Abstract
Recently, deriving candidate endophenotypes from brain imaging data has become a valuable approach to study genetic influences on schizophrenia (SZ), whose pathophysiology remains unclear. In this work we utilized a multivariate approach, parallel independent component analysis, to identify genomic risk components associated with brain function abnormalities in SZ. 5157 candidate single nucleotide polymorphisms (SNPs) were derived from genome-wide array based on their possible connections with SZ and further investigated for their associations with brain activations captured with functional magnetic resonance imaging (fMRI) during a sensorimotor task. Using data from 92 SZ patients and 116 healthy controls, we detected a significant correlation (r= 0.29; p= 2.41×10−5) between one fMRI component and one SNP component, both of which significantly differentiated patients from controls. The fMRI component mainly consisted of precentral and postcentral gyri, the major activated regions in the motor task. On average, higher activation in these regions was observed in participants with higher loadings of the linked SNP component, predominantly contributed to by 253 SNPs. 138 identified SNPs were from known coding regions of 100 unique genes. 31 identified SNPs did not differ between groups, but moderately correlated with some other group-discriminating SNPs, indicating interactions among alleles contributing towards elevated SZ susceptibility. The genes associated with the identified SNPs participated in four neurotransmitter pathways: GABA receptor signaling, dopamine receptor signaling, neuregulin signaling and glutamate receptor signaling. In summary, our work provides further evidence for the complexity of genomic risk to the functional brain abnormality in SZ and suggests a pathological role of interactions between SNPs, genes and multiple neurotransmitter pathways.
Keywords: schizophrenia, fMRI, SNP, parallel-ICA, multivariate
Introduction
Schizophrenia (SZ) is a severe psychiatric disorder demonstrating a strong genetic component with heritability estimated up to 80% based on family and twin studies (Cardno and Gottesman, 2000). In the past decade, a number of susceptibility genes have been identified from linkage and association studies (Duan et al., 2010; Harrison and Owen, 2003). However, the associations between SZ diagnosis and individual polymorphisms were often weak (Duan et al., 2010). These results suggest a polygenic model for SZ (Gottesman and Shields, 1967), hypothesizing that multiple alleles with small individual effects may interact synergistically to increase the susceptibility to the disorder. The hypothesis is supported by a recent study, demonstrating the involvement of an aggregate of common (frequency > 0.05) single nucleotide polymorphisms (SNPs), collectively accounting for a substantial proportion of variation in risk to the disorder (Purcell et al., 2009).
In response to the complex genetic architecture underlying SZ, a multivariate approach is well positioned to examine associations between SZ-related phenotypes and genetic components derived from various potential susceptibility alleles. Prata et al., examining epistasis between DAT and COMT genes, demonstrated that the nonadditive DAT-COMT interaction was associated with a SZ-altered modulation effect on cortical function during executive processing (Prata et al., 2009). Expanding the variables to 24 SNPs spanning 14 SZ putative risk genes, Meda et al. identified two genetic components (DAT and BDNF; SLC6A4, 5HTTLPR and 5HTTLPR_AG) correlating with three functional brain networks; the combined brain function-gene effects showed significant group differences in SZ (Meda et al., 2010). These positive findings have encouraged researchers to explore more genetic influences, including interactions among known risk genes and novel genes.
Simultaneously, an allied line of research has focused on defining endophenotypes given the heterogeneity of symptoms, course and outcome in SZ (Gottesman and Gould, 2003). Some identified endophenotypes are based on magnetic resonance imaging (MRI), which has demonstrated its specific value in identifying regional brain abnormalities (Rapoport et al., 2005), including structural endophenotypes (Lawrie et al., 2003; McDonald et al., 2006; Nelson et al., 1998; Sun et al., 2009) and functional networks recorded in functional MRI (fMRI) that discriminate SZ patients from healthy controls. In particular, a simple motor task has exhibited high efficacy for identifying SZ-related dysfunction in brain motor regions (Schroder et al., 1999; Schroder et al., 1995), which is of great interest given that neuromotor dysfunction is a SZ characteristic preceding the onset of the syndrome (Walker et al., 1994). In addition, the simple motor task was shown to be the most effective paradigm for extracting group-discriminating components in a comparative study with auditory oddball detection (AOD) and Sternberg item recognition paradigms (Kim et al., 2010).
The current study was designed to investigate combined effects of genetic variations on disrupted sensorimotor functional activities related to SZ. We collected genome-wide SNP data and fMRI data during a sensorimotor task and then used parallel independent component analysis (parallel-ICA) (Liu et al., 2008; Liu et al., 2009a) to examine potential associations between the two modalities. Designed based on ICA for multimodality analysis, parallel-ICA extracts components (a weighted combination of all variables) (Bell and Sejnowski, 1995) independently for each modality, and enhances the inter-modality association through a constraint on the correlation. The first application of this method for genetic association studies was on fMRI data derived during an AOD task and SNP data consisting of 384 SNPs spanning 222 genes, by Liu et al. (Liu et al., 2009b), where a brain network containing cuneus and precuneus was identified to be significantly associated with a SNP component predominantly contributed to by 10 SNPs. As a natural extension of the above work, our study broadened the range of examined SNPs to thousands of loci.
Material and Methods
Participants
Participant recruitment and data collection were conducted by The Mind Clinical Imaging consortium (MCIC), a collaborative effort of four research teams from Boston, Iowa, Minnesota and New Mexico. The institutional review board at each site approved the study and all participants provided written informed consents. Prior to inclusion in the study, all healthy participants were screened to ensure that they were free of any medical, neurological or psychiatric illnesses, including any history of substance abuse. The inclusion criteria for patients were based on a diagnosis of schizophrenia, schizophreniform or schizoaffective disorder confirmed by the Structured Clinical Interview for DSM-IV-TR Disorders (First et al., 1997) or the Comprehensive Assessment of Symptoms and History (Andreasen et al., 1992). Antipsychotic history was collected as part of the psychiatric assessment; for details, see SI Materials and Methods. 208 participants (92 patients and 116 controls) who had volunteered for both fMRI scanning and gene genotyping were included in the association study (see Table 1 for demographic information).
Table 1.
Demographic information of participants
| Demographics | SZ (92) | HC (116) | |
|---|---|---|---|
| Sex | Male | 70 | 72 |
| Female | 22 | 44 | |
| Age | Mean ± SD | 34 ± 11 | 32 ± 11 |
| Range | 18–59 | 18–58 | |
| Ethnicity | American Indian or Alaska Native | 1 | 1 |
| Asian | 6 | 5 | |
| African-American | 13 | 4 | |
| Native Hawaiian or Pacific Islander | 0 | 1 | |
| Caucasian | 72 | 105 | |
fMRI data collection and preprocessing
The original fMRI data were collected from 275 participants at four MCIC sites during a sensorimotor task, a block-design motor response to auditory stimulation. The collected fMRI data were preprocessed in SPM5 (http://www.fil.ion.ucl.ac.uk/spm). Images were realigned using INRIalign, which is a motion correction algorithm unbiased by local signal changes (Freire and Mangin, 2001; Freire et al., 2002). Data were then spatially normalized into the standard Montreal Neurological Institute space (Friston et al., 1995) and resliced to 3 × 3 × 3 mm, resulting in 53 × 63 × 46 voxels. Spatial smoothing was further applied with a 10mm Gaussian kernel. For details of data collection and preprocessing, see SI Materials and Methods. Data for each participant were then analyzed by a multiple regression incorporating regressors of the stimulus and its temporal derivative plus an intercept term, resulting in β values reflecting the relevance of regional activation to the stimulus. Finally the stimulus-on versus stimulus-off contrast images were extracted and all the voxels outside the brain or with missing measurements were excluded, resulting in a dataset of 52,322 voxels.
SNP data collection and preprocessing
DNA was extracted from a blood sample obtained from each of the 255 participants. Genotyping for all participants was performed at the Mind Research Network using the Illumina Infinium HumanOmni1-Quad assay covering 1,140,419 SNP loci. BeadStudio was used to make the final genotype calls. Next, the PLINK software package ((Purcell et al., 2007); http://pngu.mgh.harvard.edu/~purcell/plink) was used to perform a series of standard quality control procedures (Anderson et al., 2010), resulting in the final dataset spanning 777,635 SNP loci; and population stratification was assessed through principal component analysis (PCA) (Price et al., 2006); for details, see SI Materials and Methods.
Association study
To investigate genetic influence on brain function disruption in SZ, we performed a two-step analysis; the first-step was SNP filtering and the second-step was parallel-ICA. The SNP filtering was designed to reduce the number of genetic variables since parallel-ICA operates most efficiently with a lower-bound limit of approximately 0.02 for the ratio of sample size to number of SNP loci (Liu et al., 2008). To include both understudied and well-documented genetic variables, we first performed univariate ANOVA tests to select out 3318 top group-discriminating SNPs whose minor allele frequencies (MAFs) differed between patients and controls (p < 0.005, uncorrected), then 1839 SNPs residing in 61 preselected SZ-susceptibility genes from Schizophrenia Research Forum (Table S1) were also included. We further analyzed these candidate SNPs in conjunction with fMRI data using parallel-ICA to identify genetic components associated with SZ-disrupted brain functions.
Parallel-ICA was implemented with MATLAB (Fusion ICA Toolbox; http://icatb.sourceforge.net) to solve three issues simultaneously: extracting brain activation components; extracting genetic components and identifying associations between them.
The component extraction was based upon the Infomax algorithm (Bell and Sejnowski, 1995). Infomax extracts independent components through maximization of entropy, which measures uncertainty associated with a random variable. As illustrated in Eq. (1), the original datasets are first decomposed into a linear combination of underlying components. X1 (n×m1) and X2 (n×m2) denote the two data modalities, each being a two-dimensional subject × feature dataset; S1 and S2 are component matrices with each row representing an independent component; A1 and A2 are mixing matrices or loading matrices, with each column representing a loading vector associated with a specific component; r1 and r2 respectively denote the numbers of components of the two modalities; W1 and W2 are unmixing matrices. Based on the decomposition, the Infomax algorithm attempts to find the W matrix resulting in independence through maximizing an entropy function as defined in Eq. (2), where fy(Y) is the probability density function of Y; E is the expected value; H is the entropy function; W0 is the bias vector.
| (1) |
| (2) |
To enhance the inter-modality association, parallel-ICA attempts to maximize the correlation calculated between the columns of the loadings matrices A1 and A2. Thus, the two entropy terms and an additional correlation term comprise the objective function of parallel-ICA, as shown in Eq. (3), where A1i and A2j refer to the ith and jth columns of A1 and A2 respectively. This objective function will then be optimized through the gradient descent method. For a full description of the methodology and mathematical details of the algorithm, we refer readers to the original publications (Liu et al., 2008; Liu et al., 2009a).
Objective function
| (3) |
Parallel-ICA requires the component numbers (r1 and r2 in Eq. (1)) to be estimated first. In this work, the number of fMRI components was estimated to be 8 using a modified minimum description length (MDL) criterion (Li et al., 2007b). For the SNP data, due to the small difference in variance between sources and the ‘noise’ (i.e. wanted and unwanted factors), we utilized component consistency for the SNP component number estimation. With the full dataset, we fixed the number of fMRI components at 8, and investigated the consistencies of SNP components extracted by parallel-ICA with the number of SNP components ranging from 2 to 60. High overall consistency on the particular SNP component linked to fMRI was observed with the SNP component number ranging from 3 to 21, indicating a high possibility that the true component number was within this range. We further investigated the local component consistencies within a sliding window covering 5 consecutive SNP component numbers, and observed the highest consistency when SNP component number ranged from 5 to 9, where the average correlation among components was 0.97. Prompted by this, together with the fact that parallel-ICA has been shown to be less vulnerable to underestimation (Liu et al., 2008), the SNP component number was finally estimated to be 5.
Validation
To assess the fidelity of the identified association across participants, we applied a subset evaluation test, where each run of evaluation included 90% of the patients and controls for parallel-ICA analysis. We then investigated if the fMRI-SNP association identified in the full dataset was replicated in the 10 evaluations with subsets of the participants.
More informatively, we performed permutation tests to assess the validity of the identified fMRI-SNP association, that is, to investigate the possibility of the identified association occurring in random subsets of SNPs. To build a valid null distribution, for each round of permutation, we used permuted diagnostic labels to select out 3318 most group-discriminating SNPs and then combined them with the preselected 1839 SZ-related SNPs for association study. Given the estimated numbers of components, parallel-ICA extracted 40 fMRI-SNP component pairs in each of the 1000 permutation tests, resulting in the null distribution comprised of 40,000 independent tests. Then we calculated the tail probability to evaluate the significance level of the identified fMRI-SNP association.
Results
After the first-step filtering, 5157 candidate SNPs were analyzed in conjunction with fMRI contrast images by parallel-ICA. 8 fMRI components and 5 SNP components were extracted respectively, resulting in 40 fMRI-SNP component pairs. One pair exhibited a significant correlation with r of 0.29, and p-value of 2.41×10−5, passing the Bonferroni threshold of 1.25×10−3 after correction for 40 independent tests. The identified fMRI-SNP pair was replicated in subset evaluations, where we observed not only high similarities (with an average correlation of 0.82) between components derived from subsets of participants and that from the full dataset, but also stable correlations in the fMRI-SNP pairs, ranging from 0.25 to 0.38 with a median of 0.31 in the 10 subset evaluations. More importantly, within 40,000 tests from 1000 rounds of permutation, the absolute values of fMRI-SNP correlations ranged from 0.00 to 0.35 with a median of 0.05, resulting in a p-value of 3.00×10−4 for the identified association (r = 0.29), still passing the Bonferroni threshold of 1.25×10−3.
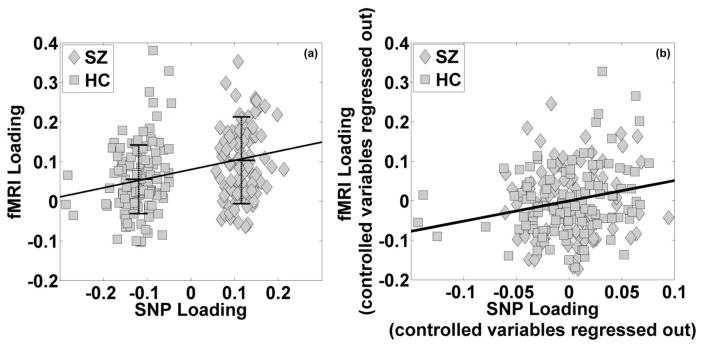
It was noted that the fMRI loading was particularly high for one participant (6-SD away from the average). After excluding this participant, the resulting correlation was 0.27 (p = 6.90×10−5), as shown in Figure 1a. We further performed a regression analysis between the fMRI and SNP loadings with controlled variables of SZ diagnosis and other factors including site, age, gender and ethnicity, as shown in Eq. (4). While these factors except for SZ diagnosis showed no significant effects on the fMRI component, the SNP component and SZ diagnosis exhibited regression effects with significance levels of 4.81×10−3 and 0.10 respectively. Correspondingly, a partial correlation analysis indicated that the SNP component uniquely explained 3.89% (r = 0.20, p = 4.31×10−3) of the fMRI variance after regressing out all the controlled variables, as shown in Figure 1b. In particular, the partial fMRI-SNP correlation was 0.21 (p = 0.02) within 116 healthy controls and 0.18 (p = 0.08) within 92 SZ patients.
Figure 1.
Scatter plots of the loading coefficients of the fMRI and SNP components (91 schizophrenia patients versus 116 healthy controls): (a) before correcting for controlling variables, mean ± SD of the fMRI loadings are marked for each group; (b) after regressing out controlling variables (SZ, site, age, gender and ethnicity).
| (4) |
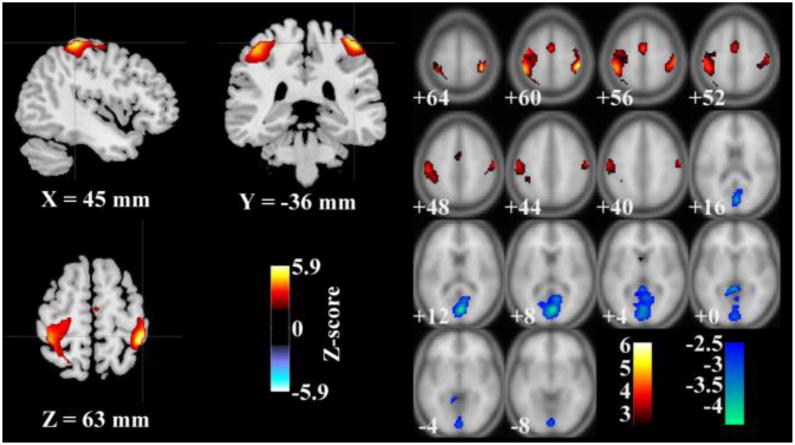
The linked fMRI component’s loadings significantly differed between groups (ANOVA, p = 4.68×10−4) with SZ patients showing higher loadings, indicating higher activations on average. The spatial map of the fMRI component thresholded at |Z| > 2.5 is shown in Figure 2, where the activated regions (red) mainly comprised the postcentral and precentral gyri and the deactivated regions (blue) mainly comprised of cuneus, posterior cingulate cortex and lingual gyrus, as listed in Table 2.
Figure 2.
Map of brain network for the identified functional magnetic resonance imaging component (|Z| > 2.5): left: regional view; right: slice view.
Table 2.
Talairach labels of brain regions of interest
| Brain region | Brodmann area | L/R Volume (cm3) | L/R random effects: max Z (x, y, z) |
|---|---|---|---|
| Positive | |||
| Postcentral Gyrus | 1, 2, 3, 5, 7, 40, 43 | 8.6/6.4 | 4.8(45, −32,54)/6.1(−45, −32,60) |
| Precentral Gyrus | 4, 6 | 5.6/2.0 | 4.1(39, −9,56)/3.9(−59, −15,45) |
| Negative | |||
| Cuneus | 17, 18, 23, 30 | 3.6/5.4 | 3.8(3, −78,12)/4.5(−3, −75,12) |
| Posterior Cingulate | 23, 29, 30, 31 | 1.3/4.2 | 3.3(6, −43,5)/3.7(−6, −69,12) |
| Lingual Gyrus | 17, 18, 19 | 1.7/2.4 | 3.4(6, −85, −8)/3.59(0, −73,6) |
The linked SNP component’s loadings, as expected, also significantly differed between groups (p < 1×10−23), with SZ patients carrying positive loadings and HC carrying negative loadings. This SNP component was predominantly contributed to by 253 SNPs (top 5% based on the absolute value of the components’ contribution weights), 138 of which were from known coding regions of 100 unique genes, while the rest were from intergenic regions. Table S2 provides a summary of the identified 253 SNPs, including the corresponding gene, Z-score of component weight, group difference in terms of MAF, and MAFs in patient and control groups. Among the 253 SNPs, 221 SNPs were from group-discriminating selection; 31 SNPs were from 16 preselected SZ-related genes; and one SNP (rs2284425 in GRIN2B) was from both. For the 31 non-group-discriminating yet SZ-related SNPs, we tested their correlations with the imaging endophenotype (the fMRI component loadings) and the group-discriminating SNPs. While exhibiting no associations with the former, all the non-group-discriminating SNPs were moderately correlated (correlation 0.28 ± 0.07, uncorrected) with some other group-discriminating SNPs, as shown in Table 3.
Table 3.
Summary of the identified non-group-discriminating single nucleotide polymorphisms
| Gene | Contributing SNPs
|
Correlation test
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| SNP | Z-score | GD (p-value) | MAF (SZ) | MAF (HC) | SNP_LD | Gene (SNP_corr) | GD (SNP_corr) | LD (r/p) | |
| BCR | rs5759636 | 2.32 | 0.57 | 0.14 | 0.15 | rs1896796 | SH3GL3 | 1.71E-04 | 0.23/6.67E-04 |
| BDNF | rs962369 | 2.08 | 0.75 | 0.27 | 0.30 | rs16855939 | C2orf48 | 4.54E-03 | 0.21/2.53E-03 |
| COMT | rs174699 | −1.91 | 0.24 | 0.05 | 0.08 | rs2297060 | STXBP6 | 2.22E-03 | 0.25/2.30E-04 |
| COMT | rs174696 | −1.92 | 0.50 | 0.27 | 0.25 | SNP2-102328778 | - | 1.73E-03 | 0.26/1.20E-04 |
| DISC1 | rs6541290 | −2.44 | 0.09 | 0.10 | 0.16 | rs9432024 | DISC1 | 1.44E-03 | −0.32/1.67E-06 |
| DNMT1 | rs721186 | 1.96 | 0.29 | 0.01 | 0.00 | rs17265803 | - | 2.74E-03 | 0.27/6.57E-05 |
| DRD3 | rs9828046 | −1.91 | 0.89 | 0.03 | 0.03 | rs4968678 | SCN4A | 3.30E-03 | 0.36/6.35E-08 |
| ERBB4 | rs11679952 | −2.28 | 0.47 | 0.30 | 0.36 | rs11639294 | - | 3.61E-03 | −0.23/8.18E-04 |
| ERBB4 | rs13392330 | 1.97 | 0.43 | 0.15 | 0.09 | rs1558789 | - | 1.86E-03 | 0.27/1.06E-04 |
| ERBB4 | rs872199 | 2.23 | 0.33 | 0.01 | 0.00 | rs12418045 | AMOTL1 | 1.43E-03 | 0.39/8.79E-09 |
| ESR1 | rs3798577 | 2.31 | 0.28 | 0.47 | 0.41 | rs7960152 | - | 2.03E-03 | −0.23/7.61E-04 |
| ESR1 | rs3798758 | −2.05 | 0.72 | 0.07 | 0.07 | rs12899474 | - | 3.53E-03 | 0.26/1.60E-04 |
| GABRA4 | rs1512130 | 2.23 | 0.27 | 0.35 | 0.30 | rs10009955 | - | 2.18E-03 | 0.27/6.86E-05 |
| GABRA4 | rs6844842 | 1.95 | 0.74 | 0.46 | 0.45 | rs10088262 | - | 2.46E-03 | −0.22/1.27E-03 |
| GAD1 | rs769406 | 2.42 | 0.27 | 0.03 | 0.01 | rs7675915 | - | 9.22E-04 | 0.27/5.95E-05 |
| GRID2 | rs6855368 | 2.00 | 0.03 | 0.54 | 0.40 | rs1748041 | PADI4 | 1.14E-03 | 0.25/2.68E-04 |
| GRID2 | rs4374594 | 1.96 | 0.01 | 0.29 | 0.19 | rs9564266 | - | 1.23E-03 | −0.27/8.32E-05 |
| HTR7 | rs17526697 | −2.09 | 0.66 | 0.10 | 0.10 | rs2088077 | WDR70 | 2.61E-03 | −0.24/6.09E-04 |
| NOTCH4 | rs8192587 | 1.96 | 0.06 | 0.02 | 0.00 | rs3846513 | - | 3.46E-03 | 0.30/7.74E-06 |
| NRG1 | rs7827456 | −1.96 | 0.05 | 0.38 | 0.40 | rs10503899 | NRG1 | 2.99E-03 | −0.50/1.32E-14 |
| NRG3 | rs10490933 | 3.06 | 0.94 | 0.01 | 0.00 | rs1520043 | - | 4.89E-03 | 0.30/1.49E-05 |
| NRG3 | rs11192642 | 2.69 | 0.20 | 0.02 | 0.00 | rs16914526 | SNTG1 | 3.18E-03 | 0.48/1.89E-13 |
| NRG3 | rs11195073 | 2.06 | 0.24 | 0.13 | 0.09 | rs7858370 | EPB41L4B | 4.94E-03 | 0.25/2.62E-04 |
| NRG3 | rs17099528 | 2.33 | 0.20 | 0.03 | 0.00 | rs2181184 | - | 1.94E-04 | −0.35/3.06E-07 |
| NRG3 | rs652183 | −2.01 | 0.74 | 0.47 | 0.53 | rs600488 | - | 3.23E-03 | 0.25/3.06E-04 |
| NRG3 | rs660464 | −2.03 | 0.73 | 0.47 | 0.53 | rs600488 | - | 3.23E-03 | 0.26/1.44E-04 |
| WNT2 | rs4727847 | 2.66 | 0.05 | 0.51 | 0.41 | rs985574 | - | 2.27E-03 | 0.23/1.02E-03 |
| WNT2 | rs2896218 | −2.33 | 0.40 | 0.39 | 0.44 | rs965281 | - | 1.85E-03 | 0.24/3.97E-04 |
| WNT2 | rs4730775 | 2.26 | 0.11 | 0.49 | 0.41 | rs13139804 | ATP10D | 2.46E-03 | −0.24/4.99E-04 |
| WNT2 | rs6947329 | 2.08 | 0.64 | 0.42 | 0.40 | rs342096 | - | 3.46E-03 | −0.25/3.14E-04 |
| WNT2 | rs916725 | −2.24 | 0.24 | 0.36 | 0.43 | rs965281 | - | 1.85E-03 | 0.21/2.03E-03 |
Note: Z-score refers to the normalized component weight and GD refers to the group difference between SZ patients and healthy controls in terms of MAF. In correlation tests, SNP_corr reports the group-discriminating SNP exhibiting the highest correlation with the tested non-group-discriminating SNP and the corresponding correlation and p-value are also included.
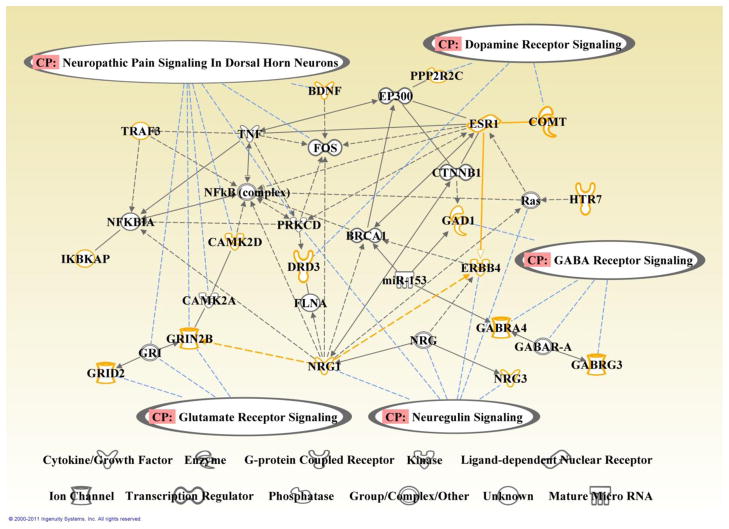
We examined the function of the identified genetic component using Ingenuity Pathways Analysis (IPA: Ingenuity® Systems, http://www.ingenuity.com), where the 100 unique contributing genes were analyzed with the whole genome as background. A number of canonical pathways were extracted including four neurotransmitter pathways: GABA receptor signaling, dopamine receptor signaling, neuregulin signaling, and glutamate receptor signaling, as illustrated in Table 4a. We further explored the interconnections among the genes involved in these pathways, and Figure 3 illustrates a function network built upon the Ingenuity pathway knowledge base, where orange nodes are the selected genes and grey nodes are bridging entities with the shortest path (nor more than 2 edges) to the selected genes. Besides the pathway analyses, the DAVID (Database for Annotation, Visualization and Integrated Discovery) bioinformatics resource (Huang et al., 2009a, b) identified the most significant cluster to be functionally related to the synapse, with a total of 10 genes involved, as summarized in Table 4b. Another marginally significant cluster was related to cell projection (a prolongation or process extending from a cell, e.g. axon, dendrite, flagellum, etc.).
Table 4.
Biological pathway analysis and functional annotation clustering
| Table 4a. Canonical pathway | Gene | p-value |
|---|---|---|
| GABA receptor signaling | GABRA4, GABRG3, GAD1 | 2.03E-03 |
| Agrin interactions at neuromuscular junctions | NRG1, NRG3, ERBB4 | 5.55E-03 |
| Dopamine receptor signaling | COMT, DRD3, PPP2R2C | 7.87E-03 |
| Neuregulin signaling | NRG1, NRG3, ERBB4 | 1.10E-02 |
| Neuropathic pain signaling in dorsal horn neurons | BDNF, CAMK2D, GRIN2B | 1.61E-02 |
| B cell activating factor signaling | IKBKAP, TRAF3 | 1.94E-02 |
| cAMP-mediated signaling | CAMK2D, DRD3, HTR7, OPRD1 | 2.71E-02 |
| Lymphotoxin β receptor signaling | IKBKAP, TRAF3 | 3.39E-02 |
| Glutamate receptor signaling | GRIN2B, GRID2 | 3.63E-02 |
| Activation of IRF by cytosolic pattern recognition receptors | IKBKAP, TRAF3 | 4.11E-01 |
| CD40 signaling | IKBKAP, TRAF3 | 4.36E-01 |
| Table 4b. Functional annotation cluster | Gene | p-value |
|---|---|---|
| Synapse | GABRA4, GABRG3, GAD1, GRIN2B, GRID2, ERBB4, SHC4, OTOF, PSD3, CTBP2 | 5.20E-03 |
| Cell projection (axon, dendrite, flagellum, etc.) | DRD3, GAD1, GRIN2B, MYCBP2, DNAH11, WNT2, ESR1, CDH13, ALCAM, MYO5A | 9.70E-02 |
Note: 4 neurotransmitter signaling pathways are extracted from Ingenuity pathway knowledge base. David bioinformatics resource identifies the most significant cluster to be functionally related to synapse, while another marginally significant cluster relating to cell projection.
Figure 3.
Function network built upon genes selected from the identified genetic component. Orange nodes: identified genes; grey nodes: employed bridging entities; black edge: interaction between two nodes (solid: direct; dashed: indirect); blue lines: the involvement of a node with a canonical pathway. Four neurotransmitter signaling pathways were identified from Ingenuity pathway knowledge base.
It is worth to note that within the patient group we did not observe any significant association between fMRI loadings and chlorpromazine (CPZ) equivalent dosages calculated from all current medications, lifetime CPZ equivalent dose years of all medications, as well as the assessment of positive/negative symptoms (SAPS/SANS) (Andreasen, 1983, 1984).
Discussion
This work was designed to analyze fMRI data in conjunction with SNP data to explore genetic risk factors underlying task-related brain function disruption in SZ. Due to the limited sample size compared to the number of SNPs, we first located 5157 candidate risk SNPs from the whole genome, including 3318 candidate SNPs exhibiting group difference in terms of MAF, and 1839 candidate SNPs residing in preselected SZ-susceptibility genes. Within these candidate risk SNPs, we further located SNPs predominantly contributing to SZ-disrupted brain functions using parallel-ICA. One fMRI-SNP pair was identified to exhibit a significant association while controlled for diagnostic labels, ethnicity, gender and age, indicating that the association was not mainly attributable to these factors. Subsequent cross-evaluation indicated that the identified association exhibited high consistency across participants. In addition, permutation tests confirmed the validity of the identified association which was significantly different from the null hypothesis of no association obtained with permuted SNPs. Overall, this validated fMRI-SNP linkage, together with the fact that both fMRI and SNP components differ between groups, suggest that genetic variations may underlie the altered brain function in SZ. It is worth to emphasize again that the goal of this study is to examine genetic influence on brain functions related to SZ, and we designed our analysis to focus on the genetic variables most relevant to SZ, so the significance level of group difference observed in the SNP component may be biased, but not the association.
fMRI component
The identified fMRI component exhibits both positive and negative weights, indicating that the brain functions exhibit positive or negative relevance to the original stimuli, denoted as activations and deactivations respectively. The associated component’s loadings are higher for patients, suggesting greater activation/deactivation in patients than in controls. The main activated regions identified in this component are postcentral and precentral gyri. The postcentral gyrus mainly comprises primary somatosensory cortex and somatosensory association cortex, responsible for receiving, processing and associating the input sensory information. The precentral gyrus mainly involves primary motor cortex and premotor cortex (supplementary motor area). These two brain regions work in association to refine, plan and execute movements based on sensory input. One would expect the precentral and postcentral gyri to be elicited by a sensorimotor task. Interestingly, adolescents who later go on to develop SZ (Mittal et al., 2008), as well as SZ patients who have never received neuroleptics (Fenton et al., 1994; Honer et al., 2005), exhibit subtle motor abnormalities that may be indicative of dysfunction of these regions. In addition, the precentral and postcentral gyri have also been reported to be altered in SZ patients, including reduction in cortical thickness and gray matter concentration (Giuliani et al., 2005; Glahn et al., 2008; Kuperberg et al., 2003; Narr et al., 2005b), as well as displaying abnormal task-related functional activation pattern (Abbott et al., 2011; Kiehl et al., 2005; Minzenberg et al., 2009; Paulus et al., 2002; Schroder et al., 1999; Shergill et al., 2000). Moreover, the hyperactivation observed in this fMRI component is consistent with a previous fMRI report that used participants partially overlapping with this study and found SZ-related hyperactivation in precentral gyrus (Kim et al., 2010). Overall, our findings are in line with the report of motor dysfunction as one of the characteristic deficits in SZ (Bilder et al., 2000).
The deactivated regions in this fMRI component, cuneus, posterior cingulate and lingual gyrus, have also been reported in SZ related studies. For instance, the posterior cingulate as part of the default mode network has been reported to be altered in SZ patients (Harrison et al., 2007). Reductions in cortical thickness and grey matter volume have been observed in both cuneus and lingual gyrus in SZ patients (Gaser et al., 1999; Narr et al., 2005a; Neckelmann et al., 2006).
SNP component
The SNP component is correlated with the fMRI component and significantly differs between patients and controls. On average, SZ patients carry higher SNP component as well as exhibit higher activation in the identified regions of the fMRI component, while healthy controls show the opposite. As for each SNP locus, a positive or negative component weight implicitly relates to an increased or decreased MAF in patients compared to controls, given that the genotype coding is based on number of minor alleles.
The SNP component is predominantly contributed to by 253 SNPs, among which 222 SNPs are from group-discriminating selection. As expected, the component weights of these SNPs strictly coincide with the MAF differences, i.e. a positive or negative weight corresponds to an increased or decreased MAF in patients, respectively (see Table S2). The remaining 31 non-group-discriminating SNPs reside in 16 out of 61 preselected susceptibility genes. Most of these SNPs still exhibit a correspondence between component weight and the trend of MAF difference, except for rs5759636 (BCR), rs9828046 (DRD3), rs174696 (COMT), and rs962369 (BDNF). Further tests show that all the non-group-discriminating SNPs moderately correlate with some other group-discriminating SNPs (Table 3), suggesting that some alleles may contribute to SZ susceptibility by interacting with others, despite weak individual effects.
Among the identified 253 SNPs, 138 SNPs are located in 100 unique genes. The remaining intergenic SNPs may affect the sequences of non-coding RNAs and regulate genes and thus they can potentially be of importance. However, these SNPs are not discussed in this manuscript given that very little is currently known about them. Instead, we focus our discussion on those participating in neurotransmitter signaling (highlighted in Table 4a), given the nature of this work to investigate fMRI-SNP associations in SZ.
GABA receptor signaling (GAD1, GABRA4 and GABRG3)
GAD1 (GAD67) has been consistently reported to show reduced mRNA levels in the dorsolateral prefrontal cortex of SZ patients (Akbarian et al., 1995; Guidotti et al., 2000; Straub et al., 2007; Volk et al., 2000). In our data, rs769406 (GAD1, minor allele type G) exhibits positive weight, suggesting a higher MAF in patients. GABRA4 and GABRG3 are expressed at high levels in brain regions. Direct connections of these genes to SZ are not yet clear, but our results indicate that the MAFs of rs6844842, rs1512130 (GABRA4, allele type C and A, respectively) and rs1029937 (GABRG3, allele type T) are all expected to be higher in patients.
Dopamine receptor signaling (DRD3, COMT and PPP2R2C)
DRD3 is shown to be over-expressed in off-antipsychotic SZ patients in postmortem studies (Gurevich et al., 1997) and COMT is involved in catabolic clearance of dopamine (Augustine, 2001). PPP2R2C, although not a common SZ susceptibility gene, is involved in the establishment and maintenance of neuronal connections (Strack et al., 1998). In our data, minor alleles in rs9828046 (DRD3, allele type A), rs174696, rs174699 (COMT, allele type C for both) and rs16838780 (PPP2R2C, allele type C) contribute to the genetic component with negative weights, implying lower MAFs in patients than in controls; minor alleles in rs4689440 and rs13434456 (PPP2R2C, allele type A for both) contribute with positive weights, indicating that corresponding MAFs are expected to be higher in patients.
Neuregulin signaling (NRG1, NRG3 and ERBB4)
The NRG1-ERBB4 cascade modulates neuronal plasticity in adult brains (Li et al., 2007a; Woo et al., 2007) and an unstable NRG3 signaling system is implicated as relating to SZ (Kao et al., 2010). In our results, minor alleles in rs17099528, rs11195073, rs11192642, rs10490933 (NRG3, allele type C, G, G and A, respectively) and rs13392330, rs872199 (ERBB4, allele type C for both) exhibit positive weights, implying higher MAFs in patients; minor alleles in rs660464, rs652183 (NRG3, allele type A and G, respectively), rs11679952 (ERBB4, allele type T) and rs7827456 (NRG1, allele type A) exhibit negative weights, suggesting lower MAFs in patients.
Glutamate receptor signaling (GRIN2B and GRID2)
Recent meta-analyses lend support to the involvement of GRIN2B in SZ (Allen et al., 2008) and GRID2 mutant mice (lacking detectable GRID2 protein in cerebellum) exhibit strong impairments of motor learning and coordination (Funabiki et al., 1995; Kashiwabuchi et al., 1995; Kishimoto et al., 2001). In our results, minor alleles in 2 SNPs in GRID2 (type A in rs6855368 and type T in rs4374594) are indicated to exhibit higher MAFs in patients, while the opposite is expected for minor allele T in rs2284425 (GRIN2B).
Besides those involved in the four neurotransmitter signaling pathways, some other genes are worth noting. For instance, CAMK2D is suggested to be present at the neuromuscular junction (Cohen et al., 2007) and participate in phosphorylation of GRIN2B (Omkumar et al., 1996). RARB encodes retinoic acid receptor beta and has been suggested as causal to SZ (Goodman, 1998). PADI4 is implicated as conferring susceptibility to rheumatoid arthritis (RA) (Suzuki et al., 2003), while the comorbidity between RA and SZ has been implied as low (Oken and Schulzer, 1999). Genes identified in functional annotation clustering (Table 4b) are also of interest, such as MYO5A, which has been implied as critical for motor learning in a recent study (Miyata et al., 2011).
In addition to the plausible connections with SZ, we emphasize the interrelations carried in this genetic component. Moderate correlations are observed in SNPs across genes or chromosomes. It is especially interesting to notice that three non-group-discriminating SNPs, rs174699 (COMT), rs9828046 (DRD3) and rs6855368 (GRID2) moderately correlate with rs2297060 (STXBP6), rs4968678 (SCN4A) and rs1748041 (PADI4), respectively, as listed in Table 3. STXBP6 may be involved in regulating SNARE complex (related to vesicle fusion in neuromediator release) formation (Scales et al., 2002); SCN4A is related to generation and propagation of action potentials and involved in neuromuscular disorder (Kullmann, 2010). Also Table 4b presents the interrelations among identified genes based on functional annotation, where the two clusters are related to synapse and cell projection. Furthermore, we explore the interactions between genes in the four pathways discussed above and show in Figure 3 that the four pathways are essentially interrelated through intermediate nodes. Different pathways also show functional influence. For instance, a hyperdopaminergic state may result in NMDA receptor hypofunction (Aalto et al., 2005; Vollenweider et al., 2000), and NRG1-ERBB4 cascade may be involved in pruning glutamate synapses (Li et al., 2007a). Overall this SNP component presents a combined effect from many SNPs, genes and pathways on altered brain function, which is associated with but not limited to SZ.
One limitation of this study lies in the inclusion of participants of various ethnicities, although we have corrected the population stratification through PCA (Price et al., 2006). To evaluate the influence, we tested the fMRI-SNP association with control for SZ diagnosis, ethnicity, gender and age and still observed a significant regression effect from the SNP component on the fMRI component (two-tailed t-test, p = 0.01). In addition, in a Caucasian-only subset (72 patients versus 105 controls), a significant regression effect from the SNP component on the fMRI component was still observed (two-tailed t-test, p = 0.02) when controlling for SZ diagnosis, gender and age. Given these observations, we have concluded that the population structure is not a major contributor to our findings.
A second limitation is the possible effect of life-long medications on fMRI activation patterns. To assess this medication influence, we first compared the fMRI component loadings of patients taking (83/91) with those not taking (8/91) antipsychotics at the time of evaluation, and oberved no group difference (ANOVA, p = 0.54). Further, after regressing out the medication effects (CPZ equivalent dosages of all current medications and lifetime CPZ equivalent dose years of all medication exposure) as well as the factors of site, age, gender and ethnicity, we still observed a similar level of correlation between the fMRI and SNP components (69 SZ patients, r = 0.20, p = 0.10). Overall, these analyses indicate that our finding on fMRI-SNP association is not majorly due to medication influence.
A third limitation might be our initial selection of SNPs. To allow parallel-ICA performing at its best condition (Liu et al., 2008), dimension reduction at the SNP array is necessary. Given that not every SNP in the genomic array is related to SZ, we believe a selection of candidate risk SNPs with a certain level of group-discriminating power is reasonable and beneficial. We also selected a number of candidate risk SNPs residing in preselected SZ-susceptibility genes, as it would be interesting to investigate whether and how these SNPs influence brain function in SZ. In the identified fMRI-SNP pair, the significance of group difference observed in the SNP component may be biased due to the inclusion of 3318 group-discriminating SNPs. However, the fMRI-SNP association has been validated through permutation, where the significance level of the association is estimated to be 0.0003. Given that permuted diagnostic labels have been used to select group-discriminating SNPs in each permutation, this significant p-value indicates a very low possibility that the identified association results from parallel-ICA being inflated by the group-discrimination selection. Furthermore, the permutation result also indicates that the identified association is not completely contributed by the preselected SNPs, considering that the same 1839 SZ-related SNPs were included in each permutation, which further confirms that the identified SNP component provides a good source of understudied SNPs with which the SZ-related SNPs may interact to disrupt the brain function. Additionally, we used the genes related to the 5157 candidate SNPs as a background that already over-represented SZ, and still observed an enrichment of SZ relevance and GABA signaling pathway in the identified SNP component through IPA, indicating that the identified component unquestionably involves SZ-related brain function. Overall, these analyses suggest that the possibility is low for our finding of SZ-related fMRI-SNP association to be a false positive resulting from the initial SNP selection.
On the other hand, it should be noted that the SNPs in high linkage disequilibrium (LD) would exhibit comparable effects in our analysis. Therefore, the SNPs identified in the parallel-ICA analysis can be in LD with true causal variants. To investigate this issue further, we calculated the correlations between each of the identified SNPs and the remaining 777,634 SNPs. The results showed that 175 out of 253 identified SNPs were in high LD (r > 0.80) with some other SNPs residing in the same genes, while one single SNP, rs872199 from ERBB4 exhibited high LD with rs7117582 from EHF. Overall, these results lead to a conclusion: the initial selection and limitation of the SNP array may result in SNPs in LD with true causal variants, however the identified genes and pathways should be unchanged.
Finally, given the difference in correlations obtained before and after controlling for a SZ diagnosis, we are aware that the group difference may play a role in the identified fMRI-SNP association. However, these three-way connections among SNP, SZ diagnosis and fMRI are rooted in the design of the experiment. Considering that we are trying to investigate the genetic factors underlying the disrupted brain functions related to SZ, we naturally expect the identified SNP and fMRI components to exhibit significant group differences, which result in the collinearity issue. In our results, the SNP component explains a total of 8.31% of the fMRI variance, among which 0.44% is shared with the factors of site, age, gender or ethnicity, and 3.98% is shared with SZ diagnosis, which is conceptually reasonable. As for the remaining 3.89% of the unique variance, although the corresponding p-value (< 5.00×10−3) does not survive Bonferroni correction (which is known to be conservative), the fMRI-SNP correlation (r ~ 0.20) is consistently observed across all participants, as well as within patients and controls. Overall, the identified fMRI-SNP association is partially shared with the group difference, which confirms the relatedness of the finding to the disorder, and the remaining major association, being consistent across groups, indicates that on average participants carrying higher SNP component also exhibit higher activations in the identified brain regions of the fMRI component.
In conclusion, we present a framework for a multivariate fMRI-SNP association study to reveal the multifaceted genetic risk factor underlying altered sensorimotor functionality in SZ. The identified fMRI component mainly comprises task-related activated regions, and shows alterations in SZ. The identified SNP component illustrates a strong contribution from multiple alleles and genes, and demonstrates interrelations among SNPs, genes and even pathways. Some identified SNPs and genes are not previously implicated in SZ risk. The fact that the fMRI component is significantly linked to the SNP component suggests that variations in these contributing alleles may underlie the disrupted brain function in SZ. This conjecture is reinforced by the observation of four relevant neurotransmitter signaling pathways.
Supplementary Material
Acknowledgments
We would like to thank Jill Fries and Marilee Morgan for processing the fMRI and genetic data. This work was supported by the National Institutes of Health; Contract grant number: 5 P20 RR 021938-03.
Footnotes
Financial disclosures. The authors declare no potential conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aalto S, Ihalainen J, Hirvonen J, Kajander J, Scheinin H, Tanila H, et al. Cortical glutamate-dopamine interaction and ketamine-induced psychotic symptoms in man. Psychopharmacology. 2005;182:375–383. doi: 10.1007/s00213-005-0092-6. [DOI] [PubMed] [Google Scholar]
- Abbott C, Juarez M, White T, Gollub RL, Pearlson GD, Bustillo J, et al. Antipsychotic dose and diminished neural modulation: a multi-site fMRI study. Progress in neuro-psychopharmacology & biological psychiatry. 2011;35:473–482. doi: 10.1016/j.pnpbp.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akbarian S, Kim JJ, Potkin SG, Hagman JO, Tafazzoli A, Bunney WE, et al. Gene-Expression for Glutamic-Acid Decarboxylase Is Reduced without Loss of Neurons in Prefrontal Cortex of Schizophrenics. Archives of General Psychiatry. 1995;52:258–266. doi: 10.1001/archpsyc.1995.03950160008002. [DOI] [PubMed] [Google Scholar]
- Allen NC, Bagade S, McQueen MB, Ioannidis JPA, Kavvoura FK, Khoury MJ, et al. Systematic meta-analyses and field synopsis of genetic association studies in schizophrenia: the SzGene database. Nature Genetics. 2008;40:827–834. doi: 10.1038/ng.171. [DOI] [PubMed] [Google Scholar]
- Anderson CA, Pettersson FH, Clarke GM, Cardon LR, Morris AP, Zondervan KT. Data quality control in genetic case-control association studies. Nature Protocols. 2010;5:1564–1573. doi: 10.1038/nprot.2010.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreasen NC. The Scale for the Assessment of Negative Symptoms (SANS) The University of Iowa; Iowa City, IA: 1983. [Google Scholar]
- Andreasen NC. The Scale for the Assessment of Negative Symptoms (SAPS) The University of Iowa; Iowa City, IA: 1984. [Google Scholar]
- Andreasen NC, Flaum M, Arndt S. The Comprehensive Assessment of Symptoms and History (Cash) - an Instrument for Assessing Diagnosis and Psychopathology. Archives of General Psychiatry. 1992;49:615–623. doi: 10.1001/archpsyc.1992.01820080023004. [DOI] [PubMed] [Google Scholar]
- Augustine GJ, editor. Neurotransmitters. 2. Sinauer; Sunderland, MA: 2001. [Google Scholar]
- Bell AJ, Sejnowski TJ. An Information Maximization Approach to Blind Separation and Blind Deconvolution. Neural Computation. 1995;7:1129–1159. doi: 10.1162/neco.1995.7.6.1129. [DOI] [PubMed] [Google Scholar]
- Bilder RM, Goldman RS, Robinson D, Reiter G, Bell L, Bates JA, et al. Neuropsychology of first-episode schizophrenia: Initial characterization and clinical correlates. American Journal of Psychiatry. 2000;157:549–559. doi: 10.1176/appi.ajp.157.4.549. [DOI] [PubMed] [Google Scholar]
- Cardno AG, Gottesman II. Twin studies of schizophrenia: From bow-and-arrow concordances to star wars mx and functional genomics. American Journal of Medical Genetics. 2000;97:12–17. [PubMed] [Google Scholar]
- Cohen TJ, Waddell DS, Barrientos T, Lu Z, Feng G, Cox GA, et al. The histone deacetylase HDAC4 connects neural activity to muscle transcriptional reprogramming. The Journal of biological chemistry. 2007;282:33752–33759. doi: 10.1074/jbc.M706268200. [DOI] [PubMed] [Google Scholar]
- Duan JB, Sanders AR, Gejman PV. Genome-wide approaches to schizophrenia. Brain Research Bulletin. 2010;83:93–102. doi: 10.1016/j.brainresbull.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenton WS, Wyatt RJ, Mcglashan TH. Risk-Factors for Spontaneous Dyskinesia in Schizophrenia. Archives of General Psychiatry. 1994;51:643–650. doi: 10.1001/archpsyc.1994.03950080055008. [DOI] [PubMed] [Google Scholar]
- First MB, Gibbon M, Spitzer RL, Williams JBW, Benjamin LS. Structured clinical interview for DSM-IV axis II personality disorders, (SCID-II) 4. American Psychiatric Press; Washington, DC: 1997. [Google Scholar]
- Freire L, Mangin JF. Motion correction algorithms may create spurious brain activations in the absence of subject motion. Neuroimage. 2001;14:709–722. doi: 10.1006/nimg.2001.0869. [DOI] [PubMed] [Google Scholar]
- Freire L, Roche A, Mangin JF. What is the best similarity measure for motion correction in fMRI time series? Ieee Transactions on Medical Imaging. 2002;21:470–484. doi: 10.1109/TMI.2002.1009383. [DOI] [PubMed] [Google Scholar]
- Friston KJ, Ashburner J, Frith CD, Poline JB, Heather JD, Frackowiak RSJ. Spatial registration and normalization of images. Human Brain Mapping. 1995;3:165–189. [Google Scholar]
- Funabiki K, Mishina M, Hirano T. Retarded vestibular compensation in mutant mice deficient in delta 2 glutamate receptor subunit. Neuroreport. 1995;7:189–192. [PubMed] [Google Scholar]
- Gaser C, Volz HP, Kiebel S, Riehemann S, Sauer H. Detecting structural changes in whole brain based on nonlinear deformations - Application to schizophrenia research. Neuroimage. 1999;10:107–113. doi: 10.1006/nimg.1999.0458. [DOI] [PubMed] [Google Scholar]
- Giuliani NR, Calhoun VD, Pearlson GD, Francis A, Buchanan RW. Voxel-based morphometry versus region of interest: a comparison of two methods for analyzing gray matter differences in schizophrenia. Schizophrenia Research. 2005;74:135–147. doi: 10.1016/j.schres.2004.08.019. [DOI] [PubMed] [Google Scholar]
- Glahn DC, Laird AR, Ellison-Wright I, Thelen SM, Robinson JL, Lancaster JL, et al. Meta-analysis of gray matter anomalies in schizophrenia: application of anatomic likelihood estimation and network analysis. Biological Psychiatry. 2008;64:774–781. doi: 10.1016/j.biopsych.2008.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman AB. Three independent lines of evidence suggest retinoids as causal to schizophrenia. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:7240–7244. doi: 10.1073/pnas.95.13.7240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman II, Gould TD. The endophenotype concept in psychiatry: Etymology and strategic intentions. American Journal of Psychiatry. 2003;160:636–645. doi: 10.1176/appi.ajp.160.4.636. [DOI] [PubMed] [Google Scholar]
- Gottesman II, Shields J. A Polygenic Theory of Schizophrenia. Proceedings of the National Academy of Sciences of the United States of America. 1967;58:199–205. doi: 10.1073/pnas.58.1.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guidotti A, Auta J, Davis JM, Gerevini VD, Dwivedi Y, Grayson DR, et al. Decrease in reelin and glutamic acid decarboxylase(67) (GAD(67)) expression in schizophrenia and bipolar disorder - A postmortem brain study. Archives of General Psychiatry. 2000;57:1061–1069. doi: 10.1001/archpsyc.57.11.1061. [DOI] [PubMed] [Google Scholar]
- Gurevich EV, Bordelon Y, Shapiro RM, Arnold SE, Gur RE, Joyce JN. Mesolimbic dopamine D-3 receptors and use of antipsychotics in patients with schizophrenia - A postmortem study. Archives of General Psychiatry. 1997;54:225–232. doi: 10.1001/archpsyc.1997.01830150047009. [DOI] [PubMed] [Google Scholar]
- Harrison BJ, Yucel M, Pujol J, Pantelis C. Task-induced deactivation of midline cortical regions in schizophrenia assessed with fMRI. Schizophrenia Research. 2007;91:82–86. doi: 10.1016/j.schres.2006.12.027. [DOI] [PubMed] [Google Scholar]
- Harrison PJ, Owen MJ. Genes for schizophrenia? Recent findings and their pathophysiological implications. Lancet. 2003;361:417–419. doi: 10.1016/S0140-6736(03)12379-3. [DOI] [PubMed] [Google Scholar]
- Honer WG, Kopala LC, Rabinowitz J. Extrapyramidal symptoms and signs in first-episode, antipsychotic exposed and non-exposed patients with schizophrenia or related psychotic illness. Journal of Psychopharmacology. 2005;19:277–285. doi: 10.1177/0269881105051539. [DOI] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Research. 2009a;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols. 2009b;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Kao WT, Wang YH, Kleinman JE, Lipska BK, Hyde TM, Weinberger DR, et al. Common genetic variation in Neuregulin 3 (NRG3) influences risk for schizophrenia and impacts NRG3 expression in human brain. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:15619–15624. doi: 10.1073/pnas.1005410107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashiwabuchi N, Ikeda K, Araki K, Hirano T, Shibuki K, Takayama C, et al. Impairment of Motor Coordination, Purkinje-Cell Synapse Formation, and Cerebellar Long-Term Depression in Glur-Delta-2 Mutant Mice. Cell. 1995;81:245–252. doi: 10.1016/0092-8674(95)90334-8. [DOI] [PubMed] [Google Scholar]
- Kiehl KA, Stevens MC, Celone K, Kurtz M, Krystal JH. Abnormal hemodynamics in schizophrenia during an auditory oddball task. Biological Psychiatry. 2005;57:1029–1040. doi: 10.1016/j.biopsych.2005.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DI, Sui J, Rachakonda S, White T, Manoach DS, Clark VP, et al. Identification of Imaging Biomarkers in Schizophrenia: A Coefficient-constrained Independent Component Analysis of the Mind Multi-site Schizophrenia Study. Neuroinformatics. 2010;8:213–229. doi: 10.1007/s12021-010-9077-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishimoto Y, Kawahara S, Suzuki M, Mori H, Mishina M, Kirino Y. Classical eyeblink conditioning in glutamate receptor subunit delta 2 mutant mice is impaired in the delay paradigm but not in the trace paradigm. European Journal of Neuroscience. 2001;13:1249–1253. doi: 10.1046/j.0953-816x.2001.01488.x. [DOI] [PubMed] [Google Scholar]
- Kullmann DN. Neurological Channelopathies. Annual Review of Neuroscience. 2010;33:151–172. doi: 10.1146/annurev-neuro-060909-153122. [DOI] [PubMed] [Google Scholar]
- Kuperberg GR, Broome MR, McGuire PK, David AS, Eddy M, Ozawa F, et al. Regionally localized thinning of the cerebral cortex in schizophrenia. Archives of General Psychiatry. 2003;60:878–888. doi: 10.1001/archpsyc.60.9.878. [DOI] [PubMed] [Google Scholar]
- Lawrie SM, Whalley HC, Job DE, Johnstone EC. Structural and functional abnormalities of the amygdala in schizophrenia. Amygdala in Brain Function: Bacic and Clinical Approaches. 2003;985:445–460. doi: 10.1111/j.1749-6632.2003.tb07099.x. [DOI] [PubMed] [Google Scholar]
- Li B, Woo RS, Mei L, Malinow R. The neuregulin-1 receptor ErbB4 controls Glutamatergic synapse maturation and plasticity. Neuron. 2007a;54:583–597. doi: 10.1016/j.neuron.2007.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YO, Adali T, Calhoun VD. Estimating the number of independent components for functional magnetic resonance imaging data. Human Brain Mapping. 2007b;28:1251–1266. doi: 10.1002/hbm.20359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Bixler JN, Calhoun VD. A multimodality ICA study - integrating genomic single nucleotide polymorphisms with functional neuroimaging data. 2008 IEEE International Conference on Bioinformatics and Biomedicine Workshops; Philadelphia, PA. 2008. pp. 151–157. [Google Scholar]
- Liu J, Pearlson G, Windemuth A, Ruano G, Perrone-Bizzozero NI, Calhoun V. Combining fMRI and SNP data to investigate connections between brain function and genetics using parallel ICA. Human Brain Mapping. 2009a;30:241–255. doi: 10.1002/hbm.20508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JY, Pearlson G, Windemuth A, Ruano G, Perrone-Bizzozero NI, Calhoun V. Combining fMRI and SNP Data to Investigate Connections Between Brain Function and Genetics Using Parallel ICA. Human Brain Mapping. 2009b;30:241–255. doi: 10.1002/hbm.20508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald C, Marshall N, Sham PC, Bullmore ET, Schulze K, Chapple B, et al. Regional brain morphometry in patients with schizophrenia or bipolar disorder and their unaffected relatives. American Journal of Psychiatry. 2006;163:478–487. doi: 10.1176/appi.ajp.163.3.478. [DOI] [PubMed] [Google Scholar]
- Meda SA, Jagannathan K, Gelernter J, Calhoun VD, Liu JY, Stevens MC, et al. A pilot multivariate parallel ICA study to investigate differential linkage between neural networks and genetic profiles in schizophrenia. Neuroimage. 2010;53:1007–1015. doi: 10.1016/j.neuroimage.2009.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minzenberg MJ, Laird AR, Thelen S, Carter CS, Glahn DC. Meta-analysis of 41 functional neuroimaging studies of executive function in schizophrenia. Archives of General Psychiatry. 2009;66:811–822. doi: 10.1001/archgenpsychiatry.2009.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittal VA, Neumann C, Saczawa M, Walker EF. Longitudinal progression of movement abnormalities in relation to psychotic symptoms in adolescents at high risk of schizophrenia. Archives of General Psychiatry. 2008;65:165–171. doi: 10.1001/archgenpsychiatry.2007.23. [DOI] [PubMed] [Google Scholar]
- Miyata M, Kishimoto Y, Tanaka M, Hashimoto K, Hirashima N, Murata Y, et al. A Role for Myosin Va in Cerebellar Plasticity and Motor Learning: A Possible Mechanism Underlying Neurological Disorder in Myosin Va Disease. Journal of Neuroscience. 2011;31:6067–6078. doi: 10.1523/JNEUROSCI.5651-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narr KL, Bilder RM, Toga AW, Woods RP, Rex DE, Szeszko PR, et al. Mapping cortical thickness and gray matter concentration in first episode schizophrenia. Cerebral Cortex. 2005a;15:708–719. doi: 10.1093/cercor/bhh172. [DOI] [PubMed] [Google Scholar]
- Narr KL, Toga AW, Szeszko P, Thompson PM, Woods RP, Robinson D, et al. Cortical thinning in cingulate and occipital cortices in first episode schizophrenia. Biological Psychiatry. 2005b;58:32–40. doi: 10.1016/j.biopsych.2005.03.043. [DOI] [PubMed] [Google Scholar]
- Neckelmann G, Specht K, Lund A, Ersland L, Smievoll AI, Neckelmann D, et al. MR morphometry analysis of grey matter volume reduction in schizophrenia: Association with hallucinations. International Journal of Neuroscience. 2006;116:9–23. doi: 10.1080/00207450690962244. [DOI] [PubMed] [Google Scholar]
- Nelson MD, Saykin AJ, Flashman LA, Riordan HJ. Hippocampal volume reduction in schizophrenia as assessed by magnetic resonance imaging - A meta-analytic study. Archives of General Psychiatry. 1998;55:433–440. doi: 10.1001/archpsyc.55.5.433. [DOI] [PubMed] [Google Scholar]
- Oken RJ, Schulzer M. At Issue: Schizophrenia and rheumatoid arthritis: The negative association revisited. Schizophrenia Bulletin. 1999;25:625–638. doi: 10.1093/oxfordjournals.schbul.a033407. [DOI] [PubMed] [Google Scholar]
- Omkumar RV, Kiely MJ, Rosenstein AJ, Min KT, Kennedy MB. Identification of a phosphorylation site for calcium/calmodulindependent protein kinase II in the NR2B subunit of the N-methyl-D-aspartate receptor. The Journal of biological chemistry. 1996;271:31670–31678. doi: 10.1074/jbc.271.49.31670. [DOI] [PubMed] [Google Scholar]
- Paulus MP, Hozack NE, Zauscher BE, Frank L, Brown GG, McDowell J, et al. Parietal dysfunction is associated with increased outcome-related decision-making in schizophrenia patients. Biological Psychiatry. 2002;51:995–1004. doi: 10.1016/s0006-3223(01)01358-0. [DOI] [PubMed] [Google Scholar]
- Prata DP, Mechelli A, Fu CHY, Picchioni M, Toulopoulou T, Bramon E, et al. Epistasis between the DAT 3′ UTR VNTR and the COMT Val158Met SNP on cortical function in healthy subjects and patients with schizophrenia. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:13600–13605. doi: 10.1073/pnas.0903007106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nature Genetics. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. American Journal of Human Genetics. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell SM, Wray NR, Stone JL, Visscher PM, O’Donovan MC, Sullivan PF, et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapoport JL, Addington AM, Frangou S, Psych M. The neurodevelopmental model of schizophrenia: update 2005. Molecular Psychiatry. 2005;10:434–449. doi: 10.1038/sj.mp.4001642. [DOI] [PubMed] [Google Scholar]
- Scales SJ, Hesser BA, Masuda ES, Scheller RH. Amisyn, a novel syntaxin-binding protein that may regulate SNARE complex assembly. Journal of Biological Chemistry. 2002;277:28271–28279. doi: 10.1074/jbc.M204929200. [DOI] [PubMed] [Google Scholar]
- Schroder J, Essig M, Baudendistel K, Jahn T, Gerdsen I, Stockert A, et al. Motor dysfunction and sensorimotor cortex activation changes in schizophrenia: A study with functional magnetic resonance imaging. Neuroimage. 1999;9:81–87. doi: 10.1006/nimg.1998.0387. [DOI] [PubMed] [Google Scholar]
- Schroder J, Wenz F, Schad LR, Baudendistel K, Knopp MV. Sensorimotor Cortex and Supplementary Motor Area Changes in Schizophrenia - a Study with Functional Magnetic-Resonance-Imaging. British Journal of Psychiatry. 1995;167:197–201. doi: 10.1192/bjp.167.2.197. [DOI] [PubMed] [Google Scholar]
- Shergill SS, Bullmore E, Simmons A, Murray R, McGuire P. Functional anatomy of auditory verbal imagery in schizophrenic patients with auditory hallucinations. American Journal of Psychiatry. 2000;157:1691–1693. doi: 10.1176/appi.ajp.157.10.1691. [DOI] [PubMed] [Google Scholar]
- Strack S, Zaucha JA, Ebner FF, Colbran RJ, Wadzinski BE. Brain protein phosphatase 2A: Developmental regulation and distinct cellular and subcellular localization by B subunits. Journal of Comparative Neurology. 1998;392:515–527. [PubMed] [Google Scholar]
- Straub RE, Lipska BK, Egan MF, Goldberg TE, Callicott JH, Mayhew MB, et al. Allelic variation in GAD1 (GAD(67)) is associated with schizophrenia and influences cortical function and gene expression. Molecular Psychiatry. 2007;12:854–869. doi: 10.1038/sj.mp.4001988. [DOI] [PubMed] [Google Scholar]
- Sun J, Maller JJ, Guo L, Fitzgerald PB. Superior temporal gyrus volume change in schizophrenia: a review on region of interest volumetric studies. Brain Research Reviews. 2009;61:14–32. doi: 10.1016/j.brainresrev.2009.03.004. [DOI] [PubMed] [Google Scholar]
- Suzuki A, Yamada R, Chang X, Tokuhiro S, Sawada T, Suzuki M, et al. Functional haplotypes of PADI4, encoding citrullinating enzyme peptidylarginine deiminase 4, are associated with rheumatoid arthritis. Nature Genetics. 2003;34:395–402. doi: 10.1038/ng1206. [DOI] [PubMed] [Google Scholar]
- Volk DW, Austin MC, Pierri JN, Sampson AR, Lewis DA. Decreased glutamic acid decarboxylase(67) messenger RNA expression in a subset of prefrontal cortical gamma-aminobutyric acid neurons in subjects with schizophrenia. Archives of General Psychiatry. 2000;57:237–245. doi: 10.1001/archpsyc.57.3.237. [DOI] [PubMed] [Google Scholar]
- Vollenweider FX, Vontobel P, Oye I, Hell D, Leenders KL. Effects of (S)-ketamine on striatal dopamine: a [11C]raclopride PET study of a model psychosis in humans. Journal of Psychiatric Research. 2000;34:35–43. doi: 10.1016/s0022-3956(99)00031-x. [DOI] [PubMed] [Google Scholar]
- Walker EF, Savoie T, Davis D. Neuromotor Precursors of Schizophrenia. Schizophrenia Bulletin. 1994;20:441–451. doi: 10.1093/schbul/20.3.441. [DOI] [PubMed] [Google Scholar]
- Woo RS, Li XM, Tao YM, Carpenter-Hyland E, Huang YZ, Weber J, et al. Neuregulin-1 enhances depolarization-induced GABA release. Neuron. 2007;54:599–610. doi: 10.1016/j.neuron.2007.04.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.