Abstract
The zinc finger transcriptional repressor Gfi-1 has been shown to play a critical role in early granulopoiesis; however, its role in late neutrophilic development is poorly understood. We report here that forced expression of a dominant negative Gfi-1 mutant, N382S, resulted in augmented mRNA levels of eosinophil major basic protein (MBP) in myeloid cells induced with G-CSF to undergo terminal neutrophilic differentiation. MBP is a cytotoxic protein that is abundantly expressed in eosinophils, but not in neutrophils. Ectopic expression of MBP inhibited the proliferation and survival of differentiating myeloid cells in response to G-CSF. Significantly, while GFI-1 is upregulated during neutrophilic differentiation, it is rapidly downregulated upon induction of eosinophilic differentiation, which was associated with increased MBP expression. Knockdown of GFI-1 in eosinophilic cells also led to increased level of MBP mRNA. These results indicate that Gfi-1 functions to inhibit the expression of MBP and aberrant expression of MBP as a result of loss of Gfi-1 function may cause premature apoptosis of differentiating neutrophils. In contrast, the rapid downregulation of Gfi-1 during eosinophilic development may allow for abundant expression of MBP, a hallmark of eosinophilic differentiation.
Keywords: Granulopoiesis, Apoptosis, Gfi-1, Eosinophil major basic protein, G-CSF
Introduction
The nuclear zinc finger protein Gfi-1 functions mainly as a transcriptional repressor that regulates the development of multiple hematopoietic lineages, including T and B cells, granulocytes, macrophages and dendritic cells (1–3). Gfi-1 has also been implicated in the regulation of hematopoietic stem cell (HSC) self-renewal and survival (4–6). The essential role of Gfi-1 in granulopoiesis was uncovered by targeted gene knockout studies showing that Gfi-1−/− mice lacked mature neutrophils and exhibited an expansion of immature monocyte-like myeloid cells in the bone marrow (BM) and peripheral blood (7–9). Myeloid precursors from Gfi-1−/− mice are unable to differentiate into mature neutrophils in vitro, but instead give rise to atypical monocytes. In contrast, overexpression of Gfi-1 accelerates neutrophilic differentiation at the expense of monocytic differentiation (10, 11). These data reveal a critical role of Gfi-1 during early myeloid development i.e., promotion of neutrophilic differentiation in an instructive manner and suppression of the monocytic development.
Consistent with its role in granulopoiesis, mutations in GFI-1 have been identified in patients with severe congenital neutropenia (SCN) (12), a condition characterized by extremely low neutrophil counts due to a maturation arrest of BM myeloid progenitor cells. One such GFI-1 mutant, N382S, carries a substitution of serine (S) for the asparagine (N) at position 382 in the fifth zinc finger (ZF) domain, which abolishes the DNA binding ability of GFI-1. The N382S mutant has been shown to act in a dominant negative (DN) manner to inhibit the function of wild type GFI-1 (10, 12). Expression of the DN N382S mutant results in premature apoptosis of differentiating myeloid 32D cells and a block in granulopoiesis with augmented monocytic development and a modest increase in cell death in moue Lin- BM cells (10, 13).
Gfi-1 contains an N-terminal SNAG domain required for transcriptional repression and 6 C-terminal ZF domains involved in DNA binding and interacting with other transcription factors. Gfi-1 represses transcription by binding to specific DNA elements containing the AATC core sequences in the promoters of its target genes (14), including Cebpe and Elane (Ela2) encoding NE (13, 15, 16). Gfi-1 also directly represses CSF1, a key positive regulator of monocytic development, such that CSF1 expression is markedly enhanced in the mouse BM cells deficient for Gfi-1 or expressing the DN N382S mutant, which appears to be responsible for the expansion of atypical monocytes in Gfi-1 −/− mice (10). However, while the expression of NE, an early marker of neutrophilic differentiation, is increased in Gfi-1−/− BM cells, the level of C/EBPε is not or barely increased in Gfi-1−/− or N382S-expressing BM cells (8, 10), likely due to the fact that Gfi-1−/− or N382S-expressing myeloid progenitors are blocked at the early stage of neutrophilic differentiation whereas C/EBPε expression is induced only at the late stage of differentiation. Thus, the role of Gfi-1 in late neutrophilic development is still poorly understood.
In the present study, we show that Gfi-1 inhibits the expression of eosinophil major basic protein (MBP), an eosinophil secondary granule protein that is expressed abundantly in eosinophils and, to a lesser extent in basophils (17). The level of MBP mRNA was dramatically induced by G-CSF in myeloid cells expressing the DN N382S mutant of Gfi-1. Overexpression of MBP in myeloid cells caused accelerated apoptosis during G-CSF-induced terminal neutrophilic differentiation. We also show that GFI-1 was rapidly downregulated upon induction of eosinophilic differentiation, which was associated with increased MBP expression. Knockdown of GFI-1 also led to increased level of MBP mRNA in eosinophilic cells. These data indicate that Gfi-1 may play an important role in late neutrophilic differentiation, but may be negatively involved in eosinophilic development.
Materials and Methods
Cells
Murine myeloid 32D/GR and L-G cells stably transfected with Gfi1 or the N382S mutant have been described (18). Cells were grown in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS), 10% WEHI-3B cell conditioned media as a crude source of interleukin-3 (IL-3), 100 μg/ml penicillin and 100 μg/ml streptomycin. The human myeloid leukemic HL-60 clone 15 cells were purchased from American Type Culture Collection (ATCC) and maintained in RPMI 1640 medium supplemented with 10% FBS and antibiotics. Cells were grown in humidified incubator with 37 °C with 5% CO2.
Expression plasmid and stable transfection
The cDNA encoding murine MBP was amplified by reverse transcriptase polymerase chain reaction (RT-PCR) from G-CSF-stimulated 32D/N382S cells. The MBP cDNA was sequenced to verify the authenticity and inserted into the GFP-RV vector, which contains an internal ribosomal entry sequence (IRES) and the cDNA encoding the humanized green fluorescent protein (GFP). The resultant expression construct (GFP-RV-MBP) were electroporated into 32D/GR cells along with pBabe-puro plasmid. Cells were selected in 1 μg/ml puromycin and the surviving cells were subcloned by limiting dilution. GFP-positive clones were expanded and expression of MBP was confirmed by RT-PCR.
For inducible expression of Gfi-1 and the N382S mutant in 32D/GR cells, the expression constructs Gfi-1-RV and N382S-RV (13) were digested with Xba I and Bam HI. The DNA fragment of approximately 2.8 kb, which contained the Gfi-1 (or N283S) cDNA, was inserted into the expression vector pTRE2hyg (Clontech). 32D/GR cells were electroporated with pUbiq.irtTA (19) encoding the glucocorticoid/tetracycline-inducible transactivator and GFP with an IRES, along with pBabe-puro plasmid. Cells were selected in puromycin and individual clones were obtained. Inducible gene expression upon treatment with 100nM dexamethasone (Dex) and 1 μg/ml doxycyclin (Dox) was evaluated by luciferase reporter assay using the pTRE2Hyg-Luc reporter construct (Clontech). A clone with strong inducible luciferase activity was transfected with pTRE2Hyg-Gfi-1 or pTRE2Hyg-N382S by electroporation. Stable transfectants were obtained after selection in 1 mg/ml hygromycin. Expression of Gfi-1 proteins in the transfectants was examined by Western blotting after Dex/Dox treatment for 24 hours.
Semi-quantitative RT-PCR
Total cellular RNA was isolated using TRIzol reagent (Invitrogen). Two μg of total RNA was reverse transcribed into cDNA using Oligo (dT)-15 primer (Promega) and AMV reverse transcriptase (Promega). Two μl of the reverse transcription reaction was then subjected to PCR amplification using the following primer pairs: 5′-CAAACTTGACAAGACCCAGGA-3′ and 5′-GGACATCTGGCAGGAAAGAA-3′ for mouse MBP; 5′-TTCACCACCATGGAGAAGGC-3′ and 5′-GGCATGGACTGTGGTCATGA-3′ for mouse Gapdh; 5′-GAAGGTCTCTGGGTGGGATA-3′ and 5′-AGGAGAGGGCAGCTCTGAAC-3′ for human MBP; 5′-CGACCACTTTGTCAAGCTCA-3′ and 5′-AGGGGAGATTCAGTGTGGTG-3′ for human GAPDH. PCR products were resolved on 1% agarose gels and visualized by ethidium bromide staining under UV light.
Western blotting analysis
Whole-cell extracts were prepared by lysing the cells in SDS lysing buffer (1% SDS, 50mM Tris-HCl [pH 7.5], 1% PMSF). Samples were boiled in SDS sample buffer, resolved by SDS/PAGE, and transfered to Immobilon membranes. The membranes were incubated with the anti-Gfi1 antibody (N-20) or anti-β-Actin antibody (Santa Cruz, CA), and the reactive proteins were visualized by enhanced chemiluminescence.
RNA interference
Short hairpin RNA (shRNA) against human GFI-1 cloned into pLKO.1 was purchased from Open Biosystems (clone TRCN0000020467). Lentiviral particles were produced by co-transfecting 293T cells with the shRNA construct along with the packaging plasmids (RRE, RSV and VSVG), using TransIT-LT1 transfection reagent (Mirus Bio). Virus-containing supernatants were harvested 24 and 48 hours later. HL-60 clone 15 cells were infected with the viral supernatant in the presence of 8 μg/ml of polybrene and selected in 2 μg/ml puromycin 48 hours after infection.
Apoptosis assay
Apoptosis was examined using the Annexin V-PE apoptosis detection kit (BD Biosciences). Briefly, 0.3 × 106 cells were collected and incubated with Annexin V-PE and 7 amino-actinomycin (7-AAD). Cells were analyzed by two-color flow cytometry using BD Cellquest Pro software (Becton Dickinson, San Jose, CA).
Results
Expression of Gfi-1 N382S in 32D/GR and L-G cells leads to up-regulation of MBP mRNA in response to G-CSF stimulation
Murine myeloid 32D/GR and L-G cells are committed to neutrophilic lineage and differentiate into mature neutrophils in response to G-CSF, thus recapitulating the G-CSF response of normal BM myeloid progenitor cells (13, 20, 21). Both cell lines have been used by others to investigate G-CSF-dependent granulopoiesis (22–26). Unlike Gfi-1−/− or N382S-expressing BM cells, which develop into atypical monocytes under the influence of CSF1 signaling, 32D/GR and L-G showed no response to CSF1 (data not shown), making them uniquely useful for addressing the role of Gfi-1 in late granulocytic development. To identify novel Gfi-1 target genes involved in G-CSF-induced neutrophilic differentiation, we compare the gene expression profiles in 32D/GR cells transfected with the empty vector (32D/Ctr) or the Gfi-1 N382S mutant (32D/N382S) using the Affymetrix oligonucleotide microarray analysis. Cells were treated with G-CSF for 24 hours prior to extraction of total RNA for gene expression profiling. The expression of a number of genes was markedly increased in 32D/N382S cells as compared with 32D/Ctr cells (Table 1).
Table 1.
Increased gene expression in 32D/N382S cells in response to G-CSF*
| Gene | Accession No. | Fold Increase (N382S vs Ctr) |
|---|---|---|
| Complement component C3, alpha and beta subunits | K02782.1 | 42.2 |
| Proteoglycan 2, bone marrow | NM_008920.1 | 21.1 |
| Ubiquitin associated domain containing 2 | BC024467.1 | 10.6 |
| Immunoglobulin superfamily, member 6 | NM_030691.1 | 9.8 |
| MHC class I cell surface glycoprotein | L22338.1 | 8.6 |
| Procollagen, type V, alpha 1 | NM_015734.1 | 5.7 |
| Microsomal glutathione S-transferase 1 | BC009155.1 | 4.9 |
| Phosphatidylinositol 3-kinase catalytic delta polypeptide | NM_008840.1 | 4.9 |
| Potassium channel beta 2 subunit | U31908.1 | 4.6 |
| Isocitrate dehydrogenase 1 (NADP+) | NM_010497.1 | 4.6 |
| Eosinophil peroxidase | NM_007946.1 | 4.3 |
Genes showing at least four-fold increase in expression are listed.
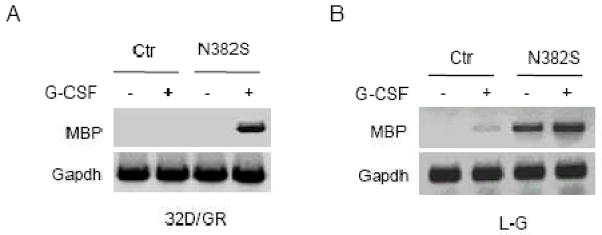
We previously showed that forced expression of the N382S mutant caused premature apoptosis and inhibited the proliferation of differentiating 32D/GR cells in response to G-CSF (13). As MBP (also called bone marrow proteoglycan 2) is a cytotoxic protein, we further examined MBP expression by semi-quantitative RT-PCR assay. As shown in Fig. 1A, MBP mRNA was not detected in 32D/Ctr and 32D/N382S cells cultured in IL-3-containing medium. Upon G-CSF stimulation, MBP mRNA was markedly induced in 32D/N382S cells, but still undetectable in 32D/Ctr cells. To further demonstrate the effect of expression of the N382S mutant, we examined MBP mRNA levels in L-G cells stably transfected with the N382S mutant (13). MBP mRNA was undetectable in L-G cells growing in IL-3-containing medium, but weakly induced by G-CSF (Fig. 1B). Significantly, MBP mRNA level was markedly augmented in L-G/N382S cells even when the cells were maintained in IL-3-containing medium and was further induced by G-CSF. Thus, suppression of Gfi-1 function by the DN N382S mutant led to aberrant expression of MBP in myeloid cells that are committed to neutrophils.
Fig. 1.
MBP is up-regulated in response to G-CSF in murine myeloid cells expressing the Gfi-1 N382S mutant. 32D/GR (A) and L-G cells (B) transfected with the empty plasmid (Ctr) or the N382S mutant were left untreated or treated with G-CSF for 24 hours. The expression of MBP was evaluated by semi-quantitative RT-PCR and compared with that of GAPDH.
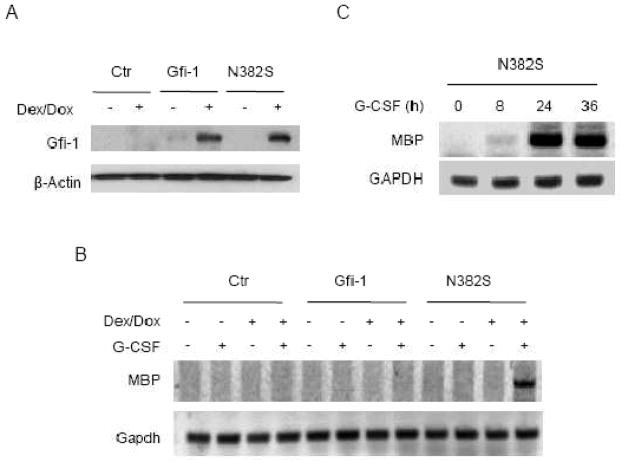
To demonstrate that the augmented levels of MBP mRNA resulted directly from the expression of the N382S mutant, we established 32D/GR cells in which the expression of Gfi-1 or the N382S was induced by addition of dexamethasone (Dex) and doxycycline (Dox). As shown in Fig. 2A, Gfi-1 proteins were barely detectable in 32D/GR cells in the absence of Dex/Dox, but induced upon Dex/Dox treatment for 24 hours. Cells were subsequently treated with G-CSF for 24 hours and then examined for MBP expression. The level of MBP mRNA was upregulated only in G-CSF-treated 32D/N382S cells that had been induced to express the N382S mutant, but not Gfi-1 (Fig. 2B). To examine the time course of MBP induction by G-CSF, we treated 32D/N382S cells with Dex/Dox for 24 hours, followed by G-CSF stimulation for different times as indicated (Fig. 2C). MBP mRNA was detected as early as 8 hours and reached maximal level 24 hours after G-CSF stimulation in 32D/N382S cells. These findings indicated that repression of G-CSF-induced MBP expression by Gfi-1 is dependent on its DNA- binding activity.
Fig. 2.
MPB is induced by G-CSF upon conditional expression of the N382S mutant. 32D/GR cells transfected with the empty vector (Ctr) or the Dex/Dox-responsive expression construct for Gfi-1 or the N382S were treated with Dex/Dox for 24 hours. (A) The expression of Gfi-1 proteins was examined by Western blot analysis. (B) Cells were subsequently stimulated with G-CSF for 24 hours prior to evaluation of MBP expression by semi-quantitative RT-PCR. (C) 32D/GR cells conditionally expressing the N382S mutant were left untreated or treated with Dex/Dox for 24 hour, followed by G-CSF stimulation for the indicated times. MBP expression was analyzed by semi-quantitative RT-PCR.
Ectopic expression of MBP inhibited cell proliferation and accelerated apoptosis in response to G-CSF
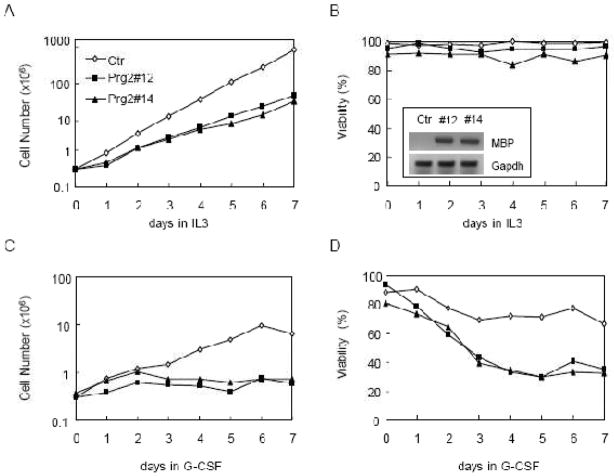
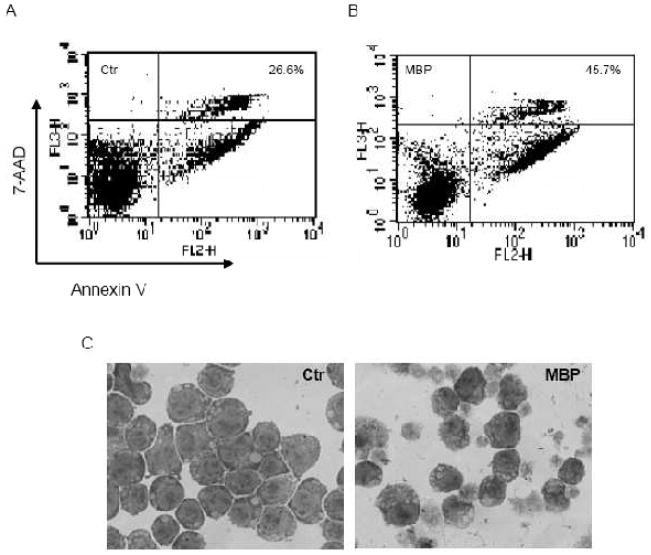
To examine the effect of MBP overexpression on cell proliferation and survival, we cloned the cDNA encoding MBP from G-CSF-treated 32D/N382S cells by RT-PCR. The MBP cDNA was inserted into GFP-RV retroviral vector and then stably transfected into 32D/GR cells. GFP-expressing cells were sorted and examined for expression of MBP mRNA by semi-quantitative RT-PCR (Fig. 3B). Control cells transfected with the empty vector and the two MBP-expressing clones (32D/MBP) were subsequently cultured in IL-3- or G-CSF-containing medium, and cell numbers and viabilities were monitored for 7 days. Ectopic expression of MBP significantly slowed down IL-3-dependent proliferation (Fig. 3A), but had no significant effect on cell viability (Fig. 3B). Notably, ectopic MBP expression blocked G-CSF-dependent proliferation and also resulted in significantly reduced cell viabilities (Fig. 3C and D). We further performed the Annexin V assays to address whether the reduced viabilities of MBP-expressing cells in response to G-CSF was due to accelerated apoptosis. Cells were treated with G-CSF for 4 days prior to the Annexin V assays. As shown in Fig. 4A and B, considerable more MBP-expressing cells stained positive for Annexin V as compared to control cells. Morphological examination of 32D/MBP cells cultured in G-CSF for 4 days revealed that a large proportion of cells displayed the features characteristic of apoptosis including shrinkage of cytoplasm and condensation of nuclei. Together, these results indicated that MBP inhibited cell proliferation and accelerated apoptosis in differentiating 32D/GR cells. Thus, aberrant expression of MBP may contribute to the premature apoptosis of 32D/N382S cells induced to differentiate with G-CSF.
Fig. 3.
Ectopic expression of MBP inhibits cell proliferation and survival. 32D/GR cells were transfected with the empty vector (Ctr) or MBP. Control cells and two independent clones (#12 and #14) were subsequently cultured in IL-3 or G-CSF as indicated. Cell numbers (A and C) and viabilities (B and D) were determined by trypan blue exclusion on the daily basis for 7 days. The expression of MBP in the two clones was confirmed by semi-quantitative RT-PCR (inset in B).
Fig. 4.
Ectopic expression of MBP results in accelerated apoptosis in response to G-CSF. Control (A) and MBP-expressing (B; pooled clones 12 and 14) 32D/GR cells were cultured in G-CSF for 4 days and then stained for annexin V and 7AAD. The percentages of annexin V-positive (early and late apoptotic) cells are indicated. (C) Morphological features of 32D/Ctr and 32D/MBP cells cultured in G-CSF for 4 days. Wright-Giemsa Stain; original magnification × 600.
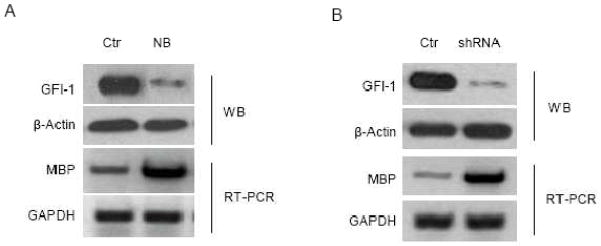
Gfi-1 negatively regulated MBP expression in eosinophilic cells
MBP is abundantly expressed in eosinophils and accounts for 50% of total granule protein (17). If Gfi-1 inhibits MBP expression, we speculated that Gfi-1 expression might be downregulated during eosinophilic differentiation. The HL-60 clone 15 cell line is an eosinophil-committed subline derived from human promyelocytic leukemia cell line HL-60 and has been shown to differentiate into mature eosinophils upon stimulation with IL-5 or n-butyrate (27, 28). Notably, the MBP protein level in HL-60 clone 15 cells increased during eosinophil differentiation (28, 29). We treated HL-60 clone 15 cells with n-butyrate to induce eosinophilic differentiation. Consistent with previous studies, MBP level was upregulated by n-butyrate treatment (Fig. 5A). Interestingly, in contrast to MBP, the expression of GFI-1 was markedly downregulated. To directly show that GFI-1 downregualtion led to increased expression of MBP, we knocked down GFI-1 expression in HL-60 clone 15 cells through lentivirus-mediated delivery of shRNAs against GFI-1. As shown in Figure 5B, GFI-1 knockdown was associated with dramatically increased MBP mRNA level in HL60 clone 15 cells. These data implicate GFI-1 as a critical negative regulator of MBP expression in eosinophils and suggest that GFI-1 downregulation during eosinophilic differentiation is required for the abundant MBP expression in mature eosinophils.
Fig. 5.
GFI-1 downregulation is associated with increased expression of MBP. (A) HL-60 clone 15 cells were left untreated (Ctr) or treated with 500 μM n-butyrate (NB) for 24 hours. (B) HL-60 clone 15 cells were infected with the empty lentivirus (Ctr) or the lentivirus containing the shRNA against GFI-1. The expression of GFI-1 and MBP was examined by Western blotting and semi-quantitative RT-PCR, respectively.
Discussion
A number of studies have demonstrated a critical role of Gfi-1 in early granulocytic differentiation. Gfi-1−/− mice lack mature neutrophils and have increased numbers of atypical myeloid cells with the characteristics of monocytic lineage (7–9). Murine BM cells deficient for Gfi-1 or transduced with the DN Gfi-1 N382S are unable to form granulocytic colonies, but instead give rise to monocytic colonies (8, 10). Monocytic development is favored over granulocytic differentiation in Gfi-1−/− and N382S-transduced cells at least in part because Gfi-1-mediaed transcriptional repression of CSF1 is lost in these cells, resulting in increased expression of CSF1 and CSF1R (8, 10). In contrast, forced expression of Gfi-1 promotes granulopoiesis and suppresses monocytic development (8, 10). These data indicate that Gfi-1 instructs neutrophil fate choice at the expense of monocytic development.
Since Gfi-1−/− BM cells fail to undergo terminal neutrophilic differentiation, the role of Gfi-1 in late granulocytic development remains poorly understood. We previously showed that forced expression of the N382S mutant resulted in increased expression of C/EBPε in 32D/GR and L-G cells induced to differentiate with G-CSF (13). Elevated levels of C/EBPε, which is normally expressed in late granulocytes, were also observed in differentiating myeloid cells derived from Gfi-1+/− mouse BM cells and in neutrophils of a patient with specific granule deficiency (SGD) that expressed low level of Gfi-1 (16). However, C/EBPε expression is barely augmented in Gfi-1−/− or N382S-expressing mouse BM cells as these cells are blocked at early stage of granulocytic differentiation and develop into atypical monocytes under the influence of enhanced CSF1 signaling (8, 10). Thus, 32D/GR and L-G cells are uniquely useful for addressing the role of Gfi-1 in late granulocytic development because they are committed to neutrophilic differentiation and show no CSF1 response.
In this paper, we have demonstrated that Gfi-1 functions to inhibit MBP expression in myeloid cells. Forced expression of the DN Gfi-1 N382S mutant, defective in DNA binding, leads to the dramatically increased level of MBP mRNA upon G-CSF stimulation in 32D/GR cells. In L-G cells, MBP expression is upregulated by the N382S mutant even in the absence of G-CSF. These data suggest that Gfi-1-mediated inhibition of MBP expression is dependent on direct DNA binding. In luciferase reporter assays, however, we observed no significant effects of expression of Gfi-1 or the N382S mutant on the activity of an approximate 2-kb promoter fragment of Prg-2, which encodes MBP (data not shown). Thus, it remains to be determined as to whether Gfi-1 directly or indirectly inhibits MBP expression.
As the most abundant eosinophil cationic protein, MBP is cytotoxic to mammalian cells (17, 30). We previously showed that the N382S mutant induced premature apoptosis of differentiating 32D/GR and L-G cells. In this paper, we have shown that ectopic expression of MBP in 32D/GR cells inhibits cell proliferation and accelerates apoptosis during the process of G-CSF-induced terminal neutrophilic differentiation. Notably, MBP also inhibits IL-3-dependent cell proliferation, but has no significant effect on survival (Fig. 3A and B). The mechanism by which MBP inhibits IL-3-stimulated proliferation remains to be determined; however, the possibility cannot be excluded that MBP overexpression may slightly inhibit cell survival leading to reduced cell number increase, but the dead cells were phagocytosed by the living cells and therefore did not significantly affect cell viability. Irrespectively, our data strongly suggest that MBP upregulation contributes to the inhibitory effect of the N382S mutant on the proliferation and survival of differentiating myeloid cells. It is also of note that the N382S mutant appears to have a more profound inhibitory effect on G-CSF-dependent cell survival than MBP does (13), which is likely because in addition to upregulation of MBP, the N382S mutant causes deregulated activation of other Gfi-1 target genes including Cebpe and Elane (12).
The expression of Gfi-1 is increased dramatically with terminal neutrophilic differentiation (13, 31). Our data suggest that an important function of Gfi-1 during late neutrophilic development is to prevent the inappropriate activation of certain myeloid-specific genes. Aberrant activation of these genes such as Prg2, Cebpe and Elane may have a detrimental effect on cell survival and proliferation in the committed neutrophilic lineage. Notably, even under the influence of enhanced CSF1 signaling, N382S-expressing mouse BM cells showed a modest increase in apoptosis after culture in G-CSF for 3 to 4 days (10). Thus, Gfi-1 not only directs neutrophil fate choice, but also may play a critical role in late neutrophilic development.
In contrast to neutrophilic differentiation, the expression of Gfi-1 was rapidly downregulated during n-butyrate-induced eosinophil differentiation in HL-60 clone 15 cells, which is associated with increased MBP expression. Furthermore, knockdown of Gfi-1 using the Gfi-1 shRNA leads to elevated expression of MBP. These data suggest that Gfi-1 downregulation may allow for MBP upregulation in differentiating eosinophils. MBP is toxic to protozoa, helminthes and bacteria and is, therefore, essential for the function of eosinophils in defense against parasitic infection and in allergic responses (32). As elevated expression of MBP is a major hallmark of eosinophilic differentiation (33, 34), it is possible that Gfi-1 downregulation may be required for normal eosinophilic differentiation. In this aspect, it is of note that Gfi-1−/− mice, while lacking mature neutrophils, have normal eosinophil counts, indicating that Gfi-1 is not required for eosinophil differentiation (8). Further studies are needed to address whether Gfi1 downregulation is essential for eosinophilic development.
Acknowledgments
This work was supported in part by grants RSG-08-307-01 (FD) from The American Cancer Society and R15HL091511 (FD) from the National Institutes of Health.
Footnotes
Conflict of interest
The authors declare that they have no conflict of interest.
References
- 1.Moroy T. The zinc finger transcription factor Growth factor independence 1 (Gfi1) Int J Biochem Cell Biol. 2005;37(3):541–6. doi: 10.1016/j.biocel.2004.08.011. [DOI] [PubMed] [Google Scholar]
- 2.Kazanjian A, Gross EA, Grimes HL. The growth factor independence-1 transcription factor: new functions and new insights. Crit Rev Oncol Hematol. 2006;59(2):85–97. doi: 10.1016/j.critrevonc.2006.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van der Meer LT, Jansen JH, van der Reijden BA. Gfi1 and Gfi1b: key regulators of hematopoiesis. Leukemia. 2010;24(11):1834–43. doi: 10.1038/leu.2010.195. [DOI] [PubMed] [Google Scholar]
- 4.Zeng H, Yucel R, Kosan C, Klein-Hitpass L, Moroy T. Transcription factor Gfi1 regulates self-renewal and engraftment of hematopoietic stem cells. EMBO J. 2004;23(20):4116–25. doi: 10.1038/sj.emboj.7600419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hock H, Hamblen MJ, Rooke HM, et al. Gfi-1 restricts proliferation and preserves functional integrity of haematopoietic stem cells. Natue. 2004;431(7011):1002–7. doi: 10.1038/nature02994. [DOI] [PubMed] [Google Scholar]
- 6.Khandanpour C, Kosan C, Gaudreau MC, et al. Growth Factor Independence 1 (Gfi1) Protects Hematopoietic Stem Cells Against Apoptosis But Also Prevents the Development of a Myeloproliferative-Like Disease. Stem Cells. 2011;29(2):376–85. doi: 10.1002/stem.575. [DOI] [PubMed] [Google Scholar]
- 7.Karsunky H, Zeng H, Schmidt T, et al. Inflammatory reactions and severe neutropenia in mice lacking the transcriptional repressor Gfi1. Nat Genet. 2002;30(3):295–300. doi: 10.1038/ng831. [DOI] [PubMed] [Google Scholar]
- 8.Hock H, Hamblen MJ, Rooke HM, et al. Intrinsic requirement for zinc finger transcription factor Gfi-1 in neutrophil differentiation. Immunity. 2003;18(1):109–20. doi: 10.1016/s1074-7613(02)00501-0. [DOI] [PubMed] [Google Scholar]
- 9.Zhu J, Jankovic D, Grinberg A, Guo L, Paul WE. Gfi-1 plays an important role in IL-2-mediated Th2 cell expansion. Proc Natl Acad Sci U S A. 2006;103(48):18214–9. doi: 10.1073/pnas.0608981103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zarebski A, Velu CS, Baktula AM, et al. Mutations in growth factor independent-1 associated with human neutropenia block murine granulopoiesis through colony stimulating factor-1. Immunity. 2008;28(3):370–80. doi: 10.1016/j.immuni.2007.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de la Luz Sierra M, Sakakibara S, Gasperini P, et al. The transcription factor Gfi1 regulates G-CSF signaling and neutrophil development through the Ras activator RasGRP1. Blood. 2010;115(19):3970–9. doi: 10.1182/blood-2009-10-246967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Person RE, Li FQ, Duan Z, et al. Mutations in proto-oncogene GFI1 cause human neutropenia and target ELA2. Nat Genet. 2003;34(3):308–12. doi: 10.1038/ng1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhuang D, Qiu Y, Kogan SC, Dong F. Increased C/EBPepsilon expression and premature apoptosis in myeloid cells expressing Gfi-1 N382S mutant associated with severe congenital neutropenia. J Biol Chem. 2006;281(16):10745–51. doi: 10.1074/jbc.M510924200. [DOI] [PubMed] [Google Scholar]
- 14.Zweidler-Mckay PA, Grimes HL, Flubacher MM, Tsichlis PN. Gfi-1 encodes a nuclear zinc finger protein that binds DNA and functions as a transcriptional repressor. Mol Cell Biol. 1996;16(8):4024–34. doi: 10.1128/mcb.16.8.4024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duan Z, Zarebski A, Montoya-Durango D, Grimes HL, Horwitz M. Gfi1 coordinates epigenetic repression of p21Cip/WAF1 by recruitment of histone lysine methyltransferase G9a and histone deacetylase 1. Mol Cell Biol. 2005;25(23):10338–51. doi: 10.1128/MCB.25.23.10338-10351.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khanna-Gupta A, Sun H, Zibello T, et al. Growth factor independence-1 (Gfi-1) plays a role in mediating specific granule deficiency (SGD) in a patient lacking a gene-inactivating mutation in the C/EBPepsilon gene. Blood. 2007;109(10):4181–90. doi: 10.1182/blood-2005-05-022004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Plager DA, Weiler DA, Loegering DA, et al. Comparative structure, proximal promoter elements, and chromosome location of the human eosinophil major basic protein genes. Genomics. 2001;71(3):271–81. doi: 10.1006/geno.2000.6391. [DOI] [PubMed] [Google Scholar]
- 18.Zhuang D, Qiu Y, Haque SJ, Dong F. Tyrosine 729 of the G-CSF receptor controls the duration of receptor signaling: involvement of SOCS3 and SOCS1. J Leukoc Biol. 2005;78(4):1008–15. doi: 10.1189/jlb.0105032. [DOI] [PubMed] [Google Scholar]
- 19.Anastassiadis K, Kim J, Daigle N, Sprengel R, Scholer HR, Stewart AF. A predictable ligand regulated expression strategy for stably integrated transgenes in mammalian cells in culture. Gene. 2002;298(2):159–72. doi: 10.1016/s0378-1119(02)00979-4. [DOI] [PubMed] [Google Scholar]
- 20.Dong F, Brynes RK, Tidow N, Welte K, Lowenberg B, Touw IP. Mutations in the gene for the granulocyte colony-stimulating-factor receptor in patients with acute myeloid leukemia preceded by severe congenital neutropenia. N Engl J Med. 1995;333(8):487–93. doi: 10.1056/NEJM199508243330804. [DOI] [PubMed] [Google Scholar]
- 21.Lee KH, Kinashi T, Tohyama K, et al. Different stromal cell lines support lineage-selective differentiation of the multipotential bone marrow stem cell clone LyD9. J Exp Med. 1991;173(5):1257–66. doi: 10.1084/jem.173.5.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ward AC, Smith L, de Koning JP, van Aesch Y, Touw IP. Multiple signals mediate proliferation, differentiation, and survival from the granulocyte colony-stimulating factor receptor in myeloid 32D cells. J Biol Chem. 1999;274(21):14956–62. doi: 10.1074/jbc.274.21.14956. [DOI] [PubMed] [Google Scholar]
- 23.Kroll SL, Barth-Baus D, Hensold JO. The carboxyl-terminal domain of the granulocyte colony-stimulating factor receptor uncouples ribosomal biogenesis from cell cycle progression in differentiating 32D myeloid cells. J Biol Chem. 2001;276(52):49410–8. doi: 10.1074/jbc.M109577200. [DOI] [PubMed] [Google Scholar]
- 24.van de Geijn GJ, Gits J, Touw IP. Distinct activities of suppressor of cytokine signaling (SOCS) proteins and involvement of the SOCS box in controlling G-CSF signaling. J Leukoc Biol. 2004;76(1):237–44. doi: 10.1189/jlb.0104041. [DOI] [PubMed] [Google Scholar]
- 25.Kitabayashi I, Yokoyama A, Shimizu K, Ohki M. Interaction and functional cooperation of the leukemia-associated factors AML1 and p300 in myeloid cell differentiation. Embo J. 1998;17(11):2994–3004. doi: 10.1093/emboj/17.11.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shimizu K, Kitabayashi I, Kamada N, et al. AML1-MTG8 leukemic protein induces the expression of granulocyte colony-stimulating factor (G-CSF) receptor through the up-regulation of CCAAT/enhancer binding protein epsilon. Blood. 2000;96(1):288–96. [PubMed] [Google Scholar]
- 27.Fischkoff SA. Graded increase in probability of eosinophilic differentiation of HL-60 promyelocytic leukemia cells induced by culture under alkaline conditions. Leuk Res. 1988;12(8):679–86. doi: 10.1016/0145-2126(88)90103-8. [DOI] [PubMed] [Google Scholar]
- 28.Ishihara K, Hong J, Zee O, Ohuchi K. Possible mechanism of action of the histone deacetylase inhibitors for the induction of differentiation of HL-60 clone 15 cells into eosinophils. Br J Pharmacol. 2004;142(6):1020–30. doi: 10.1038/sj.bjp.0705869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fischkoff SA, Pollak A, Gleich GJ, Testa JR, Misawa S, Reber TJ. Eosinophilic differentiation of the human promyelocytic leukemia cell line, HL-60. J Exp Med. 1984;160(1):179–96. doi: 10.1084/jem.160.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weyer K, Glerup S. Placental Regulation of Peptide Hormone and Growth Factor Activity by proMBP. Biol Reprod. 2011;84(6):1077–86. doi: 10.1095/biolreprod.110.090209. [DOI] [PubMed] [Google Scholar]
- 31.De La Luz Sierra M, Gasperini P, McCormick PJ, Zhu J, Tosato G. Transcription factor Gfi-1 induced by G-CSF is a negative regulator of CXCR4 in myeloid cells. Blood. 2007;110(7):2276–85. doi: 10.1182/blood-2007-03-081448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Plager DA, Adolphson CR, Gleich GJ. A novel human homolog of eosinophil major basic protein. Immunol Rev. 2001;179:192–202. doi: 10.1034/j.1600-065x.2001.790119.x. [DOI] [PubMed] [Google Scholar]
- 33.Popken-Harris P, Checkel J, Loegering D, et al. Regulation and processing of a precursor form of eosinophil granule major basic protein (ProMBP) in differentiating eosinophils. Blood. 1998;92(2):623–31. [PubMed] [Google Scholar]
- 34.Ishihara K, Satoh I, Mue S, Ohuchi K. Generation of rat eosinophils by recombinant rat interleukin-5 in vitro and in vivo. Biochim Biophys Acta. 2000;1501(1):25–32. doi: 10.1016/s0925-4439(00)00002-8. [DOI] [PubMed] [Google Scholar]