Abstract
Partial peptide sequence of a 36 kDa protein from common bean embryo axes showed 100% identity with a reported β-subunit of a heterotrimeric G protein from soybean. Analysis of the full sequence showed 96.6% identity with the reported soybean Gβ -subunit, 86% with RACK1B and C from Arabidopsis and 66% with human and mouse RACK1, at the amino acid level. In addition, it showed 85.5, 85 and 83% identities with arcA from Solanum lycopersicum, Arabidopsis (RACK1A) and Nicotiana tabacum, respectively. The amino acid sequence displayed seven WD40 domains and two sites for activated protein kinase C binding. The protein showed a constant expression level but the mRNA had a maximum at 32 h post-imbibition. Western immunoblotting showed the protein in vegetative plant tissues, and in both microsomal and soluble fractions from embryo axes. Synthetic auxin treatment during germination delayed the peak of RACK1 mRNA expression to 48 h but did not affect the protein expression level while the polar auxin transport inhibitor, naphtylphtalamic acid had no effect on either mRNA or protein expression levels. Southern blot and genomic DNA amplification revealed a small gene family with at least one member without introns in the genome. Thus, the RACK1/arcA homolog from common bean has the following features: (1) it is highly conserved; (2) it is both soluble and insoluble within the embryo axis; (3) it is encoded by a small gene family; (4) its mRNA has a peak of expression at the time point of germination stop and (5) its expression is only slightly affected by auxin but unaffected by an auxin transport blocker.
Introduction
Germination and growth in plants are natural events that involve multiple molecular reactions within complex and highly regulated biochemical cascades, which will eventually give rise to a photosynthetic, autotrophic plant. The cytoskeleton and signal and adaptor proteins play key roles during those early stages and most of the plant and animal life cycle (Kost et al. 1999, Pawson and Scott 1997). Putative adaptor proteins that can be found in the genome database of Arabidopsis are gamma adaptins, and similar adaptor proteins of the clathrin complex, proteins similar to phosphoinositol 4-phosphate adaptor protein-2, transcriptional adaptors, v-SNARE homologs and WD40 repeat proteins, among others (The Arabidopsis Information Resource, TAIR). WD40 repeat proteins have evolved to be specifically designed to interact with other partners through their WD40 domains that assemble to form an interaction platform with a characteristic propeller shape (McCahill et al. 2002). The family of WD40 repeat proteins include the β-subunit of heterotrimeric G proteins, the Tup1 transcriptional repressor (Chen et al. 2004), the Prp4 splicing factor (Dalrymple et al. 1989), cdc4 (Mallet et al. 1994), the receptor for activated C kinase (RACK; McCahill et al. 2002) and the product of the auxin-regulated gene from cultured cells (arcA; Ishida et al. 1993). In plants, the Gβ -subunit of the heterotrimeric G protein is one of the few β-propeller-type proteins that have been described and characterized, and an indirect role in auxin signaling has been reported in Arabidopsis (Ullah et al. 2003). Another protein potentially involved in signal-transduction through hormone signaling is the arcA protein of tobacco. The gene that encodes this protein was isolated from a subtraction library of cells subjected to auxin treatment (Ishida et al. 1993). Proteins with similar sequences were described as being expressed in roots, stems, cotyledons, true leaves, flowers and fruits of tomato (Kiyosue and Ryan 1999), and during nodulation in Medicago sativa (McKhann et al. 1997). This gene named Msgb1 was expressed in young embryos and in leaves, and was induced in roots after cytokinin treatment. Interestingly, the transcript was most abundant in dividing cells of nodule primordia and in the nodule meristem, suggesting a role in hormone-mediated cell division. In addition, a putative Gβ -subunit of the heterotrimeric G protein from Glycine max has also been reported (Nielsen et al. 2001), but the protein showed only three N-terminal amino acids upstream the first WD repeat. Therefore, the protein lacks the amino acids that form the α-helical sequence at the N-terminal that a true Gβ requires for interaction with the Gα -subunit (Chen et al. 2006, Nielsen et al. 2001). The sequences of the arcA, Msgb1 and G. max Gβ proteins are also more related to the receptor for activated protein kinase C (RACK1) homolog from Homo sapiens and mouse than to the canonical Gβ from Arabidopsis, and thus are more likely to have functions related to this scaffolding protein. In fact, the Arabidopsis genome contains three genes encoding RACK- or arcA-related proteins besides the canonical Gβ sequence. RACK1 proteins play key roles in signal-transduction cascades in animal cells, which range from cytoskeleton rearrangements (Osmanagic-Myers and Wiche 2004) to protein translation regulation (Nilsson et al. 2004). Recently, the function of the RACK1A gene, the homolog of arcA in Arabidopsis, was studied in loss-of-function mutants (Chen et al. 2006). These mutants showed numerous defects in developmental processes that included seed germination, leaf production and flowering. During seed germination, they had a low sensitivity to brassinosteroids and gibberellin, and hypersensitivity to abscisic acid. It was concluded that RACK1A from Arabidopsis is involved in multiple signal-transduction processes that are also related to auxin. Furthermore, in a recent study, triple RACK Arabidopsis mutants were barely able to survive at early growth stages and could not survive until maturity, although single and double mutants were viable and showed various degrees of developmental defects suggesting functional substitution (Guo and Chen 2008). As it appears that this protein plays a fundamental role in processes that lead to cell division and proliferation in both plants and animals, it is of great interest to characterize its homologs in various organisms, especially those that undergo active cell proliferation. In this work, we isolated a 36 kDa protein from common bean embryo axes whose internal partial peptide sequence showed 100% identity with a reported β-subunit of a heterotrimeric G protein. We obtained the full cDNA and genomic sequence of this protein, and identified it as the RACK1 homolog from common bean. We describe here its sequence and inherent features, cell and tissue distribution, and its expression during normal germination and under synthetic auxin treatment.
Materials and methods
Plant material
Common bean (Phaseolus vulgaris, Negro Jamapa variety) seeds were sterilized with 10% bleach for 5 min with stirring, briefly rinsed with distilled water and added with 100% ethanol and further stirred for 1 min. They were then rinsed extensively with double distilled water. The embryo axes were then excised from dry seeds with a sterile knife, dried on a paper towel and finely ground to a powder. This material was resuspended in the appropriate buffer for the isolation of the 36 kDa protein. In some cases, 15-day-old leaves or stems or 72 h post-imbibed roots were used for the preparation of extracts as described below.
Isolation and internal peptide sequence of the 36 kDa protein
All extraction procedures were carried out at 4°C. The embryo axis powder was resuspended at a ratio of 0.02 g/ml of 10 mM phosphate buffer (PB; 2.8 mM NaH2PO4, 7.2 mM Na2HPO4, pH 7.4). The suspension was stirred until completely hydrated and resuspended thoroughly. This suspension was then centrifuged at 15 000 g for 15 min and the supernatant was fractionated in a DEAE-Sephacel column (5.2 × 1 cm) previously equilibrated in PB. The column was washed extensively with PB buffer and then eluted with a gradient of 0–0.4 M NaCl in PB. The first six-eluted fractions were pooled and concentrated, and this procedure was repeated several times to obtain enough protein for sequencing. The concentrated sample was run on SDS–PAGE gels, and stained with Coomassie blue, excised from the gel and then completely destained. The stained band was cut and sent for trypsin digestion and sequencing to the Harvard Microchemistry Facility of Harvard University (Harvard, MA). Sequencing was carried out by microcapillary reverse-phase HPLC nanoelectrospray tandem mass spectrometry (μLC/MS/MS) on a Finnigan LCQ Deca quadrupole mass spectrometer.
Synthesis and antibody production against a peptide from the 36 kDa protein
A decapeptide from the amino terminal of the obtained sequences was chosen for synthesis and antibody production. The peptide with the sequence AIATPIDNSD and its corresponding antibody were custom ordered from Biosynthesis Inc. (Lewisville, TX). The decapeptide was conjugated to keyhole limpet hemocyanin for antiserum production in rabbits and the hyperimmune serum collected from the manufacturer. This antibody was termed anti-PA218. The specificity of the antibody was assessed by detecting a distinct 36 kDa band on total extracts from embryo axes extracts from both soybean and common bean, and no reaction with the pre-immune serum. In addition, the antibody specificity was tested in a peptide competition assay (see below).
cDNA sequence of RT–PCR and 3′-RACE fragments
Surface-sterilized and pre-washed common bean seeds were further washed (briefly) with diethyl pyrocarbonate (DEPC)-treated sterile water and air-dried. The embryo axes were dissected and placed immediately in a pre-chilled mortar containing liquid nitrogen in which a fine powder was prepared by grinding. Total RNA was extracted from this embryo axis powder with either the Tri™ reagent (Molecular Research Center Inc., Cincinnati, OH) or the original method of Chomczynski and Sacchi (1987), and the final RNA pellet was resuspended in DEPC-treated sterile water and stored in aliquots at −70°C until use. This RNA was used as template, and complimentary DNA was synthesized using reverse transcriptase followed by amplification by the polymerase chain reaction (PCR) with the platinum™ PCR SuperMix and platinum™ Taq DNA polymerase (Invitrogen, Carlsbad, CA, USA). The combinations of oligonucleotides for amplification were: (1) forward 5′-CTCTCATCCGACGGTCA-3′, reverse 5′-TGAAGCAGAGTGCATGGA-3′; (2) forward 5′-GCGTTTCAGCCCCAGCA-3′, reverse 5′-CTTCCAT CCGCACTCCA-3′ and (3) forward 5′-CACACCGACGTCGTGA-3′ or 5′-CACCATGGCGGAAGGACTCGTCCTGAAGG-3′, reverse 5′-ATAACGTCCAATTGCCCAAAC-3′. For 3′-rapid amplification of cDNA ends (3′-RACE), the supplied adapter primer 5′-GGCCACGCGTCGACTAGTAC(T)17-3′ was used for the cDNA synthesis, and the forward in (2) and abridged universal amplification primer 5′-GGCCACGCGTCGACTAGTAC-3′ were used for PCR according to the manufacturer’s instructions (Invitrogen). All PCR products were cloned with the TOPO TA cloning™ kit (Invitrogen) in the pCR™2.1-TOPO™ vector. Both sense and antisense chains were amplified for sequence analysis. The amplified products were analyzed on 1.4% agarose gels run in the presence of ethidium bromide. Amplified and cloned cDNA products were purified using the QuiaPrep™ Spin miniprep kit (Qiagen, Valencia, CA), and this DNA was sequenced in the sequencing facility of the Instituto de Biotecnología, UNAM (Cuernavaca, Morelos, Mexico). The total sequence of the cDNA was obtained and analyzed with the Editview program (Applied Biosystems, Foster City, CA). The sequence was reported to the Gen-Bank with accession number EU906910. For expression, amplified products were cloned in the pCR™T7/CT-TOPO™ vector (Invitrogen) and expressed in Escherichia coli cells.
Promoter sequence determination and analysis
The PvRACK1 promoter was obtained from a promoter library. Genomic DNA was digested with the enzymes EcoRV, PvuI, StuI and DraI. An adapter was placed at the 5′-end of fragments derived from the digestions to facilitate the amplification with specific primers. In order to amplify the PvRACK1 promoter, two primers at the 5′-end in reverse were used. The first reaction was done with the primer 5′-CCGTAGGTCTTGTCCTCCTTGG-3′ and the primer from the library adapter AP1 5′-GTAATACGACTCACTATAGGGC-3′. The second reaction was carried out with the primer 5′-CCACAGGATGATGGATTTGTCGC-3′ and the primer from the library adapter AP2 5′-ACTATAGGGCACGCGTGGT-3′. The amplified fragments were sequenced at the sequencing facility of the Instituto de Biotecnología, UNAM. The sequence corresponding to the promoter region was searched for regulatory regions using the FASTA format of upstream genomic sequence adjacent to the PvRACK1 coding region in a sequence comparison with the database of plant cis-acting regulatory DNA elements (www.dna.affrc.go.jp/PLACE/; Higo et al. 1999) and the database of plant PolII promoter and start of transcription regions (http://www.softberry.ru/berry.phtml).
Phylogenetic tree assembly and construction
The phylogenetic tree was constructed using Guanine Nucleotide-Binding Protein Subunit β-like amino acid sequences with the highest identity to RACK, collected from the GenBank for the following species: Aedes aegypti (AEDAE), A. gambiae (AEDGA), Apis mellifera (APIME), Arabidopsis thaliana (ARATH), Aspergillus clavatus (ASPCL), A. fumigatus (ASPFU), A. nidulans (ASPNI), A. oryzae (ASPOR), Biomphalaria glabrata (BIOGL), Blattella germanica (BLAGE), Bombyx mori (BOMMO), Brassica napus (BRANA), Canis familiaris (CANFA), Chaetomium globosum (CHAGL), Chlamydomonas incerta (CHLIN), C. reinhardtii (CHLRE), Choristoneura fumiferana (CHOFU), Coprinopsis cinerea (COPCI), Cryptococcus neoformans (CRYNE), Danio rerio (DANRE), Drosophila melanogaster (DROME), D. pseudoobscura (DROPS), Euprymna scolopes (EUPSC), Fusarium oxysporum (FUSOX), Gallus gallus (GALGA), Gibberella zeae (GIBZE), G. max (GLYMA), Heliothis virescens (HELVI), Homalodisca coagulate (HOMCO), H. sapiens (HOMSA), Hydra vulgaris (HYDVU), Ixodes scapularis (IXOSC), Lentinula edodes (LENED), Lethenteron japonicum (LETJA), Macaca mulatta (MACMU), Maconellicoccus hirsutus (MACHI), Magnaporthe grisea (MAGGR), Mamestra brassicae (MAMBR), M. sativa (MEDSA), Metarhizium anisopliae (METAN), Mus musculus (MUSMU), Mya arenaria (MYAAR), Neosartorya fischeri (NEOFI), Neurospora crassa (NEOCR), Nicotiana plumbaginifolia (NICPL), N. tabacum (NICTA), Oreochromis mossambicus (OREMO), O. niloticus (ORENI), Ostreococcus lucimarinus (OSTLU), O. tauri (OSTTA), Oryctolagus cuniculus (ORYCU), Oryza sativa (ORYSA), Pan troglodytes (PANTR), Paralichthys olivaceus (PAROL), Petromyzon marinus (PETMA), P. vulgaris (PVRACK), Plutella xylostella (PLUXY), Rattus norvegicus (RATNO), S. lycopersicum (SOLLY), S. tuberosum (SOLTU), Tetraodon nigroviridis (TETNI), Toxoptera citricida (TOXCI), Tribolium castaneum (TRICA), Ustilago maydis (USTMA), Xenopus laevis (XENLA) and X. tropicalis (XENTR). The sequences were directed to ClustalX 1.8 (Thompson et al. 1997) for joint multiple alignment. The multiple alignment was sent to the PHYLIP 3.57 package (Lim and Zhang 1999). Within this package, we input the aligned sequences into the SEQBOOT algorithm (bootstrap sequence data sets) to create 100 data sets by bootstrap resampling. These data sets were entered into PROTDIST to generate 100 protein distance matrices. These matrices were entered into the program NEIGHBOR to produce 100 phenograms of the PvRACK1 sequences using the neighbor-joining method, followed by the creation of majority-rules, strict consensus and unrooted tree with confidence intervals using CONSENSE. The resulting phylogenetic tree was displayed and edited in the SplitsTree4 program (Huson and Bryant 2006).
SDS–PAGE western blot analysis and peptide competition assay
Dissociating polyacrylamide gels were prepared at a final concentration of 12% according to Lamely (1970). Gels were run at 30 mA and Coomassie blue-or silver-stained. SDS–PAGE gels were transferred to nitrocellulose (Towbin et al. 1979). For immunoblotting, the nitrocellulose membranes were blocked for 1 h at 50°C in 3% bovine serum albumin (BSA) in PBS (2.8 mM NaH2PO4, 7.2 mM Na2HPO4, 0.14 M NaCl, pH 7.4), followed by incubation overnight at 4°C with anti-PA218 antibodies diluted 1:5000–1:10 000 in PBST (PBS added with 0.05% Triton X-100 or Tween-20). The blots were then washed three times with PBST and incubated with alkaline phosphatase-conjugated anti-rabbit IgG, diluted 1:3500 in PBST for 2 h at 25°C. The membranes were washed three times with PBST, rinsed briefly in PBS and the antigen–antibody complex was detected with bromo chloro indolyl phosphate and nitro-blue tetrazolium according to the manufacturer (Roche Laboratories, Nutley, NJ).
For the peptide competition assay, excess molar amounts of peptide:anti-PA218 antibody at 1:1, 100:1 and 200:1, were pre-incubated overnight at 4°C, followed by the usual western blot procedure.
Soluble and microsomal fraction analysis
For this preparation, embryo axis powder was resuspended at a ratio of 0.2 g/ml of PBS-C (PBS containing the commercial protease inhibitor cocktail tablet, Complete™; Roche Laboratories; according to the manufacturer’s instructions). The suspension was stirred at 4°C as described above. The suspension was centrifuged at 20 800 g for 10 min at 4°C to collect the total microsomal fraction in the supernatant. The supernatant was then centrifuged at 100 000 g for 30 min at 4°C, and the supernatant was taken as the soluble fraction whereas the pellet was considered the total microsomal fraction. Part of this pellet was resuspended in PBS and stored for further analysis. The other part was resuspended in 0.5 M KCl-PBS and incubated for 20 min at 4°C to release ionically bound proteins. This fraction was again centrifuged at 100 000 g for 30 min at 4°C. The supernatant was stored for further analysis and the pellet was resuspended in 0.5% TX-100 in PBS and incubated for 20 min at 4°C to release intrinsically membrane bound proteins. This fraction was again centrifuged at 100 000 g for 30 min at 4°C. The supernatant was stored for further analysis and the pellet was resuspended in 0.1 M Na2CO3 and incubated for 20 min at 4°C to release intrinsically bound proteins in luminal and folded membranes. This fraction was again centrifuged at 100 000 g for 30 min at 4°C. The supernatant was stored for further analysis, and the pellet was resuspended in Laemmli’s sample buffer (Lamely 1970) and heated for 5 min at 95°C to release all the proteins that remained in the cell matrix and which were not released by the previous treatments. All the fractions were standardized for protein in Coomassie blue-stained SDS–PAGE gels and then analyzed by western blot with the anti-PA218 antibody.
Western blot analyses of three plant species and various tissues of P. vulgaris
Seeds from P. vulgaris and G. max were surface-sterilized as described above, and a powder from their embryo axes was obtained with a mortar and pestle. This powder was made into an extract by hydrating and resuspending in PBS-C, followed by centrifugation as described above. Whole Pisum sativum seeds were obtained directly from pods and frozen in liquid nitrogen. Twenty-one-day-old nodules from P. vulgaris plants inoculated with the Rhizobium etli CIAT899 strain were collected from roots and frozen in liquid nitrogen. Likewise, roots, stems and leaves from 15-day-old P. vulgaris plants, and pods from various developmental stages, were also collected and frozen in liquid nitrogen. All frozen tissues were made into a powder in the presence of liquid nitrogen and resuspended in PBS-C to make a total protein extract as described above. The obtained supernatants from all extracts were quantitated for protein by the Bradford (1976) assay and then mixed with Laemmli’s sample buffer (Lamely 1970) and heated for 5 min at 95°C prior to electrophoresis and western blot analysis.
Western and northern blot analyses of P. vulgaris embryo axes and hypocotyls during germination
Surface-sterilized seeds from P. vulgaris were placed on sterile trays lined with wet paper towels for germination in the dark at 24°C ± 1. Common bean embryo axes or hypocotyls from the seedlings were dissected at various stages of germination, frozen in liquid nitrogen and homogenized to a powder as described above. For protein analysis, the powder was extracted with PBS-C and the soluble supernatants quantified for protein with the Bradford assay (Bradford 1976). The presence of protein at various post-imbibition times was detected by western blot with anti-PA218 antibodies. For RNA analysis, RNA extracted as described above from the various post-imbibition times was quantified and loaded in equal amounts on 1.4% agarose gels. After separation, the gels were rinsed sequentially in 1 × MOPS, 10× SSC (3 M NaCl, 0.3 M Na Citrate) and 20× SSC, and transferred to Hybond-N™ nylon membranes (Amersham, Piscataway, NJ). The membranes were incubated with a pre-hybridization solution containing 0.1% BSA, 7% SDS, 0.52 M NaH2PO4, 0.5 mM ethylenediaminotetraacetic acid (EDTA), pH 7.5 and incubated 1 h at 55°C. The membrane was then incubated for 24 h at 55°C with a 500 bp cDNA fragment labeled with [32P]-γ-ATP using the Rediprime™ II from Amersham according to the manufacturer’s instructions.
For auxin and auxin transport inhibitor treatment, seeds were germinated in wet paper towel-lined trays in the dark at 24 ± 1°C but these were damped and irrigated with either 25 μM 2, 4-dichlorophenoxyacetic acid (2, 4-D), 50 μM indoleacetic acid (IAA) or 25 μM naphthylphthalamic acid (NPA). The embryo axes or hypocotyls were collected at various times during the germination period and processed for western and northern blot analyses as described above.
Southern blot and genomic DNA PCR amplification
Total genomic DNA was extracted by the cetyl trimethylammonium bromide (CTAB) method (Murray and Thompson 1980), and 30 μg were digested overnight at 37°C with 30–40 U of either BglII, BanI or KpnI. The next day the digested DNA was precipitated and separated in a 1% agarose gel. The gel was transferred to a nylon membrane (Hybond-N, Amersham) by diffusion for 24 h in 0.4 M NaOH. The membrane was then washed with 2× SSC for 5 min at 25°C, placed in a tube with pre-hybridization solution [5× Denhardt’s solution (4% Ficoll, 4% polyvinylpirrolidone (PVP), 4% BSA), 5× SSC, 1% SDS and 100 μg ml−1 salmon sperm DNA], and agitated continuously for 2 h at 42°C. Then, the solution was discarded, and the radioactive cDNA probe ([32P]-γ-ATP 936 bp’s PvRACK1 DNA) was added in the pre-hybridization solution but without the salmon sperm DNA and incubated 24 h at 50°C with continuous agitation. After this time, the solution was discarded and the membrane washed extensively with 0.1% SDS in 4× SSC, and then with 0.1% SDS in 0.1× SSC until no signal was detected at the edges of the membrane. The membrane was air-dried and the radioactively labeled bands detected on a Phosphorimager (Molecular Dynamics, Sunnyvale, CA).
The genomic DNA PCR amplification was carried out using an Invitrogen AccuPrime™ SuperMix II kit (Invitrogen) with the oligonucleotides forward PvRACK5′-CACC (5′-CACCATGGCGGAAGGACTCGTCCTG-3′) and reverse PvRACK3′-TAG (5′-CTAATAACGTCCAATTGCCCAAACTCTGACC-3′). The reaction mixture was prepared with 12.5 μl AccuPrime SuperMix II, 0.5 μl 10 μM PvRACK5′-CACC, 0.5 μl 10 μM PvRACK3′-TAG, 1 μl (100 ng μl−)P. vulgaris genomic DNA, 10.5 μl H2O. The reaction was carried out by incubating 2 min at 94°C followed by 35 amplification cycles as follows: denaturing at 94°C for 15 s; alignment at 55–60°C for 15 s and extension at 68°C for 3 min. The PCR product was cloned into a TOPO™ TA cloning kit (Invitrogen) and sent for sequencing to the sequencing facility of the Instituto de Biotecnología-UNAM.
Results
A 36 kDa protein from P. vulgaris is related to proteins with β-propeller-type structure and possesses seven WD40 domains
The enriched 36 kDa protein from bean embryo axes was run on a denaturing gel and sent for internal peptide sequencing. Table 1 shows the obtained four internal sequences that, after a blast against the GenBank (Altschul et al. 1997), were 100% identical to a sequence more closely related to RACK1 but reported as a Gβ -like subunit of a heterotrimeric protein from G. max (GenBank accession numbers: Gene LOC547868; nucleotide U44850.1; protein AAB05941.1). An internal peptide from the sequence closest to the N-terminus was used to raise an antibody in rabbits (Table 1, underlined amino acids; see below). RT–PCR and 3′-RACE (rapid amplification of cDNA ends) using oligonucleotides derived from the obtained sequences allowed us to obtain the full cDNA sequence that encodes the 36 kDa protein (Fig. 1). This sequence has been deposited in the GenBank under accession number EU906910. The GenBank annotated cDNA sequence of PvRACK1 was initially obtained by RT–PCRs with primers based on the reported soybean Gβ -subunit-like sequence; for this reason, the PvRACK1 5′ cDNA had several nucleotide changes. The sequencing of PvRACK1 genomic DNA allowed us to obtain the real 5′ RACK1 sequence which had nucleotide changes at positions 6, 9, 12, 18, 21, 22, 23, 24 and 27 (G/A, A/G, A/T, C/T, G/C, A/C, A/G, G/C and A/C, respectively). However, despite these differences, there was only one change at position 8 (K to R) at the amino acid level. This corrected sequence is showed in Fig. 1. The sequence shows seven WD40 repeats (Fig. 1, boxed sequences in white and gray), and two internal sequences (Fig. 1, underlined sequences) that represent the conserved activated protein kinase C (aPKC) binding domains. Table 2 shows an analysis comparing this and other reported similar sequences from the GenBank. This analysis revealed that the closest identities were 96.6% with the G. max RACK1-related sequence reported as Gβ -like subunit of a heterotrimeric protein, 96% with M. sativa Msgb1, and 82–87% identities with various reported proteins that included the arcA homologs of A. thaliana (RACK1A; ARATH3 in Table 2) and N. tabacum, and RACK1B and C (ARATH2 and 1 in Table 2, respectively) from A. thaliana and B. napus BGB1. Lower, nonetheless significant identities (46–69%) were found with proteins from other organisms including O. tauri and C. reinhardtii (Table 2). In addition, a separate analysis yielded 66% identity of the common bean protein with RACK1 from human and mouse (data not shown). The common feature of all these proteins was the presence of WD40 domains in their sequence. In contrast, comparison with the canonical Gβ -subunit of a heterotrimeric protein from A. thaliana (At4g34460.1) showed only 25% identity at the amino acid level (data not shown). The two internal aPKC binding sequences of the common bean protein also showed a strong identity with equivalent aPKC binding sequences in other RACK homologs and 100% identity with the RACK1 sequence reported as Gβ -like protein from G. max (Table 3). The first internal aPKC binding sequence from PvRACK1 was identical to the equivalent sequence of the RACK1 sequence reported as Gβ -like protein from G. max and other reported aPKC binding sequences from H. Sapiens. It also showed 85.7 and 57% identity to equivalent sequences from Tripanosoma cruzi, and Saccharomyces cerevisiae and S. pombe, respectively (Table 3). The second internal sequence showed a close identity with plant and mammalian homologs but very low identity with unicellular eukaryotes (Table 3). These data show that the sequence from P. vulgaris as well as the G. max sequence reported as a Gβ -like subunit of a heterotrimeric protein is more closely related to RACK1 and arcA proteins. Therefore, we hereby term this protein PvRACK1.
Table 1.
Comparison and alignment of the partial amino acid sequences of the 36 kDa protein from Phaseolus vulgaris embryo axes, with the reported Gβ -like subunit of a heterotrimeric protein from Glycine max (accession number AAB05941.1).
| Species | Amino acid sequence |
|---|---|
| Sequence number 1 | |
| P. vulgaris | AHTDVVTAIATPIDNSDMIVTASR* |
| G. max | 13 AHTDVVTAIATPIDNSDMIVTASR 36 |
| Sequence number 2 | |
| P. vulgaris | LWDLAAGTSAR |
| G. max | 89 LWDLAAGTSAR 99 |
| Sequence number 3 | |
| P. vulgaris | FSPSTLQPTIVSASWDR |
| G. max | 157 FSPSTLQPTIVSASWDR 173 |
| Sequence number 4 | |
| P. vulgaris | IWDLESK |
| G. max | 261 IWDLESK 267 |
The underlined sequence corresponds to the peptide against which antibodies were raised in rabbits.
Fig. 1.
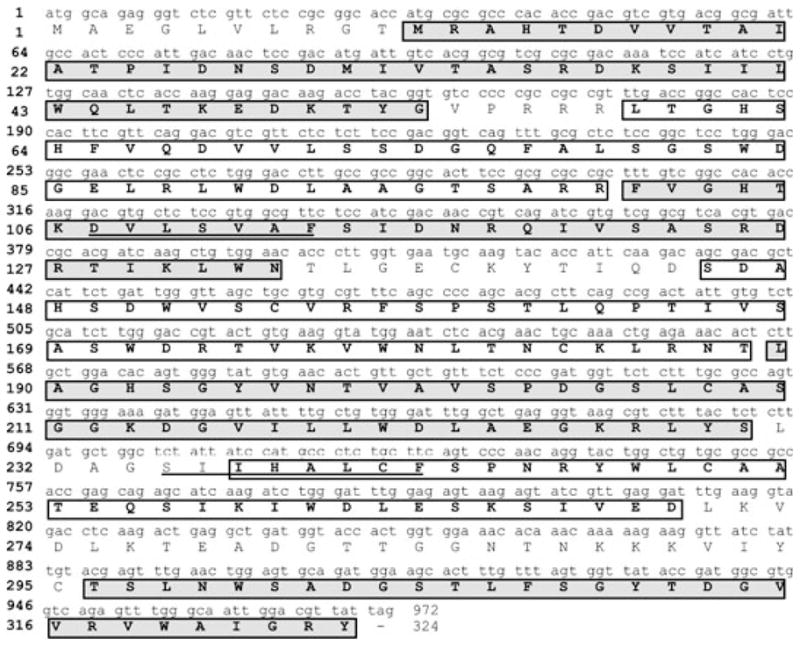
Nucleotide sequence of the cDNA that encodes PvRACK1 and its translated amino acid sequence. The 972 nucleotides translate into 324 amino acids for a protein of approximately 36 kDa. The amino acid sequence shows seven domains containing the WD40 sequence (boxed amino acids in white and gray) and two internal sequences representing the PKC binding domains (underlined amino acids).
Table 2.
Amino acid sequence identities (in percentile) among reported sequences of orthologs of PvRACK1 and other organisms, including that of the reported Gβ -like subunit of a heterotrimeric protein from Glycine max (GLYMA) which yields the highest identity score with PvRACK1. The other sequences obtained from the GenBank were those of Arabidopsis thaliana (ARATH), Brassica napus (BRANA), Chlamydomonas reinhardtii (CHLRE), Medicago sativa (MEDSA), Nicotiana plumbaginifolia (NICPL), N. tabacum (NICTA), Ostreococcus lucimarinus (OSTLU), O. tauri (OSTTA), Oryza sativa (ORYSA), Solanum lycopersicum (SOLLY), and S. tuberosum (SOLTU). Not included in the table are 66% identities of PvRACK1 to both Homo sapiens and Mus musculus RACK1. The protein alignment analysis was performed utilizing the MUSCLE and BIOEDIT software package.
| CHLRE | CHLRE | ||||||||||||||||
| OSTLU | 78.5 | OSTLU | |||||||||||||||
| OSTTA | 51.2 | 62 | OSTTA | ||||||||||||||
| ARATH3 | 67.4 | 65 | 43.1 | ARATH3 | |||||||||||||
| BRANA | 66.8 | 63.5 | 42.1 | 96 | BRANA | ||||||||||||
| ARATH2 | 67.3 | 66.7 | 44.4 | 87.4 | 87.1 | ARATH2 | |||||||||||
| ARATH1 | 67.6 | 66.4 | 44 | 87.1 | 86.5 | 92.9 | ARATH1 | ||||||||||
| MEDSA | 69.4 | 69.6 | 46.1 | 85.3 | 84.1 | 85.3 | 85.3 | MEDSA | |||||||||
| GLYMA | 68.1 | 68.1 | 45.1 | 85 | 84.4 | 85.3 | 85.3 | 95.3 | GLYMA | ||||||||
| PVRACK1 | 69.3 | 69.3 | 45.8 | 85 | 84.4 | 86.1 | 86.1 | 96 | 96.6 | PVRACK1 | |||||||
| NICPL2 | 66.4 | 66.1 | 44.3 | 78.7 | 79.3 | 79.2 | 78.6 | 83.7 | 82.5 | 83.1 | NICPL2 | ||||||
| NICTA1 | 66.4 | 66.1 | 44.3 | 78.7 | 78.7 | 79.2 | 78.6 | 83.7 | 82.8 | 83.1 | 96.3 | NICTA1 | |||||
| NICPL1 | 67.5 | 66.6 | 44.5 | 80.2 | 79.6 | 79.8 | 79.2 | 84.4 | 83.2 | 83.8 | 90.2 | 91.1 | NICPL1 | ||||
| SOLLY2 | 68.5 | 67.3 | 44.9 | 80.7 | 80.4 | 81 | 81 | 84.7 | 83.2 | 84.7 | 90.8 | 91.7 | 92.6 | SOLLY2 | |||
| SOLLY1 | 68.5 | 67.9 | 45.3 | 81.1 | 80.2 | 80.7 | 80.1 | 86.5 | 84.9 | 85.5 | 91.1 | 92 | 94.2 | 93.9 | SOLLY1 | ||
| SOLTU | 68 | 68 | 45.4 | 80.1 | 79.8 | 80.1 | 80.1 | 84.7 | 82.9 | 84 | 90.8 | 90.8 | 93.2 | 93.5 | 95.4 | SOLTU | |
| ORYSA1 | 65.1 | 65.3 | 42.6 | 66 | 66.3 | 66 | 66.3 | 66.2 | 66.5 | 66.4 | 66.2 | 67.1 | 68.1 | 67.1 | 68 | 66.6 | ORYSA1 |
| ORYSA2 | 67.6 | 66.4 | 44 | 69.6 | 69.3 | 68.7 | 69 | 71 | 70.4 | 70.9 | 69.5 | 69.5 | 71.1 | 71 | 71.5 | 71 | 81.8 |
Table 3.
Comparison of the identities of the internal aPKC binding sequences of PvRACK1 with other reported sequences, including that of the reported Gβ -like subunit of a heterotrimeric protein from Glycine max.
| Identities species | RACK1 sequence in third domain | Percentage identity with PvRACK1 | RACK1 sequence in sixth domain | Percentage identity with PvRACK1 |
|---|---|---|---|---|
| PvRACK1 (Phaseolus vulgaris) | DVLSVAF | SIIHALFC | ||
| Gβ -like (G. max) | DVLSVAF | 100 | SIIHALFC | 100 |
| arcA (Nicotiana tabacum) | DVLSVAF | 100 | SIIHSFLC | 87.5 |
| AtarcA (Arabidopsis thaliana) | DVLSVAF | 100 | SIIHSFLC | 87.5 |
| RACK1 (Homo sapiens) | DVLSVAF | 100 | DIINALCF | 75 |
| RACK1 (Mus musculus) | DVLSVAF | 100 | DIINALCF | 75 |
| RACK1 (Tripanosoma cruzi) | DVLSVTF | 85.7 | APINQICF | 37.5 |
| Asc1p/Cpc2 (Saccharomyces cerevisiae/S. pombe) | DVMSVDI | 57 | DEVFSLAF | 33.3 |
PvRACK1 is related to other WD40 domain-containing proteins from species having multigene families but is represented by a small gene family in P. vulgaris
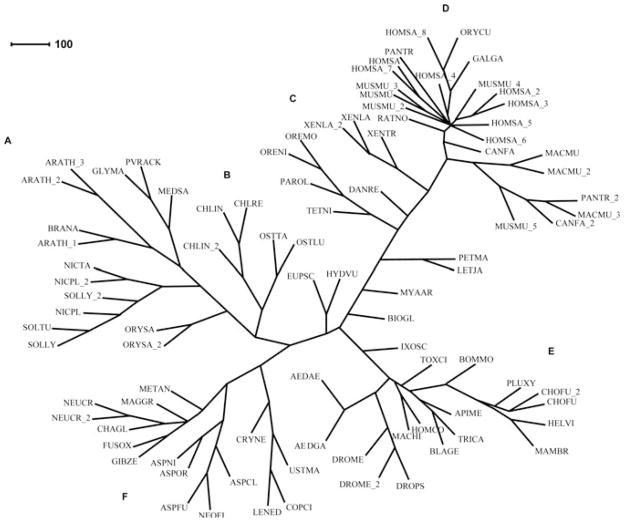
The sequence of PvRACK1 and other similar sequences from the GenBank were organized into a phylogenetic tree in order to reveal how it relates with the various orthologs in different species and kingdoms. The resulting phylogenetic tree showed that PvRACK1 (Fig. 2) clusters in a family of sequences from dicotiledoneus legume plants most closely related to G. max and M. sativa (GLYMA and MEDSA in Fig. 2, respectively) and forms a separate but closely related branch with other dicots such as A. thaliana and B. napus (ARATH and BRANA in Fig. 2, respectively), both of them appearing to share a common ancestor (Fig. 2A). Plant sequences from other non-legume dicots group in more distant but adjacent branches within the same cluster (Fig. 2A). The two most distant adjacent branches within the same cluster belong to fungi (Fig. 2F) and the unicellular algae Chlamydomonas and Ostreococcus spp (Fig. 2B). The most distant and distinct clusters in the tree, as expected, belong to sequences from the animal kingdom including humans and insects (Fig. 2D, E, respectively). This result shows that PvRACK1 is more closely related to legume dicots and they most likely evolved from a common ancestor.
Fig. 2.
Phylogenetic tree of the PvRACK1 protein. The phylogenetic tree of RACK1 protein sequences was constructed based on Guanine Nucleotide-Binding Protein Subunit β-like amino acid sequences with the highest identity to RACK, collected from the GenBank for the following species: Aedes aegypti (AEDAE), A. gambiae (AEDGA), Apis mellifera (APIME), Arabidopsis thaliana (ARATH), Aspergillus clavatus (ASPCL), A. fumigatus (ASPFU), A. nidulans (ASPNI), A. oryzae (ASPOR), Biomphalaria glabrata (BIOGL), Blattella germanica (BLAGE), Bombyx mori (BOMMO), Brassica napus (BRANA), Canis familiaris (CANFA), Chaetomium globosum (CHAGL), Chlamydomonas incerta (CHLIN), C. reinhardtii (CHLRE), Choristoneura fumiferana (CHOFU), Coprinopsis cinerea (COPCI), Cryptococcus neoformans (CRYNE), Danio rerio (DANRE), Drosophila melanogaster (DROME), D. pseudoobscura (DROPS), Euprymna scolopes (EUPSC), Fusarium oxysporum (FUSOX), Gallus gallus (GALGA), Gibberella zeae (GIBZE), Glycine max (GLYMA), Heliothis virescens (HELVI), Homalodisca coagulate (HOMCO), Homo sapiens (HOMSA), Hydra vulgaris (HYDVU), Ixodes scapularis (IXOSC), Lentinula edodes (LENED), Lethenteron japonicum (LETJA), Macaca mulatta (MACMU), Maconellicoccus hirsutus (MACHI), Magnaporthe grisea (MAGGR), Mamestra brassicae (MAMBR), Medicago sativa (MEDSA), Metarhizium anisopliae (METAN), Mus musculus (MUSMU), Mya arenaria (MYAAR), Neosartorya fischeri (NEOFI), Neurospora crassa (NEOCR), Nicotiana plumbaginifolia (NICPL), Nicotiana tabacum (NICTA), Oreochromis mossambicus (OREMO), O. niloticus (ORENI), Ostreococcus lucimarinus (OSTLU), O. tauri (OSTTA), Oryctolagus cuniculus (ORYCU), Oryza sativa (ORYSA), Pan troglodytes (PANTR), Paralichthys olivaceus (PAROL), Petromyzon marinus (PETMA), Phaseolus vulgaris (PVRACK), Plutella xylostella (PLUXY), Rattus norvegicus (RATNO), Solanum lycopersicum (SOLLY), S. tuberosum (SOLTU), Tetraodon nigroviridis (TETNI), Toxoptera citricida (TOXCI), Tribolium castaneum (TRICA), Ustilago maydis (USTMA), Xenopus laevis (XENLA) and X. tropicalis (XENTR). The tree was created as described in Materials and Methods; briefly, sequences were directed to ClustalX 1.8 (Thompson et al. 1997) and assembled with the PHYLIP 3.57 package (Lim and Zhang 1999) using the neighbor-joining method. The unrooted tree was created with CONSENSE. The resulting phylogenetic tree is displayed and edited in the SplitsTree4 program. The clusters correspond to: (A) plants; (B) unicellular algae; (C) amphibians and fishes; (D) mammals; (E) insects and (F) fungi. PvRACK1 clusters within the group of legume plant RACK proteins.
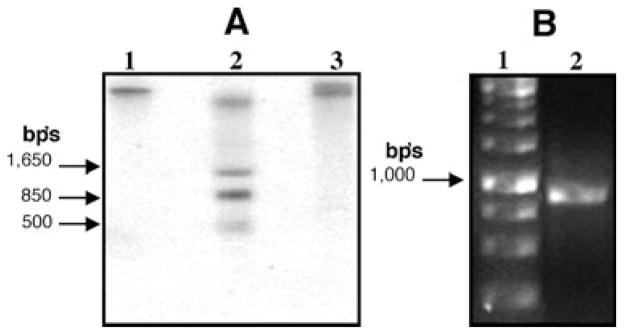
In order to gain insight toward the number of genes that code for this protein in the P. vulgaris genome, we carried out a Southern blot analysis with a 936 bp’s fragment from the sequence that includes the 3′-end. The pattern obtained with three different restriction enzymes with sites present within the sequence showed only one or very few DNA bands hybridized with the probe (Fig. 3A). Both BglII and KpnI with cut sites at nucleotides 1212 and 1284 of the reported genomic sequence FJ959041 yielded only one hybridized band (Fig. 3A, lanes 1 and 3, respectively), identical to what was observed for G. max genomic DNA cut with BglII and hybridized with the G. max RACK-related sequence probe (Nielsen et al. 2001), that had the highest identity to PvRACK1. The detection of only one band with these enzymes could arise from the fact that their cut site is very close to the 3′-end. This might result in fragments too large to enter the gel. BanI had two cut sites at nucleotides 469 and 1280, which are consistent with the pattern of hybridized bands observed (Fig. 3A, lane 2). The smaller 500 bp band observed with this enzyme is faint and probably non-specific. Further analysis by genomic DNA PCR amplification with the primers used for the complete PvRACK1 cDNA resulted in the amplification of a single band of approximately 975 bp (Fig. 3B, lane 2). The sequence obtained from this band comprised the exact nucleotide sequence previously obtained for the cDNA. These data, along with the limited number of detected hybridized bands on the Southern blot, suggest that PvRACK1 is represented by a small gene family in P. vulgaris, similar to what was observed for G. max (Nielsen et al. 2001). At least one of the representatives, the gene that was amplified by genomic PCR, presents no introns.
Fig. 3.
(A) Southern blot of Phaseolus vulgaris DNA detected with a 936 bp’s fragment from PvRACK1. The total DNA was digested with the restriction enzymes BglII, BanI and KpnI (lanes 1–3, respectively). BanI showed a limited restriction pattern of three-four bands (lane 2), whereas BglII and KpnI showed only one high molecular weight band (lanes 1 and 3, respectively). (B) Genomic PCR amplification of P. vulgaris DNA. After PCR amplification of P. vulgaris DNA with primers that anchor to the start and end of the cDNA PvRACK1 sequence, a single approximately 975 bp band was obtained (right arrow), with identical sequence to PvRACK cDNA. The arrows on the left of each gel point toward the size of some DNA molecular weight markers.
In addition to the open reading frame from the genomic sequence, which was identical to the cDNA sequence, various nucleotide regions corresponding to the 5′- and 3′-untranslated regions, which included the sequence of the promoter, were obtained. Thus, the full sequence including the promoter as well as a 3′-untranslated region was assembled and annotated in the GenBank (GenBank sequence number FJ959041). The full sequence comprised 1577 nucleotides with the 3′-UTR starting from nucleotide 1412 (GenBank sequence number FJ959041). The promoter 5′-end sequence analysis (nucleotide 1 equivalent to nucleotide 18 on reported sequence FJ959041) showed typical regions of a promoter from nucleotide 1 to 422, just prior to the ATG translation start (Fig. 4). This region showed the TATA box (Fig. 4) as well as several regulatory regions that included the response regulator sequence for the ARR1-binding element (ARRIAT Element, Fig. 4), a–300 regulatory element sequence (–300 element, Fig. 4), a telo box (telo box, Fig. 4) and several TCP domain protein binding (Site II) element regions (SITEIIATCYTC element, Fig. 4). These data suggest that PvRACK1 expression is under the control of several regulatory elements.
Fig. 4.

Promoter sequence of PvRACK1. The promoter 5′-end sequence from nucleotide 1 (nucleotide 1 equivalent to nucleotide 18 on the GenBank reported sequence number FJ959041) to 422, just prior to the ATG translation start, was analyzed for transcriptional and regulatory regions. The found regions of promoter and regulatory sequences are shown labeled and underlined. TATA: the TATA box; ARRIAT element: a response regulator sequence for the ARR1-binding element; –300 element: a–300 regulatory element sequence; telo box: a telo box and SITEIIATCYTC: several TCP domain protein binding (Site II) element regions.
Tissue distribution and cross-reactivity of PvRACK1 with an antibody raised against a decapeptide from within the 36 kDa protein sequence
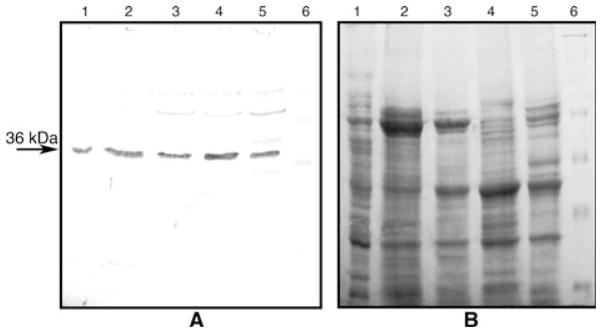
The anti-PA218 antibody, which was raised against a synthetic decapaptide from the obtained partial peptide amino acid sequence (Table 1, underlined amino acids), immunostained a 36 kDa protein present in total extracts from common bean (Fig. 5A, lane 2). In a peptide competition assay, immunostaining of the 36 kDa common bean protein was gradually inhibited by increasing excess molar amounts of the synthetic decapeptide incubated with the antibody prior to immunoblotting (Fig. 5A, lanes 5–7). Thus, the antibody was highly specific inasmuch as it could recognize the sequence of the decapeptide on the protein, against which the anti-PA218 antibody was raised. The anti-PA218 antibody also cross-reacted with a 36 kDa protein in soybean (Fig. 5A, lane 3), and pea (Fig. 5A, lane 4), indicating that similar proteins are present in other plant species. Further analysis showed that the anti-PA218 antibody was also able to recognize the 36 kDa protein in all tissue extracts at equal amounts of protein from common bean, loaded on a polyacrylamide gel and tested by western blot. All tissues showed a well detectable amount of the protein, which was found present in root nodules, roots, stems, leaves and pods (Fig. 6A, lanes 1–5, respectively), with slightly higher amounts in leaf tissue (Fig. 6A, lane 4). These data indicated that the antibody is specific, and that the protein is present in other plant species and is ubiquitous in all common bean tissues.
Fig. 5.
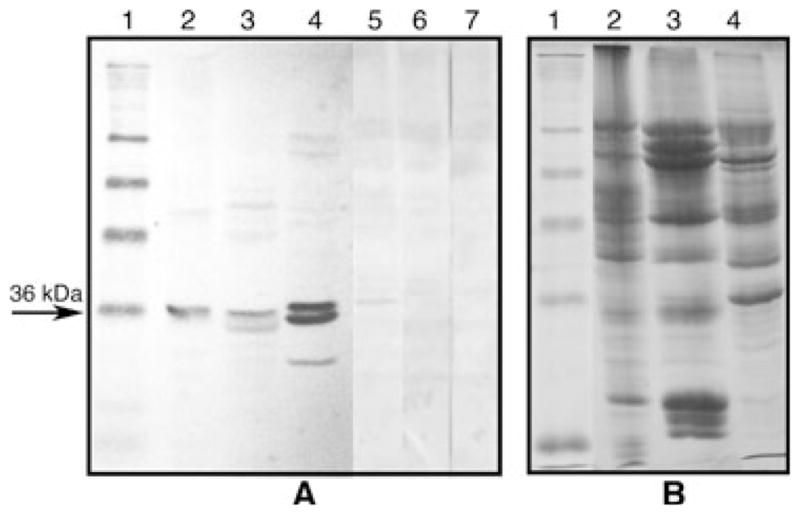
(A) Western blot of various plant homogenates with anti-PA218 antibodies. The anti-PA218 antibodies showed a strong reaction against the 36 kDa protein band and some degradation products from it in total homogenates from common bean (lane 2), soybean (lane 3) and pea (lane 4). In addition, the antibody immunostaining signal was gradually decreased when molar proportions of decapeptide:anti-PA218 of 1:1 (lane 5), 100:1 (lane 6) and 200:1 (lane 7) were incubated prior to the normal western blotting procedure. Lane 1 shows molecular weight markers of 98, 64, 50, 36, 22 and 16 kDa. (B) An equivalent denaturing 12% polyacrylamide gel stained with Coomassie blue showing the equal amounts of loaded protein (approximately 20 μg) per lane for lanes 2–4. Equal amounts of protein were also loaded in lanes 5–7.
Fig. 6.
(A) Western blot of various Phaseolus vulgaris tissues detected with anti-PA218 antibodies. The anti-PA218 antibodies showed a strong reaction against the 36 kDa protein band in root nodules (lane 1), roots (lane 2), stems (lane 3), leaves (lane 4) and pods (lane 5). Lane 6 shows BioRad prestained molecular weight markers with bands of 250, 42, 30, 22 and 15 kDa seen clearly on the gel. (B) An equivalent denaturing 12% polyacrylamide gel was stained with Coomassie blue to show the equal amounts of loaded protein (approximately 20 μg) per lane.
The PvRACK1 protein is present throughout differentially extracted fractions from common bean embryo axes
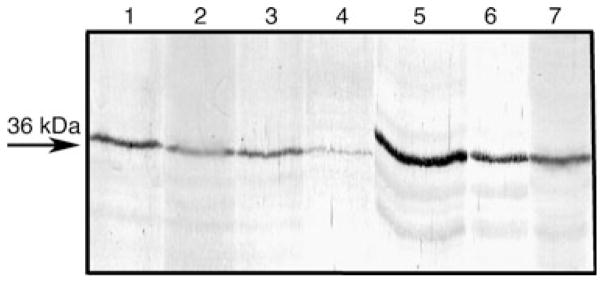
The common bean embryo axes were separated into a microsomal and a soluble fraction, and the microsomal fraction was subjected to differential extraction using salts and detergents (see section on Materials and methods). The resulting fractions were analyzed by western immunoblotting using the anti-PA218 antibody, and the analysis showed significant immunostaining in all fractions tested. First, the antibody detected the protein in both insoluble (Fig. 7, lane 1) and soluble (Fig. 7, lane 2) fractions from the microsomal preparation. Second, the soluble fraction from KCl-extracted microsomes showed a clear presence of the protein as well (Fig. 7, lane 3). Third, soluble detergent fractions from TX-100-extracted microsomes (previously extracted with KCl), also showed the presence of the protein (Fig. 7, lane 4). Next, the salt- and detergent-extracted microsomal pellet was further extracted with Na2CO3 and the carbonate-extracted soluble fraction showed a significant presence of the protein (Fig. 7, lane 5). Finally, the resulting carbonate-extracted insoluble pellet, which was further resuspended in Laemmli’s denaturing sample buffer and boiled, also showed immunostaining of the 36 kDa band (Fig. 7, lane 6). As a positive control, the total low-speed PBS supernatant from common bean embryo axes was run in parallel to show that the antibody also immunostained the 36 kDa band (Fig. 7, lane 7). These data indicate that the protein is ubiquitously distributed in the cell and it shows no special predominance in soluble, membrane-enriched, or detergent-resistant cell matrix fractions.
Fig. 7.
Distribution of PvRACK in differentially extracted microsomes and soluble fractions of embryo axes from Phaseolus vulgaris analyzed by western blot with anti-PA218 antibodies. A microsomal fraction was obtained and the insoluble membranes were differentially extracted with various salts and detergents (see Section on Materials and methods). The 36 kDa protein was detected in all fractions: insoluble 100 000 g pellet (lane 1); soluble 100 000 g supernatant (lane 2); 100 000 g supernatant from 0.5 M KCl extraction of insoluble pellet (lane 3); 100 000 g supernatant from 0.5% TX-100 extraction of KCl-extracted pellet (lane 4); 100 000 g supernatant from 0.1 M Na2CO3 extraction of KCl/TX-100-extracted pellet (lane 5); 14 000 g supernatant from Laemmli’s sample buffer extraction and heating at 95°C of KCl/TX-100/Na2CO3-extracted pellet (lane 6) and initial PBS extract (lane 7) from which the microsomal fraction was prepared. The arrow points toward the position of migration of the 36 kDa protein.
PvRACK1 protein is expressed steadily during early germination times but its mRNA is accumulated with a maximum at the end of germination
The anti-PA218 antibody was used to analyze the expression pattern of the PvRACK1 protein at various post-imbibition times. The protein was readily detectable in western blots from the extracts derived from the embryo axis at the seed stage (Fig. 8A, lane 0 h). This level of detection was more or less constant up to 40 h post-imbibition (Fig. 8A, lanes 0–40 h). In contrast, northern blot analysis of the same time points revealed that, at initial stages, the mRNA is practically undetected (Fig. 8B, lane 0 h). The mRNA remained at low levels but slightly detectable after 12 h (Fig. 8B, lane 12 h) and steadily increased to become clearly detectable at 20 h (Fig. 8B, lane 20 h). The accumulation of mRNA reached a maximum at 32 h (Fig. 8B, lane 32 h), which is the point at which germination stops in common bean (Díaz-Camino et al. 2005). From this point on, the mRNA level decreased slightly but was still clearly detectable after 40 h post-imbibition (Fig. 8B, lane 40 h). Thus, the expression of PvRACK1 mRNA appears to be induced and differentially regulated during germination of P. vulgaris.
Fig. 8.
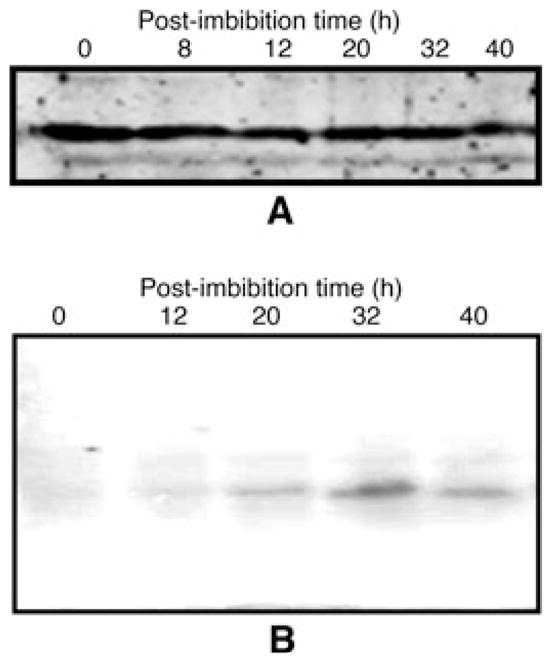
PvRACK1 protein and mRNA expression analysis at various post-imbibition times. (A) The embryo axes or hypocotyls of germinating seeds at various post-imbibition times were processed for SDS–PAGE, and the accumulated protein analyzed at various times from 0 to 40 h by western blot with anti-PA218 antibodies (see Section on Materials and methods). The level of protein remained practically unchanged throughout the various time points (lanes 0–40 h). (B) In parallel, the mRNA at equivalent time points was extracted and processed for northern blot and the membrane hybridized with a 500 bp probe from PvRACK1. Note the peak of mRNA accumulation at 32 h (B, 32 h).
A polar auxin transport blocker did not have any effect on the level of PvRACK1 mRNA accumulation but auxins delayed the point of maximum mRNA accumulation
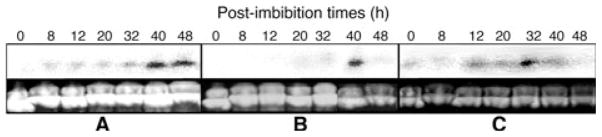
As the PvRACK1 mRNA showed a peak of accumulation in the seedlings at early germination times, PvRACK1 is also the ortholog of arcA, and auxins change significantly during germination (Bialek et al. 1992), the level of this mRNA accumulation was also analyzed under the influence of synthetic auxins at various post-imbibition times. Treatments with 25 μM 2, 4-D (Fig. 9A) and 50 μM IAA (Fig. 9B) during germination resulted in a delay of the time of maximal peak of expression of the PvRACK1 transcript, from 32 to 40 h (Fig. 9A, B; lanes 40 h, respectively), compared with the 32 h peak of mRNA accumulation in untreated seedlings (Fig. 8B, lane 32 h). In addition, treatment with the polar auxin transport blocker NPA (25 μM) had no effect on the increase of PvRACK1 transcript accumulation levels (Fig. 9C, lanes 0–48 h) and was comparable to the untreated seedlings (Fig. 8B, lanes 0–40 h). The level of protein was unchanged throughout the post-imbibition times tested in all treatments and identical to what was observed in Fig. 8A. These data indicate that, during P. vulgaris germination, the auxin treatment affected the transcript of PvRACK1 by delaying its maximal peak of accumulation, but NPA did not affect the levels of transcript or protein accumulation significantly.
Fig. 9.
Northern blots of Phaseolus vulgaris at various post-imbibition times in the presence of synthetic auxins and the polar auxin transport blocker NPA. The embryo axes or hypocotyls of germinating seeds in 25 mM 2, 4-D (A), 50 mM IAA (B) or 25 mM NPA (C) were processed for mRNA extraction and northern blot (see Section on Materials and methods). The accumulated mRNA, separated on a gel with equal amounts or RNA loaded, was hybridized at various times from 0 to 40 h with a 500 bp probe from PvRACK1. The maximal peak of mRNA accumulation was detected at 40 h in both auxin treatments (40 h in A and B, respectively), whereas in the presence of the polar auxin transport blocker NPA, it remained unchanged at 32 h (32 h in C), as in the untreated seedlings.
Discussion
The 36 kDa protein from P. vulgaris is the RACK1 and arcA homolog
A 36 kDa protein that we isolated from P. vulgaris had its closest identity to a reported Gβ -like protein from soybean and therefore, we originally believed that it was more related to a Gβ -like subunit. However, amplification of the full cDNA sequence and careful analysis of all related sequences showed that the protein is more closely related to RACK1 and arcA sequences previously annotated in the GenBank. Thus, the 36 kDa protein that we identified and sequenced is the RACK1 ortholog in P. vulgaris. The sequence showed two highly conserved aPKC binding sites with the first stretch being 100% identical to RACK1 from human and mouse (Table 3). The protozoan T. cruzi even had a 75% identity score for this sequence. The second sequence is not as conserved but it still displays a 75% identity with human and mouse aPKC binding sequences. The relatively high identity with sequences that are designed to bind aPKC in mammals contrasts sharply with the fact that in available plant genomes that have been completely sequenced, the equivalent sequences for PKC do not exist (Chen et al. 2006). Thus, either equivalent kinase-binding partners with highly divergent sequences exist, or the equivalent function for recruiting an activated kinase has been lost and they associate with other types of proteins. It is known that RACK proteins bind to proteins with C2 domains (see below), so their possible association and functional consequences in plants remain to be demonstrated. With respect to other reported similar sequences, a phylogenetic tree showed that the PvRACK1 sequence (Fig. 2) is most closely related to similar plant sequences. Within this group, distinct sub-groups can be distinctly observed. For example, one is constituted with sequences from the legumes soybean and alfalfa (GLYMA and MEDSA in Fig. 2, respectively). Another one groups with dicots such as A. thaliana and B. napus (ARATH and BRANA in Fig. 2, respectively). Other sub-group of plant sequences which fall in a more distant but adjacent branch within the same cluster are those from the Solanaceae dicots such as N. tabacum and S. tuberosum (NICTA and SOLTU in Fig. 2, respectively). As the branches of the tree diverge farther apart, the relatedness of the organisms also become more distant. Thus, distinct separations are evident from sequences that group independently together into plants (Fig. 2A), algae (Fig. 2B), amphibia and fish (Fig. 2C), mammals (Fig. 2D), insecta (Fig. 2E), and fungi (Fig. 2F) although the related sequences still share certain clusters of the tree. For example, some unicellular algae and fungi sequences occupy the two most distant but still adjacent branches to the cluster of plant sequences (Fig. 2B, F, respectively). Thus, all these sequences appear to have evolved from a common ancestor, from which they diverged into two major groups of inter-related species, one enclosing amphibians, fish, insects and mammals, and the other fungi, algae and plants. It is interesting to note that the most distant groups also differ in the availability of an aPKC binding partner (i.e. plants vs mammals).
Putative binding partners for a seven-bladed propeller structure of PvRACK1
The sequence of PvRACK1 also displays seven WD40 repeats, which in molecular models of highly identical proteins such as the G. max putative Gβ -subunit (SWISS MODEL-protein modeling service from UniProt Knowledgebase; protein GLB SOYBN Q39836) and Arabidopsis (RACK1A; Chen et al. 2006) orthologs form the seven-bladed structure of RACK1 (McCahill et al. 2002), typical of β-propeller-type proteins (Ter Haar et al. 2000). Thus, similar binding partners and interaction platforms in different parts of the molecule can be expected. The RACK1 ortholog in yeast has been proposed recently to play a role in ribosomal assembly and protein translation (Nilsson et al. 2004). In parallel, the RACK ortholog in Arabidopsis has been found associated with the 40S subunit of ribosomes in this plant (Chang et al. 2005). It is proposed that the RACK molecule would recruit aPKC to the ribosome in order to shuttle a kinase to phosphorylate initiation factor 6 and perhaps, other mRNA associated proteins (Nilsson et al. 2004). RACK has also been reported to associate in a calcium- and phosphatidylserine-dependent manner, to PKC and other proteins such as Src and β-integrin via their C2 domains. Identified plant protein orthologs containing such domains are synaptotagmin, phosphatidylinositol 3-kinase and β-integrins which have all been identified in plants (Clark et al. 2001, Craxton 2000) and could be candidates for protein–protein interaction and signal-transduction function. It has been recently reported that mutations that impair RACK1A function in Arabidopsis cause a number of defects in developmental processes such as seed germination, leaf production and flowering, as well as alterations in auxin-mediated signal-transduction mechanisms (Chen et al. 2006, Guo and Chen 2008). Those data suggest that the binding function of RACK as an adaptor protein for signal-transduction mechanisms in the plant cell is fundamental for growth and auxin-mediated differentiation and development.
RACK1 is present in other plant species, with PvRACK1 displaying a wide distribution in subcellular compartments and tissues, and a small gene family
Custom-made antibodies against a decapeptide derived from the internal PvRACK1 peptide sequence named anti-PA218 antibodies were shown to specifically immunostain a 36 kDa protein corresponding to PvRACK1 in embryo axes extracts (Fig. 5A, lanes 2, 5–7). Besides common bean, the anti-PA218 antibody detected the presence of RACK-related proteins in soybean and pea (Fig. 5A, lanes 3 and 4, respectively), which was a somewhat expected result for soybean because its related sequence shares a high identity with common bean. However, the fact that the antibody is raised against a decapeptide makes high sequence identity crucial for detecting the RACK homolog in any other plant species by immunoblotting. Thus, although the pea sequence is not reported yet, the fact that the anti-PA218 antibody is able to detect the protein implies that the RACK1 ortholog of this species is highly identical to PvRACK as well.
In addition to observing the protein in other plant species, we analyzed the tissue distribution of the protein in various common bean vegetative tissues and in differentially extracted fractions from the bean embryo axes because of its proposed roles in cell proliferation and of adaptor protein to target other proteins to different subcellular compartments. We found a ubiquitous distribution of the protein in all vegetative tissues analyzed, but we found a slightly higher amount in leaves using equal protein loads from each tissue in the gel. This was somewhat surprising because the PvRACK1 mRNA was found at lower levels in leaves than in other tissues of the plant (T. Islas-Flores, unpublished observations). These data suggest that the protein may be highly stable and perhaps being transported to this tissue. It remains to be determined how the bean plant may accomplish this transport.
Analysis of the protein distribution in differentially extracted fractions from embryo axes also showed a ubiquitous distribution within the cell. Thus, we did not find a preferential distribution of the protein in membranes but rather, it was found in the cytoplasmic soluble fraction of the cells from the embryo, as well as in the high salt extract containing the ionically bound proteins. The protein was also found as an intrinsic membrane protein because it was released with detergent from the high salt-extracted pellet. Furthermore, the luminal intrinsic proteins released with carbonate after the detergent extraction also released protein indicating that PvRACK1 was also found in Golgi and highly folded membranes. The most surprising result was that after this last extraction, a well detectable amount of protein was still released by a harsh extraction with Laemmli’s denaturing buffer and heat, indicating that a part of the protein was associated with the detergent-resistant cell matrix. The association of PvRACK1 with this matrix may indicate a functional analogy with the scaffolding action of RACK1 with plectin in the cytoskeletal matrix of animal cells where it is associated with detergent-resistant cytoskeletal fractions under normal conditions (Osmanagic-Myers and Wiche 2004). The presence of the protein in all the fractions analyzed is consistent with its proposed multifunctional role of targeting signaling proteins to different cellular compartments (including the ribosome; Nilsson et al. 2004), being its localization to these compartments critical for normal cell function.
The size of the gene family can also give some clues as to the possible existence of isovariants of the protein and ways in which it may be regulated. When we carried out a Southern blot analysis to determine the number of genes, we detected a limited number of bands after restricting the DNA with enzymes that had restriction sites present in the sequence. The hybridization pattern resembles that of the G. max ortholog, where also very few bands were observed by Southern blot analysis, and thus, it was also concluded that only one or two genes code for this protein in this species (Nielsen et al. 2001). It is also noteworthy that these two sequences are closest in the phylogenetic tree (Fig. 2). Furthermore, a genomic DNA PCR amplification assay amplified a single band of approximately 975 bp with primers used previously to amplify the whole cDNA sequence for PvRACK1 (Fig. 3B). Thus, it can be concluded that the gene family of PvRACK1 consists of only one or two genes and that at least one of these genes lacks introns. It seems a consistent pattern that some proteins such as Arabidopsis profilin have a multigene family in this species (Huang et al. 1996), whereas there are only one-two profilin genes in P. vulgaris (Guillén et al. 1999). Thus, it is not so surprising that common bean has only a few PvRACK1 representatives compared with Arabidopsis with three members (Chen et al. 2006). In addition, the lack of introns in the P. vulgaris gene contrasts sharply with the fact that all three RACK members of Arabidopsis (At3g18130.1, At3g48630.1 and At1g18080.1) have introns of 281, 310 and 592 bp, respectively (TAIR). Taken together, these data are consistent with the recent report in Arabidopsis which shows functional substitution among its RACK1 genes (Guo and Chen 2008) and suggests regulation through differential gene expression, while the common bean protein expression might be tightly regulated within the P. vulgaris untranslated regions to accommodate for its putative multifunction purposes. Indeed, the promoter sequence analysis showed several putative regulatory regions (Fig. 4). These regulatory regions included the sequence for the ARR1-binding element (ARRIAT Element, Fig. 4), which is a transcriptional activator found in Arabidopsis (Sakai et al. 2000). Another region was similar to a –300 regulatory element sequence. This sequence was reported as an enhancer of endosperm-specific transcription of the seed storage protein glutenin (Thomas and Flavell 1990). Finally, several TCP domain protein binding (Site II) element regions (SITEIIATCYTC element, Fig. 4) were also detected. These elements are implicated in the expression of genes in meristematic tissues and/or proliferating cells and, consistent with our findings, present in more than one copy and accompanied by a telo box (telo box, Fig. 4; Welchen and Gonzalez 2006). These data further suggest that PvRACK1 expression is under the control of several regulatory elements that are implicated in several fundamental functions within the plant cells that include transcriptional activation, regulated expression and proliferation.
Expression of the PvRACK1 protein is maintained constant although its mRNA appears to increase as germination proceeds and the transcript is slightly affected by auxin but unaffected by polar auxin transport blocker treatments
Western blot analysis with anti-PA218 antibodies of common bean embryo axes after the initiation and after various post-imbibition times showed a well detectable amount of the PvRACK1 protein. As we are interested in the early germination events and germination in common bean ends at 36 h (Díaz-Camino et al. 2005), we carried out our analysis from 0 to 40 h post-imbibition. The level of protein was clearly detectable from the pre-imbibed embryo axes indicating that sufficient protein is stored during the seed filling process to be utilized at the initiation and early germination events. This protein must also be quite stable because it remained at constant levels throughout the time intervals of analysis to up to 40 h, contrary to the mRNA levels, which were undetectable during the initial post-imbibition times. This suggests that the initial stored protein is important at the onset of germination, and a constant level of expression is required throughout the germination process. This is consistent with the requirement of the protein for signal-transduction processes and regulation of the protein expression per se (Nilsson et al. 2004), since the very start of germination. However, it is intriguing that the mRNA peak of accumulation is detected at 36 h, which is the time when germination ends in P. vulgaris and the radicle emerges (Díaz-Camino et al. 2005). This might suggest a de novo synthesis of this protein beginning just before the peak of germination, although this remains to be tested.
It was recently reported that stopping auxin transport through impairing the PIN2 efflux carrier that exports auxin from root cells with the chemical D’orenone severely inhibited the polarized growth of root hairs in Arabidopsis (Schlicht et al. 2008). Furthermore, since the PvRACK1 gene of P. vulgaris is also an arcA homolog (Ishida et al. 1993), auxin changes could profoundly affect its expression. Therefore, we tested the germination process under the presence of the synthetic auxins 2, 4-D and IAA, and the polar auxin transport blocker NPA. Surprisingly, neither the synthetic auxins nor the polar auxin transport blocker had any effect on the PvRACK1 protein during the post-imbibition process. However, the mRNA accumulation of PvRACK1 levels during the post-imbibition process showed a slight delay of 8 h in the peak of PvRACK mRNA accumulation when either auxin was present, suggesting that the expression of this gene during early germination in common bean is not severely affected by the level of auxin. These results also suggest that the RACK1-auxin related effects are significantly different in common bean compared with how auxins affect developmental functions in other plants such as Arabidopsis. The analysis of loss-of-function RACK mutants in these plants showed numerous defects in developmental processes that included seed germination, leaf production and flowering, and these appeared to be related to auxin-mediated signaling because of changes in the sensitivity to auxin treatments (Chen et al. 2006). Although 2, 4-D was not used in their treatments during seed germination, these mutants had a low sensitivity to brassinosteroids and gibberellin, and hypersensitivity to abscisic acid, suggesting that RACK1A from Arabidopsis is involved in multiple signal-transduction processes that are also related to auxin (Chen et al. 2006). It is also possible that due to the multifunctionality of RACK1 in common bean and, particularly during early germination events, its role in this species is more restricted to regulation of protein translation in ribosomes (Nilsson et al. 2004).
These results altogether show that PvRACK1 is the RACK1 and arcA ortholog in the genome of P. vulgaris and that it contains all the expected features (such as a seven-bladed propeller structure) of a protein with a platform for binding and interacting with other partners for multifunction purposes within the cell. Of special interest is the fact that it has two highly conserved aPKC binding sequences despite the fact that no PKC orthologs have been found in plants so far. Its small gene family and several regulatory element sequences in P. vulgaris suggest a tight regulation but its expression does not appear to be severely affected by auxins during early germination in the seedling. We are currently testing the effect of turning off its expression by taking advantage of the fact that there may be only one-two genes in the P. vulgaris genome. We are also in the search for interactive partners because finding its binding proteins will allow us to gain more insight into the various functions that PvRACK1 is proposed to serve in the plant.
Acknowledgments
This work was partially supported by a grant number 32650-B from the National Council of Science and Technology of México (CONACyT) and special Graduate Program Support Grant from CONACyT to T. Islas-Flores. A joint grant from the National Science Foundation and CONACyT to Drs E. L. B. and M. A. V. is also acknowledged. We thank the sequencing facility of the Institute of Biotechnology of UNAM for sequencing of the DNA and some oligonucleotide synthesis. We thank Arturo Ocádiz and Juan Manuel Hurtado from the Informatics Facility at the Institute of Biotechnology-UNAM for technical help.
Abbreviations
- aPKC
activated protein kinase C
- IAA
indoleacetic acid
- NPA
naphthylphthalamic acid
- PBS
phosphate-buffered saline
- RACK1
receptor for activated C kinase
- SDS–PAGE
sodium dodecyl sulfate–polyacrylamide gel electrophoresis
- 2, 4-D
2, 4-dichlorophenoxyacetic acid
Footnotes
References
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bialek K, Michalczuk L, Cohen JD. Auxin biosynthesis during seed germination in Phaseolus vulgaris. Plant Physiol. 1992;100:509–517. doi: 10.1104/pp.100.1.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. A rapid sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;22:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Chang I-F, Szick-Miranda K, Pan S, Bailey-Serres J. Proteomic characterization of evolutionarily conserved and variable proteins of Arabidopsis cytosolic ribosomes. Plant Physiol. 2005;137:848–862. doi: 10.1104/pp.104.053637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Spiegelberg BD, Lin F, Dell EJ, Hamm HE. Scaffolding and docking proteins in the heart. Interaction of Gbc with RACK1 and other WD40 repeat proteins. J Mol Cell Cardiol. 2004;37:399–406. doi: 10.1016/j.yjmcc.2004.04.019. [DOI] [PubMed] [Google Scholar]
- Chen J, Ullah H, Temple B, Liang J, Guo J, Alonso JM, Ecker JR, Jones AM. RACK1 mediates multiple hormone responsiveness and developmental processes in Arabidopsis. J Exp Bot. 2006;57:2697–2708. doi: 10.1093/jxb/erl035. [DOI] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Clark GB, Thompson G, Jr, Roux SJ. Signal transduction mechanisms in plants: an overview. Curr Sci. 2001;80:170–177. [PubMed] [Google Scholar]
- Craxton M. Genomic analysis of synaptotagmin genes. Genomics. 2000;77:43–49. doi: 10.1006/geno.2001.6619. [DOI] [PubMed] [Google Scholar]
- Dalrymple MA, Petersen Bjorn S, Friesen JD, Beggs JD. The product of the PRP4 gene of S. cerevisiae shows homology to β subunits of G proteins. Cell. 1989;58:811–812. doi: 10.1016/0092-8674(89)90930-6. [DOI] [PubMed] [Google Scholar]
- Díaz-Camino C, Conde R, Ovsenek N, Villanueva MA. Actin expression is induced and three isoforms are differentially expressed during germination in Zea mays. J Exp Bot. 2005;56:557–565. doi: 10.1093/jxb/eri034. [DOI] [PubMed] [Google Scholar]
- Guillén G, Valdés-López V, Noguez R, Olivares J, Rodríguez-Zapata LC, Pérez H, Vidali L, Villanueva MA, Sánchez F. Profilin in Phaseolus vulgaris is encoded by two genes (only one expressed in root nodules) but multiple isoforms are generated in vivo by phosphorylation on tyrosine residues. Plant J. 1999;19:497–508. doi: 10.1046/j.1365-313x.1999.00542.x. [DOI] [PubMed] [Google Scholar]
- Guo J, Chen J-G. RACK1 genes regulate plant development with unequal genetic redundancy in Arabidopsis. BMC Plant Biol. 2008;8:108. doi: 10.1186/1471-2229-8-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higo K, Ugawa Y, Iwamoto M, Korenaga T. Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 1999;27:297–300. doi: 10.1093/nar/27.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, McDowell JM, Weise MJ, Meagher RB. The Arabidopsis profilin gene family (Evidence for an ancient split between constitutive and pollen-specific profilin genes) Plant Physiol. 1996;111:115–126. doi: 10.1104/pp.111.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson DH, Bryant D. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol. 2006;23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- Ishida S, Takahashi Y, Nagata T. Isolation of a cDNA of an auxin-regulated gene encoding a G protein β-subunit-like protein from tobacco BY-2 cells. Proc Natl Acad Sci USA. 1993;90:11152–11156. doi: 10.1073/pnas.90.23.11152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyosue T, Ryan CA. Molecular cloning of two cDNAs encoding G protein beta subunit-like proteins from tomato (Accession Nos. AB022686 and AB022687). Plant Gene Register (PGR99-045) Plant Physiol. 1999;119:1567–1568. [Google Scholar]
- Kost B, Mathur J, Chua NH. Cytoskeleton in plant development. Curr Opin Plant Biol. 1999;2:462–470. doi: 10.1016/s1369-5266(99)00024-2. [DOI] [PubMed] [Google Scholar]
- Lamely UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lim A, Zhang L. WebPHYLIP: a web interface to PHYLIP. Bioinformatics. 1999;15:1068–1069. doi: 10.1093/bioinformatics/15.12.1068. [DOI] [PubMed] [Google Scholar]
- Mallet L, Bussereau F, Jacquet M. Nucleotide sequence analysis of an 11.7 kb fragment of yeast chromosome II including BEM1, a new gene of the WD-40 repeat family and a new member of the KRE2/MNT1 family. Yeast. 1994;10:819–831. doi: 10.1002/yea.320100612. [DOI] [PubMed] [Google Scholar]
- McCahill A, Warwicker J, Bolger GB, Houslay MD, Yarwood SJ. The RACK1 scaffold protein: a dynamic cog in cell response mechanisms. Mol Pharmacol. 2002;62:1261–1273. doi: 10.1124/mol.62.6.1261. [DOI] [PubMed] [Google Scholar]
- McKhann HI, Frugier F, Petrovics G, Coba de la Peña T, Jurkevitch E, Brown S, Kondorosi E, Kondorosi A, Crespi M. Cloning of a WD-repeat-containing gene from alfalfa (Medicago sativa): a role in hormone-mediated cell division? Plant Mol Biol. 1997;34:771–780. doi: 10.1023/a:1005899410389. [DOI] [PubMed] [Google Scholar]
- Murray MG, Thompson WF. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980;8:4321–4326. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen NC, Beilinson V, Bassüner R, Reverdatto S. A Gβ -like protein from soybean. Physiol Plant. 2001;111:75–82. [Google Scholar]
- Nilsson J, Sengupta J, Frank J, Nissen P. Regulation of eukaryotic translation by the RACK1 protein: a platform for signalling molecules on the ribosome. EMBO Rep. 2004;5:1137–1141. doi: 10.1038/sj.embor.7400291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osmanagic-Myers S, Wiche G. Plectin-RACK1 (receptor for activated C kinase 1) scaffolding. J Biol Chem. 2004;279:18701–18710. doi: 10.1074/jbc.M312382200. [DOI] [PubMed] [Google Scholar]
- Pawson T, Scott JD. Signaling through scaffold, anchoring, and adaptor proteins. Science. 1997;278:2075–2080. doi: 10.1126/science.278.5346.2075. [DOI] [PubMed] [Google Scholar]
- Sakai H, Aoyama T, Oka A. Arabidopsis ARR1 and ARR2 response regulators operate as transcriptional activators. Plant J. 2000;24:703–711. doi: 10.1046/j.1365-313x.2000.00909.x. [DOI] [PubMed] [Google Scholar]
- Schlicht M, Samajová O, Schachtschabel D, Mancuso S, Menzel D, Boland W, Balu ska F. D’orenone blocks polarized tip growth of root hairs by interfering with the PIN2-mediated auxin transport network in the root apex. Plant J. 2008;55:709–717. doi: 10.1111/j.1365-313X.2008.03543.x. [DOI] [PubMed] [Google Scholar]
- Ter Haar E, Harrison SC, Kirchhausen T. Peptide-in-groove interactions link target proteins to the β-propeller of clathrin. Proc Natl Acad Sci USA. 2000;97:1096–1100. doi: 10.1073/pnas.97.3.1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas MS, Flavell RB. Identification of an enhancer element for the endosperm-specific expression of high molecular weight glutenin. Plant Cell. 1990;2:1171–1180. doi: 10.1105/tpc.2.12.1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin J, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullah H, Chen J, Temple B, Boyes DC, Alonso JM, Davis KR, Eckerd JR, Jones AM. The β-subunit of the Arabidopsis G protein negatively regulates auxin-induced cell division and affects multiple developmental processes. Plant Cell. 2003;15:393–409. doi: 10.1105/tpc.006148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welchen E, Gonzalez DH. Overrepresentation of elements recognized by TCP-domain transcription factors in the upstream regions of nuclear genes encoding components of the mitochondrial oxidative phosphorylation machinery. Plant Physiol. 2006;141:540–545. doi: 10.1104/pp.105.075366. [DOI] [PMC free article] [PubMed] [Google Scholar]