Abstract
The general transcription initiation factor TFIID was originally identified, purified, and characterized with a biochemical assay in which accurate transcription initiation is reconstituted with multiple, chromatographically separable activities. Biochemical analyses have demonstrated that TFIID is a multiprotein complex that directs preinitiation complex assembly on both TATA box-containing and TATA-less promoters, and some TFIID subunits have been shown to be molecular targets for activation domains in DNA-binding regulatory proteins. These findings have most commonly been interpreted to support the view that transcriptional activation by upstream factors is the result of enhanced TFIID recruitment to the core promoter. Recent insights into the architecture and cell-cycle regulation of the multiprotein TFIID complex prompt both a reassessment of the functional role of TFIID in gene activation and a review of some of the less well-appreciated literature on TFIID. We present a speculative model for diverse functional roles of TFIID in the cell, explore the merits of the model in the context of published data, and suggest experimental approaches to resolve unanswered questions. Finally, we point out how the proposed functional roles of TFIID in eukaryotic class II transcription fit into a model for promoter recognition and activation that applies to both eubacteria and eukaryotes.
TFIID and Transcription Activation
The initial description of TFIID (1) was followed by early biochemical studies that identified it as the “TATA box-recognition factor” and focused on its interactions with the core promoter (2, 3). Nuclease and chemical footprinting techniques revealed two different types of DNA interaction patterns by a highly purified human TFIID preparation. One (on the Ad2ML promoter) extends over a broad region from nucleotide positions −47 to +35, whereas the other (on the human hsp70 promoter) is restricted to a narrow region over the TATA element (4). While the TATA box is thought to be the primary site of specific DNA binding by TFIID, downstream interactions have been shown to be sequence-dependent as well (5–9). Such interactions could contribute to TATA-independent modes of TFIID binding to core promoters (reviewed in ref. 10). This latter notion is further substantiated by the demonstration that TATA-binding protein (TBP)-associated factors (TAFs) are required for basal transcription from TATA-less promoters (11) and, further, that the TATA-specific DNA-binding activity of TBP is dispensable for transcription initiation from TATA-less promoters (12).
The first studies implicating TFIID as a target for transcriptional activators employed partially purified natural TFIID and demonstrated that activator proteins could have both quantitative and qualitative effects on TFIID-promoter binding (3, 13–15). Comparison of recombinant TBP with natural TFIID preparations in functional assays provided strong evidence that a multiprotein TFIID complex, but not TBP alone, can mediate activator-dependent transcription in vitro and suggested coactivator functions for TAFs (16–18). Furthermore, the availability of TAFs in recombinant form led to the in vitro demonstration of selective physical interactions with specific activators (reviewed in ref. 19). The functional significance of these interactions is supported by in vitro correlations of activator function with the capability to interact with TAFs (based on activator mutagenesis and anti-TAF antibody inhibition studies) and with the presence of the interacting TAF(s) in functional TFIID complexes (20–25). Recent genetic experiments similarly demonstrate the importance of particular activator–TAF interactions in transcriptional enhancement of specific genes in the Drosophila embryo (26), whereas studies in yeast have suggested that TAFs are not generally required for the activation of many genes (refs. 27 and 28; reviewed in ref. 29).
Although the ability to reconstitute partial and complete recombinant TFIID complexes (23) provides the means for examining mechanistic possibilities of TAFII coactivator functions in vitro, a high degree of transcriptional activation in vitro also requires additional coactivators that are not tightly associated with TFIID and that are only in part biochemically defined (30). In addition, the absence in purified transcription systems of natural restrictions, such as TBP-interacting negative cofactors (reviewed in ref. 30) and packaging of DNA within chromatin, may give rise to in vitro phenomena, such as activator-independent basal transcription, that have no in vivo correlates (10).
Activation Mechanisms: Direct vs. Indirect
Transcriptional activators appear to function both in vitro and in vivo by increasing the rate of transcription initiation, elongation or both (31, 32). On the basis of eubacterial studies (33) it is thought that initiation rates can be modulated at several steps that include preinitiation complex (PIC) formation, isomerization of the resulting complex, and promoter clearance.
In eukaryotes the assembly of a functional class II PIC involves the binding of at least six well-characterized general transcription factors (GTFs), RNA polymerase II and other cofactors to the core promoter region in an ordered fashion. This may involve multiple, sequential steps that have been characterized by in vitro binding experiments (reviewed in ref. 10) or the recruitment of a preassembled holo-RNA polymerase that may contain a subset of GTFs (reviewed in ref. 34). Activators may enhance the formation of a functional PIC by a “direct” mechanism that involves interactions with free GTFs (including TFIID) and/or a holoenzyme complex that effectively recruit these components to the core promoter. Alternatively, they may act by an “indirect” mechanism that involves interactions with, and modifications of, a preexisting TFIID–core promoter complex that in turn mediate recruitment of the other components. (Both mechanisms may be facilitated or mediated by coactivator proteins.)
The idea that activators could function “directly” by recruiting TFIID to the core promoter originated in the observation that the upstream activator USF and partially purified TFIID can mutually stabilize their respective DNA interactions (3) and was supported by studies with the pseudorabies activator IE (13, 14). A later demonstration that the herpes virus activator VP16 can interact with the TBP subunit of TFIID (35) inaugurated a host of similar studies, some of which indeed indicated TBP-mediated activator function (refs. 36 and 37; reviewed in ref. 38). More recently, studies employing partially reconstituted TFIID complexes have provided evidence that the synergistic action of multiply bound activators (39) can be explained by recruitment of TFIID through specific activator–TAFII interactions in vitro (23, 40, 41).
Curiously, however, an activation mechanism based solely on TFIID recruitment seems at odds with the observed high stability of TFIID–DNA complexes that (at least on several promoters) are refractory to both template challenge (42–44) and nucleosomal repression (45). It is worth emphasizing that in vitro assays that employ nonsaturating concentrations of TFIID may exaggerate the effects of activator-dependent recruitment of TFIID and, consequently, overshadow alternative TFIID-mediated activator effects. Thus the operative in vitro activation mechanism may depend crucially on the particular experimental design.
One alternative activation mechanism posits TFIID as a mediator for activator-dependent GTF/holoenzyme recruitment. An early mutagenesis analysis of the Ad EIIa late promoter, for example, showed that upstream elements do not necessarily act merely to overcome a rate-limiting step imposed by an inefficient TATA element (46). Similarly, a mechanistic study on the Ad2 E1B promoter failed to show any effect of Sp1 binding on the stability of the human TFIID–E1b promoter complex, but argued instead that a qualitative difference led to increased initiation rates (47). The same conclusion was drawn in a study that utilized specific monoclonal antibodies and the detergent sarkosyl to show that an acidic activator could act through template-committed complexes containing TFIID and TFIIA to increase the number of productive PICs (48).
A possible mechanism for such a qualitative difference in template-committed complexes was suggested by footprinting analyses on the Ad5E4 promoter. These studies demonstrated that activators can induce a downstream extension of the normally restricted TFIID footprint, an event that correlated with increased recruitment of other GTFs and RNA polymerase II and increased transcription initiation (15, 49, 50). These observations indicated an “indirect” activation mechanism involving an activator-induced conformational change of a preexisting TFIID–promoter complex that in turn facilitates productive PIC formation by incoming RNA polymerase II and other GTFs.
In this context, it is noteworthy that some mechanistic studies have revealed TFIIB incorporation into the PIC as a limiting step in transcription initiation (51), confirming the relevance of a so-called “rapid start complex” containing both TFIIB and RNA polymerase II as an intermediate within the PIC assembly pathway (43). Importantly, one TFIID subunit, Drosophila TAFII40 (homologue of human TAFII31), was shown to be capable of interacting with both acidic activation surfaces and with TFIIB (20). Recent analyses of activator function on the Ad5E4 promoter have indeed demonstrated a correlation between the aforementioned activator-induced isomerization of promoter-bound TFIID and functional TFIIB recruitment (52, 53).
Recent Insights in TFIID Structure and Function
Toward a detailed understanding of the role of TFIID in promoter recognition and PIC formation, crystallographic studies have shown that specific binding of TBP to the TATA element induces dramatic distortions of the DNA helix (54, 55). As revealed by subsequent structural studies of TBP–TATA–TFIIB (56) and TBP–TATA–TFIIA (57, 58) ternary complexes, the unique TBP–TATA structure facilitates stable interactions of TFIIA and TFIIB that in turn may allow formation of the complete PIC. More recent biochemical and biophysical studies have demonstrated that the TFIID complex contains a histone octamer-like structure consisting of hTAF80, hTAF31, and hTAF20/15 (or dTAF62, dTAF42, dTAF28/22) (59, 60). Studies examining interactions of recombinant factors and partially disrupted native human (in vitro) and yeast (in vivo) TFIID complexes indicate that components of the presumptive TAF octamer are central to the architecture of the TFIID multiprotein complex (28, 61). The location of several TFIID subunits was recently mapped relative to Ad2ML promoter DNA sequences by a site-specific photocrosslinking study (62). The same study also showed that TFIID binding to the Ad2ML promoter induces negative supercoiling that is mediated by TAFs, consistent with the earlier observation that TFIID binding to the Ad2ML promoter DNA results in a DNase I footprinting pattern reminiscent of nucleosomal DNA wrapping (3, 63).
Interestingly, a number of TFIID-interacting transcriptional cofactors are related to chromatin components. For example, the negative cofactor NC2 (Dr1/DRAP1), which can regulate TFIIB access to the TFIID–promoter complex through binding to TBP in competition with TFIIA (64–66), is a hetero-dimer that is composed of histone H2A- and H2B-related subunits and capable of DNA binding (67–69). It is thus the most recent member of a growing class of transcription cofactors that were originally described as, or bear structural relationships to, chromatin-associated proteins; these include topoisomerase I (PC3/Dr2) (70–72), HMG1 (NC1) (73), HMG2 (74, 75), HMG17 (76), HMG I(Y) (77), LEF-1 (78), and DSP1 (79). Although the mechanism of coactivator function for some of these proteins remains to be elucidated, these observations emphasize the close link between the structural organization of chromatin around a given start site and the mechanisms responsible for the precise regulation of transcriptional initiation at that site. Furthermore, a functional connection has long been suggested by a yeast genetic screen for utilization of a cryptic transcription initiation site; it not only yielded mutations in TBP itself but also in histones and what appear to be regulators of chromatin structure (e.g., see ref. 80).
Within this context it is of particular interest that an initial cell–biological study of TFIID revealed that transcriptionally inactive mitotic chromosomes contain significant amounts of TFIID (81), although mitotic phosphorylation of the DNA-binding domains of some activator proteins correlates with their DNA dissociation (82). Mitotic phosphorylation also seems to regulate the activity of the TFIID complex, but not necessarily via its ability to bind to core promoter elements (discussed further below). Instead, biochemical data indicate that multiple serine/threonine phosphorylations of TBP and TAFs selectively inhibit the ability of TFIID to mediate transcriptional activation (81), thereby providing an indication that TFIID activity can be regulated in ways other than through stoichiometric positive or negative cofactor interactions. Finally, these observations suggest that some class II promoters may bind TFIID constitutively, and that transcription initiation may be regulated during the cell cycle by reversible modifications of TFIID subunits.
A regulatory function of TFIID within the chromatin context is also suggested by the provocative findings that the largest TFIID subunit, TAFII250, contains protein kinase (83) and histone acetyltransferase (84) activities. The latter finding follows the discovery that yeast and human coactivators such as GCN5 (85) and CBP/p300 (86) contain acetyl-transferase activities that are thought to be involved in modulating DNA accessibility in chromatin. Given the existence of a histone-related octamer within TFIID, it is of immediate interest to characterize the specificity and regulation of these enzymatic activities.
A New Conceptual Framework for TFIID Function
Components involved in packaging of DNA into chromatin have coevolved with the factors that constitute the transcription machineries to fulfill their respective functional requirements in the eukaryotic cell. Significant progress in our understanding of transcriptional regulation thus requires consideration—both in experimental design and in interpretation—of the topological state and organization of the physiological promoter-containing template. It thus is not surprising that several transcriptional cofactors have been identified as chromatin components, their evolutionary relatives, or their regulators. Finally, recent studies have also established a structural and biochemical relationship of the GTF TFIID with protein components of the nucleosome. Here, we attempt to accommodate these findings in a novel model to describe a possible physiological mode of function for TFIID in transcriptional regulation. It may be stated as follows:
The TFIID complex bound to certain class II core promoters may be regarded as a specialized chromatin component that fulfills the topological requirements necessary to mediate and maintain the inducibility of genes. As such, the TFIID–core promoter complex is capable of conformational changes that allow it to switch from a transcriptionally inactive state to an active state in a process that is effected by gene- and cell type-specific activators and that can be modulated by cofactors or covalent modifications.
In the following sections, we discuss the experimental evidence for this proposal, indicate developmental and evolutionary aspects, and suggest experimental approaches to test additional predictions.
(i) In Vivo, TFIID Is Present on Some Promoters That Are Transcriptionally Inactive But Inducible.
Early in vitro studies showed that TFIID binding to core promoter DNA is mutually exclusive with nucleosome formation (45). Consistent with this observation, positioned nucleosomes have been found to repress class II gene transcription by inhibiting access of the general transcription machinery to core promoter sequences in vivo. As an example, permanent inactivation of the cell type-specific STE2 gene in yeast alpha cells correlates with the positioning of a nucleosome over the TATA box region (87).
However, consistent with the remarkable stability of TFIID–DNA complexes demonstrated in the earliest characterization of TFIID in vitro (3, 44), an above-mentioned study (81) demonstrated that TFIID, potentially in promoter complexes, can persist on transcriptionally inactive mitotic chromosomes in vivo. Evidence for TFIID binding to the core promoter regions of genes that are transcriptionally inactive is provided by a number of in vivo footprinting studies. The yeast HSP82 promoter, for example, exhibits both constitutive occupancy of its core promoter and a markedly distorted helix that is indicative of TBP binding to the TATA box (88). Similarly, genomic footprinting studies on the transcriptionally inactive Drosophila H3 (89) and yeast cyc1 (90) genes are indicative of the presence of promoter-bound TFIID.
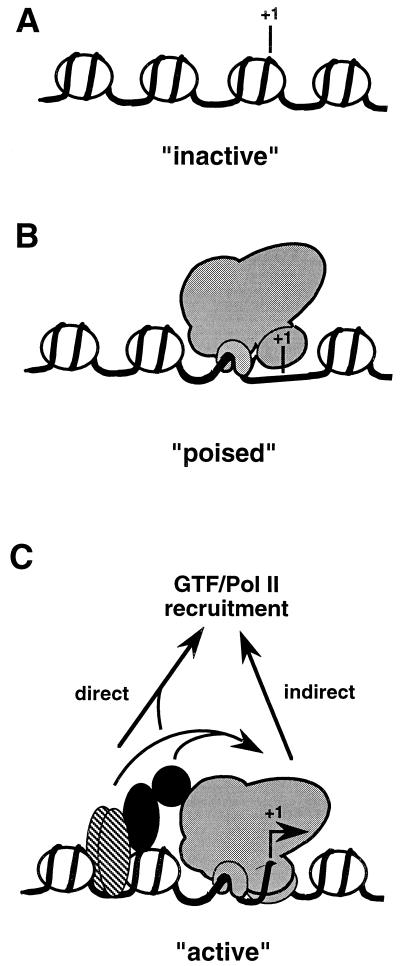
Taking these data into account, we propose that three states of class II gene activity can be distinguished, as outlined in Fig. 1. First, promoters not containing TFIID are transcriptionally inactive (Fig. 1A) and cannot be activated without chromatin remodeling, a process that, in some cases, could require DNA synthesis and cell division. Second, so-called “poised genes” are present in an inactive state but are rapidly inducible; they contain TFIID but lack activators that could provide the inducing stimulus (Fig. 1B). Third, actively transcribing genes contain TFIID as well as activators bound to their respective promoter sites (Fig. 1C).
Figure 1.
States of gene expression. Within physiological chromatin, each class II gene may be present in any one of three states that thereby determines its capacity to be transcribed. (A) “Inactive” genes are packaged in nucleosomes and inaccessible to the transcription machinery. PIC assembly and initiation must be preceded by major chromatin remodeling that, in some cases, may require DNA synthesis and mitosis. (B) “Poised” genes contain TFIID bound to the core promoter region and thus are rapidly inducible though otherwise inactive. The conformation of this complex, in the absence of an inducing stimulus (activator) renders the promoter inaccessible to RNA polymerase II and other GTFs (or the holoenzyme). (C) “Active” genes contain promoter-bound activators that recruit RNA polymerase II and GTFs (or the holoenzyme) either (i) “indirectly,” by inducing a conformational change in the TFIID–core promoter complex that renders the initiation region accessible or (ii) “directly,” via protein–protein interactions with these components.
These proposed states of gene activity are illustrated by studies on the interleukin 2 (IL-2) promoter, which is only active in activated T lymphocytes. It was demonstrated (91) by in vivo footprinting experiments that the expression of IL-2 correlates with the occupancy of cis sequences for specific upstream factors (e.g., NF-AT). In contrast, the TATA box region and downstream core promoter sequences were found to be occupied both in resting and activated T cells, with only minor changes in the in vivo footprinting pattern around the initiation site in response to gene activation by ionomycin and phorbol ester. Importantly, protein binding at the core promoter region could not be detected in pro-myelocytic HL-60 cells that are incapable of synthesizing IL-2 (91).
Taken together, these observations suggest that the presence or absence of TFIID at core promoter sequences may determine the expression capabilities of certain genes, but may not necessarily be indicative of ongoing initiation. Studying in vivo promoter occupancy by TFIID on tightly regulated promoters may therefore serve to distinguish “poised and rapidly inducible” from “inactive” genes. Which genes are “poised” is likely to be controlled in a tissue and cell type-specific manner, and may indeed be characteristic (and therefore diagnostic) for particular cell lineages. This issue is of particular interest in studies that are aimed at understanding the molecular basis for cell fate restriction/commitment in development, as well as the imprinting of gene expression patterns to successive generations. Thus, just as stably bound TFIIIA was originally proposed to be responsible for persistent and preferential expression of somatic 5S ribosomal RNA genes (relative to oocyte-type genes) during Xenopus development (92), stable TFIID–core promoter complexes on a subset of class II genes might similarly determine an inheritable gene expression program.
As argued above, an analysis of core promoter occupancy of a given gene may be indicative of the relevant mechanism of gene regulation. Although the operative mechanism could be cell type-dependent, core promoter occupancy data from a large number of genes may nonetheless allow for a promoter classification scheme that is based on functional criteria (i.e., the mechanism of regulation), rather than the presence or absence of poorly conserved (i.e., poorly identifiable) core promoter elements such as TATA and initiator elements. We speculate that such a classification scheme of genes may more reliably correlate promoter regulation with transcriptional cofactor requirement (e.g., TAFII250) and, possibly, the function of the gene product (e.g., a cell cycle regulator, see below).
(ii) The TFIID–Core Promoter Complex Is Capable of Conformational Changes That Can Be Effected by Activators: An “Active” Conformation Allows Efficient PIC Assembly, While an “Inactive” Conformation Does Not.
In addition to studies supporting the idea that activators can function on preassembled core promoter complexes as summarized above, there is accumulating evidence for an important role of activator-mediated changes in the topology of the TFIID–promoter complex during the activation process.
Activator-induced isomerization of the TFIID nucleoprotein complex in response to either a natural (15) or an artificial activator (50) was originally discovered in studies of the Ad5E4 promoter. Importantly and as mentioned above, the activator-induced conformational change of the TFIID–Ad5E4 core promoter complex was shown to correlate with enhanced binding of remaining GTFs and RNA polymerase II (15, 49). More recently, detailed studies using highly purified components have further emphasized activator-induced isomerization of the TFIID–TFIIA–core promoter complex as an important step in transcription activation (52, 53, 93). Whether topological changes in the human TFIID–Ad2ML core promoter complex that are induced by the general coactivator TFIIA, recently demonstrated by site-specific crosslinking (62), are necessary or sufficient for enhanced PIC assembly remains to be investigated; but the fact that such conformational changes are not easily discernible by simple nuclease protection assays implies that activator- or coactivator-induced isomerization of the TFIID nucleoprotein complex may be more common than previously assumed.
The functional analysis of distinct conformational states of TFIID would clearly be facilitated if the TFIID–core promoter complex could be locked in a particular conformation. Covalent modification of TFIID subunits or cofactor binding to the complex (as discussed further in iv) may have such an effect, without compromising DNA binding per se, and would thus prove immensely useful as experimental tools. Likewise, in vitro studies with partially assembled TFIID complexes (23) may be used to shed light on the roles of individual TFIID subunits in core promoter interactions and conformational changes of the complex in response to transcriptional activators.
(iii) Isomerization of the TFIID–Core Promoter Complex in Response to Activators Is Affected by the Core Promoter Sequence as Well as by the Interaction Characteristics of Individual TFIID Subunits to DNA and to Each Other.
A number of studies have noted the role of the core promoter sequence in determining the extent and selectivity of activator function in vitro and in vivo (see references in refs. 9 and 11). Differential binding of TFIID to different core promoters (4–7, 94) and the ability of promoter-bound TFIID to undergo conformational changes in response to activator or coactivator interactions (15, 52, 53, 62, 93) argue strongly for a role of TAFs in the function of core promoter sequences. Our model implies that the DNA sequence and topological characteristics of a given core promoter will also determine the mechanistic consequences of activator interactions with TFIID subunits—for example, by affecting the ability of the TFIID–core promoter complex to undergo conformational changes.
Topological aspects of promoter DNA have been shown in vitro to affect both basal promoter activity (95, 96) and transcriptional activation (97), as well as TFIID binding (98, 99). Furthermore, promoter topology may affect GTF requirements in a core promoter sequence-specific manner (100, 101).
Given that the sequence of a promoter may dictate its capacity to be activated through conformational changes of the TFIID–core promoter complex, the architecture of the TFIID complex and its DNA interaction surfaces should similarly determine the extent and mechanism of activation. Indeed, mutations that affect activator-dependent (but not basal) transcription have been mapped to the DNA interaction surface of yeast TBP (102, 103), implying that these residues are important for DNA contacts predominantly in an activator-driven pathway. Similarly, it may be possible to identify specific TAF mutations that affect either direct DNA interactions or the stereospecific arrangement of the TFIID nucleoprotein complex and that selectively affect the function either of specific activators or of specific core promoters per se. First indications that this may be a valid prediction come from studies of a temperature-sensitive allele of human TAFII250 that selectively affects the function of specific activators on the cyclin D1 promoter (104) and from studies in yeast indicating that TAF requirements for the activation of specific genes are determined by core promoter sequences (refs. 27 and 127; reviewed in refs. 29 and 105).
(iv) There May Be General Cofactors That Affect the Conformation of the TFIID–DNA Complex and/or Facilitate or Inhibit Activator-Induced Conformational Changes.
There are at least two possible mechanisms, covalent modification by specific enzymatic activities and stoichiometric binding of cofactors, that may reversibly affect the ability of the TFIID–core promoter complex to undergo conformational change.
As mentioned above, it was shown recently that TBP and associated TAFIIs are phosphorylated in HeLa cells during mitosis, an event that coincides with the loss of activator-dependent TFIID function in vitro (81). Importantly, the activator-independent (basal) transcription activity of TFIID was not significantly reduced, suggesting that phosphorylation of TFIID subunits does not inhibit TFIID promoter binding per se but rather affects productive activator interactions or the mechanistic consequences thereof (81).
Repressors bound to the TFIID–promoter complex could inhibit binding of the remaining GTFs either directly by steric hindrance or indirectly by altering the topology of the TFIID–promoter complex. Thus, the repressor NC2 (Dr1/DRAP1) binds directly to TFIIA interaction sites on TBP to inhibit TBP–TFIIA–DNA complex formation (64, 66). This interaction in turn results in significant conformational changes within the TBP–DNA complex that inhibit efficient recruitment of TFIIB (67, 68).
Conversely, TFIIA, originally characterized as a GTF, has more recently been implicated as an important coactivator molecule (52, 53, 64, 93, 106). In addition, TFIIA recruitment to the TFIID–DNA complex is accompanied by a dramatic conformational change in the complex (62, 107). Thus, specific mutations in TFIIA (106, 108) may be used to delineate the functional role of TFIIA in activator-induced isomerization of the TFIID–core promoter complex and to determine whether these conformational changes are intrinsic to TFIIA’s coactivator function.
Although TFIID can interact with, and function through, downstream core promoter DNA, with preference for the initiator element consensus (109), a number of additional initiator-binding proteins have been identified (10). While their specific requirements in accurate initiation remain unclear, we imagine that they could in fact function as coactivators by affecting the conformation of the TFIID–core promoter complex.
Finally, by analogy to recently described factors (SWI/SNF, NURF, RSC) that affect nucleosome positioning and stability (110, 111), coactivators (e.g., GCN5, P/CAF, CBP/p300, or indeed TFIID’s own TAFII250) that exhibit histone-acetyltransferase activity (112) could also modulate activator-dependent conformational changes of the TFIID–promoter complex or subsequent steps of activator function. At present, it cannot be ruled out that the as yet biochemically poorly defined coactivators PC2 and PC5 may contain similar enzymatic activities. Order-of-addition experiments indicate that they act at a step subsequent to TFIID–TFIIA–DNA complex formation (113).
(v) Considering TFIID as a Stable Component of the Chromatin Template Reveals Striking Parallels Between Eukaryotic and Eubacterial Initiation Mechanisms.
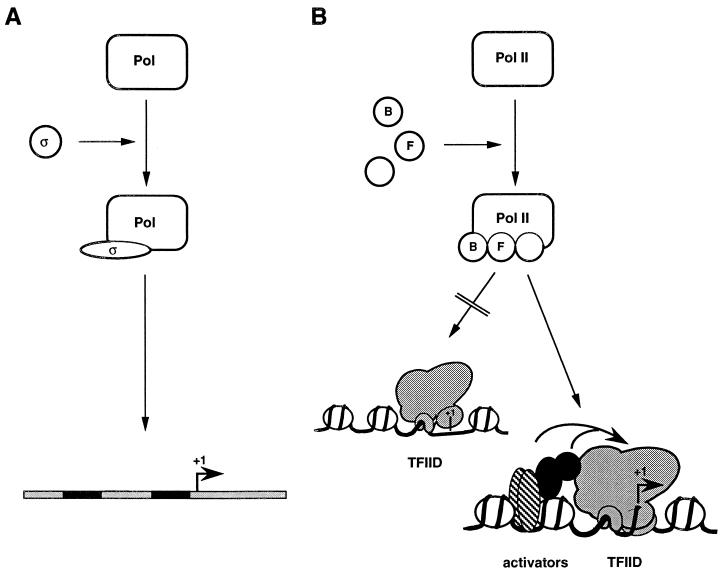
The sheer complexity of the eukaryotic general transcription machinery has discouraged comparisons to eubacterial initiation mechanisms. In eubacteria (Fig. 2A), initiation requires association of the core RNA polymerase with a single sigma factor; this interaction triggers a conformational change in sigma that enables the holoenzyme to recognize the −10 and −35 elements of the promoter (114). Promoter binding or function of the holoenzyme may be aided by activator proteins via specific interactions with different holoenzyme subunits (reviewed in refs. 105 and 115). Following initiation of transcription, sigma factor dissociates from the elongating polymerase and is free to associate with free core RNA polymerase molecules for subsequent rounds of promoter recognition and transcription initiation.
Figure 2.
Universal aspects in transcription initiation mechanisms. (A) Initiation in eubacteria requires the binding of a sigma factor to the RNA polymerase (Pol) to form the holoenzyme. This event induces a conformational change in the sigma factor, enabling it to recognize specific sequences of proximal promoter elements, leading to transcription initiation. Activators may facilitate this recognition step through interactions with holoenzyme components. (B) Initiation on class II promoters may involve analogous steps of holoenzyme assembly with GTFs binding to RNA polymerase II (Pol II), depicted here to emphasize parallels to the eubacterial paradigm. This holoenzyme (or RNA polymerase II and unbound GTFs) is able to recognize the initiation region via TFIID, but only when TFIID is bound to the core promoter DNA in a particular conformation(s) and not in another(s). The transcriptionally active conformation of TFIID can be induced by a large variety of upstream-bound activators via the coactivator function of certain TAFs and soluble cofactors.
In eukaryotes, it was originally speculated that TFIID might play a sigma-like role in class II transcription by virtue of its recognition of a T/A-rich promoter element. However, TFIID remains associated with the core promoter after transcription initiation, at least in vitro (44, 63, 116), and the crystallographic structures of TBP and sigma rule out any evolutionary relatedness between these proteins (38, 117). On the other hand, certain regions of TFIIF and sigma are related in sequence and function (118, 119), and studies of promoter recognition and start site selection by the core RNA polymerase, as well as cycling in reinitiation, argue for a sigma-like role of TFIIB (120). Interestingly, recent genetic and biochemical studies have suggested that TFIIB and TFIIF, as well as other GTFs, may associate with RNA polymerase II prior to PIC formation (121); however, the exact composition of an eukaryotic holoenzyme complex remains controversial (122, 123). Despite the observations that TFIIF contains a cryptic sigma-like DNA-binding domain (119) and that template-bound activators can interact with holoenzyme components (124), the nucleosomal organization of chromatin may still inhibit access of GTFs/RNA polymerase II or the holoenzyme complex to some promoters.
Within the more physiological context of a chromatin template, a constitutive TFIID–core promoter complex may function on some genes to regulate promoter access and recognition by eukaryotic RNA polymerase and cognate GTFs (Fig. 2B). In this model, the TFIID–core promoter complex within the chromatin template does not allow TFIIB-mediated binding of GTFs/RNA polymerase II in the absence of activators, whereas an activator-effected conformational change renders the complex capable of supporting subsequent steps of the initiation pathway. These steps may also be modulated by additional activator–GTF interactions through a direct recruitment mechanism.
Based on these considerations, certain parallels and differences between transcription mechanisms in eubacteria and eukaryotes are evident. Thus, whereas eubacterial holoenzymes are capable of binding the naked promoter DNA (Fig. 2A), eukaryotic GTFs and RNA polymerase II (or preassembled holoenzymes) require TFIID bound to the promoter to access the template (Fig. 2B)—such that certain stable TFIID–promoter complexes (within the chromatin context) may be formally equivalent to the eubacterial DNA template. Further, whereas recruitment of the eubacterial holoenzyme to cognate promoters may or may not require activators, the physiological assembly of functional PICs in eukaryotic cells generally requires activators both for “direct” and “indirect” (via the TFIID–core promoter complex) recruitment mechanisms.
Summary
We have proposed that on certain promoters the TFIID–promoter complex may serve as a specialized nucleoprotein complex that allows recruitment of RNA polymerase II and downstream GTFs, or a preassembled holoenzyme, to a chromatin template. Such a notion is remarkably consistent with interpretations of the earliest successful attempt to reconstitute accurate transcription initiation on a eukaryotic gene in a cell-free system. In this case, a purified RNA polymerase III was shown to support accurate initiation on specific (5S RNA) genes in a natural chromatin template but not on a purified template DNA (125). Similarly, RNA polymerase II and GTFs were shown to be capable of mutual association and subsequent PIC assembly on a template that was “committed” by prior binding of TFIID (63, 126).
Alongside chromatin packaging components such as nucleosomes, whose own regulatory functions in gene expression are increasingly recognized, TFIID has evolved in eukaryotes to provide mechanisms for promoter recognition by the class II transcription machinery. In this context, a stable TFIID–core promoter complex regulating promoter accessibility through conformational changes, as proposed by our model, may be regarded as a specialized “nucleosome-like” protein–DNA complex. Thus, such a structure may contribute to packaging of promoter DNA within chromatin and provide a means for regulated access of RNA polymerase and GTFs to the template. This is achieved both by excluding nucleosomes from the core promoter and, more importantly, by virtue of alternative TFIID–promoter conformations that can be reversibly affected by activators, transcriptional cofactors and posttranslational modifications.
What role does activator-effected recruitment of TFIID, as observed in vitro, have in the proposed scenario of TFIID as a constitutively bound chromatin component? While we have argued that inducible gene regulation and fine control of gene activity is not necessarily the result of regulating TFIID access to the DNA, the apparent heterogeneity of core promoters and gene- and cell type-specific regulatory proteins suggests a large diversity of regulatory mechanisms. Moreover, even for those promoters showing stably bound TFIID in specific cell types, certain activators may still effectively recruit TFIID to the core promoter during differentiation or at a specific point in the cell cycle (e.g., during the establishment of a nascent chromatin structure in S phase).
As studies of the transcription field have expanded from determining in vitro factor requirements on model templates to include an understanding of their functions in the physiological context, we believe that experimental techniques, interpretations and working hypotheses must reflect the shifted priorities. It is with this intention that we have put forward the present model. Its potential value will depend upon its ability to encourage and to provide a framework for diverse experimental efforts directed to further our understanding of TFIID’s physiological roles and mechanisms of function.
Acknowledgments
We are grateful to many of our colleagues and collaborators, particularly Drs. A. J. Koleske, E. Martinez, N. Segil, S. K. Burley, N. Heintz, and J. E. Darnell, Jr. for stimulating discussions and/or critical reading of the manuscript. A.H. thanks D. Baltimore for his encouragement and support.
ABBREVIATIONS
- TBP
TATA binding protein
- TAF
TBP-associated factor
- PIC
preinitiation complex
- GTF
general transcription factor
- Ad
adenovirus
References
- 1.Matsui T, Segall J, Weil P A, Roeder R G. J Biol Chem. 1980;255:11992–11996. [PubMed] [Google Scholar]
- 2.Parker C S, Topol J. Cell. 1984;36:357–369. doi: 10.1016/0092-8674(84)90229-0. [DOI] [PubMed] [Google Scholar]
- 3.Sawadogo M, Roeder R G. Cell. 1985;43:165–175. doi: 10.1016/0092-8674(85)90021-2. [DOI] [PubMed] [Google Scholar]
- 4.Nakajima N, Horikoshi M, Roeder R G. Mol Cell Biol. 1988;8:4028–4040. doi: 10.1128/mcb.8.10.4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nakatani Y, Horikoshi M, Brenner M, Yamamoto T, Besnard F, Roeder R G, Freese E. Nature (London) 1990;348:86–88. doi: 10.1038/348086a0. [DOI] [PubMed] [Google Scholar]
- 6.Verrijzer C P, Chen J-L, Yokomori K, Tjian R. Cell. 1995;81:1115–1125. doi: 10.1016/s0092-8674(05)80016-9. [DOI] [PubMed] [Google Scholar]
- 7.Burke T W, Kadonaga J T. Genes Dev. 1996;10:711–724. doi: 10.1101/gad.10.6.711. [DOI] [PubMed] [Google Scholar]
- 8.Wang J C, van Dyke M W. Biochim Biophys Acta. 1993;1216:73–80. doi: 10.1016/0167-4781(93)90039-g. [DOI] [PubMed] [Google Scholar]
- 9.Kaufmann J, Smale S T. Genes Dev. 1994;8:821–829. doi: 10.1101/gad.8.7.821. [DOI] [PubMed] [Google Scholar]
- 10.Roeder R G. Trends Biochem Sci. 1996;21:327–335. [PubMed] [Google Scholar]
- 11.Martinez E, Chiang C-M, Ge H, Roeder R G. EMBO J. 1994;13:3115–3126. doi: 10.1002/j.1460-2075.1994.tb06610.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martinez E, Zhou Q, L’Etoile N, Oelgeschläger T, Berk A J, Roeder R G. Proc Natl Acad Sci USA. 1995;92:11864–11868. doi: 10.1073/pnas.92.25.11864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abmayr S M, Workman J L, Roeder R G. Genes Dev. 1988;2:542–553. doi: 10.1101/gad.2.5.542. [DOI] [PubMed] [Google Scholar]
- 14.Workman J L, Abmayr S M, Cromlish W A, Roeder R G. Cell. 1988;55:211–219. doi: 10.1016/0092-8674(88)90044-x. [DOI] [PubMed] [Google Scholar]
- 15.Horikoshi M, Hai T, Lin Y-S, Green M R, Roeder R G. Cell. 1988;54:1033–1042. doi: 10.1016/0092-8674(88)90118-3. [DOI] [PubMed] [Google Scholar]
- 16.Hoffmann A, Sinn E, Yamamoto T, Wang J, Roy A, Horikoshi M, Roeder R G. Nature (London) 1990;346:387–390. doi: 10.1038/346387a0. [DOI] [PubMed] [Google Scholar]
- 17.Peterson M G, Tanese N, Pugh B F, Tjian R. Science. 1990;248:1625–1630. doi: 10.1126/science.2363050. [DOI] [PubMed] [Google Scholar]
- 18.Pugh B F, Tjian R. Cell. 1990;61:1187–1197. doi: 10.1016/0092-8674(90)90683-6. [DOI] [PubMed] [Google Scholar]
- 19.Burley S K, Roeder R G. Annu Rev Biochem. 1996;65:769–799. doi: 10.1146/annurev.bi.65.070196.004005. [DOI] [PubMed] [Google Scholar]
- 20.Goodrich J A, Hoey T, Thut C J, Admon A, Tjian R. Cell. 1993;75:519–530. doi: 10.1016/0092-8674(93)90386-5. [DOI] [PubMed] [Google Scholar]
- 21.Hoey T, Weinzierl R O J, Gill G, Chen J-L, Dynlacht B D, Tjian R. Cell. 1993;72:247–260. doi: 10.1016/0092-8674(93)90664-c. [DOI] [PubMed] [Google Scholar]
- 22.Chiang C-M, Roeder R G. Science. 1995;267:531–536. doi: 10.1126/science.7824954. [DOI] [PubMed] [Google Scholar]
- 23.Chen J-L, Attardi L D, Verrijzer C P, Yokomori K, Tjian R. Cell. 1994;79:93–105. doi: 10.1016/0092-8674(94)90403-0. [DOI] [PubMed] [Google Scholar]
- 24.Jacq X, Brou C, Lutz Y, Davidson I, Chambon P, Tora L. Cell. 1994;79:107–117. doi: 10.1016/0092-8674(94)90404-9. [DOI] [PubMed] [Google Scholar]
- 25.Gill G, Pascal E, Tseng Z H, Tjian R. Proc Natl Acad Sci USA. 1994;91:192–196. doi: 10.1073/pnas.91.1.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sauer F, Wassarman D A, Rubin G M, Tjian R. Cell. 1996;87:1271–1284. doi: 10.1016/s0092-8674(00)81822-x. [DOI] [PubMed] [Google Scholar]
- 27.Moqtaderi Z, Bai Z, Poon D, Weil P A, Struhl K. Nature (London) 1996;383:188–191. doi: 10.1038/383188a0. [DOI] [PubMed] [Google Scholar]
- 28.Walker S S, Reese J C, Apone L M, Green M R. Nature (London) 1996;383:185–188. doi: 10.1038/383185a0. [DOI] [PubMed] [Google Scholar]
- 29.Tansey W P, Herr W. Cell. 1997;88:729–732. doi: 10.1016/s0092-8674(00)81916-9. [DOI] [PubMed] [Google Scholar]
- 30.Kaiser K, Meisterernst M. Trends Biochem Sci. 1996;21:342–345. [PubMed] [Google Scholar]
- 31.Blair W S, Fridall R A, Cullen B R. EMBO J. 1996;15:1658–1665. [PMC free article] [PubMed] [Google Scholar]
- 32.Blau J, Xiao H, McCracken S, O’Hare P, Greenblatt J, Bentley D L. Mol Cell Biol. 1996;16:2044–2055. doi: 10.1128/mcb.16.5.2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McClure W R. Annu Rev Biochem. 1985;54:171–204. doi: 10.1146/annurev.bi.54.070185.001131. [DOI] [PubMed] [Google Scholar]
- 34.Koleske A J, Young R A. Trends Biochem Sci. 1995;20:113–116. doi: 10.1016/s0968-0004(00)88977-x. [DOI] [PubMed] [Google Scholar]
- 35.Stringer K F, Ingles C J, Greenblatt J. Nature (London) 1990;345:783–786. doi: 10.1038/345783a0. [DOI] [PubMed] [Google Scholar]
- 36.Ingles C J, Shales M, Cress W D, Triezenberg S J, Greenblatt J. Nature (London) 1991;351:588–590. doi: 10.1038/351588a0. [DOI] [PubMed] [Google Scholar]
- 37.Wu Y, Reece R J, Ptashne M. EMBO J. 1996;16:3951–3963. [PMC free article] [PubMed] [Google Scholar]
- 38.Nikolov D B, Burley S K. Nat Struct Biol. 1994;1:621–637. doi: 10.1038/nsb0994-621. [DOI] [PubMed] [Google Scholar]
- 39.Lin Y S, Carey M, Ptashne M, Green M R. Nature (London) 1990;345:359–361. doi: 10.1038/345359a0. [DOI] [PubMed] [Google Scholar]
- 40.Sauer F, Hansen S K, Tjian R. Science. 1995;270:1783–1788. doi: 10.1126/science.270.5243.1783. [DOI] [PubMed] [Google Scholar]
- 41.Sauer F, Hansen S K, Tjian R. Science. 1995;270:1825–1828. doi: 10.1126/science.270.5243.1825. [DOI] [PubMed] [Google Scholar]
- 42.Fire A, Samuels M, Sharp P A. J Biol Chem. 1984;259:2509–2516. [PubMed] [Google Scholar]
- 43.Hawley D K, Roeder R G. J Biol Chem. 1985;260:8163–8172. [PubMed] [Google Scholar]
- 44.van Dyke M W, Sawadogo M, Roeder R G. Mol Cell Biol. 1989;9:342–344. doi: 10.1128/mcb.9.1.342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Workman J L, Roeder R G. Cell. 1987;51:613–622. doi: 10.1016/0092-8674(87)90130-9. [DOI] [PubMed] [Google Scholar]
- 46.Huang D-H, Horikoshi M, Roeder R G. J Biol Chem. 1988;263:12596–12601. [PubMed] [Google Scholar]
- 47.Schmidt M C, Zhou Q, Berk A J. Mol Cell Biol. 1989;9:3299–3307. doi: 10.1128/mcb.9.8.3299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.White J, Brou C, Wu J, Lutz Y, Moncollin V, Chambon P. EMBO J. 1992;11:2229–2240. doi: 10.1002/j.1460-2075.1992.tb05282.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hai T, Horikoshi M, Roeder R G, Green M. Cell. 1988;54:1043–1051. doi: 10.1016/0092-8674(88)90119-5. [DOI] [PubMed] [Google Scholar]
- 50.Horikoshi M, Carey M F, Kakidani H, Roeder R G. Cell. 1988;54:665–669. doi: 10.1016/s0092-8674(88)80011-4. [DOI] [PubMed] [Google Scholar]
- 51.Lin Y S, Green M R. Cell. 1991;64:971–981. doi: 10.1016/0092-8674(91)90321-o. [DOI] [PubMed] [Google Scholar]
- 52.Chi T, Lieberman P, Ellwood K, Carey M. Nature (London) 1995;377:254–257. doi: 10.1038/377254a0. [DOI] [PubMed] [Google Scholar]
- 53.Chi T, Carey M. Genes Dev. 1996;10:2540–2550. doi: 10.1101/gad.10.20.2540. [DOI] [PubMed] [Google Scholar]
- 54.Kim Y, Geiger J H, Hahn S, Sigler P B. Nature (London) 1993;365:512–520. doi: 10.1038/365512a0. [DOI] [PubMed] [Google Scholar]
- 55.Kim J L, Nikolov D B, Burley S K. Nature (London) 1993;365:520–527. doi: 10.1038/365520a0. [DOI] [PubMed] [Google Scholar]
- 56.Nikolov D B, Chen H, Halay E D, Usheva A A, Hisatake K, Lee D K, Roeder R G, Burley S K. Nature (London) 1995;377:119–128. doi: 10.1038/377119a0. [DOI] [PubMed] [Google Scholar]
- 57.Geiger J H, Hahn S, Lee S, Sigler P B. Science. 1996;272:830–836. doi: 10.1126/science.272.5263.830. [DOI] [PubMed] [Google Scholar]
- 58.Tan S, Hunzikar Y, Sargent D F, Richmond T J. Nature (London) 1996;381:127–134. doi: 10.1038/381127a0. [DOI] [PubMed] [Google Scholar]
- 59.Hoffmann A, Chiang C-M, Oelgeschläger T, Xie X, Burley S K, Nakatani Y, Roeder R G. Nature (London) 1996;380:356–359. doi: 10.1038/380356a0. [DOI] [PubMed] [Google Scholar]
- 60.Xie X, Kokubo T, Cohen S L, Mirza U A, Hoffmann A, Chait B T, Roeder R G, Nakatani Y, Burley S K. Nature (London) 1996;380:316–322. doi: 10.1038/380316a0. [DOI] [PubMed] [Google Scholar]
- 61.Hoffmann A, Roeder R G. J Biol Chem. 1996;271:18194–18202. doi: 10.1074/jbc.271.30.18194. [DOI] [PubMed] [Google Scholar]
- 62.Oelgeschläger T, Chiang C-M, Roeder R G. Nature (London) 1996;382:735–738. doi: 10.1038/382735a0. [DOI] [PubMed] [Google Scholar]
- 63.van Dyke M W, Roeder R G, Sawadogo M. Science. 1988;241:1335–1338. doi: 10.1126/science.3413495. [DOI] [PubMed] [Google Scholar]
- 64.Meisterernst M, Roeder R G. Cell. 1991;67:557–567. doi: 10.1016/0092-8674(91)90530-c. [DOI] [PubMed] [Google Scholar]
- 65.Inostroza J A, Mermelstein F H, Ha I, Lane W S, Reinberg D. Cell. 1992;70:477–489. doi: 10.1016/0092-8674(92)90172-9. [DOI] [PubMed] [Google Scholar]
- 66.Kim T K, Zhao Y, Ge H, Bernstein R, Roeder R G. J Biol Chem. 1995;270:10976–10981. doi: 10.1074/jbc.270.18.10976. [DOI] [PubMed] [Google Scholar]
- 67.Goppelt A, Stelzer G, Lottspeich F, Meisterernst M. EMBO J. 1996;15:3105–3116. [PMC free article] [PubMed] [Google Scholar]
- 68.Mermelstein F, Yeung K, Cao J, Inostroza J A, Erdjument-Bromage H, Eagelson K, Landsman D, Levitt P, Tempst P, Reinberg D. Genes Dev. 1996;10:1033–1048. doi: 10.1101/gad.10.8.1033. [DOI] [PubMed] [Google Scholar]
- 69.Gadbois E L, Chao D M, Reese J C, Green M R, Young R A. Proc Natl Acad Sci USA. 1997;94:3145–3150. doi: 10.1073/pnas.94.7.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kretzschmar M, Meisterernst M, Roeder R G. Proc Natl Acad Sci USA. 1993;90:11508–11512. doi: 10.1073/pnas.90.24.11508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Merino A, Madden K R, Lane W S, Champoux J J, Reinberg D. Nature (London) 1993;365:227–232. doi: 10.1038/365227a0. [DOI] [PubMed] [Google Scholar]
- 72.Shykind B M, Kim J, Stance L, Champoux J J, Sharp P A. Genes Dev. 1997;11:397–407. doi: 10.1101/gad.11.3.397. [DOI] [PubMed] [Google Scholar]
- 73.Ge H, Roeder R G. J Biol Chem. 1994;269:17136–17140. [PubMed] [Google Scholar]
- 74.Shykind B M, Kim J, Sharp P A. Genes Dev. 1995;9:1354–1365. doi: 10.1101/gad.9.11.1354. [DOI] [PubMed] [Google Scholar]
- 75.Stelzer G, Goppelt A, Lottspeich F, Meisterernst M. Mol Cell Biol. 1994;14:4712–4721. doi: 10.1128/mcb.14.7.4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Paranjape S M, Krumm A, Kadonaga J T. Genes Dev. 1995;9:1978–1991. doi: 10.1101/gad.9.16.1978. [DOI] [PubMed] [Google Scholar]
- 77.Thanos D, Maniatis T. Cell. 1992;71:777–789. doi: 10.1016/0092-8674(92)90554-p. [DOI] [PubMed] [Google Scholar]
- 78.Love J J, Li X, Case D A, Giese K, Grosschedl R, Wright P E. Nature (London) 1995;376:791–795. doi: 10.1038/376791a0. [DOI] [PubMed] [Google Scholar]
- 79.Lehming N, Thanos D, Brickman J M, Ma J, Maniatis T, Ptashne M. Nature (London) 1994;371:175–179. doi: 10.1038/371175a0. [DOI] [PubMed] [Google Scholar]
- 80.Bortvin A, Winston F. J Biol Chem. 1996;272:1473–1476. doi: 10.1126/science.272.5267.1473. [DOI] [PubMed] [Google Scholar]
- 81.Segil N, Guermah M, Hoffmann A, Roeder R G, Heintz N. Genes Dev. 1996;10:2389–2400. doi: 10.1101/gad.10.19.2389. [DOI] [PubMed] [Google Scholar]
- 82.Segil N, Roberts S B, Heintz N. Science. 1991;254:1814–1816. doi: 10.1126/science.1684878. [DOI] [PubMed] [Google Scholar]
- 83.Dikstein R, Ruppert S, Tjian R. Cell. 1996;84:781–790. doi: 10.1016/s0092-8674(00)81055-7. [DOI] [PubMed] [Google Scholar]
- 84.Mizzen C A, Yang X-J, Kokubo T, Brownell J E, Bannister A J, Owen-Hughes T, Workman J, Wang L, Berger S L, Kazourides T, Nakatani Y, Allis C D. Cell. 1996;87:1261–1270. doi: 10.1016/s0092-8674(00)81821-8. [DOI] [PubMed] [Google Scholar]
- 85.Kuo M-H, Brownell J E, Sobel R E, Ranalli T A, Cook R G, Edmonson D G, Roth S Y, Allis C D. Nature (London) 1996;383:269–272. doi: 10.1038/383269a0. [DOI] [PubMed] [Google Scholar]
- 86.Ogryzko V V, Schiltz R L, Russanova V, Howard B H, Nakatani Y. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 87.Ganter B, Tan S, Richmond T J. J Mol Biol. 1993;234:975–987. doi: 10.1006/jmbi.1993.1652. [DOI] [PubMed] [Google Scholar]
- 88.Gross D S, English K E, Collins K W, Lee S W. J Mol Biol. 1990;216:611–631. doi: 10.1016/0022-2836(90)90387-2. [DOI] [PubMed] [Google Scholar]
- 89.Weber J A, Gilmour D S. Nucleic Acids Res. 1995;23:3327–3334. doi: 10.1093/nar/23.16.3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chen J, Ding M, Pederson D S. Proc Natl Acad Sci USA. 1994;91:11909–11913. doi: 10.1073/pnas.91.25.11909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Brunvand M W, Krumm A, Groudine M. Nucleic Acids Res. 1993;21:4824–4829. doi: 10.1093/nar/21.20.4824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wolffe A P, Brown D D. Science. 1988;241:1626–1632. doi: 10.1126/science.241.4873.1626. [DOI] [PubMed] [Google Scholar]
- 93.Lieberman P M, Berk A J. Genes Dev. 1994;8:995–1006. doi: 10.1101/gad.8.9.995. [DOI] [PubMed] [Google Scholar]
- 94.Chiang C-M, Ge H, Wang Z, Hoffmann A, Roeder R G. EMBO J. 1993;12:2749–2762. doi: 10.1002/j.1460-2075.1993.tb05936.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mizutani M, Ura K, Hirose S. Nucleic Acids Res. 1991;19:2907–2911. doi: 10.1093/nar/19.11.2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hirose S, Suzuki Y. Proc Natl Acad Sci USA. 1988;85:718–722. doi: 10.1073/pnas.85.3.718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mizushima-Sugano J, Roeder R G. Proc Natl Acad Sci USA. 1986;83:8511–8515. doi: 10.1073/pnas.83.22.8511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mizutani M, Ohta T, Watanabe H, Handa H, Hirose S. Proc Natl Acad Sci USA. 1990;88:718–722. doi: 10.1073/pnas.88.3.718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Parvin J D, McCormick R J, Sharp P A, Fisher D E. Nature (London) 1995;373:724–727. doi: 10.1038/373724a0. [DOI] [PubMed] [Google Scholar]
- 100.Parvin J D, Timmers H T, Sharp P A. Cell. 1992;68:1135–1144. doi: 10.1016/0092-8674(92)90084-p. [DOI] [PubMed] [Google Scholar]
- 101.Holstege F C, Tantin D, Carey M, van der Vliet P C, Timmers H T. EMBO J. 1995;14:810–819. doi: 10.1002/j.1460-2075.1995.tb07059.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kim T K, Hashimoto S, Kelleher R J I, Flanagan P M, Kornberg R D, Horikoshi M, Roeder R G. Nature (London) 1994;369:252–255. doi: 10.1038/369252a0. [DOI] [PubMed] [Google Scholar]
- 103.Lee M, Struhl K. Mol Cell Biol. 1996;15:5461–5469. doi: 10.1128/mcb.15.10.5461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Suzuki-Yagawa Y, Guermah M, Roeder R G. Mol Cell Biol. 1997;17:3284–3294. doi: 10.1128/mcb.17.6.3284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ptashne M, Gann A. Nature (London) 1997;386:569–577. doi: 10.1038/386569a0. [DOI] [PubMed] [Google Scholar]
- 106.Ma D, Olave I, Merino A, Reinberg D. Proc Natl Acad Sci USA. 1996;93:6583–6588. doi: 10.1073/pnas.93.13.6583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wang W, Gralla J D, Carey M. Genes Dev. 1992;6:1716–1727. doi: 10.1101/gad.6.9.1716. [DOI] [PubMed] [Google Scholar]
- 108.Ozer J, Bolden A H, Lieberman P M. J Biol Chem. 1996;271:11182–11190. doi: 10.1074/jbc.271.19.11182. [DOI] [PubMed] [Google Scholar]
- 109.Purnell B A, Gilmour D S. Mol Cell Biol. 1993;13:2593–2603. doi: 10.1128/mcb.13.4.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kingston R E, Bunker C A, Imbalzano A N. Genes Dev. 1996;10:905–920. doi: 10.1101/gad.10.8.905. [DOI] [PubMed] [Google Scholar]
- 111.Cairns B R, Lorch Y, Li Y, Zhang M, Lacomis L, Erdjument-Bromage H, Tempst P, Du J, Laurent B, Kornberg R D. Cell. 1996;87:1249–1260. doi: 10.1016/s0092-8674(00)81820-6. [DOI] [PubMed] [Google Scholar]
- 112.Roth S Y, Allis C D. Cell. 1996;87:5–8. doi: 10.1016/s0092-8674(00)81316-1. [DOI] [PubMed] [Google Scholar]
- 113.Halle J P, Stelzer G, Goppelt A, Meisterernst M. J Biol Chem. 1995;270:21307–21311. doi: 10.1074/jbc.270.36.21307. [DOI] [PubMed] [Google Scholar]
- 114.Helmann J D, Chamberlin M J A. Annu Rev Biochem. 1988;57:839–872. doi: 10.1146/annurev.bi.57.070188.004203. [DOI] [PubMed] [Google Scholar]
- 115.Busby S, Ebright R H. Cell. 1994;79:743–746. doi: 10.1016/0092-8674(94)90063-9. [DOI] [PubMed] [Google Scholar]
- 116.Zawel L, Kumar K P, Reinberg D. Genes Dev. 1995;9:1479–1490. doi: 10.1101/gad.9.12.1479. [DOI] [PubMed] [Google Scholar]
- 117.Malhotra A, Severinova E, Darst S A. Cell. 1996;87:127–136. doi: 10.1016/s0092-8674(00)81329-x. [DOI] [PubMed] [Google Scholar]
- 118.McCracken S, Greenblatt J. Science. 1991;253:900–902. doi: 10.1126/science.1652156. [DOI] [PubMed] [Google Scholar]
- 119.Tan S, Garrett K P, Conaway R C, Conaway J W. Proc Natl Acad Sci USA. 1994;91:9808–9812. doi: 10.1073/pnas.91.21.9808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Killeen M, Coulombe B, Greenblatt J. J Biol Chem. 1992;267:9463–9466. [PubMed] [Google Scholar]
- 121.Koleske A J, Young R A. Nature (London) 1994;368:466–469. doi: 10.1038/368466a0. [DOI] [PubMed] [Google Scholar]
- 122.Berk A J. Proc Natl Acad Sci USA. 1995;92:11952–11954. doi: 10.1073/pnas.92.26.11952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Björklund S, Kim Y-J. Trends Biochem Sci. 1996;21:335–337. doi: 10.1016/s0968-0004(96)10051-7. [DOI] [PubMed] [Google Scholar]
- 124.Barberis A, Pearlberg J, Simkovich N, Farrell S, Reinagel P, Bamdad C, Sigal G, Ptashne M. Cell. 1995;81:359–368. doi: 10.1016/0092-8674(95)90389-5. [DOI] [PubMed] [Google Scholar]
- 125.Parker C S, Roeder R G. Proc Natl Acad Sci USA. 1977;74:44–48. doi: 10.1073/pnas.74.1.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Reinberg D, Roeder R G. J Biol Chem. 1987;262:3310–3321. [PubMed] [Google Scholar]
- 127.Shen, W.-C. & Green, M. R. (1997) Cell, in press.