Abstract
The human blood platelet circulates in the blood as a non-adherent disk. Upon receiving signals of blood vessel damage, the platelet reorganizes its actin cytoskeleton which transforms it into a spiky dynamic adherent glue. This transformation involves a temporal sequence of four morphologically distinct steps that can be reproducible in vitro. The actin dynamics that underlie these shape changes depend on a large number of actin-binding proteins. Maintenance of the discoid shape requires actin-binding proteins that inhibit these reorganizations, whereas transformation involves other proteins, some to disassemble old filaments and others to polymerize new ones. F-actin-affinity chromatography identified a large set of actin-binding proteins including VASP, Arp2 and 2E4/kaptin. Recent discoveries show that VASP inhibits filament disassembly and Arp2/3 is required to polymerize new filaments. Morphological analysis of the distribution of these actin-binding proteins in spread platelets together with biochemical measurements of their interactions with actin lead to a model of interactions with actin that mediate shape change.
Keywords: Platelet activation, Shape change, Actin polymerization, Cytoskeleton, Actin-binding proteins, Arp2/3, VASP, 2E4/kaptin, Gelsolin, ADF/cofilin, Lamellipodia, Filopodia
I. Introduction
Blood clotting is essential for life in multicellular organisms with circulatory systems. The clotting mechanism in humans depends on soluble clotting factors and the blood platelet. Signals from a damaged vessel wall activate the circulating platelet to change shape. Shape change is a reproducible temporal sequence of morphological steps that transform the non-sticky discoid platelet into spiky, sticky, flattened glue that spreads over small tears in the vessel wall, adheres to other cells, and recruits more platelets to the site of damage. After the clot has formed, it contracts, an event that pulls the edges of the wound together.
Platelet shape change depends on actin. Actin is a highly conserved 42-kDa protein found in all eukaryotic cells. Actin polymerizes to form filaments. Inside platelets, actin exists in a dynamic equilibrium between the monomeric or globular form (G-actin) and the polymeric filamentous form (F-actin). Actin serves as a building block that is built into a variety of higher order structures with the help of a large number of different actin-binding proteins. In the platelet after spreading there are four morphologically and functionally distinct actin structures which are analogous to similar structures in other cells. Each of these structures contains a different complement of actin binding proteins which are involved in their formation, maintenance, and physiologic function within the platelet. Thus, shape change is a very complex process, involving a large number of actin-binding proteins.
The central question is how actin dynamics are regulated by these actin-binding proteins. In the platelet, shape change must involve the dissociation of existing actin structures and the re-formation of new ones. Previous models proposed that actin filament severing and barbed end uncapping combine to produce these two effects (Stossel, 1994; Stossel et al., 1999). Proteins performing severing and capping have been identified as gelsolin and capping protein, but until recently it was not known how de novo nucleation is achieved. New discoveries point to the Arp2/3 complex as the nucleator (Li, Kim & Bearer, 2002).
Recently, using Listeria bacteria as a model for actin polymerization, a set of proteins has been discovered that is minimally required for regulated actin polymerization in vitro (Loisel et al., 1999). F-actin affinity chromatography had previously identified these same set of proteins in platelets, which include Arp2/3, cofilin, and capping protein, as well as 2E4/kaptin, gelsolin, VASP, and profilin.
In this review, we describe the morphological events of shape change and provide the experimental evidence that links these events with actin. We then provide a synthesis of how the many individual actin-binding proteins (ABP) interact with actin, where they are located in the platelet, and what specific role they might play in the complex choreography of shape change. We close with a model invoking novel mechanisms for platelet actin dynamics.
II. Platelets and Actin
A. Morphology of Shape Change
Platelets are anucleate fragments derived from the cortical cytoplasm of the bone marrow megakaryocyte. As such, platelets are analogous to a biochemical separation of cellular cortex, enriched in signaling machinery and membrane-associated cytoskeletal components but lacking nuclei, and depleted in Golgi apparatus and rough endoplasmic reticulum. After release from the megakaryocyte, platelets circulate in the blood in a discoid shape with a mean diameter of 3.1 ± 0.3 μm an average thickness of 1.0 ± 0.2 μm, and a calculated volume of approximately 7 fm3 (David-Ferreira, 1974; O'Brien and Woodhouse, 1968). Platelets circulating in the blood range in age from 1 to 120 days old and are thus somewhat heterogeneous both in size and responsiveness to agonists.
The discoid platelet in the blood stream is “at rest” (Fig. 1). The resting platelet does not stick to other cells or proteins in the blood. Upon stimulation by any one of a large number of agonists, platelets change shape and become sticky. This shape change involves a sequence of temporally reproducible morphological events. This series of event was best described using video microscopy under Nomarski optics to follow individual platelets as they spread on glass (Allen et al., 1979).
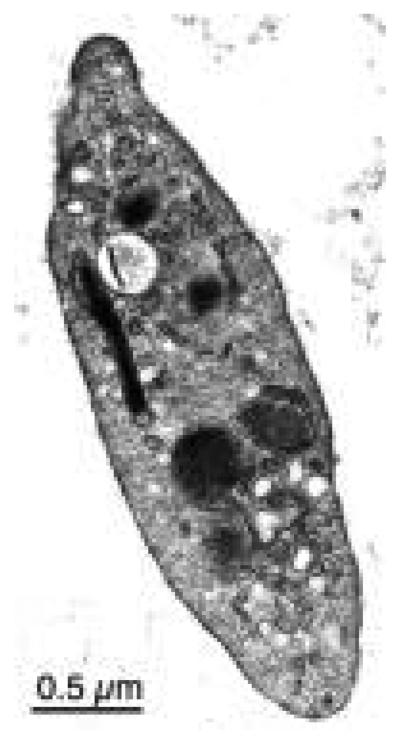
FIG. 1.
Electron microscopy of a resting platelet. (Bearer, unpublished)
Video microscopy reveals that platelets undergo four successive stages as they activate and spread (Fig. 2). Because of the difficulty of imaging platelets in suspension, direct observation of shape change in living platelets was performed on platelets activated on a flat surface, glass coverslips. Shape change begins with a contractile event, rounding, whereupon the platelet loses its discoid shape, becoming spherical and slightly smaller. This rounding is followed immediately (< 1 min) by a burst of dynamic protrusions from the surface of structures termed “pseudopodia”. After the platelet adheres to the flat surface, it begins to spread lamellipodia. Lamellipodia can arise from the lateral membrane between pseudopodia or from the sides of pseudopodia. By observation with Nomarski optics, the platelet achieves a fully spread morphology as quickly as 10 min after contact with a glass surface, and seldom takes longer than 30 minutes.
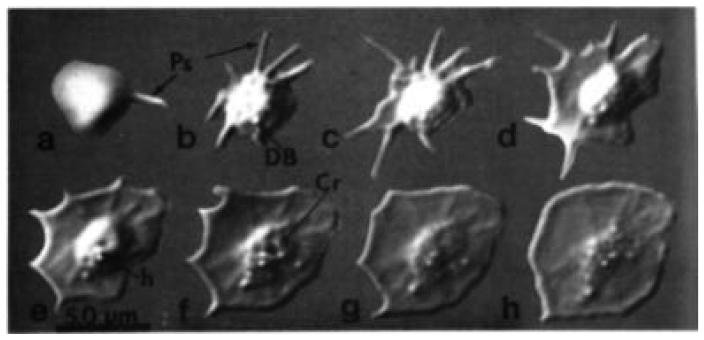
FIG. 2.
Video microscopy of the same platelet as it spreads on glass, transforming from spheroid (a) to fully spread (h). Photographs were taken at 1-min intervals until the last, which was after a 5-min interval. Structures visible include pseudopodia (Ps), dense bodies (DB), and crater (Cr). Scale bar = 0.5 μm. (Reproduced from Allen et al., 1979, by permission.)
Pseudopodia extend and retract. Extension velocity varies from 0.75 to 7.5 μm/min, whereas retraction is less common and slower, at −1.9 μm/min. Abrupt changes in either direction or velocity also occur. During any given time period, different pseudopodia from a single platelet can be extending or retracting at different rates and in opposite directions independently of each other. Thus, the mechanism governing pseudopodial behavior is not coordinated throughout the platelet, but must be locally regulated.
In contrast, lamellipodia extend much more slowly (1.5–150-fold more slowly than pseudopodial extension), and spreading is often not continuous—there are frequent pauses, but retractions are uncommon. Initial rates are fast (~0.5 μm/min). Typically, as measured for 10 different platelets, 1.5–2 μm is accomplished in the first 8–10 min, but only very slow to imperceptible extension is seen after that. Only rarely is any retraction detected, and never more than a few tenths of a micron.
Platelet granules can also be detected by video microscopy with Nomarski optics. Granules are aggregated in the dome of central cytoplasm, termed the “hyalomere.” As defined by electron-microscopic analysis, platelets contain two types of granules, dense granules and alpha granules, and two types of cannalicular systems, an open cannalicular system (OCS) and a dense cannalicular system (DCS). The DCS contains the calcium sequestration system and the enzymes responsible for synthesizing arachidonic acid and other lipid mediators.
Both types of granules are visible by video studies (Allen et al., 1979). These, together with immunogold decoration for granule contents (Stenberg, et al., 1984) have demonstrated that degranulation occurs most often into the OCS, with granule contents subsequently released through the pores into the extracellular space. Degranulation is often observed just at the point when spreading activity slows, although it can occur any time during the activation process.
Further reorganization of the cytoskeleton occurs after spreading is accomplished, as can be detected by immunofluoresence using antibodies to adhesion plaque proteins such as vinculin or VASP, (Nachmias and Golla, 1991; Reinhard et al., 1992). Adhesion plaques continue to mature for at least an hour after contact.
Finally, for the platelet to mediate the subsequent contraction of the clot, which is complete one hour after activation (Pollard et al., 1977), further structural rearrangements must also occur to produce antiparallel alignments of the polar actin filaments necessary for contractile forces. These later steps in platelet behavior have not been studied by real-time video microscopy of individual platelets.
B. Actin in Platelets
1. Role of Actin
It is currently believed that actin reorganization is fundamental to the process of shape change. Platelets contain a large amount of actin (0.5 mM, estimated at 15–20% of the total protein) (Fox, 1993; Hartwig, 1999). During activation, there is a dramatic increase in the proportion of actin that is polymerized with increases in the length and changes in the organization of the actin filaments.
Unstimulated platelets have 40–50% of their total actin as filaments (Fox and Phillips, 1981, 1983). This increases to 70% within 20 sec after thrombin stimulation, as determined using the DNase I inhibition assay to determine the concentration of monomeric actin in platelet extracts lysed with Triton-X 100 before and after thrombin stimulation (Fox and Phillips, 1981). This was barbed-end elongation, since cytochalasin treatment abolished the effect entirely in a dose-dependent manner. It is estimated that there are ~2000 filaments in each platelet with an average filament length of ~1.1 μm (Hartwig, 1992, 1999).
2. Actin Structures in the Platelet after Spreading
In the glass-activated fully spread platelet, actin filaments form four structures that are distinct in their molecular composition and in their function (Fig. 3) (Bearer, 1995). The use of phalloidin to stain for actin allowed the determination of actin structures quickly by fluorescence microscopy. Thus, the actin structures in the spread platelet could be determined without the requirement for detergent extraction, which is necessary for electron-microscopy of whole mounts.
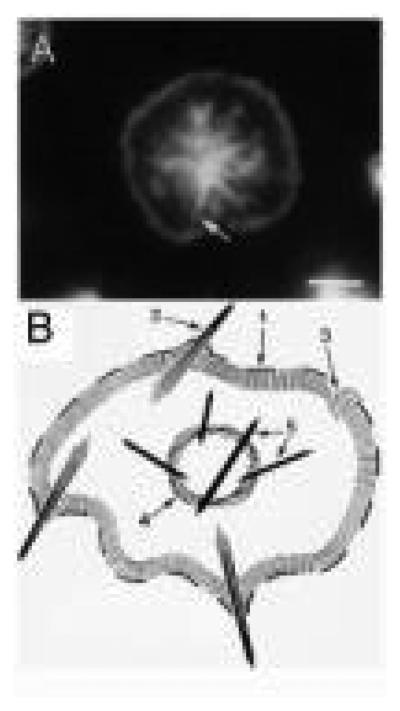
FIG. 3.
Actin structures by fluorescence. (A) An example of a platelet 15 min after spreading on glass imaged by fluorescence microscopy of F-actin as stained with phalloidin. Arrow indicates the position of a former filopodia. (B) Diagram of the actin structures in the spread platelets: (1) leading edge of the lamellipodium; (2) filopodia; (3) lamellipodium; (4) contractile ring; (5) stress fibers. (Reproduced from Bearer, 1995, by permission.)
The four actin structures in the fully spread platelet are filopodia, lamellipodia, stress-like fibers, and a contractile ring (Fig. 3). These structures appear analogous to actin filament structures formed in fibroblasts and other cells in culture (Karlsson et al., 1984). These different actin structures must form in the 10 min during which the platelet transforms from discoid to fully spread. The contractile ring appears to form first during the rounding stage, probably by myosin-mediated contraction of actin filaments from the membrane skeleton of the resting cell (Stark et al., 1991). Filopodia projection follows rounding. Some of these pseudopodia remain as filopodia. After attachment to the glass surface via both the hyalomere and the filopodia, lamellipodia form. Finally, adhesion plaques develop (Nachmias and Golla, 1991; Reinhard et al., 1992). These plaques form more quickly when platelets are spread on fibronectin coated coverslips.
To explain what is known and not known about the formation of these structures, we first describe key features of the biochemical behavior of actin relevant to platelet physiology. Next, we review information about the structure of actin in the resting platelet. Then, we discuss key actin binding proteins and their relationship to the formation of these structures.
C. Biochemistry of Actin
Actin is a 42 kDa, highly conserved protein that is found inside cells in either the globular (G-actin) or filamentous (F-actin) homopolymer states. In humans the known actin genes include six functional genes that encode three types of actin: α, β and γ (Pollard, 2001). Platelet actin is composed of β and γ isoforms, as is the case for most non-muscle cells. The three α isoforms are exclusively expressed in muscle (Kaitlina, 2001). All isoforms are very similar at the sequence level, and functional differences between them are not well defined. Each isoform will apparently co-polymerize with the other isoforms although there are differences in rate constants (Kaitlina, 2001). The β isoform has been implicated in initiating actin filament nucleation, but this activity may also be possible for the γ isoform.
That actin is so widely conserved across species and ubiquitously expressed has produced two experimental advantages for the study of platelets. Actin dynamics in other cells, even yeast, can usually be extrapolated to platelets; and platelets can be used as the source for proteins or as the model in which to investigate actin behavior likely to be universally applicable to other cells and other species.
The biochemistry of actin polymerization has been the focus of intense study for many years. It would not be possible to cover all the details in this review. Among the key features of actin relevant to platelets are some of the details of its molecular structure and several points regarding its polymerization.
Actin contains an ATP pocket that must retain either ATP or ADP for the actin molecule to retain its tertiary conformation. This pocket also harbors a divalent cation, either Mg2+ or Ca2+. The contents of this nucleoside pocket affect the rate of actin polymerization in vitro. Actin monomers have four functional surfaces, three that interface with other actin monomers in the filament, and one that is free and can bind other proteins.
In cultured cells, G-actin is in a dynamic equilibrium with F-actin. The amount of G-actin in cell extracts can be quantified using the DNase I inhibition assay (Fox et al., 1981), as DNase I binds actin monomers with 1:1 stoichiometry. Binding to actin inhibits DNase I activity and thus nucleoside hydrolysis provides an accurate measure of actin monomer concentration in the sample (Fox and Phillips, 1983). To study polymerization rates, actin covalently coupled to the fluorochrome pyrene via cysteine 373 is most commonly used (Kouyama and Mihashi, 1980). The pyrenyl actin fluoresces at a different wave length as a polymer than as a monomer, and this shift in wavelength is directly proportional to the number of actin molecules in filaments. Other biochemical methods include sedimentation amounts and rates, viscosity measurements, and light scattering. (Most traditional methods are described in detail in four publications, Methods in Enzymology, volumes 96, 134 and 186, and Methods in Molecular Biology: The Cytoskeleton.)
In addition to biochemical methods to measure various parameters of actin behavior, observation by electron microscopy has revealed elongation rates at the two ends (Pollard, 1986; Pollard and Cooper, 1986; Pollard and Mooseker, 1981).
Direct observation of filaments is also possible at the light level using phalloidin labeled filaments (Fig. 4). This was pioneered by Spudich for the study of myosin interaction with actin (Kron and Spudich, 1986) and later adapted for the imaging of filaments during severing (Fig. 4; Bearer, 1991) and branching (Blanchoin et al., 2000). Unlike microtubules, actin is too small for detection by DIC-video microscopy. Direct conjugation of a fluorochrome to actin monomers provides information about filament behavior independent of phalloidin (Bearer, 1992b). Such labeling is not bright enough to image the monomer, dimers, and trimers that nucleate assembly. Thus, we have yet to image actin polymerization directly in real time.
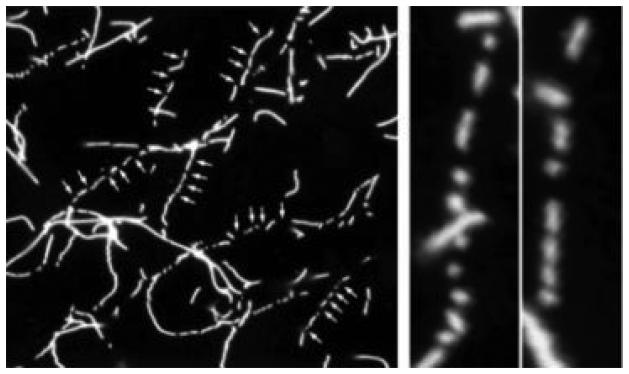
FIG. 4.
Direct observation of actin filament severing. Rhodamine-phalloidin allows individual actin filaments to be imaged by fluorescence microscopy. Such imaging revealed for the first time that gelsolin severed filaments and was phalloidin-blind. In addition, microscopy of the effects of proteins on actin filaments can be used as a biochemical assay to follow proteins through purification strategies. (Reproduced from Bearer, 1991, by permission.)
The actin filament is a bipolar double helix with a fast-growing “barbed” end and a slow growing “pointed” end (Korn et al., 1987; Pollard, 1986). The affinity of monomer for the barbed end (~1 μM) is ten-fold higher than that for the pointed end (~0.1 μM) in the presence of ATP. With ADP, both ends add monomer with the same low affinity as the pointed end with ATP. Cytochalasin, a fungal toxin, binds actin filaments at the barbed end and prevents filament growth (Cooper, 1987). It is generally accepted that most if not all actin filament growth occurs from the barbed ends inside cells.
Actin polymerization occurs readily in vitro in the absence of other proteins at actin concentrations above 1 μM and in the presence of physiologic salt (Korn et al., 1987). The amount of polymerized actin depends on monomer concentration. Thus, the proportion of actin in the filamentous form inside the cell can be regulated by decreasing the available monomer concentration. This is accomplished by monomer-binding proteins that bind G-actin and decrease the effective monomer concentration.
The initial step in actin polymerization is slow but can be accelerated by the addition of short actin filaments (Lal et al., 1984). Such filaments are thought to “seed” the crystallization process that underlies filamentation. It is believed that this initial slow, “lag”, phase is due to the kinetics of actin dimer and trimer formation. These small polymers are unstable, with estimated Kd equal to Ka. It has long been hypothesized that stabilization of dimers and trimers would accelerate polymerization. Such a stabilizer (often referred to as “the nucleator”) has long been sought, and a number of candidates proposed (Higgs and Pollard, 2001; Machesky and Insall, 1999; Pantaloni et al., 2001; Stossel, 1994; Stossel et al., 1999; Welch, 1999). All of these candidates have been found in platelets. Arp2/3, the recently discovered strongest contender for this nucleator, is described in more detail below (Section V.B.1).
To maintain stable filaments over time, the cell appears to use several strategies. One is continuous replacement of actin monomers in a filament by a process dubbed “treadmilling.” The working model of actin dynamics in quiescent cells predicts that turnover of existing filaments occurs by addition of monomer to the barbed end and loss of monomer from the pointed end. Indeed, such treadmilling of filaments has been observed in cells in culture (Wang, 1985). Another strategy to maintain stable filaments is to complex the filaments with other proteins that slow dissociation of monomers. In this case, monomers may be replaced at break points along the filament length.
In addition to maintaining filaments, cells must depolymerize filaments rapidly when changing shape. Actin is very slow to depolymerize in vitro. Therefore, cells must use a variety of strategies to get rid of inconvenient filaments, including severing them, blocking the barbed end to growth, accelerating pointed-end off rates, or removing side-binding proteins. Again, proteins with each of these functions have been found in platelets. More detail about a representative example of each type of protein is provided below (Section IV).
D. A Proteomics Approach to the study of Platelet Cytoskeleton: F-Actin Affinity Chromatography to Identify All of the Proteins that Bind to Actin
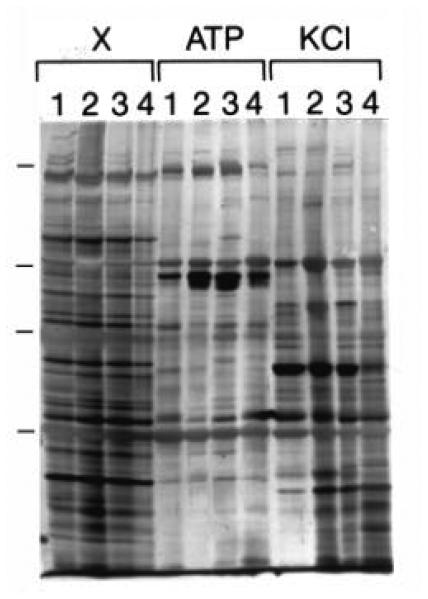
In an early effort to obtain a global picture of all the possible proteins involved in platelet actin dynamics, we used F-actin-affinity chromatography (Fig. 5; Bearer, 1995). With this approach, we identified 33 different proteins from extracts of ADP-activated human platelets that specifically bound to actin. This abundance was not surprising, but it did leave a logistical challenge: how to select among these 33 those that were most significant for platelet physiology.
FIG. 5.
F-actin affinity chromatography identifies a large number of F-actin-binding proteins in platelets. Four individual experiments are shown (lanes labeled 1–4). Platelets were activated for 1 min with ADP, protein solubilized by sonication in a low ionic strength buffer containing detergent, and the lysate clarified by high-speed centrifugation. Extracts (lanes labeled X) were loaded on filamentous actin affinity columns and eluted sequentially with 5 mM ATP (lanes labeled ATP) and 1.0 M KCl (lanes labeled KCl). Note that a large number of different protein species reproducibly elute with each of the elution buffers. (Reproduced from Bearer, 1995, by permission).
Our selection process was to generate antibodies to all those proteins that we could not identify as having been previously discovered in platelets or in other systems. This left us with 14 potentially novel proteins. We next used the antibodies to screen these proteins for their localization in platelets spread on glass. These antibodies were also useful to follow interesting proteins through more standard purification protocols by western blot, to screen expression libraries, and to determine location and expression of these proteins in other cells by immunofluorescence.
The 14 different proteins fell into nine categories based on their molecular weights and location in spread platelets, cultured fibroblasts, and human skeletal muscle (Bearer, 1995; Table 1). Some proteins were concentrated at all sites of actin polymerization, while others were restricted to filopodia, lamellipodia, stress fibers, or the contractile ring. Some antibodies stained skeletal muscle, indicating a possible function in contraction rather than polymerization. Some stained fibroblasts at the leading edge, while others stained stress fibers. Some stained diffusely in the cytoplasm, and some gave nuclear staining as well.
TABLE I.
Characteristics of Actin-Binding Proteins Identified by F-Actin-Affinity Chromatography from ADP-Activated Human Platelets
| Category | Cell type | Staining pattern | Ab no. | Molecular-weight (kDa) |
|---|---|---|---|---|
| 1 | Platelets Fibroblasts Muscle |
Contractile ring Perinuclear Nonreactive |
6 18 |
65 65 |
| 2 | Platelets Fibroblasts Muscle |
Leading edge Leading edge and perinuclear Z-band in muscle |
7 | 95 |
| 3 | Platelets Fibroblasts Muscle |
Contractile ring Stress fibers Both Z and A bands |
14 16 |
68 70 |
| 4 | Platelets Fibroblasts Muscle |
Leading edge and filopodia Filopodia Nonreactive |
19 20 |
110 110 |
| 5 | Platelets Fibroblasts Muscle |
Leading edge Diffuse Nonreactive |
21 | Many bands |
| 6 | Platelets Fibroblasts Muscle |
Lamellipodia/diffuse Nonreactive Z-band/diffuse |
26 28 |
75 75 |
| 7 | Platelets Fibroblasts Muscle |
Filopodia Nonreactive Diffuse | 30 | 72 |
| 8 | Platelets Fibroblasts Muscle |
Lamellipodium Lamellipodium A-band (myosin-like) |
32 | 43 |
| 9 | Platelets Fibroblasts Muscle |
Leading edge Diffuse Actin-like (I-band) |
33 | 130 |
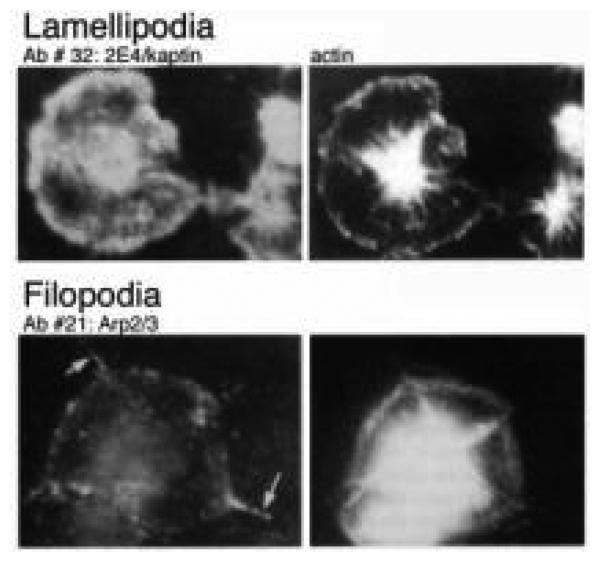
Three of these proteins were selected for further study: Kaptin/2E4, VASP, and Arp2, subsequently found to be a member of the Arp2/3 complex. These proteins were chosen as representative of three of the more interesting classes from among the nine different patterns. Antibodies against kaptin/2E4 stained at the edge of the spread cell, antibodies to VASP stained focal adhesions, and antibodies to Arp2 stained filopodia (Fig. 6). Each of these proteins is discussed below in the context of their participation in specific aspects of the actin dynamics occurring during shape change.
FIG. 6.
Immunofluorescence of actin-binding proteins. Two examples of patterns obtained using antibodies against platelet proteins eluting from the actin columns. Ab #32 (2E4/kaptin) stains lamellipodia and the cytoplasm, whereas Ab #21 (Arp2/3) stains the filopodia at this stage of spreading. (Adapted from Bearer, 1995, by permission.)
Proteins that interact with actin to regulate the polymerization/depolymerization of individual filaments, or the organization of groups of filaments into higher order structures may play several roles during different stages of platelet shape change. Although most proteins have only one specific type of interaction with actin, the state of the actin and the presence of other proteins can influence how this specific activity affects the cytoskeleton of the cell as a whole. Thus a single activity can produce many different outcomes when viewed from the perspective of the whole platelet during its many morphological changes.
In this review, specific proteins are described in the section pertaining to a particular stage in shape change when their activity is most likely to be functionally significant. However, several of these proteins are likely to mediate events that occur during more than one stage.
III. Actin Filaments in the Resting Platelet
A. Two Pools of Actin: Monomeric and Polymeric
The resting platelet has two pools of actin filaments: those associated with the membrane skeleton, and those that course through the cytoplasm. In addition to two filamentous pools, resting platelets have a large pool of monomeric actin (300–350 μM) (Fox and Phillips, 1983; Nachmias and Yoshida, 1988). This is hundreds of times larger than the critical concentration (0.2 and 1.0 μM for barbed and pointed ends, respectively) (Pollard and Cooper, 1986). Thus, mechanisms exist that prevent polymerization and inhibit actin dynamics. These mechanisms must be switched off for shape change to occur.
B. Two Structures Containing Filamentous Actin
1. The Membrane Skeleton
Early electron microscopy had revealed that platelets contain microtubules as well as microfilaments–6–8-nm filaments presumably composed of actin (Nachmias and Yoshida, 1988; White, 1969, 1984; White and Clawson, 1980). By thin section transmission electron-microscopy (TEM), microtubules were found in a coil at the periphery of the disc in the resting cell (White, 1969). Although the microtubules are not the focus of this review, it should be mentioned that interactions between actin and microtubules, detected in TEM but as yet poorly understood biochemically, may be involved in some aspects of actin structure in the resting platelet and/or in actin dynamics during shape change (White and Krumweide, 1976; White and Rao, 1998).
Microfilaments were best observed in TEM after partial lysis with digitonin or polyethylene glycol, which extracted some of the dense cytoplasmic material. In these extracted platelets, microfilaments were predominantly found in the area between the membrane and the microtubule coil. A subset of these filaments was observed attached to the plasma membrane, some in evenly spaced arrays perpendicular to the membrane (Zucker-Franklin, 1970). A lysine-based fixative was developed that better preserved these sparse filaments in resting platelets against the effects of osmium for TEM (Boyles et al., 1985). This lysine fixation also confirmed the presence of a microfilamentous network in the deep cytoplasm of resting platelets which had been previous imaged in negatively stained whole mounts (Nachmias, 1980).
While these were elegant images of the platelet, their preparation required denaturing steps that could have perturbed the normal architecture of the living platelet. Hence, application of quick-freeze techniques to capture unfixed, living specimens and image them as a whole rather than after sectioning was needed (Bearer, 1983, 1990; Nakata and Hirokawa, 1987). Such quick-freeze studies mainly revealed information about the membrane skeleton because the central cytoplasm in whole platelets was too dense to permit imaging of filamentous structures.
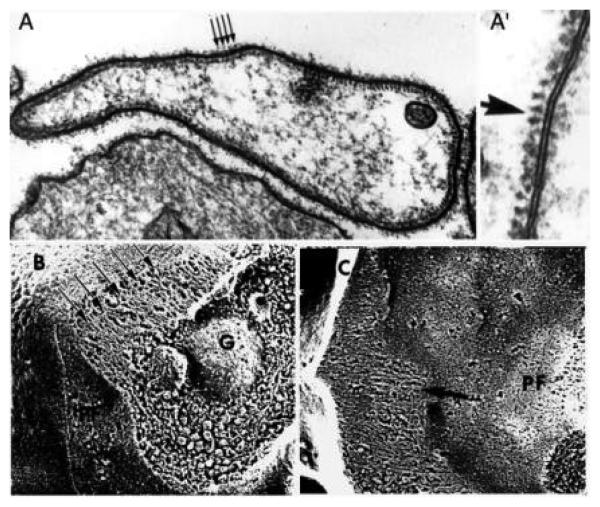
In platelets captured live by quick freezing, the membrane skeleton was found to be composed of a regular array of the submembranous filaments, as observed in quick-frozen deep-etched platelets (Fig. 7) (Bearer, 1983, 1990). Quick-freezing avoided chemical fixation while fracturing of frozen specimens followed by etching of the water from the surface revealed submembranous filaments in their natural state. In these intact specimens, the submembranous filament system was a continuous shell of apparently parallel filaments lying just beneath the plasma membrane of the resting platelet (Fig. 7A). The regular spacing of these filaments was confirmed in freeze-substituted platelets stained with tannic acid and observed by TEM (Fig. 7A′). These filaments had a diameter of ~9 nm and were separated from each other with a periodicity of 15 nm and from the plasma membrane by a similar distance (15 nm). In these preparations, the membrane and its skeleton appeared to have three layers–the lipid bilayer, a fuzzy coating just beneath it, and a cage of filaments inside that.
FIG. 7.
The platelet membrane skeleton is composed of three layers. By thin section, the platelet membrane is lined by a periodic array of filaments that appear as dots when transected in sectioning for thin-section electron microscopy (A and A′) and as a linear array in quick-freeze deep-etch replicas of living platelets frozen in suspension (B and C). In A′ the cytoplasmic surface is to the left of the membrane. Note the rough texture of the P-face of the plasma membrane (PF) which is embossed by the underlying net of the membrane skeleton to which the sheet of actin filaments attaches. The three layers are best appreciated in (C), where the extracellular surface (EF), the membrane skeleton (PF) and the filaments (arrows) are shown. A granule is seen lying beneath the membrane (G). (Adapted from Bearer, 1990, with permission.)
This membrane skeleton was more complex than that described for the red blood cell (Shen et al., 1986). The submembranous filament network was predicted to contain spectrin and significantly more actin than the red cell membrane skeleton (Bearer, 1983, 1990). It was proposed that the platelet membrane skeleton must be dissociated for shape change to occur.
The structure of the resting platelet membrane skeleton has also been imaged by metal replicas of platelets extracted with 7.5% Triton, and quick-frozen freeze-dried (Hartwig and DeSisto, 1991). This technique allowed immunogold labeling for the determination of its molecular composition. In these extracted specimens, the membrane skeleton appeared as a tightly woven planar sheet. The membrane skeleton was further simplified after Triton extraction by sedimentation onto a coverslip, which sheared it from the underlying filament core. This simplified membrane skeleton appeared as a uniform porous sheet, composed of thin (4–5 nm diameter) filaments (240 nm maximal length) connected at intervals by globular particles. As previously predicted, spectrin was identified as the 4–5-nm filaments, and actin-binding protein (ABP) was found at the interstices of the network by immunogold labeling. Double-label immunogold for ABP and GPIb demonstrated colocalization in the globular particles at the interstices of the spectrin net. In addition to spectrin, myosin II was also found in the membrane skeleton. These structural results confirmed and extended previous biochemical data describing the molecular composition of the membrane skeleton as obtained by differential centrifugation (Boyles et al., 1985; Fox, 1985, 1993; Fox et al., 1988).
It is not clear whether this extracted membrane-skeleton represents the submembranous filaments seen in thin sections and quick-frozen living platelets, since the extracted, sheared cytoskeleton had none of the regular spacing of linear elements seen in the other preparations and the measurements of the structures were also different between intact and extracted preparations. Images of quick-frozen deep-etched platelets captured live by freezing suggest that the membrane skeleton has two layers (Fig. 7). The first is a tightly woven sheet directly linked to integral membrane proteins such as GPIb which coats the undersurface of the plasma membrane and contains spectrin, ABP, and some short actin filaments. The second is a deeper layer containing a regularly spaced cage of actin filaments thickened by side-binding proteins and attached along their sides to the spectrin-ABP web. Thus, one interpretation is that different layers are imaged more effective in each type of preparation.
2. The Filamentous Core
Triton-extracted platelets centrifuged onto coverslips have a second component in addition to the membrane skeleton which appeared to be the filamentous core of the resting platelet cytoplasm. This structure was composed of a dense aggregate from which long filaments, 0.5–1.0 μm in length, extended (Hartwig and DeSisto, 1991). Labeling with the S1 fragment of myosin identified virtually all these filaments as actin. These radiating actin filaments were attached to the membrane skeleton along their sides. Based on these observations, it was proposed that curved bundles of actin filaments extend in the living cell from the central core to contact the membrane skeleton. These filaments may be continuous with the regularly spaced filaments that line the membrane.
Attachment between these filaments and the membrane skeleton must be very strong. Despite extraction and sedimentation, actin filaments from the central core retained their attachment to the membrane skeleton. The sturdiness of this architecture argues for its role in the maintenance of the discoid shape of the platelet during circulation, although this idea has not yet been directly tested.
3. The Biochemistry of Attachment of Membrane Skeleton to the Surface Membrane
The attachment of the membrane skeleton to the surface plasma membrane of the platelet is at least partly mediated through the binding of ABP to the cytoplasmic tail of GPIb, a subunit of the von Willebrand factor (vWF) receptor (Cunningham et al., 1996; Ezzell et al., 1988; Fox, 1985; Fox et al., 1988) When vWF attaches to subendothelial basement membrane, it undergoes a conformational change that renders it sticky to platelets (Englund et al., 2001; Jackson et al., 2000). Biochemical analysis of the membrane cytoskeleton was initially performed using differential centrifugation of resting platelets after Triton solubilization. Platelets are first treated with detergent to permeabilize the membrane and release cytosolic proteins and other molecules. The actin cytoskeleton and all its attached components can be separated by sedimentation at low g force (10,000 × g for 15 min). This low-speed pellet, termed the “Triton-insoluble cytoskeleton,” is useful for the study of actin in activated platelets. But in the resting platelet, very little material sediments in the Triton-insoluble low-speed pellet. The membrane skeleton of the resting platelet is collected by high speed centrifugation (100,000 × g for 1 hr) of the low speed supernatant (Fox, 1985, 1993). Proteins in the high-speed pellet were identified by Western blot. Presence in this fraction was indicative of association with macromolecular structures, such as the membrane skeleton, as was subsequently confirmed by immunogold labeling of membrane skeletons discussed above.
The dense mesh of the resting platelet membrane skeleton probably plays a significant role in maintaining the smooth surface contours of the discoid platelet and in restraining the platelet from projecting pseudopodia while in circulation. For shape change to proceed, this membrane skeleton must be dismantled. Mechanisms to do that involve depolymerization and proteolysis of the membrane skeleton as discussed below.
IV. Inhibition of Actin Polymerization in Resting Platelets
In addition to physical constraints to shape change contributed by the membrane skeleton, other factors must prevent actin polymerization in the resting platelet, since the actin monomer concentration in the resting platelet, estimated at 0.3 mM, is significantly above the critical concentration for polymerization (0.2–1.0 μM). Some of the biochemical restraints have been elucidated in the form of proteins with three different effects on actin. These three effects—monomer sequestration, barbed-end capping, and filament stabilization—synergize to inhibit polymerization and stabilize existing filaments.
Monomer binding proteins sequester actin monomer. This lowers the free monomer concentration and thereby raises the amount of actin required for polymerization to occur. When the barbed ends of actin filaments are capped, monomer addition is possible only on the pointed end. This further raises the amount of monomer required for filament elongation because the pointed end has a 10-fold lower affinity for monomer than the barbed end. Stabilization of filaments by proteins that bind along their length inhibits depolymerization from the pointed end, which would otherwise occur for barbed-end capped filaments at monomer concentrations below the critical concentration for pointed-end stability.
A. Monomer-Binding Proteins
Platelets have two major monomer-binding proteins: profilin and thymosin β4 (Tβ4) (Table II). Several other proteins found in platelets also bind monomer in vitro, including cofilin and gelsolin, but these have additional effects on actin filaments which are likely more important in the overall physiology.
TABLE II.
Examples of Actin-Binding Proteins in Platelets
| Type of Activity | Concentration | Cofactors |
|---|---|---|
| Monomer-binding proteins | ||
| Profilin | 0.5 μM | Binds polyproline repeats, inhibited by PIP2 |
| Thymosin β4 | 500 μM | |
| Depolymerizing factors | ||
| Cofilin/ADF | 50 μM | Activated by dephosphorylation, inactivated by phosphorylation by LIM kinase |
| Barbed-end capping proteins | ||
| Gelsolin | 12 μM | Calcium, PIP2 |
| Capping protein | 2–5 μM | PIP2 |
| gCap 39 | 5 μM | |
| Side-binding proteins | ||
| VASP | 5 μM | Phosphorylation by cGMP/cAMP kinases |
| Actin-binding protein | Proteolysis | |
| Spectrin | Proteolysis | |
| Nucleator | ||
| Arp2/3 | ND | N-WASp/Scar |
| Kaptin (2E4) | ND |
1. Thymosin β4 (Tβ4)
Soluble actin in the platelet is found in a 1:1 complex with Tβ4 (Safer et al., 1991). Tβ4 is a 5-kDa protein that binds only to actin monomers. Its only function detected so far inside cells is actin monomer binding (Weber et al., 1992). When actin monomer is bound to Tβ4, it cannot polymerize. Tβ4 has a Kd for monomeric actin of 0.4–0.7 μM, and is present in platelets at ~560 μM. Mathematical calculations assuming that most (95%) of the barbed ends in platelet are capped and, based on these in vitro binding constants, predict that most of the monomeric actin in platelets is sequestered in the form of Tβ4–actin dimers.
Profilin facilitates the transfer of monomer from Tβ4 to the free barbed ends of actin filaments (Goldschmidt-Clermont et al., 1992; Pantaloni and Carlier, 1993). Free barbed ends have a higher affinity for actin monomer than Tβ4. Tβ4 releases actin monomer in the presence of free barbed ends. There may be other mechanisms that inhibit binding of Tβ4 to actin and inactive it, since release of monomer is a necessary step in the formation of new filaments.
2. Profilin
Profilin, a 14-kDa protein initially isolated from platelets (Carlsson et al., 1977), was first thought to be the monomer-binding protein principally responsible for monomer sequestration in platelets, but there is not a high enough concentration for profilin (0.5 μM) to account for all of the sequestration. Profilin's affinity for actin is affected by PIP2 (Lassing and Lindberg, 1985), which dissociates it from actin. Profilin acts as an ATP exchange factor for actin—profilin-stabilized actin monomers are “recharged” by exchanging hydrolyzed ADP for fresh ATP (Goldschmidt-Clermont et al., 1992; Goldschmidt-Clermont et al., 1991a). Since ATP-actin polymerizes from the barbed end ten times more rapidly than ADP-actin, such exchange can have a profound impact on the critical concentration of actin required for filaments to form. Consistent with this model are results from platelets in which the amount of profilin bound to actin increases during the first minute after thrombin activation (Lind et al., 1987).
There are two known human profilin genes that produce three profilin isoforms, profilin I and profilins IIa and IIb ((Di Nardo et al., 2000; Lambrechts et al., 2000a), but only profilin I has been detected in platelets (Kwiatkowski and Bruns, 1988). The two isoforms have 62.1% sequence identity (Honore et al., 1993) but all three bind actin monomers and binding is regulated by PIP2, although the affinities differ (Gieselmann et al., 1995; Machesky et al., 1990).
Profilin probably has other roles besides monomer binding, including participation in signal transduction. Profilin interacts with phospholipase C, inactivating it (Goldschmidt-Clermont et al., 1991b). Actual binding to PLC occurred after activation, when PLC is phosphorylated and other cytoplasmic proteins dissociate from it. In yeast, profilin deficiency slows growth (Haarer et al., 1993), suggesting that in yeast, profilin may play a role in signaling and cell cycle.
B. Capping Proteins
1. Gelsolin
Gelsolin was initially believed to prevent actin polymerization in the resting platelet by binding to the barbed ends and blocking their elongation. However, only about 10% of the platelet gelsolin is associated with actin in the resting cell (Lind et al., 1987), and this is not enough to account for capping of 2000 barbed ends. In addition, in gelsolin-minus mouse platelets, there is only a small, 10%, increase in actin filaments in the resting cell, which demonstrates that if the filaments are capped, then some other protein must be involved (Barkalow et al., 1996). Barbed-end capping proteins other than gelsolin found in platelets include CapZ (also known as capping protein, cap32/34), flightless 1 and gCAP39.
2. Capping Protein
CapZ/capping protein has been considered the most likely to be responsible for most of the capping of filaments in the resting cell (Barkalow et al., 1996; Nachmias et al., 1996). There is 2–5 μM capping protein in platelets. Reports are conflicting about its association with the Triton-insoluble cytoskeleton during activation. In the resting platelet, most (75–80%) of the total capping protein pellets is in the high-speed supernatant which contains both the core and membrane skeletal filaments (Nachmias et al., 1996). After thrombin stimulation (10 sec), 15% of this capping protein leaves the high speed pellet and becomes soluble. In another report (Barkalow et al., 1996), 35% of the capping protein is found in the low-speed Triton-insoluble pellet, which primarily composed of the central core filaments of the resting platelets. After thrombin stimulation (20 sec), the amount of capping protein in the Triton-insoluble low-speed pellet increases to 60%. Thus, while some capping protein dissociates from actin altogether in the early stages of thrombin stimulation, that associated with the low-speed pellet increases. This increase in the low-speed pellet likely represents a tighter association between the filaments of the core and those in the membrane, possible a consequence of the contractile events occurring during rounding, which is the morphological correlate of this time course. Indeed, in both studies immunomicroscopy localizes capping protein to the membrane of the lamellipodia after spreading.
Taken together, these two studies suggest that most of the capping protein is associated with the membrane-bound filaments in the resting platelet, since these are soluble at low g force after Triton extraction and would not be found in the low-speed pellet. After thrombin stimulation, the membrane skeleton is not solubilized and becomes more firmly attached to the core filaments, which results in the appearance of membrane-associated proteins such as capping protein in the low-speed pellet. Variability of detection of capping protein in the supernatant probably reflects a dynamic equilibrium during the process of polymerization, when capping protein comes on and off filaments in a time course too rapid to capture by these biochemical techniques.
It has been postulated that uncapping of the barbed ends by capping protein accompanies activation and be responsible for the rapid polymerization of filaments occurring (Hartwig, 1999; Kwiatkowski, 1999; Stossel, 1994; Stossel et al., 1999). Thus it was expected that capping protein would shift from an insoluble filament-associated state, to a soluble unattached state in a single event after stimulation. However, this model now appears too simplistic. Instead of a single uncapping event, reversible dynamic capping and uncapping of growing filaments probably continues throughout the process of shape change. This focuses elongation on those particular filaments that are not capped (Pantaloni et al., 2001). The formation of complex multi-filament networks such as those formed during platelet spreading likely involves continuous rounds of polymerization, severing, and capping. In the resting cell, uncapping is inhibited and this prevents actin remodeling.
C. Side-binding Proteins Involved in Maintaining Discoid Shape
1. Vasoactive-Stimulated Phosphoprotein (VASP)
Dismantling of the actin in the resting platelet with inactivation of inhibitory proteins is a necessary first stage of shape change, rounding. Vasoactive substances, such as nitroprusside, block platelet shape change and activate cGMP & cAMP-dependent protein kinases. VASP was originally identified as the major substrate of cAMP and cGMP kinases in human platelets (Halbrugge et al., 1990; Halbrugge and Walter, 1989). VASP was considered an important inhibitor of platelet activation which could serve as a target for antithrombotic drugs. Knockout experiments have confirmed this idea (Aszodi et al., 1999; Hauser et al., 1999). Inhibition of platelet aggregation by low doses of cyclic nucleotides is impaired in VASP minus platelets (Aszodi et al., 1999). VASP-minus platelets also show an increase in aggregation in response to collagen, increased thrombin-induced adhesion to fibrinogen, and enhanced expression of P-Selectin (Hauser et al., 1999). Additional details about VASP that are not specifically related to platelets can be found in a recent review (Rienhard et al., 2001).
VASP was the first member of the VASP/Ena/Evl family of proteins to be identified. Ena, a gene that enhanced the phenotypic defect of Ableson mutations in Drosophila (Gertler et al., 1995) is more ubiquitously expressed, although platelets apparently have only one member of this family of proteins, VASP. Ena/VASP proteins have three structural domains. The amino and carboxy domains are termed Ena–VASP homology domains 1 and 2 (EVH1 and 2) while the middle domain contains a polyproline-rich stretch (Reinhard et al., 1992).
VASP binds via the polyproline repeats directly to the Listeria bacteria protein, ActA, which is the bacterial protein required for actin polymerization activity (Niebuhr et al., 1997). However, VASP was not found to be required for the reconstitution of bacterial actin-based motility in vitro (Loisel et al., 1999). In vitro, VASP binds polyproline (Bearer et al., 2000a) and profilin (Reinhard et al., 1995). VASP also interacts with vinculin, and this regulation appears to be regulated by phosphotidylinositide 4,5-bisphosphate (Huttelmaier et al., 1998).
In vitro, VASP both bundles filaments and nucleates actin polymerization (Fig. 8; (Bachmann et al., 1999; Bearer et al., 1996, 2000a; Dunaway et al., 1997; Huttelmaier et al., 1999; Lambrechts et al., 2000b; Manchester et al., 1998). It is controversial whether bundling of filaments by VASP actually occurs in vivo, although the actin bundles imaged by electron microscopy in platelets (Gonnella and Nachmias, 1981; Nachmias, 1980) are very similar to those produced by VASP in vitro (E. L. Bearer, unpublished observations). VASP is localized along the length of the filopodia that form in the initial stages of shape change, and thus is likely to be involved in the loose bundling of those filaments (Bearer et al., 2000a). Differences in the location of VASP in platelet filopodia and in the filopodia of cultured cells raise the question of whether platelet filopodia are formed by the same mechanisms that form filopodia in other cell types. Our working hypothesis is that the filopodial-like projections of platelets are in fact a heterogeneous mix of actin-based structures, some of which are identical to filopodia produced by cultured cells and others of which are produced by alternate processes. Indeed, microtubules have been found in some filopodial-like protrusions in platelets but are not detected in filopodia in other cell types (White and Krumweide, 1976; White and Rao, 1998).
FIG. 8.
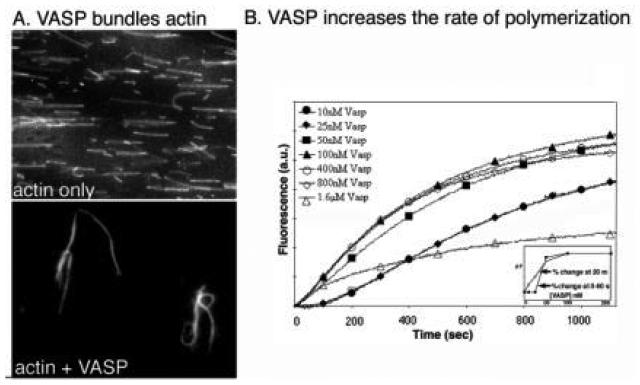
VASP bundles actin filaments and nucleates polymerization. (A) Fluorescently labeled filaments (top panel) mixed with VASP (lower panel) display thick bundles proportional to the amount of VASP added. (B) Polymerization of pyrene-actin(2 μM) is accelerated in the presence of VASP. Calcium-actin is more sensitive to VASP (inset, open squares) than magnesium-actin (inset closed squares). (Reproduced by Bearer et al., 2000, by permission.)
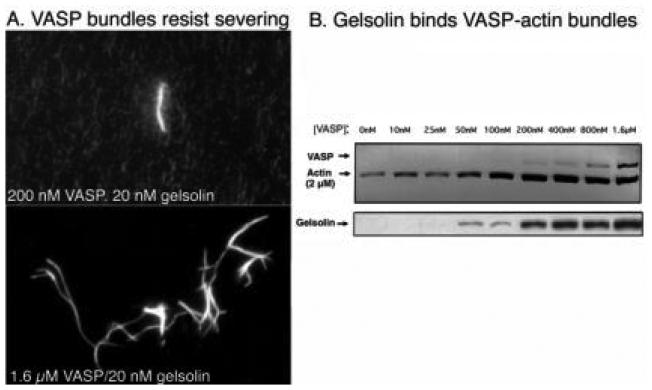
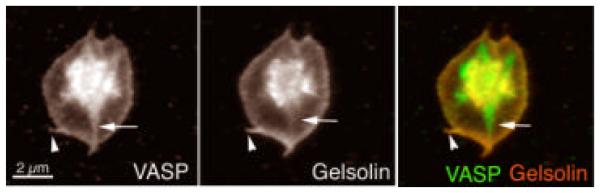
VASP-bundled filaments resist gelsolin severing (Fig. 9; Bearer et al., 2000a). Direct observation of labeled actin allows visualization of gelsolin severing (Bearer, 1991). VASP bundles these filaments, with increasing numbers of bundles seen at increasing VASP concentrations. Addition of gelsolin causes severing of individual filaments but not of the bundles (Fig. 9). At high VASP:actin ratios (1:5) all filaments are in bundles and no severing is detected. That gelsolin could bind to VASP-actin bundles was shown by co-sedimentation of both proteins with actin filaments. In fact, the amount of gelsolin bound to the bundles increases as a function of VASP. The amount of actin that sediments also increases with VASP concentration, confirming biochemically the results obtained by microscopy (Fig. 9B). Thus, VASP inhibits gelsolin severing but not its binding to actin filaments. Inside platelets, VASP and gelsolin colocalized, particularly at the leading edge of the lamellipodia during the later stages of spreading (Fig. 10). VASP was concentrated in focal adhesions where no gelsolin was detected.
FIG. 9.
Gelsolin binds but does not sever VASP-bundled filaments. (A) By fluorescence microscopy, VASP bundles filaments even at low concentrations. Only the bundles resist the severing effects of gelsolin. The shower of dots in the background (top panel) indicates short pieces left after gelsolin severing of individual filaments not part of a bundle. At higher VASP concentrations, all filaments are grouped into bundles, and no severing is detected (lower panel). (B) Sedimentation of VASP-actin bundles demonstrates that VASP protects filaments from solubilization by gelsolin, but gelsolin binds the filaments. Increasing amounts of gelsolin are found in the sediment, directly proportional to the amount of VASP. (Reproduced from Bearer et al., 2000, by permission.)
FIG. 10.
Immunofluorescence of VASP and gelsolin in a spread platelet. VASP and gelsolin colocalize in the lamellipodia and filopodia (arrowheads) but not in stress fibers, where only VASP is found (arrows). (See also color insert.)
In addition to side binding and bundling, VASP also nucleates filament assembly both with Mg2+-actin and with Ca2+-actin. With Ca2+-actin, nucleation activity is greatest, with maximal effects at 1:50 actin:VASP ratio, while with Mg2+-actin, a 1:1 ratio is required for maximum activity (Bearer et al., 2000a).
Stabilization of filaments by VASP explains all of its cellular behaviors. The resistance to severing of VASP-bound filaments could serve to stabilize filaments in the resting platelet. Conversely, during activation, VASP stabilization of new filaments would potentiate de novo polymerization. Thus, VASP could have one activity, stabilization, which would produce opposite results (prevention of remodeling and potentiation of polymerization) inside the platelet. These two activities could appear paradoxical when viewed from the context of the whole cell.
Phosphorylation of VASP apparently regulates its affinity for actin, its ability to bundle filaments and it nucleating activity (Harbeck et al., 2000; Horstrup et al., 1994; Lambrechts et al., 2000b). Phosphorylation occurs at three serine/threonine residues and is mediated in vitro and in vivo by cAMP- and cGMP-dependent protein kinases (Butt et al., 1994). Phosphorylation does not affect VASP binding to profilin (Reinhard et al., 1995). Which phosphorylation site(s) is required for this activity has not yet been defined, although phosphorylation of serine 157 is the most likely regulatory site.
In fibroblasts, overexpression of VASP slows migration, while VASP/Ena knockouts have accelerated motility (Bear et al., 2000). Thus, VASP/Ena family members play a physiological role in cell adhesion and shape change required for motility. Whether VASP plays an inhibitory role exclusively, or also participates in filament formation inside the cell remains to be determined.
VASP could be a “switch,” playing both an inhibitory and a promotional role in actin dynamics. By releasing filaments, it could permit severing, and by binding it could potentiate elongation by stabilizing nascent polymers. That VASP is phosphorylated on three sites further supports its role as a switch. Regulation of VASP phosphorylation state involves kinases of the cGMP/cAMP type (Butt et al., 1994; Halbrugge et al., 1990) and serine/threonin phosphatases (Abel et al., 1995). Hence, VASP is phosphorylated in platelets in response to signals that inhibit platelet activation. VASP's stabilization of filaments and its location in adhesion plaques suggest a second physiologic role later in thrombus formation: formation of stress fibers. this is discussed further below (Section VI.B.1).
D. Other Proteins: Actin Biding Protein (ABP), Tropomyosin, and Spectrin
Stabilization of the actin filaments in the resting platelet is also mediated by three well known actin-binding proteins, actin-binding protein (filamin), tropomyosin, and spectrin (Hartwig, 1999). Significantly, in the platelet, two of these proteins appear to be major substrates for the calcium-dependent protease activated by calcium influx upon activation (Fox et al., 1985, 1987). Not content with merely disassociation, the platelet digests a number of proteins during activation. Calpain, a calcium-dependent protease, is apparently responsible for this proteolysis (Croce et al. 1999) and is discussed below.
V. Actin Filaments in Activated Platelets
Upon activation, existing filaments are disassembled and new filaments form. The first step, rounding, apparently depends on depolymerization of existing filaments. Pseudopodial protrusion may occur coincident with disassembly, but filopodia appear to form after rounding. Lamellipodia that appear between filopodia when platelets spread on glass are later steps. Each of these events is likely to depend on different actin-binding proteins with unique activities.
A. Rounding: Disassembly and Contraction
The first morphologically recognizable stage in platelet activation is rounding, when the platelet loses its discoid form and becomes a sphere. All agonists that have been studied produce this initial result. Thus, it appears that rounding can be activated through any of a number of signaling pathways. Because rounding is a very brief step in the sequence of shape change, it has been difficult to study biochemically.
Rounding induced by agonists is thought to be secondary to two main events: (1) severing of existing actin filaments; (2) chemical alteration of actin-filament binding proteins, including phosphorylation, dephosphorylation, and proteolysis, as well as inhibition or activation of their interaction with actin by small G proteins and small metabolites, such as Ca+ and PIP2. During rounding, the connections between the core actin filaments and the membrane skeleton appear to be dissociated, the membrane skeleton is dismantled, and the core filaments contract into a microfilamentous shell, the contractile ring (Stark et al., 1991). This process is probably necessary for subsequent protrusive activity at the membrane surface.
1. Severing
a. Gelsolin
Gelsolin (82 kDa) was initially considered responsible for most of the actin dynamics detected in platelets because of its many different effects on actin (reviewed in Kwiatkowski, 1999; Sun et al., 1999). The abundance of gelsolin in platelets (5 μM) and its pronounced effect on actin filaments in vitro argue that it plays an important role in platelet shape change. Studies in gelsolin knockout mice suggest that gelsolin's main role is to sever existing actin filaments, thereby allowing reorganization of the cytoskeleton.
Gelsolin severs actin filaments in the presence of calcium (Bearer, 1991) which causes a rapid measurable decrease in actin polymers (Yin et al., 1981). In the platelet, calcium levels rise from 10–20 nM to 3–5 μM upon activation (Brass, 1984; Davies et al., 1989). After severing a filament, gelsolin remains bound to the barbed end, preventing elongation. This barbed-end binding would be expected to potentiate depolymerization from the pointed ends. Gelsolin may bind as many as three monomers (Bryan, 1988). Two apposite functions have been proposed for this binding: monomer sequestration, which depresses filament formation, or nucleation from the pointed end, which would enhance polymerization.
Uncapping of gelsolin-capped filaments has been proposed as a mechanism for the reorganization of actin filaments in platelets (Hartwig, 1992; Hartwig et al., 1995). Gelsolin releases actin slowly in the presence of PIP2, a metabolite of phospholipase Cγ (Yin et al., 1988). Uncapping of the severed ends is mediated by a rise in PIP2, which also occurs upon platelet activation. If gelsolin were first to sever and then release the barbed ends, rapid elongation of new filaments could ensue.
Gelsolin-severed filaments would be expected to depolymerize from the pointed ends. This would provide substantial increase in monomeric actin, which would accelerate elongation of new filaments nucleated at their pointed ends by other proteins (Bearer et al., 2000a).
Gelsolin-null mice have no overt bleeding problems, but when measured in vitro, bleeding time is prolonged to twice that of normal mice (Witke et al., 1995). Resting platelets from gelsolin-minus mice have an increased actin filament concentration (10–33% more than wild type) as measured by phalloidin staining followed by fluorescence cell sorting. While contact with glass causes a calcium-dependent severing of the long filaments in the normal platelet, this fragmentation is not observed in gelsolin-minus platelets. Thus, prolonged bleeding time is likely due to a loss in the first step of shape change during which the existing filaments are severed. When filaments are not severed, depolymerization would be slower, and monomer pool would not rise rapidly. Thus, slowed actin polymerization could be a result of a decrease in monomer availability.
That gelsolin-minus platelets still have actin polymerization has been explained by the presence of other proteins whose functions replace those of gelsolin. Indeed, capping protein appears to be coordinately regulated with gelsolin (Barkalow et al., 1996). In gelsolin-minus mouse platelets, an increased amount of capping protein associates with the cytoskeleton of detergent-permeabilized platelets after low-speed centrifugation, which corresponds to the increased F-actin. After thrombin stimulation, the amount of capping protein in the cytoskeleton increases, but this behavior is not significantly different in gelsolin-minus platelets as compared to wild type.
b. Cofilin/Actin Depolymerizing Factor (ADF)
Cofilin/ADF was first identified in platelets as an 18–19-kDa phosphoprotein dephosphorylated by a thrombin-activated process (Imaoka et al., 1983). This phosphoprotein was subsequently identified as cofilin (Davidson and Haslam, 1994). The molar concentration of cofilin/ADF in platelets is very high, up to 10% that of actin, or 50 μM. In platelets, dephosphorylation of cofilin/ADF is activated by calcium, independent of protein kinase C, and by GTP S, which activates G proteins. Cofilin/ADF is phosphorylated in resting platelets exclusively on the serine 3 at the amino terminus. Phosphorylation on serine of cofilin/ADF inhibits its activity and dephosphorylation activates it. In platelets, ADP- and thrombin-induced dephosphorylation of cofilin/ADF is inhibited by 1-napthylphosphate but not by okadaic acid.
In vitro, cofilin/ADF appears to act as an “actin dynamizing factor,” increasing by 25-fold the rate of treadmilling, via increasing monomer loss from the pointed end without effecting barbed ends (Carlier et al., 1997). Cofilin coats actin filaments, possibly preferentially binding to ADP-actin subunits, and produces a subtle change in the filament structure (McGough, 1998). Addition of recombinant plant cofilin/ADF to platelet extracts increases the rate of actin-based motility of the Listeria bacteria (Carlier et al., 1997). Although cofilin/ADF was reported to sever filaments (MacIver et al., 1991), the filaments in this study were imaged with phalloidin, which inhibits cofilin's ability to increase depolymerization. A mechanism invoking severing is now considered incompatible with results obtained using other methods to study ADF/cofilin effects on actin.
In cultured cells, cofilin/ADF dephosphorylation is stimulated by EGF (Chan et al., 2000). Injection of anti-cofilin antibodies inhibits EGF-induced actin polymerization at the cell cortex in metastatic mammary adenocarcinoma cells (MTLn3). Cofilin/ADF is phosphorylated by the LIM-kinase family (Arber et al., 1998; Yang et al., 1998). Overexpression of the kinase domain of LIM kinase results in near total phosphorylation of cofilin/ADF and is sufficient to completely inhibit the appearance of barbed ends and lamellipodial protrusions in response to EGF (Zebda et al., 2000). In high-resolution fluorescence and electron-microscopy, cofilin/ADF is located inside the leading edge of the lamellipodium (Svitkina and Borisy, 1999)
Cofilin is likely to synergize with gelsolin to produce the depolymerizing activity occurring in the rounding stage of platelet shape change. As discussed above for capping protein, cofilin's activity is likely to be continuously regulated throughout shape change and does not, as formerly assumed, occur in one event upon activation. Given that cofilin/ADF is now known to be required for actin-based Listeria motility (Loisel et al., 1999), it will be important to analyze the location and timing of activation of cofilin in platelets.
2. Contractile Proteins
Myosin II has also been identified in platelets (Pollard et al., 1977) and found associated with the membrane cytoskeleton (Hartwig and DeSisto, 1991). Myosin II appears to regulate the clustering of the integrin receptor (Kovacsovics and Hartwig, 1996). Myosin II is regulated by phosphorylation of the light chain during activation which increases its association with the Triton-insoluble cytoskeleton (Fox and Phillips, 1982). Inhibition of phosphorylation by the calmodulin inhibitor trifluoperazine blocked this effect, as did the platelet activation inhibitors prostacylcin and prostaglandin E1. Activation of myosin therefore accompanies shape change, and is likely to be involved in mediating a contraction of actin filaments at the cortex during rounding and the subsequent formation of a ring of actin around the hyalomere that centralizes the granules as the platelet spreads (Stark et al., 1991). There are likely to be other members of the myosin family of proteins in platelets as well, but these remain undiscovered.
B. Formation of Filopodia and Lamellipodia: Re-assembly and Polymerization
Disassembly of filaments produces two synergistic effects: (1) The rigid cytoskeleton of the discoid cell is broken down, allowing new structures to form inside the platelet and permitting deformation of the membrane to accommodate them as various types of protrusions; (2) actin monomer concentration rises as monomers are released from the pointed ends of severed filaments. In addition, monomer-binding proteins such as thymosin 4 may also lose their affinity and release the monomer pool already existing in the resting cell.
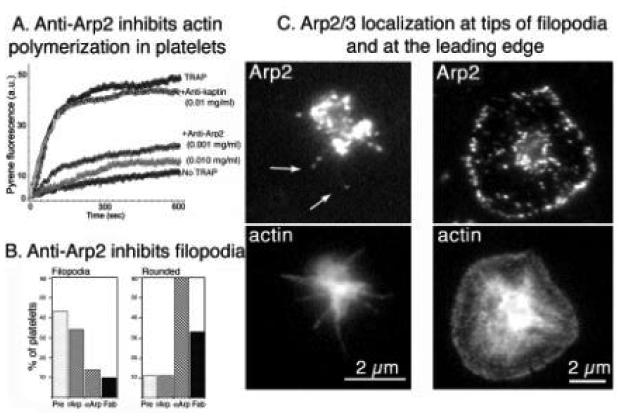
Formation of new filaments in platelets occurs through barbed-end elongation. These barbed ends could be a consequence of uncapping of filaments initially severed by gelsolin or those capped by capping protein. Very recent evidence suggests that nucleation of new filaments by Arp2/3 is the major event responsible for de novo formation of filaments after activation (Li, Kim and Bearer, 2002).
1. Arp2/3: Background
We have recently demonstrated that Arp2/3 is required for all steps of shape change following rounding (Li, Kim and Bearer, 2002). During the writing of this review, two new in-depth reviews have appeared (Higgs and Pollard, 2001; Pantaloni et al., 2001). Here, we first summarize key biochemical features of Arp2/3 and then report recent information about its presence and activity in platelets. (See Note added in proof.)
We initially isolated Arp2 as a 44-kDa protein, one of the 33 proteins that bind to F-actin affinity columns and present in platelet filopodia (Bearer and Alberts, 1988; Bearer, 1995). The Arp2/3 complex was also discovered using polyproline affinity columns to isolate potential profilin-binding proteins from the soil amoeba Acanthamoeba castellani (Machesky et al., 1994). Subsequently, the full seven-member Arp2/3 complex was isolated from platelet cytoskeletons (Welch et al., 1997). To describe how Arp2/3 was discovered in platelets, we need to review some background information about the actin-based motility of intracellular pathogens.
Several intracellular pathogens, including the bacteria Listeria, Shigella, Salmonella, and Rickettsia as well as viruses (Cossart, 2000), induce the polymerization of actin filaments in the cell cytoplasm. Actin polymerization drives the bacteria through the cytoplasm at 2–4 μm/min (Loisel et al., 1999). This motility can be reconstituted in extracts, including extracts from platelets (Theriot et al., 1994). However, bacteria outside a host cell cannot induce actin polymerization with purified actin (Tilney et al., 1992)—additional cellular proteins are needed.
Because Listeria are motile in platelet extracts (Egile et al., 1999; Laurent and Carlier, 1997; Laurent et al., 1999; Theriot et al., 1994), it was assumed that platelets contained the cellular actin nucleation factor that this bacteria co-opts for the induction of actin polymerization. Thus, platelet extracts were used as a source of proteins for the isolation of this factor (Welch et al., 1997). The platelet Arp2/3 complex was biochemically isolated using a novel assay to test fractions from platelet extracts for actin nucleation activity. The assay involves observing the ability of various fractions to induce a cloud of actin filaments around Listeria bacteria. Actin covalently labeled with rhodamine allows individual filaments to be imaged by fluorescence microscopy (Bearer, 1992b).
Listeria swim in platelet extracts, but Shigella do not (Egile et al., 1999), although both bacteria swim in brain extracts and both recruit Arp2/3. The basis for this difference appears to be the mechanism of recruitment of Arp2/3 and its activation. While the ActA protein of Listeria recruits and activates Arp2/3 directly, the IgsA protein of Shigella recruits Arp2/3 indirectly, using the N-WASp protein as an intermediary and activator. Platelets do not contain N-WASp; they contain only WASp (Scherbina et al., 2001). These results have implications concerning the activation of Arp2/3 in platelets as discussed further below.
Arp2/3 is a seven peptide complex (Table III), with two members of the actin-related protein family, Arp2 and Arp3, as its name implies. Additionally, it contains five other subunits of smaller molecular weight which are novel. All of the subunits are highly conserved, with sequences similar across phyla from the soil amoeba, to yeast, to human (Higgs and Pollard, 2001). In vitro, Arp2/3 induces the pointed-end nucleation of actin polymerization (Mullins et al., 1998; Welch et al., 1998a). This nucleation activity may be enhanced through binding to the sides of pre-existing polymers (Amann and Pollard, 2001), which could explain the branching of actin filaments nucleated by Arp2/3. Alternatively, branching could be occasioned by Arp2/3 adding to the barbed end of existing polymers and acting as a Y adaptor, with growth of filaments from each arm of the Y (Pantaloni et al., 2000). This latter hypothesis would produce two filaments in place of one. In soil amoeba and in tissue culture cells, Arp2/3 is located at the leading edge by immunofluoresence (Machesky et al., 1994; Welch and Mitchison, 1998b). By immunogold electron microscopy, Arp2/3 has been found at 70° branch points in the meshwork of actin in the lamellipodia (Svitkina and Borisy, 1999).
TABLE III.
Arp2/3 Subunitsa
| Protein | Molecular weight (kDa) |
Protein family | Interaction with actin |
Interaction with other Arp2/3 subunits |
Activity |
|---|---|---|---|---|---|
| Arp3 | 47 | Actin-related protein |
Pointed-end binding |
ARC2 ARC3 ARC4 |
NA |
| Arp2 | 44 | Actin-related protein |
Pointed-end binding |
ARC1 ARC2 |
Antibody blocks polymerization in vitro; antibody blocks platelet shape change morphologically |
| ARC1 | 40 | 7 WD repeats | Filament side binding |
Arp2 ARC2 |
— |
| ARC2 | 34 | — | Branching, side binding |
Arp2 Arp3 ARC1 ARC4 |
Antibody blocks branching in vitro; antibody blocks propulsion but not polymerization in cells |
| ARC3 | 21 | — | Arp3 ARC4 |
Binds WASp/SCAR; mediate activation of Arp2/3 | |
| ARC4 | 20 | — | Arp3 ARC2 |
— | |
| ARC5 | 16 | — | ARC3 ARC4 |
— — |
References as in text and as reported in Higgs and Pollard (2001).
Antibody inhibition of Arp2/3 in Acanthamoeba extracts using anti-Arp2 but not anti-Arp3 antibodies eliminates actin polymerization activity (Mullins and Pollard, 1999). In contrast, injection of anti-ARC2 (p34) peptide antibodies blocks propulsion at the leading edge but not polymerization (Bailly et al., 2001). This suggests that individual subunits are effectors for different aspects of Arp2/3-mediated actin polymerization.
2. Regulation of Arp2/3
Studies on activation of Arp2/3 in systems other than platelets demonstrates at least two mechanisms. First, WASp family members can greatly increase Arp2/3 activity in vitro. In elegant two-hybrid screens, it was found that the p21 subunit of Arp2/3 (ARC3) bound SCAR (Machesky and Insall, 1999), a member of the Wiskott-Aldritch protein (WASp) family (Ochs, 1998). In addition to the ARC3 subunit, other Arp2/3 complex members may also bind or contact WASp/SCAR family members (Zalevsky et al., 2001). The founding member of this family is the hematopoietic protein WASp, mutated in the Wiskott-Aldritch Syndrome. SCAR was initially identified in Dictyostelium where it is a genetic suppressor of cAMP receptor mutant phenotypes (Bear et al., 1998). Binding to an activated WASp family member in turn activates Arp2/3. WASp family members are likely to be activated by cdc42, but other Rho-family G proteins, PIP2, and proteins containing SH3 domains may also contribute to activation. Second, short actin filaments may also increase the nucleation ability of Arp2/3. binding to the sides of existing filaments appears to activate Arp2/3 (Amann et al., 2001). Tropomyosin (and possibly other side-binding proteins) block this activation (Blanchoin et al., 2001).
WASp itself does not play a role in the physiological activation of actin polymerization in human platelets (Rengan et al., 2000). Platelets from Wiskott-Aldritch Syndrome platelets have no detectable defect in actin nucleating activity (Rengan et al., 2000). Shigella bacteria that induce actin tails by recruiting Arp2/3 via N-WASp in brain extracts cannot do so in platelet extracts (Egile et al., 1999). Human WASp has only one WD repeat and is exclusively expressed in hematopoietic cells (Oda and Ochs, 2000). In contrast, N-WASp, an isoform of WASp, contains two WD repeats and N-WASp activates Arp2/3 in vitro (reviewed in Higgs and Pollard, 2001). However, although N-WASp is more ubiquitously expressed than WASp, it is not expressed in platelet (Oda and Ochs, 2000; Rengan et al., 2000). This explains the failure of Shigella to swim in platelet extracts, since Shigella recruit Arp2/3 through N-WASp. But it also leaves open the question of how Arp2/3 is activated in platelets. Other members of the WASp/SCAR family are being actively sought (A. Oda, personal communication).
Activation of Arp2/3 by preexisting actin filaments offers an attractive hypothesis to explain activation in platelets. As discussed above, the severing of existing filaments underlies the transition from discoid to rounding, the first step in activation. Such severing is mediated by calcium activation of gelsolin (Barkalow et al., 1996; Hartwig, 1992, 1999; Hartwig et al., 1995; Witke et al., 1995), which then remains bound to the barbed ends, capping them and preventing repolymerization from the barbed end. Hypotheses invoking the uncapping of these filaments have been proposed, although the mechanism for uncapping, PIP2 inhibition, has proven less than robust in vitro.
Short filaments produced by gelsolin severing at the membrane surface would serve as excellent sites for the Arp2/3-mediated polymerization of actin that follows rounding. Indeed, the idea that Arp2/3 can nucleate off the sides of filaments is particularly seductive, since prying gelsolin off the barbed ends of severed filaments has proven to be extremely difficult in vitro. The controversial uncapping of filaments by capping proteins also appears insufficiently robust to produce the explosive polymerization required for the rapid and dramatic morphological changes of platelet activation.
One likely scenario is that gelsolin severing, together with disassociation of side-binding proteins, would produce short filaments for Arp2/3 nucleation sites in the platelet cortex. In the resting platelet VASP or other side-binding proteins could inhibit the activation of Arp2/3 by blocking its binding sites on filament sides. This suggests another mechanism by which discoid platelets are maintained quiescent.
In the Listeria model, Arp2/3-mediated motility also requires capping protein and cofilin. Both of these proteins are present in platelets. Models describing the complex interactions between these proteins and actin in the formation of actin networks and in producing force have been proposed (Pantaloni et al., 2001). These models are likely to be applicable to the platelet lamellipodial formation as well.
3. Kaptin/2E4
Kaptin/2E4 is a ~43-kDa protein initially discovered in our F-actin-affinity chromatography screen for platelet actin-binding proteins. Kaptin/2E4 is located at sites of actin polymerization in virtually all cells (Bearer, 1992a, 1992c, 1993, 1995; Bearer and Abraham, 1999). In platelets, kaptin/2E4 is located in the 1-μm edge of the lamellipodia (Fig. 11); and in fibroblasts, antibodies to kaptin/2E4 label the outer 2/3 of the actin filaments in the lamellipodia as well as in microspikes and filopodia. Kaptin/2E4 may be an ATP-sensitive actin-binding protein, as it elutes from F-actin columns with ATP and is similarly extracted from platelets and cultured fibroblasts. In vitro, kaptin/2E4 binds the ends of actin filaments, but instead of inhibiting elongation as capping proteins do, it potentiates polymerization slightly (Bearer, 1992a). Kaptin/2E4 is upregulated in the embryonic mesoderm prior to the early movements of gastrulation in Xenopus, suggesting it plays a crucial role in cell motility (Bearer, 1992c).
FIG. 11.
Kaptin/2E4 localizes to the leading edge of platelets. Platelets spread on glass were stained with antibodies generated against the recombinant 2E4/kaptin protein. As seen with the original antibody (#32) generate against the human protein isolated by F-actin-affinity chromatography, staining was throughout the lamellipodia and in the dome of cytoplasm (the hyalomere). (Reproduced from Bearer and Abraham, 1999, by permission.)
A specific role for kaptin/2E4 in platelet shape change has not been identified yet, but its uniform location at the leading edge suggests that it may be involved in regulating growth of filaments at the barbed ends. It is also found at the tips of stereocilia in the hair cells of the inner ear (Bearer et al., 2000b). Its location on human chromosome 19q13.3 is within a deafness locus, DFNA 4. In one family with this deafness syndrome, no coding region mutations were found, however. This family also has no known platelet defects; this could mean that there are promoter sequences specific for cochlear expression mutated in DFNA4 that do not effect expression of kaptin/2E4 in other tissues, such as the megakaryocyte.
VI. Stress Fibers and Adhesion Plaques
A. Adhesion
The final stages of spreading involve the maturation of adhesions with the consequent formation of actin-based stress fibers. These elements are important for the final stages of thrombosis contraction. Maturation of adhesion involves a complex series of steps: (1) activation of the attachment system, either via the platelet receptor or its ligand; (2) signaling into the cell which produces a cascade of events; (3) recruitment of cytoplasmic and membrane proteins to the attachments site; (4) polymorization of actin and organization of filaments into higher order structures such as stress fibers.
Platelets have a large number of glycoprotein receptors for extracellular matrix, a full reviewing of which would far exceed the scope of the chapter. However, some key concepts are emerging from studies on three of these receptors which may prove to be paradigms for transmembrane links between the extracellular matrix and the cytoskeleton in platelets, and for extracellular matrix adhesion in other cells as well. We have selected features of these for discussion here.
1. Glycoprotein Ib
Adhesion to extracellular matrix must be coordinated with attachments through the membrane to the underlying cytoskeleton. In the case of GP1 IX, the receptor is already tightly bound to the membrane skeleton, as discussed above. GP1b/IX is sticky for ligand, vWF, when the platelet is in circulation, but GPIb does not bind vWF until vWF is activated by interaction with exposed endothelial basement membrane. Thus, exposure of basement membrane can cause a cascade of events in which the platelets are pulled out of circulation by confrontation with an activated ligand. Tight coupling of the receptor to the membrane skeleton may help the platelet withstand this interaction. Washed platelets adhere and spread on glass coated with ristocetin-activated vWF. Such spreading appears to result in the formation of the four distinct actin structures (Zaffran et al., 2000). Thus, activated vWF can act as an agonist and produce all of the actin dynamics required for shape change.
Binding to activated vWF results in trans-activation of the integrin receptor, and possibly could lead to activation of other extracellular receptors on the platelet surface as well (Zaffran et al., 2000). The signaling molecules that mediate this cross talk between GP1b and the integrin receptor are not known but may involve 14-3-3, a signaling molecule found associated with the GIb-IX receptor complex. However, signaling from GP1b to the integrin receptor could be reproduced in CHO cells which lack the thrombin receptor pathway; thus it appears that GPIb signaling involves a different pathway from that which activates integrin receptors in response to thrombin.
In contrast, the platelet integrin receptor, αIIbβ3 is not sticky in resting platelets; it must first be activated by an internal mechanism, termed “inside-out” activation (Hughes and Pfaff, 1998). Once activated, it binds to fibronectin and this in turn activates a second signaling pathway, termed “outside-in” activation.
2. The Integrin Receptor
The integrin receptor, a heterodimer composed of two glycoprotein subunits, is activated to bind fibronectin by thrombin and other agonists (Calderwood et al., 2000). Upon binding its ligand, it transmits a signal back into the cytoplasm, which results in the formation of adhesion plaques (Phillips et al., 2001). In platelets, these adhesion plaques contain a set of structural proteins, including VASP, talin, APB, α-actinin, zyxin, paxcillin, skelemin and vinculin. The subunits of the integrin receptors appear to bind directly to a number of these actin-binding proteins, including-actinin, skelemin, and vinculin. The activated integrin receptor may also recruit a macromolecular cluster of proteins containing signaling enzymes such as G-proteins, kinases, proteases, and phosphatases. This conglomerate could recruit WASp/SCAR family members and other downstream effectors of G proteins and activate Arp2/3 as described above.
After activation, the integrin receptor becomes clustered, possibly through direct interactions with myosin II (Kovacsovics and Hartwig, 1996) Concomitant with this clustering, actin filaments accumulate and the focal adhesion plaque forms with its full complement of enzymes and structural proteins. It remains speculative how the activation of the integrin receptor triggers the formation of stress fibers in platelets or in any cell. The process involves a series of back-and-forth communication between the receptors and the cytoskeleton. While phosphorylation may activate actin polymerization, formation of filaments appears to be necessary for full phosphorylations to occur. Thus, while interaction of integrin receptors with immobilized ligands triggers tyrosine phosphorylation of the receptor, cytochalasin D, which blocks barbed-end elongation of actin filaments, inhibits spreading and also decreases receptor phosphorylation (Haimovich et al., 1993).
3. Calcium-Dependent Proteases
The calcium-dependent proteases including calpain comprise another class of the enzymes that concentrates in adhesion plaques. It has been known for some time that spectrin, ABP, and several other platelet actin-binding proteins are hydrolyzed during activation, as mentioned above. Recently, the physiological significance of this hydrolysis has been demonstrated in two sets of experiments. First, integrin clusters are dependent on calpain activity. Formation of these clusters is important for activation of the small G proteins, Rac and Rho, which can also trigger focal complexes and focal adhesions (Bialkowska et al., 2000; Kulkarni et al., 1999). Dominant negative μ-calpain expressed in cultured cells inhibited cluster formation induced by integrin activation but not those induced by Rac and Rho. This suggests that in the normal sequence of events, integrin clusters induce Rac and Rho secondarily, and these G proteins in turn precipitate the series of events that produces actin polymerization. These experiments support earlier work demonstrating decreased receptor phosphorylation when actin polymerization is blocked with cytochalasin. Second, in calpain knockout mice, platelet clotting is significantly impaired, demonstrating a role for this enzyme in normal platelet physiology as well (Azam et al., 2001). Introduction of calpastatin into human platelets demonstrates that calpain activation is required for secretion, thrombin-induced aggregation, and full spreading on glass (Croce et al., 1999).
4. Glycoprotein VI, the Collagen Receptor
Collagen is a powerful agonist for platelets. One of the integrin receptors, GPVI, which binds collagen, appears to mediate a rapid phosphorylation of WASp in platelets (Oda et al., 1998). Since this member of the WASp/SCAR family has not been shown to be involved in Arp2/3-mediated actin polymerization, it is not clear yet whether this pathway impacts the cytoskeleton directly. However, WASp shifts from the supernatant to the Triton-insoluble cytoskeleton upon activation, and is an endogenous substrate for calpain, implicating it in actin dynamics in platelets. WASp in platelets is likely to play a more significant role in megakaryocyte platelet production than in platelet activation, since WASp platelets are abnormal in size and shape but not in activation (Oda and Ochs, 2000).
Other signaling proteins found in platelets also may be more functionally significant for platelet production during megakaryocyte maturation than for platelet activation. This includes protein kinase Cα, which colocalizes with actin filaments at the cortex of the megakaryocyte during proplatelet formation (Rojnuckarin and Kaushansky, 2001). The recent identification of thrombopoieitin, which allows megakaryocyte-enriched cultures to be grown in vitro will allow further dissection of the role of these molecules in platelet formation.
B. Actin-Binding Proteins in Adhesion Plaques
Platelet adhesion plaques appear to be analogous to those formed in every other mammalian cell studied. A large number of proteins assemble onto the intracellular surface of adhesion plaques regardless of the extracellular receptor activated. Cytoskeletal attachment proteins include zyxin, vinculin, talin, α-actinin, and VASP. Recent information links VASP and α-actinin to the signaling cascade that results in recruitment of actin in maturing adhesion plaques.
1. VASP's Second Role: Adhesion Plaques and Stress Fiber Formation
The carboxy terminus of VASP targets it to adhesion plaques (Reinhard et al., 1992), as do its polyproline repeats which may bind zixin in cells (Bear et al., 2000). The polyproline repeats of ActA can compete VASP off of adhesion (Bear et al., 2000) but not off the leading edge, suggesting there are two molecular mechanisms for the recruitment of VASP.
Adhesion to fibrinogen stimulates association of SLP-76 with SLAP-130 adaptor and with VASP (Obergfell et al., 2001). SLAP-130, SLP-76 and VASP colocalize in the lamellipodia of PMA-potentiated glass-activated platelets. That SLP-76 is essential for platelet function has been confirmed in knockout mice, where SLP-76 deficiency results in hemorrhage. Platelets from SLP-76 knockouts bind fibrinogen normally but fail to spread and have decreased phosphotyrosine (Judd et al., 2000).
2. α-Actinin
α-Actinin is a 95-kDa protein that forms homodimers linked in an antiparellel orientation (Matsudaira, 1991). In platelets, α-actinin colocalizes with actin in adhesion plaques and stabilizes the filaments (Lazarides and Burridge, 1975). Of the many cytoskeletal-associated adhesion plaque proteins, α-actinin is the one that binds both the cytoplasmic tail of matrix receptors and to actin (Otey et al., 1990). Thus α-actinin appears to be the bridge cementing surface attachment to the cytoskeleton. Signaling molecules also bind α-actinin, suggesting that it plays a role as a scaffolding protein that transmits information about cell attachment into the cell interior. Interactions between α-actinin and its ligands is differentially regulated by calcium, PIP2, and PIP3 (Fukami et al., 1992; Greenwood et al., 2000).
In human platelets, α-actinin is tyrosine phosphorylated on Y12 by focal adhesion kinase (FAK) (Izaguire et al., 1999, 2001). Tyrosine phosphorylation decreases α-actinin affinity for F-actin, suggesting a role for phosphorylation in regulating recruitment of actin to adhesion plaques by α-actinin.
VII. A Model of Actin Dynamics in Platelet Activation
A hypothetical model of the first stages in platelet shape change is shown in Figure 12. The cortex of the discoid circulating platelet has at least three tightly opposed layers: (1) the outer layer, a lipid-glycoprotein membrane supported by a second layer; (2) a dense net of spectrin, actin-binding protein, and myosin firmly attached to the cytoplasmic tail of the transmembrane glycoprotein, Ib-IX; (3) an innermost layer, a cage of regularly spaced actin filaments running parallel to the plane of the membrane. The actin filaments that attach along their sides to the membrane skeleton probably arise from bundles of actin that radiate out from a central filamentous core. In this “resting” stage gelsolin is soluble, not attached to the F-actin, and VASP is involved in bundling the filaments radiating from the central core.
FIG 12.
Proposed model of actin reorganization in platelet activation and spreading. The discoid platelet has a trilaminar membrane shield composed of the outer membrane, the spectrin-ABP-rich inner membrane skeleton and a cage of closely aligned actin filaments that connect to the central core via radiating spokes. VASP bundles the filaments radiating from the central core. Most filaments are long and their barbed ends are capped. Monomer is sequestered by thymosin β4.
Upon activation, the platelet rounds and contracts, the membrane skeleton is dissociated, at least in part through proteolysis. Side-binding proteins such as VASP, are released from the radiating bundles and gelsolin is activated by a calcium influx to sever these filaments and the membrane-associated cage. Activation of myosin results in condensation of the radiating filaments, severed from their membrane anchors, into the central core and formation of the contractile ring.
Disassociation and severing are rapidly followed by rounds of polymerization of new actin from activated nucleators like Arp2/3 and 2E4/kaptin and from uncapping of severed barbed ends. Arp2/3 is activated at the membrane surface and on the sides of the severed filaments. Cofilin regulates filament length and recycles monomers that are recharged by profilin with ATP. Capping protein regulates which filaments are free to elongate, and hence the site of lamellipodia formation. VASP and other side-binding proteins restructure the new filaments into bundles. (See also color insert.)
Upon stimulation, the platelet rounds. Calcium enters, activating gelsolin which severs the long filaments radiating from the core and those lining the membrane. Activation of myosin pulls the proximal lengths of severed filaments into the central core where the contractile ring forms. Concomitant with filament severing, activation of calcium-dependent proteases digests spectrin and actin-binding protein in the membrane skeleton, which is loosened and becomes distensible.
Polymerization of new filaments begins and involves two processes:
Nucleation of new filaments by Arp2/3, 2E4/kaptin, and/or other nucleating systems: Activation of Arp2/3 at the surface and by the denuded sides of existing filaments sparks pointed-end nucleation. Filaments grow outwards towards the membrane. Capping protein regulates the sites of barbed-end elongation and cofilin controls pointed end off rates, maintaining the growing filaments at a constant length. 2E4/kaptin could be involved in regulating Arp2/3 activity, controlling barbed-end elongation of Arp2/3-nucleated filaments, or nucleating new filaments on its own.
Elongation of uncapped filaments: Gelsolin releases barbed ends, which elongate. VASP and other cross-linking proteins reassociate with new filaments, organizing them into filopodia. As the concentration of side-binding proteins drops, the lamellipodia extends as sheet of a short actin filaments.
This model is a simplification of a more complicated choreography. Many more proteins must be involved in order to achieve the organization of the new actin into higher order structures such as filopodia and stress fibers, which mediate the adhesion and contraction of the platelet. What is needed is a logical multivariate model in which the combinatorial affinities and activities of each protein can be studied in the context of the whole set of interacting molecules.
Platelet activation and clot formation must be tightly controlled so that hemostasis is maintained without intravascular thrombosis and consequent ischemia. Abnormal clotting is responsible for major morbidity and mortality in the developed world, including heart attack and stroke. Now that details of the molecular mechanisms of actin dynamics in the platelet are identified, these cytoskeletal regulators are strong candidates for targets of drug intervention for abnormal clot prevention.
VII. Summary
Platelet shape change is the morphological equivalent of activation. Shape change is a temporal sequence of structural change: rounding, filopodial projection, spreading, and contraction. Actin dynamics underlie these changes. Actin is a ubiquitously expressed eukaryotic protein whose biochemical and structural features in platelets are similar to those in other cells. Thus, platelets can serve as an experimental model for the discovery of universal mechanisms regulating actin dynamics. In the discoid resting platelet, there are two pools of actin filaments: those that line the membrane and those in the central core. In the first stage of activation, rounding, these structures are disassembled by severing and proteolysis. New filaments form by elongation of severed ends and nucleation of new filaments as the platelet spreads. The spread platelet has four distinct actin structures that differ in composition and function: filopodia, lamellipodia, stress fibers, and the contractile ring. The formation and function of these structures is mediated by a large number of actin-binding proteins, the full complement of which were identified by F-actin affinity chromatography. Our recent discovery demonstrates that Arp2/3 is necessary for all actin polymerization in the platelet upon thrombin stimulation and is required for formation of filopodia and lamellipodia during spreading on glass. The Arp2/3 complex, cofilin, and capping protein are sufficient to reconstitute actin-based motility of the intracellular bacteria, Listeria, in vitro. Cofilin and capping protein are also found in platelets. Other proteins involved in platelet actin dynamics include gelsolin (a calcium-dependent severing protein), VASP (a phosphoprotein that bundles and nucleates filament assembly), and 2E4/kaptin (an ATP-dependent end-binding protein). In addition to mediating shape change, actin dynamics also play a role in integrin signaling and platelet adhesion. Future challenges will be to determine how these many different proteins and pathways interweave to produce the physiologically significant event of clot formation and retraction.
Note added in proof
Arp/23 is the key regulator of actin polymerization during agonist activation of platelets: A key question in platelet actin dynamics has been the identification of the “nucleator”—i.e., the protein(s) responsible for initiating the formation of actin filaments. Because of its activity in vitro, the Arp2/3 complex was considered the best candidate. However, although Arp2/3 was known to be present in platelets, its contribution to actin dynamics was not known, nor has a physiological role for Arp2/3 been experimentally determined in any other cell.
By two new approaches, it has now been shown that Arp2/3 is the major nucleator of actin polymerization in human platelets during shape change (Li, Kim and Bearer, 2002). We generated inhibitory antibodies against recombinant Arp2, and developed a novel method to load them into platelets without activating them. TRAP stimulation causes a five-fold increase in actin polymerization in permeabilized platelets which is decreased by anti-Arp2 to that of un-stimulated platelets (Fig. 13A). Arp2/3 activity is also required for platelets to spread on glass (Fig. 13B). when permeabilized platelets are loaded with pre-immune antiserum, 45% form filopodia whereas loading with anti-Arp2 or its Fab fragments decrease this by 4-fold. Instead of spreading, platelets loaded with anti-Arp2 or its Fab fragments remain in the rounded stage or form blebs. Recombinant Arp2 reverses this anti-Arp2 effect. Immunofluorescence of platelets spread on glass shows that the Arp2/3 complex is at the tips of filopodia and in the lamellipodium (Fig. 13C). this location together with the date from the inhibition studies demonstrates that Arp2/3 activity is directly responsible for the formation of actin structures in the platelet during either activation in suspension or while spreading on glass.
FIG. 13.
Arp2/3 is required for actin polymerization and for shape change in platelets. (A) Pyrene-actin assay showing the effects of anti-Arp2 on actin polymerization in TRAP-stimulated permeabilized platelets. (B) Histogram of the morphological effects of Arp2/3 inhibition on platelets loaded with pre-immune-antibodies (Pre), anti-Arp2 antibody (αArp) or its Fab fragments (Fab). Anti-Arp2 decreases filopodial formation, and freezes platelets at the rounded. Recombinant Arp2 (rArp) blocks this effect. (C) Arp2/3 is located at the tips of filopodia (arrows) and in the lamellipodia in two platelets spread on glass and double labeled for Arp2 (top) and actin (bottom) (Modified from Li, Kim and Bearer, 2002, with permission).
References
- Abel K, Mieskes G, et al. Dephosphorylation of the focal adhesion protein VASP in vitro and in intact human platelets. FEBS Lett. 1995;370(3):184–8. doi: 10.1016/0014-5793(95)00817-s. [DOI] [PubMed] [Google Scholar]
- Allen RD, Zacharski LR, et al. Transformation and motility of human platelets: details of the shape change and release reaction observed by optical and electron microscopy. J. Cell Biol. 1979;83(1):126–42. doi: 10.1083/jcb.83.1.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amann KJ, Pollard TD. The Arp2/3 complex nucleates actin filament branches from the sides of pre-existing filaments. Nat. Cell Biol. 2001;3(3):306–10. doi: 10.1038/35060104. [DOI] [PubMed] [Google Scholar]
- Arber S, Barbayannis FA, et al. Regulation of actin dynamics through phosphorylation of cofilin by LIM-kinase. Nature. 1998;393(6687):805–9. doi: 10.1038/31729. [DOI] [PubMed] [Google Scholar]
- Aszodi A, Pfeifer A, et al. The vasodilator-stimulated phosphoprotein (VASP) is involved in cGMP- and cAMP-mediated inhibition of agonist-induced platelet aggregation, but is dispensable for smooth muscle function. EMBO J. 1999;18(1):37–48. doi: 10.1093/emboj/18.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azam M, Andrabi SS, et al. Disruption of the mouse mu-calpain gene reveals an essential role in platelet function. Mol. Cell Biol. 2001;21(6):2213–20. doi: 10.1128/MCB.21.6.2213-2220.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann C, Fischer L, et al. The EVH2 domain of the vasodilator-stimulated phosphoprotein mediates tetramerization, F-actin binding, and actin bundle formation. J. Biol. Chem. 1999;274(33):23549–57. doi: 10.1074/jbc.274.33.23549. [DOI] [PubMed] [Google Scholar]
- Bailly M, Ichetovkin I, et al. The F-actin side binding activity of the Arp2/3 complex is essential for actin nucleation and lamellipod extension. Curr. Biol. 2001;11(8):620–5. doi: 10.1016/s0960-9822(01)00152-x. [DOI] [PubMed] [Google Scholar]
- Barkalow K, Witke W, et al. Coordinated regulation of platelet actin filament barbed ends by gelsolin and capping protein. J. Cell Biol. 1996;134 doi: 10.1083/jcb.134.2.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bear JE, Loureiro JJ, et al. Negative regulation of fibroblast motility by Ena/VASP proteins. Cell. 2000;101(7):717–28. doi: 10.1016/s0092-8674(00)80884-3. [DOI] [PubMed] [Google Scholar]
- Bear JE, Rawls JF, et al. SCAR, a WASP-related protein, isolated as a suppressor of receptor defects in late Dictyostelium development. J. Cell Biol. 1998;142(5):1325–35. doi: 10.1083/jcb.142.5.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bearer EL. Anionic lipid domains in cell membranes. University of California; San Francisco: 1983. Dept. of Biochemistry and Biophysics. San Francisco. [Google Scholar]
- Bearer EL. Platelet membrane skeleton revealed by quick-freeze deep-etch. Anat. Rec. 1990;227(1):1–11. doi: 10.1002/ar.1092270102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bearer EL. Direct observation of actin filament severing by gelsolin and binding by gCap39 and CapZ. J. Cell. Biol. 1991;115(6):1629–38. doi: 10.1083/jcb.115.6.1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bearer EL. An actin-associated protein present in the microtubule organizing center and the growth cones of PC-12 cells. J. Neurosci. 1992a;12(3):750–61. doi: 10.1523/JNEUROSCI.12-03-00750.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bearer EL. Fluorescence microscopy of single actin filaments labeled by conjugation to rhodamine. Biol. Bull. 1992b;183:361–362. doi: 10.1086/BBLv183n2p361. [DOI] [PubMed] [Google Scholar]
- Bearer EL. Actin and actin-associated proteins in Xenopus embryos: relationship to cytoarchitecture and gastrulation. In: Pederson R, editor. Cytoskeleton in Development. Academic Press; New York: 1992c. p. 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bearer EL. Role of actin polymerization in cell locomotion: molecules and models. Am. J. Respir. Cell Mol. Biol. 1993;8(6):582–91. doi: 10.1165/ajrcmb/8.6.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bearer EL. Cytoskeletal domains in the activated platelet. Cell Motil. Cytoskeleton. 1995;30(1):50–66. doi: 10.1002/cm.970300107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bearer EL, Abraham MT. 2E4 (kaptin): a novel actin-associated protein from human blood platelets found in lamellipodia and the tips of the stereocilia of the inner ear. Eur. J. Cell Biol. 1999;78(2):117–26. doi: 10.1016/S0171-9335(99)80013-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bearer EL, Chen AF, et al. 2E4/Kaptin (KPTN) - a candidate gene for the hearing loss locus, DFNA4. Ann. Hum. Genet. 2000b;64(3):189–19. doi: 10.1046/j.1469-1809.2000.6430189.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bearer EL, Frackelton R, et al. A complex of platelet proteins that bind profilin and affect actin polymerization. Molec. Biol. Cell. 1996;7 [Google Scholar]
- Bearer EL, Prakash JM, et al. VASP protects actin filaments from gelsolin: an in vitro study with implications for platelet actin reorganizations. Cell Motil. Cytoskeleton. 2000a;47(4):351–64. doi: 10.1002/1097-0169(200012)47:4<351::AID-CM8>3.0.CO;2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bialkowska K, Kulkarni S, et al. Evidence that beta3 integrin-induced Rac activation involves the calpain-dependent formation of integrin clusters that are distinct from the focal complexes and focal adhesions that form as Rac and RhoA become active. J. Cell. Biol. 2000;151(3):685–96. doi: 10.1083/jcb.151.3.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchoin L, Pollard TD, et al. Interactions of ADF/cofilin, Arp2/3 complex, capping protein and profilin in remodeling of branched actin filament networks. Curr. Biol. 2000;10(20):1273–82. doi: 10.1016/s0960-9822(00)00749-1. [DOI] [PubMed] [Google Scholar]
- Blanchoin L, Pollard TD, Hitchcock-DeGregori SE. Inhibition of the Arp2/3 complex-nucleated actin polymerization and branch formation by tropomyosin. Curr. Biol. 2001;11:1300–1304. doi: 10.1016/s0960-9822(01)00395-5. [DOI] [PubMed] [Google Scholar]
- Boyles J, Fox JEB, et al. Organization of the cytoskeleton in resting, discoid platelets: preservation of actin filaments by a modified fixation that prevents osmium damage. J. Cell Biol. 1985;101:1463. doi: 10.1083/jcb.101.4.1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brass LF. Ca2+ hemostasis in unstimulated platelets. J. Biol. Chem. 1984;259:12563–12570. [PubMed] [Google Scholar]
- Bryan J. Gelsolin has three actin-binding sites. J. Cell Biol. 1988;106(5):1553–62. doi: 10.1083/jcb.106.5.1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butt E, Abel K, et al. cAMP- and cGMP-dependent protein kinase phosphorylation sites of the focal adhesion vasodilator-stimulated phosphoprotein (VASP) in vitro and in intact human platelets. J. Biol. Chem. 1994;269(20):14509–17. [PubMed] [Google Scholar]
- Calderwood DA, Shattil SJ, et al. Integrins and actin filaments: reciprocal regulation of cell adhesion and signaling. J. Biol. Chem. 2000;275(30):22607–10. doi: 10.1074/jbc.R900037199. [DOI] [PubMed] [Google Scholar]
- Carlier M-F, Laurent V, et al. Actin depolymerizing factor (ADF/cofilin) enhances the rate of filament turnover: implication in actin-based motility. J. Cell Biol. 1997;136(6):1307–22. doi: 10.1083/jcb.136.6.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsson L, Nystrom LE, et al. Actin polymerizability is influenced by profilin, a low molecular weight protein in non-muscle cells. J. Mol. Biol. 1977;115(3):465–83. doi: 10.1016/0022-2836(77)90166-8. [DOI] [PubMed] [Google Scholar]
- Chan AY, Bailly M, et al. Role of cofilin in epidermal growth factor-stimulated actin polymerization and lamellipod protrusion. J. Cell Biol. 2000;148(3):531–42. doi: 10.1083/jcb.148.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper JA. Effects of cytochalasin and phalloidin on actin. J. Cell Biol. 1987;105(4):1473–8. doi: 10.1083/jcb.105.4.1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cossart P. Actin-based motility of pathogens: the Arp2/3 complex is a central player. Cell Microbiol. 2000;2(3):195–205. doi: 10.1046/j.1462-5822.2000.00053.x. [DOI] [PubMed] [Google Scholar]
- Cunningham JG, Meyer SC, et al. The cytoplasmic domain of the a-subunit of glycoprotein (GP) Ib mediates attachment of the entire GP Ib-IX complex to the cytoskeleton and regulates von Willebrand factor-induced changes in cell morphology. J. Biol. Chem. 1996;271:11581–11587. doi: 10.1074/jbc.271.19.11581. [DOI] [PubMed] [Google Scholar]
- David-Ferreira JF. The blood platelet: Electron microscopic studies. Int. Rev. Cytol. 1974;17:99–148. doi: 10.1016/s0074-7696(08)60406-4. [DOI] [PubMed] [Google Scholar]
- Davidson MM, Haslam RJ. Dephosphorylation of cofilin in stimulated platelets: roles for a GTP-binding protein and Ca2+ Biochem. J. 1994;301(Pt 1):41–7. doi: 10.1042/bj3010041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies T, Drotts D, et al. Cytoplasmic calcium is necessary for thrombin induced platelet secretion. J. Biol. Chem. 1989;264:19600–1960. [PubMed] [Google Scholar]
- Di Nardo A, Gareus R, et al. Alternative splicing of the mouse profilin II gene generates functionally different profilin isoforms. J. Cell Sci. 2000;113(Pt 21):3795–803. doi: 10.1242/jcs.113.21.3795. [DOI] [PubMed] [Google Scholar]
- Dunaway S, Allen P, et al. Association between polyproline and Grb-2, VASP, and gelsolin from human platelets. Mol. Cell Biol. 1997;8(255a) [Google Scholar]
- Egile C, Loisel TP, et al. Activation of the CDC42 effector N-WASP by the Shigella flexneri IcsA protein promotes actin nucleation by Arp2/3 complex and bacterial actin-based motility. J. Cell Biol. 1999;146(6):1319–32. doi: 10.1083/jcb.146.6.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englund GD, Bodnar RJ, et al. Regulation of von Willebrand Factor Binding to the Platelet Glycoprotein Ib-IX by a Membrane Skeleton-dependent Inside-out Signal. J. Biol. Chem. 2001;276(20):16952–9. doi: 10.1074/jbc.M008048200. [DOI] [PubMed] [Google Scholar]
- Ezzell RM, Kenney DM, et al. Localization of the domain of actin-binding protein that binds to membrane glycoprotein Ib and actin in human platelets. J. Biol. Chem. 1988;263(26):13303–9. [PubMed] [Google Scholar]
- Fox JE. Identification of actin-binding protein as the protein linking the membrane skeleton to glycoproteins on platelet plasma membrane. J. Biol. Chem. 1985;260:11970–11977. [PubMed] [Google Scholar]
- Fox JE. The platelet cytoskeleton. Thromb. And Haemos. 1993;70:884–893. [PubMed] [Google Scholar]
- Fox JE, Boyles JK, et al. Identification of a membrane skeleton in platelets. J. Cell Biol. 1988;102:1748–1757. doi: 10.1083/jcb.106.5.1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox JE, Dockter ME, et al. An improved method for determining the actin filament content of nonmuscle cells by the DNase I. inhibition assay. Anal Biochem. 1981;117:170–7. doi: 10.1016/0003-2697(81)90707-7. [DOI] [PubMed] [Google Scholar]
- Fox JE, Goll DE, et al. Identification of two proteins (actin-binding protein and P235) that are hydrolyzed by endogenous Ca2+-dependent protease during platelet aggregation. J. Biol. Chem. 1985;260(2):1060–6. [PubMed] [Google Scholar]
- Fox JE, Phillips DR. Inhibition of actin polymerization in blood platelets by cytochalasins. Nature. 1981;292:650–652. doi: 10.1038/292650a0. [DOI] [PubMed] [Google Scholar]
- Fox JE, Phillips DR. Role of phosphorylation in mediating the association of myosin with the cytoskeletal structures of human platelets. J. Biol. Chem. 1982;257:4120–4126. [PubMed] [Google Scholar]
- Fox JE, Phillips DR. Polymerization and organization of actin filaments within platelets. Sem. Heme. 1983;20:243–260. [PubMed] [Google Scholar]
- Fox JE, Reynolds CC, et al. Spectrin is associated with membrane-bound actin filaments in platelets and is hydrolyzed by the Ca2+-dependent protease during platelet activation. Blood. 1987;69(2):537–4. [PubMed] [Google Scholar]
- Fukami K, Furuhashi K, et al. Requirement of phosphatidylinositol 4,5-bisphosphate for alpha-actinin function. Nature. 1992;359(6391):150–2. doi: 10.1038/359150a0. [DOI] [PubMed] [Google Scholar]
- Gertler FB, Comer AR, et al. enabled, a dosage-sensitive suppressor of mutations in the Drosophila Abl tyrosine kinase, encodes an Abl substrate with SH3 domain-binding properties. Genes Dev. 1995;9(5):521–33. doi: 10.1101/gad.9.5.521. [DOI] [PubMed] [Google Scholar]
- Gieselmann R, Kwiatkowski DJ, et al. Distinct biochemical characteristics of the two human profilin isoforms. Eur. J. Biochem. 1995;229(3):621–8. doi: 10.1111/j.1432-1033.1995.tb20506.x. [DOI] [PubMed] [Google Scholar]
- Goldschmidt-Clermont MF, Furman MI, et al. The control of actin nucleotide exchange by thymosin B4 and profilin. A potential regulatory mechanism for actin polymerization in cells. Mol. Biol. Cell. 1992;3:1015–1024. doi: 10.1091/mbc.3.9.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldschmidt-Clermont PJ, Kim JW, et al. Regulation of phospholipase C-gamma 1 by profilin and tyrosine phosphorylation. Science. 1991b;251(4998):1231–3. doi: 10.1126/science.1848725. [DOI] [PubMed] [Google Scholar]
- Goldschmidt-Clermont PJ, Machesky LM, et al. Mechanism of the interaction of human platelet profilin with actin. J. Cell Biol. 1991a;113(5):1081–9. doi: 10.1083/jcb.113.5.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonnella PA, Nachmias VT. Platelet activation and microfilament bundling. J. Cell Biol. 1981;89(1):146–51. doi: 10.1083/jcb.89.1.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwood JA, Theibert AB, et al. Restructuring of focal adhesion plaques by PI 3-kinase. Regulation by PtdIns (3,4,5)-p(3) binding to alpha-actinin. J. Cell Biol. 2000;150(3):627–42. doi: 10.1083/jcb.150.3.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haarer BK, Petzold AS, et al. Mutational analysis of yeast profilin. Mol. Cell Biol. 1993;13(12):7864–73. doi: 10.1128/mcb.13.12.7864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haimovich B, Lipfert L, et al. Tyrosine phosphorylation and cytoskeletal reorganization in platelets are triggered by interaction with integrin receptors with their immobilized ligands. J. Biol. Chem. 1993;268:15868–15877. [PubMed] [Google Scholar]
- Halbrugge M, Friedrich C, et al. Stoichiometric and reversible phosphorylation of a 46-kDa protein in human platelets in response to cGMP- and cAMP-elevating vasodilators. J. Biol. Chem. 1990;265(6):3088–93. [PubMed] [Google Scholar]
- Halbrugge M, Walter U. Purification of a vasodilator-regulated phosphoprotein from human platelets. Eur. J. Biochem. 1989;185(1):41–50. doi: 10.1111/j.1432-1033.1989.tb15079.x. [DOI] [PubMed] [Google Scholar]
- Harbeck B, Huttelmaier S, et al. Phosphorylation of the vasodilator-stimulated phosphoprotein regulates its interaction with actin. J. Biol. Chem. 2000;275(40):30817–25. doi: 10.1074/jbc.M005066200. [DOI] [PubMed] [Google Scholar]
- Hartwig JH. Mechanisms of actin rearrangements mediating platelet activation. J. Cell Biol. 1992;118(6):1421–42. doi: 10.1083/jcb.118.6.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwig JH. Platelet morphology. In: Loscalzo J, Schafer AI, editors. Thrombosis and Hemorrhage. 2nd edition Williams and Wilkins; Baltimore: 1999. pp. 207–227. [Google Scholar]
- Hartwig JH, Bokoch GM, et al. Thrombin receptor ligation and activated Rac uncap actin filament barbed ends through phosphoinositidesynthesis in permeabilized human platelets. Cell. 1995;82(4):643–53. doi: 10.1016/0092-8674(95)90036-5. [DOI] [PubMed] [Google Scholar]
- Hartwig JH, DeSisto M. The cytoskeleton of the resting human blood platelet: structure of the membrane skeleton and its attachment to actin filaments. J. Cell Biol. 1991;112(3):407–25. doi: 10.1083/jcb.112.3.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser W, Knobeloch KP, et al. Megakaryocte hyperplasia and enhanced agonist-induced platelet activation in vasodilator-stimulate phosphoprotein knockout mice. Proc. Natl. Acad. Sci. U.S.A. 1999;96(14):8120–5. doi: 10.1073/pnas.96.14.8120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgs HN, Pollard TD. Regulation of actin filament networks through the Arp2/3 complex: Activation by a Diverse Array of Proteins. Annu. Rev. Biochem. 2001;70:649–676. doi: 10.1146/annurev.biochem.70.1.649. [DOI] [PubMed] [Google Scholar]
- Honore B, Madsen P, Andersen AH, Leffers H. Regulation of actin filament networks through the Arp2/3 complex: Activation by a Diverse Array of Proteins. Annu. Rev. Biochem. 2001;70:649–676. doi: 10.1146/annurev.biochem.70.1.649. [DOI] [PubMed] [Google Scholar]
- Horstrup K, Jablonka B, et al. Phosphorylation of focal adhesion vasodilator-stimulated phosphoprotein at Ser157 in intact human platelets correlates with fibrinogen receptor inhibition. Eur. J. Biochem. 1994;225(1):21–7. doi: 10.1111/j.1432-1033.1994.00021.x. [DOI] [PubMed] [Google Scholar]
- Hughes PE, Pfaff M. Integrin affinity modulation. Trends Cell Biol. 1998;8(9):359–64. doi: 10.1016/s0962-8924(98)01339-7. [DOI] [PubMed] [Google Scholar]
- Huttelmaier S, et al. The interaction of the cell contact proteins VASP and vinculin is regulated by phosphotidylinositide 4,5 bisphosphate. Curr. Biol. 1998;8:479–488. doi: 10.1016/s0960-9822(98)70199-x. [DOI] [PubMed] [Google Scholar]
- Huttelmaier S, Harbeck B, et al. Characterization of the actin binding properties of the vasodilator-stimulated phosphoprotein VASP. FEBS Lett. 1999;451(1):68–74. doi: 10.1016/s0014-5793(99)00546-3. [DOI] [PubMed] [Google Scholar]
- Imaoka T, Lynham JA, et al. Purification and characterization of the 47,000-dalton protein phosphorylated during degranulation of human platelets. J. Biol. Chem. 1983;258(18):11404–14. [PubMed] [Google Scholar]
- Izaguire G, Aguirre L, et al. The cytoskeltal/nonmuscle isoform of a-actinin is phosphorylated on its actin-binding domain by focal adhesion kinase. J. Biol. Chem. 2001 doi: 10.1074/jbc.M101678200. in press. [DOI] [PubMed] [Google Scholar]
- Izaguire G, Aguirre L, et al. Tyrosine phosphorylation of a-actinin in activated platelets. J. Biol. Chem. 1999;274:3712–37020. doi: 10.1074/jbc.274.52.37012. [DOI] [PubMed] [Google Scholar]
- Jackson SP, Mistry N, et al. Platelets and the injured vessel wall-- “rolling into action”: focus on glycoprotein Ib/V/IX and the platelet cytoskeleton. Trends Cardiovasc. Med. 2000;10(5):192–7. doi: 10.1016/s1050-1738(00)00062-1. Review. [DOI] [PubMed] [Google Scholar]
- Judd BA, Myung PS, et al. Hematopoietic reconstitution of SLP-76 corrects hemostasis and platelet signaling through alpha IIb beta3 and collagen receptors. Proc. Natl. Acad. Sci. U. S. A. 2000;97(22):12056–61. doi: 10.1073/pnas.97.22.12056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaitlina SY. Functional specificity fo actin isoforms. Int. Rev. Cytol. 2001;202:35–98. doi: 10.1016/s0074-7696(01)02003-4. [DOI] [PubMed] [Google Scholar]
- Karlsson R, Lassing I, et al. The organization of microfilaments in spreading platelets: a comparison with fibrovblasts and glial cells. J. Cell Physiol. 1984;121:96–113. doi: 10.1002/jcp.1041210113. [DOI] [PubMed] [Google Scholar]
- Korn ED, Carlier M-F, et al. Actin polymerization and ATP hydrolysis. Science. 1987;238(4827):638–44. doi: 10.1126/science.3672117. [DOI] [PubMed] [Google Scholar]
- Kouyama T, Mihashi K. Pulse-fluorometry study on actin and heavy meromyosin using F-actin labelled with N-(1-pyrene)maleimide. Eur. J. Biochem. 1980;105:279–87. doi: 10.1111/j.1432-1033.1980.tb04499.x. [DOI] [PubMed] [Google Scholar]
- Kovacsovics TJ, Hartwig JH. Thrombin-induced GPIb centralization on the platelet surface requires actin assembly and myosin II activation. Blood. 1996;87:618–629. [PubMed] [Google Scholar]
- Kron SJ, Spudich JA. Fluorescent actin filaments move on myosin fixed to a glass surface. Proc. Natl. Acad. Sci. U. S. A. 1986;83(17):6272–6. doi: 10.1073/pnas.83.17.6272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni S, Saido TC, et al. Calpain mediates integrin-induced signaling at a point upstream of Rho family members. J. Biol. Chem. 1999;274(30):21265–75. doi: 10.1074/jbc.274.30.21265. [DOI] [PubMed] [Google Scholar]
- Kwiatkowski DJ. Functions of gelsolin: motility, signaling, apoptosis, cancer. Curr. Opin. Cell Biol. 1999;11(1):103–8. doi: 10.1016/s0955-0674(99)80012-x. [DOI] [PubMed] [Google Scholar]
- Kwiatkowski DJ, Bruns GA. Human profilin. Molecular cloning, sequence comparison, and chromosomal analysis. J. Biol. Chem. 1988;263(12):5910–5. [PubMed] [Google Scholar]
- Lal AA, Korn ED, et al. Rate constants for actin polymerization in ATP determined using cross-linked actin trimers as nuclei. J. Biol. Chem. 1984;259(14):8794–800. [PubMed] [Google Scholar]
- Lambrechts A, Braun A, et al. Profilin II is alternatively spliced, resulting in profilin isoforms that are differentially expressed and have distinct biochemical properties. Mol. Cell Biol. 2000a;20(21):8209–19. doi: 10.1128/mcb.20.21.8209-8219.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambrechts A, Kwiatkowski AV, et al. cAMP-dependent protein kinase phosphorylation of EVL, a Mena/VASP relative, regulates its interaction with actin and SH3 domains. J. Biol. Chem. 2000b;275(46):36143–51. doi: 10.1074/jbc.M006274200. [DOI] [PubMed] [Google Scholar]
- Lassing I, Lindberg U. Specific interaction between phosphatidylinositol 4,5-bisphosphate and profilactin. Nature. 1985;314(6010):472–4. doi: 10.1038/314472a0. [DOI] [PubMed] [Google Scholar]
- Laurent V, Carlier M-F. Use of platelet extracts for actin-based motility of Listeria monocytogenes. In: Celis JE, editor. Cell Biology: A laboratory Handbook. Vol. 2. Academic Press; San Diego, CA: 1997. pp. 359–365. [Google Scholar]
- Laurent V, Loisel TP, et al. Role of proteins of the Ena/VASP family in actin-based motility of Listeria monocytogenes. J. Cell Biol. 1999;144(6):1245–58. doi: 10.1083/jcb.144.6.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarides E, Burridge K. Alpha-actinin: immunofluorescent localization of a muscle structural protein in nonmuscle cells. Cell. 1975;6(3):289–98. doi: 10.1016/0092-8674(75)90180-4. [DOI] [PubMed] [Google Scholar]
- Li Z, Kim ES, Bearer EL. The Arp2/3 complex is required for actin polymerization during platelet shape change. Blood. 2002 doi: 10.1182/blood.v99.12.4466. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lind SE, Janmey PA, et al. Reversible binding of actin to gelsolin and profilin in human platelet extracts. J. Cell Biol. 1987;105(2):833–42. doi: 10.1083/jcb.105.2.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loisel TP, Boujemaa R, et al. Reconstitution of actin-based motility of Listeria and Shigella using pure proteins. Nature. 1999;401(6753):613–6. doi: 10.1038/44183. [DOI] [PubMed] [Google Scholar]
- Machesky LM, Atkinson SJ, et al. Purification of a cortical complex containing two unconventional actins from Acanthamoeba by affinity chromatography on profilin-agarose. J. Cell Biol. 1994;127(1):107–15. doi: 10.1083/jcb.127.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machesky LM, Goldschmidt-Clermont PJ, et al. The affinities of human platelet and Acanthamoeba profilin isoforms for polyphosphoinositides account for their relative abilities to inhibit phospholipase C. Cell Regul. 1990;1(12):937–50. doi: 10.1091/mbc.1.12.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machesky LM, Insall RH. Signaling to actin dynamics. J. Cell Biol. 1999;146(2):267–72. doi: 10.1083/jcb.146.2.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacIver SK, Zot HG, et al. Characterization of actin filament severing by actophorin from Acanthamoeba castellanii. J. Cell Biol. 1991;115(6):1611–20. doi: 10.1083/jcb.115.6.1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manchester R, Allen PG, et al. Affects of VASP and profilin on actin polymerization. Mol. Biol. Cell. 1998;9:141a. [Google Scholar]
- Matsudaira P. Modular organization of actin crosslinking proteins. Trends Biochem. Sci. 1991;16(3):87–92. doi: 10.1016/0968-0004(91)90039-x. [DOI] [PubMed] [Google Scholar]
- McGough A. F-actin-binding proteins. Curr. Opin. Struct. Biol. 1998;8(2):166–76. doi: 10.1016/s0959-440x(98)80034-1. [DOI] [PubMed] [Google Scholar]
- Mullins RD, Heuser JA, et al. The interaction of Arp2/3 complex with actin: nucleation, high affinity pointed end capping, and formation of branching networks of filaments. Proc. Natl. Acad. Sci. U. S. A. 1998;95(11):6181–6. doi: 10.1073/pnas.95.11.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins RD, Pollard TD. Rho-family GTPases require the Arp2/3 complex to stimulate actin polymerization in Acanthamoeba extracts. Curr. Biol. 1999;9(8):405–15. doi: 10.1016/s0960-9822(99)80187-0. [DOI] [PubMed] [Google Scholar]
- Nachmias VT. Cytoskeleton of human platelets at rest and after spreading. J. Cell Biol. 1980;86:795–802. doi: 10.1083/jcb.86.3.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachmias VT, Golla R. Vinculin in relation to stress fibers in spread platelets. Cell Motil. and Cytoskel. 1991;20:190–202. doi: 10.1002/cm.970200303. [DOI] [PubMed] [Google Scholar]
- Nachmias VT, Golla R, et al. Cap Z, a calcium insensitive capping protein in resting and activated platelets. FEBS Lett. 1996;378(3):258–62. doi: 10.1016/0014-5793(95)01474-8. [DOI] [PubMed] [Google Scholar]
- Nachmias VYT, Yoshida K. The cytoskeleton of the blood platelet. Adv. Cell Bio. 1988;2:181–211. [Google Scholar]
- Nakata T, Hirokawa N. Cytoskeletal reorganization of human platelets after stimulation revealed by quick-freeze deep-etch technique. J. Cell Biol. 1987;105:1771–1780. doi: 10.1083/jcb.105.4.1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niebuhr K, Ebel F, et al. A novel proline-rich motif present in ActA of Listeria monocytogenes and cytoskeletal proteins is the ligand for the EVH1 domain, a protein module present in the Ena/VASP family. EMBO J. 1997;16(17):5433–44. doi: 10.1093/emboj/16.17.5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien JR, Woodhouse MA. Platelets. Their size, shape and stickiness in vitro: Degranulation and propinquity. Exp. Biol. Med. 1968;3:90–102. [Google Scholar]
- Obergfell A, Judd BA, et al. The molecular adapter SLP-76 relays signals from platelet integrin alphaIIbbeta3 to the actin cytoskeleton. J. Biol. Chem. 2001;276(8):5916–23. doi: 10.1074/jbc.M010639200. [DOI] [PubMed] [Google Scholar]
- Ochs HD. The Wiskott-Aldrich syndrome. Smin. Hematol. 1998;35(4):332–45. Review. [PubMed] [Google Scholar]
- Oda A, Ochs HD. Wiskott-Aldrich syndrome protein and platelets. Immunol Rev. 2000;178:111–7. doi: 10.1034/j.1600-065x.2000.17808.x. Review. [DOI] [PubMed] [Google Scholar]
- Oda A, Ochs HD, et al. Collagen induces tyrosine phosphorylation of Wiskott-Aldrich syndrome protein in human platelets. Blood. 1998;92(6):1852–8. [PubMed] [Google Scholar]
- Otey CA, Pavalko FM, et al. An interaction between a-actinin and B1 integrin subunit. J. Cell Biol. 1990;111:721–729. doi: 10.1083/jcb.111.2.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantaloni D, Boujemaa R, et al. The Arp2/3 complex branches filament barbed ends: functional antagonism with capping proteins. Nat. Cell Biol. 2000;2(7):385–91. doi: 10.1038/35017011. [DOI] [PubMed] [Google Scholar]
- Pantaloni D, Carlier M-F. How profilin promotes actin filament assembly in the presence of thymosin B4. Cell. 1993;75:1007–1014. doi: 10.1016/0092-8674(93)90544-z. [DOI] [PubMed] [Google Scholar]
- Pantaloni D, Le Clainche C, et al. Mechanism of actin-based motility. Science. 2001;292(5521):1502–6. doi: 10.1126/science.1059975. [DOI] [PubMed] [Google Scholar]
- Phillips DR, Prasad KS, Mangenello J, Bao M, Nannizzi-Alaimo L. Integrin tyrosine phosphorylation in platelet signaling. Curr. Opin. Cell. Biol. 2001;13:546–554. doi: 10.1016/s0955-0674(00)00250-7. [DOI] [PubMed] [Google Scholar]
- Pollard T, Fujiwara K, et al. Contractile proteins in platelet activation and contraction. Ann. NY Acad. Sci. 1977;283:218–236. [Google Scholar]
- Pollard TD. Rate constants for the reactions of ATP- and ADP-actin with the ends of actin filaments. J. Cell Biol. 1986;103(6 Pt 2):2747–54. doi: 10.1083/jcb.103.6.2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard TD. Genomics, the cytoskeleton and motility. Nature. 2001;409:842–843. doi: 10.1038/35057029. [DOI] [PubMed] [Google Scholar]
- Pollard TD, Cooper JA. Actin and actin-binding proteins: a critical review. Ann. Rev. Biochem. 1986;55:987–1035. doi: 10.1146/annurev.bi.55.070186.005011. [DOI] [PubMed] [Google Scholar]
- Pollard TD, Mooseker MS. Direct measurement of actin polymerization rate constants by electron microscopy of actin filaments nucleated by isolated microvillus cores. J. Cell Biol. 1981;88(3):654–9. doi: 10.1083/jcb.88.3.654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhard M, Giehl K, et al. The proline-rich focal adhesion and microfilament protein VASP is a ligand for profilins. EMBO J. 1995;14(8):1583–9. doi: 10.1002/j.1460-2075.1995.tb07146.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhard M, Halbrugge M, et al. The 46/50 kDa phosphoprotein VASP purified from human platelets is a novel protein associated with actin filaments and focal contacts. EMBO J. 1992;11(6):2063–70. doi: 10.1002/j.1460-2075.1992.tb05264.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rengan R, Ochs HD, et al. Actin cytoskeletal function is spared, but apoptosis is increased, in WAS patient hematopoietic cells. Blood. 2000;95(4):1283–92. [PubMed] [Google Scholar]
- Rienhard M, Jarchau T, et al. Actin-based motility: Stop and go with Ena./VASP proteins. TBS. 2001;26:243–249. doi: 10.1016/s0968-0004(00)01785-0. [DOI] [PubMed] [Google Scholar]
- Rojnuckarin P, Kaushansky K. Actin reorganization and proplatelet formation in murine megakaryocytes: the role of protein kinase calpha. Blood. 2001;97(1):154–61. doi: 10.1182/blood.v97.1.154. [DOI] [PubMed] [Google Scholar]
- Safer D, Elzinga M, et al. Thymosin beta 4 and Fx, an actin-sequestering peptide, are indistinguishable. J. Biol. Chem. 1991;266(7):4029–32. [PubMed] [Google Scholar]
- Scherbina A, Miki H, Kenney DM, Rosen FS, Takenawa T, Remold-O'Donnell E. WASp and N-WASp in human platelets differ in sensitivity to protease calpain. Blood. 2001;98:2988–2991. doi: 10.1182/blood.v98.10.2988. [DOI] [PubMed] [Google Scholar]
- Shen BW, Josephs R, et al. Ultrastructure of the intact skeleton of human erythrocyte membrane. J. Cell Biol. 1986;102:997–1006. doi: 10.1083/jcb.102.3.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark F, Golla R, et al. Formation and contraction of a microfilamentous shell in saponin-permeabilized platelets. J. Cell Biol. 1991;112(5):903–13. doi: 10.1083/jcb.112.5.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenberg PE, Shuman MA, et al. Redistribution of alpha-granules and their contents in thrombin-stimulated platelets. J. Cell Biol. 1984;98(2):748–60S. doi: 10.1083/jcb.98.2.748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stossel TP. The machinery of cell crawling. Sci. Am. 1994;271(3):54–5. doi: 10.1038/scientificamerican0994-54. 58-63. [DOI] [PubMed] [Google Scholar]
- Stossel TP, Hartwig JH, et al. Cell crawling two decades after Abercrombie. Biochem. Soc. Symp. 1999;65:267–80. [PubMed] [Google Scholar]
- Sun HQ, Yamamoto M, et al. Gelsolin, a multifunctional actin regulatory protein. J. Biol. Chem. 1999;274(47):33179–82. doi: 10.1074/jbc.274.47.33179. [DOI] [PubMed] [Google Scholar]
- Svitkina TM, Borisy GG. Arp2/3 complex and actin depolymerizing factor/cofilin in dendritic organization and treadmilling of actin filament array in lamellipodia. J. Cell Biol. 1999;145(5):1009–26. doi: 10.1083/jcb.145.5.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theriot JA, Rosenblatt J, et al. Involvement of profilin in the actin-based motility of L. monocytogenes in cells and in cell-free extracts. Cell. 1994;76(3):505–17. doi: 10.1016/0092-8674(94)90114-7. [DOI] [PubMed] [Google Scholar]
- Tilney LG, DeRosier DJ, et al. How Listeria exploits host cell actin to form its own cytoskeleton. II. Nucleation, actin filament polarity, filament assembly, and evidence for a pointed end capper. J. Cell Biol. 1992;118(1):83–93. doi: 10.1083/jcb.118.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YL. Exchange of actin subunits at the leading edge of living fibroblasts: possible role of treadmilling. J. Cell Biol. 1985;101(2):597–602. doi: 10.1083/jcb.101.2.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber A, Nachmias VT, et al. Interaction of Thymosin B4 with muscle and platelet actin: Implications for actin sequestration in resting platelets. Biochem. 1992;31:6179–6185. doi: 10.1021/bi00142a002. [DOI] [PubMed] [Google Scholar]
- Welch MD. The world according to Arp: regulation of actin nucleation by the Arp2/3 complex. Trends Cell Biol. 1999;9(11):423–7. doi: 10.1016/s0962-8924(99)01651-7. Review. [DOI] [PubMed] [Google Scholar]
- Welch MD, Iwamatsu A, et al. Actin polymerization is induced by Arp2/3 protein complex at the surface of Listeria monocytogenes. Nature. 1997;385(6613):265–9. doi: 10.1038/385265a0. [DOI] [PubMed] [Google Scholar]
- Welch MD, Mitchison TJ. Purification and assay of the platelet Arp2/3 complex. Methods Enzymology. 1998b;298:52–61. doi: 10.1016/s0076-6879(98)98008-9. [DOI] [PubMed] [Google Scholar]
- Welch MD, Rosenblat J, et al. Interaction of human Arp2/3 complex and the Listeria monocytogenes ActA protein in actin filament nucleation. Science. 1998a;128(5373):105–8. doi: 10.1126/science.281.5373.105. [DOI] [PubMed] [Google Scholar]
- White JB. The submembrane filaments of blood platelets. Am. J. Path. 1969;56:267–277. [PMC free article] [PubMed] [Google Scholar]
- White JB. Arrangements of actin filaments in the cytoskeleton of human platelets. Am. J. Path. 1984;117:207–216. [PMC free article] [PubMed] [Google Scholar]
- White JB, Clawson CC. Biostructure of blood platelets. Ultrastruc. Path. 1980;1:533–558. doi: 10.3109/01913128009140561. [DOI] [PubMed] [Google Scholar]
- White JB, Krumweide M. An ultrastructural basis for shape change induced in platelets by chilling. Blood. 1976;30:625–635. Overview article. [PubMed] [Google Scholar]
- White JB, Rao GHR. Microtubule coil versus surface membrane cytoskeleton in maintenance and restoration of platelet discoid shape. Am. J. Path. 1998;152:597–609. [PMC free article] [PubMed] [Google Scholar]
- Witke W, Sharpe AH, et al. Hemostatic, inflammatory, and fibroblast responses are blunted in mice lacking gelsolin. Cell. 1995;81(1):41–51. doi: 10.1016/0092-8674(95)90369-0. [DOI] [PubMed] [Google Scholar]
- Yang N, Higuchi O, et al. Cofilin phosphorylation by LIM-kinase 1 and its role in Rac-mediated actin reorganization. Nature. 1998;393(6687):809–12. doi: 10.1038/31735. [DOI] [PubMed] [Google Scholar]
- Yin HL, Hartwig JH, et al. Ca2+ control of actin filament length. Effects of macrophage gelsolin on actin polymerization. J. Biol. Chem. 1981;256(18):9693–7. [PubMed] [Google Scholar]
- Yin HL, Iida K, et al. Identification of a polyphosphoinositide-modulated domain in gelsolin which binds to the sides of actin filaments. J. Cell Biol. 1988;106(3):805–12. doi: 10.1083/jcb.106.3.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaffran Y, Meyer SC, et al. Signaling across the platelet adhesion receptor glycoprotein Ib-IX induces alpha IIbbeta 3 activation both in platelets and a transfected Chinese hamster ovary cell system. J. Biol. Chem. 2000;275(22):16779–87. doi: 10.1074/jbc.275.22.16779. [DOI] [PubMed] [Google Scholar]
- Zalevsky J, Grigorova I, et al. Activation of the Arp2/3 complex by the Listeria acta protein. Acta binds two actin monomers and threesubunits of the Arp2/3 complex. J. Biol. Chem. 2001;276(5):3468–75. doi: 10.1074/jbc.M006407200. [DOI] [PubMed] [Google Scholar]
- Zebda N, Bernard O, et al. Phosphorylation of ADF/cofilin abolishes EGF-induced actin nucleation at the leading edge and subsequent lamellipod extension. J. Cell Biol. 2000;151(5):1119–28. doi: 10.1083/jcb.151.5.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zucker-Franklin D. The submembranous fibrils of human blood platelets. J. Cell Biol. 1970;47:293–299. doi: 10.1083/jcb.47.1.293. [DOI] [PMC free article] [PubMed] [Google Scholar]