Abstract
The increasing power and popularity of cryo-electron (cryo-EM) microscopy in structural biology brought about the development of so-called hybrid methods, which permit the interpretation of cryo-EM density maps beyond their nominal resolution in terms of atomic models. The Cryo-EM Modeling Challenge 2010 is the first community effort to bring together developers of hybrid methods as well as cryo-EM experimentalists. Participating in the challenge, the molecular dynamics flexible fitting (MDFF) method was applied to a number of cryo-EM density maps. The results are described here with special emphasis on the use of symmetry-based restraints to improve the quality of atomic models derived from density maps of symmetric complexes; on a comparison of the stereochemical quality of atomic models resulting from different hybrid methods; and on application of MDFF to electron crystallography data.
Introduction
Cryo-electron microscopy (cryo-EM) is furnishing images of biological complexes for over 30 years.1 Continuous progress in instrumentation and methodology has consolidated cryo-EM as a major biomolecular structure determination technique, especially for large macromolecular complexes, the crystallization of which is typically too challenging. Apart from obviating a need for crystallization, the fact that structures can be solved in solution means that conditions closer to physiological ones can be employed, and even significant conformational changes can be imaged by means of image classification (see Fischer et al.2 for a striking example).
Cryo-EM single-particle reconstruction yields now routinely structures at sub-nanometer resolution, in some cases approaching atomic resolution.3–10 Even though de novo modeling of atomic structures is now possible in exceptional cases, interpretation of cryo-EM maps typically leverages available crystallographic structures. Approaches to merge structural information of different modalities are collectively called hybrid methods, and are in fact analogous to methods employed for X-ray structure determination. Electron density maps solved by X-ray crystallography at ~3 Å resolution, for instance, clearly do not feature atomic resolution per se, but since the structure of the building blocks (mainly amino acid residues and nucleotides) is known at very high resolution, complete atomic models can be built given such electron density maps.
A number of hybrid methods that combine structural information from X-ray crystallography and cryo-EM have been proposed in the last few years (see Trabuco et al.11). The authors' group has developed the molecular dynamics flexible fitting (MDFF) method, in which an initial atomic model is subject to a molecular dynamics (MD) simulation with a modified potential energy function that includes a term derived from the cryo-EM density map.11,12 Through this term, atoms experience steering forces that locally drive them toward high-density regions. In addition to the steering forces derived from the EM data, structural restraints are applied to preserve secondary structure of proteins and nucleic acids,11,12 as well as to ensure stereochemical correctness,13 thus avoiding the so-called over-fitting problem. Moreover, symmetry-based restraints can be introduced.14 Restraints that ensure stereochemical correctness are widely used in crystallography.15–18 In combination with hybrid methods, restraints are adapted to preserve the stereochemistry of the starting model. The first applications of MDFF employed in vacuo MD simulations,12,19 but now MDFF simulations are usually performed in explicit or implicit solvent.11,20 MDFF has been successfully applied to several macromolecular complexes.14,19,21–45 For a practical guide on applying MDFF, see Trabuco et al.11 as well as a tutorial available on the MDFF website (http://www.ks.uiuc.edu/Research/mdff).
As part of the Cryo-EM Modeling Challenge 2010 (http://ncmi.bcm.edu/challenge), we have applied MDFF to a number of density maps, namely: GroEL (4Å), GroEL-GroES complex (7.7 Å), Mm-cpn in its closed state (4.3Å), lidless Mm-cpn in its open state (8Å), aquaporin-0 (2.5Å, electron crystallography), VP6 component of rotavirus (3.8Å), and a bacterial ribosome (6.4Å). Below we describe the atomic models obtained with special emphasis on: symmetry-based restraints, tools to detect and prevent stereochemical errors, and application of MDFF to crystallographic data.
Results
Atomic models were obtained by applying MDFF to seven density maps provided in the Cryo-EM Modeling Challenge 2010 (Fig. 1). In order to assess the quality of the fit for each case, the cross-correlation coefficient (CCC) between initial/final structures and the corresponding maps was calculated, showing an improvement in all cases (Table 1). The classical CCC suffers from an important limitation: all volume within the box encompassing the density map is taken into account in the calculation, such that a significant portion of the data points used does not actually correspond to the macromolecule.11,12 In fact, a CCC coefficient close to one can be obtained simply by defining a sufficiently large bounding box. Furthermore, in cases where only a subset of the macromolecular components imaged can be modeled, the unoccupied regions in the map lead to a lower global CCC. Thus, it is usually preferable to use a local CCC measure, in which only regions around the macromolecule are used in the calculation. Initial and final local CCCs presented in Table 1 show more clearly the improvement of the fit obtained by employing MDFF simulations.
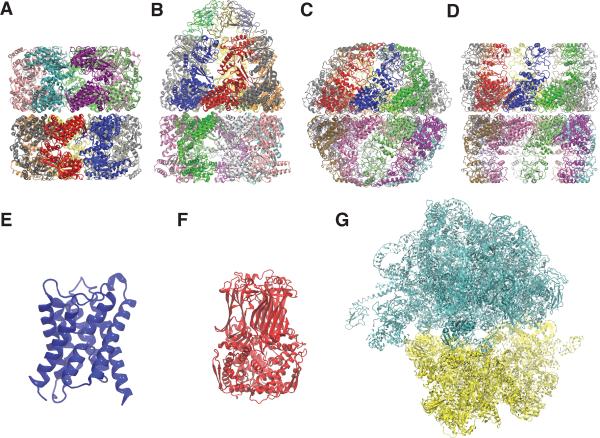
Figure 1.
Atomic models obtained by applying MDFF to density maps from the Cryo-EM Modeling Challenge 2010: (A) GroEL; (B) GroEL-GroES complex in the ATP-bound state; (C) Mm-cpn in the closed state; (D) lidless Mm-cpn in the open state; (E) aquaporin 0; (F) VP6 component of rotavirus; and (G) a bacterial ribosome.
Table 1.
Local and global cross-correlation coefficients (CCC) between atomic models and density maps. Local CCCs were calculated using a density threshold of 1 and 0.2 standard deviation above the mean for maps with resolution higher and lower than 5 Å, respectively. Global CCCs are given in parentheses.
| System | Resolution | Initial | Final |
|---|---|---|---|
| Aquaporin | 3 Å | 0.58 (0.34) | 0.69 (0.39) |
| GroEL | 4 Å | 0.32 (0.74) | 0.57 (0.86) |
| GroEL-GroES | 7.7 Å | 0.44 (0.84) | 0.71 (0.90) |
| Rotavirus | 3.8 Å | 0.68 (0.56) | 0.75 (0.61) |
| Mm-cpn in closed state | 4.3 Å | 0.51 (0.74) | 0.59 (0.76) |
| Mm-cpn in open state | 8 Å | 0.15 (0.39) | 0.77 (0.88) |
| Ribosome | 6.4 Å | 0.30 (0.75) | 0.46 (0.80) |
Symmetry-restrained MDFF
Many biological systems possess structural symmetry. Examples are large macromolecules such as the GroEL-GroES and Mm-cpn complexes, which were included in the Cryo-EM Modeling Challenge 2010. During the 3D single-particle reconstruction process, EM maps of such systems are often symmetrized to improve resolution.46–48 Similarly, incorporation of symmetry information into hybrid methods can increase the quality of the resulting models. For this purpose, support for enforcing a given symmetry has been implemented into MDFF.14 During a symmetry-restrained MDFF simulation, a harmonic potential energy term is imposed in addition to the MDFF restraints described in Methods and Materials, minimizing the root-mean-squared deviation (RMSD) between the current fitted structure, which is usually not symmetric due to thermal fluctuations in MD simulations, and a perfectly symmetric structure. The latter structure is calculated iteratively by averaging over all symmetric subunits of the current structure.
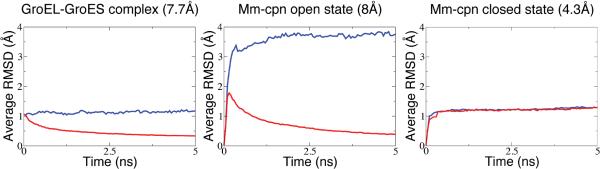
Symmetry-restrained MDFF was applied to three symmetric EM maps from the Cryo-EM Modeling Challenge 2010, namely a GroEL-GroES complex with 7-fold rotational symmetry (7.7Å resolution) and two Mm-cpn complexes with 8-fold rotational symmetry and 2-fold reflection symmetry, one in a closed state (4.3Å) and the other a lidless variant in an open state (8Å). For the sake of comparison, an MDFF simulation without symmetry-based restraints was also performed in each case. As expected, fitting simulations with symmetry restraints yield more symmetric structures, reflected in the lower average RMSD values (Fig. 2), defined by ; here RMSDi(t) is the RMSD between the current structure and the calculated symmetric structure of the i-th symmetric unit and N is the total number of symmetric units. As can be seen in Fig. 2, symmetry restraints have little effect in case of the 4.3-Å Mm-cpn map; at this resolution range, map features are detailed enough that equivalent subunits adopt similar structures in a regular MDFF simulation. However, in case of the 8-Å Mm-cpn map the effect of the symmetry restraints is strong (<RMSDi(t)> of ~0.5Å versus ~4 Å).
Figure 2.
Average Cα RMSD values between subunits and idealized symmetric structure throughout MDFF trajectories. The lower the average RMSD value, the more symmetric the structure is. Traces for simulations with and without restraints are shown in red and blue, respectively. Based on data previously presented in Chan et al.14
In addition to maintaining structural symmetry and, hence, improving quality of fit for low-resolution data, the use of symmetry restraints can avoid the so-called edge distortion effect. Assume, for instance, that a cryo-EM reconstruction is available for a helical macromolecular complex composed of a very large number of subunits, and a crystal structure is available for the monomer. In order to obtain an atomic model, one would naturally fit a few copies of the monomeric structure into a representative region of the density map. The edge distortion effect refers to the fact that, in a fitting simulation, monomers at the edges can be dragged into adjacent unoccupied densities. For more details on benefits and usage of symmetry-restrained MDFF see Chan et al.14
Stereochemical errors
Force fields used in MD simulations do not contain any energy term designed to enforce a given chirality or cis/trans peptide bond configuration. In general, both enantiomers or peptide bond isomers can arise, although the two forms are separated by a large energy barrier. Thus, for equilibrium MD simulations starting with error-free atomic structures, stereochemical errors are not expected to arise. In simulations such as MDFF employing external forces, however, it is possible for artifactual chirality or peptide bond configurations to arise, resulting in an atomic model with stereochemical errors. Hybrid methods do apply forces to atoms to drive them into the EM density map and, thus, may introduce stereochemical errors, unless additional measures are taken. To ensure stereochemical integrity of molecular models, software tools were developed to identify, visualize, and interactively correct stereochemical errors in biomolecular structures.13 These tools are implemented as two plugins (Chirality and Cispeptide) to the molecular visualization and analysis package VMD.49 Given a stereochemically correct starting structure, the plugins can also be used to generate harmonic restraints designed to prevent stereochemical errors from arising. Such stereochemical restraints are included indeed in standard MDFF protocols. Practical details about use of the Chirality and Cispeptide plugins are covered in Schreiner et al.,13 and in a tutorial available on the MDFF website (http://www.ks.uiuc.edu/Research/mdff). Since the data of these stereochemical restraints are written out as files for input to the simulation package NAMD,50 they can also be utilized by other MD simulation packages that support user-defined internal-coordinate restraints after adaptation of the data files.
During the Cryo-EM Modeling Challenge 2010, atomic models obtained by different flexible fitting methods were deposited and made publicly available. Analysis of such models can help answer the question of whether hybrid methods not based on MD simulations are also prone to introducing stereochemical errors in the generated atomic models. Table 2 presents the number of chirality errors and cis peptide bonds identified by the Chirality and Cispeptide VMD plugins, respectively, for each deposited atomic model obtained with a flexible fitting method, covering a total of five different methods. Structures derived with MDFF or Rosetta51 were stereochemically correct, whereas the other three investigated methods (DireX,52 Gorgon,6,53 and Froda54,55) introduced stereochemical errors. The different stereochemical quality of models may stem from the different definition of allowed configurations and the usage of additional restraints in the various methods.
Table 2.
Chirality errors and cis peptides identified in atomic models obtained by various flexible fitting methods, deposited as part of the Cryo-EM Modeling Challenge 2010. The number of chirality errors is given first, followed by the number of cis peptide bonds. For rotavirus and ribosome, 3 and 6 cis peptides, respectively, involving Pro present in the original structures were not considered. Mm-cpn models obtained by Gorgon contain only a single unit, so the total number stereochemical errors can be obtained by multiplying the given values by 16. Cases for which a model was not deposited are identified with n/a.
| System | Resolution | MDFF | DireX | Rosetta | Gorgon | Froda |
|---|---|---|---|---|---|---|
| Aquaporin | 3 Å | 0, 0 | 0, 0 | 0, 0 | n/a | n/a |
| GroEL | 4 Å | 0, 0 | 0, 4 | 0, 0 | n/a | n/a |
| GroEL-GroES | 7.7 Å | 0, 0 | 194, 60 | 0, 0 | n/a | 14, 11 |
| Rotavirus | 3.8 Å | 0, 0 | 0, 1 | n/a | n/a | n/a |
| Mm-cpn closed state | 4.3 Å | 0, 0 | 0, 0 | 0, 0 | 244, 202 | n/a |
| Mm-cpn open state | 8 Å | 0, 0 | n/a | 0, 0 | 267, 257 | 694, 115 |
| Ribosome | 6.4 Å | 0, 0 | n/a | n/a | n/a | n/a |
Apart from peptide bond configuration and chirality, the stereochemical quality of structures can be addressed by comparison with available crystal structure data and structure validation tools like Molprobity.56 Here, we chose the 3.8-Å map of the VP6 component of rotavirus to compare with the crystal structure and analyze the comparison with Molprobity. The crystal structure used for comparison is the same as the starting structure used for the fitting (PDB 1QHD5). The RMSD between the heavy atoms of the crystal structure and the MDFF model is 1.17Å, showing close resemblance of the MDFF model to the high-resolution (1.95Å) crystal structure. Some degree of deviation should be expected due to different molecular environments of the models, i.e., a crystal and a solution environment. Another reason for a significant RMSD value may be a superposition of different conformers present in the cryo-EM map.
An alternative comparison between MDFF model and crystal structure is furnished by backbone dihedrals. The main advantage of using the RMSD between these internal coordinates is that it does not rely on any alignment. Taking all backbone dihedrals into account, the RMSD for the Φ and Ψ angles are both about 21 degrees. Excluding flexible loop regions from the comparison yields RMSDs of 2 and 2.6 degrees for the Φ and Ψ angles, respectively. The reduced RMSD clearly shows that most of the differences stem from the loop regions. Apart from the fact that secondary structure elements were kept restrained during MDFF, a dominant uncertainty in the positions of loop regions should be expected since these regions are less resolved in the EM data and very flexible in MD simulations.
Although no stereochemical errors were detected in the final fitted structure of the VP6 component of rotavirus, the Molprobity server detected differences relative to the crystal structure. Overall, the MDFF-derived model shows a larger degree of flexibility than observed for the crystal structure (Table 3). The origin of the differences is of the same nature as already discussed for the comparison of RMSD in real space. The obtained differences are observed even after a short structure optimization (see Methods and Materials for the simulation protocol), suggesting that the variation in structural parameters is compatible with the map and moreover is within a reasonable range as guaranteed by the force field. A more aggressive optimization using tighter convergence criteria and not taking into account the EM data would be able to reduce even more deviations from the ideal values (Table 3). This means, however, that the effect of the temperature, which is present in the EM map, will be eliminated.
Table 3.
Comparison of stereochemistry of initial structure, final structure and structure after further minimization without EM data for the map of VP6 component of rotavirus (3.8 Å) as analyzed by the Molprobity web server.56 Definitions of the stereochemistry indicators are covered in Chen et al.56
| Stereochemistry indicators | Initial | Final | Minimized |
|---|---|---|---|
| Clashscore | 0.95 | 6.01 | 0.48 |
| Poor rotamers | 4.43% | 9.49% | 1.27% |
| Ramachandran outliers | 0% | 3.85% | 0.26% |
| Ramachandran favored | 97.44% | 87.44% | 96.67% |
| Cβ violations | 9 | 101 | 2 |
| Residues with bad bonds | 0% | 16.33% | 0% |
| Residues with bad angles | 0% | 11.48% | 0.26% |
Application to electron crystallography
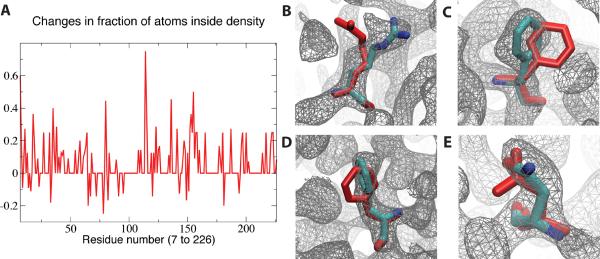
The MDFF method had been developed and validated initially as a real-space refinement method for cryo-EM density maps.12 Since the Cryo-EM Modeling Challenge 2010 provided a density map obtained by electron crystallography (aquaporin-0 resolved at 2.5Å), the applicability of MDFF to such data source was tested. The originally solved structure (PDB 3M9I57) was thus refined with MDFF. For the purpose of the challenge, which focuses on obtaining structural models based on cryo-EM data, the density map was treated only as an EM map and kept unchanged throughout the refinement process. As a proof-of-principle, a very simple measure was defined to determine if MDFF could potentially provide qualitative improvements. A molecular envelope was defined using a density threshold and, for each residue in the atomic model, the fraction of heavy atoms inside the envelope was calculated before and after MDFF refinement (Fig. 3A). Overall, MDFF lead to an increase in the fraction of atoms inside the molecular envelope. Focusing on a few peaks from the plot in Fig. 3A, one can visually inspect the effect of MDFF on the local structure at the side chain level (Fig. 3B–E). The images show that, in principle, MDFF refinement has the potential to improve the quality of structures solved by electron crystallography.
Figure 3.
MDFF applied to a 2.5-Å electron crystallography map of aquaporin-0. (A) Per-residue fraction of heavy atoms inside a molecular envelope defined by a density threshold of one standard deviation above the mean. Examples of residues with improved placement of side chains within the density, as identified by peaks in the plot, with initial structure colored in red: (B) Arg-11; (C) Phe-18; (D) Phe-136; and (E) Asn-197.
It should be noted, however, that in crystallographic refinement the phases and, hence, the density map are updated iteratively. Furthermore, one could utilize information like beta factors or even the original diffraction data in the MDFF fitting process. Despite the stated shortcomings, improvement of structure by refinement to a static map showcased here suggests the possibility to develop an extension of MDFF to fit crystallographic data.
Discussion
MDFF was applied to obtain atomic models for several density maps determined by the Cryo-EM Modeling Challenge 2010, covering a range of system sizes (single proteins to large complexes) and resolutions (2.5–8Å). Symmetry-restrained MDFF14 was applied to three different cryo-EM maps, showing that symmetry restraints are required to produce symmetric models at ~8Å resolution, but not at ~4Å. Furthermore, a comparative analysis of atomic models obtained using different flexible fitting methods revealed that certain approaches are prone to introducing stereochemical errors. MDFF protocols include extra harmonic restraints designed to prevent such errors from arising.13
MDFF benefits from the flexibility of NAMD, its underlying MD package,50 as highlighted by the aforementioned examples. As the MDFF method matures and is applied to a larger range of systems, limitations can often be addressed by introducing special-purpose restraints, such as the ones designed to preserve symmetry or stereochemical correctness. Being part of the very efficient NAMD software, MDFF also benefits from NAMD's high scalability, which is almost linear with system size and number of CPUs. For example, the solvated VP6 component of the rotavirus in the challenge is comprised of 105,537 atoms and the MDFF simulation speed was about 0.66 days per ns using 48 CPUs, while the larger GroEL-GroES system in solvent consists of 594,845 atoms and the speed was about 3.79 days per ns using 48 CPUs, showing almost linear scaling with system sizes. Speed and number of cores required by systems of other sizes should be approximately equal to linear interpolation of these numbers. For instance from linear interpolation of the above numbers one would expect to get about 3.8 days per ns on an eight-core machine for a system of 100,000 atoms. This scalability makes MDFF applicable even to very large systems. In light of ever increasing computational power, computational cost should not be a barrier to the use of MDFF.
Since MDFF is based on MD simulations, any molecule that has been parameterized in MD force fields can be modeled by MDFF. Macromolecules can also be modeled in a realistic environment, namely solvated by water molecules and ions, or even embedded in a lipid membrane.11,21,22 The current release of NAMD 2.8 supports the generalized Born implicit solvent model20. Calculations using implicit solvent models reduce the computational cost by avoiding computations associated with water, which comprises often the largest part of the system. MDFF results obtained with the Born model were shown to provide better agreement with the corresponding results in explicit solvent than in-vacuo fittings.20 Since the implicit solvent feature was not yet available at the time of the challenge, the benefits cannot be showcased here. Readers are referred to Tanner et al.20 for more information on implicit-solvent MDFF.
The Cryo-EM Modeling Challenge 2010 was the first community effort to evaluate the performance of different hybrid methods in a systematic way. Structural biologists wishing to apply hybrid methods now have more information to decide which approach is most appropriate to their particular problem. Although concrete conclusions are not made in this first attempt of the challenge, it is expected that the next challenge will provide more information for potential users of hybrid methods. Such efforts also motivate researchers to push the boundaries of their methods by applying them to new kinds of problems. As an example, the first proof-of-principle application of MDFF to crystallographic data resulted from the challenge. The hybrid modeling field as a whole stands to benefit greatly from such community efforts. Method developers can learn from each other and further improve their own methods in an iterative way, and opportunities arise for combining multiple methods to tackle modeling tasks of ever increasing difficulty.
Materials and Methods
The following structures were used as the starting point for the MDFF simulations presented in this paper: PDB 3E7658 was fitted into the 4-Å GroEL map4; PDB 2C7D59 was fitted into the 7.7-Å GroEL-GroES map59; PDB 3LOS3 was fitted into the 4.3-Å map of Mm-cpn in a closed state3; a homology model of a lidless Mm-cpn complex was built with Modeller60 using PDB 3LOS as a template and fitted into the 8-Å map of lidless Mm-cpn in an open state3; PDB 3M9I57 was fitted into the 2.5-Å aquaporin 0 map57; PDB 1QHD61 was fitted into the 3.8-Å map of a VP6 component of rotavirus5; and PDB 2WDG/2WDI62 was fitted into the 6.4-Å ribosome map63 after removing bound factors. Each structure was first rigid-body docked into the corresponding density maps using colores from the Situs package64.
Aquaporin-0 was embedded in a POPE lipid bilayer of size 100 Å× 100 Å, with TIP3P65 water molecules added with a 15-Å padding in the direction orthogonal to the membrane. The ribosome system was simulated in vacuo, whereas the remaining structures were solvated in a TIP3P water box with a 10-Å padding in all directions. All systems simulated in explicit solvent had their total charge neutralized by adding Na+ or Cl− ions.
MD simulations were performed with a development version of NAMD 2.8.50 The CHARMM27 force field with CMAP corrections66,67 was used for all systems except for the ribosome, which was simulated with the AMBER99 force field68 including the SB69 and BSC070 corrections, and converted to CHARMM format27 to allow for system building with VMD.49 The dielectric constant was set to 80 for the ribosome simulation in vacuo and to 1 for the remaining systems. Temperature was maintained at 300 K using Langevin dynamics with a damping constant of 5 ps−1. For the aquaporin system (embedded in a membrane), pressure was maintained at 1 atm employing a Nosé-Hoover-Langevin piston with a decay period of 200 fs and time constant of 100 fs. The RESPA71,72 multiple-time-stepping algorithm was used with an integration time step of 1 fs, short-range forces calculated every 2 fs, and long-range electrostatics calculated every 4 fs. Nonbonded interactions were calculated with a 10-Å cut-off. Long-range electrostatic forces were computed by the particle-mesh Ewald summation method using a grid spacing smaller than 1 Å. All systems simulated in explicit solvent were subjected to restrained MD simulations prior to flexible fitting, with harmonic restraints first applied to all protein atoms, followed by restraints applied only to backbone atoms, thus allowing water, ions, lipids (in case of aquaporin), and side chains to equilibrate.
The grid scaling, an MDFF parameter that controls the balance between the map-derived potential energy term and the normal MD force field, was set to 0.3.12 Harmonic restraints were applied to enforce correct chirality and peptide bond configuration.13 Each MDFF simulation was performed until convergence of the protein RMSD, followed by 3,000 steps of energy minimization in the presence of the density-derived MDFF energy term and a grid scaling of 10. For symmetry-restrained MDFF simulations,14 only Cα atoms experienced symmetry restraints, with the force constant linearly increased from 0 to 10 kcal/mol/Å2 throughout the simulations.
Acknowledgments
This work was supported by grants from the National Institutes of Health (P41-RR005969) and the National Science Foundation (PHY0822613). L.G.T. and E.S. were supported by fellowships from the European Molecular Biology Organization and the Humboldt Foundation, respectively. The authors acknowledge the Texas Advanced Computing Center (TACC) at the University of Texas at Austin for providing HPC resources under grant number MCA93S028 and the Extreme Science and Engineering Discovery Environment (XSEDE) supported by National Science Foundation for providing computing resources under grant number OCI-1053575. All-atom MD simulations discussed here were performed using the package NAMD.50 Molecular images in this article were rendered using the molecular visualization software VMD.49
References
- [1].Frank J. Quart Rev Biophys. 2009;42:139–158. doi: 10.1017/S0033583509990059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Fischer N, Konevega AL, Wintermeyer W, Rodnina MV, Stark H. Nature. 2010;466:329–333. doi: 10.1038/nature09206. [DOI] [PubMed] [Google Scholar]
- [3].Zhang J, Baker ML, Schröder GF, Douglas NR, Reissmann S, Jakana J, Dougherty M, Fu CJ, Levitt M, Ludtke SJ, Frydman J, Chiu W. Nature. 2010;463:379–383. doi: 10.1038/nature08701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Ludtke SJ, Baker ML, Chen D-H, Song J-L, Chuang DT, Chiu W. Structure. 2008;16:441–448. doi: 10.1016/j.str.2008.02.007. [DOI] [PubMed] [Google Scholar]
- [5].Zhang X, Settembre E, Xu C, Dormitzer PR, Bellamy R, Harrison SC, Grigorieff N. Proc Natl Acad Sci USA. 2008;105:1867–1872. doi: 10.1073/pnas.0711623105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Baker ML, Zhang J, Ludtke SJ, Chiu W. Nat Protoc. 2010;5:1697–1708. doi: 10.1038/nprot.2010.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Hryc CF, Chen D-H, Chiu W. Curr Opin Virol. 2011;1:110–117. doi: 10.1016/j.coviro.2011.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Zhang R, Hryc CF, Cong Y, Liu X, Jakana J, Gorchakov R, Baker ML, Weaver SC, Chiu W. EMBO J. 2011;30:3854–3863. doi: 10.1038/emboj.2011.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Maki-Yonekura S, Yonekura K, Namba K. Nat Struct Mol Biol. 2010;17:417–422. doi: 10.1038/nsmb.1774. [DOI] [PubMed] [Google Scholar]
- [10].Cheng L, Zhu J, Hui WH, Zhang X, Honig B, Fang Q, Zhou ZH. J Mol Biol. 2010;397:852–863. doi: 10.1016/j.jmb.2009.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Trabuco LG, Villa E, Schreiner E, Harrison CB, Schulten K. Methods. 2009;49:174–180. doi: 10.1016/j.ymeth.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Trabuco LG, Villa E, Mitra K, Frank J, Schulten K. Structure. 2008;16:673–683. doi: 10.1016/j.str.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Schreiner E, Trabuco LG, Freddolino PL, Schulten K. BMC Bioinform. 2011;12:190. doi: 10.1186/1471-2105-12-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Chan K-Y, Gumbart J, McGreevy R, Watermeyer JM, Sewell BT, Schulten K. Structure. 2011;19:1211–1218. doi: 10.1016/j.str.2011.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Petrova T, Podjarny A. Rep Progr Phys. 2004;67:1565–1605. [Google Scholar]
- [16].Tronrud DE. Acta Cryst D. 2004;60:2165–2168. doi: 10.1107/S090744490402356X. [DOI] [PubMed] [Google Scholar]
- [17].Kleywegt GJ. Acta Cryst D. 1996;52:842–857. doi: 10.1107/S0907444995016477. [DOI] [PubMed] [Google Scholar]
- [18].Hendrickson WA. Meth Enzym. 1985;115:252–270. doi: 10.1016/0076-6879(85)15021-4. [DOI] [PubMed] [Google Scholar]
- [19].Villa E, Sengupta J, Trabuco LG, LeBarron J, Baxter WT, Shaikh TR, Grassucci RA, Nissen P, Ehrenberg M, Schulten K, Frank J. Proc Natl Acad Sci USA. 2009;106:1063–1068. doi: 10.1073/pnas.0811370106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Tanner DE, Chan K-Y, Phillips J, Schulten K. J Chem Theor Comp. 2011 doi: 10.1021/ct200563j. Article ASAP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Sener MK, Hsin J, Trabuco LG, Villa E, Qian P, Hunter CN, Schulten K. Chem Phys. 2009;357:188–197. doi: 10.1016/j.chemphys.2009.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Hsin J, Gumbart J, Trabuco LG, Villa E, Qian P, Hunter CN, Schulten K. Biophys J. 2009;97:321–329. doi: 10.1016/j.bpj.2009.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Gumbart J, Trabuco LG, Schreiner E, Villa E, Schulten K. Structure. 2009;17:1453–1464. doi: 10.1016/j.str.2009.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Seidelt B, Innis CA, Wilson DN, Gartmann M, Armache J-P, Villa E, Trabuco LG, Becker T, Mielke T, Schulten K, Steitz TA, Beckmann R. Science. 2009;326:1412–1415. doi: 10.1126/science.1177662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Becker T, Bhushan S, Jarasch A, Armache J-P, Funes S, Jossinet F, Gumbart J, Mielke T, Berninghausen O, Schulten K, Westhof E, Gilmore R, Mandon EC, Beckmann R. Science. 2009;326:1369–1373. doi: 10.1126/science.1178535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Trabuco LG, Harrison CB, Schreiner E, Schulten K. Structure. 2010;18:627–637. doi: 10.1016/j.str.2010.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Trabuco LG, Schreiner E, Eargle J, Cornish P, Ha T, Luthey-Schulten Z, Schulten K. J Mol Biol. 2010;402:741–760. doi: 10.1016/j.jmb.2010.07.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Kim H, Hsin J, Liu Y, Selvin PR, Schulten K. Structure. 2010;18:1443–1449. doi: 10.1016/j.str.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Bhushan S, Gartmann M, Halic M, Armache JP, Jarasch A, Mielke T, Berninghausen O, Wilson DN, Beckmann R. Nat Struct Mol Biol. 2010;17:313–317. doi: 10.1038/nsmb.1756. [DOI] [PubMed] [Google Scholar]
- [30].Lorenz M, Holmes KC. Proc Natl Acad Sci USA. 2010;107:12529–12534. doi: 10.1073/pnas.1003604107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Bhushan S, Meyer H, Starosta AL, Becker T, Mielke T, Berninghausen O, Sattler M, Wilson DN, Beckmann R. Mol Cell. 2010;40:138–146. doi: 10.1016/j.molcel.2010.09.009. [DOI] [PubMed] [Google Scholar]
- [32].Fu J, Hashem Y, Wower I, Lei J, Liao HY, Zwieb C, Wower J, Frank J. EMBO J. 2010;29:3819–3825. doi: 10.1038/emboj.2010.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Wei F, Bro P, Giudic E, Rollan J-P, Thoma D, Felde B, Gillet R. EMBO J. 2010;29:3810–3818. doi: 10.1038/emboj.2010.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Li W, Trabuco LG, Schulten K, Frank J. Proteins Struct Func Bioinf. 2011;79:1478–1486. doi: 10.1002/prot.22976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Agirrezabala X, Schreiner E, Trabuco LG, Lei J, Ortiz-Meoz RF, Schulten K, Green R, Frank J. EMBO J. 2011;30:1497–1507. doi: 10.1038/emboj.2011.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Frauenfeld J, Gumbart J, van der Sluis EO, Funes S, Gartmann M, Beatrix B, Mielke T, Berninghausen O, Becker T, Schulten K, Beckmann R. Nat Struct Mol Biol. 2011;18:614–621. doi: 10.1038/nsmb.2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Armache J-P, Jarasch A, Anger AM, Villa E, Becker T, Bhushan S, Jossinet F, Habeck M, Dinda G, Franckenber S, Marquz V, Mielk T, Thomm M, Berninghausen O, Beatrix B, Södina J, Westho E, Wilso DN, Beckman R. Proc Natl Acad Sci USA. 2011;107:19754–19759. doi: 10.1073/pnas.1010005107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Ghosh-Kumar M, Alam TI, Draper B, Stack JD, Rao VB. Nucleic Acids Res. 2011;39:2742–2755. doi: 10.1093/nar/gkq1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Becker T, Armache J-P, Jarasch A, Anger AM, Villa E, Sieber H, Motaal BA, Mielke T, Berninghausen O, Beckmann R. Nat Struct Mol Biol. 2011;18:715–720. doi: 10.1038/nsmb.2057. [DOI] [PubMed] [Google Scholar]
- [40].Yassin AS, Agrawal RK, Banavali NK. PLoS One. 2011;6:e21771. doi: 10.1371/journal.pone.0021871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Wollmann P, Cui S, Viswanathan R, Berninghausen O, Wells MN, Moldt M, Witte G, Butryn A, Wendler P, Beckmann R, Auble DT, Hopfner K-P. Nature. 2011;475:403–407. doi: 10.1038/nature10215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Kostyuchenko VA, Jakana J, Liu X, Haddow AD, Aung M, Weaver SC, Chiu W, Lok S-M. J Virol. 2011;85:9327–9333. doi: 10.1128/JVI.05015-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Guo Q, Yuan Y, Xu Y, Feng B, Liu L, Chen K, Sun M, Yang Z, Lei J, Gao N. Proc Natl Acad Sci USA. 2011;108:13100–13105. doi: 10.1073/pnas.1104645108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Bai X-C, Pan X-J, Wang X-J, Ye Y-Y, Chang L-F, Leng D, Lei J, Sui S-F. Structure. 2011;19:1328–1337. doi: 10.1016/j.str.2011.06.013. [DOI] [PubMed] [Google Scholar]
- [45].Strunk BS, Loucks CR, Su M, Vashisth H, Cheng S, Schilling J, Brooks CL, III, Karbstein K, Skiniotis G. Science. 2011;333:1449–1453. doi: 10.1126/science.1208245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Ludtke SJ, Baldwin PR, Chiu W. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- [47].Egelman EH. Ultramicroscopy. 2000;85:225–234. doi: 10.1016/s0304-3991(00)00062-0. [DOI] [PubMed] [Google Scholar]
- [48].van Heel M, Harauz G, Oriova EV, Schmidt R, Schatz M. J Struct Biol. 1996;116:17–24. doi: 10.1006/jsbi.1996.0004. [DOI] [PubMed] [Google Scholar]
- [49].Humphrey W, Dalke A, Schulten K. J Mol Graphics. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- [50].Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. J Comp Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Das R, Baker D. Annu Rev Biochem. 2008;77:363–382. doi: 10.1146/annurev.biochem.77.062906.171838. [DOI] [PubMed] [Google Scholar]
- [52].Schröder GF, Brunger AT, Levitt M. Structure. 2007;15:1630–1641. doi: 10.1016/j.str.2007.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Baker ML, Zhang J, Ludtke SJ, Chiu W. Nat Protoc. 2010;5:1697–1708. doi: 10.1038/nprot.2010.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Baker ML, Baker MR, Hryc CF, DiMaio F. Meth Enzym. 2010;483:1–29. doi: 10.1016/S0076-6879(10)83001-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Jolley CC, Wells SA, Fromme P, Thorpe MF. Biophys J. 2008;94:1613–1621. doi: 10.1529/biophysj.107.115949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Farrell DW, Speranskiy K, Thorpe MF. Proteins Struct Func Bioinf. 2010;78:2908–2921. doi: 10.1002/prot.22810. [DOI] [PubMed] [Google Scholar]
- [57].Hite RK, Li Z, Walz T. EMBO J. 2010;29:1652–1658. doi: 10.1038/emboj.2010.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Kiser PD, Lorimer GH, Palczewski K. Acta Cryst F. 2009;65:967–971. doi: 10.1107/S1744309109032928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Ranson NA, Clare DK, Farr GW, Houldershaw D, Horwich AL, Saibil HR. Nat Struct Mol Biol. 2006;13:147–152. doi: 10.1038/nsmb1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Sali A, Blundell TL. J Mol Biol. 1993;234:779. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- [61].Mathieu M, Petitpas I, Navaza J, Lepault J, Kohli E, Pothier P, Prasad B, Cohen J, Rey FA. EMBO J. 2001;20:1485–1497. doi: 10.1093/emboj/20.7.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Voorhees RM, Weixlbaumer A, Loakes D, Kelley AC, Ramakrishnan V. Nat Struct Mol Biol. 2009;16:528–533. doi: 10.1038/nsmb.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Schuette J-C, Murphy FV, Kelley AC, Weir JR, Giesebrecht J, Connell SR, Loerke J, Mielke T, Zhang W, Penczek PA, Ramakrishnan V, Spahn CMT. EMBO J. 2009;28(6):755–65. doi: 10.1038/emboj.2009.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Wriggers W, Milligan RA, McCammon JA. J Struct Biol. 1999;125:185–195. doi: 10.1006/jsbi.1998.4080. [DOI] [PubMed] [Google Scholar]
- [65].Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. J Chem Phys. 1983;79:926–935. [Google Scholar]
- [66].MacKerell AD, Jr., Bashford D, Bellott M, Dunbrack RL, Jr., Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, Joseph D, Kuchnir L, Kuczera K, Lau FTK, Mattos C, Michnick S, Ngo T, Nguyen DT, Prodhom B, Reiher IWE, Roux B, Schlenkrich M, Smith J, Stote R, Straub J, Watanabe M, Wiorkiewicz-Kuczera J, Yin D, Karplus M. J Phys Chem B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- [67].MacKerell AD, Jr., Feig M, Brooks CL., III J Comp Chem. 2004;25:1400–1415. doi: 10.1002/jcc.20065. [DOI] [PubMed] [Google Scholar]
- [68].Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KM, Jr., Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA. J Am Chem Soc. 1995;117:5179–5197. [Google Scholar]
- [69].Hornak V, Abel R, Okur A, Strockbine B, Roitberg A, Simmerling C. Proteins. 2006;65:712–725. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Perez A, Marchán I, Svozil D, Sponer J, Cheatham TE, Laughton CA, Orozco M. Biophys J. 2007;92:3817–3829. doi: 10.1529/biophysj.106.097782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Grubmüller H, Heller H, Windemuth A, Schulten K. Mol Sim. 1991;6:121–142. [Google Scholar]
- [72].Tuckerman M, Berne BJ, Martyna GJ. J Chem Phys. 1992;97:1990–2001. [Google Scholar]