Abstract
Most flagellated green algae swim toward or away from the light source, i.e., display positive or negative phototaxis. Usually, the sign of phototaxis, positive or negative, is variable depending on various factors, but the mechanism that controls it has been unclear. Using Chlamydomonas reinhardtii, an excellent model organism for studying phototaxis, we have recently shown that cellular redox poise plays a key role; cells show positive phototaxis when treated with reactive oxygen species (ROS), whereas they show negative phototaxis when treated with ROS quenching reagents. Here we discuss the possible mechanism of the redox regulation of phototactic sign, questions to be clarified in the future and its possible application.
Keywords: Chlamydomonas, cilia, flagella, green algae, phototaxis, redox
Phototaxis is a behavior of organisms to move toward or away from the incident light (called positive or negative phototaxis, respectively). Phototrophic microorganisms switch between positive and negative phototaxis to inhabit under proper light conditions for photosynthesis, like the chloroplasts in higher plant leaves move to optimize their photosynthesis. A unicellular green alga Chlamydomonas reinhardtii is an excellent model organism to study phototaxis because cells quickly show phototactic turning upon light perception, and a number of mutants defective in phototaxis have been isolated. Molecular mechanisms of photoreception at the eyespot and the regulation of flagellar beating for phototactic turning have been extensively studied in Chlamydomonas.1 The switching mechanism of the phototactic sign has also been studied. Factors that have been shown to affect it include light intensity, circadian rhythms and ionic strength of the extracellular solution.2-4 However, the intrinsic factor that determines the phototactic sign has been an open question. In our recent study, we focused on the cytoplasmic redox state as a candidate determinant of the phototactic sign.5
The cytoplasm is kept in a moderately reduced redox state by several mechanisms that include glutathione and thioredoxin (called redox homeostasis).6 However, it can transiently change to relatively oxidized or overly reduced states upon alterations in activities of photosynthesis or respiration, or upon exposure to external factors. Cellular reactions against such changes in the redox state are called redox regulation. In this decade, the redox regulation has become very important in the cell biology field, as many kinds of cellular activities have been shown to be regulated by redox poise: for example, the regulation of intracellular Ca2+ concentration and chemoattraction of leukocytes for wound detection.7,8
Previous studies showed that (1) photosynthetic activity affects the phototactic sign; (2) photosynthetic activity affects the cellular redox poise; and (3) redox poise modulates flagellar motility.9-11 Hence, we surmised that the redox poise could be a good candidate for the determinant of the phototactic sign. After treatments with reactive oxygen species (ROS) reagents to oxidize the cytoplasm, Chlamydomonas cells showed positive phototaxis, while they showed negative phototaxis after treatments with ROS-quenching reagents (Fig. 1). Use of a chlorophyll-deficient mutant assured that the effects of those ROS and ROS-quenching reagents (called “redox reagents”) were not through any changes in the photosynthetic activity.12 This was further confirmed by our direct measurements of photosynthetic activity in Chlamydomonas cells in the presence and absence of the redox agents (data not shown). These data suggest that the cytoplasmic redox poise determines the phototactic sign: when it becomes oxidized, the cells show positive phototaxis, and vice versa.
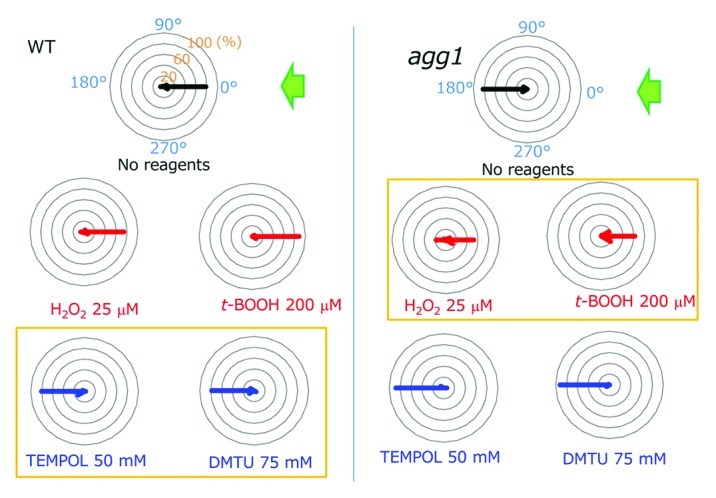
Figure 1. Summary of the effects of ROS or ROS scavengers on the phototactic sign. Polar histograms depict the percentage of cells moving in a particular direction relative to light illuminated from the right, with or without treatment with redox reagents (12 bins of 30°, n = 50 cells per condition). Wild type cells exhibited positive whereas agg1 mutant cells exhibited negative phototaxis without treatment with any reagents. Note that wild type cells exhibited negative phototaxis after treatment with ROS scavengers (blue), whereas agg1 cells showed positive phototaxis after treatment with ROS (red) (histograms in yellow boxes). Modified from Figure 2 in Wakabayashi et al.5
Redox reagents clearly switched the phototactic sign of Chlamydomonas cells. However, several questions remain to be answered. First, how much do those reagents affect the cytoplasmic redox potential? Since some ROS-quenching reagents not only quench ROS but also produce different types of ROS during itsvquenching reaction, it is necessary to monitor the change in the cellular redox potential after treatment with redox reagents to elucidate the true effects of the redox reagents.13 Several techniques have been developed to quantify the cellular redox potential in vivo, such as “redox-sensitive GFP” (roGFP, a GFP variant which functions as a ratiometric dual emission redox potential sensor) and “Redoxflour” (tandemly linked CFP and YFP, which show fluorescence resonance energy transfer in a redox-dependent manner).14-16 Though it may be difficult to directly utilize these fluorescence-based techniques in phototrophic organisms that have strong chloroplast auto-fluorescence, quantification of the cytoplasmic redox potential is clearly the next challenge. Second, how does the redox regulation of phototactic sign contribute to cell viability in nature? One possibility is that it helps cells to maintain the redox homeostasis of the cytoplasm: when the cytoplasm is oxidized, cells migrate toward light and increase photosynthesis activity, and thus the cytoplasm becomes reduced because of the reducing equivalents brought from chloroplast. However, in the chloroplast, stronger light may produce greater amounts of ROS, which may make the cellular redox state more oxidized if diffused into the cytoplasm. To clarify the overall effect resulting from the two apparently opposing effects, further understanding of the mechanisms of redox homeostasis and the relationship between cytoplasmic and chloroplast redox states is necessary.
With our present discovery of redox-dependent phototactic sign, we can now control the swimming direction of Chlamydomonas cells using redox reagents and light. An obvious application of this finding would be to concentrate algae at will. When dimethylthiourea (DMTU), a ROS-quenching reagent, is added to Chlamydomonas wild-type cell culture and green light is applied from above, cells quickly moved to the bottom of the bottle (Fig. 2). By discarding the upper phase, we can easily obtain ~10 fold denser cell culture without using pumps or centrifuges, which is harmful for cells. This method should contribute to industrial use of algae as bioenergy source, as well as to basic science of motile phototrophic algae.

Figure 2. Concentration of Chlamydomonas cells based on the redox regulation of phototaxis. About 1-liter culture of wild type cells were treated with 75 mM dimethylthiourea (DMTU) and illuminated with a green LED from the top.
Acknowledgments
We thank Drs Ichiro Terashima and Ko Noguchi (University of Tokyo) for the measurement of photosynthetic activity of Chlamydomonas cells after treatment with the redox reagents and for their helpful discussion and Dr Ritsu Kamiya (University of Tokyo) for his critical reading of the manuscript. This work was supported by a grant from Japan Society for Promotion of Sciences (#22770189) to K.W. and a grant from the Asahi Glass Foundation to K.W.
Footnotes
Previously published online: www.landesbioscience.com/journals/cib/article/18890
References
- 1.Hegemann P, Berthold P. Sensory photoreceptors and light control of flagellar activity. The Chlamydomonas Sourcebook Second Edition. Waltham, MA: Academic Press, 2009:395-430. [Google Scholar]
- 2.Feinleib MEH, Curry GM. The relationship between stimulus intensity and oriented phototactic response (topotaxis) in Chlamydomonas. Physiol Plant. 1971;25:346–52. doi: 10.1111/j.1399-3054.1971.tb01453.x. [DOI] [Google Scholar]
- 3.Morel-Laurens N. Calcium control of phototactic orientation in Chlamydomonas reinhardtii: sign and strength of response. Photochem Photobiol. 1987;45:119–28. doi: 10.1111/j.1751-1097.1987.tb08412.x. [DOI] [PubMed] [Google Scholar]
- 4.Kondo T, Johnson CH, Hastings JW. Action spectrum for resetting the circadian phototaxis rhythm in the CW15 strain of Chlamydomonas 1. Cells in darkness. Plant Physiol. 1991;97:197–205. doi: 10.1104/pp.95.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wakabayashi K, Misawa Y, Mochiji S, Kamiya R. Reduction-oxidation poise regulates the sign of phototaxis in Chlamydomonas reinhardtii. Proc Natl Acad Sci U S A. 2011;108:11280–4. doi: 10.1073/pnas.1100592108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wheeler GL, Grant CM. Regulation of redox homeostasis in the yeast Saccharomyces cerevisiae. Physiol Plant. 2004;120:12–20. doi: 10.1111/j.0031-9317.2004.0193.x. [DOI] [PubMed] [Google Scholar]
- 7.Higo T, Hattori M, Nakamura T, Natsume T, Michikawa T, Mikoshiba K. Subtype-specific and ER lumenal environment-dependent regulation of inositol 1,4,5-trisphosphate receptor type 1 by ERp44. Cell. 2005;120:85–98. doi: 10.1016/j.cell.2004.11.048. [DOI] [PubMed] [Google Scholar]
- 8.Niethammer P, Grabher C, Look AT, Mitchison TJ. A tissue-scale gradient of hydrogen peroxide mediates rapid wound detection in zebrafish. Nature. 2009;459:996–9. doi: 10.1038/nature08119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takahashi T, Watanabe M. Photosynthesis modulates the sign of phototaxis of wild-type Chlamydomonas reinhardtii. Effects of red background illumination and 3-(3′,4′-dichlorophenyl)-1,1-dimethylurea. FEBS Lett. 1993;336:516–20. doi: 10.1016/0014-5793(93)80867-T. [DOI] [PubMed] [Google Scholar]
- 10.Forti G, Furia A, Bombelli P, Finazzi G. In vivo changes of the oxidation-reduction state of NADP and of the ATP/ADP cellular ratio linked to the photosynthetic activity in Chlamydomonas reinhardtii. Plant Physiol. 2003;132:1464–74. doi: 10.1104/pp.102.018861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wakabayashi K, King SM. Modulation of Chlamydomonas reinhardtii flagellar motility by redox poise. J Cell Biol. 2006;173:743–54. doi: 10.1083/jcb.200603019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang WY, Boynton JE, Gillham NW. Genetic control of chlorophyll biosynthesis: effect of increased delta-aminolevulinic acid synthesis on the phenotype of the y-1 mutant of Chlamydomonas. Mol Gen Genet. 1977;152:7–12. doi: 10.1007/BF00264933. [DOI] [Google Scholar]
- 13.Mochizuki M, Yamazaki S, Kano K, Ikeda T. Kinetic analysis and mechanistic aspects of autoxidation of catechins. Biochim Biophys Acta. 2002;1569:35–44. doi: 10.1016/S0304-4165(01)00230-6. [DOI] [PubMed] [Google Scholar]
- 14.Hanson GT, Aggeler R, Oglesbee D, Cannon M, Capaldi RA, Tsien RY, et al. Investigating mitochondrial redox potential with redox-sensitive green fluorescent protein indicators. J Biol Chem. 2004;279:13044–53. doi: 10.1074/jbc.M312846200. [DOI] [PubMed] [Google Scholar]
- 15.Dooley CT, Dore TM, Hanson GT, Jackson WC, Remington SJ, Tsien RY. Imaging dynamic redox changes in mammalian cells with green fluorescent protein indicators. J Biol Chem. 2004;279:22284–93. doi: 10.1074/jbc.M312847200. [DOI] [PubMed] [Google Scholar]
- 16.Yano T, Oku M, Akeyama N, Itoyama A, Yurimoto H, Kuge S, et al. A novel fluorescent sensor protein for visualization of redox states in the cytoplasm and in peroxisomes. Mol Cell Biol. 2010;30:3758–66. doi: 10.1128/MCB.00121-10. [DOI] [PMC free article] [PubMed] [Google Scholar]


