Abstract
Nicotine metabolism and genetic variation have an impact on nicotine addiction and smoking abstinence, but further research is required. The nicotine metabolite ratio (NMR) is a robust biomarker of nicotine metabolism used to categorize slow and normal nicotine metabolizers (lower 25th quartile cutoff). In two randomized clinical trials of smoking abstinence treatments, we conducted NMR-stratified analyses on smoking abstinence across 13 regions coding for nicotinic acetylcholine receptors and proteins involved in the dopamine reward system. Gene × NMR interaction P-values were adjusted for multiple correlated tests, and we used a Bonferroni-corrected α-level of 0.004 to determine system-wide significance. Three SNPs in DRD1 (rs11746641, rs2168631, rs11749035) had significant interactions (0.001 ≤ adjusted P-values ≤ 0.004), with increased odds of abstinence within slow metabolizers (ORs=3.1–3.5, 95% CI 1.7–6.7). Our findings support the role of DRD1 in nicotine dependence, and identify genetic and nicotine metabolism profiles that may interact to impact nicotine dependence.
Keywords: Genetic association studies, heterogeneity, smoking abstinence, nicotine metabolism, nicotine metabolite ratio, DRD1
Introduction
Nicotine addiction is a persistent global public health issue with long-term quitting success achieved by only a small percentage of smokers [1]. Research continues to identify and characterize genetic markers and biological processes that impact nicotine addiction and smoking abstinence.
The cytochrome P450 enzyme (CYP2A6) converts 80–90% of nicotine to cotinine. CYP2A6 subsequently metabolizes cotinine to 3-hydroxycotinine (3-HC). Nicotine metabolite ratio (NMR), the ratio of 3-HC to cotinine, is a stable phenotypic marker of nicotine metabolism [2] and variation of NMR has been shown to be related to genetic variation in the CYP2A6 gene [2] and to factors such as sex and hormone levels that alter nicotine metabolic rate [3].
The nicotinic acetylcholine receptor (nAChR) gene regions and genes involved in the dopamine reward system have been associated with nicotine dependence [4–6]. The nAChRs influence the dopamine pathways [7], and nicotine stimulates dopamine release in the brain reward circuits [8]. A previous study did not find an association between smoking rate and the interaction between the chr15q25.1 nAChR region and NMR [9]. However, their independent associations make the interplay between nicotine metabolism and genetic variants involved in nAChR signaling and dopamine transmission on smoking abstinence of particular interest.
In this study, we assessed the interaction between strata of NMR status (slow metabolizers, lower 25th quartile) and a priori candidate genes in the nicotinic receptor and dopaminergic pathways on smoking abstinence.
Study Sample
We pooled subjects enrolled in two smoking abstinence pharmacogenetic effectiveness trials conducted by the University of Pennsylvania Transdisciplinary Tobacco Use Research Center assessing the efficacy of alternate forms of nicotine replacement (NRT) and bupropion therapy (BUP) [5]. Smokers in the NRT trial were randomized (open-label) to transdermal nicotine (patch) or nicotine nasal spray. The BUP trial was double-blind and randomized, where smokers received placebo or bupropion. The studies had similar designs with subjects recruited using identical methods, making them directly comparable for analysis [5].
After applying exclusion criteria, 1,111 subjects consented to treatment and provided a blood sample for genotyping and NMR measurement. We limited analyses to self-identified Caucasians with phenotype and genotype data (N=626) to avoid potential confounding and heterogeneity of effect estimates. Females comprised 51% percent of participants, 46% were college graduates, the mean age was 45 years old (SD=11), the average cigarettes smoked per day was 23 (SD=10), and the mean Fagerström Test for Nicotine Dependence (FTND) [10] score was 5.5 (SD=2.2). Of 626 participants: 164 were slow metabolizers (lower 25th NMR quartile; NMR≤0.28); 462 were normal metabolizers (upper three NMR quartiles; NMR>0.28).
The NRT and BUP trials provided medication and group behavioral smoking abstinence counseling. NRT participants (N=298) began assigned treatments at target quit date (TQD) and continued for 8 weeks. BUP participants (N=318) initiated assigned treatments two weeks prior to TQD for a total of 10 weeks. The primary outcome was biochemically confirmed seven-day point-prevalence abstinence at the end of treatment (EOT), assessed eight weeks post-TQD. Per convention [11], non-abstinent participants reported smoking within seven days prior to EOT, failed to provide a saliva sample, or had carbon monoxide levels >10ppm (NRT) or cotinine levels >15ng/ml (BUP). Among 626 participants, 183 (29%) were abstinent at EOT.
Genotyping procedures
Within a larger candidate gene study, we focused on 13 gene regions: six coding for nicotinic acetylcholine receptors (nAChRs – CHRNA2, CHRNA4, CHRNA5-CHRNA3-CHRNB4, CHRNA7, CHRNB2, CHRNB3-CHRNA6) and seven involved in the dopamine reward system (the dopamine receptor gene family – DRD1, DRD2, DRD3, DRD4, DRD5; catechol-o-methyltransferase COMT, DRD1 interacting protein gene CALCYON). We genotyped 281 SNPs across these genes (44 with a priori putative function, 237 to capture underlying genetic structure) at the University of Southern California Epigenetics Center using the GoldenGate® assay (Illumina, San Diego, CA, USA). Among the 281 SNPs, 12 with genotype call rates <95% were excluded from analyses. One additional SNP deviated significantly from Hardy-Weinberg equilibrium (adjusted threshold P=1.9×10−4) (Supplementary Table 1). Complete SNP selection and quality control procedures have been described previously [4].
SNP Analysis
To estimate NMR-stratified genetic associations with abstinence at EOT, we used logistic regression to obtain odds ratios for marginal SNP effects within strata of slow and normal nicotine metabolizers. For each SNP we tested an additive or dominant genetic model consistent with previously reported analyses. The most common genotype served as the referent. All models were adjusted for gender, age, treatment and FTND. We performed a 1-df likelihood ratio test (LRT) on SNP × NMR interaction terms. Analyses were performed using the R Statistical Program [12].
Correlated tests adjustment and system-level significance
Interaction 1-df LRT P-values were adjusted to account for the correlation and number of tests performed across SNPs within a gene region. Test statistics were modeled as asymptotically distributed multivariate normal with a co-variance structure estimated from the correlation of SNPs [4]. Final observed and adjusted P-values are reported. Overall significance was determined using an additional Bonferroni correction across the 13 gene regions, giving a system-wide α-level of 0.05/13=0.004 [4].
Results
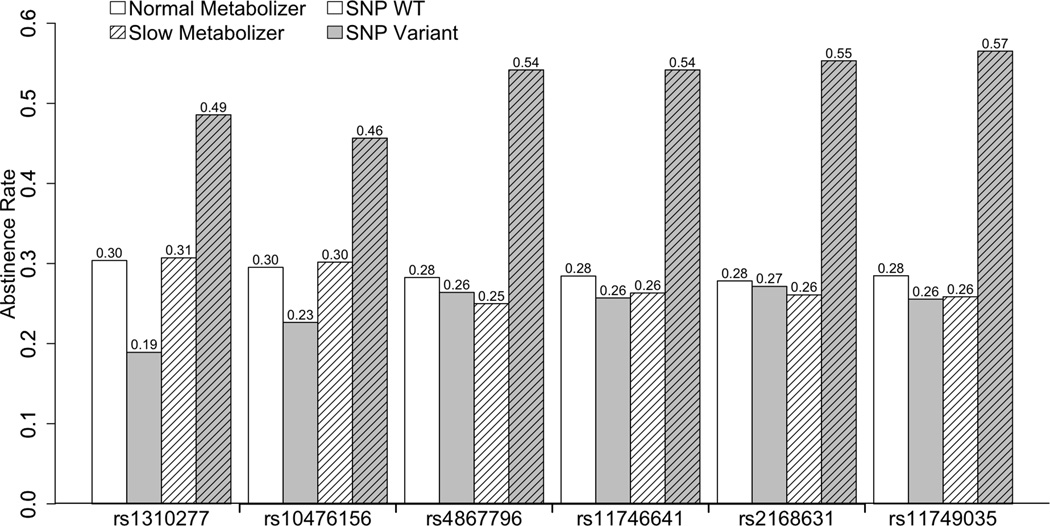
In our study, six SNPs located in DRD1 had significant gene × NMR interactions on abstinence (Table 1). One SNP was located in haplotype block 1 (rs1310277 [merged into dbSNP rs266001], one SNP in block 3 (rs10476156), one SNP between blocks 4 and 5 (rs4867796), and three SNPs in block 5 (rs11746641, rs2168631, rs11749035; r2≥0.9) (Supplementary Figure 1). The interactions for the three SNPs located in block 5 achieved system-wide significance (0.001≤adjusted P-values≤0.004). Within slow metabolizers, the minor allele was associated with increased odds of abstinence (OR=3.1–3.5, 95% CI 1.7–6.7), but that association was null within normal metabolizers (OR=0.8–0.9, 95% CI 0.5–1.3). Abstinence rates (Figure 1) reflect these associations, and were higher for slow metabolizers carrying the minor allele for each of these SNPs (46–57%) compared to the other three metabolizer/genotype groups (19–30%).
Table 1.
Interaction between DRD1 and CHRNA5-CHRNA3-CHRNB4 Chromosome 15 nAChR region SNPs and nicotine metabolism rate (slow vs. normal metabolizers) on abstinence at end of treatment.
| Haplotype | Slow Metabolizer | Normal Metabolizer | Interaction P-value | ||
|---|---|---|---|---|---|
| SNP | Block | OR (95% CI)a | OR (95% CI)a | Observed | Adjustedb |
| DRD1 | |||||
| rs1310277 | 1 | 1.98 (1.03 – 3.83) | 0.53 (0.32 – 0.87) | 1.32 × 10−3 | 2.26 × 10−2 |
| rs10476156 | 3 | 2.05 (1.10 – 3.80) | 0.64 (0.41 – 1.00) | 1.25 × 10−3 | 2.22 × 10−2 |
| rs4867796 | b/w 4–5 | 2.75 (1.53 – 4.94) | 0.91 (0.61 – 1.37) | 1.48 × 10−3 | 2.40 × 10−2 |
| rs11746641 | 5 | 3.10 (1.67 – 5.79) | 0.82 (0.53 – 1.25) | 2.06 × 10−4 | 4.05 × 10−3 |
| rs2168631 | 5 | 3.30 (1.75 – 6.22) | 0.87 (0.58 – 1.30) | 1.96 × 10−4 | 3.96 × 10−3 |
| rs11749035 | 5 | 3.49 (1.84 – 6.67) | 0.79 (0.52 – 1.21) | 5.95 × 10−5 | 1.25 × 10−3 |
| CHRNA5-CHRNA3-CHRNB4 | |||||
| rs2036527 | 1 | 1.52 (0.94 – 2.45) | 0.81 (0.59 – 1.11) | 3.88 × 10−2 | 2.85 × 10−1 |
| rs1051730 | 2 | 1.54 (0.95 – 2.48) | 0.85 (0.62 – 1.16) | 4.23 × 10−2 | 2.99 × 10−1 |
| rs1317286 | 2 | 1.50 (0.92 – 2.44) | 0.85 (0.62 – 1.16) | 4.81 × 10−2 | 3.24 × 10−1 |
| rs7178270 | 3 | 0.46 (0.23 – 0.91) | 1.07 (0.68 – 1.68) | 3.37 × 10−2 | 2.57 × 10−1 |
Odds Ratio of quitting associated with minor allele at SNP in specific NMR group
P-value corrected for the correlation structure within respective gene region
Figure 1.
Abstinence rates across different DRD1 genotype groups within slow and normal nicotine metabolizers (N = 164 and 462, respectively).
-
-Normal metabolizers, carrying two major alleles (i.e., WT) – Unlined, White bars
-
-Normal metabolizers, carrying at least one minor allele (i.e., Variant) – Unlined, Grey bars
-
-Slow metabolizers, carrying two major alleles (i.e., WT) – Lined, White bars
-
-Slow metabolizers, carrying at least one minor allele (i.e., Variant) – Lined, Grey bars
Of note are gene × NMR interactions (unadjusted P-values=0.03–0.05) for four SNPs (rs7178270, rs2036527, rs1051730, rs1317286) in the chr15q25.1 CHRNA5-CHRNA3-CHRNB4 nAChR region (Table 1), three of which are in strong LD (rs2036527, rs1051730, rs1317286; r2≥0.9). Although they do not achieve region-wide significance after adjustment for correlated tests, two have strong a priori associations with nicotine dependence (rs1051730 [6], rs1317286 [13]). Within slow metabolizers, the minor alleles for these SNPs are associated with suggestive increases in odds of abstinence (OR=1.5, 95% CI 0.9–2.5), but slight decreases within normal metabolizers (OR=0.85, 95% CI 0.6–1.2). For rs7178270, the minor allele is associated with decreased odds of abstinence within slow metabolizers (OR=0.46, 95% CI 0.2–0.9), but null within normal metabolizers (OR=1.1, 95% CI 0.7–1.7). For the three SNPs in strong LD, the minor allele is associated with a suggestive increase in odds of abstinence within slow metabolizers (OR=1.5, 95% CI 1.0–2.5), but null within normal metabolizers (OR=0.9, 95% CI 0.6–1.2).
Discussion
In summary, six SNPs in DRD1 have significant gene × nicotine metabolism ratio interactions with smoking abstinence at EOT. The minor alleles for these SNPs were associated with significantly increased abstinence rates within slow metabolizers. We find that gene × nicotine metabolism interactions are more strongly associated with smoking abstinence than unstratified gene effects. Prior studies have also shown associations between DRD1 polymorphisms and nicotine dependence [14]. The SNPs in DRD1 that interacted with NMR in our study are neither found in prior nicotine dependence studies nor in LD with SNPs reported in previous studies, but they may be in LD with an undiscovered functional variant for nicotine dependence and D1 dopamine receptor expression.
In the absence of information on the functional consequences of the relevant DRD1 variants, the mechanism of the interaction with the rate of nicotine metabolism is unknown. Nicotine exposure has been shown to upregulate D1 dopamine receptor expression and activity in key brain regions important for nicotine reward [15]. Such upregulation may contribute to the level of nicotine dependence, and the extent of upregulation may be influenced by the rate of nicotine metabolism.
The association for rs1051730 in the chr15q25.1 nAChR region replicates previous findings between this SNP and nicotine dependence. An interaction was reported between rs1051730 and CYP2A6, where cigarette consumption and FTND both increased for those in increasing risk categories (homozygous for the rs1051730 minor allele and/or normal nicotine metabolizers as determined by CYP2A6 genotype) [6]. This is consistent with our finding, where normal nicotine metabolizers as assessed by NMR carrying the rs1051730 minor allele are more likely to relapse.
Strengths and limitations have been described previously [4,5]. Strengths include bias reduction through baseline biomarker measurements and the prospective assessment of abstinence. Also, our P-value adjustment for multiple correlated test is less conservative than a Bonferroni adjustment. However, while gene effects have been shown to differ across treatments [4,5], we lack a sufficient sample to detect small gene × NMR × treatment interaction effects.
In summary, we observe significant gene × NMR interactions in which six DRD1 SNPs are associated with increased odds of smoking abstinence with slow nicotine metabolizers. We also replicate previous findings for an interaction between rs1051730 in the chr15q25.1 nAChR region and the rate of nicotine metabolism. Independent validation of our results is necessary before more conclusions can be made from these findings.
Supplementary Material
Acknowledgements
This research was supported by grants from NIDA, NCI, NIGMS and NHGRI U01 DA020830, NIDA R01 DA002277, and NCI P50 CA084735.
Conflicts of Interest
Dr. Ray has received grant funding from Pfizer and is currently an employee of GlaxoSmithKline Biologicals. Dr. Swan has served as a consultant for Pfrzer. Dr. Lerman has served as a consultant for and/or received research supporl from Pfizer, AstraZeneca, Novartis, Targacept, and Glaxo SmithKline. Dr. Tyndale owns shares and participates in Nicogen Research Inc., a company focused on novel smoking cessation treatment approaches. No Nicogen funds were used in this work and no other Nicogen parlicipants reviewed the manuscript. Dr. Tyndale has also consulted for Novartis and McNeil. Dr. Benowitz is a paid consultant for several pharmaceutical companies that market smoking cessation medications and has been a paid expert witness against tobacco companies in litigation related to nicotine addiction. This research was not supported by industry funds.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lerman CE, Schnoll RA, Munafo MR. Genetics and smoking cessation improving outcomes in smokers at risk. Am J Prev Med. 2007;33:S398–S405. doi: 10.1016/j.amepre.2007.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dempsey D, Tutka P, Jacob P, 3rd, Allen F, Schoedel K, Tyndale RF, et al. Nicotine metabolite ratio as an index of cytochrome P450 2A6 metabolic activity. Clin Pharmacol Ther. 2004;76:64–72. doi: 10.1016/j.clpt.2004.02.011. [DOI] [PubMed] [Google Scholar]
- 3.Benowitz NL, Lessov-Schlaggar CN, Swan GE, Jacob P., 3rd Female sex and oral contraceptive use accelerate nicotine metabolism. Clin Pharmacol Ther. 2006;79:480–488. doi: 10.1016/j.clpt.2006.01.008. [DOI] [PubMed] [Google Scholar]
- 4.Conti DV, Lee W, Li D, Liu J, Van Den Berg D, Thomas PD, et al. Nicotinic acetylcholine receptor beta2 subunit gene implicated in a systems-based candidate gene study of smoking cessation. Hum Mol Genet. 2008;17:2834–2848. doi: 10.1093/hmg/ddn181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lerman C, Jepson C, Wileyto EP, Epstein LH, Rukstalis M, Patterson F, et al. Role of functional genetic variation in the dopamine D2 receptor (DRD2) in response to bupropion and nicotine replacement therapy for tobacco dependence: results of two randomized clinical trials. Neuropsychopharmacology. 2006;31:231–242. doi: 10.1038/sj.npp.1300861. [DOI] [PubMed] [Google Scholar]
- 6.Wassenaar CA, Dong Q, Wei Q, Amos CI, Spitz MR, Tyndale RF. Relationship between CYP2A6 and CHRNA5-CHRNA3-CHRNB4 variation and smoking behaviors and lung cancer risk. J Natl Cancer Inst. 2011;103:1342–1346. doi: 10.1093/jnci/djr237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Livingstone PD, Wonnacott S. Nicotinic acetylcholine receptors and the ascending dopamine pathways. Biochem Pharmacol. 2009;78:744–755. doi: 10.1016/j.bcp.2009.06.004. [DOI] [PubMed] [Google Scholar]
- 8.Hong LE, Gu H, Yang Y, Ross TJ, Salmeron BJ, Buchholz B, et al. Association of nicotine addiction and nicotine's actions with separate cingulate cortex functional circuits. Arch Gen Psychiatry. 2009;66:431–441. doi: 10.1001/archgenpsychiatry.2009.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Falcone M, Jepson C, Benowitz N, Bergen AW, Pinto A, Wileyto EP, et al. Association of the nicotine metabolite ratio and CHRNA5/CHRNA3 polymorphisms with smoking rate among treatment-seeking smokers. Nicotine Tob Res. 2011;13:498–503. doi: 10.1093/ntr/ntr012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heatherton TF, Kozlowski LT, Frecker RC, Fagerstrom KO. The Fagerstrom Test for Nicotine Dependence: a revision of the Fagerstrom Tolerance Questionnaire. Br J Addict. 1991;86:1119–1127. doi: 10.1111/j.1360-0443.1991.tb01879.x. [DOI] [PubMed] [Google Scholar]
- 11.SRNT. Biochemical verification of tobacco use and cessation. Nicotine Tob Res. 2002;4:149–159. doi: 10.1080/14622200210123581. [DOI] [PubMed] [Google Scholar]
- 12.R Development Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing. 2010 [Google Scholar]
- 13.Li MD, Xu Q, Lou XY, Payne TJ, Niu T, Ma JZ. Association and interaction analysis of variants in CHRNA5/CHRNA3/CHRNB4 gene cluster with nicotine dependence in African and European Americans. Am J Med Genet B Neuropsychiatr Genet. 2010;153B:745–756. doi: 10.1002/ajmg.b.31043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang W, Ma JZ, Payne TJ, Beuten J, Dupont RT, Li MD. Significant association of DRD1 with nicotine dependence. Hum Genet. 2008;123:133–140. doi: 10.1007/s00439-007-0453-9. [DOI] [PubMed] [Google Scholar]
- 15.Bahk JY, Li S, Park MS, Kim MO. Dopamine D1 and D2 receptor mRNA up-regulation in the caudate-putamen and nucleus accumbens of rat brains by smoking. Prog Neuropsychopharmacol Biol Psychiatry. 2002;26:1095–1104. doi: 10.1016/s0278-5846(02)00243-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.