Abstract
Spatial normalization reshapes an individual’s brain to match the shape and size of a template image. This is a crucial step required for group-level statistical analyses. The most popular standard templates are derived from MRI scans of young adults. We introduce specialized templates that allow normalization algorithms to be applied to stroke-aged populations. First, we developed a CT template: while this is the dominant modality for many clinical situations, there are no modern CT templates and popular algorithms fail to successfully normalize CT scans. Importantly, our template was based on healthy individuals with ages similar to what is commonly seen in stroke (mean 65 years old). This template allows studies where only CT scans are available. Second, we derived a MRI template that approximately matches the shape of our CT template as well as processing steps that aid the normalization of scans from older individuals (including lesion masking and the ability to generate high quality cortical renderings despite brain injury). The benefit of this strategy is that the resulting templates can be used in studies where mixed modalities are present. We have integrated these templates and processing algorithms into a simple SPM toolbox (http://www.mccauslandcenter.sc.edu/CRNL/tools/spm8-scripts).
Keywords: voxel-based lesion symptom mapping, MRI, CT, stroke, aging, human
Introduction
Spatial normalization refers to an automated process for warping the orientation, size and shape of an individual’s brain scan to match a standard stereotaxic space. This process is useful for conducting group statistics (required for making inferences regarding a population), where tests can be applied to a group of images registered in uniform space. This application is common for functional neuroimaging studies (e.g. functional magnetic resonance imaging; fMRI) and lesion mapping studies (e.g. voxelwise lesion symptom mapping [VLSM, Bates et al. 2003; Rorden et al. 2007]). Contemporary spatial normalization algorithms are guided by template images derived from a population of neurologically healthy individuals, such that the normalization warps each individual’s brain to approximately match the shape and size of the template image. The majority of these templates are derived from healthy young adults, though specialized templates have been created for pediatric studies (Wilke et al., 2008) and healthy aging (Lemaître et al., 2005; Grabner et al., 2006). Our aim was to develop templates designed for common clinical imaging applications, i.e. for senior-adult brain injury applications, where the current templates may not be appropriate. Specifically, we developed a computerized axial tomography [CT] template that allows robust normalization for this modality, based on a population with ages comparable to typical stroke patients. CT is the dominant modality for many clinical situations yet there are no modern CT templates and popular algorithms fail to successfully normalize CT scans. Second, we created a Magnetic Resonance Imaging (MRI) template of aging adults designed to match the size and shape of our CT template. These templates allow popular neuroimaging normalization algorithms to be applied to a wider range of clinical populations.
CT remains the modality of choice for many clinical studies, with advantages typically including speed, cost, and reduced exclusion criteria relative to MRI. For example, Karnath et al. (2004) noted that CT was the preferred modality for 72 out of 140 stroke patients at admission in their university center of neurology. Likewise, it is the only modality suitable for many clinical and research situations, for example observing the consequences of improvised explosive devices (IEDs) or other penetrating head wounds which often leave metal fragments (e.g. Koenigs et al., 2008). It would be a dramatic loss of information and of representativeness if such cases would simply be neglected in anatomical studies, leaving researchers to only include those individuals who (for various clinical reasons) get an MRI brain scan during their hospital stay. Surprisingly, we know of no modern CT templates for normalization. SPM versions since the 1990’s have included the template ‘transm’ that is based on low-resolution transmission images from 11 individuals scanned with a Siemens Ecat Exact HR+, a system that was designed for Positron Emission Tomography rather than CT, and presumably the effective resolution was poor relative to modern dedicated CT scanners as the resulting template does not reveal major features such as the ventricles (Figure 1, left column). Similar to virtually all of the popular templates, this transmission template is normalized to match the MNI templates (from the Montreal Neurological Institute, Collins et al., 1994) allowing inferences to be made across studies that use different modalities. Solomon et al. (2007) described a sophisticated method to normalize CT scans to MNI space using the professional ABLe software. In this method, scans are thresholded to hide bone (bright values such as bone and the choroid plexus are set to dark intensities), the image is scalp-stripped using BET (Smith, 2002), and the resulting images are normalized using AIR (Woods et al., 1998). Despite this advanced processing, this method relies on an unsmoothed scalp-stripped template from a single young adult male acquired in the 1990s, and the quality of this image is inferior to modern CT images (Figure 1, middle column). Developing a modern open-source CT template thus could aid analysis of datasets where MRI scans are not available or not appropriate, for example the Vietnam Head Injury Study (VHIS) registry which includes 199 individuals (Koenigs et al., 2010).
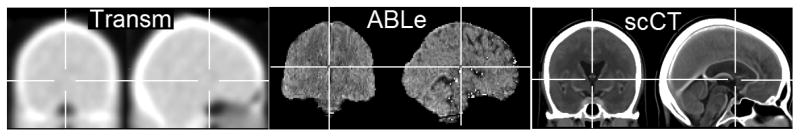
Figure 1.
CT template images. Left column: The ‘Transm’ission scan that is distributed with SPM is based on PET transmission scans from 11 young adults, note poor subcortical definition. Middle column: ABLe’s template is based on a skull-stripped low-resolution CT scan from a single healthy young adult. Right column: Our stroke-control CT (‘scCT’) template based on 30 individuals without brain injury who had a high resolution CT scan. Note that for normalization this image is blurred with a 8mm FWHM.
We suspect there are two major reasons why no modern CT templates exist. First, while CT scans have great clinical utility, they do expose participants to a dose of ionizing radiation, making it difficult to justify exposure for healthy volunteers. This is important, as template images should be generated from neurologically healthy individuals, as inclusion of participants with brain injury would lead to templates that are influenced by the deformations of shape and appearance resulting from the injury. We solved this by collecting data from individuals where a CT scan was ordered but where subsequent inspection was able to rule out major structural brain abnormalities. Second, image intensity in CT scans is calibrated (using Hounsfield units), but these units provide very little dynamic range for the CSF and soft tissue, components that are important for accurate brain alignment. Specifically, for a calibrated CT scan, approximate intensities of air are −1000, cerebral spinal fluid (CSF) 0, white matter 25, gray matter 35, blood 60, blood with gadolinium (Gd) contrast ~150, and bone 1000. Whereas MRI scans are not calibrated like CT scans, a much larger proportion of the full-contrast range is assigned to different intracranial tissue types. Therefore, normalization using popular cost-functions applied to a raw CT scan will be primarily driven by the contrast of air and bone, and features such as ventricles will not align well (an artifact that is obvious from first principles, but which we provide evidence for later). This problem is obvious for cost functions such as least-squares (as air and bone have the most relative contrast), but can also be derived from first priniciples for other cost functions such as histogram-based mutual information (where soft tissues have relatively equivalent similarity metrics). However, as commonly observed by radiologists, while the magnitude of the difference between water and parenchyma in Hounsfield units is numerically small (relative to air and bone), there is robust difference in terms of signal to noise (so radiologists can choose brightness and contrast values to see this definition). To solve this, we developed an invertible intensity transform that allows us to use the popular cost functions during normalization while emphasizing the tissue contrast between CSF and parenchyma.
Our second aim was to create a MRI template that would match the approximate size and shape of our CT template. While participants in studies of adult stroke have a mean age in their 60s (e.g. Karnath et al., 2004, 2011), most lesion mapping studies normalize data to the same MNI152 template that is distributed with popular packages such as SPM and FSL (Collins et al., 1994). According to the SPM source code, this template is based on 152 individuals (86 male) with a mean age of 25 (median=24, stdev=4.9). Therefore, the MNI152 template shows less age-related atrophy (wide sulci, large ventricles) than the average adult stroke patient. On one hand, using the MNI152 template is very useful, as it does provide a close correspondence between stroke studies and the majority of neuroimaging studies that are conducted on young individuals. On the other hand, the nonlinear functions employed creating an age-specific template for stroke patients will provide a more representative image of the actual extent and location of injury. In any case, as our CT template was drawn from a clinical aging adult population, the CT template necessarily had larger ventricles than previous templates based on young adults. Therefore, our aim was to create a MRI template that matched our CT template. The benefit of this strategy is that the resulting templates can be used in studies where mixed modalities are present (though it should be noted that the precise shape of the templates will vary slightly and that the extent of apparent injury varies across modality, requiring caution for mixed modality studies).
For our MRI template, we decided to use SPM’s unified normalization-segmentation routines (Ashburner and Friston, 2005) that have proved very robust for normalizing scans from individuals with brain lesions (Crinion et al., 2007; Andersen et al., 2010). This method works by having a priori maps of gray matter, white matter and CSF based on a population of healthy adults. This normalization creates a virtuous cycle, where improved normalization leads to improved segmentation which in turn improves the normalization.
Unfortunately, regardless of the template used, automated normalization algorithms that work well for scans from healthy individuals can perform poorly when provided with images from individuals with brain injury. For example, the non-linear components often employed in normalization can act to shrink the size of a brain lesion (Brett et al., 2001; Andersen et al., 2010), and distort the local healthy tissue (as the abnormal tissue does not match the corresponding location on the template which was derived from healthy individuals). Brett and colleagues (2001) introduced the concept of lesion cost function masking, where regions identified as unusual do not contribute to the normalization transforms. However, this technique has historically been somewhat laborious, requiring the dilation and thresholding of the mask, and the reslicing and binarization of the lesion map to standard space. Here we create a SPM toolbox that incorporates all these steps for both our CT and MRI templates, easing the use of this method.
In summary, our objective was to develop CT and MRI templates and algorithms that can aid normalizing data from older (stroke-aged) individuals. We also conducted a careful validation to demonstrate that these methods can work reliably. Specifically, our CT normalization should outperform the usage of the decades old SPM transmission scan template as well as the proprietary method included with ABLe. Likewise, our validation was designed to establish that our MRI template is competitive with previously described techniques that have proved robust for normalizing T1-wegithed images from individuals with brain injury (Crinion et al., 2007; Andersen et al., 2010)
Methods
Participants
Thirty-five high-resolution CT scans were acquired to create the CT template. These CT scans were obtained from individuals who presented emergently with specific neurological deficits (such as decreased level of consciousness or slurred speech) and were suspected to have a stroke. In all these individuals, these neurological deficits were later found to be a consequence of metabolic abnormality rather than stroke. This conclusion was based on their normal imaging findings and evidence of metabolic disorder on subsequent comprehensive medical evaluation. Two individuals were excluded for being younger than 40 years old, one was excluded for being over 90 years old, one was excluded due to a shunt that was visible on the CT, and one was excluded because the scan’s field of view did not include complete coverage of the head. Of the remaining 30 individuals, 17 were men and the mean age was 61.3 years (median = 64.8; stdev = 18.38). All CT scans used in template creation were acquired on the same Siemens Sensation 64, peak 120 kV, 348mA X-ray Tube Current. The reconstructed resolution of all the images ranged from 0.69×0.69×0.5mm to 0.45×0.45×0.5mm with full brain coverage. A representative scan is shown in Figure 2 (left column) Scans included a bolus of Gd enhancement, which does tend to make blood appear brighter than unenhanced images.
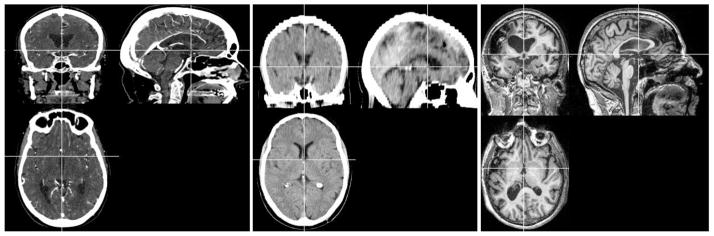
Figure 2.
Examples of the individual participant images described here. Left column: High resolution enhanced CT scan used to generate our new CT template image. Middle column: example of a low-resolution unenhanced image used to validate the CT templates. Right column: example of a T1-weighted image used for creating an aging template (neurologically healthy participants) and to validate the aging MRI template (based on individuals with chronic stroke, as shown in this example image).
Also, our desire was to have a MRI template that closely matches our CT template, though note that we do not have MRI scans from the individuals who had CT images. Therefore, data for our MRI template were drawn from previous work by our team (Bonilha et al., 2009) based on 99 individuals. Our initial selection criteria were designed to match the age of the CT template. However, using these values we observed that the MRI template exhibited dramatically smaller ventricles than the CT template previously described. We suggest this may reflect sampling error, with the CTs drawn from people who experienced an event that prompted clinical observation, whereas the participants for the MRI study volunteered and probably reflect individuals with unusually good brain health relative to their age. In addition, we excluded individuals where the scans’ field of view did not show the full range of the head covered by standard templates. Therefore we ended up selecting a subset of 50 individuals with a mean age of 72.9 years old (median = 74; stdev = 7.63), with 18 men. Note that there was a trend for a higher ratio of women in for our MRI template than our CT template (Chi-squared χ(1)=3.254, p < 0.071), potentially reflecting the population at the retirement home where we did our recruiting. These data were acquired using a Siemens Trio 3T scanner equipped with a 12-element head coil and parallel imaging located at the University of South Carolina as previously described (Bonilha et al., 2009). These images were utilized a T1-weighted MP-RAGE sequence with 1 mm isotropic voxels, TR=2250 ms, TI=900ms, TE=4.52 ms, matrix size=256×256, flip angle=9°, 160 sagittal slices.
Template creation
All CT images were manually clipped to remove excess signal from the neck and sides of the head, and the header transform was manually translated and rotated to roughly match the MNI template by an experienced neuroimager [CR]. The image intensity was then transformed from Hounsfield units using an invertible formula. The conversion from Hounsfield units (assuming the darkest voxel in the source image has an intensity of −1000) is as follows: values −1000..−100 are translated to be from 0..900, values from −99..100 are linearly scaled to the range 911…3100, and values where i>100 receive the value [i+3000]. We then used SPM8’s coregister function to linearly align each image to match the MNI152 T1 template using 12 parameters (rotations, translations, zooms and shears, each in 3 dimensions), and resliced to an isotropic 1mm resolution using 3rd-degree b-spline interpolation with an unusually large bounding box (XYZ min; max [−90 −126 −82; 90 90 108]) – we intentionally included an extra 10mm in the inferior dimension so that subsequent nonlinear functions would not be sampling beyond the edge of our final template space. Advanced Normalization Tools (ANTS) diffeomorphic normalization (Avants and Gee, 2004) was then run for four iterations using the default cost function and the affine average of all the images as a starting estimate. The output of this first ANTS run was then coregistered to MNI space using SPM, now with a standard template bounding box ([−90 −126 −72; 90 90 108]). ANTS was then re-run with the same source images but now using this new coregistered image as the template. The resulting image was now in asymmetric MNI space. To create a symmetrical template we first created two images: one based on the left and mirror-left hemisphere and the other based on the right and mirror-right hemisphere. These two images were again processed with ANTS, resulting in a symmetrical image where the left and right hemispheres contributed equally.
Our aim was to develop a MRI-based template from healthy aging adults that can be used for SPM’s unified segmentation-normalization (Ashburner and Friston, 2005). All images were manually clipped to remove excess signal from the neck and sides of the head, and the header transform was manually translated and rotated to roughly match the MNI template by an experienced neuroimager [LB]. The data were then processed with SPM8’s New Segment functions to segment gray matter, white matter and CSF tissue maps and files for DARTEL (‘Diffeomorphic Anatomical Registration using Exponentiated Lie algebra’, Ashburner, 2007). For this step we used an in-house template with standard bounding box based on healthy adults imaged using a Siemens Trio. During this stage, we also generated bias corrected images in native space. Then, SPM8’s “DARTEL create template” function was applied and followed by SPM8’s affine normalization to MNI space to create a template (Ashburner and Friston, 2009). Left and right hemispheres were averaged to yield a symmetrical template. To create an image for display purposes, we normalized each bias-corrected T1-weighted scan to this template image; as the resulting image was slightly asymmetrical we made a new image composed of a left and mirror left hemisphere and another composed of a right and mirror right hemisphere and used ANTS align these two images into a symmetrical template.
SPM Toolbox
In addition to developing new templates, we have developed a set of SPM8 scripts in the form of a plugin toolbox (http://www.mccauslandcenter.sc.edu/CRNL/tools/spm8-scripts) that allow easy implementation and batch processing using our templates. Here we describe the features of these toolboxes. As with the rest of SPM8’s code, all scripts and the images use the General Public License (either version 2, or as an option, any later version).
Our toolbox has a ‘CT normalize’ feature for dealing with computerized axial tomography. This converts a CT scan from Hounsfield units to the image brightness range of our template (described above) and then normalizes the data using SPM’s standard normalization function. After normalization the image is converted back to Hounsfield units. If a lesion image is included, this script automatically generates a dilated lesion mask that is used to ensure normalization is driven by intact brain tissue (Brett et al., 2001), and outputs a normalized binary lesion map as well as the normalized CT.
Our ‘MR segment-normalize’ function allows the user to normalize magnetic resonance imaging data. The user can select whether to use SPM’s default MNI152 template (based on young adults) or our new stroke control template based on healthy older participants. The user can specify three types of inputs: T1 only, T1 plus lesion map, or T1 with lesion map and pathological scan (e.g. FLAIR, T2, DWI). In all cases, a unified segmentation-normalization (Ashburner and Friston, 2005) will be performed on the anatomical scan. When only provided with a T1 scan, the software computes a standard unified normalization. If provided with both a T1 and lesion map the following steps are run: smooth the lesion with 3mm FWHM and 0.5 threshold (to remove jagged edges created during drawing), create dilated binary lesion mask using 8mm FWMHM and 0.001 threshold, conduct normalization with lesion masked cost function, reslice normalized T1 scan and 3mm smoothed lesion, and binarize the normalized lesion (mimicking the steps suggested by Brett et al., 2001). The resulting normalized lesion map can then be analyzed with ABLe (Solomon et al., 2007), BrainVox (Frank et al., 1997; Rudrauf et al., 2008), NPM (Rorden et al., 2007) or VLSM (Bates et al., 2003) software packages. If a pathological scan is provided, it is assumed that the lesion was drawn on that scan rather than the T1, and the pathological scan is used to coregister the lesion map to the T1 image before the standard lesion plus T1 processing steps are conducted. The toolbox also generates a normalized brain-extracted T1 image, for example with the default 0.1 threshold a binary image with where voxels have an intensity of zero unless they have at least 10% combined gray and white matter (or if the voxel was marked in the lesion map) in which case they are set to have a value of one. This binary map is smoothed with a 1mm FWHM (effectively feathering the image, which improves volume rendering surface gradient estimation) and then the T1 map is modulated using this smoothed mask. The resulting brain extracted image is useful for verifying the quality of normalization and rendering the cortical surface (as seen in Figure 5).
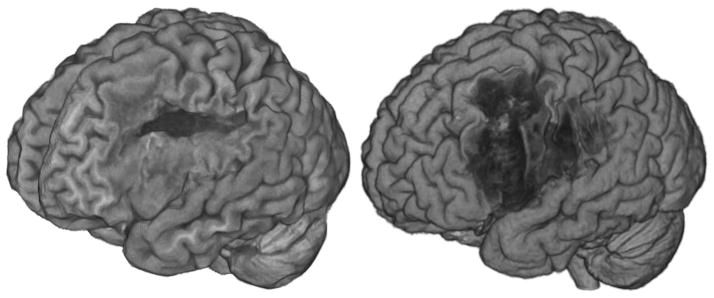
Figure 5.
Volume rendering MRI scans can be a good way to visualize the extent of brain injury. The left image reveals the performance of FSL’s Brain Extraction Technique (Smith, 2002) when provided with an image from a stroke patient. BET is not designed for brain lesions, and the quality of the extraction is not optimal. The right image shows the same participant’s image extracted using our routines and SPM8’s unified segmentation normalization. Note that when using segmentation normalization these renderings also give insight into the quality of the normalization, as misclassified tissue will be clearly visible.
Validation
In order to evaluate the performance of our CT normalization template and method, we normalized the images from twenty neurologically healthy individuals. These data came from a different neurology center, and were low-resolution, unenhanced scans. Our intention was to validate our normalization on a set of average-quality images, rather than the high-quality images used for template creation. The mean age was 56.7 years (range = 42–76, median = 56, stdev = 10.12). All CT scans used in template validation images were acquired on a Siemens Sensation 16 scanner with an in-plane matrix of 512×512 pixels, X-ray tube voltage of 120 kV, and an effective tube current time product of 285 mAs. Sequential axial sections with a thickness of 0.6 mm and an in-plane resolution of 0.43×0.43 mm2 were acquired and reconstructed to 30–40 axial sections covering the whole head, with a thickness of 4.43 – 4.75 mm. A sample image is shown in Figure 2 (middle column). All images were normalized with five methods: a clone of the proprietary functions used by the professional ABLe software, SPM8 coregistration to the T1-weighted template based on 152 individuals from the MNI, standard SPM8 normalization to the ‘transm’ template that is distributed with SPM8, normalization to our stroke-control CT template, and a hybrid of the ABLe algorithm and our own algorithm and template. Note that SPM8’s coregistration uses a normalized mutual information cost function that is suitable across modalities (adapting methods described by Maes F, Collignon et al., 1997) whereas the other SPM8 normalization routines rely on the intramodal least-squares cost function. To emulate the proprietary ABLe method, we used BET (Smith, 2002) and AIR 3.08 (Woods et al., 1998) to replicate the ABLe technique (Solomon et al., 2007). We started by setting all voxels with a Hounsfield brightness greater than 100 (centers of bone and choroid plexus) to match the dark intensity of air. Next, we manually adjusted the intensity of each image to provide a clear 8-bit grayscale image of the soft-tissue and then scalp-stripped the images using BET with a fractional intensity threshold of 0.35. For healthy control participants, the resulting images were normalized using the same AIR parameters used by ABLe (12 parameter affine normalization with the defaults as well as 25 iterations, 1e-05 convergence threshold, 5mm FWHM smoothing, standard deviation of ratio images cost function). Since we utilized the transformation matrices for our clinical CT validation, we used SPM’s normalize function instead of AIR for these scans (restricted to 12 parameter affine transform, with default settings). For the second and third methods, we tested the benefits of using the raw Hounsfield units versus image intensity rescaling using our reversible transform. For our own method (where the template is already scaled using our transform) we compared normalization with and without a brain mask. SPM8’s normalization has three stages: a coarse affine stage, a precise affine stage and finally a precise nonlinear normalization stage. The brain mask was used to weight the template image after the first stage (so that the template and source image were approximately aligned, allowing the mask to influence brain regions). For completeness, we also examined a hybrid of our own template and the technique used by ABLe where the template and source images are explicitly scalp-stripped prior to normalization (whereas our technique uses an implicit brain mask after the initial coarse affine normalization). To do this, we scalp stripped our template and the participant scans, and then conducted our SPM-based nonlinear normalization (without specifying a brain mask).
One potential criticism of the validation outlined above is that it examines neurologically healthy adults, while our ultimate aim is to provide a tool that can be applied to clinical populations with neurological injury. Therefore, we conducted a second validation using CT scans from 21 individuals with acute stroke, all of whom were right handed and 7 were men. The mean age was 68 years (range = 50–81, median = 69, stdev = 9.13), with images acquired within seven days of injury (mean 2.19). All injuries were unilateral, with 13 influencing the right hemisphere (17 were ischemic, 4 were hemorrhagic). These scans were from the same system and used the same parameters as the neurologically healthy CT validation set (see above). We normalized each of these scans using the ‘transm’ template (with our Hounsfield transform), with our clone of the ABLe method and with our new toolbox, template and lesion masking method. Next, we asked four individuals with expertise in neuroimaging (LB, JF, HOK, CR) to conduct a simple behavioral judgment task where they rated which normalization technique was most successful. On each trial, they observed two images: the left image showed a column of the middle axial, coronal and sagittal slice for an individual with the Montreal Neuroimaging Institute brain mask (as distributed with FSL, Smith et al., 2004) overlayed as a translucent red; the right column showed a similar image from the same individual except using a different normalization method. On each trial, the participant was asked to make an unspeeded button response regarding which normalization was more successful. Each participant completed 42 trials (21 trials ‘transm’ versus new; 21 trials ABLe versus new). The location of the pair of images was randomized to eliminate response biases.
The previously described validation compares alignment with the cortical surface of the MNI template. It is logically possible that normalization to a different template might lead to very consistent normalization, though not precisely in MNI space. Another concern is that a method that performed well in the previous validation could in theory be generally more accurate than a poorer-scoring method, yet it may perform poorly for certain regions of the brain (e.g. a well scoring method may be very inconsistent in one region of the brain, such as the frontal cortex). To directly address this criticism, we adapted Crinion et al.’s (2007) MRI anatomical landmark validation to our CT dataset. With this method, an observer identifies local anatomical features across a group of indivdiuals prior to normalization. The images are then normalized with different methods and analyses are then conducted to determine which method yields a tighter cluster of normalized landmarks. In other words, one of the prime assumptions of normalization is that anatomical features are aligned across individuals, and this measure tests the consistency of this alignment. However, several of the landmarks described by Crinion et al. are not easily identified in the CT modality. Therefore, we selected 19 landmarks: 9 bilateral locations (anterior, posterior and temporal horns of the lateral ventricles; quadrigeminal bodies; genu of the internal capsule; caudal end of the posterior limb of the internal capsule; limen insulae; medial end of the central sulcus; medial aspect of the amygdala) as well as the center of the splenium of the corpus callosum. For each of the clincial CT scans a neurologist with neuroimaging expertise (HOK) identified the location of the 19 landmarks using MRIcron’s “Landmark” tool (www.mricro.com). Not all landmarks were clearly identifiable or spatially discrete in each subject, and therefore we did not assign 11 of the 399 possible (19 landmarks multipled by 21 individuals; each individual contributed at least 16 landmarks and each landmark was observed in at least 17 individuals).
To validate our MRI template, we examined normalization performance on images from 29 patients with chronic left hemisphere stroke (6–350 months post stroke, lesion volume 7.7–278.1cc), as described by Fridriksson et al. (2012). MRI images were acquired using a 3T Siemens Trio equipped with a twelve-channel head coil, with both a T2-weighted and high-resolution (1×1×1mm, 256×256mm FOV, 160 sagittal slices) T1-weighted images were acquired. Lesions were drawn on the T2 image using MRIcron software (www.mricro.com), with the lesion maps coregistered to match the T1 scans using SPM8. SPM8’s display function was used to manually rotate and translate the T1 image and corresponding lesion map to roughly match the SPM template images (e.g. translations to set the anterior commissure as the origin). The T1 image and lesion were then aligned to standard space using SPM’s linear normalization features and resliced to an isotropic 1mm. This step is not typically required, but was done in this case so that all images were in roughly the same orientation for the subsequent mapping of anatomical landmarks. For each patient, an experienced neuroimager (CR) attempted to mark the 24 anatomical landmarks described by Crinion et al. (2007). Note that while Crinion et al. (2007) identified these locations in the images of healthy adults (prior to inclusion of simulated lesions), we identified these landmarks in the scans of neurological patients. Crinion et al. (2007) do not report the age of the healthy controls (nor Brett et al., 2001; who used the same dataset), however by mapping these landmarks directly onto images from older adults with injury provides a realistic measure of anatomical variability due to aging and brain injury induced shifts in anatomical location. This addresses concerns raised by Andersen et al. (2010) regarding reliance on images that are atypical of stroke research. However, as a necessary consequence of this decision, it was impossible to localize some of these landmarks in some patients (where the injury decimated the landmark). Furthermore, we found it difficult to consistently identify four of the landmarks described by Crinion et al. in our population, and therefore our analysis is based on 20 landmarks. Specifically, we found substantial variability in the occipital horn (which is bulbous in some individuals and very elongated in others, a feature that is exacerbated following stroke) and found it difficult to consistently identify the cortical occipital landmark ‘OC’ described by Crinion et al. (2007). Therefore, these two features were omitted bilaterally. In total we identified 15–20 landmarks per participant (mean 18.0). We used the location of these landmarks to compare performance of four different techniques: linear-only normalization, default unified segmentation-normalization (as espoused by Crinion et al., 2007), lesion-masked unified segmentation normalization (advocated by Andersen et al., 2010), and lesion-masked unified segmentation normalization using our new age-matched template. All normalization segmentations were performed with medium regularization. While our primary aim in developing an age-matched MRI template is to have a correspondence with our CT template, it is important to demonstrate that this template does not degrade normalization performance. Therefore, our hypothesis is that the aged-matched template would perform similarly to lesion-masked normalization using the default template (based on young adults). Based on previous work (Brett et al., 2001; Crinion et al., 2007) we expected all nonlinear methods to outperform linear-only normalization. We did not have a clear hypothesis regarding the performance of lesion masked normalization using our template versus unmasked nonlinear normalization using the default template: it is possible that unmasked normalization provides more accurate alignment of markers, but this may be at the expense of lesion volume (a criticism raised by Andersen et al., 2010).
Results
CT Validation
As suggested by Solomon et al. (2007), we used two methods to check registration of CT scans from healthy volunteers. First, we visually inspected the images to see if there was approximate registration between each source image and the template. If this succeeded for all images, we created two new images for each technique: a mean image and a standard deviation image for the group. These descriptive images were scaled in Hounsfield units (i.e. if registration was computed on transformed data, the aligned data was reconverted back to Hounsfield units). For our statistical analysis, we computed a repeated-measures t-test for the standard deviation images, comparing corresponding voxels across methods being contrasted. As we wanted to test alignment within the brain, this test was only computed for voxels that have a 50% or greater probability of being brain as indexed on the ‘brainmask’ image that is distributed with SPM8. As all images were in Hounsfield units, this measures how well tissues are aligned, providing a direct index of how well normalization is matching features across the group. For this statistical analysis we added one extra step to the ABLe algorithm: the transforms computed by AIR for the scalp-stripped image were also applied to the original unstripped images (such that any minor BET errors near the brain surface would not cause errors when computing the group standard deviation).
Visual inspection suggested that ABLe provides a reliable method for normalization. Coregistration of CT scans to the MNI 152 T1-weighted MRI template consistently failed, regardless of whether the transforms were computed for Hounsfield or transformed intensities (see ‘to T1’ row in Figure 3). Specifically, the CT brains were scaled to be smaller than the template brains, suggesting that different tissues were being coregistered across the modalities. Normalization of the raw Hounsfield CT scans to the ‘transm’ template consistently failed, perhaps because air has negative values in the CT scans, but SPM fills edges with zeros. On the other hand, the intensity transformed CT scans did normalize to the ‘transm’ template (see ‘to Transm’ row of Fig. 3). Finally, visual inspection suggests that images also successfully normalized to our new CT template, regardless of whether the brain mask was used (row ‘to scCT’ in Figure 3 shows performance when a brain mask is used). Visual inspection suggests sharper edges in the mean image when the new CT template is used relative to the ‘transm’ template (see Fig. 4). Repeated measures t-tests confirm that the CT template with brain mask outperforms the ABLe template and technique (t[238624]=109; mean SD = 19 versus 30 Hounsfield units), the ‘transm’ template (t[238624]=396; mean SD = 19 versus 195 Hounsfield units), and also outperforms the CT template without the brain mask (t[238624]=158; mean SD = 19 versus 31 Hounsfield units). Finally, our implicit mask outperformed the hybrid method (an explicit scalp stripped version of our template and the source images) (t[238624]=105; mean SD = 19 versus 29 Hounsfield units), . These analysis survive a p<0.001 corrected for multiple comparisons using the Bonferroni correction.
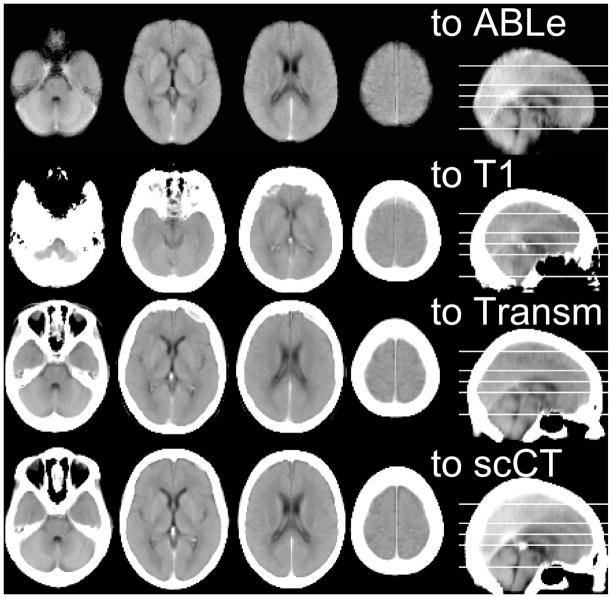
Figure 3.
Mean images for the same 20 neurologically healthy individuals normalized using four different normalization techniques. Poor normalization can be observed as blurriness (poor alignment of individuals) and incorrect overall alignment to MNI coordinates (with axial slices corresponding to −32, 0, 24 and 60mm respectively). The top row (‘to ABLe’) shows our simulation of ABLe’s proprietary method (which uses explicit brain extraction, removing scalp and bone signals). The second row (‘to T1’) shows coregistration to SPM8’s default T1 template image, note that the overall size of the brain is smaller than other methods demonstrating misregistration of tissues. The third row (‘to Transm’) shows normalization to SPM8’s transmission template (using our novel image intensity transform, otherwise this method consistently and dramatically failed, not shown), some blurriness is visible relative to other methods. The bottom row (to scCT) shows normalization using our new template and intensity transform, note relatively crisp alignment of tissues.
Figure 4.
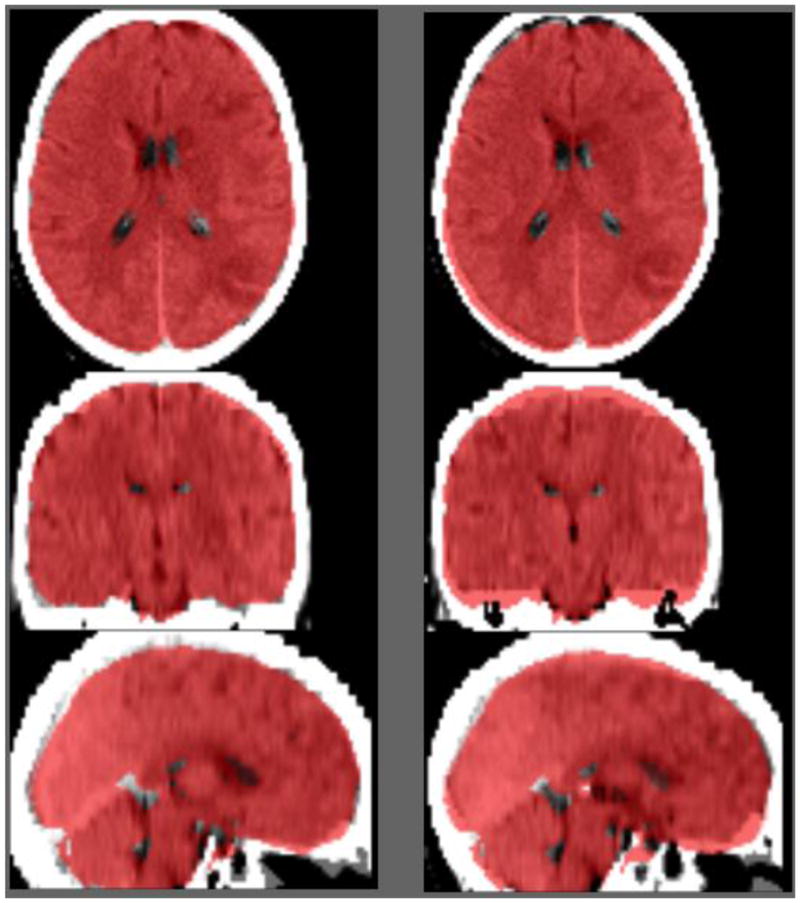
Example trial in our two-alternative forced choice trial. On each trial the observer examined the normalization of an acute stroke patient’s CT scan using two different normalization methods. The MNI brain mask (as distributed with FSL) is displayed overlayed on top of each image (red). The task is to report whether the left or right normalization was more successful. In this example, the normalization of the image using our new template is shown on the left and the performance of our ABLe clone is shown on the right.
We also conducted a validation based on individuals with acute stroke. Since the lesions can lead to changes in image intensity, the validation method outlined for healthy individuals could not be applied to this group. Therefore, we developed the visual inspection task described in the methods section, and illustrated in Figure 4. The results of this two-alternative forced choice task were compared using binomial probability (testing the null hypothesis that participants could not distinguish between normalization methods, and hence the probability of preferring one of the two methods displayed was 50%). All four participants preferred the new method to the ABLe method in 21 of 21 trials (p < 0.00000048), whereas three preferred the new method over ‘transm’ in 21 of 21 trials and one of the individuals preferred the new method over ‘transm’ in 17 of 21 trials (0.0036). All results survive Bonferroni correction for multiple comparisons.
Our final CT validation test compared the performance of different techniques when normalizing the location of 19 anatomical landmarks. To accomplish this we used SPM8’s deformation toolbox and Ged Ridgway’s map_coords function (www.cs.ucl.ac.uk/staff/g.ridgway/vbm/) to transform landmark coordinates from native to normalized space. For each method, we calculated the mean location for each landmark across the group and then determined the distance of each individual’s landmark from the group mean (using Pythagorean theorem, providing our measure of error). We then used OpenStat (www.statprograms4u.com) to conduct a two-factor repeated-measures ANOVA with three levels for technique (our new method, ABLe, and our intensity transform followed by normalization to the Transm template) and nineteen levels for anatomical landmarks. We found a main effect for technique (F(2,40)=3.607 p < 0.036) and landmark location (F(18,360)=4.394 p< 0.0005), but no evidence for an interaction between technique and landmark location (F(36,720)=0.761 p< 0.844). Numerically, our new method (mean error 4.48mm, root mean square error 4.96mm) outperformed ABLe (4.79 and 5.43mm) and the Transm normalization (5.19 and 5.66mm). To understand the main effect of technique, we conducted planned paired comparisons using the repeated measures t-test and found our new method outperformed ABLe (t(387)=3.03, p<0.0026) as well as the Transm normalization (t(387)=10.37, p<0.00005), while ABLe outperformed the Transm method (t(387)=3.728, p<0.0002). The main effect of anatomical locations is not particularly interesting as it reflects that in all three techniques (our new method, ABLe, and Transm) the location of some anatomical landmarks are more variable across participants than are others. While it is always difficult to interpret null results, the failure to observe a clear interaction between technique and location suggests that no method is atypically inconsistent across regions.
MRI Validation
For each method, we computed the normalized location of each anatomical landmark and for each individual measured the distance from that location to the group mean landmark location. For each individual, we computed the root-mean-square error for all available landmarks. We then conducted a repeated measure single factor Analysis of Variance (ANOVA) with four levels: linear-only normalization (affine), default unified normalization-segmentation (unified), lesion-masked normalization-segmentation (masked) and lesion-masked normalization-segmentation using our aging-adult template (aging). We found a main effect of normalization type (F(3,84) = 28.3, Greenhouse-Geisser-Corrected {0.3719} p<0.0000045). Numerically, the mean performance (in units of mm root-mean-square error) from worst to best was affine (6.08), unified (5.01), masked (4.96), aging (4.89). Planned pair-wise comparisons (reporting two-tailed p-values, not corrected for multiple comparisons, all 28 degrees of freedom) showed that the aging method outperformed the affine (t=5.27 p < 0.0001), unified (t=2.3, p < 0.028) and had similar performance to the masked method (t=1.48, p <0.148). For completeness, is should be noted that linear performed poorer than masked (t=5.41 p< 0.0001) and unified (t=5.27 p< 0.0001), while there was a trend for masked to outperform unified (t=1.73 p< 0.0949).
Discussion
There has been a rapid advance in statistical methods for correlating brain injury with subsequent neurological symptoms including stroke, brain resection and tumors (Frank et al., 1997; Rorden and Brett, 2000; Bates et al., 2003; Bonilha et al., 2007; Kimberg et al., 2007; Rorden et al., 2007; Solomon et al., 2007; Rudrauf et al., 2008; Wick et al., 2008). These methods allow us to see how brain injury disrupts brain function, complementing findings based on other methods that examine functional brain activation. Despite these advances, there are no modern templates to aid the normalization of CT scans, which remain one of the popular clinical modalities for visualizing brain injury. Our primary aim thus was to develop a robust method for normalizing CT scans, which can work with both high-quality and standard quality images.
During validation we found that a popular open source method for MRI normalization consistently fails when applied to CT scans. While a simple (and invertible) intensity transform allow reasonable normalization using the Transm template distributed with SPM (landmark alignment for clinical quality CT scans where brain injury is present yielded a root mean square error of 5.66mm), we found that our new template yielded reliably better normalization (root mean square error of 4.96mm). However, as with all normalization routines, we emphasize the importance of manually setting the origin and angulation of the source image to approximately match the template. For most MRI scans, the origin recorded in the DICOM header refers to the scanner’s isocenter, and is therefore often somewhat near the template origin (which for SPM is the anterior commissure). However, we found that many CT scanners appear to list the table center as the origin, and therefore the origin recorded in the DICOM headers of CT scans are often dramatically different from the template origin. Also, it should be emphasized that the popular neuroimaging tools (and the NIfTI format that they use) assume that interslice spacing is equidistant, whereas clinical scans are sometimes taken with thinner slices near the posterior fossa and thicker slices through the vertex. Therefore, equidistant acquisitions are preferable, and care should be taken when converting and interpolating non-equidistant acquisitions. Nevertheless, our validation analysis suggests that our template and associated normalization toolbox provides a robust method for normalizing clinical CT scans. As a result, we believe our CT template and algorithm can be applied to a wide range of clinical applications.
We also developed an MRI template that is quite similar in size and shape to our CT template. As this new template is based on older adults, it may be more appropriate for studies including stroke patients than templates based on younger adults. In theory, the CT template and MRI template from older adults could be combined in a study where one does not have either MRI or CT scans from all individuals but a mixture of both (c.f. Dronkers, 1996; Bates et al., 2003; Karnath et al., 2004, 2011). This is particularly useful in studies aiming to describe the fundamental brain areas related to a certain behavioral disorder in a representative sample of brain-injured individuals by including each and every adequate stroke patient admitted. By using the new CT and MRI templates, no subjects need to be excluded simply due to the type of scanning they obtained at admission; biases related to clinical constellations that direct initial CT scanning or initial MRI scanning can be avoided and the rich data set from a typical clinical population admitted to a neurology center can be used to address the scientific question. On the other hand we note that each modality (even within MRI, e.g. T1, T2, DWI, perfusion) is sensitive to different properties, and therefore mixed modality studies will probably have greater variance (and therefore lower statistical power). Further, it is crucial that modality is orthogonal to the dependent variables. Our validation demonstrated that this template performed statistically similar and numerically better than when using the default templates based on young adults (as suggested by Andersen et al., 2010) and outperforming unmasked normalization to a template based on young adults (as suggested by Crinion et al., 2007). Our results provide corroboration for Andersen et al.’s (2010) contention that lesion masking remains important for many real world (versus simulated) lesions, in contrast to the claims of Crinion et al. (2007). Future users should use their discretion when selecting between our older-adult template and the default SPM-young adult template: our aging MRI template is more similar to our CT template and demands less ventricle deformation when applied to older adults, though the young adult template yields coordinates more similar to functional imaging studies conducted on young adults.
As mentioned previously, our toolbox employs SPM’s unified segmentation normalization in order to provide a robust registration of anatomical images. One consequence of this approach is that one necessarily estimates the location of gray and white matter during the normalization. Our toolbox uses these estimates to generate a scalp-stripped image of the normalized scan (removing all tissue that is not classified as gray matter, white matter, or part of a lesion mask). Visual inspection of a rendering of this image is useful for detecting poor normalization, as misclassified tissue will be clearly visible. While there are robust tools for brain extraction of images from healthy adults (Shattuck et al., 2009), these routines can fail when a brain injury is present, whereas our tool explicitly preserves lesioned tissue. Therefore, our toolbox provides an easy method for visualizing cortical abnormalities. Figure 5 demonstrates the performance of a popular scalp-stripping tool that was not designed for use with clinical populations (Smith, 2002) and the extraction method we describe here. The resulting renderings can help visualize the extent of brain injury in stroke, or aid in surgical planning when tumors or depth electrodes are present.
There is one important caveat that applies to our templates relative to the default templates distributed with SPM. The default templates distributed with SPM are asymmetric, whereas our templates are symmetrical. The human brain has functional and anatomical asymmetries (Toga and Thompson, 2002), and the templates typically used with popular software exhibit subtle asymmetries. While our templates were carefully aligned to match the MNI templates, our final images are intentionally made symmetrical. Our decision was biased by the small number of healthy high-resolution CT scans we had available for creating our CT template, and therefore by averaging hemispheres we hoped to reduce any idiosyncratic features. However, we also feel there is a clear rationale for creating symmetrical templates for analyzing brain injury. First of all, many studies of brain injury intentionally flip lesions to collapse across lesion side (Donchin et al., 2012; Zeller et al., 2011; Küper et al., 2011). Asymmetrical templates will pose challenges if the flipping is done after normalization, and can lead to differential biases if flipping occurs prior to normalization (e.g. individuals with unflipped brains will have slightly stronger correspondence to the template than unflipped). In addition, mirror flipping is a method to examine the fate of a lesions’ homologue (Richardson et al., 2011), which assumes hemispheric symmetry. Finally, if one wants to make comparisons regarding the difference between the left and right hemisphere it is critical to start with an unbiased template, such that any differences observed reflect the observations rather than differences inherent in the template. In any case, we emphasize that the symmetry differences are subtle and our templates are smoothed. Indeed, it is worth noting that our two-alternative forced choice validation of the CT template used the asymmetrical MNI brain map overlay as a reference, yet observers reliably judged that our symmetrical template outperformed other methods that used asymmetrical templates.
While the most popular templates are tuned for work with young adults (the population where most current theoretical neuroimaging research is focused), our aim was to develop high-quality templates for common clinical applications. Our templates are based on healthy aging adults that can help scientists and clinicians understand the consequences and treatment of brain injury that is common in this age group (tumors, stroke). In particular, the CT template combined with algorithmic changes (reversible transformation of image brightness) provides a robust, open source method for normalizing this modality. As neuroimaging matures, there is a pressing need to translate robust methods to tackle clinically relevant problems. We believe the templates and techniques described here will enhance such endeavors.
Acknowledgments
This work was supported by the National Institutes of Health (NS054266; DC009571), and the Deutsche Forschungsgemeinschaft (KA 1258/10-1). We thank Johannes Rennig, Julia Suchan and Urszula Mihulowicz, University of Tuebingen, for their help with data screening and organization in order to test the new CT template. Jeff Solomon helped us clone the normalization routines of ABLe.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Andersen SM, Rapcsak SZ, Beeson PM. Cost function masking during normalization of brains with focal lesions: still a necessity? Neuroimage. 2010;53(1):78–84. doi: 10.1016/j.neuroimage.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner J, Friston KJ. Unified segmentation. NeuroImage. 2005;26:839–851. doi: 10.1016/j.neuroimage.2005.02.018. [DOI] [PubMed] [Google Scholar]
- Ashburner J. A fast diffeomorphic image registration algorithm. NeuroImage. 2007;38(1):95–113. doi: 10.1016/j.neuroimage.2007.07.007. [DOI] [PubMed] [Google Scholar]
- Ashburner J, Friston KJ. Computing average shaped tissue probability templates. Neuroimage. 2009;45(2):333–41. doi: 10.1016/j.neuroimage.2008.12.008. [DOI] [PubMed] [Google Scholar]
- Avants B, Gee JC. Geodesic estimation for large deformation anatomical shape averaging and interpolation. Neuroimage Suppl. 2004;1:S139–50. doi: 10.1016/j.neuroimage.2004.07.010. [DOI] [PubMed] [Google Scholar]
- Bates E, Wilson SM, Saygin AP, Dick F, Sereno MI, Knight RT, Dronkers NF. Voxel-based lesion-symptom mapping. Nat Neurosci. 2003;6(5):448–50. doi: 10.1038/nn1050. [DOI] [PubMed] [Google Scholar]
- Bonilha L, Yasuda CL, Rorden C, Li LM, Tedeschi H, de Oliveira E, Cendes F. Does resection of the medial temporal lobe improve the outcome of temporal lobe epilepsy surgery? Epilepsia. 2007;48(3):571–8. doi: 10.1111/j.1528-1167.2006.00958.x. [DOI] [PubMed] [Google Scholar]
- Bonilha L, Eckert MA, Fridriksson J, Hirth VA, Moser D, Morgan PS, Rorden C. Age-related relative volume preservation of the dominant hand cortical region. Brain Res. 2009;1305:14–9. doi: 10.1016/j.brainres.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brett M, Leff AP, Rorden C, Ashburner J. Spatial normalization of brain images with focal lesions using cost function masking. Neuroimage. 2001;14(2):486–500. doi: 10.1006/nimg.2001.0845. [DOI] [PubMed] [Google Scholar]
- Maes F, Collignon A, Vandermeulen D, Marchal G, Suetens P. Multimodality image registration by maximization of mutual information. IEEE Trans Med Imaging. 1997;16(2):187–98. doi: 10.1109/42.563664. [DOI] [PubMed] [Google Scholar]
- Collins DL, Neelin P, Peters TM, Evans AC. Automatic 3D intersubject registration of MR volumetric data in standardized Talairach space. J Comput Assist Tomogr. 1994;18:192–205. [PubMed] [Google Scholar]
- Crinion J, Ashburner J, Leff A, Brett M, Price C, Friston K. Spatial normalization of lesioned brains: performance evaluation and impact on fMRI analyses. Neuroimage. 2007;37(3):866–75. doi: 10.1016/j.neuroimage.2007.04.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donchin O, Rabe K, Diedrichsen J, Lally N, Schoch B, Gizewski ER, Timmann D. Cerebellar regions involved in adaptation to force field and visuomotor perturbation. J Neurophysiol. 2012;107(1):134–47. doi: 10.1152/jn.00007.2011. [DOI] [PubMed] [Google Scholar]
- Frank RJ, Damasio H, Grabowski TJ. Brainvox: an interactive, multimodal visualization and analysis system for neuroanatomical imaging. Neuroimage. 1997;5(1):13–30. doi: 10.1006/nimg.1996.0250. [DOI] [PubMed] [Google Scholar]
- Fridriksson J, Richardson JD, Fillmore P, Cai B. Left hemisphere plasticity and aphasia recovery. NeuroImage. 2012 doi: 10.1016/j.neuroimage.2011.12.057. [YNIMG-09034] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabner G, Janke AL, Budge MM, Smith D, Pruessner J, Collins DL. Symmetric atlasing and model based segmentation: an application to the hippocampus in older adults. Med Image Comput Comput Assist Interv. 2006;9(2):58–66. doi: 10.1007/11866763_8. [DOI] [PubMed] [Google Scholar]
- Hartley SW, Scher AI, Korf ES, White LR, Launer LJ. Analysis and validation of automated skull stripping tools: a validation study based on 296 MR images from the Honolulu Asia aging study. Neuroimage. 2006;30(4):1179–86. doi: 10.1016/j.neuroimage.2005.10.043. [DOI] [PubMed] [Google Scholar]
- Karnath HO, Fruhmann Berger M, Küker W, Rorden C. The anatomy of spatial neglect based on voxelwise statistical analysis: a study of 140 patients. Cereb Cortex. 2004;14:1164–72. doi: 10.1093/cercor/bhh076. [DOI] [PubMed] [Google Scholar]
- Karnath HO, Rennig J, Johannsen L, Rorden C. The anatomy underlying acute versus chronic spatial neglect: a longitudinal study. Brain. 2011;134:903–912. doi: 10.1093/brain/awq355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimberg DY, Coslett HB, Schwartz MF. Power in Voxel-based lesion-symptom mapping. J Cogn Neurosci. 2007;19(7):1067–80. doi: 10.1162/jocn.2007.19.7.1067. [DOI] [PubMed] [Google Scholar]
- Koenigs M, Huey ED, Raymont V, Cheon B, Solomon J, Wassermann EM, Grafman J. Focal brain damage protects against post-traumatic stress disorder in combat veterans. Nat Neurosci. 2008;11(2):232–7. doi: 10.1038/nn2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenigs M, Holliday J, Solomon J, Grafman J. Left dorsomedial frontal brain damage is associated with insomnia. J Neurosci. 2010;30(47):16041–3. doi: 10.1523/JNEUROSCI.3745-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Küper M, Brandauer B, Thürling M, Schoch B, Gizewski ER, Timmann D, Hermsdörfer J. Impaired prehension is associated with lesions of the superior and inferior hand representation within the human cerebellum. J Neurophysiol. 2011;105(5):2018–29. doi: 10.1152/jn.00834.2010. [DOI] [PubMed] [Google Scholar]
- Lemaître H, Crivello F, Grassiot B, Alpérovitch A, Tzourio C, Mazoyer B. Age- and sex-related effects on the neuroanatomy of healthy elderly. Neuroimage. 2005;26(3):900–11. doi: 10.1016/j.neuroimage.2005.02.042. [DOI] [PubMed] [Google Scholar]
- Richardson JD, Baker JM, Morgan PS, Rorden C, Bonilha L, Fridriksson J. Cerebral perfusion in chronic stroke: implications for lesion-symptom mapping and functional MRI. Behav Neurol. 2011 Jan 1;24(2):117–22. doi: 10.3233/BEN-2011-0283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rorden C, Brett M. Stereotaxic display of brain lesions. Behav Neurol. 2000;12(4):191–200. doi: 10.1155/2000/421719. [DOI] [PubMed] [Google Scholar]
- Rorden C, Karnath HO. Using human brain lesions to infer function: a relic from a past era in the fMRI age? Nat Rev Neurosci. 2004;5(10):813–9. doi: 10.1038/nrn1521. [DOI] [PubMed] [Google Scholar]
- Rorden C, Karnath HO, Bonilha L. Improving lesion-symptom mapping. J Cogn Neurosci. 2007;19(7):1081–8. doi: 10.1162/jocn.2007.19.7.1081. [DOI] [PubMed] [Google Scholar]
- Rudrauf D, Mehta S, Bruss J, Tranel D, Damasio H, Grabowski TJ. Thresholding lesion overlap difference maps: application to category-related naming and recognition deficits. Neuroimage. 2008;41(3):970–84. doi: 10.1016/j.neuroimage.2007.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shattuck DW, Prasad G, Mirza M, Narr KL, Toga AW. Online resource for validation of brain segmentation methods. Neuroimage. 2009;5(2):431–9. doi: 10.1016/j.neuroimage.2008.10.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon J, Raymont V, Braun A, Butman JA, Grafman J. User friendly software for the analysis of brain lesions (ABLe) Comput Methods Programs Biomed. 2007;86:245–254. doi: 10.1016/j.cmpb.2007.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM. Fast robust automated brain extraction. Human Brain Mapping. 2002;17(3):143–55. doi: 10.1002/hbm.10062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM, Jenkinson M, Woolrich MW, Beckmann CF, Behrens TE, Johansen-Berg H, Bannister PR, De Luca M, Drobnjak I, Flitney DE, Niazy RK, Saunders J, Vickers J, Zhang Y, De Stefano N, Brady JM, Matthews PM. Advances in functional and structural MR image analysis and implementation as FSL. Neuroimage. 2004;23:S208–S219. doi: 10.1016/j.neuroimage.2004.07.051. [DOI] [PubMed] [Google Scholar]
- Toga AW, Thompson PM. Mapping brain asymmetry. Nat Rev Neurosci. 2003 Jan;4(1):37–48. doi: 10.1038/nrn1009. [DOI] [PubMed] [Google Scholar]
- Wick W, Stupp R, Beule AC, Bromberg J, Wick A, Ernemann U, Platten M, Marosi C, Mason WP, van den Bent M, Weller M, Rorden C, Karnath HO. A novel tool to analyze MRI recurrence patterns in glioblastoma. Neuro Oncol. 2008;10(6):1019–24. doi: 10.1215/15228517-2008-058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilke M, Holland SK, Altaye M, Gaser C. Template-O-Matic: a toolbox for creating customized pediatric templates. Neuroimage. 2008;41(3):903–13. doi: 10.1016/j.neuroimage.2008.02.056. [DOI] [PubMed] [Google Scholar]
- Woods RP, Grafton ST, Watson JD, Sicotte NL, Mazziotta JC. Automated image registration: II. Intersubject validation of linear and nonlinear models. J Comput Assist Tomogr. 1998;22(1):153–65. doi: 10.1097/00004728-199801000-00028. [DOI] [PubMed] [Google Scholar]
- Zeller D, Gross C, Bartsch A, Johansen-Berg H, Classen J. Ventral premotor cortex may be required for dynamic changes in the feeling of limb ownership: a lesion study. J Neurosci. 2011;31(13):4852–7. doi: 10.1523/JNEUROSCI.5154-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]