Abstract
Neuroanatomists posit that the central nucleus of the amygdala (Ce) and bed nucleus of the stria terminalis (BST) comprise two major nodes of a macrostructural forebrain entity termed the extended amygdala. The extended amygdala is thought to play a critical role in adaptive motivational behavior and is implicated in the pathophysiology of maladaptive fear and anxiety. Resting functional connectivity of the Ce was examined in 107 young anesthetized rhesus monkeys and 105 young humans using standard resting-state functional magnetic resonance imaging (fMRI) methods to assess temporal correlations across the brain. The data expand the neuroanatomical concept of the extended amygdala by finding, in both species, highly significant functional coupling between the Ce and the BST. These results support the use of in vivo functional imaging methods in nonhuman and human primates to probe the functional anatomy of major brain networks such as the extended amygdala.
Keywords: connectivity, resting-state fMRI, rhesus macaque, human, bed nucleus of the stria terminalis, central nucleus
Introduction
The amygdala occupies an important position in contemporary neural models of emotion, and mounting evidence suggests that amygdalar circuits plays a key role in the pathophysiology of anxiety, mood disorders and substance abuse (Aggleton, 2000; LeDoux, 2007; Shinnick-Gallagher et al., 2003; Whalen and Phelps, 2009). As research into the function, hodology and clinical significance of the amygdala progresses, the organization of this forebrain region continues to be redefined (Roy et al., 2009; Solano-Castiella et al., 2010) and some have called into question whether the conceptualization of the amygdala as a unitary entity remains relevant (Cassell, 1998; McDonald, 2003; Swanson and Petrovitch, 1998). The amygdala is hypothesized to have four major supranuclear divisions: a superficial cortical-like nuclear group, a basolateral nuclear complex, an unclassified cell group (e.g. the intercalated cell islands), and an extended amygdala (Heimer et al., 1999; Paxinos and Mai, 2004). Considerable focus has recently been placed on the clinical relevance of the extended amygdala (Luyten et al., 2012; Somerville et al., 2012), an anatomical construct that was originally described by Johnson in the early part of the last century (Johnson, 1923) and elaborated more recently by de Olmos, Alheid, Heimer and colleagues (Alheid and Heimer, 1988; De Olmos and Ingram, 1972; Heimer, 2003). The extended amygdala is a basal forebrain continuum of striatal-like medium spiney neurons that run from the dorsal amygdala, through the substantia innominata (SI), to the bed nuclei of the stria terminalis (BST) and the shell of the nucleus accumbens (Alheid, 2003; de Olmos and Heimer, 1999; Martin et al., 1991; McDonald, 1992). Heimer and colleagues have proposed that two parallel columns of extended amygdala neurons exist within the basal forebrain, a central extended amygdala that includes the central nucleus of the amygdala (Ce) and lateral portions of the BST (BSTL), and a medial extended amygdala including the medial nucleus of the amygdala (Me) and medial BST (see Figures 1a,b).
Fig 1. The extended amygdala.
(A) The human basal forebrain depicted in a series of coronal sections through the right hemisphere from the level of the ventral striatum (1) to the level of the caudal amygdala (4). The central extended amygdala is depicted in yellow, the medial extended amygdala in green. The images were modified (with permission) from Heimer et al. 1999, Art by Medical Scientific Illustration, Crozet, Virginia. See original reference for details. (B) Schematic pseudo-3D illustration of the right amygdala within the medial temporal lobe (modified with permission). For continuity, the central extended amygdala is depicted in yellow, the medial extended amygdala in green. The extended amygdala is depicted as a continuous bridge of neurons running from the Ce and Me, through the SI region, to the BST and the VS (shell of the nucleus accumbens). (C) 3D rendering of the rhesus monkey brain showing the approximate location and angle of the enlarged photo on the right. The autoradiograph (dark-field illumination) depicts axon transport in the rhesus monkey following an injection of 3H-amino acids into the region of the Ce. Note the dense labeling of axons and terminals through the sublenticular area of the extended amygdala (reprinted with permission). Abbreviations: ac: anterior commissure; BLA: basolateral complex; BSTL: lateral bed nucleus of stria terminalis; BSTM: medial bed nucleus of stria terminalis; BSTS/st: bed nucleus of stria terminialis, supra-capsular part/stria terminalis; Ce: central nucleus of the amygdala; f: fornix; ic: internal capsule; LV: lateral ventricle; Me: medial nucleus of the amygdala; opt: optic tract; ox: optic chiasm; SI; substantia innominata; SLEA: sublenticular part of extended amygdala; VS: ventral striatum.
Neuroanatomical studies in rodents and nonhuman primates support the central extended amygdala concept by demonstrating that the Ce and BSTL are strongly connected and share many efferent targets (de Olmos and Heimer, 1999; Dong et al., 2001; Heimer and Van Hoesen, 2006; Nagy and Pare, 2008); see Figure 1c). Additionally, although first noted by Johnson (1923), recent developmental data suggest that some of the constituent neurons of the Ce and BSTL are derived from similar embryological origins (Bupesh et al., 2011). Despite the fact that these neuroanatomical data support the concept of the extended amygdala, little data addresses the degree to which activity in the Ce and BST regions comprise a functional network.
Components of the extended amygdala play pivotal roles mediating diverse behavioral, emotional, and physiological responses associated with stress, anxiety, reproduction and other motivational states (Duvarci et al., 2009; Park et al., 2012; Regev et al., 2011). Animal work suggests altered amygdala and BST function may be important in the pathophysiology of psychiatric disorders (Davis et al., 2009; Fox et al., 2008; Heimer, 2003; Oler et al., 2010; Sink et al., 2012). To examine whether the neuroanatomical concept of the extended amygdala pertains to measures of temporal functional connectivity, we used well-established functional magnetic resonance imaging (fMRI) methods in a large group of young anesthetized rhesus monkeys to assess whether intrinsic activity in the Ce region is linked and coordinated with that in the BST. Resting-state fMRI reflects synchronized variations in the neuronal activity of a network, providing a window into the interaction, or connection, between brain areas. Rhesus monkeys were used since the cytoarchitecture of the macaque amygdala is similar to that in the human (Freese and Amaral, 2009), and this species provides optimal translational models for understanding brain mechanisms that underlie human psychopathology (Kalin and Shelton, 2003; Machado and Bachevalier, 2003).
fMRI scans were performed in a large sample of peri-adolescent monkeys using methods modified from prior work demonstrating the reliability of collecting resting fMRI data in anesthetized rhesus monkeys (Vincent et al., 2007). Blood oxygenation level dependent (BOLD) time-series data were extracted from a right hemisphere Ce region of interest (ROI) that served as a seed cluster for later connectivity analyses. To assess the relevance of monkey Ce resting-state connectivity to humans, we used a similar strategy to examine the functional connectivity of the dorsal amygdala in an equivalently sized sample of children and adolescents in which resting-state fMRI data were collected.
Methods
Non-human primate imaging analyses
Participants
107 rhesus macaque monkeys (57 female; mean (± s.d.) age = 2.59 (± 1.02) years) were used in this study. All animals were mother-reared, and pair-housed at the Harlow Primate Laboratory or the Wisconsin National Primate Research Center. All study procedures were performed in accordance with the guidelines set forth by the University of Wisconsin-Madison Animal Care and Use Committee (IACUC).
MRI data acquisition
Magnetic resonance imaging (MRI) data were collected using a General Electric Signa 3T scanner (General Electric Medical Systems, Waukesha, WI) with a quadrature birdcage extremity coil. Structural MRI data were acquired using an axial 3D T1-weighted inversion-recovery fast gradient echo sequence (repetition time, 9.4 ms; echo time, 2.1 ms; field of view, 14 cm; flip angle, 10°; number of excitations, 2; in-plane resolution, 0.2734 mm; number of slices, 248; slice thickness, 1 mm; −0.05 mm interslice gap). Before undergoing MRI acquisition, the monkeys were anesthetized with an intramuscular injection of ketamine (15 mg/kg). Structural MRI data were collected immediately prior to functional scans.
Functional MRI data acquisition
Functional data were acquired using a series of coronal T2*-weighted echo-planar images (EPI) with the following parameters: TR = 2.5s; TE = 25ms; Flip angle = 90 degrees; FOV = 140mm; 26 slices; matrix 64×64, voxel size 2.19×2.19×3.1 mm; 360 volumes; and duration = 15 min.
Resting state data analysis -Preprocessing
All preprocessing steps were carried out in AFNI (Cox, 1996). EPI data were corrected for slice timing differences and volumes were registered using rigid-body image registration to correct for subject motion. Field-map correction was performed to correct for echo-planar image warping induced by B0-field inhomogenetities. For each subject, EPI volumes were aligned to that subject’s anatomical image (using AFNI’s 3dAllineate) and then aligned to a standard anatomical template (derived from the average of 238 high-resolution anatomical images, see below) using a 12-parameter affine transformation, and re-sampled to 0.625×0.625×0.625mm resolution. Data were then spatially smoothed (using a Gaussian kernel with FWHM 3mm), and temporally band-pass filtered to 0.01–0.1Hz (using 3dFourier). The first three imaging volumes for each resting run were ignored to allow the magnetization to reach equilibrium.
Rhesus MRI brain template
The anatomical template used in this study was created using standard methods, and was transformed to the stereotaxic space of Paxinos, Huang, Petrides & Toga (Paxinos et al., 2009). First, each subject’s T1-MRI image was manually striped of non-brain tissue using SPAMALIZE; http://brainimaging.waisman.wisc.edu/~oakes/spam/spam_frames.htm. Brain extracted MRI images were originally registered to a 34-brain template in standard space, using a 12-parameter linear transformation with FMRIB Software Library’s “flirt” tool (FSL; http://www.fmrib.ox.ac.uk/fsl/) (Jenkinson et al., 2002). Images were manually verified, and averaged to create a study-specific 238-brain template in standard space. The brain-extracted MRI images in original space were then transformed to match this study-specific template using non-linear transformation tool in FSL (“fnirt”).
5-HTT Availability Map and region of interest (ROI) creation
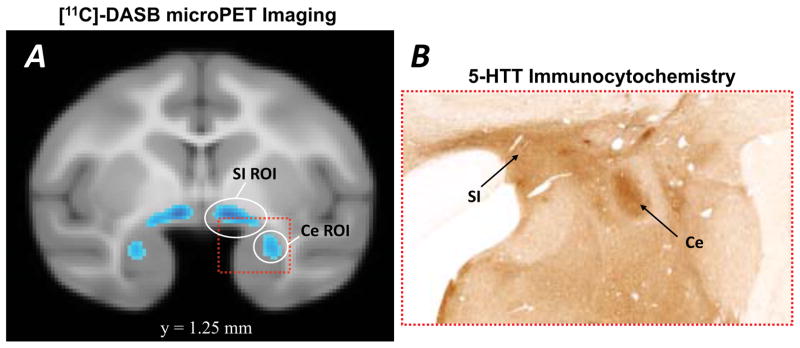
The [11C]DASB-PET methods are detailed elsewhere (Christian et al., 2009), and are only briefly described here. DASB is a high-affinity ligand of the serotonin transporter (5-HTT), and the carbon-11 for the radiolabeling was produced with a National Electrostatics 9SDH 6 MeV Van de Graff tandem accelerator (Middleton, WI). [11C]DASB-PET data were acquired in an independent sample of 34 rhesus monkeys (mean age = 4.4 years; 12 male, 22 female) using a Concorde microPET P4 scanner (Tai et al., 2001). The dynamic PET time series were transformed into parametric images with each voxel representing the distribution volume ratio (DVR) serving as an index of receptor binding (Innis et al., 2007). The cerebellum was used as a reference region, and all voxels were divided by the mean cerebellar binding values. Each subject’s DVR image was transformed (based on the corresponding MRI transformation) into the same standard space that the fMRI data were. This 5-HTT availability map, thresholded at 250x background binding, precisely localizes the Ce because compared with other dorsal amygdalar regions the lateral division of the Ce has the highest levels of 5-HTT binding (Bauman and Amaral, 2005; Freedman and Shi, 2001; O’Rourke and Fudge, 2006) (Figure 2b). The right Ce ROI used in the present study was 40 mm3 and can be seen in Figure 2a.
Fig 2. Serotonin transporter (5-HTT) labeling in the central nucleus (Ce) region of rhesus monkey amygdala used to delineate the seed region for monkey fMRI analysis.
(A) In vivo PET image demonstrating 5-HTT binding availability in the Ce and dorsomedially adjacent substantia innominata (SI) region, adapted from data first reported in Christian et al., (2009). The image is thresholded at 250X the background binding level, and the circles indicate the regions of interest (ROIs) used in the present study as seed clusters for analysis of the monkey functional connectivity data. (B) Comparable low-power photomicrograph showing the dense and selective expression of 5-HTT in the lateral division of the Ce (image provided by Dr. Julie Fudge, University of Rochester School of Medicine and reprinted with permission of the publisher).
Resting state connectivity analysis
Functional connectivity was computed using a seed-region based approach. The amygdala ROI was defined using the 5-HTT availability, as described in the section above. The resting-state EPI signal intensity time courses were averaged over the seed ROI. Single-subject connectivity maps were generated by computing a voxelwise general linear model (GLM) with the mean seed time-series as the explanatory variable. The resulting single-subject correlation coefficient (“connectivity”) maps were normalized using Fisher’s r-to-z transformation. In order to account for physiological noise, average signal intensity time courses from the white matter (WM) and cerebral spinal fluid (CSF), were included as nuisance regressors (Jo et al., 2010). These nuisance regressors were obtained in the following manner. First, each subject’s T1-weighted anatomical image was segmented into WM, gray matter, and CSF using the FAST routine from the FMRIB Software Library (FSL) (Zhang et al., 2001). WM masks were eroded by 2 voxels in each dimension in order to avoid partial volume contributions from gray matter and CSF. To isolate the ventricular CSF, the CSF mask was multiplied by a template-defined mask of the lateral ventricles. The ventricular CSF and WM masks were then transformed to template space and the average EPI signal over the ventricular CSF and white matter were extracted. For group level analyses, a main effect t-test (against zero) was performed on the Fisher-Z transformed correlation coefficients for the seed time-series (using 3dttest).
Calculating the 95% Spatial Confidence Intervals
Intrinsic fluctuations in the Ce signal were significantly correlated with a number of brain regions (see Table 1). Connectivity maps were stringently thresholded (t = 10.0, and a minimum spatial extent of 10.0 mm3), allowing us to isolate the regions with the strongest functional connectivity with the Ce seed. Only clusters > 10 mm3 that were located outside of the Ce seed region were further examined. Within clusters, local maxima were identified using code adapted from FMRISTAT (http://www.math.mcgill.ca/keith/fmristat/). Spatial confidence intervals (CI) around local maxima in the connectivity maps were calculated using FMRISTAT (95% CI ≈ tneighbor ≥ ((tmax)2 − χ(3)2(.05))1/2);(Ma et al., 1999).
Table 1.
Locations of peaks in the functional connectivity data are presented above with the hemisphere, brain region and the volume of each Ce-connected cluster. All correlations were positive and only clusters larger that 10 mm3 are reported. Also presented are the t-values and the location (in millimeters relative to the anterior commissure, ac) of the peak voxel within each macroscopic region in the cluster.
| monkey | cluster | volume (mm3) | local maxima | coordinate of peak in mm relative to the ac | |||
|---|---|---|---|---|---|---|---|
| hemisphere | t | x | y | z | |||
| L | anterior temporal lobe | 551 | amygdala | 17.31 | −12.500 | 1.250 | −5.000 |
| R | basal forebrain | 88 | BST region | 13.98 | 3.750 | 0.625 | 2.500 |
| L | basal forebrain | 73 | BST region | 12.08 | −5.625 | 0.000 | 3.125 |
| R | lateral occipital cortex | 47 | V4/V5 | 11.11 | 18.750 | −28.125 | 8.125 |
| R | anterior temporal lobe | 1680 | inferotemporal cortex (TE) | 10.86 | 21.875 | 0.625 | −13.125 |
| L | lateral occipital cortex | 13 | V4/V5 | 10.75 | −22.500 | −28.125 | 9.375 |
| human | cluster | volume (mm3) | local maxima | coordinate of peak in mm relative to the ac | |||
|---|---|---|---|---|---|---|---|
| hemisphere | t | x | y | z | |||
| L | anterior temporal lobe/striatum/basal forebrain | 10963 | amygdala | 27.29 | 22 | 7 | −11 |
| nucleus accumbens | 17.67 | 9 | −7 | −1 | |||
| putamen | 17.34 | 30 | 5 | 6 | |||
| subiculum/hippocampus | 16.06 | 24 | 30 | −8 | |||
| R | anterior temporal lobe/striatum/basal forebrain/posterior insula | 34893 | hippocampus | 23.42 | −26 | 16 | −15 |
| putamen/claustrum | 23.08 | −31 | 0 | 0 | |||
| posterior thalamus | 18.82 | −18 | 34 | −1 | |||
| parietal operculum | 18.20 | −47 | 22 | 18 | |||
| insula | 17.84 | −40 | 17 | 20 | |||
| nucleus accumbens/BST region | 17.11 | −6 | −3 | −1 | |||
| cerebellum | 16.70 | −25 | 34 | −20 | |||
| nucleus accumbens | 16.45 | −8 | −7 | 1 | |||
| dorsal temporopolar region | 16.15 | −35 | −11 | −24 | |||
| R | temporal lobe | 284 | superior temporal sulcus | 17.55 | −51 | 4 | −11 |
| R | temporal lobe | 58 | lateral temporopolar region | 16.51 | −42 | −14 | −10 |
| R | cingulate gyrus | 35 | posterior midcingulate cortex | 16.19 | −10 | 10 | 42 |
Human imaging analyses
Participants
To generate a comparable sample size, subjects from three independent datasets were combined. Data from two different publicly available datasets were downloaded from the NITRC (http://www.nitrc.org/projects/fcon_1000/) and combined with fMRI datasets collected in our lab. The two sets of images downloaded from the online database came from New York University (NYU, Milham, M.P./Castellanos, F.X.) and the Nathan Kline Institute (NKI/Rockland sample, Castellanos, F.X., Leventhal, B., Milham, M.P., Nooner, K.). Thus for the present study, we analyzed data from 105 subjects. Sixty-six of the subjects (sixty-three percent of the total sample) were selected from the Wisconsin Study of Family and Work, a prospective longitudinal study of child and adolescent development (Essex et al., 2011). The mean (± s.d.) age of the 66 adolescents was 18.44 (± 0.19) years, 34 were male, and 10% were racial or ethnic minorities. All study procedures were performed in accordance with the guidelines set forth by the University of Wisconsin-Madison Human Subjects IRB. Eighteen child and adolescent subjects (eleven males, seventeen percent of the total sample size) were selected from the NYU data available online on the NITRC database. The mean (± s.d.) age was 12.14 (± 2.49) years, with the youngest participant being 7.88 years of age and the oldest 15.58 years old. More details on the subjects are available in previous published studies (Di Martino et al., 2008; Kelly et al., 2010; Margulies et al., 2007; Shehzad et al., 2009). Finally, twenty-one subjects (11 males, twenty percent of the total number of participants) with a mean (± s.d.) age of 12.25 (± 3.67) years, were selected from the NKI dataset, the youngest participant being 4 years of age and the oldest 17 years old.
fMRI data collection
For the NYU dataset, A 3-Tesla Siemens Allegra scanner was used to obtain the EPI and anatomical images. A gradient echo sequence with the following parameters was used for the EPI: TR = 2s; TE = 25ms; Flip angle = 90 degrees; FOV = 192mm; 39 axial slices; matrix 64×64, voxel size 3×3×3 mm; 197 volumes; and duration = 6 min 38 s. The anatomical images were acquired using a T1-weighted MP-RAGE sequence with the following parameters: TR = 2.5s; TE = 4.35ms; TI = 900ms; Flip angle = 8 degrees; FOV = 256mm; 176 slices; and voxel size 1×1×1 mm). For the NKI dataset a 3T Siemens Magnetom TrioTim scanner was used. The parameters for the EPI scans were as follows: TR = 2.5s; TE = 30ms; Flip angle = 80 degrees; FOV = 216mm; 38 axial slices; voxel size 3×3×3 mm; and duration = 10 min 55 s. The anatomical images were acquired with the following properties: T1-weighted; MP-RAGE; TR = 2.5s; TE = 3.5ms; TI = 1200ms; Flip angle = 8 degrees; FOV = 256mm; 192 slices; and voxel size 1×1×1 mm). Finally, the brain data collected at the Waisman Laboratory for Brain Imaging and Behavior were acquired on a 3-Tesla Discovery MR750 with the following EPI scan parameters: TR = 2s; TE = 25ms; Flip angle = 60 degrees; FOV = 24cm; Slice thickness = 5mm; Matrix = 64×64; 30 sagittal slices.
Resting state data analysis: Preprocessing
All preprocessing steps were similar to those performed on the monkey data, and were carried out in AFNI (Cox, 1996). Every subject from each dataset was analyzed separately with the same steps. Slice timing correction, motion correction (using Fourier interpolation), spatial smoothing (using a Gaussian kernel with FWHM 6mm), and temporal band-pass filtering to 0.01–0.1Hz (using 3dFourier) were performed. The first three imaging volumes for each resting run were ignored to allow the magnetization to reach equilibrium.
Image Alignment and Registration
Each anatomical MR image was skull-stripped and aligned to the EPI in native space using AFNI. Functional and structural brain data were then registered using affine transformation to a standard template in AFNI that was centered relative to the posterior edge of the anterior commissure. Taking the average of all 105 brain scans generated a mean anatomical image.
Resting-state connectivity analysis
Functional connectivity was computed using a seed-region based approach. An ROI corresponding to the Ce region was manually created in the right amygdala on a standard 152-brain MRI template (see figure 3d, inset). This was accomplished using the ROI drawing tool in AFNI and was based on the detailed human brain atlas of Mai (Mai et al., 2003), which is in the same standard space to which the brain data were aligned. The right Ce seed ROI was then inflated by 2mm in each cardinal (A–P, R–L, I–S) direction in order to account for slight differences in registration across subjects, resulting in a Ce ROI cluster of 944 mm3. The seed ROIs were mapped back to each subject’s EPI data, and the resting-state signal intensity time courses were averaged over the ROI. Connectivity maps were generated for each subject by regressing the seed time-series against all other voxels in the brain. In order to account for physiological processes and motion, average signal intensity time courses from the white matter and CSF, along with six parameters of estimated subject movement (3 translation, 3 rotation), were included as nuisance regressors in this regression. Average CSF and white matter signals were obtained by segmenting each subject’s T1-weighted anatomical image (using FSL’s FAST routine), multiplying these masks by tissue priors to localize CSF signal to the lateral ventricles and to reduce partial volume contributions of gray matter and CSF to the white matter, resampling these masks to the EPI data, and averaging the signal over the CSF and white matter. For group level analyses, a main effect t-test (against zero) was performed on the Fisher-Z transformed correlation coefficients for the seed time-series (using 3dttest). A t = 16.0 was used to stringently threshold the human connectivity map, allowing us to isolate regions with the strongest functional connectivity with the Ce seed.
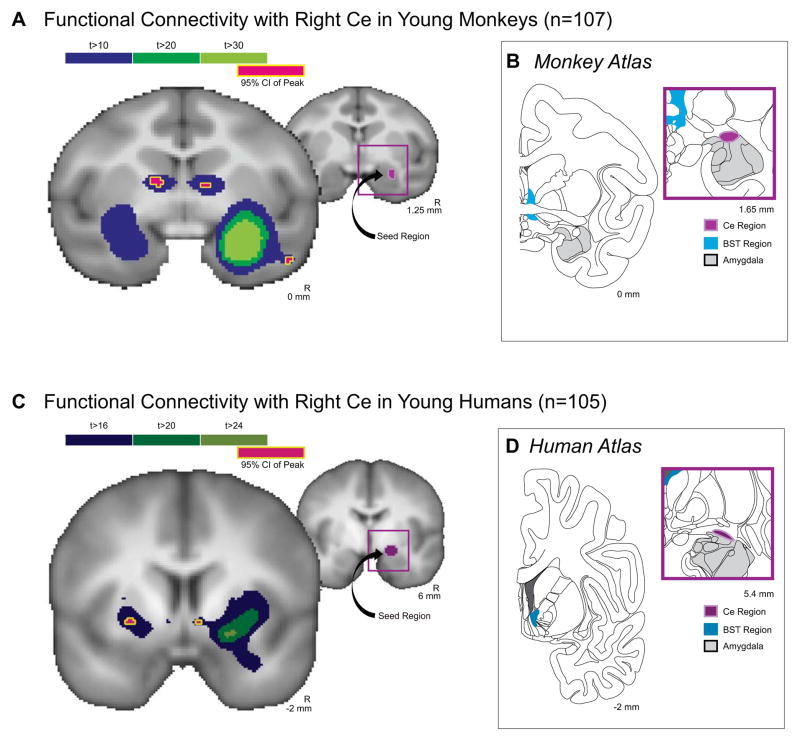
Fig 3. Ce and BST are functionally connected in the monkey and human.
(A) Monkey and (C) human brain regions where the fMRI time-series were highly correlated (monkey: dark purple: t = 10.0; dark green: t = 20; light green: t = 30; human: dark purple: t = 16.0; dark green: t = 20; light green: t = 24). The Ce seed region is depicted in the adjacent section for both monkey and human (light purple). Red with yellow outline represents the 95% spatial confidence intervals around the peaks of the Ce-correlated activity within each cluster. Sections modified (and reprinted with permission) from stereotaxic atlases of the (B) rhesus monkey brain (Paxinos et al., 2009) and (D) human brain (Mai et al., 2003) corresponding to the MRI slices shown in A and C, respectively. The BST is depicted in light blue.
Results
In anesthetized peri-adolescent monkeys, spontaneous fluctuations in the right Ce BOLD signal were highly significantly correlated with BOLD signal fluctuations within six clusters identified using a stringent statistical threshold (t > 10.0; p < 6.0e–17). The six clusters consisted of bilateral anterior temporal lobe, bilateral basal forebrain regions corresponding to the BST (Figure 3a) and bilateral parieto-occipital cortex (V4/V5). The locations of the peaks functionally connected to the Ce (Table 1) were determined within each cluster by calculating the 95% spatial confidence intervals around the voxels containing the maximum t-values. Outside of the right temporal lobe cluster containing the seeded region, the cluster demonstrating the most significant functional connectivity with the right Ce seed was in the contralateral (left-hemisphere) amygdala, while the clusters displaying the third and fourth highest temporal correlation with the Ce time-series contained the right and left BST, respectively. We formally tested the extent to which connectivity with the BST was greater than connectivity with V4/V5, the region exhibiting the next highest peak correlated with Ce (see Table 1). Connectivity data were extracted from the BST containing clusters, and connectivity data were also extracted from the V4/V5 cluster in the right hemisphere. To compare the extracted correlation coefficients, the Pearson’s r-values were converted to standardized scores with Fisher’s z-transform, and a paired-sample t-test of the difference between Ce-BST vs. Ce-V4/V5 connectivity was conducted. This analysis revealed that Ce-BST connectivity was significantly greater than Ce-V4/V5 connectivity (t = 4.629, p < 0.00001).
To understand the extent to which Ce function is linked to components of the extended amygdala other than BST, we examined Ce-SI connectivity. While significant connectivity was observed between Ce and SI (t = 6.373, p < 1.0e–8), it is important to note that the level of connectivity between these regions was below the statistical threshold that was used to identify the initial six Ce connected clusters reported in Table 1. To examine whether the Ce-BST connectivity was significantly greater than that observed for Ce-SI, we tested the difference between Ce-BST and Ce-SI connectivity. As performed previously to identify the Ce seed, the [11C]-DASB 5-HTT map was used to generate a SI seed. This SI seed region can be seen in Figure 2a, dorsal and medial to the Ce seed. Ce connectivity data were extracted from the SI region in the right hemisphere, the correlation coefficients were standardized with Fisher’s z transform, and a paired-sample t-test of the difference between Ce-BST vs Ce-SI connectivity was conducted. The analysis revealed that Ce-BST connectivity was significantly greater than Ce-SI connectivity (t = 2.213, p < 0.03).
We next sought to assess whether the tight functional coupling between Ce and BST we observed in anesthetized adolescent monkeys is present in awake young humans. As can be seen in Figure 3c, the thresholded Ce-connectivity map of the human brain revealed discrete clusters of voxels with significant functional connectivity in the BST region (t > 16.0; p < 8.0e–30). The locations of the peaks in the human Ce functional connectivity data are presented Table 1. While differences in Ce functional connectivity exist between the monkey and human, these potential disparities should be interpreted with caution due to necessary methodological differences in acquiring data from the two species (e.g., anesthesia). The possible confound to cross-species comparison posed by the use of the 5-HTT map in the monkeys versus manually drawn Ce ROI for the humans, should also be noted.
Discussion
The present study expands the anatomical concept of the extended amygdala to a functional level by providing evidence in 2 primate species for evolutionarily conserved strong temporal coupling between the Ce and BST regions. This finding is consistent with anatomical and neurochemical data linking the Ce and BST, and supports the hypothesis that these structures are components of a coordinated functional network. Rodent studies suggest an important dissociation between the Ce and BST with respect to defensive behaviors, such that the Ce is involved in acute fear-related responding, and the BST is thought to mediate anxiety-like responses to sustained or ambiguous threats (Walker and Davis, 2008). In previous rhesus monkey studies, we reported that individual differences in BST (Fox et al., 2008; Kalin et al., 2005) and Ce (Oler et al., 2010) metabolic activity, as assessed with PET imaging, were predictive of individual differences in trait-like anxiety in young monkeys. Additionally, recent human imaging studies have associated these regions with vigilance, threat monitoring and anticipatory anxiety (Alvarez et al. 2010; Mobbs et al., 2010; Somerville et al. 2010; Straube et al., 2007). The current findings present novel evidence that Ce and BST functional activations, as assessed with the BOLD signal, are highly coordinated. These data support the hypothesis that coordinated actions between key components of the extended amygdala are the substrate for the adaptive interplay between immediate responses to threat and longer-term, sustained vigilance for potential future threats.
The present fMRI data do not address the directionality of connectivity between these structures. It is important to note that the monkey and human amygdala share a similar pattern of connections with cortical and sub-cortical regions (Amaral et al., 1992; Freese and Amaral, 2009). In primates the BST receives inputs from almost all of the amygdalar nuclei (Price and Amaral, 1981; Price et al., 1987), including the Ce (see Fig 1c). Preliminary work in rhesus monkeys has examined the extent to which neurons in the BST project to the Ce. Using retrograde tracer injections into the Ce, few retrogradely labeled cells are observed in the BST (personal communication, Julie Fudge, University of Rochester). It is noteworthy that injections of retrograde tracers into the Ce reveal dense projections originating in other amygdala nuclei (Fudge and Tucker, 2009). In the rat, the species most extensively studied with tract tracing techniques, only a few reports describe light structural connections projecting from BST to Ce (Pitkänen, 2000). Thus, the marked preponderance of efferent projections from Ce to BST support similar directionality underlying the functional connectivity observed in the present study between Ce and BST. It is also unclear whether the source of the Ce-BST functional connectivity documented here is via monosynaptic or polysynaptic pathways. It is possible that the temporal correlation between Ce and BST signal is due to direct monosynaptic Ce→ BST connections via the stria terminalis and/or the ventral amygdalofugal pathway/sublenticular bundle (Amaral et al., 1992; Klingler and Gloor, 1960). Alternatively, it is possible that Ce-BST functional connectivity could result from shared afferent input (i.e., from the basolateral complex) driving activity in both the Ce and BST.
It is important to emphasize that the extended amygdala concept involves regions other than the Ce and BST. The SI, which is interposed between the Ce and BST, is hypothesized to contain extended amygdala neurons, as extensive histochemical studies demonstrate islands of cell groups in the SI region that appear to be continuous with cells in the Ce and BST (Heimer et al., 1999; Heimer et al., 1997). To understand the extent to which Ce function is linked to components of the extended amygdala other than the BST, Ce-SI connectivity was examined. Although temporal fluctuations in the SI signal were significantly correlated with the Ce time-series, analyses revealed that Ce-BST connectivity was significantly greater than Ce-SI connectivity (p < 0.03). Perhaps, the relatively decreased connectivity found between Ce and SI is secondary to the fact that SI extended amygdala cells are interdigitated with other cell types such as magnocellular cholinergic cells. Within the extended amygdala, the data support a particularly robust functional connectivity between Ce and BST.
Taken together the data support the use of in vivo functional imaging methods in nonhuman and human primates to quantify the degree of coupling between components of the extended amygdala. Future studies using functional connectivity to assess the degree of spontaneous and task-evoked coupling between the Ce and BST have the potential to provide new insights into the pathophysiology of anxiety, affective, and stress-related disorders. Finally, these data provide an fMRI-based demonstration that the extended amygdala is conserved across primate species, and that, at rest, the Ce and BST function as a coordinated circuit, supporting further investigation of the role of the BST in anxiety-like processes in primates.
Acknowledgments
This work was supported by the HealthEmotions Research Institute and the National Institute of Mental Health: MH046729 (to N.H.K.), MH081884 (to N.H.K.), MH084051 (to R.J.D, M.J.E. & N.H.K.). We thank the staff of the Harlow Center for Biological Psychology, HealthEmotions Research Institute, Waisman Laboratory for Brain Imaging and Behavior, Wisconsin National Primate Center, J. M. Armstrong, B. Christian, M. Jesson, E. Larson, K. Myer, M. Riedel, A. J. Shackman, J. Storey and H. Van Valkenberg.
References
- Aggleton JP. The Amygdala. A Functional Analysis 2000 [Google Scholar]
- Alheid GF. Extended Amygdala and Basal Forebrain. Ann NY Acad Sci. 2003;985:185–205. doi: 10.1111/j.1749-6632.2003.tb07082.x. [DOI] [PubMed] [Google Scholar]
- Alheid GF, Heimer L. New perspectives in basal forebrain organization of special relevance for neuropsychiatric disorders: the striatopallidal, amygdaloid, and corticopetal components of substantia innominata. Neuroscience. 1988;27:1–39. doi: 10.1016/0306-4522(88)90217-5. [DOI] [PubMed] [Google Scholar]
- Alvarez RP, Chen G, Bodurka J, Kaplan R, Grillon C. Phasic and sustained fear in humans elicits distinct patterns of brain activity. Neuroimage. 2010;55:389–400. doi: 10.1016/j.neuroimage.2010.11.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaral DG, Price JL, Pitkänen A, Carmichael ST. Anatomical organization of the primate amygdaloid complex. The Amygdala: Neurobiological Aspects of Emotion, Memory, and mental Dysfunction. 1992:1–66. [Google Scholar]
- Bauman MD, Amaral DG. The distribution of serotonergic fibers in the macaque monkey amygdala: an immunohistochemical study using antisera to 5-hydroxytryptamine. Neuroscience. 2005;136:193–203. doi: 10.1016/j.neuroscience.2005.07.040. [DOI] [PubMed] [Google Scholar]
- Bupesh M, Abellan A, Medina L. Genetic and experimental evidence supports the continuum of the central extended amygdala and a mutiple embryonic origin of its principal neurons. J Comp Neurol. 2011;519:3507–3531. doi: 10.1002/cne.22719. [DOI] [PubMed] [Google Scholar]
- Cassell MD. The amygdala: myth or monolith? Trends Neurosci. 1998;21:200–201. doi: 10.1016/s0166-2236(97)01207-1. [DOI] [PubMed] [Google Scholar]
- Christian BT, Fox AS, Oler JA, Vandehey NT, Murali D, Rogers J, Oakes TR, Shelton SE, Davidson RJ, Kalin NH. Serotonin transporter binding and genotype in the nonhuman primate brain using [C-11]DASB PET. Neuroimage. 2009;47:1230–1236. doi: 10.1016/j.neuroimage.2009.05.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox RW. AFNI: software for analysis and visualization of functional magnetic resonance neuroimages. Comput Biomed Res. 1996;29:162–173. doi: 10.1006/cbmr.1996.0014. [DOI] [PubMed] [Google Scholar]
- Davis M, Walker DL, Miles L, Grillon C. Phasic vs Sustained Fear in Rats and Humans: Role of the Extended Amygdala in Fear vs Anxiety. Neuropsychopharmacology. 2009 doi: 10.1038/npp.2009.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Olmos JS, Heimer L. The concepts of the ventral striatopallidal system and extended amygdala. In: McGinty JF, editor. Advancing from the ventral striatum to the extended amygdala: Implications for neuropsychiatry and drug abuse. The New York Academy of Sciences; New York: 1999. pp. 1–32. [DOI] [PubMed] [Google Scholar]
- De Olmos JS, Ingram WR. The projection field of the stria terminalis in the rat brain. An experimental study. J Comp Neurol. 1972;146:303–334. doi: 10.1002/cne.901460303. [DOI] [PubMed] [Google Scholar]
- Di Martino A, Scheres A, Margulies DS, Kelly AM, Uddin LQ, Shehzad Z, Biswal B, Walters JR, Castellanos FX, Milham MP. Functional connectivity of human striatum: a resting state FMRI study. Cereb Cortex. 2008;18:2735–2747. doi: 10.1093/cercor/bhn041. [DOI] [PubMed] [Google Scholar]
- Dong HW, Petrovich GD, Swanson LW. Topography of projections from amygdala to bed nuclei of the stria terminalis. Brain Res Brain Res Rev. 2001;38:192–246. doi: 10.1016/s0165-0173(01)00079-0. [DOI] [PubMed] [Google Scholar]
- Duvarci S, Bauer EP, Pare D. The bed nucleus of the stria terminalis mediates inter-individual variations in anxiety and fear. J Neurosci. 2009;29:10357–10361. doi: 10.1523/JNEUROSCI.2119-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essex MJ, Shirtcliff EA, Burk LR, Ruttle PL, Klein MH, Slattery MJ, Kalin NH, Armstrong JM. Influence of early life stress on later hypothalamic-pituitary-adrenal axis functioning and its covariation with mental health symptoms: a study of the allostatic process from childhood into adolescence. Dev Psychopathol. 2011;23:1039–1058. doi: 10.1017/S0954579411000484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox AS, Shelton SE, Oakes TR, Davidson RJ, Kalin NH. Trait-like brain activity during adolescence predicts anxious temperament in primates. PLoS ONE. 2008;3:e2570. doi: 10.1371/journal.pone.0002570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freedman LJ, Shi C. Monoaminergic innervation of the macaque extended amygdala. Neuroscience. 2001;104:1067–1084. doi: 10.1016/s0306-4522(01)00157-9. [DOI] [PubMed] [Google Scholar]
- Freese JL, Amaral DG. Neuroanatomy of the Primate Amygdala. In: Whalen PJ, Phelps EA, editors. The Human Amygdala. Guilford; New York: 2009. pp. 3–42. [Google Scholar]
- Fudge JL, Tucker T. Amygdala projections to central amygdaloid nucleus subdivisions and transition zones in the primate. Neuroscience. 2009;159:819–841. doi: 10.1016/j.neuroscience.2009.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heimer L. A new anatomical framework for neuropsychiatric disorders and drug abuse. Am J Psychiatry. 2003;160:1726–1739. doi: 10.1176/appi.ajp.160.10.1726. [DOI] [PubMed] [Google Scholar]
- Heimer L, de Olmos JS, Alheid GF, Pearson J, Sakamoto N, Shinoda K, Marksteiner J, Switzer RC., Iii . Chapter II The human basal forebrain. Part II. In: Bloom FE, ABr, Hökfelt T, editors. Handbook of Chemical Neuroanatomy. Elsevier; 1999. pp. 57–226. [Google Scholar]
- Heimer L, Harlan RE, Alheid GF, Garcia MM, de Olmos J. Substantia innominata: a notion which impedes clinical-anatomical correlations in neuropsychiatric disorders. Neuroscience. 1997;76:957–1006. doi: 10.1016/s0306-4522(96)00405-8. [DOI] [PubMed] [Google Scholar]
- Heimer L, Van Hoesen GW. The limbic lobe and its output channels: implications for emotional functions and adaptive behavior. Neurosci Biobehav Rev. 2006;30:126–147. doi: 10.1016/j.neubiorev.2005.06.006. [DOI] [PubMed] [Google Scholar]
- Innis RB, Cunningham VJ, Delforge J, Fujita M, Gjedde A, Gunn RN, Holden J, Houle S, Huang SC, Ichise M, Iida H, Ito H, Kimura Y, Koeppe RA, Knudsen GM, Knuuti J, Lammertsma AA, Laruelle M, Logan J, Maguire RP, Mintun MA, Morris ED, Parsey R, Price JC, Slifstein M, Sossi V, Suhara T, Votaw JR, Wong DF, Carson RE. Consensus nomenclature for in vivo imaging of reversibly binding radioligands. J Cereb Blood Flow Metab. 2007;27:1533–1539. doi: 10.1038/sj.jcbfm.9600493. [DOI] [PubMed] [Google Scholar]
- Jenkinson M, Bannister P, Brady M, Smith S. Improved optimization for the robust and accurate linear registration and motion correction of brain images. Neuroimage. 2002;17:825–841. doi: 10.1016/s1053-8119(02)91132-8. [DOI] [PubMed] [Google Scholar]
- Jo HJ, Saad ZS, Simmons WK, Milbury LA, Cox RW. Mapping sources of correlation in resting state FMRI, with artifact detection and removal. Neuroimage. 2010;52:571–582. doi: 10.1016/j.neuroimage.2010.04.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JB. Further contributions to the study of the evolution of the forebrain. Journal of Comparative Neurology. 1923;35:337–481. [Google Scholar]
- Kalin NH, Shelton SE. Nonhuman primate models to study anxiety, emotion regulation, and psychopathology. Ann N Y Acad Sci. 2003;1008:189–200. doi: 10.1196/annals.1301.021. [DOI] [PubMed] [Google Scholar]
- Kalin NH, Shelton SE, Fox AS, Oakes TR, Davidson RJ. Brain regions associated with the expression and contextual regulation of anxiety in primates. Biol Psychiatry. 2005;58:796–804. doi: 10.1016/j.biopsych.2005.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly C, Uddin LQ, Shehzad Z, Margulies DS, Castellanos FX, Milham MP, Petrides M. Broca’s region: linking human brain functional connectivity data and non-human primate tracing anatomy studies. Eur J Neurosci. 2010;32:383–398. doi: 10.1111/j.1460-9568.2010.07279.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klingler J, Gloor P. The connections of the amygdala and of the anterior temporal cortex in the human brain. J Comp Neurol. 1960;115:333–369. doi: 10.1002/cne.901150305. [DOI] [PubMed] [Google Scholar]
- LeDoux J. The amygdala. Curr Biol. 2007;17:R868–874. doi: 10.1016/j.cub.2007.08.005. [DOI] [PubMed] [Google Scholar]
- Luyten L, Casteels C, Vansteenwegen D, van Kuyck K, Koole M, Van Laere K, Nuttin B. Micro-Positron Emission Tomography Imaging of Rat Brain Metabolism during Expression of Contextual Conditioning. The Journal of Neuroscience. 2012;32:254–263. doi: 10.1523/JNEUROSCI.3701-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Worsley KJ, Evans AC. Variability of spatial location of activation in fMRI and PET CBF images. Neuroimage. 1999;9:S178. [Google Scholar]
- Machado CJ, Bachevalier J. Non-human primate models of childhood psychopathology: the promise and the limitations. J Child Psychol Psychiatry. 2003;44:64–87. doi: 10.1111/1469-7610.00103. [DOI] [PubMed] [Google Scholar]
- Mai JK, Assheuer J, Paxinos G. Atlas of the Human Brain. 2. Elsevier Academic Press; San Diego: 2003. [Google Scholar]
- Margulies DS, Kelly AM, Uddin LQ, Biswal BB, Castellanos FX, Milham MP. Mapping the functional connectivity of anterior cingulate cortex. Neuroimage. 2007;37:579–588. doi: 10.1016/j.neuroimage.2007.05.019. [DOI] [PubMed] [Google Scholar]
- Martin LJ, Powers RE, Dellovade TL, Price DL. The bed nucleus-amygdala continuum in human and monkey. J Comp Neurol. 1991;309:445–485. doi: 10.1002/cne.903090404. [DOI] [PubMed] [Google Scholar]
- McDonald AJ. Cell types and intrinsic connections of the amygdala. In: Aggelton JP, editor. The Amygdala: Neurobiological Aspects of Emotion, Memory, and mental Dysfunction. Wiley-Liss; New York: 1992. pp. 67–96. [Google Scholar]
- McDonald AJ. Is There an Amygdala and How Far Does It Extend?: An Anatomical Perspective. Ann NY Acad Sci. 2003;985:1–21. doi: 10.1111/j.1749-6632.2003.tb07067.x. [DOI] [PubMed] [Google Scholar]
- Mobbs D, Yu R, Rowe JB, Eich H, FeldmanHall O, Dalgleish T. Neural activity associated with monitoring the oscillating threat value of a tarantula. Proc Natl Acad Sci U S A. 2010;107:20582–20586. doi: 10.1073/pnas.1009076107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy FZ, Pare D. Timing of impulses from the central amygdala and bed nucleus of the stria terminalis to the brain stem. J Neurophysiol. 2008;100:3429–3436. doi: 10.1152/jn.90936.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Rourke H, Fudge JL. Distribution of serotonin transporter labeled fibers in amygdaloid subregions: implications for mood disorders. Biol Psychiatry. 2006;60:479–490. doi: 10.1016/j.biopsych.2005.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oler JA, Fox AS, Shelton SE, Rogers J, Dyer TD, Davidson RJ, Shelledy W, Oakes TR, Blangero J, Kalin NH. Amygdalar and hippocampal substrates of anxious temperament differ in their heritability. Nature. 2010;466:864–868. doi: 10.1038/nature09282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Wheeler RA, Fontillas K, Keithley RB, Carelli RM, Wightman RM. Catecholamines in the bed nucleus of the stria terminalis reciprocally respond to reward and aversion. Biol Psychiatry. 2012;71:327–334. doi: 10.1016/j.biopsych.2011.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paxinos G, Huang X, Petrides M, Toga A. The rhesus monkey brain in stereotaxic coordinates. 2. Academic Press; San Diego: 2009. [Google Scholar]
- Paxinos G, Mai JK. The human nervous system. 2. Elsevier Science; Amsterdam: 2004. [Google Scholar]
- Pitkänen A. Connectivity of the rat amygdaloid complex. In: Aggleton JP, editor. The Amygdala. A Functional Analysis. Oxford University Press; New York: 2000. pp. 31–117. [Google Scholar]
- Price JL, Amaral DG. An autoradiographic study of the projections of the central nucleus of the monkey amygdala. J Neurosci. 1981;1:1242–1259. doi: 10.1523/JNEUROSCI.01-11-01242.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price JL, Russchen FT, Amaral DG. The limbic region II: The amygdaloid complex. In: Björkland A, Hökfelt T, Swanson LW, editors. Handbook of chemical neuroanatomy. Elsevier; Amsterdam: 1987. pp. 279–388. [Google Scholar]
- Regev L, Neufeld-Cohen A, Tsoory M, Kuperman Y, Getselter D, Gil S, Chen A. Prolonged and site-specific over-expression of corticotropin-releasing factor reveals differential roles for extended amygdala nuclei in emotional regulation. Mol Psychiatry. 2011;16:714–728. doi: 10.1038/mp.2010.64. [DOI] [PubMed] [Google Scholar]
- Roy AK, Shehzad Z, Margulies DS, Kelly AM, Uddin LQ, Gotimer K, Biswal BB, Castellanos FX, Milham MP. Functional connectivity of the human amygdala using resting state fMRI. Neuroimage. 2009;45:614–626. doi: 10.1016/j.neuroimage.2008.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shehzad Z, Kelly AM, Reiss PT, Gee DG, Gotimer K, Uddin LQ, Lee SH, Margulies DS, Roy AK, Biswal BB, Petkova E, Castellanos FX, Milham MP. The resting brain: unconstrained yet reliable. Cereb Cortex. 2009;19:2209–2229. doi: 10.1093/cercor/bhn256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinnick-Gallagher P, Pitkanen A, Shekhar A, Cahill L, editors. The Amygdala in Brain Function: Basic and Clinical Applications. The New York Academy of Sciences; New York: 2003. [Google Scholar]
- Sink KS, Walker DL, Freeman SM, Flandreau EI, Ressler KJ, Davis M. Effects of continuously enhanced corticotropin releasing factor expression within the bed nucleus of the stria terminalis on conditioned and unconditioned anxiety. Mol Psychiatry. 2012 doi: 10.1038/mp.2011.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solano-Castiella E, Anwander A, Lohmann G, Weiss M, Docherty C, Geyer S, Reimer E, Friederici AD, Turner R. Diffusion tensor imaging segments the human amygdala in vivo. Neuroimage. 2010;49:2958–2965. doi: 10.1016/j.neuroimage.2009.11.027. [DOI] [PubMed] [Google Scholar]
- Somerville LH, Wagner DD, Wig GS, Moran JM, Whalen PJ, Kelley WM. Interactions Between Transient and Sustained Neural Signals Support the Generation and Regulation of Anxious Emotion. Cerebral Cortex. 2012 doi: 10.1093/cercor/bhr373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somerville LH, Whalen PJ, Kelley WM. Human bed nucleus of the stria terminalis indexes hypervigilant threat monitoring. Biol Psychiatry. 2010;68:416–424. doi: 10.1016/j.biopsych.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straube T, Mentzel HJ, Miltner WH. Waiting for spiders: brain activation during anticipatory anxiety in spider phobics. Neuroimage. 2007;37:1427–1436. doi: 10.1016/j.neuroimage.2007.06.023. [DOI] [PubMed] [Google Scholar]
- Swanson LW, Petrovitch GD. What is the amygdala? Trends in Neurosciences. 1998;21:323–330. doi: 10.1016/s0166-2236(98)01265-x. [DOI] [PubMed] [Google Scholar]
- Tai C, Chatziioannou A, Siegel S, Young J, Newport D, Goble RN, Nutt RE, Cherry SR. Performance evaluation of the microPET P4: a PET system dedicated to animal imaging. Phys Med Biol. 2001;46:1845–1862. doi: 10.1088/0031-9155/46/7/308. [DOI] [PubMed] [Google Scholar]
- Vincent JL, Patel GH, Fox MD, Snyder AZ, Baker JT, Van Essen DC, Zempel JM, Snyder LH, Corbetta M, Raichle ME. Intrinsic functional architecture in the anaesthetized monkey brain. Nature. 2007;447:83–86. doi: 10.1038/nature05758. [DOI] [PubMed] [Google Scholar]
- Walker DL, Davis M. Role of the extended amygdala in short-duration versus sustained fear: a tribute to Dr. Lennart Heimer. Brain Struct Funct. 2008;213:29–42. doi: 10.1007/s00429-008-0183-3. [DOI] [PubMed] [Google Scholar]
- Whalen PJ, Phelps EA, editors. The Human Amygdala. Guilford; New York: 2009. [Google Scholar]
- Zhang Y, Brady M, Smith S. Segmentation of brain MR images through a hidden Markov random field model and the expectation-maximization algorithm. IEEE Trans Med Imaging. 2001;20:45–57. doi: 10.1109/42.906424. [DOI] [PubMed] [Google Scholar]