hydrolysate was dried in a Speed-Vac concentrator and resuspended in 20 µL of water containing 1 mg mL−1 of each of the phospho-amino acid markers: phospho-Ser, phospho-Thr, and phospho-Tyr (Sigma). Two microliters of the hydrolysate was analyzed by ascending silica-TLC (Merck) using a mixture of ethanol and ammonia (3.5:1.6, v/v) as a solvent system (Muñoz and Marshall, 1990). The positions of the phospho-amino acid markers were detected by ninhydrin staining silica-TLC plates. The plates were then exposed for autoradiography to locate the positions of the 32P-labeled amino acids.
Site-Directed Mutagenesis
Wild-type OLE3 (pRSET C-OLE3) templates (80 ng) and sense and antisense primers (25 pmol) were added to PCR tubes containing 0.2 mm deoxyribonucleotide triphosphates, 1 mm MgSO4, 2.5 units of Pfu polymerase, and 1× reaction buffer. Amplification was carried out under the following conditions: denaturation of the template at 95°C for 4 min followed by 20 cycles at 94°C for 45 s (denaturation), 52°C for 1 min (annealing), and 72°C for 9 min (extension). The reaction was continued for 20 min at 72°C for complete extension. The primers that anneal to the sense strand were used on the pRSET C vector harboring the OLE3 gene to perform site-directed mutagenesis. The PCR-amplified mixture was treated with DpnI (10 units) for 1 h at 37°C to digest the methylated template, and the resulting mixture was transformed into DH5α-competent cells. The presence of mutations was confirmed by sequencing the plasmid DNA. The following primers were used: S14A, 5′-GGAGGAGGAGGGGCCTATGGATCATCC-3′; S17A, 5′-GGGTCCTATGGAGCATCCTATGGTGGA-3′; S18A, 5′-TCCTATGGATCAGCCTATGGTGGAGGA-3′; S27A, 5′-GGCACCTATGGTGCATCTTATGGAACC-3′; and S28A, 5′-ACCTATGGTTCAGCTTATGGAACCTCC-3′.
Preparation of Phospho-OLE3
Microsomes from S. cerevisiae (50 µg) overexpressing OLE3 were incubated with 25 µg of recombinant AhSTYK with 50 µm ATP in 50 mm Tris-HCl (pH 7.5) and 10 mm MgCl2 for 30 min at 30°C. OLE3 phosphorylation was confirmed by a radiolabeled kinase assay. The phosphorylated sample mixture was dialyzed, and the resulting proteins were quantified.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AY027437 (AhSTY kinase), AY722696 (AhOleosin 3), and NM127998 (AtSTY kinase).
Acknowledgments
We are indebted to Director C.S. Nautiyal and Dr. Samir Sawant (Council of Scientific and Industrial Research, National Botanical Research Institute, Lucknow) for allowing us to carry out confocal microscopy in their facility. We thank Dr. Dinesh Nagagowda (Council of Scientific and Industrial Research, Central Institute of Medicinal and Aromatic Plants, Lucknow) for helpful discussion and a generous gift of positive control plasmid constructs for the protein-protein interaction study.
Glossary
- TAG
triacylglycerol
- MGAT
monoacylglycerol acyltransferase
- MAG
monoacylglycerol
- DAG
diacylglycerol
- STYK
serine/threonine/tyrosine protein kinase
- BiFC
bimolecular fluorescence complementation
- EYFP
enhanced yellow fluorescent protein
- YFP
yellow fluorescent protein
- LPC
lysophosphatidylcholine
- PC
phosphatidylcholine
- PLA2
phospholipase A2
- LPA
lysophosphatidic acid
- TLC
thin-layer chromatography
References
- Abdel-Ghany SE, Müller-Moulé P, Niyogi KK, Pilon M, Shikanai T. (2005) Two P-type ATPases are required for copper delivery in Arabidopsis thaliana chloroplasts. Plant Cell 17: 1233–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abell BM, Hahn M, Holbrook LA, Moloney MM. (2004) Membrane topology and sequence requirements for oil body targeting of oleosin. Plant J 37: 461–470 [DOI] [PubMed] [Google Scholar]
- Ali N, Halfter U, Chua NH. (1994) Cloning and biochemical characterization of a plant protein kinase that phosphorylates serine, threonine, and tyrosine. J Biol Chem 269: 31626–31629 [PubMed] [Google Scholar]
- Austin JR, II, Frost E, Vidi P-A, Kessler F, Staehelin LA. (2006) Plastoglobules are lipoprotein subcompartments of the chloroplast that are permanently coupled to thylakoid membranes and contain biosynthetic enzymes. Plant Cell 18: 1693–1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudoin F, Napier JA. (2000) The targeting and accumulation of ectopically expressed oleosin in non-seed tissues of Arabidopsis thaliana. Planta 210: 439–445 [DOI] [PubMed] [Google Scholar]
- Beisson F, Ferté N, Bruley S, Voultoury R, Verger R, Arondel V. (2001) Oil-bodies as substrates for lipolytic enzymes. Biochim Biophys Acta 1531: 47–58 [DOI] [PubMed] [Google Scholar]
- Binder BM, Harper JF, Sussman MR. (1994) Characterization of an Arabidopsis calmodulin-like domain protein kinase purified from Escherichia coli using an affinity sandwich technique. Biochemistry 33: 2033–2041 [DOI] [PubMed] [Google Scholar]
- Bréhélin C, Kessler F, van Wijk KJ. (2007) Plastoglobules: versatile lipoprotein particles in plastids. Trends Plant Sci 12: 260–266 [DOI] [PubMed] [Google Scholar]
- Chen JCF, Tsai CCY, Tzen JTC. (1999) Cloning and secondary structure analysis of caleosin, a unique calcium-binding protein in oil bodies of plant seeds. Plant Cell Physiol 40: 1079–1086 [DOI] [PubMed] [Google Scholar]
- Citovsky V, Gafni Y, Tzfira T. (2008) Localizing protein-protein interactions by bimolecular fluorescence complementation in planta. Methods 45: 196–206 [DOI] [PubMed] [Google Scholar]
- Citovsky V, Lee LY, Vyas S, Glick E, Chen MH, Vainstein A, Gafni Y, Gelvin SB, Tzfira T. (2006) Subcellular localization of interacting proteins by bimolecular fluorescence complementation in planta. J Mol Biol 362: 1120–1131 [DOI] [PubMed] [Google Scholar]
- De Domenico S, Bonsegna S, Lenucci MS, Poltronieri P, Di Sansebastiano GP, Santino A. (2011) Localization of seed oil body proteins in tobacco protoplasts reveals specific mechanisms of protein targeting to leaf lipid droplets. J Integr Plant Biol 53: 858–868 [DOI] [PubMed] [Google Scholar]
- Ellis RH, Hong TD, Roberts EH. (1991) An intermediate category of seed storage behaviour? I. COFFEE. J Exp Bot 41: 1167–1174 [Google Scholar]
- Granneman JG, Moore HP, Krishnamoorthy R, Rathod M. (2009) Perilipin controls lipolysis by regulating the interactions of AB-hydrolase containing 5 (Abhd5) and adipose triglyceride lipase (Atgl). J Biol Chem 284: 34538–34544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirayama T, Oka A. (1992) Novel protein kinase of Arabidopsis thaliana (APK1) that phosphorylates tyrosine, serine and threonine. Plant Mol Biol 20: 653–662 [DOI] [PubMed] [Google Scholar]
- Huang AHC. (1992) Oil bodies and oleosins in seeds. Annu Rev Plant Physiol Plant Mol Biol 43: 177–200 [Google Scholar]
- Huang AHC. (1996) Oleosins and oil bodies in seeds and other organs. Plant Physiol 110: 1055–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CY, Chung CI, Lin YC, Hsing YI, Huang AH. (2009) Oil bodies and oleosins in Physcomitrella possess characteristics representative of early trends in evolution. Plant Physiol 150: 1192–1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamberti G, Gügel IL, Meurer J, Soll J, Schwenkert S. (2011) The cytosolic kinases STY8, STY17, and STY46 are involved in chloroplast differentiation in Arabidopsis. Plant Physiol 157: 70–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leprince O, van Aelst AC, Pritchard HW, Murphy DJ. (1998) Oleosins prevent oil-body coalescence during seed imbibition as suggested by a low-temperature scanning electron microscope study of desiccation-tolerant and -sensitive oilseeds. Planta 204: 109–119 [Google Scholar]
- Martin BA, Wilson RF. (1984) Subcellular localization of TAG synthesis in spinach leaves. Lipids 19: 117–121 [DOI] [PubMed] [Google Scholar]
- Martin T, Sharma R, Sippel C, Waegemann K, Soll J, Vothknecht UC. (2006) A protein kinase family in Arabidopsis phosphorylates chloroplast precursor proteins. J Biol Chem 281: 40216–40223 [DOI] [PubMed] [Google Scholar]
- Muñoz G, Marshall SH. (1990) An alternative method for a fast separation of phosphotyrosine. Anal Biochem 190: 233–237 [DOI] [PubMed] [Google Scholar]
- Murphy DJ. (October 15, 2011) The dynamic roles of intracellular lipid droplets: from archaea to mammals. Protoplasma http://dx.doi.org/10.1007/s00709-011-0329-7 [DOI] [PubMed] [Google Scholar]
- Orlova I, Nagegowda DA, Kish CM, Gutensohn M, Maeda H, Varbanova M, Fridman E, Yamaguchi S, Hanada A, Kamiya Y, et al. (2009) The small subunit of snapdragon geranyl diphosphate synthase modifies the chain length specificity of tobacco geranylgeranyl diphosphate synthase in planta. Plant Cell 21: 4002–4017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napier JA, Stobart AK, Shewry PR. (1996) The structure and biogenesis of plant oil bodies: the role of the ER membrane and the oleosin class of proteins. Plant Mol Biol 31: 945–956 [DOI] [PubMed] [Google Scholar]
- Parthibane V, Rajakumari S, Venkateshwari V, Iyappan R, Rajasekharan R. (2012) Oleosin is bifunctional enzyme that has both monoacylglycerol acyltransferase and phospholipase activities. J Biol Chem 287: 1946–1954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng FY, Weselake RJ. (2011) Gene coexpression clusters and putative regulatory elements underlying seed storage reserve accumulation in Arabidopsis. BMC Genomics 12: 286–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy MM, Rajasekharan R. (2007) Serine/threonine/tyrosine protein kinase from Arabidopsis thaliana is dependent on serine residues for its activity. Arch Biochem Biophys 460: 122–128 [DOI] [PubMed] [Google Scholar]
- Rudrabhatla P, Rajasekharan R. (2002) Developmentally regulated dual-specificity kinase from peanut that is induced by abiotic stresses. Plant Physiol 130: 380–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudrabhatla P, Rajasekharan R. (2003) Mutational analysis of stress-responsive peanut dual specificity protein kinase: identification of tyrosine residues involved in regulation of protein kinase activity. J Biol Chem 278: 17328–17335 [DOI] [PubMed] [Google Scholar]
- Rudrabhatla P, Reddy MM, Rajasekharan R. (2006) Genome-wide analysis and experimentation of plant serine/threonine/tyrosine-specific protein kinases. Plant Mol Biol 60: 293–319 [DOI] [PubMed] [Google Scholar]
- Sanders D, Pelloux J, Brownlee C, Harper JF. (2002) Calcium at the crossroads of signaling. Plant Cell (Suppl) 14: S401–S417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiestl RH, Gietz RD. (1989) High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr Genet 16: 339–346 [DOI] [PubMed] [Google Scholar]
- Schmidt MA, Herman EM. (2008) Suppression of soybean oleosin produces micro-oil bodies that aggregate into oil body/ER complexes. Mol Plant 1: 910–924 [DOI] [PubMed] [Google Scholar]
- Sessa G, Raz V, Savaldi S, Fluhr R. (1996) PK12, a plant dual-specificity protein kinase of the LAMMER family, is regulated by the hormone ethylene. Plant Cell 8: 2223–2234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shekar S, Tumaney AW, Rao TJVS, Rajasekharan R. (2002) Isolation of lysophosphatidic acid phosphatase from developing peanut cotyledons. Plant Physiol 128: 988–996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada TL, Shimada T, Takahashi H, Fukao Y, Hara-Nishimura I. (2008) A novel role for oleosins in freezing tolerance of oilseeds in Arabidopsis thaliana. Plant J 55: 798–809 [DOI] [PubMed] [Google Scholar]
- Siloto RMP, Findlay K, Lopez-Villalobos A, Yeung EC, Nykiforuk CL, Moloney MM. (2006) The accumulation of oleosins determines the size of seed oilbodies in Arabidopsis. Plant Cell 18: 1961–1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczegielniak J, Klimecka M, Liwosz A, Ciesielski A, Kaczanowski S, Dobrowolska G, Harmon AC, Muszyńska G. (2005) A wound-responsive and phospholipid-regulated maize calcium-dependent protein kinase. Plant Physiol 139: 1970–1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taybi T, Patil S, Chollet R, Cushman JC. (2000) A minimal serine/threonine protein kinase circadianly regulates phosphoenolpyruvate carboxylase activity in Crassulacean acid metabolism-induced leaves of the common ice plant. Plant Physiol 123: 1471–1482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tregear JW, Jouannic S, Schwebel-Dugue N, Kreis M. (1996) An unusual protein kinase displaying characteristics of both the serine/threonine kinase and tyrosine families is encoded by the Arabidopsis thaliana gene ATN1. Plant Sci 117: 107–119 [Google Scholar]
- Wahlroos T, Soukka J, Denesyuk A, Wahlroos R, Korpela T, Kilby NJ. (2003) Oleosin expression and trafficking during oil body biogenesis in tobacco leaf cells. Genesis 35: 125–132 [DOI] [PubMed] [Google Scholar]
- Wan B, Lin Y, Mou T. (2007) Expression of rice Ca2+-dependent protein kinases (CDPKs) genes under different environmental stresses. FEBS Lett 581: 1179–1189 [DOI] [PubMed] [Google Scholar]
- Wang TW, Balsamo RA, Ratnayake C, Platt KA, Ting JT, Huang AH. (1997) Identification, subcellular localization, and developmental studies of oleosins in the anther of Brassica napus. Plant J 11: 475–487 [DOI] [PubMed] [Google Scholar]
- Xu F, Meng T, Li P, Yu Y, Cui Y, Wang Y, Gong Q, Wang NN. (2011) A soybean dual-specificity kinase, GmSARK, and its Arabidopsis homolog, AtSARK, regulate leaf senescence through synergistic actions of auxin and ethylene. Plant Physiol 157: 2131–2153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Klessig DF. (2001) MAPK cascades in plant defense signaling. Trends Plant Sci 6: 520–527 [DOI] [PubMed] [Google Scholar]