It is probably fair to say that DNA structure holds center stage in many processes of genetic recombination. Understanding the conformation of branched DNA intermediates and their recognition and manipulation by proteins is the key to deducing the underlying mechanisms. In the original Holliday (1) model for homologous genetic recombination, the central intermediate is a four-way junction where four DNA helices are linked by the covalent continuity of the individual strands. The three-dimensional structure of this species has been much investigated in the last decade, and a general consensus has emerged from a variety of experiments. It is important, however, not to be seduced by too static a view of the structure, and new experiments by David Millar, Walter Chazin, and their coworkers (2) have highlighted the dynamic character of the junction.
In the absence of added metal ions, the four-way junction is extended with an open central region (3, 4). However, upon addition of magnesium or other metal ions the junction folds by pairwise coaxial stacking of helices into the stacked X-structure (reviewed in refs. 5 and 6) (Fig. 1). This structure is a substrate for a variety of junction-selective enzymes, most of which alter the global structure on binding (reviewed in ref. 7). In free solution, the DNA junction folds to create an antiparallel structure, probably governed by the favorable juxtaposition of backbones and grooves when the small angle is approximately 60°. The lowering of symmetry on formation of the stacked X-structure creates two inequivalent kinds of strands; the two continuous strands turn about the pseudo-continuous axes that pass through the stacked helical pairs, while the exchanging strands pass between the stacked helices at the crossover. The general features of the stacked X-structure have been confirmed by a variety of experimental methods (3, 8–11), including more recent NMR studies (12–14).
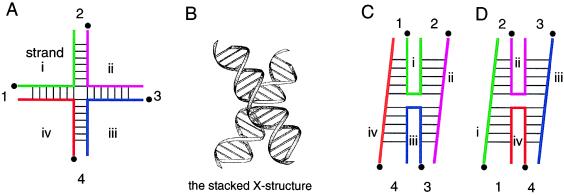
Figure 1.
The four-way DNA junction. (A) A four-way junction may be considered as four helices held together by the strands that exchange from one to the next. The strands are labeled i, ii, iii, and iv, and the helical arms 1, 2, 3, and 4. The 5′ ends of the strands are denoted by the •. (B) In the presence of magnesium ions, the junction folds by pairwise coaxial stacking into an antiparallel X-shaped structure. Two possible conformers of this structure differ in the nature of the stacking partners. In C, helices 1 and 4 are stacked, as are helices 2 and 3, while in D, helix 1 is stacked with helix 2, and helix 3 with helix 4. Note that the nature of all strands is changed by interconversion between stacking conformers; e.g., strands i and iii are exchanging strands in conformer C, but become continuous strands in D. The view represented by C and D is rotated 90° with respect to that in B.
Two possible alternative forms of the stacked X-structure exist. They frequently are called isomers, but Altona (15) has pointed out that these are more properly termed conformers. These species correspond to the two alternative choices of stacking partners (Fig. 1 C and D). It could be envisaged that a given junction might exist in both forms. Given free interconversion, the relative populations would be determined by the difference in free energy between the two conformers. The interconversion of stacking conformers changes the character of every strand; thus exchanging strands become continuous strands and vice versa. It also may change the neighboring stacked base pairs. It is these interactions that are likely to be primary determinants of relative conformer stability. Perhaps by good fortune, most of the junctions first studied were found to exist predominantly in one stacked conformation (3, 11). However, the methods used were not very sensitive, and less than 10% of the alternative conformer probably would not have been detected. Thus even relatively small free energy differences between the forms would have been sufficient to give this result. Nevertheless, there were indications that not all junctions behaved in this way. We studied one particularly G+C-rich junction that gave results consistent with a mixture of equally populated conformers (D. R. Duckett and D.M.J.L., unpublished data). Moreover, when methods that could detect minor populations of alternative stacking forms were used, these were found even for the well behaved junctions. Thus when the restriction enzyme MboII was used to probe the structure of one well studied junction, it was found that a low level of the alternative stacking conformer was present, and that this must be in a freely interconverting equilibrium with the major conformer (16). It is this aspect that has been addressed directly in the new study.
Structural studies of four-way junctions frequently have been a stimulus for the application of new approaches to the study of nucleic acid conformation, and this is true of the new work. The groups of Millar and Chazin (2) have combined their expertise to generate a two-pronged approach to examine the conformational equilibrium for three DNA junctions. One technique used is time resolved fluorescence resonance energy transfer (trFRET) to gain long-range distance information between the ends of designated arms of the junctions. The other is an NMR approach that provides local information on the environment of a base present at the point of strand exchange.
In a FRET approach, two different fluorophores are chemically attached to the ends of selected helical arms, and the efficiency of energy transfer is measured spectroscopically. Because the dipolar coupling is inversely related to the sixth power of the distance between the dyes, the efficiency is higher when the dyes are closer together. This method originally was used in the steady state to demonstrate the antiparallel character of the four-way DNA junction (17). Millar and Chazin, et al. (2) have placed fluorescent labels onto two arms such that the distance between them will be either long or short, depending on the choice of stacking partners (Fig. 2). In the time-resolved mode the decay of the excited state of the fluorescein donor is measured (18), and the decay has been deconvoluted in terms of distances corresponding to the two stacking conformers.
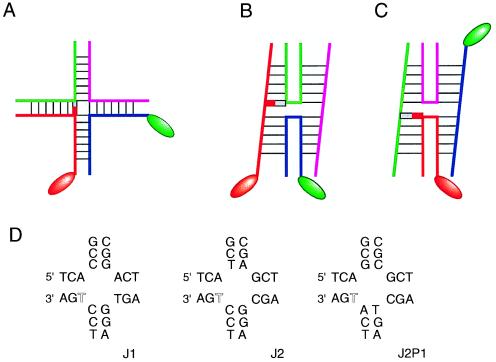
Figure 2.
The experimental approach of Millar, Chazin, et al. (2). (A) The modifications made to the junction in its extended form. In the FRET experiments, donor and acceptor fluorophore pairs were attached to the ends of two different helical arms (indicated by the oval shapes). In one stacking conformer (B) these are relatively close together across the narrow angle of the stacked X-structure, while in the other they are at opposite ends of a stacked pair of helices (C). For the NMR experiments a thymine base at the point of strand exchange (indicated by the red block) was modified by substitution of ring nitrogen atoms by the spin 1/2 15N. In conformer B the substituted thymine is located on the continuous strand, whereas in C it is on the exchanging strand. The central sequences of the three junctions studied are shown in D with the modified thymine highlighted in open type.
The NMR method used in this study depends critically on the selective introduction of a single 1,3 15N-labeled thymine base at the point of strand exchange. The local magnetic environment can be probed either by the 15N nucleus itself (using two-dimensional heteronuclear multiple quantum correlation spectra) or by examining the attached hydrogen-bonded imino proton (by means of a one-dimensional 15N-filtered 1H-detected NMR experiment). The labeled thymine base is attached to a continuous strand in one stacking conformer (Fig. 2), whereas exchange of stacking moves it to an exchanging strand (and also may change the nature of the neighboring base stacked on one side).
Three different four-way DNA junctions were compared in these experiments, differing only in arrangement of the central base pairs. In the first junction (J1) examined it was found that the trFRET data could be accounted for by a single (>95%) end-to-end distance, and only one environment was detected for the imino proton at 13 ppm. Although these results indicated that J1 existed predominantly as a single conformer with a unique choice of stacking partners, this contrasted markedly with the other two junctions examined. J2 can be obtained from J1 by the exchange of two central base pairs. It was found that the altered sequence gave trFRET data corresponding to two inter-fluorophore distances, and two 15N resonances were detected, indicative of similar populations of the two stacking conformers. A further junction, J2P1, was constructed by effectively rotating the four central base pairs by 90°, while keeping the remaining junction constant; thus any changes induced would have to result from nearest neighbor effects. Interestingly, both the trFRET and the NMR experiments indicated that the isomer bias was changed to about 1:4 for this junction, showing the influence of the neighboring base pairs on the conformational equilibrium.
These experiments provide convincing evidence that a four-way junction of particular sequence can populate both possible stacking conformers simultaneously, and that the relative populations are determined by base sequence extending at least as far as the next position from the center. In fact, in recent comparative gel electrophoresis experiments we have found the conformer population can be influenced by the third position from the point of strand exchange (R. Grainger and D.M.J.L., unpublished data). Experiments using MboII cleavage across four-way DNA junctions indicated that the stacking conformers undergo continuous interconversion (16), and recent experiments from Li, et al. (19) provide further evidence for this interconversion. Some limits can be set on the rate of the process, because the interconversion is clearly slow compared with the NMR timescale.
These conclusions may hold particular significance when we come to think about models for integrase family site-specific recombination events. Such reactions proceed via Holliday junction intermediates (20–24) that are created in one set of strand exchanges and resolved in a second set. Simone Nunes-Düby, Art Landy, and coworkers suggested a new model for integrative recombination in phage λ (25). In contrast to previous models that required branch migration of the junction formed after the first strand exchanges, this involved “peeling back” the strands after cleavage by integrase, locating the junction in the middle of the overlap region between cleavage sites (Fig. 3). The key event was then an isomerization of the junction, i.e., an exchange of stacking partners of exactly the kind discussed above, such that the second set of strand exchanges are carried out in the same stereochemical environment as the first set. This model requires no rotation of the DNA helices and is satisfyingly symmetrical.
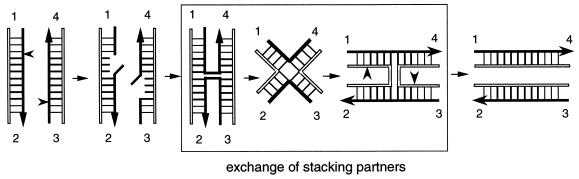
Figure 3.
A model for integrase site-specific recombination according to Nunes-Düby et al. (25). This mechanism involves little or no branch migration, and the key event is the exchange of helical stacking partners in the four-way junction formed between the two sets of strand exchanges.
A critical aspect of this model is that the reaction is DNA structure-driven. In the light of the demonstrated distortion of junction structure by cellular and phage resolving enzymes (7) perhaps this might be surprising, but persuasive evidence supporting this has been presented in two recent papers. Landy and Azaro (26) have shown that by subtly altering the sequence around the region of strand exchange for phage λ, the direction of resolution could be changed. Thus the resolution bias seems to be governed by the intrinsic DNA structure, and the data provide evidence for an conformational transition interconverting stacking conformers between the two strand exchange reactions. Although both sets of strand exchange are brought about by integrase in the case of phage λ, they are catalyzed by separate proteins, XerC and XerD, in the case of Xer site-specific recombination. Sherratt and coworkers (27) have used junctions that are constrained into one or other stacking conformers by means of tethering to examine the dependence of XerC and XerD cleavages on the junction structure. The result is striking. XerD is active only when the junction adopts the conformer that places its target cleavage sites on the exchanging strands. XerC is less fussy about the structure of its target, but also exhibits a preference for exchanging strands.
Both of these studies illustrate that the intrinsic structural preferences of the DNA junctions appear to exert a strong influence on the outcome of the recombination reactions, and that an exchange of stacking partners in the junction is a critical event in the proposed mechanism. Although physical studies of the isolated DNA structures may seem oversimplistic in some cases, it is clear that such data can provide important raw material for thinking about biologically important mechanisms.
References
- 1.Holliday R. Genet Res. 1964;5:282–304. [Google Scholar]
- 2.Miick S M, Fee R S, Millar D P, Chazin W J. Proc Natl Acad Sci USA. 1997;94:9080–9084. doi: 10.1073/pnas.94.17.9080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Duckett D R, Murchie A I H, Diekmann S, von Kitzing E, Kemper B, Lilley D M J. Cell. 1988;55:79–89. doi: 10.1016/0092-8674(88)90011-6. [DOI] [PubMed] [Google Scholar]
- 4.Clegg R M, Murchie A I H, Zechel A, Lilley D M J. Biophys J. 1994;66:99–109. doi: 10.1016/S0006-3495(94)80765-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lilley D M J, Clegg R M. Quart Rev Biophys. 1993;26:131–175. doi: 10.1017/s0033583500004054. [DOI] [PubMed] [Google Scholar]
- 6.Seeman N C, Kallenbach N R. Annu Rev Biophys Biomol Struc. 1994;23:53–86. doi: 10.1146/annurev.bb.23.060194.000413. [DOI] [PubMed] [Google Scholar]
- 7.White M F, Giraud-Panis M-J E, Pöhler J R G, Lilley D M J. J Mol Biol. 1997;269:647–664. doi: 10.1006/jmbi.1997.1097. [DOI] [PubMed] [Google Scholar]
- 8.Clegg R M, Murchie A I H, Zechel A, Carlberg C, Diekmann S, Lilley D M J. Biochemistry. 1992;31:4846–4856. doi: 10.1021/bi00135a016. [DOI] [PubMed] [Google Scholar]
- 9.Cooper J P, Hagerman P J. J Mol Biol. 1987;198:711–719. doi: 10.1016/0022-2836(87)90212-9. [DOI] [PubMed] [Google Scholar]
- 10.Cooper J P, Hagerman P J. Proc Natl Acad Sci USA. 1989;86:7336–7340. doi: 10.1073/pnas.86.19.7336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Churchill M E, Tullius T D, Kallenbach N R, Seeman N C. Proc Natl Acad Sci USA. 1988;85:4653–4656. doi: 10.1073/pnas.85.13.4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen S M, Heffron F, Chazin W J. Biochemistry. 1993;32:319–326. doi: 10.1021/bi00052a040. [DOI] [PubMed] [Google Scholar]
- 13.Carlstrom G, Chazin W J. Biochemistry. 1996;35:3534–3544. doi: 10.1021/bi952571n. [DOI] [PubMed] [Google Scholar]
- 14.Pikkemaat J A, van den Elst H, van Boom J H, Altona C. Biochemistry. 1994;33:14896–14907. doi: 10.1021/bi00253a029. [DOI] [PubMed] [Google Scholar]
- 15.Altona C. J Mol Biol. 1996;263:568–581. doi: 10.1006/jmbi.1996.0599. [DOI] [PubMed] [Google Scholar]
- 16.Murchie A I H, Portugal J, Lilley D M J. EMBO J. 1991;10:713–718. doi: 10.1002/j.1460-2075.1991.tb08001.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murchie A I H, Clegg R M, von Kitzing E, Duckett D R, Diekmann S, Lilley D M J. Nature (London) 1989;341:763–766. doi: 10.1038/341763a0. [DOI] [PubMed] [Google Scholar]
- 18.Eis P S, Millar D P. Biochemistry. 1993;32:13852–13860. doi: 10.1021/bi00213a014. [DOI] [PubMed] [Google Scholar]
- 19.Li X, Wang H, Seeman N C. Biochemistry. 1997;36:4240–4247. doi: 10.1021/bi9629586. [DOI] [PubMed] [Google Scholar]
- 20.Kitts P A, Nash H A. Nature (London) 1987;329:346–348. doi: 10.1038/329346a0. [DOI] [PubMed] [Google Scholar]
- 21.Nunes-Düby S E, Matsomoto L, Landy A. Cell. 1987;50:779–788. doi: 10.1016/0092-8674(87)90336-9. [DOI] [PubMed] [Google Scholar]
- 22.Hoess R, Wierzbicki A, Abremski K. Proc Natl Acad Sci USA. 1987;84:6840–6844. doi: 10.1073/pnas.84.19.6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jayaram M, Crain K L, Parsons R L, Harshey R M. Proc Natl Acad Sci USA. 1988;85:7902–7906. doi: 10.1073/pnas.85.21.7902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCulloch R, Coggins L W, Colloms S D, Sherratt D J. EMBO J. 1994;13:1844–1855. doi: 10.1002/j.1460-2075.1994.tb06453.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nunes-Düby S E, Azaro M A, Landy A. Curr Biol. 1995;5:139–148. doi: 10.1016/s0960-9822(95)00035-2. [DOI] [PubMed] [Google Scholar]
- 26.Azaro M A, Landy A. EMBO J. 1997;16:3744–3755. doi: 10.1093/emboj/16.12.3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arciszewska L K, Grainge I, Sherratt D J. EMBO J. 1997;16:3731–3743. doi: 10.1093/emboj/16.12.3731. [DOI] [PMC free article] [PubMed] [Google Scholar]