Abstract
Background
Sepsis is the leading cause of death in Intensive Care Units. Novel sepsis biomarkers and targets for treatment are needed to improve mortality from sepsis. MicroRNAs (miRNAs) have recently been used as finger prints for sepsis, and our goal in this prospective study was to investigate if serum miRNAs identified in genome-wide scans could predict sepsis mortality.
Methodology/Principal Findings
We enrolled 214 sepsis patients (117 survivors and 97 non-survivors based on 28-day mortality). Solexa sequencing followed by quantitative reverse transcriptase polymerase chain reaction assays was used to test for differences in the levels of miRNAs between survivors and non-survivors. miR-223, miR-15a, miR-16, miR-122, miR-193*, and miR-483-5p were significantly differentially expressed. Receiver operating characteristic curves were generated and the areas under the curve (AUC) for these six miRNAs for predicting sepsis mortality ranged from 0.610 (95%CI: 0.523–0.697) to 0.790 (95%CI: 0.719–0.861). Logistic regression analysis showed that sepsis stage, Sequential Organ Failure Assessment scores, Acute Physiology and Chronic Health Evaluation II scores, miR-15a, miR-16, miR-193b*, and miR-483-5p were associated with death from sepsis. An analysis was done using these seven variables combined. The AUC for these combined variables’ predictive probability was 0.953 (95% CI: 0.923–0.983), which was much higher than the AUCs for Acute Physiology and Chronic Health Evaluation II scores (0.782; 95% CI: 0.712–0.851), Sequential Organ Failure Assessment scores (0.752; 95% CI: 0.672–0.832), and procalcitonin levels (0.689; 95% CI: 0.611–0.784). With a cut-off point of 0.550, the predictive value of the seven variables had a sensitivity of 88.5% and a specificity of 90.4%. Additionally, miR-193b* had the highest odds ratio for sepsis mortality of 9.23 (95% CI: 1.20–71.16).
Conclusion/Significance
Six serum miRNA’s were identified as prognostic predictors for sepsis patients.
Trial Registration
ClinicalTrials.gov NCT01207531
Introduction
Sepsis is a state of sustained infection that results in a severe systemic inflammatory response and, possibly, shock [1]. Despite significant improvements in critical care medicine during recent decades, sepsis remains as one of the leading causes of mortality in the intensive care unit (ICU) [2]. Numerous studies have attempted to identify biomarkers to predict sepsis mortality, including serum C-reactive protein (CRP), Procalcitonin (PCT) levels [3], and Acute Physiology and Chronic Health Evaluation II scores (APACHE II) and Sequential Organ Failure Assessment scores (SOFA) [4].
However, CRP levels on the day of sepsis diagnosis have proved to have poor predictive value for mortality [5]. PCT, APACHE II scores, and SOFA scores have shown better predictive values compared to CRP levels; however, their areas under receiver operating characteristic (AUROC) curves were only 0.75 (95% CI: 0.66–0.85) [6], 0.75 (95% CI: 0.67–0.83), and 0.8 (95% CI: 0.72–0.88), respectively [7]. Thus, the sensitivities and specificities of these four biomarkers, which are widely used in clinical practice, are not sufficiently high to predict sepsis mortality.
MicroRNAs (miRNAs) are endogenous RNAs of ∼22 nt that play important regulatory roles by targeting mRNAs for cleavage or translational repression [8]. Recently, serum or plasma miRNAs levels have been shown to be stable and reproducible [9], [10], and the unique expression patterns of serum miRNAs can be used as finger prints for evaluating the prognoses of various diseases [9], [11]. To our knowledge, only miR-223 and miR-146a have shown diagnostic value for sepsis and only one plasma miRNA, miR-150, was identified as a potential biomarker for sepsis prognosis [12]. As is well known, sepsis is a complex syndrome that involves multiple organs and tissues, and not only leukocytes, and a single biomarker is not sufficient to reflect the severity of sepsis. Thus, a panel of biomarkers is needed to evaluate sepsis prognosis.
There have been no studies that used genome-wide screening of serum or plasma for sepsis prognosis or mortality from sepsis. Numerous methods have been used to detect the genome-wide expression profiles of miRNAs, such as Exqion miRCURY LNA array technology, the Illumina BeadChip platform, the Febit automated Geniom Real Time Analyzer platform, and Affymetrix GeneChip miRNA array technology [13]–[15]. However, all of these microarrays are limited due to the possibility of false positive results. Next generation sequencing is now being used to detect miRNA expression profiles in the sera of patients with different diseases [16], [17]. Compared to microarrays, these have shown better detection capabilities both in terms of the quality and quantity of miRNAs. These results showed that next generation sequencing is a promising method for genome-wide miRNA screening [18].
In this study, we first used Solexa sequencing to screen for serum miRNAs among 9 surviving and 9 non-surviving sepsis patients. Then, qRT-PCR was used to validate the Solexa sequencing results in a cohort of 166 sepsis patients. After validation, the predictive values of the screened miRNAs were compared to those of currently used clinical biomarkers. A multivariable logistic regression analysis was also used to evaluate the predictive values and odds ratios of these miRNAs.
Results
Clinical Characterizations of the Sepsis Patients
From July 2010 to March 2011, a total of 214 sepsis patients were screened. Based on 28-day mortality, they included 117 survivors and 97 non-survivors who were matched by sex (p = 0.406) and age (p = 0.730). Pulmonary infection was the leading cause of sepsis in these patients. There were no significant differences in the causes of sepsis between the two groups (pulmonary infection, p = 0.179; post-surgery, p = 0.591; acute pancreatitis, p = 0.981; multiple trauma, p = 0.491). However, APACHE II scores and SOFA scores were significantly different between sepsis survivors and non-survivors ( Table 1 ).
Table 1. Clinical data for 214 sepsis patients.
| Variables | Survivors (n = 117) | Non-survivors (n = 97) | P-value |
| Male/female | 72/36 | 58/37 | 0.406A |
| Ages (years) | 60.7±18.9 | 59.6±18.8 | 0.730B |
| Cause of sepsis (%) | |||
| Pulmonary infection | 39(33.3%) | 41(42.3%) | 0.179A |
| Post-surgery | 30(25.6%) | 23(23.7%) | 0.591A |
| Acute pancreatitis | 10(8.5%) | 7(7.2%) | 0.981A |
| Multiple trauma | 17(14.6%) | 11(11.3%) | 0.491A |
| Others | 21(18.0%) | 15(15.5%) | 0.629A |
| Intercurrent conditions | |||
| CHD# | 15(12.8%) | 10(10.3%) | 0.569A |
| Cancer | 9(7.7%) | 13(13.4%) | 0.171A |
| Hypertension | 21(17.9%) | 17(17.5) | 0.936A |
| Diabetes | 7(6.0%) | 5(5.2%) | 0.793A |
| COPD* | 25(21.4%) | 20(17.1%) | 0.894A |
| SOFA score | 5.80±3.33 | 9.54±3.33 | <0.001B |
| APACHE II score | 15.75±6.46 | 22.72±6.62 | <0.001B |
| CRP (mg/dl) | 3.5(0.2,6.2) | 12.15(6.7,32.8) | 0.687C |
| PCT (ng/ml) | 1.97(0.05,3.78) | 8.29(3.91,191.44) | <0.001C |
A: Chi Square tests; B: Student t test; C: Mann-Whitney U tests; #: Coronary heart disease;
: Chronic obstructive pulmonary disease.
Genome-wide Screening for Serum miRNAs by Solexa Sequencing
We selected 9 survivors and 9 non-survivors who were matched by sex, age, and sepsis severity for initial Solexa sequencing ( Table 2 ). This sequencing detected 153 known miRNAs in the non-surviving group and 173 known miRNAs in the surviving group. The expression profiles of these miRNAs are shown in Table S1 and Table S2. Then, we compared the expressions of these known miRNA between the two groups; a large number of miRNAs were differentially expressed (Table S3).
Table 2. Clinical characteristics of the sepsis patients used for Solexa sequencing.
| Variables | Survivors(n = 9) | Non-survivors(n = 9) | p-value |
| Male/female | 6/3 | 7/2 | 0.599A |
| Ages (years) | 62.3±20.8 | 60.9±21.5 | 0.887B |
| Type | |||
| Sepsis | 3 | 3 | - |
| Severe sepsis | 3 | 3 | - |
| Septic shock | 3 | 3 | - |
| SOFA scores | 6.0±3.7 | 9.6±3.6 | 0.057B |
| APACHE II | 14.5±6.5 | 20.9±6.0 | 0.060B |
| CRP (mg/dl) | 13.8 (6.1, 20.6) | 14.15 (0.5, 22.2) | 0.116B |
| PCT (ng/ml) | 4.87(0.37,10.36) | 10.18(0.45,32.76) | 0.404B |
| WBC (×109/L) | 11.84(3.01,23.29) | 10.08(7.62,29.6) | 0.559B |
A: Chi Square tests; B: Mann-Whitney U tests.
The level of an miRNA was considered to be significantly differentially expressed only if two criteria were both satisfied: (1) there were at least 20 copies in either the surviving or non-surviving group [16]; and (2) there was at least a two-fold different expression level between the two groups [16], [19]. After screening, 11 miRNAs were identified. These included 6 up-regulated miRNAs (miR-378, miR-483-5p, miR-193b*, miR-122, miR-499-5p, miR-206) and 5 down-regulated miRNAs (miR-451, miR-16, miR-15b, miR-15a, miR-486-5p) in the non-surviving group compared to the surviving group; these are shown in Table 3 . Despite its low fold-change of 1.8, miR-223 was also chosen because miR-223 had been previously proven to be a biomarker of sepsis [20]. Thus, 12 miRNAs were selected for a small sample size validation by qRT-PCR.
Table 3. Fold-changes of selected miRNAs in Non-survivors (D) and Survivors (S).
| miR-names | Fold-change* | p-value |
| Down-regulated innon-survivors’ sera | ||
| hsa-miR-451 | –3.27 | 0 |
| hsa-miR-16 | –3.21 | 0 |
| hsa-miR-15b | –3.07 | 0 |
| hsa-miR-15a | –2.28 | <0.001 |
| hsa-miR-486-5p | –2.19 | 0 |
| hsa-miR-223 | –1.81 | <0.001 |
| Up-regulated innon-survivors’ sera | ||
| hsa-miR-378 | 2.38 | <0.001 |
| hsa-miR-483-5p | 2.05 | 0 |
| hsa-miR-193b* | 2.77 | 0 |
| hsa-miR-122 | 3.14 | 0 |
| hsa-miR-499-5p | 4.40 | 0 |
| hsa-miR-206 | 4.88 | 0 |
Fold change formula: Fold-change = log2 (D/S).
qRT-PCR Small Sample Size Validation and Large Sample Size Confirmation
24 healthy controls were used to normalize the expression levels of the miRNAs measured in our study and all of these miRNAs were detectable in normal controls’ sera. Based on previous results [21], [22], U6 snRNA was selected as an internal normalization control for miRNA qPCR. In our study, there were no significant differences in raw Ct values for U6 snRNA among normal controls, non-survivors, and survivors (p = 0.376; Kruskal-Wallis H).
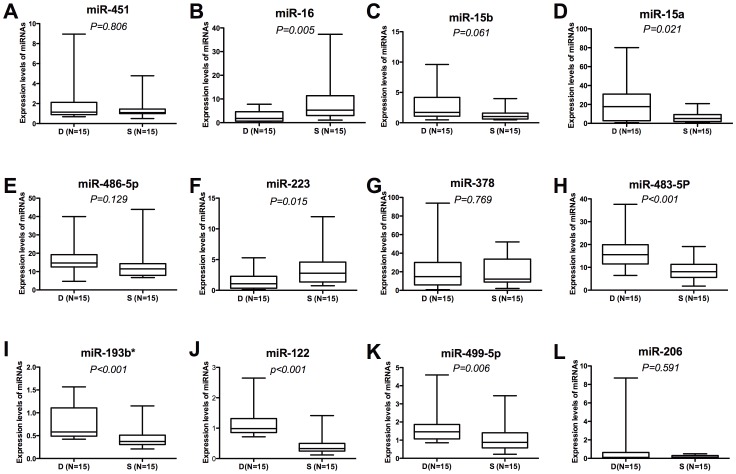
We used qRT-PCR to validate the altered expressions of the 12 candidate miRNAs for 15 survivors and 15 non-survivors. After validation, seven miRNAs were identified (associated p-values in parentheses): miR-223 (p = 0.015); miR-15a (p = 0.005); miR-16 (p = 0.021); miR-499-5p (p = 0.006); miR-122 (p<0.001), miR-193b*(p<0.001) and miR-483-5p (p<0.001). Compared to the surviving group, miR-16 and miR-223 were up-regulated in the non-surviving group and the levels of miR-15a, miR-499-5p, miR-193b*, miR-122, and miR-483-5p were up-regulated ( Figure 1 ). Although levels of miR-15b were not significantly different in the validation set, previous study has proved that levels of miR-15b were significantly higher than other expressed miRNAs in T cell subsets [23]. Hence, miR-15b was also selected for further confirmation.
Figure 1. Comparison of 12 miRNAs expression levels between sepsis non-survivors and survivors in a validation set.
Differentially expressed miRNAs identified by Solexa sequencing were validated by qRT-PCR in sepsis survivors (S; N = 15) compared to non-survivors (D; N = 15). MiR-16 (p = 0.005), miR-15a (p = 0.021), miR-223 (p = 0.015), miR-483-5p (p<0.001), miR-193b* (p<0.001), miR-122 (p<0.001), and miR-499-5p (p = 0.006) were significantly different after validation with qRT-PCR. Expression levels of the 12 miRNAs were normalized to U6 snRNA above normal controls and given as fold-changes (2–ΔΔCt ). △△Ct = (CtmiRNA-CtU6 snRNA) patients-(CtmiRNA-CtU6 snRNA)controls. Mann-Whitney U-test was used for statistical comparisons.
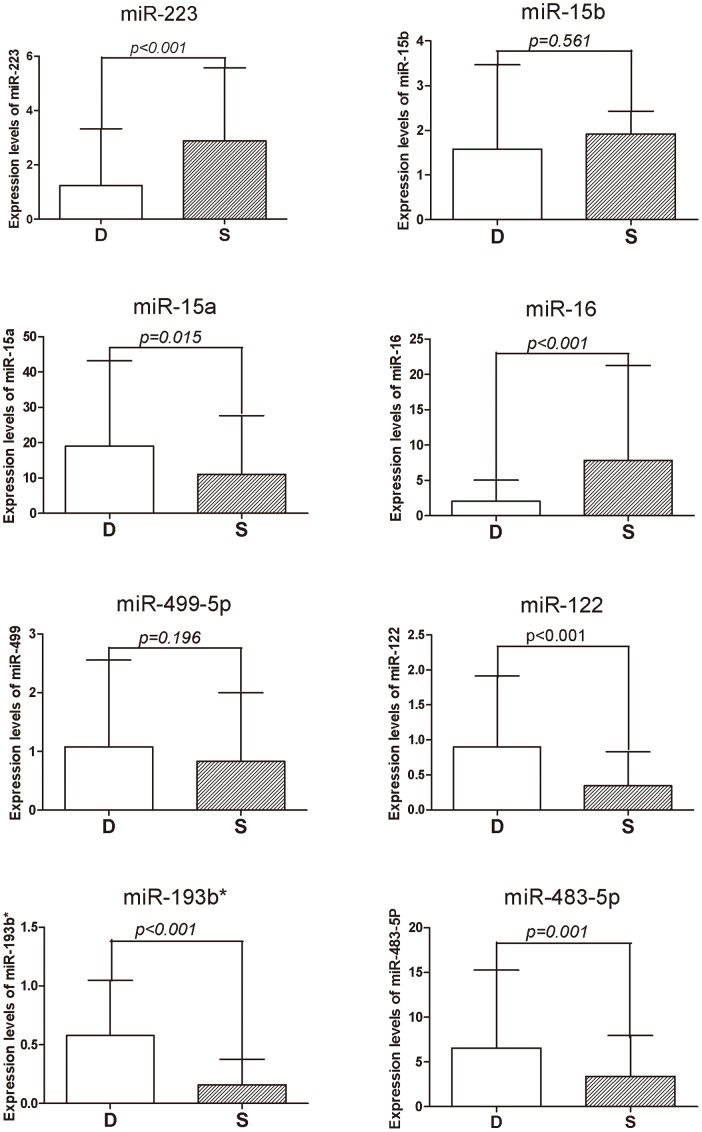
Next, the eight differentially expressed miRNAs were further validated in a confirmation set comprised of 93 survivors and 73 non-survivors. As a result, two of the eight miRNAs’ levels (miR-15b and miR-499-5p) were not significantly different. Six miRNAs were confirmed to be significantly differentially expressed and these miRNA expression pattern alterations in the confirmation set were consistent with those of the validation set. The expression levels of miR-15a (p = 0.015), miR-193b* (p<0.001), miR-122 (p<0.001), and miR-483-5p (p<0.001) in non-survivors were significantly higher than in survivors, and the levels of miR-223(p<0.001) and miR-16 (p<0.001) were significantly lower ( Figure 2 ).
Figure 2. Expression levels of 8 miRNAs in sepsis non-survivors and survivors in a confirmation set.
These 8 miRNAs were significantly differentially expressed between sepsis non-survivors and survivors after qRT-PCR validation in a smaller study sample. Then, these were checked by qRT-PCR in a larger study sample size (Non-survivors = D, n = 73; Survivors = S, n = 93). Only 6 of the 8 miRNAs remained as significantly different between the D group and S groups. Expression levels of these 8 miRNAs were normalized to U6 snRNA U6 snRNA above normal controls and given as fold-changes (2–ΔΔCt ), △△Ct = (CtmiRNA-CtU6 snRNA)patients-(CtmiRNA-CtU6 snRNA)controls. Mann-Whitney U-test or student t-test was used for statistical comparisons.
Comparisons of the Predictive Mortality Values of the Six miRNAs with SOFA Scores, APACHE II Scores, and PCT Levels
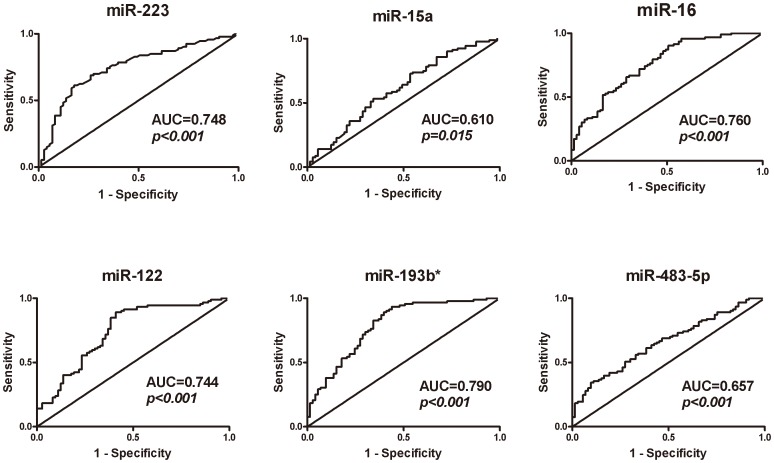
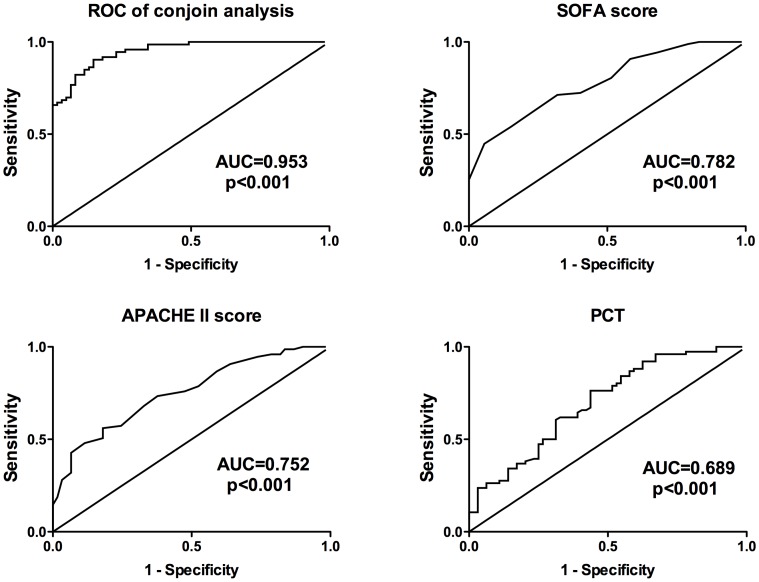
To investigate the predictive mortality value of the six confirmed miRNAs, receiver operating characteristic (ROC) curves were generated and the areas under the curves (AUCs) were calculated. The AUCs for the six miRNAs ranged from 0.610 (95%CI: 0.523–0.697) to 0.790 (95%CI: 0.719–0.861) ( Figure 3 ). The AUCs for SOFA scores, APACHE II scores, and PCT levels were, respectively, 0.782 (95%CI: 0.712–0.851), 0.752 (95%CI: 0.672–0.832), and 0.689 (95%CI: 0.611–0.784) ( Figure 4 ). The AUC for miR-193b* had the highest value, followed by those for SOFA scores and miR-16. AUCs for miR-223 and miR-122 were nearly identical to the AUC for APACHE II scores.
Figure 3. Receiver operating characteristic (ROC) curves for serum miRNAs for sepsis non-survivors (n = 73) and survivors (n = 93).
These six miRNAs were finally confirmed by qRT-PCR. Areas under the ROC curves are also shown.
Figure 4. Receiver operating characteristic (ROC) curves for serum miRNAs and clinically used indicators for sepsis non-survivors (n = 73) and survivors (n = 93).
Serum levels of miRNAs were quantified using real time qPCR. Each qPCR was done in triplicate in 96-well plates. Expression levels of the selected miRNAs were normalized to U6 snRNA and presented as fold-changes (2-ΔΔCt) above normal controls.
Four Serum miRNAs Identified as Risk Factors for Sepsis Patients’ Prognosis
Logistic regression analysis was used to evaluate possible associations between the six miRNAs and sepsis patients’ mortality. First, univariate logistic regression was used for the six miRNAs along with sepsis stage (sepsis = 1, severe sepsis = 2, septic shock = 3), age, sex, SOFA scores, APACHE II scores, CRP and PCT levels, white blood cell counts (WBC), coronary heart diseases (CHD), diabetes, hypotension, and COPD. The sepsis stage, six serum miRNAs, SOFA scores, APACHE II scores, PCT levels and diabetes were significantly different between survivors and non-survivors (all p<0.05). These variables were used in multivariable logistic regression analysis.
Only the sepsis stage, SOFA scores, APACHE II scores, miR-15a, miR-16, miR193b*, and miR-483-5p were entered into the final regression equation ( Table 4 ). The predictive values of these seven variables were evaluated by ROC analysis. The AUC of these combined seven variables was 0.953 (95%CI: 0.923–0.985), which was much higher than SOFA scores, APACHE II scores, or any of the miRNAs ( Figure 4 ). When the cut-off point was set at 0.550, the predictive value of the seven variables had a sensitivity of 88.5% and a specificity of 90.4%. Then, we calculated the predictive value when only SOFA and APACHE II scores were combined. The AUC was 0.891 (95%CI: 0.843–0.939), which was lower than when the miRNAs results were added.
Table 4. Multivariate logistic regression analysis of the variables (Forward conditional).
| Variables | Univariate | Multivariate | ||
| ORs | p-value | ORs | p-value | |
| Sepsis stage | 3.15(1.94, 5.10) | <0.001 | 6.96(2.11,22.99) | 0.001 |
| Sex | 1.51(0.78, 2.92) | 0.23 | - | - |
| Ages | 0.99(0.98, 1.00) | 0.73 | - | - |
| SOFA score | 1.37(1.23, 1.53) | <0.001 | 1.25(1.01, 1.54) | 0.039 |
| APACHE II | 1.18(1.10, 1.26) | <0.001 | 1.14(1.02,1.27) | 0.026 |
| CRP | 1.02(0.98, 1.07) | 0.33 | - | - |
| PCT | 1.06(1.03, 1.10) | 0.001 | - | - |
| WBC | 1.01(0.96, 1.06) | 0.817 | - | - |
| Serum miR-223 | 0.67(0.54, 0.82) | <0.001 | - | - |
| Serum miR-15a | 1.02(1.00, 1.04) | 0.022 | 1.03(1.00, 1.07) | 0.037 |
| Serum miR-16 | 0.81(0.72, 0.91) | <0.001 | 0.60(0.45,0.81) | 0.001 |
| Serum miR-122 | 5.23(2.40,11.41) | 0.02 | - | - |
| Serum miR-193b* | 37.88(10.64,134.90) | <0.001 | 9.07(1.32,62.42) | 0.025 |
| Serum miR-483-5p | 1.08(1.01, 1.15) | 0.007 | 1.29(1.08,1.55) | 0.006 |
| CHD | 1.31(0.55, 3.11) | 0.548 | _ | _ |
| Diabetes | 3.74(1.14, 12.31) | 0.030 | _ | _ |
| Hypotension | 1,70(0.70, 4.14) | 0.245 | _ | _ |
| COPD | 1.37(0.52, 3.58) | 0.525 | _ | _ |
Among these seven factors, miR-193b* had the highest odds ratio (OR) of 9.07 (95% CI: 1.32–62.42), followed by sepsis stage, miR-483-5p, SOFA scores, APACHE II scores, miR-15a, and miR-16 with ORs, respectively, of 6.96 (95% CI: 2.11–22.99), 1.29 (95% CI: 1.08–1.55), 1.25 (95%CI: 1.01, 1.54), 1.14 (95% CI: 1.02–1.27), 1.03 (95% CI: 1.00–1.07), and 0.60 (95% CI: 0.45–0.81). These results indicated that sepsis stage, miR-193b*, miR-483-5p, APACHE II scores, and miR-15a were independent risk factors for sepsis patients’ death. miR-16 was identified as a factor associated with decreased risk of death for sepsis patients.
Discussion
Several circulating miRNAs have recently been reported to be biomarkers for sepsis diagnosis and prognosis [12], [20]. In our study, a genome-wide Solexa method was first used to screen 18 sepsis patients’ sera for miRNAs. Then, qRT-PCR was used for 196 sepsis patients to confirm the results of Solexa sequencing. Two methods for validation and a large sample size made our results more convincing. As a result, six miRNAs were confirmed to be significantly differentially expressed between sepsis survivors and non-survivors.
Among these six miRNAs, the predictive value of miR-193b* for sepsis mortality was better than SOFA scores and APACHE II scores, which are both composites that integrate numerous sepsis indicators. However, serum miR-15a and miR-483-5p had poor predictive values for sepsis mortality. Thus, we used a logistic regression analysis to select those miRNAs that were associated with death from sepsis. These results showed that a combination of miR-15a, miR-16, miR-193b*, miR-483-5p, SOFA scores, APACHE II scores, and sepsis stage had a much better predictive value for mortality. Therefore, if we were to integrate these six miRNAs into a composite indicator in clinical practice, this composite indicator would have a much better predictive value for sepsis mortality than SOFA scores and APACHE II scores.
These six miRNAs were differentially expressed even when survivors and non-survivors were matched by sepsis severity. Hence, these biomarkers may be genetic markers and may not necessarily be suitable for characterizing sepsis progression. Functional studies involving these biomarkers will be needed, which may provide additional valuable information for treating physicians.
Previous studies of miRNAs in sepsis patients primarily focused on the miRNA expression profiles of WBCs. miR-146b, miR-150, miR-342, and miR-let-7 g were found to be differentially expressed by WBCs from healthy donors after treatment with E. Coli lipopolysaccharide (LPS) infusion for 4 hours [24], and miR-150, miR-182, miR-342-5p, and miR-486 were identified in WBCs from sepsis patients’ peripheral blood [12]. However, none of these miRNAs were present in the expression profiles of our six miRNAs. These differences may be due to the following reasons.
In peripheral blood serum, circulating miRNAs are found in microvesicles (MVs), which have been identified as carriers of genetic information exchange between cells [25]. The majority of peripheral blood MVs derive from platelets [26], [27] and a second, large population of MVs derives from mononuclear phagocyte cell lineages. Only a small percentage of MVs are derived from T-cells and neutrophils [27]. In sepsis patients, WBCs are not the only blood cells that are involved in sepsis pathogenesis. Thus, the serum miRNAs screened for using a genome-wide method might derive mostly from other sources given the small percentage of miRNAs from white cells.
Functional studies of these screened miRNAs in our study have already been performed by other investigators. miR-223, miR-15a, and miR-16 have been found to be associated with the innate immune system by targeting IKKα mRNA, which is involved in the NF-κB signaling pathway [28]. Functional studies of miR-193b* have mostly focused on cancer [29], [30]. miR-122, a liver-specific miRNA, was found to be associated with hepatocellular carcinoma [31] and chronic hepatitis [32]. Abnormal expression of miR-483-5p was indicative of a poor prognosis for adrenocortical carcinomas [33]. To date, no direct functional study has shown that these miRNAs were associated with sepsis. Hence, much work needs to be done.
This study was novel in several respects. First, we addressed a significant medical condition-sepsis-from a new perspective: the involvement of serum miRNAs. Second, Solexa sequencing was first used for genome-wide screening of sepsis patients’ sera to identify differentially expressed miRNAs. Third, this was the first time that a high- throughput method was used to screen for serum miRNAs to evaluate sepsis prognosis. Finally, this is the first report to show that miR-16, miR-15a, miR-193b*, and miR-483-5p were associated with sepsis prognosis.
However, this study had some limitations. We only used 9 survivors and 9 non-survivors in our discovery set. Although six miRNAs were found to be valuable for predicting mortality of sepsis patients, a number of miRNAs with low expression levels might have been left out during the initial screening by Solexa sequencing. The 9 samples in each group were comprised of 3 sepsis patients, 3 severe sepsis patients, and 3 septic shock patients. Solexa sequencing performed only for single subgroups may discover some specific miRNAs in these subgroups. Another limitation was that miR-499 were significantly differentially expressed in the validation set with p value of 0.006 but not in confirmation set with value of 0.196. This result meant that relatively small numbers in sample groups could readily give misleading results and these results of our study were still needed to be validated in a much larger sample group. In addition, conditions of the patients and comorbidity influences may also affect the results. In addition, in our study, we only evaluated the levels of these miRNAs in patients who were admitted to ICU within 24 hours. Whether there is a progression or increase of these biomarkers when these patients approach death is still unknown. Hence, evaluation of the dynamic changes of these miRNAs during the sepsis process was essential to our further study.
Regardless of these limitations, the six miRNAs were found to be valuable predictors of sepsis mortality. In future studies, a focus on the trends of the expression level changes of these six miRNAs during hospitalization in the ICU would be even more valuable. In addition, predicting the target genes of these six miRNAs and post-experimental validation of these target genes may provide new targets for the treatment of sepsis.
In conclusion, six serum miRNAs identified in our study can be used to predict sepsis patients’ mortality.
Materials and Methods
The protocol for this trial and supporting CONSORT checklist are available as supporting information; see Checklist S1 and Protocol S1.
Ethics
Written informed consent was obtained from all sepsis patients and normal controls. This study was approved by the Committee on Ethics of the Chinese PLA General Hospital.
Study Subjects
All patients who were diagnosed with sepsis met the definitions of the American College of Chest Physicians/Society of Critical Care Medicine (ACCP/SCCM) [34]. Patients younger than 18 years of age or those who were immunosuppressed were excluded from this study. Patients who did not receive adequate treatment because of economic hardship were also excluded. A total of 214 sepsis patients (117 survivors and 97 non-survivors) were enrolled from July 2010 to March 2011. Healthy controls (n = 24) were recruited from the Health Screening Center of the Chinese PLA General Hospital and they did not have any type of known infection or known medical condition.
Based on 28-day mortality, we divided sepsis patients into a surviving group and a non-surviving group. First, to detect differently expressed miRNAs between the surviving and non-surviving groups, 9 survivors and 9 non-survivors who were matched by sex (p = 0.902), age (p = 0.887), and sepsis severity (SOFA scores, p = 0.057; APACHE II scores, p = 0.060) were screened by Solexa sequencing (miRBase 15.0). Next, qRT-PCR was used in a validation set (15 survivors and 15 non-survivors) who were also matched by sex (p = 0.821), age (p = 0.329), and sepsis severity (SOFA scores, p = 0.089; APACHE II scores, p = 0.071) to validate the Solexa sequencing results in a small sample size. Finally, the validated serum miRNAs in the small sample size were confirmed in a confirmation set (93 survivors and 73 non-survivors).
Blood Samples and Serum Total RNA Isolation
Blood samples of all the patients were drawn within 24 hours after a diagnosis of sepsis was made. Coagulated blood samples were collected in tubes containing a separating gel and clot activator and centrifuged at 3000 rpm for 15 min at room temperature. The supernatant was transferred to Eppendorf tubes. These samples were centrifuged at 15,000 rpm for 30 min to precipitate cell debris, and the supernatants were stored at -80°C until RNA extraction. Serum total RNA was isolated using a mirVana PARIS kit (Ambion, #1556) according to the manufacturer ‘s instructions. All serum RNA preparations were pre-treated with RNase-free DNase I (Promega) to eliminate potential DNA contamination.
Solexa Sequencing and Bioinformatics Analysis
The serum samples of 9 survivors and 9 non-survivors were pooled for their respective groups. Total RNA was purified directly for Solexa sequencing analysis using an Illumina Genome Analyzer (Illumina, San Diego, CA, USA) according to the manufacturer's instructions [35]. Briefly, a pair of Solexa adaptors were ligated to the 5' and 3' ends of total RNA, and small RNA molecules were amplified by reverse transcription PCR (RT-PCR) using a RT-PCR kit (Invitrogen). The adaptor primers for 17 cycles and fragments around 30 bp (small RNA + adaptors) were isolated from an agarose gel. Purified RNA was used directly for cluster generation and sequencing analysis using an Illumina Genome Analyzer (Illumina, San Diego, CA, USA) according to the manufacturer's instructions. After a bioinformatics analysis [36], the remaining miRNAs were searched against the Sanger miRBase (version15.0) [37] to identify conserved, known miRNAs. Then, the fold-changes of the known miRNAs between the two groups were determined by: Log2(copies of non-survivors/copies of survivors). This work was done at the Beijing Genomics Institute (BGI).
qRT-PCR Validation
After total RNA isolation, reverse transcription (RT) was done in a reaction volume of 7.5 µL, which contained 2.08 µL of water, 0.75 µL of 10× reverse transcription buffer, 0.095 µL of RNase inhibitor, 0.075 µL of dNTPs, 0.5 µL of Multiscribe reverse transcriptase, 1.5 µL of miRNA-specific stem-loop RT primer (Applied Biosystems), and 2.5 µL (5 ng) of digested RNA preparations. RT used a Mastercycler Ep Gradient at 16°C for 30 min, 42°C for 30 min, and 85°C for 5 min. Real-time qPCR used a TaqMan MicroRNA assay (Applied Biosystems). Briefly, 4.5 µL of 5∶28 diluted RT product was combined with 5.0 µL of TaqMan gene expression Master Mix and 0.5 µL of Taqman miRNA assay. qPCR used an ABI PRISM 7300 detection system at 95°C for 5 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. All qPCR reactions were performed in triplicate and the raw Ct (threshold cycle) of each sample was the mean value of three Ct values. The data were analyzed by the 2-ΔΔCT method.
CRP and PCT Determinations
Serum CRP levels were determined using scattering turbidimetry (CardioPhase hsCRP, Siemens, Germany) and PCT was determined using an enzyme-linked fluorescence analysis kit (ELFA, VIDAS BRAHMS PCT kit, bioMerieux SA, France). All assays were done according to the manufacturer’s instructions and duplicate measurements were made.
Statistical Methods
Expression levels of selected miRNAs detected by qRT-PCR were normalized to U6 snRNA and presented as the fold-change (2-ΔΔCt) above the normal control: △△Ct = (CtmiRNA-CtU6 snRNA)patients-(CtmiRNA-CtU6 snRNA)controls. Results for normally distributed continuous variables are given as means (±standard errors of the mean) and compared between groups by Student’s t-tests. Results for non-normally distributed continuous variables are summarized as medians (interquartile ranges) and were compared by Mann-Whitney U tests. To evaluate the predictive values of selected miRNAs, ROC curves were generated and the areas under the curves were determined. Multivariable logistic regression analysis was used to screen for independent risk or protective factors for sepsis. Then, an analysis of a combination of these screened variables was done and areas under the curve for the predictive probabilities of these variables were also determined. P-values <0.05 were considered statistically significant. All statistical analyses used SPSS software (version 16.0).
Supporting Information
CONSORT Checklist.
(DOC)
Trial Protocol.
(PDF)
Expression profiles of serum miRNAs in non-surviving sepsis patients (n = 9) detected by Solexa sequencing.
(DOC)
Expression profiles of serum miRNAs in surviving sepsis patients (n = 9) detected by Solexa sequencing.
(DOC)
Comparisons of miRNAs between 9 survivors (S) and 9 non-survivors (D) detected by Solexa sequencing.
(XLS)
Acknowledgments
We wish to thank all of the doctors and nurses in the Department of Respiratory Diseases, the Emergency Department, and the Department of Surgery’s ICU for their support and assistance.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by the Chinese National Science & Technology Pillar Program (2009BAI86B03). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Rivers E, Nguyen B, Havstad S, Ressler J, Muzzin A, et al. Early goal-directed therapy in the treatment of severe sepsis and septic shock. N Engl J Med. 2001;345:1368–1377. doi: 10.1056/NEJMoa010307. [DOI] [PubMed] [Google Scholar]
- 2.Kampmeier TG, Rehberg S, Westphal M, Lange M. Vasopressin in sepsis and septic shock. Minerva Anestesiol. 2010;76:844–850. [PubMed] [Google Scholar]
- 3.Hillas G, Vassilakopoulos T, Plantza P, Rasidakis A, Bakakos P. C-reactive protein and procalcitonin as predictors of survival and septic shock in ventilator-associated pneumonia. Eur Respir J. 2010;35:805–811. doi: 10.1183/09031936.00051309. [DOI] [PubMed] [Google Scholar]
- 4.Chen SJ, Chao TF, Chiang MC, Kuo SC, Chen LY, et al. Prediction of patient outcome from Acinetobacter baumannii bacteremia with Sequential Organ Failure Assessment (SOFA) and Acute Physiology and Chronic Health Evaluation (APACHE) II scores. Intern Med. 2011;50:871–877. doi: 10.2169/internalmedicine.50.4312. [DOI] [PubMed] [Google Scholar]
- 5.Silvestre J, Povoa P, Coelho L, Almeida E, Moreira P, et al. Is C-reactive protein a good prognostic marker in septic patients? Intensive Care Med. 2009;35:909–913. doi: 10.1007/s00134-009-1402-y. [DOI] [PubMed] [Google Scholar]
- 6.Fritz HG, Brandes H, Bredle DL, Bitterlich A, Vollandt R, et al. Post-operative hypoalbuminaemia and procalcitonin elevation for prediction of outcome in cardiopulmonary bypass surgery. Acta Anaesthesiol Scand. 2003;47:1276–1283. doi: 10.1046/j.1399-6576.2003.00239.x. [DOI] [PubMed] [Google Scholar]
- 7.Bellon M, Lepelletier Y, Hermine O, Nicot C. Deregulation of microRNA involved in hematopoiesis and the immune response in HTLV-I adult T-cell leukemia. Blood. 2009;113:4914–4917. doi: 10.1182/blood-2008-11-189845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 9.Chen X, Ba Y, Ma L, Cai X, Yin Y, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 10.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hu Z, Chen X, Zhao Y, Tian T, Jin G, et al. Serum microRNA signatures identified in a genome-wide serum microRNA expression profiling predict survival of non-small-cell lung cancer. J Clin Oncol. 2010;28:1721–1726. doi: 10.1200/JCO.2009.24.9342. [DOI] [PubMed] [Google Scholar]
- 12.Vasilescu C, Rossi S, Shimizu M, Tudor S, Veronese A, et al. MicroRNA fingerprints identify miR-150 as a plasma prognostic marker in patients with sepsis. PLoS One. 2009;4:e7405. doi: 10.1371/journal.pone.0007405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li J, Wan Y, Guo Q, Zou L, Zhang J, et al. Altered microRNA expression profile with miR-146a upregulation in CD4+ T cells from patients with rheumatoid arthritis. Arthritis Res Ther. 2010;12:R81. doi: 10.1186/ar3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schmid R, Baum P, Ittrich C, Fundel-Clemens K, Huber W, et al. Comparison of normalization methods for Illumina BeadChip HumanHT-12 v3. BMC Genomics. 2010;11:349. doi: 10.1186/1471-2164-11-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keller A, Leidinger P, Borries A, Wendschlag A, Wucherpfennig F, et al. miRNAs in lung cancer - studying complex fingerprints in patient's blood cells by microarray experiments. BMC Cancer. 2009;9:353. doi: 10.1186/1471-2407-9-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu R, Zhang C, Hu Z, Li G, Wang C, et al. A five-microRNA signature identified from genome-wide serum microRNA expression profiling serves as a fingerprint for gastric cancer diagnosis. Eur J Cancer. 2011;47:784–791. doi: 10.1016/j.ejca.2010.10.025. [DOI] [PubMed] [Google Scholar]
- 17.Wu Q, Lu Z, Li H, Lu J, Guo L, et al. Next-Generation Sequencing of MicroRNAs for Breast Cancer Detection. J Biomed Biotechnol. 2011;2011:597145. doi: 10.1155/2011/597145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Git A, Dvinge H, Salmon-Divon M, Osborne M, Kutter C, et al. Systematic comparison of microarray profiling, real-time PCR, and next-generation sequencing technologies for measuring differential microRNA expression. RNA. 2010;16:991–1006. doi: 10.1261/rna.1947110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu E, Zhao F, Xu G, Hou H, Zhou L, et al. mirTools: microRNA profiling and discovery based on high-throughput sequencing. Nucleic Acids Res. 2010;38:W392–397. doi: 10.1093/nar/gkq393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang F, Wang J, Liu D, Su Y. Normalizing genes for real-time polymerase chain reaction in epithelial and nonepithelial cells of mouse small intestine. Anal Biochem. 2010;399:211–217. doi: 10.1016/j.ab.2009.12.029. [DOI] [PubMed] [Google Scholar]
- 21.Gui J, Tian Y, Wen X, Zhang W, Zhang P, et al. Serum microRNA characterization identifies miR-885–5p as a potential marker for detecting liver pathologies. Clin Sci (Lond) 2011;120:183–193. doi: 10.1042/CS20100297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang ZX, Bian HB, Wang JR, Cheng ZX, Wang KM, et al. Prognostic significance of serum miRNA-21 expression in human non-small cell lung cancer. J Surg Oncol. 2011. [DOI] [PubMed]
- 23.Wu H, Neilson JR, Kumar P, Manocha M, Shankar P, et al. miRNA profiling of naive, effector and memory CD8 T cells. PLoS One. 2007;2:e1020. doi: 10.1371/journal.pone.0001020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schmidt WM, Spiel AO, Jilma B, Wolzt M, Muller M. In vivo profile of the human leukocyte microRNA response to endotoxemia. Biochem Biophys Res Commun. 2009;380:437–441. doi: 10.1016/j.bbrc.2008.12.190. [DOI] [PubMed] [Google Scholar]
- 25.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 26.Janowska-Wieczorek A, Wysoczynski M, Kijowski J, Marquez-Curtis L, Machalinski B, et al. Microvesicles derived from activated platelets induce metastasis and angiogenesis in lung cancer. Int J Cancer. 2005;113:752–760. doi: 10.1002/ijc.20657. [DOI] [PubMed] [Google Scholar]
- 27.Hunter MP, Ismail N, Zhang X, Aguda BD, Lee EJ, et al. Detection of microRNA expression in human peripheral blood microvesicles. PLoS One. 2008;3:e3694. doi: 10.1371/journal.pone.0003694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li T, Morgan MJ, Choksi S, Zhang Y, Kim YS, et al. MicroRNAs modulate the noncanonical transcription factor NF-kappaB pathway by regulating expression of the kinase IKKalpha during macrophage differentiation. Nat Immunol. 2010;11:799–805. doi: 10.1038/ni.1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen J, Zhang X, Lentz C, Abi-Daoud M, Pare GC, et al. miR-193b Regulates Mcl-1 in Melanoma. Am J Pathol. 2011. [DOI] [PMC free article] [PubMed]
- 30.Chen J, Feilotter HE, Pare GC, Zhang X, Pemberton JG, et al. MicroRNA-193b represses cell proliferation and regulates cyclin D1 in melanoma. Am J Pathol. 2010;176:2520–2529. doi: 10.2353/ajpath.2010.091061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mizuguchi Y, Mishima T, Yokomuro S, Arima Y, Kawahigashi Y, et al. Sequencing and bioinformatics-based analyses of the microRNA transcriptome in hepatitis B-related hepatocellular carcinoma. PLoS One. 2011;6:e15304. doi: 10.1371/journal.pone.0015304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bihrer V, Friedrich-Rust M, Kronenberger B, Forestier N, Haupenthal J, et al. Serum miR-122 as a biomarker of necroinflammation in patients with chronic hepatitis C virus infection. Am J Gastroenterol. 2011;106:1663–1669. doi: 10.1038/ajg.2011.161. [DOI] [PubMed] [Google Scholar]
- 33.Soon PS, Tacon LJ, Gill AJ, Bambach CP, Sywak MS, et al. miR-195 and miR-483–5p Identified as Predictors of Poor Prognosis in Adrenocortical Cancer. Clin Cancer Res. 2009;15:7684–7692. doi: 10.1158/1078-0432.CCR-09-1587. [DOI] [PubMed] [Google Scholar]
- 34.Bone RC, Balk RA, Cerra FB, Dellinger RP, Fein AM, et al. Definitions for sepsis and organ failure and guidelines for the use of innovative therapies in sepsis. The ACCP/SCCM Consensus Conference Committee. American College of Chest Physicians/Society of Critical Care Medicine. Chest. 1992;101:1644–1655. doi: 10.1378/chest.101.6.1644. [DOI] [PubMed] [Google Scholar]
- 35.Hafner M, Landgraf P, Ludwig J, Rice A, Ojo T, et al. Identification of microRNAs and other small regulatory RNAs using cDNA library sequencing. Methods. 2008;44:3–12. doi: 10.1016/j.ymeth.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Calabrese JM, Seila AC, Yeo GW, Sharp PA. RNA sequence analysis defines Dicer's role in mouse embryonic stem cells. Proc Natl Acad Sci U S A. 2007;104:18097–18102. doi: 10.1073/pnas.0709193104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ruby JG, Jan C, Player C, Axtell MJ, Lee W, et al. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell. 2006;127:1193–1207. doi: 10.1016/j.cell.2006.10.040. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
CONSORT Checklist.
(DOC)
Trial Protocol.
(PDF)
Expression profiles of serum miRNAs in non-surviving sepsis patients (n = 9) detected by Solexa sequencing.
(DOC)
Expression profiles of serum miRNAs in surviving sepsis patients (n = 9) detected by Solexa sequencing.
(DOC)
Comparisons of miRNAs between 9 survivors (S) and 9 non-survivors (D) detected by Solexa sequencing.
(XLS)