Abstract
BACKGROUND
Interleukin-12 (IL-12) and related cytokines induce activation and differentiation of T cells. Our aim was to investigate the associations between genetic differences in IL-12-family cytokines and the pathogenesis of chlamydial disease.
METHODS
The final study population consisted of 100 women with Chlamydia trachomatis-induced tubal factor infertility (TFI) and 125 pregnant women as controls. Three single nucleotide polymorphisms (SNPs) of IL12A and seven SNPs of IL12B genes were determined from isolated DNA using the Sequenom system with matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry.
RESULTS
We found that the IL12B SNP rs3212227 was associated with both susceptibility and severity of TFI. The minor allele C was rare and only one CC homozygote was found among the controls. AC heterozygotes were more common among TFI cases than among controls (P = 0.009) and were associated with increased risk of TFI [odds ratios (OR) = 2.44, 95% confidence intervals (CI) = 1.23–4.87]. Carrying the minor allele C was also associated with disease severity (P for trend = 0.008) and moderate (OR = 2.51, 95% CI = 1.06–5.95) and severe tubal damage (OR = 2.73, 95% CI = 1.15–6.52).
CONCLUSIONS
The results suggest that variation in the IL12B gene partly explains inter-individual differences in disease susceptibility and severity.
Keywords: IL-12, disease susceptibility, disease severity, chlamydial infection, C. trachomatis
Introduction
Persistent or repeat Chlamydia trachomatis infection and consequent inflammation can damage Fallopian tubes and are associated with reproductive disorders, including pelvic inflammatory disease, infertility and ectopic pregnancy (Paavonen and Eggert-Kruse, 1999). Host immunogenetic factors may partly explain why some women are more susceptible to the development of tissue damage during infection while most infected women do not develop clinical complications (Morré et al., 2009; Öhman et al., 2009).
Interferon-gamma (IFN-γ) is critical in the activation of cell-mediated immune responses against viral and intracellular infections (Boehm et al., 1997), and also against C. trachomatis, an obligatorily intracellular pathogen (Rottenberg et al., 2002). In a previous study, we found an association between the IFNG +874 single nucleotide polymorphism (SNP) and the intensity of the C. trachomatis-specific cell-mediated immune response, although the differences between genotype groups were small (Öhman et al., 2011). However, associations between the studied SNP and disease susceptibility or severity were not found. Despite this, the genotype associated with a lower lymphocyte proliferation (LP) response was more common among infertile women with severe tubal damage than among cases with minor or moderate tubal damage (Öhman et al., 2009). These results suggest that other factors that regulate IFN-γ production, such as interleukin-12 (IL-12), may be of interest.
Production of IFN-γ from NK cells and lymphocytes is induced by IL-12 which acts as an antagonist to IL-10 (Boehm et al., 1997). IL-12 and IL-10 are both mainly produced by monocytes, macrophages and dendritic cells, which are the first leukocytes to interact with pathogens. Through antigen presentation and cytokine expression, they initiate and coordinate the development of antigen-specific cell-mediated immune response (O'Garra and Murphy, 2009). Monocytes have an important role in the regulation of the cell-mediated immune response during C. trachomatis infection by secreting IL-10. SNPs in the promoter region of the IL10 gene are associated with inter-individual variation in IL-10 expression, intensity of the cell-mediated response and susceptibility to severe tubal damage (Öhman et al., 2006, 2009).
Endocervical IL-12 levels are increased in women infected with C. trachomatis (Wang et al., 2005). We have previously found an association between the intronic IL12B SNP rs2853694 and the intensity of the C. trachomatis-specific cell-mediated immune response (Öhman et al., 2010). This suggests that the cytokine has an active role in the immune cascade in chlamydial infection and that polymorphisms within or flanking its coding gene can affect the outcome. IL-12 is a heterodimeric cytokine, encoded by two separate genes; i.e. IL12A which encodes subunit p35 and IL12B which encodes subunit p40. Subunit p40 is a common building block of IL-12 and IL-23 cytokines (Trinchieri, 2003). SNPs of both IL12A and IL12B genes have been identified and some of them have been associated with infectious diseases. In particular, the SNP marker rs3212227 of IL12B has been linked with recurrent chlamydial infections (Geisler et al., 2010) and with susceptibility to psoriasis (Cargill et al., 2007; Nair et al., 2008).
The role of variation in IL-12 coding genes in the pathogenesis of C. trachomatis-induced tubal damage is not known. We therefore selected three SNPs of the IL12A gene and seven of the IL12B gene to study the associations between these polymorphisms and susceptibility to, and the severity of, C. trachomatis-induced tubal damage.
Materials and Methods
The study population
The study population consisted of 163 women (median age 33, range: 23–40 years) who attended the Infertility Clinic of the Department of Obstetrics and Gynecology, Helsinki University Hospital during 1990–2005 and had accurate laparoscopic evaluation of tubal factor infertility (TFI). A history of past C. trachomatis infection was analysed in these cases by assessing both cell-mediated immunity and antibody responses to C. trachomatis elementary bodies (EB) and to CHSP60 antigens. Serum for antibody analysis was available from all 163 TFI cases and cell samples for LP testing were available from 72 of these cases. However, 26 TFI cases did not have an immune marker of past C. trachomatis infection and were excluded. The control group consisted of 179 pregnant women (median age 36, range: 27–44) from the Helsinki area, whose samples were provided by the Finnish Maternity Cohort (FMC) serum bank collected in 2006.
Due to a low quantity of DNA for some of the samples, the genotyping failures accumulated to some subjects. The subjects who had three or more missing SNP genotypes (54 controls and 37 TFI cases) were excluded from further analysis. The study population for genetic association analysis therefore contained 125 controls and 100 TFI cases.
The TFI cases were grouped into three categories according to the severity of tubal damage, following the classification of Hull and Rutherford (Rutherford and Jenkins, 2002). Minor damage with proximal or distal tubal occlusion but no tubal distension and at most flimsy adhesions was found in 24 of the 100 cases with TFI, moderate damage with unilateral sactosalpinx or moderate tubal adhesions was found in 41 cases and severe damage with bilateral sactosalpinx or extensive adhesions was found in 35 cases.
Genotyping of cytokine polymorphism
Leukocytes, blood clots and serum were used as DNA source material. The DNA was extracted from leukocytes by using a guanidine hydrochloride method. A MagNA Pure LC instrument (Roche Diagnostics) was used for DNA extraction from blood clots and serum. For blood clots, a DNA Isolation Kit—Large Volume (Roche Diagnostics) was used, following the DNA LV Cells Protocol, and for serum a Total Nucleic Acid Isolation Kit—Large Volume (Roche Diagnostics) was used, following the Total NA/LV Serum Plasma protocol. The quantity of DNA in serum samples was low and contributed to genotyping failure in some samples.
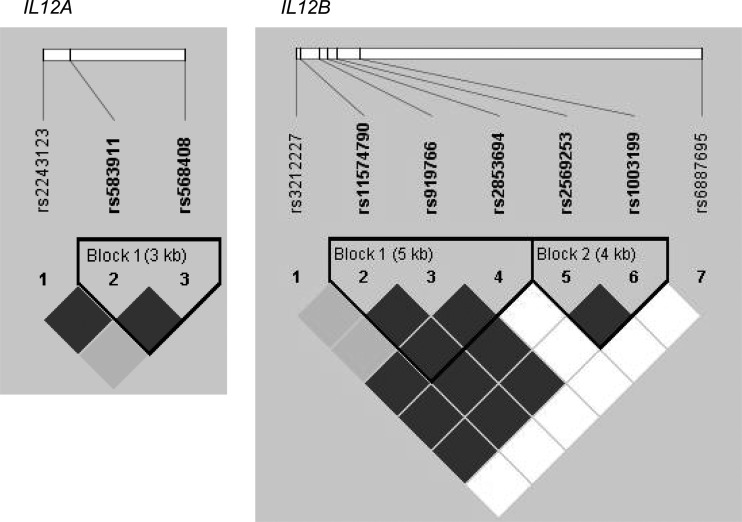
IL12A and IL12B SNPs were determined using the Sequenom system with matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry. Three SNPs of IL12A (rs2243123, rs583911, rs568408) and seven of IL12B (rs3212227, rs1003199, rs11574790, rs919766, rs2569253, rs2853694 and rs6887695) genes were studied (Table I and Fig. 1).
Table I.
The sequences for the PCR primers, the extension primers and the PCR product length for the SNPs used are shown.
| SNP ID | Second PCR primer | First PCR primer | Amplicon (bp) | Extension primer |
|---|---|---|---|---|
| rs2243123 | ACGTTGGATGAGTCTTTCTCATGCTGCTCC | ACGTTGGATGGGTGAATCCAGTGTAAGCAG | 100 | ctCTGCTCCCTCTGGAC |
| rs583911 | ACGTTGGATGAGCTTGTCTTAAGGGTTTGC | ACGTTGGATGCAAGTATAACTTCTAAAGGG | 100 | GCATGTTTGTTATATCCATCA |
| rs568408 | ACGTTGGATGGTCAAAAATACTTGATCAG | ACGTTGGATGGGATTAAGAACTAGGGAGGG | 97 | GTCAAAAATACTTGATCAGAGGTAT |
| rs3212227 | ACGTTGGATGGGATCACAATGATATCTTTGC | ACGTTGGATGCTGATTGTTTCAATGAGC | 85 | cccTATCTTTGCTGTATTTGTATAGTT |
| rs1003199 | ACGTTGGATGGCCTAGAATGACTTTCTTGAC | ACGTTGGATGAACCTAGAGGAAGAGGTAAG | 99 | CCATGATATATAAAACACAGATACC |
| rs11574790 | ACGTTGGATGCAGCTTACCCTGTGACTATG | ACGTTGGATGCCGTGAAGACTCTATCTTTC | 98 | GCCAAGGGGTCTTCA |
| rs919766 | ACGTTGGATGGCTACAATCACTAGGAACTC | ACGTTGGATGGGGTCAGAAGAGCTGAAGT | 97 | AGGAACTCTCTCCCCAA |
| rs2569253 | ACGTTGGATGAGTTTCTCTGTACAGTTGGC | ACGTTGGATGCTGCCACACAGTAAATTCGG | 94 | tGTTGGCTGACTCCTC |
| rs2853694 | ACGTTGGATGCAAGGTGCAATTTCAGCAAG | ACGTTGGATGTTCCTGAAGCCTCATAGCAC | 100 | TTGTAGCTTTGAATTCTCC |
| rs6887695 | ACGTTGGATGGTTTGAGAGAAGCAGTGTAG | ACGTTGGATGGTCACAAGCGTAGTAAATGG | 98 | GCAGTGTAGTGTAGTGGT |
Figure 1.
LD maps for the markers genotyped in the IL12A and IL12B genes. Black squares represent high LD; white squares represent low LD. Locations of SNPs are shown.
A candidate gene approach was used and the selected SNPs were chosen because of their potential role effect on IL-12 in antichlamydial immunity and chlamydial pathogenesis. The study included particularly those SNPs for which evidence of functionality or an association with other diseases existed, and a few that were involved in more speculative areas.
Chlamydia trachomatis-specific immune responses
A history of previous C. trachomatis infection was evaluated by assessing C. trachomatis-specific humoral and cell-mediated immunity, as described earlier (Tiitinen et al., 2006). Chlamydia trachomatis- and chlamydial heat shock protein 60 (CHSP60)-specific IgG serum antibodies were assayed by ELISA kits (Medac Diagnostika, Hamburg, Germany). The antibody results were recorded as mean absorbance of duplicate samples at 450 nm. The threshold for a positive antibody level [mean optical density (OD) value of negative controls 0.350] was OD > 0.4. Peripheral blood mononuclear cells (PBMCs) were isolated and cell-mediated immunity was assessed by measuring LP responses to C. trachomatis EB strains E and F and to CHSP60 (kindly provided by Professor Richard Morrison), as previously described (Kinnunen et al., 2002). The LP responses were measured as counts per minute (cpm) of incorporated [3H]-methyl thymidine, using a liquid scintillation counter (Wallac, Turku, Finland), and the results were expressed as median stimulation indices (SI = median cpm in the presence of antigen divided by median cpm in its absence) of triplicate cultures. The viability and reactivity of the cultured PBMCs were controlled in each experiment by requiring SI of >10 in response to the control Pokeweed mitogen.
Statistical analyses
A χ2 test for trend was used to test genotype distribution trends. Genotypes conferring susceptibility are presented in terms of odds ratios (ORs) with 95% confidence intervals (95% CIs) (SPSS 18.0 software). The two-sample test of proportions was performed to compare genotype frequencies between cases and controls (Stata 5.0 statistical software). Haploview 4.2 software (Barrett et al., 2005) was used to analyse the linkage disequilibrium (LD) between SNPs in IL12A and IL12B genes.
Sample size calculations in Stata and Epi Info suggested, for example, that a study of 100 cases and controls would have 80% power to detect a marker which is present in 30% of cases and 10% of controls, with 99% confidence, i.e. allowing for five multiple comparisons. If the marker were present in 40% of cases, this could be detected with 80% power and 99.9% confidence, i.e. allowing for 50 independent comparisons. For a stronger effect than this, even more comparisons are allowed, for example, a marker present in 50% of cases gives adequate power, even if 500 independent comparisons were made. So we concluded that the study had adequate power to detect polymorphisms that were strongly associated with disease, even allowing for multiple comparisons in a conservative way.
Results
Genotyping
Overall genotyping performance in the final study population was 83.6–99.6% depending on the SNP.
In the control group, all 10 studied polymorphisms were in Hardy–Weinberg equilibrium (HWE). Two of the studied IL12B SNPs, rs11574790 (P = 0.001) and rs919766 (P = 0.004), which exhibited strong LD (Fig. 1), were not in HWE in the TFI cases.
Immune markers
Serum for antibody analysis was available from all 163 TFI cases and from 179 controls. Cells for LP analysis were available from 72 TFI cases. Cells from controls were not available. The immune responses are presented in Table II. We found that 26 of the 163 (16%) TFI cases were not reactive to any of the studied markers and these were excluded from the genetic susceptibility analysis because of lack of C. trachomatis attributable evidence.
Table II.
Humoral and cell-mediated reactivity to C. trachomatis-specific antigens in cases of TFI and control women.
| Controls, n (%) | TFI cases, n (%) | OR (95% CI) | |
|---|---|---|---|
| IgG C. trachomatis | |||
| Negative | 145 (86.8) | 69 (42.3) | 1 |
| Positive | 22 (13.2) | 94 (57.7) | 8.98 (5.20–15.49) |
| IgG CHSP60 | |||
| Negative | 135 (80.8) | 73 (44.8) | 1 |
| Positive | 32 (19.2) | 90 (55.2) | 5.20 (3.17–8.52) |
| SI C. trachomatis E | |||
| Negative | N/A | 19 (26.4) | N/A |
| Positive | 53 (73.6) | ||
| SI C. trachomatis F | |||
| Negative | N/A | 16 (30.8) | N/A |
| Positive | 36 (69.2) | ||
| SI CHSP60 | |||
| Positive | N/A | 39 (54.2) | N/A |
| Negative | 33 (45.8) | ||
The risk of TFI among C. trachomatis- and CHSP60-IgG-positives was estimated using logistic regression. ORs with 95% CIs are presented.
SI, stimulation index.
The prevalence of C. trachomatis-specific IgG antibodies was 4.4-fold higher and the prevalence of CHSP60-specific IgG antibodies was 2.9-fold higher in TFI cases than in the controls. Presence of chlamydial antibodies was associated with an increased risk for TFI (Table II).
Genetic associations with disease susceptibility
Genotype distributions in the TFI cases and controls were compared to analyse the possible risk factors of disease susceptibility (Table III). For the IL12B gene, a divergent genotype distribution was found in connection with the SNP rs3212227. The minor allele C was rare; among TFI cases no CC homozygotes were found. The AC genotype was more common in the TFI women than in controls (P = 0.009) and was associated with an increased risk for TFI (OR = 2.44, 95% CI = 1.23–4.87).
Table III.
Distribution of cytokine genotypes in cases with chlamydial TFI and controls.
| Gene, SNP |
n (%) |
P-test of proportion | χ2 for trend (P-value) | OR (95% CI) | |
|---|---|---|---|---|---|
| TFI women | Controls | ||||
| IL12A, rs2243123 | |||||
| TT | 53 (55.2) | 64 (54.2) | 0.887 | 0.868 | Reference |
| CT | 35 (36.5) | 47 (39.8) | 0.614 | 0.90 (0.51–1.59) | |
| CC | 8 (8.3) | 7 (5.9) | 0.494 | 1.38 (0.47–4.06) | |
| IL12A, rs583911 | |||||
| AA | 33 (38.8) | 34 (32.1) | 0.331 | 0.468 | Reference |
| AG | 33 (38.8) | 47 (44.3) | 0.443 | 0.72 (0.38–1.39) | |
| GG | 19 (22.4) | 25 (23.6) | 0.841 | 0.78 (0.36–1.68) | |
| IL12A, rs568408 | |||||
| GG | 73 (76.8) | 89 (73.6) | 0.58 | 0.664 | Reference |
| AG | 18 (18.9) | 27 (22.3) | 0.545 | 0.81 (0.42–1.59) | |
| AA | 4 (4.2) | 5 (4.1) | 0.977 | 0.98 (0.25–3.77) | |
| IL12B, rs3212227 | |||||
| AA | 66 (69.5) | 89 (84.0) | 0.015 | 0.028 | Reference |
| AC | 29 (30.5) | 16 (15.1) | 0.009 | 2.44 (1.23–4.87) | |
| CC | 0 (0) | 1 (0.9) | 0.343 | ||
| IL12B rs11574790 | |||||
| CC | 86 (86.9) | 106 (84.8) | 0.66 | 0.858 | Reference |
| CT | 10 (10.1) | 19 (15.2) | 0.259 | 0.65 (0.29–1.47) | |
| TT | 3 (3.0) | 0 (0) | 0.05 | ||
| IL12B rs919766 | |||||
| AA | 85 (85.9) | 106 (84.8) | 0.824 | 0.716 | Reference |
| AC | 11 (11.1) | 19 (15.2) | 0.372 | 0.72 (0.33–1.60) | |
| CC | 3 (3.0) | 0 (0) | 0.05 | ||
| IL12B rs2853694 | |||||
| CC | 31 (32.6) | 36 (31.0) | 0.804 | 0.782 | Reference |
| AC | 41 (43.2) | 57 (49.1) | 0.386 | 0.84 (0.45–1.56) | |
| AA | 23 (24.2) | 23 (19.8) | 0.443 | 1.16 (0.55–2.46) | |
| IL12B rs2569253 | |||||
| CC | 28 (34.1) | 48 (45.3) | 0.123 | 0.095 | Reference |
| CT | 41 (50.0) | 47 (44.3) | 0.441 | 1.50 (0.80–2.80) | |
| TT | 13 (15.9) | 11 (10.4) | 0.265 | 2.03 (0.80–5.13) | |
| IL12B rs1003199 | |||||
| TT | 31 (31.6) | 48 (41.4) | 0.141 | 0.315 | Reference |
| CT | 52 (53.1) | 50 (43.1) | 0.146 | 1.61 (0.89–2.92) | |
| CC | 15 (15.3) | 18 (15.5) | 0.966 | 1.29 (0.57–2.93) | |
| IL12B rs6887695 | |||||
| GG | 58 (58.6) | 70 (56.5) | 0.749 | 0.856 | Reference |
| CG | 34 (34.3) | 46 (37.1) | 0.67 | 0.89 (0.51–1.57) | |
| CC | 7 (7.1) | 8 (6.5) | 0.855 | 1.06 (0.36–3.09) | |
The two-sample test of proportions was performed to compare genotype frequencies in cases and controls. A χ2 test for trend was used to analyse a possible trend between disease and genotype. ORs with 95% CIs are shown.
Genetic associations with disease severity
The relationships between IL12A and IL12B genotypes and the degree of tubal damage were studied. Only the IL12B SNP rs3212227 was significantly associated with disease severity (P for trend = 0.008). Data on SNP rs3212227 are shown in Table IV. The AC and CC genotypes were combined because of the small number of CC homozygotes. Subjects carrying the C allele had an increased risk of moderate (OR = 2.51, 95% CI = 1.06–5.95) and severe tubal damage (OR = 2.73, 95% CI = 1.15–6.52).
Table IV.
Comparison of genotype distribution and risk estimates of different degrees of tubal damage.
| IL12B, rs3212227 | Controls, n (%) | Severity of tubal damage (Hull and Rutherford) |
χ2 for trend (P–value) | Minor damage | Moderate damage | Severe damage | ||
|---|---|---|---|---|---|---|---|---|
| Minor, n (%) | Moderate, n (%) | Severe, n (%) | OR (95% CI) | OR (95% CI) | OR (95% CI) | |||
| AA | 89 (84.0) | 18 (78.3) | 25 (67.6) | 23 (65.7) | 0.008 | 1 | 1 | 1 |
| AC or CC | 17 (16.0) | 5 (21.7) | 12 (32.4) | 12 (34.3) | 1.45 (0.48–4.45) | 2.51 (1.06–5.95) | 2.73 (1.15–6.52) | |
A χ2 test for trend was used to analyse the relationship between genotype and disease severity. ORs with 95% CIs are shown.
Discussion
In the present study, we investigated the role of IL12A and IL12B polymorphisms in the pathogenesis of C. trachomatis-induced TFI in a unique and well-characterised population that included accurate laparoscopic evaluation of the cases and uniform classification of the degree of tubal damage. This setting enabled us to study the association between SNPs and the severity of disease manifestations, in addition to disease susceptibility and gave an excellent opportunity to study the role of genes in pathogenesis.
We found that one of the studied SNPs, rs3212227 of IL12B, was associated with TFI and the severity of tubal damage. This SNP is recognized as a susceptibility correlate in many inflammatory conditions from infections to autoimmune diseases (Cargill et al., 2007; Nair et al., 2008; McGovern et al., 2009; Phawong et al., 2010; Wang et al., 2010). The other SNPs studied were not associated with susceptibility to TFI or severity of tubal damage.
The immune response to genital C. trachomatis infection is complex (Loomis and Starnbach, 2002). It seems that a delicate balance between pro- and anti-inflammatory cytokines is required for infection clearance and at the same time to avoid immune-mediated pathology (Debattista et al., 2003). Chlamydia trachomatis infection increases endocervical IL-12 production in vivo (Wang et al., 2005) indicating that IL-12 has an active role in the immune cascade provoked by C. trachomatis. The SNP rs3212227 has also been associated with recurrent C. trachomatis infections (Geisler et al., 2010). Particularly, the minor allele C, which in our data increased the risk and severity of TFI, was associated with recurrent C. trachomatis infections. As repeated infections and continuous inflammation increase the risk of tubal damage, our findings are in agreement with those by Geisler et al. In contrast, for inflammatory autoimmune illnesses, such as psoriasis and Crohn's disease, the common genotype AA has been linked to disease susceptibility (Cargill et al., 2007; Nair et al., 2008; McGovern et al., 2009).
The results of this study suggest that IL12B is involved in the disease process, but more data are needed to investigate the mechanism behind the genotype association. Differences in IL12B may be reflected by individual variation in levels of IL-12 and IL-23 cytokines, which are composed of a common IL-12p40 and distinct IL-12p35 or IL-23p19 subunits, respectively. The IL-12 effect is mediated through the IFN-γ pathway necessary to defend against C. trachomatis. The effects of IL-23 are less well known, but it seems to play a key role in the induction of Th17 cells. These cells induce production of proinflammatory cytokines and might be responsible for the inflammation-driven pathogenesis instead of the Th1 subset (Goriely et al., 2009). Besides effects mediated through IL-12 and IL-23, it is also possible that IL-12p40 has a biological activity on its own, as it has been reported that IL-12p40 can act as an antagonist of IL-12 in vitro (Zhang and Wang, 2008) and thus mediate an anti-inflammatory effect. This hypothesis is also supported by Müller-Berghaus et al. (2004) who showed that elevated levels of IL-12p40 were associated with lower levels of IL-12p70.
Although the mechanism behind the connection is still unsolved, the disease associations and the literature (Cargill et al., 2007; Nair et al., 2008; McGovern et al., 2009; Geisler et al., 2010) suggest that the IL12B rs3212227 allele C is linked with a weaker, and allele A with a stronger, inflammatory response. Subjects with the AA genotype seem to cope with C. trachomatis infection better than subjects who carry the allele C. This might be a result of more efficient Chlamydia clearance and also protective immunity.
In conclusion, we showed evidence here that a transcribed marker in the IL12B gene is associated with susceptibility to C. trachomatis-induced TFI and with severe disease manifestation. The results may explain some individual variations in the manifestations of C. trachomatis disease. Further studies dealing with the IL12B polymorphism and the immune response are still needed to reveal the mechanism behind the disease association and to increase understanding of the pathogenesis of the disease, as this will be needed in the design of new therapeutic strategies.
Authors' roles
The study was designed by H.Ö. and H.-M.S. The study population was collected and laparoscopic evaluations were performed by A.T., M.H. and J.P. The selection of SNPs and design of their analysis was done by A.N., R.B., J.R. and L.-L.J. The SNPs were genotyped by L.-L.J. and H.Ö. The data were analysed by H.Ö. The manuscript was drafted by H.Ö., H.-M.S. and R.B. All authors contributed to the final version of the manuscript. The final version was seen and approved by all authors.
Funding
This study was supported by EpiGenChlamydia Consortium (EU FP6: LSHG-CT-2007-037637) and the Wellcome Trust grant 075491/Z/04 (J.R and L.L.J).
Conflict of interest
None declared.
References
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Boehm U, Klamp T, Groot M, Howard JC. Cellular responses to interferon-gamma. Annu Rev Immunol. 1997;15:749–795. doi: 10.1146/annurev.immunol.15.1.749. [DOI] [PubMed] [Google Scholar]
- Cargill M, Schrodi SJ, Chang M, Garcia VE, Brandon R, Callis KP, Matsunami N, Ardlie KG, Civello D, Catanese JJ, et al. A large-scale genetic association study confirms IL12B and leads to the identification of IL23R as psoriasis-risk genes. Am J Hum Genet. 2007;80:273–290. doi: 10.1086/511051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debattista J, Timms P, Allan J. Immunopathogenesis of Chlamydia trachomatis infections in women. Fertil Steril. 2003;79:1273–1287. doi: 10.1016/s0015-0282(03)00396-0. [DOI] [PubMed] [Google Scholar]
- Geisler WM, Jiang B, Song W, Shrestha S, Tang J. Immune response gene variants as correlates of genital Chlamydia in adolescents. Proceedings of the Twelfth International Symposium on Human Chlamydial Infections, Hof bei Salzburg, Austria; San Francisco: International Chlamydia Symposium; 2010. pp. 181–184. [Google Scholar]
- Goriely S, Cavoy R, Goldman M. Interleukin-12 family members and type I interferons in Th17-mediated inflammatory disorders. Allergy. 2009;64:702–709. doi: 10.1111/j.1398-9995.2009.02039.x. [DOI] [PubMed] [Google Scholar]
- Kinnunen AH, Surcel HM, Lehtinen M, Karhukorpi J, Tiitinen A, Halttunen M, Bloigu A, Morrison RP, Karttunen R, Paavonen J. HLA DQ alleles and interleukin-10 polymorphism associated with Chlamydia trachomatis-related tubal factor infertility: a case–control study. Hum Reprod. 2002;17:2073–2078. doi: 10.1093/humrep/17.8.2073. [DOI] [PubMed] [Google Scholar]
- Loomis WP, Starnbach MN. T cell responses to Chlamydia trachomatis. Curr Opin Microbiol. 2002;5:87–91. doi: 10.1016/s1369-5274(02)00291-6. [DOI] [PubMed] [Google Scholar]
- McGovern DP, Rotter JI, Mei L, Haritunians T, Landers C, Derkowski C, Dutridge D, Dubinsky M, Ippoliti A, Vasiliauskas E, et al. Genetic epistasis of IL23/IL17 pathway genes in Crohn's disease. Inflamm Bowel Dis. 2009;15:883–889. doi: 10.1002/ibd.20855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morré SA, Karimi O, Ouburg S. Chlamydia trachomatis: identification of susceptibility markers for ocular and sexually transmitted infection by immunogenetics. FEMS Immunol Med Microbiol. 2009;55:140–153. doi: 10.1111/j.1574-695X.2009.00536.x. [DOI] [PubMed] [Google Scholar]
- Müller-Berghaus J, Kern K, Paschen A, Nguyen XD, Kluter H, Morahan G, Schadendorf D. Deficient IL-12p70 secretion by dendritic cells based on IL12B promoter genotype. Genes Immun. 2004;5:431–434. doi: 10.1038/sj.gene.6364102. [DOI] [PubMed] [Google Scholar]
- Nair RP, Ruether A, Stuart PE, Jenisch S, Tejasvi T, Hiremagalore R, Schreiber S, Kabelitz D, Lim HW, Voorhees JJ, et al. Polymorphisms of the IL12B and IL23R genes are associated with psoriasis. J Invest Dermatol. 2008;128:1653–1661. doi: 10.1038/sj.jid.5701255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Garra A, Murphy KM. From IL-10 to IL-12: how pathogens and their products stimulate APCs to induce T(H)1 development. Nat Immunol. 2009;10:929–932. doi: 10.1038/ni0909-929. [DOI] [PubMed] [Google Scholar]
- Öhman H, Tiitinen A, Halttunen M, Birkelund S, Christiansen G, Koskela P, Lehtinen M, Paavonen J, Surcel HM. IL-10 polymorphism and cell-mediated immune response to Chlamydia trachomatis. Genes Immun. 2006;7:243–249. doi: 10.1038/sj.gene.6364293. [DOI] [PubMed] [Google Scholar]
- Öhman H, Tiitinen A, Halttunen M, Lehtinen M, Paavonen J, Surcel HM. Cytokine polymorphisms and severity of tubal damage in women with Chlamydia-associated infertility. J Infect Dis. 2009;199:1353–1359. doi: 10.1086/597620. [DOI] [PubMed] [Google Scholar]
- Öhman H, Bailey R, Natividad A, Tiitinen A, Halttunen M, Paavonen J, Surcel H-M. IL-12p40 polymorphisms and C. trachomatis-specific lymphocyte proliferative responses. Proceedings of the Twelfth International Symposium on Human Chlamydial Infections, Hof bei Salzburg, Austria; San Francisco: International Chlamydia Symposium; 2010. pp. 169–172. [Google Scholar]
- Öhman H, Tiitinen A, Halttunen M, Paavonen J, Surcel HM. Cytokine gene polymorphism and Chlamydia trachomatis-specific immune responses. Hum Immunol. 2011;72:278–282. doi: 10.1016/j.humimm.2010.12.012. [DOI] [PubMed] [Google Scholar]
- Paavonen J, Eggert-Kruse W. Chlamydia trachomatis: impact on human reproduction. Hum Reprod Update. 1999;5:433–447. doi: 10.1093/humupd/5.5.433. [DOI] [PubMed] [Google Scholar]
- Phawong C, Ouma C, Tangteerawatana P, Thongshoob J, Were T, Mahakunkijcharoen Y, Wattanasirichaigoon D, Perkins DJ, Khusmith S. Haplotypes of IL12B promoter polymorphisms condition susceptibility to severe malaria and functional changes in cytokine levels in Thai adults. Immunogenetics. 2010;62:345–356. doi: 10.1007/s00251-010-0439-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottenberg ME, Gigliotti-Rothfuchs A, Wigzell H. The role of IFN-gamma in the outcome of chlamydial infection. Curr Opin Immunol. 2002;14:444–451. doi: 10.1016/s0952-7915(02)00361-8. [DOI] [PubMed] [Google Scholar]
- Rutherford AJ, Jenkins JM. Hull and Rutherford classification of infertility. Hum Fertil (Camb) 2002;5:S41–45. doi: 10.1080/1464727022000199911. [DOI] [PubMed] [Google Scholar]
- Tiitinen A, Surcel HM, Halttunen M, Birkelund S, Bloigu A, Christiansen G, Koskela P, Morrison SG, Morrison RP, Paavonen J. Chlamydia trachomatis and chlamydial heat shock protein 60-specific antibody and cell-mediated responses predict tubal factor infertility. Hum Reprod. 2006;21:1533–1538. doi: 10.1093/humrep/del014. [DOI] [PubMed] [Google Scholar]
- Trinchieri G. Interleukin-12 and the regulation of innate resistance and adaptive immunity. Nat Rev Immunol. 2003;3:133–146. doi: 10.1038/nri1001. [DOI] [PubMed] [Google Scholar]
- Wang C, Tang J, Crowley-Nowick PA, Wilson CM, Kaslow RA, Geisler WM. Interleukin (IL)-2 and IL-12 responses to Chlamydia trachomatis infection in adolescents. Clin Exp Immunol. 2005;142:548–554. doi: 10.1111/j.1365-2249.2005.02946.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Tang S, Shen H. Association of genetic polymorphisms in the IL12-IFNG pathway with susceptibility to and prognosis of pulmonary tuberculosis in a Chinese population. Eur J Clin Microbiol Infect Dis. 2010;29:1291–1295. doi: 10.1007/s10096-010-0985-0. [DOI] [PubMed] [Google Scholar]
- Zhang S, Wang Q. Factors determining the formation and release of bioactive IL-12: regulatory mechanisms for IL-12p70 synthesis and inhibition. Biochem Biophys Res Commun. 2008;372:509–512. doi: 10.1016/j.bbrc.2008.05.081. [DOI] [PubMed] [Google Scholar]