Abstract
The structure activity relationship study of a small molecule Rev-erbα agonist is reported. The potency and efficacy of the agonists in a cell-based assay were optimized as compared to the initial lead. Modest mouse pharmacokinetics coupled with an improved in vitro profile make 12e a suitable in vivo probe to interrogate the functions of Rev-erbα in animal models of disease.
Rev-erbα was originally identified as an orphan nuclear hormone receptor based on its canonical domain structure.1 Rev-erbβ was identified based on its homology to other nuclear receptors (NR) and has an overlapping pattern of expression with Rev-erbα. Rev-erbs have particularly high expression in the liver, adipose tissue, skeletal muscle and brain2–4 and are expressed in a circadian manner in these tissues.5–8 The Rev-erbs are unique within the NR superfamily in that they lack the typical C-terminal AF2 domain (helix 12), which is required for coactivator protein binding. Although these receptors lack the ability to activate transcription of target genes due to their inability to recruit transcriptional coactivator proteins, both have been shown to be effective repressors of transcription due to their ability to recruit transcriptional corepressor proteins such as NCoR and HDAC3.9, 10 It has been recently demonstrated that the porphyrin heme functions as a ligand for Rev-erbα and Rev-erbβ.9–2 Heme binds reversibly and specifically to the ligand binding domain (LBD) of Rev-erb. Binding induces a conformational change in the LBD that results in the ability of the receptor to recruit NCoR and thus repress target gene transcription. The nuclear hormone receptors, Rev-erbα and Rev-erbβ, regulate a number of physiological functions including the circadian rhythm, glucose and lipid metabolism, adipogenesis, and cellular differentiation.13, 14 The observation that these NRs are ligand regulated suggests that development of synthetic ligands may be possible.
Recently, the first nonporphyrin synthetic ligand for Rev-erbα, GSK4112/SR6452/1 (Figure 1) was identified.15, 16 This ligand acts as an agonist, mimicking the action of heme and resets the circadian rhythm in a phasic manner. It also represses expression of gluconeogenic genes in liver cells and reduces glucose output in primary hepatocytes. 1 was identified in a fluorescence resonance energy transfer (FRET) assay that significantly and specifically enhances the Rev-erbα-NCoR interaction with an EC50 value of 0.40 μM. We recapitulated this data showing that SR6452 was able to modulate the interaction of either Rev-erbα or Rev-erbβ with an NCoR CoRNR box peptide using Luminex technology.17, 18 SR6452 dose-dependently increased the interaction of both Rev-erbα and Rev-erbβ with the NCoR peptide, indicating that the ligand modulates the activity of both Rer-erb subtypes. Direct binding of an analog (12e) to Rev-erbα was also confirmed by circular dichroism analysis.18
Figure 1.
GSK4112/SR6452 lead
The compound was reported to show no activity on related nuclear hormone receptors (LRH1, SF1, FXR, or RORα) using the same FRET assay and no activity on LXRα or LXRβ in reporter-gene assays. Unfortunately, the pharmacokinetic profile of 1 in rodents was poor hampering its use as an in vivo tool. Additionally, 1 had modest potency and limited efficacy in a cellular assay (in-house data). With the goal of interrogating the function of Rev-erbα in animal models of disease, we needed a more potent compound with improved potency and efficacy and an adequate in vivo profile. Based on trisubstituted amine 2, we initiated the structure-activity relationships (SAR) study described herein.
Based on the lead structure 1, the three portions of the molecule were individually modified in a step-wise fashion investigating R, R1, and R2 as in 2. The analogues 4–11 were synthesized in straightforward fashion starting from commercially available starting materials (Scheme 1). In one instance, reductive amination of t-butyl glycine (3) with p-chlorobenzaldehyde afforded secondary amine 4. Functionalization of the third amine substituent was carried out by a second reductive amination or sulfonylation or acylation as described to give products 5a–e. Alternatively, reductive amination of t-butyl glycine and 5-nitrothiophenecarboxaldehyde (6) uneventfully afforded secondary amine 7. This could then be converted to final products 8 in a similar fashion. Lastly, the 5-nitrothiophenecarboxaldehyde and p-chlorobenzylamine (9) could be condensed to yield amine 10, which was converted to final products 11.
Scheme 1.
Reagents and conditions: a. NaBH(OAc)3, HOAc, Cl(CH2)2Cl, 4-Cl-PhCHO; b. RCHO, NaBH(OAc)3, HOAc, Cl(CH2)2Cl; c. RCOCl, TEA; d. RSO2Cl, TEA; e. NaBH(OAc)3, HOAc, Cl(CH2)2Cl, H2NCH2CO2tBu; f. 5-nitrothiophenecarboxaldehyde, NaBH(OAc)3, HOAc, Cl(CH2)2Cl.
Compounds were screened in a cell-based luciferase assay in a two-step format.18–20 Cells were co-transfected with an expression plasmid harboring full-length Rev-erbα and a luciferase reporter driven by the Bmal1 promoter. Compounds were first screened at two concentrations (1 μM and 10 μM) to determine the effect on repression of Bmal1 transcription. Rev-erb is a transcriptional repressor. Rev-erb agonists lead to recruitment of co-repressors, which leads to repression of transcription. Maximum inhibition at 10 μM is reported.21 The lower the value, the more efficacious the agonist is at repressing transcription. A value of 1.0 effectively means no repression. Compounds that appeared efficacious at 10 μM were then fully titrated in an eleven-point dose response format to generate EC50 values. In our in-house cell-based assay, GSK4112/SR6452/1 showed only modest potency and minimal efficacy (Table 1).
Table 1.
Nitrothiophene Analogs
| Compound | R | aMax Inh | EC50 (μM) |
|---|---|---|---|
| 1 |

|
0.82 | 2.3 |
| 4 | H | 1.2 | bNT |
| 5a |

|
0.81 | NT |
| 5b |

|
1.1 | NT |
| 5c |

|
0.82 | NT |
| 5d |

|
0.93 | NT |
| 5w |

|
0.88 | NT |
results are average of 2 or more experiments.
Value = fold change relative to DMSO control at 10 μM compound;
NT = not tested.
All standard deviations ≤ 25%.
Given the potential for issues with the nitrothiophene residue in vivo, we assessed replacement of this group first. Several small heterocycles and carbocycles were tried as nitrothiophene isosteres, however none of them showed any improvement with regards to efficacy (Table 1). One might argue that the 4-pyridyl analog (5a) and the benzothiazole analog (5c) were equally efficacious as 1, however these early compounds were not fully titrated. Compounds 4, 5b and 5d showed no repression. Temporarily unsuccessful in replacing the nitrothiophene ring, we moved on to investigate the other two portions of the molecule.
Efforts were then focused on replacing the p-chlorobenzyl group (Table 2). The compounds shown are only a subset of those actually made however they are representative of the group. Substitutions on the benzyl group had modest effects on efficacy (8a–d) as did the naphthyl analogs (8e–f), however this did not translate into an improved EC50. Converting the amine to an amide or sulfonamide showed improved efficacy, and 8i was the first compound synthesized with an EC50 <1 μM. This represented a nice improvement over 1. Unfortunately, we were unable to assess the in vivo characteristics of 8i as this analog could not be detected in the mass spectrometer due to poor ionization under a number of conditions.
Table 2.
p-Cl-Benzyl Analogs
| Compound | R1 | aMax Inh | EC50 (μM) |
|---|---|---|---|
| 1 |

|
0.82 | 2.3 |
| 8a |

|
0.58 | 2.5 |
| 8b |

|
1.1 | bNT |
| 8c |

|
0.68 | 2.9 |
| 8d |

|
0.77 | 2.0 |
| 8e |

|
0.60 | 2.5 |
| 8f |

|
0.60 | NT |
| 8g |

|
0.50 | 3.0 |
| 8h |

|
0.48 | 2.6 |
| 8i |

|
0.37 | 0.8 |
results are average of 2 or more experiments.
Value = fold change relative to DMSO control at 10 μM compound;
NT = not tested. .
All standard deviations ≤ 25%.
Finally, we began to modify the third segment of 1 and looked to modify the acetic ester side chain (Table 3). We found that the t-butyl ester residue was not important for activity, as the corresponding methyl ester (11a), primary amide (11c), and nitrile (11d) were all equipotent. Attempts to replace the ester group with aryl and heteroaryl residues (11e–g) were slightly misleading as improved efficacy at 10 μM did not translate into an improved EC50. Saturated ring systems were accommodated (11i–k), however only one showed improved cellular EC50 (11k). Amides and sulfonamides (11l–m) showed nice improvements in efficacy, however these analogs also displayed only modest EC50's. The most potent and efficacious analog identified was carbamate 11k. In an effort to further improve the activity of 11k, we investigated ring size and linker length (Table 4).
Table 3.
Ester Analogs
| Compound | R2 | aMax Inh | EC50 (μM) |
|---|---|---|---|
| 1 |
|
0.82 | 2.3 |
| 11a |
|
0.80 | 3.9 |
| 11b |
|
0.85 | bNT |
| 11c |
|
0.75 | 3.0 |
| 11d |

|
0.79 | 2.5 |
| 11e |

|
0.70 | 8.8 |
| 11f |
|
0.45 | 5.5 |
| 11g |

|
0.50 | 4.0 |
| 11h |
|
1.0 | NT |
| 11i |

|
0.40 | 7.8 |
| 11j |

|
0.30 | 7.4 |
| 11k |
|
0.33 | 1.8 |
| 11l |

|
0.40 | 2.6 |
| 11m |

|
0.50 | 2.5 |
results are average of 2 or more experiments.
Value = fold change relative to DMSO control at 10 μM compound;
NT = not tested. .
All standard deviations ≤ 25%.
Table 4.
Pipendine and Pyrrolidine Analogs
| Compound | R2 | aMax Inh | EC50 (μM) |
|---|---|---|---|
| 11k |
|
0.33 | 1.8 |
| 11n |
|
0.80 | bNT |
| 11o |

|
0.90 | NT |
| 11p |

|
0.31 | NT |
| 11q |

|
0.14 | NT |
| 11r |

|
0.24 | 1.6 |
results are average of 2 or more experiments.
Value = fold change relative to DMSO control at 10 μM compound;
NT = not tested. .
All standard deviations ≤ 25%.
The first aspect investigated was to see if the methyl pyrrolidine side chain was optimal. We examined other 5-and 6-membered ring isomers (11n–r) with and without the methylene linker between the nitrogen atom and the ring. These analogs were synthesized simply via reductive amination with the corresponding ketone or aldehydes. All products are racemic at this stage, however, they could be made in enantiomeric fashion if desired. This might also lead to improvements in potency. The methyl pyrrolidine group in 11k was certainly better than either isomer of the piperidines (11n–o), however the analogs lacking the methylene spacer appeared to be equipotent (11p–r).
We next considered modifications to the carbamate group in 11k (Table 5). Deprotection of the t-butoxycarbonyl group (BOC) in 11k by exposure to acid was uneventful (Scheme 2). Installation of the R3-substituent via standard chemistry afforded products 12. Slightly smaller carbamates (12e,f) showed similar efficacy at 10 μM, but with nearly 3-fold improvement in EC50's. The corresponding ureas (12g–i) and sulfonamides (12j,l–m) were also equally efficacious and considerably more potent than 11k. Urea 12i and sulfonamide 12m were the most potent analogs synthesized. Amide 12c was equipotent to 11k. Removal of the BOC group and substitution with alkyl groups (12a–b) led to a substantial drop in efficacy. Clearly the additional hydrogen bond acceptors are important for activity.
Table 5.
Carbamates, amides, ureas, and sulfonamides
| Compound | R3 | aMax Inh | EC50 (μM) |
|---|---|---|---|
| 11k |
|
0.33 | 1.8 |
| 12a |

|
0.95 | bNT |
| 12b |
|
1.00 | NT |
| 12c |
|
0.35 | 1.6 |
| 12d |
|
0.55 | NT |
| 12e |
|
0.35 | 0.70 |
| 12f |
|
0.35 | 0.67 |
| 12g |
|
0.33 | 0.62 |
| 12h |
|
0.35 | 0.67 |
| 12i |
|
0.37 | 0.40 |
| 12j |
|
0.30 | 0.57 |
| 12k |
|
>1 | NT |
| 12l |
|
0.40 | 0.63 |
| 12m |
|
0.45 | 0.45 |
results are average of 2 or more experiments.
Value = fold change relative to DMSO control at 10 μM compound;
NT = not tested. .
All standard deviations ≤ 25%.
Scheme 2.
Reagents and conditions: a. TFA, CH2Cl2; b. NaBH(OAc)3, HOAc, Cl(CH2)2Cl, R3CHO; c. R3COCl, TEA; d. R3SO2Cl, TEA; e. R3NCO; f. R3OCOCl, TEA, CH2Cl2.
As a secondary screen of in vitro activity, select compounds were tested in a Gal4-Rev-erb LBD cotransfection assay. 12e dose-dependently increased the Rev-erb-dependent repressor activity assessed in HEK293 cells expressing a chimaeric Gal4 DNA binding domain (DBD): REV-ERB ligand binding domain (LBD) α or β and a Gal4-responsive luciferase reporter. It's half-maximum inhibitory concentration (IC50) against Rev-erbα was 670 nM, in good correlation with its BMAL data.18
The in vivo properties of several analogs were examined in mouse (Table 6)22. As Rev-erbα is highly expressed in the central nervous system (CNS), brain penetration was also evaluated. Mice were given a 10 mg/kg IP dose of drug, and plasma and brain levels of drug were determined 2h later. The hydrophobic lead 1 had limited exposure in plasma, although CNS penetration was good. Urea 12h had somewhat better plasma exposure, with reduced brain penetration. The reduced brain penetration is not surprising given the increased polar surface area of the urea. Carbamate 12e had slightly better plasma and brain exposure as 1. Most surprising was sulfonamide 12j which displayed the best CNS exposure. These trisubstituted amines have several metabolic soft spots which may contribute to their poor exposure and it's not clear yet the liability of the nitrothiophene ring. Interestingly, upon increasing the dose to 50 mg/kg IP for 12e, plasma exposure and brain exposure increase significantly, although drug formulation was also modified. With an EC50 = 0.7 μM, given a 50 mg/kg dose, there is over 10-fold concentration of drug in brain at t=8h.
Table 6.
In vivo properties of selected Rev-erbα agonists
| Compound | a[Plasma] (μM) | [Brain] (μM) | eb.p. (%) |
|---|---|---|---|
| b 1 | 0.25 | 0.35 | 140 |
| b 12h | 0.53 | 0.24 | 53 |
| b 12j | 0.54 | 1.3 | 242 |
| b 12e | 0.53 | 0.53 | 100 |
| c 12e | 6.7 | d8.7 | 100 |
Mice sacrificed at t = 2h. Brain and plasma levels of drug determined;
Mice dosed 10 mg/kg IP in 10:10:80 DMSO:Tween:water;
Mice dosed 50 mg/kg IP in 15% cremaphor EL;
brain levels at 8h;
b.p. = brain penetration.
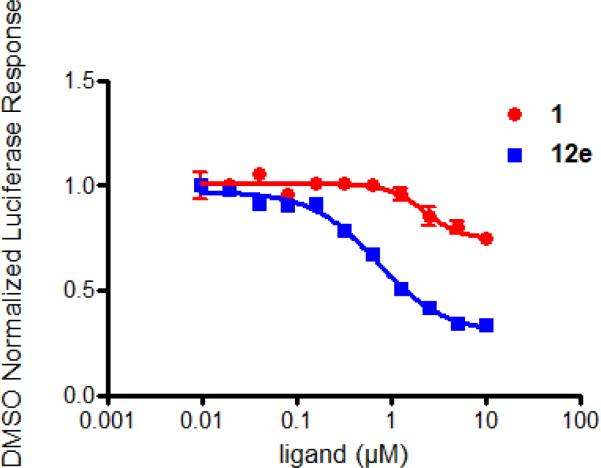
As can be seen from the curves in Figure 2, we have greatly improved the overall efficacy and potency of compounds in this series when compared to the lead GSK4112/SR6452/1. Compounds like 12e, 12h, and 12j also have good plasma and brain exposure such they might represent useful tools to study the function of Rev-erbα in vivo in models of disease. Progress in this area is on-going and will be reported in due course.
Figure 2.
Cell-based comparison between lead 1 and optimized analog 12e.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lazar MA, Hodin RA, Darling DS, Chin WW. Molecular and Cellular Biology. 1989;9(3):1128–1136. doi: 10.1128/mcb.9.3.1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dumas B, Harding HP, Choi HS, Lehmann KA, Chung M, Lazar MA, Moore DD. Molecular Endocrinology. 1994;8(8):996–1005. doi: 10.1210/mend.8.8.7997240. [DOI] [PubMed] [Google Scholar]
- 3.Forman BM, Chen J, Blumberg B, Kliewer SA, Henshaw R, Ong ES, Evans RM. Molecular Endocrinology. 1994;8(9):1253–1261. doi: 10.1210/mend.8.9.7838158. [DOI] [PubMed] [Google Scholar]
- 4.Bonnelye E, Vanacker JM, Desbiens X, Begue A, Stehelin D, Laudet V. Cell Growth & Differentiation. 1994;5(12):1357–1365. [PubMed] [Google Scholar]
- 5.Zvonic S, Ptitsyn AA, Conrad SA, Scott LK, Floyd ZE, Kilroy G, Wu XY, Goh BC, Mynatt RL, Gimble JM. Diabetes. 2006;55(4):962–970. doi: 10.2337/diabetes.55.04.06.db05-0873. [DOI] [PubMed] [Google Scholar]
- 6.Balsalobre A, Damiola F, Schibler U. Cell. 1998;93(6):929–937. doi: 10.1016/s0092-8674(00)81199-x. [DOI] [PubMed] [Google Scholar]
- 7.Guillaumond F, Dardente H, Giguere V, Cermakian N. Journal of Biological Rhythms. 2005;20(5):391–403. doi: 10.1177/0748730405277232. [DOI] [PubMed] [Google Scholar]
- 8.Torra IP, Tsibulsky V, Delaunay F, Saladin R, Laudet V, Fruchart JC, Kosykh V, Staels B. Endocrinology. 2000;141(10):3799–3806. doi: 10.1210/endo.141.10.7708. [DOI] [PubMed] [Google Scholar]
- 9.Raghuram S, Stayrook KR, Huang P, Rogers PM, Nosie AK, McClure DB, Burris LL, Khorasanizadeh S, Burris TP, Rastinejad F. Nat Struct Mol Biol. 2007;14(12):1207–13. doi: 10.1038/nsmb1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yin L, Wu N, Curtin JC, Qatanani M, Szwergold NR, Reid RA, Waitt GM, Parks DJ, Pearce KH, Wisely GB, Lazar MA. Science. 2007;318(5857):1786–1789. doi: 10.1126/science.1150179. [DOI] [PubMed] [Google Scholar]
- 11.Burris TP. Mol Endocrinol. 2008;22(7):1509–20. doi: 10.1210/me.2007-0519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kumar N, Solt LA, Wang Y, Rogers PM, Bhattacharyya G, Kamenecka TM, Stayrook KR, Crumbley C, Floyd ZE, Gimble JM, Griffin PR, Burris TP. Endocrinology. 2010;151:3015–3025. doi: 10.1210/en.2009-0800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feng D, Liu T, Sun Z, Bugge A, Mullican SE, Alenghat T, Liu XS, Lazar MA. Science. 2011;331(6022):1315–9. doi: 10.1126/science.1198125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Solt LA, Kojetin DJ, Burris TP. Future Med Chem. 2011;3(5):623–38. doi: 10.4155/fmc.11.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meng QJ, McMaster A, Beesley S, Lu WQ, Gibbs J, Parks D, Collins J, Farrow S, Donn R, Ray D, Loudon A. J Cell Sci. 2008;121(Pt 21):3629–35. doi: 10.1242/jcs.035048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grant D, Yin L, Collins JL, Parks DJ, Orband-Miller LA, Wisely GB, Joshi S, Lazar MA, Willson TM, Zuercher WJ. ACS Chem Biol. 2010;5(10):925–32. doi: 10.1021/cb100141y. [DOI] [PubMed] [Google Scholar]
- 17.Kumar N, Solt LA, Wang Y, Rogers PM, Bhattacharyya G, Kamenecka TM, Stayrook KR, Crumbley C, Floyd ZE, Gimble JM, Griffin PR, Burris TP. Regulation of adipogenesis by natural and synthetic REV-ERB ligands. Endocrinology. 2010;151(7):3015–25. doi: 10.1210/en.2009-0800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Solt LA, Wang Y, Banerjee S, Hughes T, Kojetin DJ, Lundasen T, Shin Y, Liu J, Cameron MD, Noel R, Yoo SH, Takahashi JS, Butler AA, Kamenecka TM, Burris TP. Regulation of circadian behaviour and metabolism by synthetic REV-ERB agonists. Nature. 2012 doi: 10.1038/nature11030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kojetin D, Wang YJ, Kamenecka TM, Burris TP. Identification of SR8278, a Synthetic Antagonist of the Nuclear Heme Receptor REV-ERB. Acs Chemical Biology. 2011;6(2):131–134. doi: 10.1021/cb1002575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Raghuram S, Stayrook KR, Huang P, Rogers PM, Nosie AK, McClure DB, Burris LL, Khorasanizadeh S, Burris TP, Rastinejad F. Identification of heme as the ligand for the orphan nuclear receptors REV-ERBalpha and REV-ERBbeta. Nat Struct Mol Biol. 2007;14(12):1207–13. doi: 10.1038/nsmb1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.All assays are performed as contransfections with the dual glow luciferase and data are normalized to constitutively active renilla luciferase reporter activity. This provides normalization for any non-specific effects of the compounds and we also monitor renilla luciferase activity directly to determine potential toxicity.
- 22.CNS exposure was evaluated in C57Bl6 mice (n = 3). Compounds were dosed at 10 mg/kg intraperitoneally and after 2 h blood and brain were collected. Plasma was generated and the samples were frozen at −80°C. The plasma and brain were mixed with acetonitrile (1:5 v:v or 1:5 w:v, respectively). The brain sample was sonicated with a probe tip sonicator to break up the tissue, and samples were analyzed for drug levels by LCMS/MS. Plasma drug levels were determined against standards made in plasma and brain levels against standards made in blank brain matrix. All procedures were approved by the Scripps Florida IACUC.