Abstract
Gap junction channels provide a conduit for communication between neighboring cells. The function of gap junction channels is regulated by posttranslational modifications of connexins, the proteins that comprise these channels. Ubiquitination of connexins has increasingly been viewed as one mechanism by which cells regulate the level of connexins present in cells, as well as the corresponding intercellular communication. Here we review the current knowledge of connexin ubiquitination and the effects this may have on gap junctional communication.
Keywords: connexin, ubiquitination, degradation, proteasome, lysosome
INTRODUCTION
Gap junctions are comprised of connexin hexamers, or connexons, that dock across the intercellular space with connexons on adjacent cells [1]. These plasma membrane channels mediate direct cell to cell communication, allowing the transfer of molecules less than 1000 daltons, such as small metabolites and secondary messengers. Gap junctional regulation and therefore, intercellular communication is critical for the proper maintenance of normal cell and tissue function [2–4].
Connexins are four-pass transmembrane proteins, with two extracellular loops, an intracellular loop, and N- and C-terminal cytoplasmic regions. Particularly for transmembrane proteins, connexins have remarkably short half-lives, ranging from 1.5 to 5 hours [5–9]. Since the amount of connexin protein correlates directly to the level of gap junctional intercellular communication (GJIC) 1, an understanding of how connexin levels are regulated is critical. There has been a great deal of work contributing to the current understanding of the processes that regulate GJIC and connexin turnover.
Connexins are co-translationally inserted into the endoplasmic reticulum (ER), then trafficked through the Golgi. At some point during the secretory pathway, connexins oligomerize into hexameric connexons. Depending on the connexin isoform, oligomerization has been observed in different locations from the ER, to the ER-Golgi intermediate compartment, to the trans-Golgi network [10–15]. These connexons, that will comprise one-half of the complete gap junction channel, are transported to the plasma membrane to form gap junctions and large accretions of gap junctions, known as gap junction plaques. Recently, undocked connexons at the plasma membrane have been demonstrated to have channel activity as well. These hemichannels can be opened under conditions such as mechanical shear stress [16, 17], membrane depolarization [18], and changes in ionic concentrations [19, 20], and can regulate the passage of ions and metabolites [16, 17, 21, 22]. Hemichannels may also have important roles in tissue remodeling [17, 23] and in cell death [24–27]. From the plasma membrane, connexins are internalized as double membrane annular gap junctions, or connexosomes, or possibly, as undocked individual connexons, then degraded [28]. It is clear that connexins undergo degradation through both the lysosomal and proteasomal degradation pathways [9, 29–37], with an additional pathway of autophagy recently revealed [38, 39] (Figure 1). However, the precise mechanisms that regulate connexin trafficking to and from the plasma membrane and subsequently through to degradation are still being studied and debated. Posttranslational modifications of connexins are thought to contribute to the regulation of connexin function and trafficking. Connexin phosphorylation, which is known to regulate gap junction channel closure and connexon membrane stability (reviewed in [40–42]), has been studied intensely. More recently, other modifications have been identified such as hydroxylation, methylation, and acetylation [43–45], the last of which has been reported to affect connexin43 (Cx43) localization in the heart [43]. This review will focus on a different type of posttranslational modification that may also have a significant impact on the connexin life cycle. This modification, which has been increasingly studied by a number of groups, is the ubiquitination of connexins, and we will discuss what effect this modification has on connexin trafficking and turnover (relevant studies summarized in Table 1).
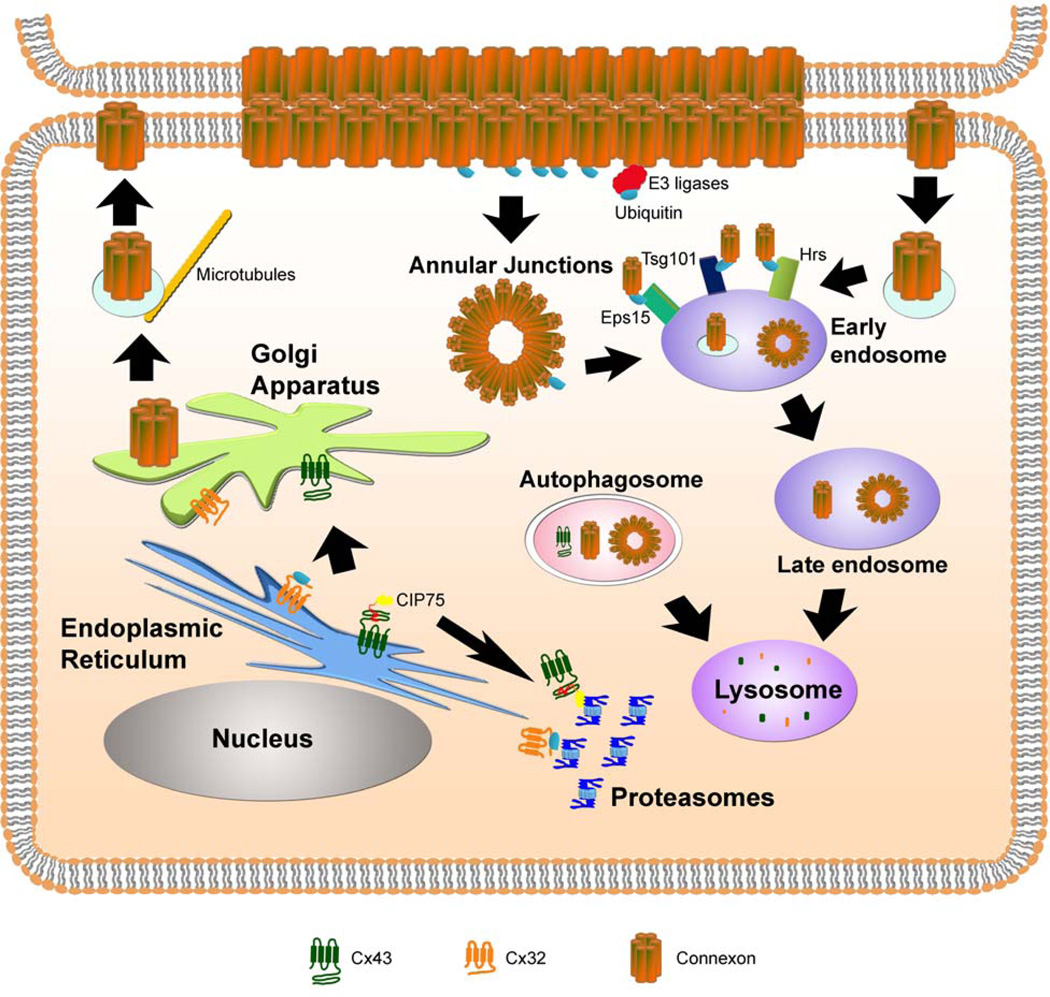
Figure 1. Regulation of the degradation of connexin proteins via proteasomal, lysosomal, and autophagic pathways.
During protein synthesis, connexins are co-translationally inserted into the ER membrane. Upon proper folding, connexins are subsequently trafficked to the Golgi apparatus and trans Golgi network where the individual proteins oligomerize into hexameric connexons. Misfolded connexins can be ubiquitinated and degraded by the proteasome through ERAD, as in the case of Cx32. Alternatively, non-ubiquitinated Cx43 interacts with the accessory factor, CIP75, which facilitates Cx43 ERAD. Connexons are trafficked to the plasma membrane on microtubules, where the hexamers dock with connexons on neighboring cells, across the intercellular space. Intact gap junctions are internalized as the double membrane annular junctions. Individual connexons may also undergo endocytosis. Annular junctions or internalized connexons may enter the endo-lysosomal pathway leading to the degradation of the connexin proteins. Ubiquitination of connexins by E3 ligases may occur either at the plasma membrane or after internalization from the membrane. The ubiquitin-interacting proteins (Eps15, Hrs, and Tsg101) may act to sort the ubiquitinated connexins through the endocytic pathway, until the connexins are ultimately degraded in the lysosome. Autophagy of connexins or connexons is another emerging mechanism for degradation.
Table 1.
Summary of factors involved in connexin ubiquitination and degradation
| Nedd4 family of E3 ligases | Ubiquitin ligases possibly responsible for connexin ubiquitination at the plasma membrane [49, 52] |
| Hrs Tsg101 Eps15 | Ubiquitin-binding proteins required for Cx43 internalization and downregulation of intercellular communication [49, 56] |
| EGF TPA | Activates MAPK signaling which results in increased Cx43 ubiquitination, internalization, and degradation [33, 34, 54–56] |
| Akt | Serine protein kinase that phosphorylates Cx43 to stabilize membrane-localized Cx43 [58] |
| CIP75 | Mediates ubiquitin-independent proteasomal degradation of Cx43 [79–81] |
Ubiquitination (or ubiquitylation) is a process that had primarily been studied as having a role in marking substrates for degradation by the 26S proteasome, with additional functions subsequently revealed. Ubiquitin is a 76 amino acid polypeptide that is highly conserved and expressed in all eukaryotes, from yeast to humans. In ubiquitination, an ubiquitin moiety is covalently attached to target substrates through a series of enzymatic events (reviewed in [46]), which begins with the E1 ubiquitin-activating enzyme forming a high-energy thioester bond with ubiquitin, activating ubiquitin. The activated ubiquitin is then loaded onto an E2 ubiquitin-conjugating enzyme, which will associate with an E3 ubiquitin ligase. The E3 ligase facilitates the covalent linkage of ubiquitin with either the target substrate at lysine residues, or with another ubiquitin forming a polyubiquitin modification. The creation of the polyubiquitin chains has been observed in vivo to attach at the lysine29, lysine48, and lysine63 residues in ubiquitin [46]. While lysine48 linkages are believed to function as the proteasomal degradation tag, lysine63 linkages are proposed to act as internalization signals during endocytosis [47, 48]. This review will focus primarily on the studies of the ubiquitination of Cx43, the most studied connexin regarding connexin ubiquitination.
Cx43 Ubiquitination
The earliest reports of Cx43 ubiquitination arose from studies of Cx43 degradation. The first report of Cx43 ubiquitination was from Laing and Beyer in 1995 [30]. A cell line expressing thermo-labile E1 enzyme was used to study Cx43 ubiquitination and degradation in the absence of E1 activity. Loss of the E1 ubiquitin activation at the restrictive temperature resulted in the stabilization of Cx43 protein, which was presumed to be caused by the loss of ubiquitination and thus, the targeting of Cx43 to the proteasome for degradation. Additionally, sequential immunoprecipitations (first of Cx43, followed by ubiquitin) detected higher molecular weight Cx43 protein that was not detected in the presence of competing free ubiquitin [30]. These data suggested that Cx43 exists in cells with covalently-attached ubiquitin moieties, and perhaps, that ubiquitination is important for regulating Cx43 protein levels.
Subsequent studies reported that Cx43 ubiquitination could result from the cellular exposure to epidermal growth factor (EGF) and the phorbol ester 12-O-tetradecanoylphorbol 13-acetate (TPA) [33, 34]. Use of ubiquitin antibodies that had different specificities to mono- and polyubiquitin indicated that Cx43 was monoubiquitinated at multiple lysine sites in response to TPA [33]. Cx43 multi-monoubiquitination was confirmed in untreated conditions [49], which is significant because monoubiquitination is believed to be involved in proteasome-independent processes, such as internalization and localization [48, 50]. A decrease in the amount of Cx43 at the plasma membrane was also observed in response to both EGF and TPA treatments [33, 34].
The presence of ubiquitinated Cx43 in actual gap junction plaques at areas of cell-cell contact was captured in an immunoelectron microscopy study using freeze-fracture followed by immunogold labeling [51]. Using the fungal metabolite brefeldin A (BFA) to block the transport of Cx43 from the Golgi to the plasma membrane, a high amount of plaques were observed to be ubiquitinated (50%), which dropped to only 14% after a one hour wash-out of BFA. A reduction in GJIC was also observed with the BFA block that was reversed upon BFA wash-out [51]. These data indicated that ubiquitination might occur as a natural part of the Cx43 life cycle, where it may have a role in the trafficking of older gap junctions from the plasma membrane.
One key, but unknown, component in the process of Cx43 ubiquitination was the identity of the E3 ligase that was responsible for the ubiquitination. Nedd4 was the first E3 ligase identified initially as a Cx43 binding partner that appeared to be required for Cx43 internalization, as loss of Nedd4 by siRNA resulted in the accumulation of Cx43 gap junction plaques at the plasma membrane [52]. Further studies on Nedd4 reported that the presence of Nedd4 was required for Cx43 ubiquitination [49]. Other groups have observed the interaction of Cx43 with additional E3 ligases, another member of the Nedd4 family WWP1, and the RING E3 ligase, TRIM21. TRIM21 and WWP1 appear to have active roles in Cx43 ubiquitination (V. Chen, personal communication and L. Matesic, personal communication, respectively).
Internalization, endocytic trafficking, and lysosomal degradation
Cx43 internalization from the plasma membrane and subsequent trafficking through the endocytic pathway to the lysosome for degradation has been extensively studied. TPA and EGF cause a reduction in Cx43 levels at the plasma membrane [33, 34, 53], and are known to induce Cx43 internalization through the MAPK signaling pathway [33, 34, 54, 55]. Cx43 turnover following TPA treatment appeared to be due to both lysosomal and proteasomal degradation [33]. Since Cx43 monoubiquitination, and not polyubiquitination, was detected after TPA treatment, the proteasomal degradation data was surprising as monoubiquitination is not normally observed in proteasomally-degraded substrates. However, it was thought that the proteasomal inhibitors might have an indirect effect on Cx43 levels in the TPA response [33, 56]. Further work found that the subset of Cx43 localized to plaques in the plasma membrane was ubiquitinated in response to TPA treatment [56]. EGF treatment also causes Cx43 internalization and ubiquitination [34]. The type of Cx43 ubiquitination that occurs in response to EGF has not been analyzed and, therefore, it is unclear whether Cx43 is polyubiquitinated (such as by lysine48 linkages) for proteasomal degradation after EGF treatment or monoubiquitinated, as in the case of TPA treatment. In addition, a recent study of ubiquitinated Cx43 proposed that Cx43 may undergo a non-canonical internalization that is mediated by an unidentified ubiquitin modification that is not dependent on the YXXØ tyrosine-sorting signal previously identified to be required for membrane Cx43 internalization [57]. These data suggest that Cx43 ubiquitination is a determinant for the specific cellular internalization mechanism or pathway.
The previous studies of EGF and TPA-induced Cx43 internalization and ubiquitination also provided evidence of proteasomal involvement in the mechanism of Cx43 turnover [33, 34]. However, TPA induced the monoubiquitination of Cx43, which is not typically the ubiquitin tag that leads to proteasomal degradation [33]. In addition, it appeared that EGF stimulated Cx43 proteasomal degradation, because treatment with proteasomal inhibitors, but not lysosomal inhibitors, alleviated the EGF-induced reduction of Cx43 [34]. One possible explanation for these results is that another protein was more directly responsible for Cx43 internalization and turnover, and that the activity of this protein was somehow regulated by proteasomal degradation. A recent study using a Cx43 mutant that cannot be ubiquitinated suggested that proteasomal degradation is an indirect stimulator of Cx43 membrane localization [58]. This Cx43 ubiquitination mutant contained a series of point mutations where all the lysine residues (ubiquitin acceptor sites) were mutated to arginine residues. In the absence of ubiquitination, Cx43 was still trafficked to the plasma membrane and formed functional channels, although initial studies suggested the possibility that the gap junctional communication might not be efficient as gap junctions formed with wild-type Cx43. Akt/protein kinase B (PKB) was identified as the link between ubiquitination and the effect of proteasomal inhibition on Cx43 membrane stabilization. Akt has previously been demonstrated to phosphorylate Cx43 [59], and Akt phosphorylation of Cx43 in the cell membrane stabilized Cx43 in the membrane. Akt ubiquitination and subsequent degradation is one mechanism to regulate its activities, so disruption of Akt turnover by blocking its proteasomal degradation increases the amount of active kinase available to stabilize Cx43. Thus, Akt ubiquitination and degradation by the proteasome indirectly affects Cx43 levels at the plasma membrane [58]. However, the effects of complete loss of Cx43 ubiquitination on internalization and subsequent intracellular trafficking is unclear.
Biochemical fractionation identified the presence of ubiquitinated Cx43 in both the double membrane plaques (previously described as the TritonX-100 insoluble fraction [60]) and the soluble protein fraction, suggesting that the intracellular endocytosed Cx43 was also or still ubiquitinated [33]. Ubiquitination has been proposed to modulate the endocytic trafficking of Cx43 from gap junction plaques. Interestingly, presumably ubiquitinated Cx43 was observed to colocalize with two known ubiquitin-binding proteins, hepatocyte growth factor-regulated tyrosine kinase substrate (Hrs) and tumor susceptibility gene 101 (Tsg101) [56]. Both proteins are involved in the ESCRT (endosomal sorting complex required for transport) machinery for endocytic trafficking and reportedly mediate the trafficking of ubiquitinated growth factor receptors from the endosome to lysosomes [61]. Upon first glance, depletion of Hrs and/or Tsg101 by siRNA appeared to have little effect on Cx43, as membrane localization and GJIC are not significantly affected [56]. However, prolonged induction of internalization with TPA in concert with a block of protein synthesis in the siRNA-treated cells resulted in increased Cx43 levels at the plasma membrane and enhanced GJIC. This study proposes that Hrs and Tsg101 are responsible for directing Cx43 towards lysosomal degradation by sorting Cx43 at the early endosome, instead of possibly being recycled back to the plasma membrane. Cx43 recycling has previously been demonstrated during cellular stress situations [37] or mitosis [62]. Furthermore, prolonged (3 hours or more) induction of Cx43 internalization by TPA treatment in cells depleted for Hrs and Tsg101 by siRNA resulted in increased Cx43 localization at the plasma membrane, increased amounts of phosphorylated Cx43, and increased levels of GJIC compared to a shorter time point of 1.5 hours [56]. This strongly suggested that the Hrs and Tsg101 ubiquitin-binding proteins were necessary to dictate the movement of Cx43 towards lysosomal degradation, instead of being recycled back to the plasma membrane to again contribute to GJIC. Additionally, the loss of both Hrs and Tsg101 resulted in the increased amounts of ubiquitinated Cx43 and the ubiquitination of Cx43 at the membrane was observed to be dependent on the proteasome. This effect was thought to result from the depletion of intracellular pools of ubiquitin when proteasomal degradation is blocked, which would result in a reduction in the amount of ubiquitin available for conjugation [56]. The Hrs/Tsg101 study was followed shortly by the report of an interaction between ubiquitinated Cx43 and epidermal growth factor receptor substrate 15 (Eps15), another ubiquitin-binding domain-containing protein that is a part of the ESCRT machinery [49]. These studies suggested that a highly-regulated mechanism mediates Cx43 internalization from the plasma membrane, trafficking through the endo-lysosomal pathway, and ending with Cx43 degradation. The internalization and trafficking of the previously discussed lysine mutant form of Cx43 from the plasma membrane has not yet been extensively studied so it is not known how the loss of Cx43 ubiquitination affects the trafficking of Cx43 through the endo-lysosomal pathways as the interactions with the ubiquitin-binding proteins Hrs, Tsg101, and Eps15 are likely to be affected.
Autophagy
Besides direct lysosomal degradation, autophagy is a degradation pathway that utilizes lysosomes for the ultimate step of proteolysis, but is distinct from the endo-lysosomal degradation pathway (reviewed in [63]). Ubiquitination has not been demonstrated to be a requirement for autophagy; however, an ubiquitin-binding protein, p62/SQSTM1, is a substrate that is degraded via autophagy. p62 contains the ubiquitin-associated (UBA) ubiquitin-binding domain which has been demonstrated to regulate the turnover of ubiquitinated proteins (reviewed in [64]). A growing body of work suggests that p62 is a receptor for ubiquitinated proteins that can contribute to the autophagic degradation of its substrates (reviewed in [65]).
The involvement of autophagy in regulating Cx43 was not recognized until recently, when two studies identified Cx43-containing autophagosome structures [38, 39]. In the heart, Cx43 is normally localized to the intercalated disc regions of the plasma membrane in cardiomyocytes where functional gap junctions are responsible for propagating the electrical signal required for the normal cardiac rhythm. In heart disease, Cx43 is relocalized away from the intercalated discs to the lateral membranes of the cardiomyocyte cells [66–69]. Electron microscopy studies of failing canine heart myocardium showed Cx43-containing multilamellar membrane structures near the lateral membranes that morphologically resembled autophagosomes [39]. To further test the theory that internalized Cx43 gap junctions were in, or associated with, autophagosomes, the colocalization of Cx43 with a marker for autophagosomes, GFP-LC3 (microtubule-associated protein light chain 3), was examined in transfected HeLa cells. Colocalization of Cx43 and LC-3 was observed. This study also analyzed the targeting of Cx43 to lipid rafts, which has been previously reported [70–72]. It was suggested that the observed multilamellar structures might contain Cx43 targeted to lipid rafts [39]. Using TritonX-100 solubility fractions, two fractions of Cx43 were identified: one fraction that also contained lipid raft markers such as the sphingolipid GM1 and caveolin-3, and a denser, less buoyant fraction. There was an increase in the amount of Cx43 in the lipid raft fractions in the failing heart tissues, as well as an increase in the amount of the autophagy marker LC3-II, which is a cleaved form of LC3-I that results with the onset of autophagy [39]. One suggestion is that in diseased heart tissue, Cx43 is removed from the intercalated disc area, processed through lipid raft areas of the plasma membrane as a part of the degradation machinery that would involve autophagosomes [39]. A relationship between lipid rafts and autophagosomes has not yet been established and thus, will require more evidence to confirm. In a subsequent study, using cultured cells, the involvement of autophagy in Cx43 degradation was observed in cells subjected to starvation conditions, which induces autophagy [38]. Cx43 was found to localize to cytoplasmic structures surrounded by LC3, and blocking autophagic degradation with a lysosomal inhibitor or siRNA of the autophagy-related Atg proteins (Atg5) prevented the starvation-induced loss of Cx43 protein as well as increased the colocalization of Cx43 with GFP-LC3. Electron microscopy detected the double membrane structures that are characteristic of autophagosomes. p62, which has been implicated in the autophagy of ubiquitinated proteins, also colocalized with Cx43 [38]. However, it is still unclear what role the ubiquitination of Cx43 has on Cx43 autophagy. Since p62 has the UBA ubiquitin-binding domain, this may serve as the mechanism that targets ubiquitinated Cx43 to the autophagosome.
The involvement of autophagy in the turnover of gap junction plaques from the plasma membrane has also been recently reported where internalized intact annular gap junctions colocalized with LC3, p62, and other autophagy-related proteins. In addition, inhibition of autophagy by siRNA knock-down of the Atg proteins, Atg6 (beclin-1), or Atg8 (LC3) reduced Cx43 turnover, resulting in the accumulation of cytoplasmic annular gap junctions (M. Falk, personal communication). In light of the recent data suggesting that ubiquitination of Cx43 has an effect on the post-internalization trafficking of Cx43 from the plasma membrane, it would be interesting to determine whether the p62 ubiquitin-binding protein is part of the mechanism that determines the fate of internalized and ubiquitinated Cx43 gap junctions.
Proteasomal degradation
Like lysosomal degradation, there is extensive evidence of a role for proteasomal degradation in the Cx43 life cycle. The proteasomal degradation pathway is regulated by a multitude of proteins that result in the trafficking of target substrates to the 26S proteasome holoenzyme complex. This complex is comprised of two subunits, the 20S core particle (CP) and the 19S regulatory particle (RP) (reviewed in [73]). The 20S CP consists of four rings containing seven subunits that are stacked on one another. The β-type subunits comprise the inner two rings and are proteolytically active, while the α-type subunits comprise the outermost rings. The 19S RP, or cap complex, consists of a base and lid, and flank the core, with one RP on each end of the 20S CP. Several proteins associated with the 19S RP, such as Rpn1 and Rpn10, are able to bind ubiquitinated proteins [74–78].
The study that initially identified Cx43 ubiquitination pointed to a role for ubiquitin-mediated proteasomal degradation as a major process of Cx43 turnover [30]. Additional studies using proteasomal inhibitors further implicated the proteasome in regulating Cx43 levels at the plasma membrane and the corresponding effect on GJIC [9, 32, 36]. Two reasons for this effect have been proposed: first, by blocking proteasomal degradation, Cx43 in the ER has more time to fold properly and to traffic to the membrane, resulting in more Cx43 gap junctions. Or, alternatively, the proteasome has an immediate effect on membrane-localized Cx43, either by facilitating the degradation of that subset of Cx43 directly, or indirectly, by facilitating the degradation of another protein involved in Cx43 localization or trafficking.
The degradation of Cx43 through the process of ER-associated degradation (ERAD) has been demonstrated through a series of studies. Increasing the amount of Cx43 in the ER by blocking trafficking to the cell membrane using BFA, plus a concurrent block of proteasomal degradation, elevated Cx43 protein levels [32], which was suggestive of ERAD. Further studies using ER stress inducers, such as DTT, which prevents the disulfide bond formation in mature Cx43 protein, revealed enhanced dislocation from the ER into the cytoplasm and elevated proteasomal degradation in response to the ER stress [9, 36], again supporting a role for ERAD in Cx43 degradation. While proteasomal degradation and ERAD are typically associated with ubiquitinated substrates, the ubiquitination state of Cx43 undergoing ERAD has not been studied in depth.
Our laboratory identified a novel Cx43-interacting protein through a yeast two-hybrid screen that we called CIP75 for Cx43-interacting protein of 75 kDa [79]. CIP75 is a member of the ubiquitin-like (UbL)-UBA domain family of ubiquitin-binding proteins. With an N-terminal UbL domain, and one or more UBA domains at the C-terminus, this family of proteins has been demonstrated to bind to ubiquitin and ubiquitinated proteins via the UBA domain, as well as subunits of the proteasome via the UbL domain (reviewed in [64]). Our initial studies suggested that CIP75 was involved in facilitating Cx43 proteasomal degradation, as overexpression of CIP75 reduced the half-life of Cx43 and the knock-down of CIP75 by siRNA had the opposite effect of significantly increasing the Cx43 half-life [79]. Use of a proteasomal inhibitor indicated that this effect was via the proteasomal degradation pathway. Proteins that are targeted for degradation by the proteasome are typically marked for degradation by the covalently-attached ubiquitin tag so the ubiquitination state of the subset of Cx43 that specifically interacted with CIP75 was examined. Prior reports of Cx43 ubiquitination and ubiquitination of proteasome substrates suggested that the Cx43 interacting with CIP75 would be ubiquitinated [30, 33, 34, 46]. Surprisingly, extensive work demonstrated that while CIP75 is indeed an ubiquitin-binding protein, which was capable of binding both monoubiquitin and lysine48-linked tetraubiquitin, as well as ubiquitinated proteins, the subset of Cx43 that interacted with CIP75 did not appear to be ubiquitinated [80]. Our initial experiments included biochemical immunoprecipitations that failed to detect interacting ubiquitinated Cx43. We subsequently generated a series of three lysine to arginine Cx43 point mutants in an attempt to eliminate any covalent ubiquitin modification. The specific lysines that are ubiquitinated in Cx43 have not been identified so we mutated the lysines in or near the CIP75 binding domain in mutant 1, all the lysines in the C-terminal tail, which contains the CIP75 binding region, in mutant 2, and, all 27 lysines in Cx43 in mutant 3. In all cases, CIP75 was able to interact with Cx43, indicating that Cx43 ubiquitination is not a requirement for interaction with CIP75 [80]. While this result was unexpected, structural NMR data supported this conclusion [81]. An analysis of the binding region of the CIP75 UBA domain for the C-terminal tail of Cx43, as well as for ubiquitin, determined that both Cx43 and ubiquitin interact with overlapping regions of CIP75. This suggested a mechanism involving the competition between ubiquitin or ubiquitinated proteins and the non-ubiquitinated Cx43 C-terminus, which may be indicative of the regulation of CIP75 function and activity. Significantly, these data point to an uncommon situation of a non-ubiquitinated substrate that undergoes proteasomal degradation by the 26S proteasome. While many substrates have been identified as undergoing ubiquitin-independent proteasomal degradation [82], it should be noted that the bulk of these substrates have been demonstrated to undergo degradation by the 20S core proteasomal subunit, not the 26S proteasome holoenzyme complex. For example, extensive work has conclusively demonstrated that oxidized proteins do not require ubiquitination and are degraded specifically by the 20S CP [83–87]. This is thought to occur because oxidation may cause some conformational changes in the proteins that allow them to directly enter the 20S CP, instead of requiring the ATP-dependent activity of the 19S regulatory particle that typically is given credit for unfolding proteins to be able to enter the 20S CP barrel. Only a limited number of substrates have been identified that also have ubiquitin-independent degradation through the 26S proteasome [82, 88], the best known and most conclusively studied substrate being the enzyme ornithine decarboxylase [89–91].
Cx32 ERAD
Because Cx32 is linked to the causation of the X-linked Charcot-Marie Tooth disease (CMTX), a human peripheral neuropathy [92–94], there has been much interest in elucidating the regulation of Cx32, including its ubiquitination, as it may provide insight into the basis of CMTX. As for Cx43, Cx32 degradation is also mediated by both lysosomal and proteasomal degradation, although the bulk of the studies have focused on the degradation of Cx32 by the proteasome. One of the mutants that has been identified in CMTX patients is the Cx32 E208K point mutant [95], which exhibits an intracellular trafficking defect such that it remains in the ER. Use of this mutant has been particularly useful in studying Cx32 ubiquitination and ERAD. The Cx32 E208K mutant does not oligomerize into connexons and is not transported to the plasma membrane to form functional gap junction channels [35]. ER stress, induced by DTT treatment, triggers the dislocation of wild-type Cx32 from the ER into the cytoplasm where it undergoes proteasomal degradation, as was seen with Cx43 [29, 36]. The ER-localized Cx32 E208K mutant was observed in both non-ubiquitinated and polyubiquitinated states [29]. There was significantly more polyubiquitinated Cx32 E208K protein versus non-ubiquitinated in the cytoplasm than in the ER membrane, which could only be observed upon inhibition of the proteasome. The extent of polyubiquitination was decreased in response to cellular stresses such as a heat shock or oxidative stress. After cellular stress, there was a decrease in Cx32 polyubiquitination, particularly in the cytoplasm [29]. This indicated that, unlike Cx43, Cx32, which is dislocated from the ER into the cytoplasm for ERAD, is polyubiquitinated, as is the case for almost all proteins that undergo 26S proteasomal degradation.
Cx43 degradation and disease
The regulation of connexins and their function is known to be crucial for normal cell homeostasis. An increasing body of work argues for the importance of connexins in human diseases as well. As previously mentioned, Cx32 has been linked to the human CMTX disease. Connexin defects have also been discovered in other diseases including oculodentaldigital dysplasia (ODDD), deafness, cataracts, and skin disorders (reviewed in [28, 93]). A role for connexins in cancer has also been pursued (reviewed in [96]). A recent study analyzed the mechanisms by which proteasomal inhibition effects cell death in cancer cells [97], which is the reason for the use of the proteasomal inhibitor, bortezomib (commercially marketed as Velcade), as a cancer therapeutic in multiple myeloma and more. Using the proteasomal inhibitor, MG132, the study demonstrated that a long-term block of proteasomal degradation triggered cell apoptosis after induction of ER stress with tunicamycin or thapsigargin. Interestingly, overexpression of Cx43 can increase the apoptotic response to MG132, as well as sensitizing cells to apoptosis brought on by ER stress. While inhibition of proteasomal degradation has consistently been proven to increase GJIC, this study reported that the increased apoptotic sensitivity due to Cx43 overexpression did not appear to be dependent on the ability of Cx43 to create functional gap junctions [97]. Other studies have also implicated a GJIC-independent function for connexins [98, 99]. Thus, the regulation of connexins, either dependent or independent of their communication abilities, may have important consequences for a vast array of cellular and tissue processes and pathways.
Concluding Remarks
It is evident from a growing body of work that connexins have critical functions not only in normal cell and tissue function and homeostasis, but also in human diseases under aberrant conditions. Altered gene and protein expression, and protein trafficking/localization have been observed in a number of disease states. These observations clearly demonstrate the necessity for a better understanding of the regulation of the connexin life cycle. One component that has commanded attention is the mechanisms that regulate connexin turnover. With such a short half-life of 1.5 to 5 hours, the cell must tightly regulate the processes that mediate connexin degradation. Lysosomal, and perhaps, autophagic actions appear to have an important role in regulating the turnover of ubiquitinated connexins that are internalized from gap junctions at the cell membrane. Proteasomal degradation, mediated by ERAD, acts in the turnover of both ubiquitinated and non-ubiquitinated connexins from the ER and may function as part of the cellular stress response which determines cell survival. With new tools and innovative techniques available to examine the role of ubiquitination in mediating connexin degradation, the processes that contribute to the regulation of connexin protein levels and GJIC will be elucidated, which will hopefully provide better insight into the mechanisms underlying disease pathologies.
Highlights.
Connexin43 ubiquitin modification affects intracellular trafficking and degradation
Monoubiquitination affects connexin43 internalization and endosomal trafficking
Ubiquitination is involved in connexin32 proteasomal degradation
Ubiquitination is not required for CIP75-mediated connexin43 proteasomal degradation
ACKNOWLEDGEMENTS
This work was supported by awards from the American Heart Association (11POST5460028) to VS, the Hawaii Community Foundation (11ADVC-49235) to VS and AFL, and the National Institutes of Health to AFL (CA052098-18).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Abbreviations used: GJIC, gap junctional intercellular communication; ER, endoplasmic reticulum; Cx43, connexin43; EGF, epidermal growth factor; TPA, 12-O-tetradecanoylphorbol 13-acetate; BFA, brefeldin A; UBA, ubiquitin-associated domain; LC3, light chain 3; ERAD, ER-associated degradation; Cx32, connexin32; CMTX, X-linked Charcot-Marie Tooth disease; ESCRT, endosomal sorting complex require for transport; Hrs, hepatocyte growth-factor regulated tyrosine kinase substrate; Tsg101, tumor susceptibility gene 101; Eps15, epidermal growth factor substrate 15.
REFERENCES
- 1.Goodenough DA, Goliger JA, Paul DL. Annu Rev Biochem. 1996;65:475–502. doi: 10.1146/annurev.bi.65.070196.002355. [DOI] [PubMed] [Google Scholar]
- 2.Vinken M, Vanhaecke T, Papeleu P, Snykers S, Henkens T, Rogiers V. Cell Signal. 2006;18:592–600. doi: 10.1016/j.cellsig.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 3.Wei CJ, Xu X, Lo CW. Annu Rev Cell Dev Biol. 2004;20:811–838. doi: 10.1146/annurev.cellbio.19.111301.144309. [DOI] [PubMed] [Google Scholar]
- 4.White TW, Paul DL. Annu Rev Physiol. 1999;61:283–310. doi: 10.1146/annurev.physiol.61.1.283. [DOI] [PubMed] [Google Scholar]
- 5.Beardslee MA, Laing JG, Beyer EC, Saffitz JE. Circ Res. 1998;83:629–635. doi: 10.1161/01.res.83.6.629. [DOI] [PubMed] [Google Scholar]
- 6.Darrow BJ, Laing JG, Lampe PD, Saffitz JE, Beyer EC. Circ Res. 1995;76:381–387. doi: 10.1161/01.res.76.3.381. [DOI] [PubMed] [Google Scholar]
- 7.Fallon RF, Goodenough DA. J Cell Biol. 1981;90:521–526. doi: 10.1083/jcb.90.2.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Laird DW, Puranam KL, Revel JP. Biochem J. 1991;273(Pt 1):67–72. doi: 10.1042/bj2730067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Musil LS, Le AC, VanSlyke JK, Roberts LM. J Biol Chem. 2000;275:25207–25215. doi: 10.1074/jbc.275.33.25207. [DOI] [PubMed] [Google Scholar]
- 10.Das S, Smith TD, Sarma JD, Ritzenthaler JD, Maza J, Kaplan BE, Cunningham LA, Suaud L, Hubbard MJ, Rubenstein RC, Koval M. Mol Biol Cell. 2009;20:2593–2604. doi: 10.1091/mbc.E08-07-0790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Das Sarma J, Wang F, Koval M. J Biol Chem. 2002;277:20911–20918. doi: 10.1074/jbc.M111498200. [DOI] [PubMed] [Google Scholar]
- 12.Diez JA, Ahmad S, Evans WH. Eur J Biochem. 1999;262:142–148. doi: 10.1046/j.1432-1327.1999.00343.x. [DOI] [PubMed] [Google Scholar]
- 13.Koval M. Trends Cell Biol. 2006;16:159–166. doi: 10.1016/j.tcb.2006.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maza J, Das Sarma J, Koval M. J Biol Chem. 2005;280:21115–21121. doi: 10.1074/jbc.M412612200. [DOI] [PubMed] [Google Scholar]
- 15.Maza J, Mateescu M, Das Sarma J, Koval M. Cell Commun Adhes. 2003;10:319–322. doi: 10.1080/cac.10.4-6.319.322. [DOI] [PubMed] [Google Scholar]
- 16.Cherian PP, Siller-Jackson AJ, Gu S, Wang X, Bonewald LF, Sprague E, Jiang JX. Mol Biol Cell. 2005;16:3100–3106. doi: 10.1091/mbc.E04-10-0912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Siller-Jackson AJ, Burra S, Gu S, Xia X, Bonewald LF, Sprague E, Jiang JX. J Biol Chem. 2008;283:26374–26382. doi: 10.1074/jbc.M803136200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Contreras JE, Saez JC, Bukauskas FF, Bennett MV. Cell Commun Adhes. 2003;10:245–249. doi: 10.1080/cac.10.4-6.245.249. [DOI] [PubMed] [Google Scholar]
- 19.Srinivas M, Calderon DP, Kronengold J, Verselis VK. J Gen Physiol. 2006;127:67–75. doi: 10.1085/jgp.200509397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gomez-Hernandez JM, de Miguel M, Larrosa B, Gonzalez D, Barrio LC. Proc Natl Acad Sci U S A. 2003;100:16030–16035. doi: 10.1073/pnas.2530348100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garre JM, Retamal MA, Cassina P, Barbeito L, Bukauskas FF, Saez JC, Bennett MV, Abudara V. Proc Natl Acad Sci U S A. 2010;107:22659–22664. doi: 10.1073/pnas.1013793107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anselmi F, Hernandez VH, Crispino G, Seydel A, Ortolano S, Roper SD, Kessaris N, Richardson W, Rickheit G, Filippov MA, Monyer H, Mammano F. Proc Natl Acad Sci U S A. 2008;105:18770–18775. doi: 10.1073/pnas.0800793105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Knight MM, McGlashan SR, Garcia M, Jensen CG, Poole CA. J Anat. 2009;214:275–283. doi: 10.1111/j.1469-7580.2008.01021.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bargiotas P, Monyer H, Schwaninger M. Curr Mol Med. 2009;9:186–194. doi: 10.2174/156652409787581646. [DOI] [PubMed] [Google Scholar]
- 25.Goodenough DA, Paul DL. Nat Rev Mol Cell Biol. 2003;4:285–294. doi: 10.1038/nrm1072. [DOI] [PubMed] [Google Scholar]
- 26.Sato M, Jiao Q, Honda T, Kurotani R, Toyota E, Okumura S, Takeya T, Minamisawa S, Lanier SM, Ishikawa Y. J Biol Chem. 2009;284:31431–31440. doi: 10.1074/jbc.M109.014068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stout C, Goodenough DA, Paul DL. Curr Opin Cell Biol. 2004;16:507–512. doi: 10.1016/j.ceb.2004.07.014. [DOI] [PubMed] [Google Scholar]
- 28.Laird DW. Biochem J. 2006;394:527–543. doi: 10.1042/BJ20051922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kelly SM, Vanslyke JK, Musil LS. Mol Biol Cell. 2007;18:4279–4291. doi: 10.1091/mbc.E07-05-0487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laing JG, Beyer EC. J Biol Chem. 1995;270:26399–26403. doi: 10.1074/jbc.270.44.26399. [DOI] [PubMed] [Google Scholar]
- 31.Laing JG, Tadros PN, Green K, Saffitz JE, Beyer EC. Cardiovasc Res. 1998;38:711–718. doi: 10.1016/s0008-6363(98)00060-1. [DOI] [PubMed] [Google Scholar]
- 32.Laing JG, Tadros PN, Westphale EM, Beyer EC. Exp Cell Res. 1997;236:482–492. doi: 10.1006/excr.1997.3747. [DOI] [PubMed] [Google Scholar]
- 33.Leithe E, Rivedal E. J Biol Chem. 2004;279:50089–50096. doi: 10.1074/jbc.M402006200. [DOI] [PubMed] [Google Scholar]
- 34.Leithe E, Rivedal E. J Cell Sci. 2004;117:1211–1220. doi: 10.1242/jcs.00951. [DOI] [PubMed] [Google Scholar]
- 35.VanSlyke JK, Deschenes SM, Musil LS. Mol Biol Cell. 2000;11:1933–1946. doi: 10.1091/mbc.11.6.1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.VanSlyke JK, Musil LS. J Cell Biol. 2002;157:381–394. doi: 10.1083/jcb.200111045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.VanSlyke JK, Musil LS. Mol Biol Cell. 2005;16:5247–5257. doi: 10.1091/mbc.E05-05-0415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lichtenstein A, Minogue PJ, Beyer EC, Berthoud VM. J Cell Sci. 2011;124:910–920. doi: 10.1242/jcs.073072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hesketh GG, Shah MH, Halperin VL, Cooke CA, Akar FG, Yen TE, Kass DA, Machamer CE, Van Eyk JE, Tomaselli GF. Circ Res. 2010;106:1153–1163. doi: 10.1161/CIRCRESAHA.108.182147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Solan JL, Lampe PD. Biochem J. 2009;419:261–272. doi: 10.1042/BJ20082319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Solan JL, Lampe PD. J Membr Biol. 2007;217:35–41. doi: 10.1007/s00232-007-9035-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Solan JL, Lampe PD. Biochim Biophys Acta. 2005;1711:154–163. doi: 10.1016/j.bbamem.2004.09.013. [DOI] [PubMed] [Google Scholar]
- 43.Colussi C, Rosati J, Straino S, Spallotta F, Berni R, Stilli D, Rossi S, Musso E, Macchi E, Mai A, Sbardella G, Castellano S, Chimenti C, Frustaci A, Nebbioso A, Altucci L, Capogrossi MC, Gaetano C. Proc Natl Acad Sci U S A. 2011;108:2795–2800. doi: 10.1073/pnas.1013124108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Locke D, Bian S, Li H, Harris AL. Biochem J. 2009;424:385–398. doi: 10.1042/BJ20091140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shearer D, Ens W, Standing K, Valdimarsson G. Invest Ophthalmol Vis Sci. 2008;49:1553–1562. doi: 10.1167/iovs.07-1193. [DOI] [PubMed] [Google Scholar]
- 46.Fang S, Weissman AM. Cell Mol Life Sci. 2004;61:1546–1561. doi: 10.1007/s00018-004-4129-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Clague MJ, Urbe S. Cell. 2010;143:682–685. doi: 10.1016/j.cell.2010.11.012. [DOI] [PubMed] [Google Scholar]
- 48.Hicke L, Dunn R. Annu Rev Cell Dev Biol. 2003;19:141–172. doi: 10.1146/annurev.cellbio.19.110701.154617. [DOI] [PubMed] [Google Scholar]
- 49.Girao H, Catarino S, Pereira P. Exp Cell Res. 2009;315:3587–3597. doi: 10.1016/j.yexcr.2009.10.003. [DOI] [PubMed] [Google Scholar]
- 50.Mosesson Y, Yarden Y. Isr Med Assoc J. 2006;8:233–237. [PubMed] [Google Scholar]
- 51.Rutz ML, Hulser DF. Eur J Cell Biol. 2001;80:20–30. doi: 10.1078/0171-9335-00140. [DOI] [PubMed] [Google Scholar]
- 52.Leykauf K, Salek M, Bomke J, Frech M, Lehmann WD, Durst M, Alonso A. J Cell Sci. 2006;119:3634–3642. doi: 10.1242/jcs.03149. [DOI] [PubMed] [Google Scholar]
- 53.Leithe E, Brech A, Rivedal E. Biochem J. 2006;393:59–67. doi: 10.1042/BJ20050674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sirnes S, Kjenseth A, Leithe E, Rivedal E. Biochem Biophys Res Commun. 2009;382:41–45. doi: 10.1016/j.bbrc.2009.02.141. [DOI] [PubMed] [Google Scholar]
- 55.Sirnes S, Leithe E, Rivedal E. Biochem Biophys Res Commun. 2008;373:597–601. doi: 10.1016/j.bbrc.2008.06.095. [DOI] [PubMed] [Google Scholar]
- 56.Leithe E, Kjenseth A, Sirnes S, Stenmark H, Brech A, Rivedal E. J Cell Sci. 2009;122:3883–3893. doi: 10.1242/jcs.053801. [DOI] [PubMed] [Google Scholar]
- 57.Catarino S, Ramalho JS, Marques C, Pereira P, Girao H. Biochem J. 2011;437:255–267. doi: 10.1042/BJ20102059. [DOI] [PubMed] [Google Scholar]
- 58.Dunn CA, Su V, Lau AF, Lampe PD. J Biol Chem. 2012 doi: 10.1074/jbc.M111.276261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Park DJ, Wallick CJ, Martyn KD, Lau AF, Jin C, Warn-Cramer BJ. Cell Commun Adhes. 2007;14:211–226. doi: 10.1080/15419060701755958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Musil LS, Goodenough DA. J Cell Biol. 1991;115:1357–1374. doi: 10.1083/jcb.115.5.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Raiborg C, Stenmark H. Nature. 2009;458:445–452. doi: 10.1038/nature07961. [DOI] [PubMed] [Google Scholar]
- 62.Boassa D, Solan JL, Papas A, Thornton P, Lampe PD, Sosinsky GE. Traffic. 2010;11:1471–1486. doi: 10.1111/j.1600-0854.2010.01109.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.He C, Klionsky DJ. Annu Rev Genet. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Su V, Lau AF. Cell Mol Life Sci. 2009;66:2819–2833. doi: 10.1007/s00018-009-0048-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Komatsu M, Ichimura Y. FEBS Lett. 2010;584:1374–1378. doi: 10.1016/j.febslet.2010.02.017. [DOI] [PubMed] [Google Scholar]
- 66.Beardslee MA, Lerner DL, Tadros PN, Laing JG, Beyer EC, Yamada KA, Kleber AG, Schuessler RB, Saffitz JE. Circ Res. 2000;87:656–662. doi: 10.1161/01.res.87.8.656. [DOI] [PubMed] [Google Scholar]
- 67.Peters NS, Green CR, Poole-Wilson PA, Severs NJ. Circulation. 1993;88:864–875. doi: 10.1161/01.cir.88.3.864. [DOI] [PubMed] [Google Scholar]
- 68.Severs NJ, Bruce AF, Dupont E, Rothery S. Cardiovasc Res. 2008;80:9–19. doi: 10.1093/cvr/cvn133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Smith JH, Green CR, Peters NS, Rothery S, Severs NJ. Am J Pathol. 1991;139:801–821. [PMC free article] [PubMed] [Google Scholar]
- 70.Locke D, Liu J, Harris AL. Biochemistry. 2005;44:13027–13042. doi: 10.1021/bi050495a. [DOI] [PubMed] [Google Scholar]
- 71.Schubert AL, Schubert W, Spray DC, Lisanti MP. Biochemistry. 2002;41:5754–5764. doi: 10.1021/bi0121656. [DOI] [PubMed] [Google Scholar]
- 72.Langlois S, Cowan KN, Shao Q, Cowan BJ, Laird DW. Mol Biol Cell. 2008;19:912–928. doi: 10.1091/mbc.E07-06-0596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Voges D, Zwickl P, Baumeister W. Annu Rev Biochem. 1999;68:1015–1068. doi: 10.1146/annurev.biochem.68.1.1015. [DOI] [PubMed] [Google Scholar]
- 74.Deveraux Q, Ustrell V, Pickart C, Rechsteiner M. J Biol Chem. 1994;269:7059–7061. [PubMed] [Google Scholar]
- 75.Elsasser S, Gali RR, Schwickart M, Larsen CN, Leggett DS, Muller B, Feng MT, Tubing F, Dittmar GA, Finley D. Nat Cell Biol. 2002;4:725–730. doi: 10.1038/ncb845. [DOI] [PubMed] [Google Scholar]
- 76.Husnjak K, Elsasser S, Zhang N, Chen X, Randles L, Shi Y, Hofmann K, Walters KJ, Finley D, Dikic I. Nature. 2008;453:481–488. doi: 10.1038/nature06926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.van Nocker S, Sadis S, Rubin DM, Glickman M, Fu H, Coux O, Wefes I, Finley D, Vierstra RD. Mol Cell Biol. 1996;16:6020–6028. doi: 10.1128/mcb.16.11.6020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Young P, Deveraux Q, Beal RE, Pickart CM, Rechsteiner M. J Biol Chem. 1998;273:5461–5467. doi: 10.1074/jbc.273.10.5461. [DOI] [PubMed] [Google Scholar]
- 79.Li X, Su V, Kurata WE, Jin C, Lau AF. J Biol Chem. 2008;283:5748–5759. doi: 10.1074/jbc.M709288200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Su V, Nakagawa R, Koval M, Lau AF. J Biol Chem. 2010;285:40979–40990. doi: 10.1074/jbc.M110.170753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kieken F, Spagnol G, Su V, Lau AF, Sorgen PL. Journal of Biological NMR. 2010 doi: 10.1007/s10858-010-9397-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jariel-Encontre I, Bossis G, Piechaczyk M. Biochim Biophys Acta. 2008;1786:153–177. doi: 10.1016/j.bbcan.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 83.Davies KJ. Biochimie. 2001;83:301–310. doi: 10.1016/s0300-9084(01)01250-0. [DOI] [PubMed] [Google Scholar]
- 84.Ferrington DA, Sun H, Murray KK, Costa J, Williams TD, Bigelow DJ, Squier TC. J Biol Chem. 2001;276:937–943. doi: 10.1074/jbc.M005356200. [DOI] [PubMed] [Google Scholar]
- 85.Grune T, Reinheckel T, Davies KJ. FASEB J. 1997;11:526–534. [PubMed] [Google Scholar]
- 86.Orlowski M, Wilk S. Arch Biochem Biophys. 2003;415:1–5. doi: 10.1016/s0003-9861(03)00197-8. [DOI] [PubMed] [Google Scholar]
- 87.Shringarpure R, Grune T, Mehlhase J, Davies KJ. J Biol Chem. 2003;278:311–318. doi: 10.1074/jbc.M206279200. [DOI] [PubMed] [Google Scholar]
- 88.Jariel-Encontre I, Pariat M, Martin F, Carillo S, Salvat C, Piechaczyk M. J Biol Chem. 1995;270:11623–11627. doi: 10.1074/jbc.270.19.11623. [DOI] [PubMed] [Google Scholar]
- 89.Bercovich Z, Rosenberg-Hasson Y, Ciechanover A, Kahana C. J Biol Chem. 1989;264:15949–15952. [PubMed] [Google Scholar]
- 90.Glass JR, Gerner EW. J Cell Physiol. 1987;130:133–141. doi: 10.1002/jcp.1041300119. [DOI] [PubMed] [Google Scholar]
- 91.Murakami Y, Matsufuji S, Kameji T, Hayashi S, Igarashi K, Tamura T, Tanaka K, Ichihara A. Nature. 1992;360:597–599. doi: 10.1038/360597a0. [DOI] [PubMed] [Google Scholar]
- 92.Bergoffen J, Scherer SS, Wang S, Scott MO, Bone LJ, Paul DL, Chen K, Lensch MW, Chance PF, Fischbeck KH. Science. 1993;262:2039–2042. doi: 10.1126/science.8266101. [DOI] [PubMed] [Google Scholar]
- 93.Krutovskikh V, Yamasaki H. Mutat Res. 2000;462:197–207. doi: 10.1016/s1383-5742(00)00037-5. [DOI] [PubMed] [Google Scholar]
- 94.Zhou L, Griffin JW. Curr Opin Neurol. 2003;16:307–313. doi: 10.1097/01.wco.0000073931.19076.52. [DOI] [PubMed] [Google Scholar]
- 95.Fairweather N, Bell C, Cochrane S, Chelly J, Wang S, Mostacciuolo ML, Monaco AP, Haites NE. Hum Mol Genet. 1994;3:29–34. doi: 10.1093/hmg/3.1.29. [DOI] [PubMed] [Google Scholar]
- 96.Naus CC, Laird DW. Nat Rev Cancer. 2010;10:435–441. doi: 10.1038/nrc2841. [DOI] [PubMed] [Google Scholar]
- 97.Huang T, Zhu Y, Fang X, Chi Y, Kitamura M, Yao J. Cancer Sci. 2010;101:713–721. doi: 10.1111/j.1349-7006.2009.01421.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kalra J, Shao Q, Qin H, Thomas T, Alaoui-Jamali MA, Laird DW. Carcinogenesis. 2006;27:2528–2537. doi: 10.1093/carcin/bgl110. [DOI] [PubMed] [Google Scholar]
- 99.Zhang YW, Kaneda M, Morita I. J Biol Chem. 2003;278:44852–44856. doi: 10.1074/jbc.M305072200. [DOI] [PubMed] [Google Scholar]