Abstract
Environmental stresses are manifold and so are the responses they elicit. This is particularly true for higher eukaryotes where various tissues and cell types are differentially affected by the insult. Type and scope of the stress response can therefore differ greatly among cell types. Given the importance of the Ubiquitin Proteasome System (UPS) for most cellular processes, it comes as no surprise that the UPR plays a pivotal role in counteracting the effects of stressors. Here we outline contributions of the UPS to stress sensing, signaling, and response pathways. We make no claim to comprehensiveness but choose selected examples to illustrate concepts and mechanisms by which protein modification with ubiquitin and proteasomal degradation of key regulators ensures cellular integrity during stress situations.
Keywords: Ubiquitin ligases, stress sensors, stress signaling, ubiquitin proteasome pathway
1. Introduction
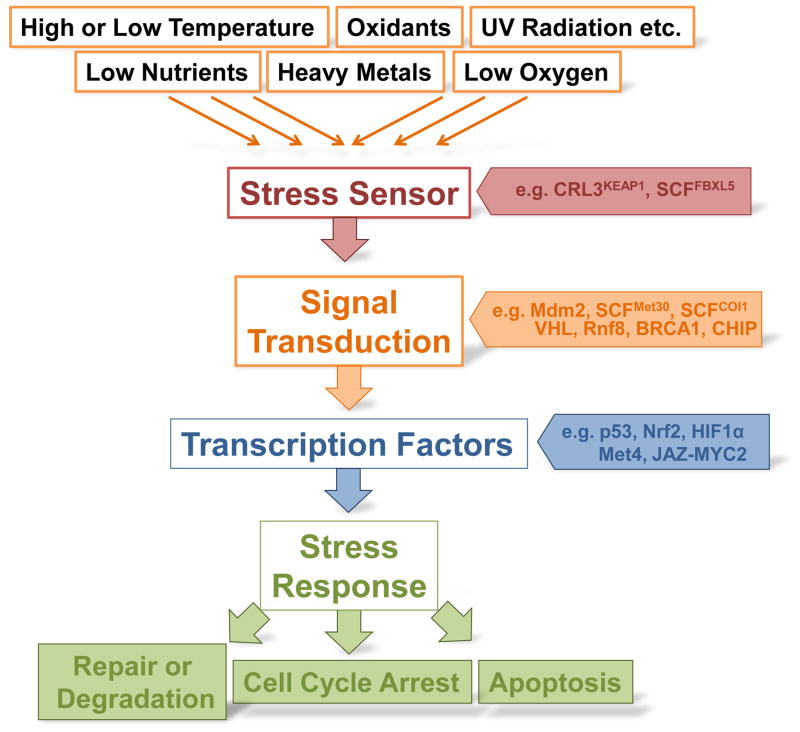
Cells constantly have to battle diverse situations that endanger cellular and genomic integrity. Products of metabolic processes cause some of these stresses internally; a changing environment causes others. Regardless of the cause, cells need to have processes in place to sense the stress, transduce the information, and induce proper counter measures, including adaptation to the new environment to guarantee the survival of the cell or organism (Figure 1). Cellular integrity may be preserved by repair of the stress-induced damage or by degradation and replacement of heavily damaged macromolecules. When these counter measures are insufficient, cells choose more radical actions such as apoptosis or macroautophagy to protect the organism. The major targets of insults are macromolecules like DNA, mRNAs, proteins, and lipids. Damage to these structural components and important regulators can compromise cellular and genomic integrity. Stress response pathways therefore often evoke cell cycle arrest to avoid distribution of damaged macromolecules, particularly damaged DNA, to daughter cells.
Fig. 1. The stress response.
Stress response requires sensing of stress, and transduction of the signal to response elements that induce gene expression programs to counteract stressors. A few selected examples for ubiquitin ligases functioning at distinct steps of the stress response are indicated.
Proteolysis by the proteasome plays an important role in stress response pathways. One important function is the removal of damaged proteins to avoid accumulation as potentially harmful aggregates and to eliminate proteins with compromised activity. In many cases this degradation pathway is not protein specific but recognizes features characteristic for damaged proteins (for example oxidized or misfolded). In some cases this proteasome pathway may be partly independent of ubiquitylation (e.g. oxidative stress). In addition, selective ubiquitylation and degradation of regulatory factors coordinate stress response processes. A common theme here is that a normally unstable transcription factor is stabilized by stress signals, and then induces a transcriptional program to coordinate removal of damaged components, cell cycle arrest, and adaptation (e.g. p53, Nrf2, HIF1).
This review will give a short overview about the ubiquitylation process and discuss concepts of the ubiquitin proteasome system (UPS) in stress sensing, signaling and response pathway activation. A major stress response pathway with extensive involvement of the UPS is the reaction to various kinds of DNA damage. A review article in this issue is devoted to the DNA damage response [***insert a cross-reference to the accompanying manuscript from A Peyroche***]. Similarly, protein quality control pathways in different cellular compartments that play essential roles in survival of many if not most stress situations are discussed in a separate review article [***insert a cross-reference to the accompanying manuscript from R Gardner***]. We will therefore only very briefly examine these important stress response pathways here, and refer the interested reader to the accompanying reviews in this issue [***insert again cross-references to the articles of R Gardner and A Peyroche] or other recent review articles [1, 2]. However, we will briefly describe the role of the UPS in other common stress response pathways such as heat shock and hypoxia, as well as less known stress situations induced for example by aneuploidy. Finally we will briefly examine the important role of stress response and proteolysis for human health as they counteract stress as a potent inducer of cancer, aging, and neurological disorders.
2. Ubiquitylation
The small 76 amino acid protein ubiquitin is covalently attached to typically lysine residues, or the N-terminus in target proteins to form a posttranslational modification that regulates most cellular and developmental processes. Ubiquitylation can come in the form of mono, poly, or multiubiquitylation [3–7]. This complexity of the ubiquitin signal is further increased because there are 7 lysine residues (K6, K11, K27, K29, K33, K48 and K63) as well as the amino-terminus in ubiquitin that can be used to form distinct polyubiquitin chains [8–11]. The various chain topologies are structurally diverse and can define different functions of the ubiquitin chain, such as targeting proteins for proteasome-dependent proteolysis, or modulation of protein function, structure, assembly, and localization.
Ubiquitin is attached to substrates by the E1-E2-E3 cascade of enzymes, consisting of ubiquitin activating enzymes (E1s), ubiquitin conjugating enzymes (E2s), and finally the ubiquitin ligases (E3s) [7]. Ubiquitin activation is an energy dependent process, which initiates the cascade that culminates in formation of an isopeptide linkage, typically between a lysine residue of the substrate and the terminal carboxyl group of ubiquitin. This step is usually achieved via ubiquitin ligases, which are responsible for substrate recognition and stimulation of E2s activity. E3s are therefore central to the ubiquitylation process and govern most processes in cells. This is accentuated by the fact that there are more ubiquitin ligases encoded in the human genome (estimated 600–1000) than there are protein kinases (518) [12].
3. Concepts of Cellular Stress Response
Cellular strategies to battle various stresses vary, but we can distinguish three general phases: Stress sensing, signaling, and response (Figure 1). Typically cells don’t sense the stressors themselves but rather their consequences such as damaged macromolecules or the reduction in ATP production. It is however crucial that the stress situation is detected before major damage occurs so that cells have time to activate response and adaptation mechanisms. Once the stress has been detected, cells transduce the signal to ensure that the appropriate countermeasures are induced. This usually involves cascades of posttranslational protein modifications, especially phosphorylation. The stress signal ultimately reaches transcription factors to activate a stress response program by modulation of gene expression profiles. The subsequent increase in repair and defense capacities may be sufficient for adaptation to the stress condition, but depending on the severity of the damage, cells may trigger more drastic measures like apoptosis or senescence to protect the tissue or organism. Most stressors induce not just a single response pathway, but a network of usually integrated pathways to deal with multiple different types of damages. This is an important feature of effective stress response because a single stress situation can lead to different damages. For example oxidative stress not only leads to oxidized proteins and lipids that need to be removed, but also induces DNA damage.
The ubiquitin proteasome pathway plays important roles at all levels of stress response. The increased need for protein modification by ubiquitin is evident from dedicated stress-inducible ubiquitin genes in all eukaryotes. Among the 4 ubiquitin genes in yeast (UBI1-4), UBI1-3 generate ubiquitin during normal growth, and the polyubiquitin gene UBI4 (five head-to-tail repeats) compensates for the increased need during stress [13, 14]. Mammals also have 4 ubiquitin genes (UbA52, UbA80, UbB, UbC), and the two polyubiquitin genes UbB and UbC are stress induced [15]. Even though ubiquitin is mostly recycled before ubiquitylated substrates enter the proteasome, basal turnover of ubiquitin results in approximate half-lives of 2 and 30hrs in yeast and mammals, respectively [16, 17]. Increased protein ubiquitylation during stress accelerates ubiquitin turnover. Interestingly, low levels of free ubiquitin present a cellular stress situation known as ubiquitin stress that is counteracted by increased transcription and enhanced disassembly of ubiquitin conjugates [18, 19]. In addition, ubiquitin stress in yeast triggers a change in proteasome composition through increased association of the deubiquitylating enzyme Ubp6 and subsequent enhanced ubiquitin recycling [20]. The human Ubp6 ortholog Usp14 may have similar functions in mammals. Limiting the availability of ubiquitin by deletion of stress induced polyubiquitin genes or reduced recycling at the proteasome renders cells hypersensitive to most stress situations, which further underscores the role of the UPS in coordination of stress response.
3.1. Stress Sensing and the UPS
Detecting stress situations for preemptive induction of stress response programs before cells are significantly harmed is critical. Such early detection can be achieved by stress sensors that are hypersensitive to the particular stress situation. Please note that the term stress sensor is often used for key factors in stress response pathways, but here we use its narrower meaning for describing the factors that directly sense the stressor. One example is the ubiquitin ligase CRL3KEAP1, a major regulator of oxidative stress response that presents a thiol-based redox sensor. Keap1 is the substrate receptor of the Cullin-3-RING ubiquitin ligase CRL3KEAP1. As such it controls ubiquitylation and degradation of the Nrf2 (Nuclear factor erythroid 2-related factor 2) transcription factor, which coordinates stress response transcription programs. Mutational analyses identified three critical cysteine residues (Cys 151, Cys 273, and Cys 288) in Keap1 that are sensitive to electrophiles or reactive oxygen species, and reaction of these cysteine residues blocks CRL3KEAP1 mediated Nrf2 ubiquitylation [21, 22]. Details about molecular consequences resulting from Cys 151, Cys 273, and Cys 288 oxidation or electrophile adduction remain somewhat controversial [21–24], but it is widely accepted that the ubiquitin ligase CRL3KEAP1 is the major sensor for oxidative stress.
The budding yeast E3 SCFMET30 is another Cullin-RING ubiquitin ligase that may directly function as stress sensor [25]. The SCFMET30 pathway specifically responds to nutrient stress and exposure to the heavy metals cadmium or arsenic, but does not sense general oxidative stress [26, 27]. Cadmium stress induces selective dissociation of the substrate receptor Met30 from the core ligase resulting in activation of the transcription factor Met4 and initiation of stress response programs.
The cellular response to iron limitation is also an example for an SCF ubiquitin ligase functioning as a putative stress sensor. Iron regulatory protein 2 (IRP2) promotes translation of several mRNAs important for iron homeostasis [28]. The ubiquitin ligase SCFFBXL5 ubiquitylates IRP2 and induces its degradation when intracellular iron levels are high, but not during iron limitation [29]. SCFFBXL5 appears to sense iron levels by two distinct mechanisms. First, the N-terminal region of Fbxl5 contains a hemerythrin-like domain [29], which is a oxygen-carrying domain with di-iron metal centers [30]. A loaded hemerythrin-like domain is important for stability of Fbxl5, because iron limitation or hypoxia induces proteasomal Fbxl5 degradation, and consequently stabilization of the SCFFBXL5 substrate IRP2. In addition, binding of IRP2 to Fbxl5 requires iron, leading to further stabilization IRP2 during conditions of iron limitation [29]. SCFFBXL5 is therefore a putative stress sensor responding to both iron limitation and hypoxia.
These examples demonstrate that core components of the ubiquitin system can function as stress sensors. However, less direct involvement of UPS components in stress sensing is more common. Ubiquitin ligase activity may be regulated directly by stress sensors and stress sensors themselves can be controlled by ubiquitylation. Stress sensors may also modify targets of ubiquitin ligases to induce or prevent substrate ubiquitylation. These events following stress detection transduce the signal to ultimately activate the response program and are discussed below.
3.2. Stress Signal Transduction and the UPS
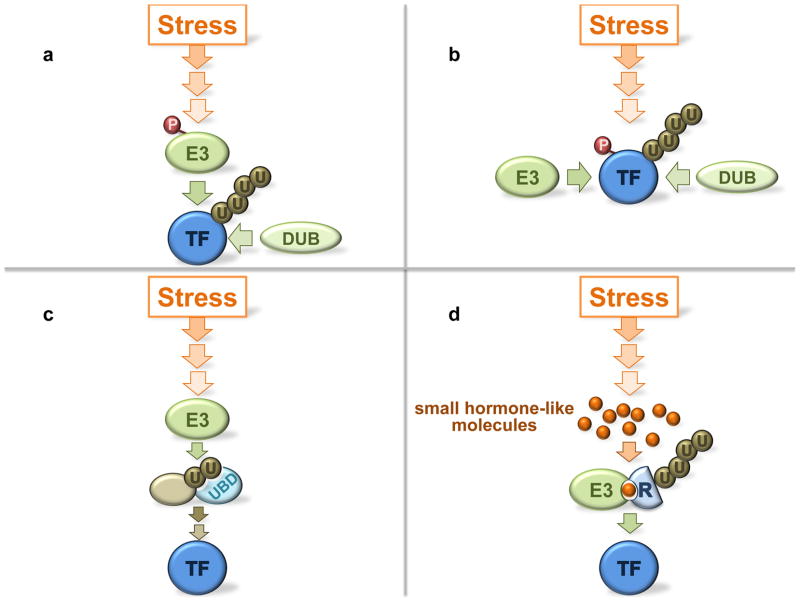
The ubiquitin proteasome system is integral to most if not all signal transduction pathways [***insert a cross-reference to the accompanying manuscript from S Urbe***]. Stress signaling is no exception. Both proteasome dependent and independent regulation by ubiquitylation are important to connect stress sensing with activation of response pathways. Components of the UPS, in particular ubiquitin ligases, can be integral parts of the core signaling cascade (Figure 2a), or act at the periphery by controlling abundance of signaling components (Figure 2b). For example, the RING-type ubiquitin ligase Mdm2 controls ubiquitylation and stability of the tumor suppressor protein p53 [31], which is activated in response to a large variety of stress situations [32]. Regulation of the p53 pathway is extraordinarily complex and depending on the stress situation Mdm2 can be the target of regulation or function as reader of stress induced p53 modifications.
Fig. 2. Concepts of ubiquitin ligase involvement in stress signal transduction.
Four examples for how ubiquitin ligases transmit and control stress signals are depicted. (a) Ubiquitin ligases are part of the core signal flow and are directly controlled by upstream signaling components. Examples are Mdm2 phosphorylation or inhibition of Mdm2 by protein binding. Deubiquitylating enzymes (DUBs) may oppose ubiquitin ligase activity. (b) An E3 substrate is posttranslationally modified by the stress signal cascade and the modification prevents ubiquitin ligase binding, or stimulates binding. An example for the former is ATM mediated p53 phosphorylation. Protein modifications, particularly phosphorylation, as substrate recruitment signals are widespread and typical for SCF-type ligases. (c) Ubiquitylation of a signaling component creates a binding site for a downstream component that binds via an ubiquitin binding domain (UBD) and transmits the stress signal. Such ubiquitin signals can be monoubiquitin modifications or polyubiquitin chains, are typically non-proteolytic signals, and often involve specific polyubiquitin chain topologies such as K63-linked chains. Examples are abundant in DNA damage response pathways. (d) Plants frequently use small hormone like molecules for signaling and feature several cullin-RING ubiquitin ligases that recognize their substrates only in the presence of the hormone. The hormone acts as “molecular glue”, initiates substrate binding to the E3, and controls ubiquitylation and subsequent degradation. Examples are plant stress response pathways mediated by jasmonate. TF: transcription factor.
Oncogenic stress, the inappropriate activation of oncogenes or proto-oncogenes, leads to a response that activates p53 by preventing its ubiquitylation and degradation [33]. One of the important mechanisms is activation of the tumor suppressor ARF by oncogenic stress. ARF binds Mdm2 and blocks its ubiquitin ligase activity towards p53, leading to activation of oncogenic stress response pathway [34].
A similar example for Mdm2 as part of the core signaling cascade is the response to ribosomal stress. Ribosomal stress is induced by disruption of rRNA synthesis or mutations in some ribosomal subunits as observed in the human Diamond-Blackfan syndrome [35]. In either case excess ribosomal subunits are released from the nucleoli and binding of the large subunit L11 to Mdm2 was shown to activate ubiquitylation and subsequent degradation of its binding partner Mdm4 resulting in p53 activation [36].
Activation of p53 in response to ionizing radiation follows a somewhat different mechanism that relies on modification of p53 to control its ubiquitylation by Mdm2. The kinase ATM is the major signaling component for the response to ionizing radiation and phosphorylates p53 on serine 15 and other residues, which reduces binding of Mdm2 resulting in p53 stabilization and activation of the stress response pathway [37]. The signal is reinforced by phosphorylation of Mdm2 and Mdm4, the major negative regulators of p53. These phosphorylation events activate Mdm2 autoubiquitylation as well as Mdm2-mediated Mdm4 ubiquitylation resulting in their degradation. The deubiquitylating enzyme HAUSP further adds to the complexity of this pathway. HAUSP normally counteracts Mdm4 degradation by trimming the ubiquitin chain on Mdm4, but its access to Mdm4 is restricted during DNA damage response [38].
Many stress situations lead to activation of p53. Stress-specific mechanisms of p53 stabilization as outlined with a few examples above allow a tailored stress response. For example, ribosomal stress launches much of the p53 response program, but uses the ribosomal subunit L11 to regulate p53 stability, thus circumvents ATM activation to stabilize p53, which would otherwise activate the full range of the ATM-mediated DNA damage response.
Stress signaling pathways with similar complexity and multiple roles for ubiquitylation are activated in response to the various forms of DNA damage. We will use selected examples from this pathway to illustrate non-proteolytic roles of ubiquitylation during stress signaling (Fig. 2c) and refer the interested reader to several excellent reviews for comprehensive discussion of this topic [2, 39]. Signaling at DNA double strand breaks is initiated through phosphorylation of the histone variant H2AX by ATM. The resulting γ-H2AX is recognized by Mdc1 and initiates an ubiquitylation cascade that ultimately amplifies the signal and leads to assembly of repair complexes. Mdc1 itself is phosphorylated by ATM forming a binding site for the ubiquitin ligase Rnf8 and its E2 Ubc13, which monoubiquitylates H2AX and γ-H2AX around the damage site [40–42]. Next, the ubiquitin ligase Rnf168 docks to the ubiquitylated histones via its two MIU-type ubiquitin binding domains [43, 44], amplifies the ubiquitin signal, possibly by synthesis of K63-linked polyubiquitin chains that are subsequently recognized by the two UIM-type ubiquitin binding domains in Rap80. Yet another ubiquitin ligase, BRCA1, bound to a number of additional proteins is recruited to the damage site by Rap80 [45–48]. The molecular details of signaling events following the recruitment of the BRCA1-A complex are less clear, but result in cell cycle delay and DNA repair.
A similar interplay of non-proteolytic ubiquitylation and recruitment of signaling factors via their ubiquitin binding domains (UBDs) is likely fundamental in other stress signaling pathways. The critical role of UBDs in these pathways is evident, and given the non-proteolytic nature of some of the ubiquitin modifications involved in signaling, the importance of deubiquitylating enzymes to fine-tune and terminate signaling is apparent.
Plants have particularly well developed stress response pathways because they are stationary and cannot escape environmental stress situations. Frequently plants employ small signaling molecules similar to hormones to convey stress signals throughout the plant. One particularly interesting mechanism of stress signaling emerged from studies of the Arabidopsis jasmonate signaling [49] (Figure 2d). Jasmonates are small molecules including jasmonic acid and other oxylipin derivatives that are conjugated to isoleucine to form the active hormone. They induce stress response pathways during ozone exposure, wounding, water deficit, or pathogen and pest attack [50]. The jasmonate induced stress response is activated by the transcription factor MYC2, which is usually repressed by JAZ proteins. Jasmonate triggers ubiquitylation of JAZ proteins by the ubiquitin ligase SCFCOI1, relieving MYC2 repression to launch the stress response program. Remarkably, the complex of SCFCOI1 and its substrates the JAZ proteins serves as the jasmonate receptor [51–54].
Specifically, jasmonate binds to a hormone binding pocket in the F-box subunit COI1 and a loop region in JAZ proteins traps it there, forming a highly selective and hormone-dependent interaction between ubiquitin ligase and substrate, which initiates JAZ ubiquitylation and degradation [54]. Similar functions for ubiquitin ligases as receptors for other hormone-like small molecules have been described in plants but remain to be demonstrated for E3s in animal cells [55].
The few examples outlined above offer a glimpse into the importance of the UPS in stress signal transduction. The roles of ubiquitylation in these pathways go beyond protein degradation. Monoubiquitylation or specific ubiquitin chain architectures such as K63-linked chains or linear ubiquitin chains generate protein signals that are recognized by ubiquitin binding domains in other signaling proteins to assemble signaling hubs. In addition, modification with ubiquitin-like proteins such as SUMO contributes significantly to transduction of stress signals and ultimately ensures efficient stress response.
3.3. Activation of Stress Response Programs and the UPS
Stress signals ultimately reach transcription factors that when activated launch the stress response program. The general concept is that stress-related transcription factors are maintained in a repressed state by ubiquitin ligases through ubiquitylation and subsequent degradation. The stress signal prevents transcription factor degradation leading to its activation and induction of transcription programs to cope with the stressors. Mechanistically transcription factor stabilization is achieved by either inhibition of the ubiquitin ligase, or by changes in posttranslational modifications on the transcription factor to prevent recognition by the E3. A combination of both mechanisms often reinforces the stress response. Alternatively, stress signals can trigger degradation of repressor proteins that under normal conditions inactivate stress-controlled transcription factors, as described above for the jasmonate response in plants. A typical example for a transcription factor that is targeted by a stress controlled E3 is Nrf2. As outlined above, Nrf2 is ubiquitylated and inactivated by the ubiquitin ligase CRL3KEAP1, which is also the sensor for oxidative stress. During non-stress conditions CRL3KEAP1 maintains low Nrf2 levels, but stress induced oxidation of several reactive cysteine residues in Keap1 inactivate CRL3KEAP1 allowing Nrf2 accumulation in the nucleus and subsequent activation of the stress response program [56].
Conceptually this mechanism of stress induced stabilization of a transcription factor is related to p53 activation through inhibition of the Mdm2 ubiquitin ligase by association with inhibitory proteins such as ARF or the ribosomal subunit L11 as described above. An alternative pathway to p53 stabilization by ATM-mediated phosphorylation that blocks the Mdm2 binding site on p53 has also been described above.
A variation of this modification-controlled E3 binding governs the response to hypoxia. HIF1α, which controls transcriptional response to low oxygen, is constitutively ubiquitylated and degraded by the VHL ubiquitin ligase during normoxia. Hydroxylation of proline 402 and 564 in the oxygen-dependent degradation domain of HIF1α forms the recognition site for VHL binding and is thus the signal for HIF1α degradation. Proline hydroxylation is dramatically reduced during hypoxia, resulting in HIF1α stabilization and activation of the stress response program [57]. Additional ubiquitin ligases that regulate prolyl hydroxylases to reinforce the signal are discussed below.
In budding yeast the response to heavy metal stress is coordinated by the transcriptional activator Met4 [25]. The ubiquitin ligase SCFMET30 polyubiquitylates Met4 to suppress the heavy metal stress response during normal conditions. Ubiquitylation does not however, induce Met4 degradation but maintains it in a dormant state standing by for rapid activation [58]. Heavy metal stress induces disassembly of SCFMET30 preventing ubiquitylation of Met4, but deubiquitylation by so far unknown deubiquitylating enzymes is required to rapidly convert the pool of ubiquitylated Met4 into its transcriptionally active form [26, 27].
The interplay between ubiquitin ligases and stress-responsive transcription factors is a recurring theme, and many pathways follow this concept with various variations. However, other mechanisms for stress-induced transcription programs that are not directly linked to transcription factor ubiquitylation are also well documented as described below for activation of heat shock factor 1.
3.4. Stress Response and the UPS
Cells under stress need to prevent further damage by launching stress defense measures, repair damaged components, or remove them when irreparable. Cells also often induce cell cycle arrest to prevent distribution of damaged macromolecules to daughter cells and to allow time for repair. These cell cycle checkpoint arrests are particular important if the stress situation leads to DNA damage. Checkpoints are tightly controlled by the UPS at multiple levels. Due to space limitation we will not discuss these important aspects of stress response here and refer the interested reader to the accompanying manuscript of J Benanti [***insert a cross-reference to the accompanying manuscript from J Benanti***] and other excellent reviews [59–62].
Stress defense measures depend on the type of stressor. Examples are induction of DNA repair enzymes in response to DNA damaging agents, and increased synthesis of glutathione or other scavengers to counteract oxidative stress or detoxify heavy metals.
Although most stressors cause specific damage that evokes selective countermeasures, a number of different stress conditions lead to protein damage and thus misfolded proteins. Consequently protein quality control pathways are induced by various adverse conditions such as heat, osmotic, oxidative, and heavy metal stress. As a first response, different chaperone systems attempt to refold proteins. However, when refolding is unsuccessful the UPS degrades proteins. The challenge for the ubiquitin system is to recognize terminally unfolded or damaged proteins. The chaperones HSP70 and HSP90 play a particularly important role in both refolding and selection of proteins for ubiquitylation by the ubiquitin ligase CHIP [63]. How CHIP differentiates between refolding intermediates and its irreparably damaged substrates is not completely understood, but contribution of a “timer” mechanism that is based on the duration a damaged protein spends on HSP70 or HSP90 seems likely [1]. In addition, protein damage generally reduces global translation and increases expression of quality control proteins, which presents an adaptive response observed for many stress situations.
Stress response pathways aim to protect from damage and their activation helps cells and organisms to adapt to the changed environment.
3.5. Coordination of Stress Response by the UPS
Stress elicits many different cellular responses and their coordination is critical for effective stress protection and survival. A simple concept for integration of cellular responses to a stressor is the coordinated control by a single transcription factor. For example, p53 directly induces expression of both DNA repair factors (e.g. DDB2, GADD45A) and cell cycle inhibitors (e.g. p21) to synchronize DNA repair with cell cycle arrest. One stressor often also activates several signaling pathways to integrate the response. Hypoxia leads to p53 and HIF1α stabilization to coordinate both downstream responses [64]. An interesting example for an ubiquitin ligase as the coordinator of stress responses is yeast SCFMET30. The two main substrates of this E3 are the transcription factor Met4, which induces glutathione biosynthesis [65], and a cell cycle inhibitory protein Met32 [66]. Heavy metal stress inactivates SCFMET30 leading to simultaneous stabilization of Met32 to induce cell cycle arrest, and activation of Met4 to induce glutathione synthesis for detoxification [67].
As with all signaling pathways, there is a myriad of connections, cross talk, and cross-activation in stress signaling to integrate individual response pathways into an effective defense against the stress situation.
4. The UPS in Selected Stress Response Pathways
4.1. Hypoxia
Energy production in aerobic organisms requires oxygen. When the availability of oxygen is too low to match the demand it creates significant stress, which elicits a rapid adaptive response. This response includes a temporary cell cycle arrest, reduction in energy production as well as secretion of factors propagating survival and vascularization. Three major signaling pathway act to choreograph this response: The protein quality control pathways induce degradation of proteins that cannot be repaired by refolding, the mTOR pathway alters translation to minimize further damage, and signaling o the hypoxia-Inducible factors (HIF) induces transcriptional programs to increase expression of proteins to coordinate and execute response to hypoxia stress.
HIFs are basic helix-loop-helix/PAS proteins consisting of one α and one β subunit. They activate transcription by binding hypoxia-responsive elements (HREs) in the promoter of genes regulating cell cycle, cell death, cell migration, metabolism, angiogenesis, and erythropoiesis [68]. Many aspects of the hypoxia response are thus controlled by HIFs, and particularly by the HIF1α/HIF1β dimer making it the key factor for cell survival during hypoxia.
Ubiquitylation is a key factor in HIF1 regulation. Proline hydroxylation generates a protein binding site on HIF1α that is specifically recognized by the von Hippel-Lindau protein, which is the substrate receptor of a cullin-RING ubiquitin ligase [57]. The VHL ubiquitin ligase consists of Cullin-2, the RING finger protein Rbx1, the substrate receptor pVHL, and Elongins B and C [69]. HIF1α ubiquitylation induces its degradation by the proteasome via a process involving the AAA-ATPase p97 [70]. The HIF1β subunit remains stable but is inactive as a transcription factor in the absence of HIF1α.
Proline hydroxylation serves as an elegant means for linking HIF1α ubiquitylation to the intracellular oxygen tension. The prolyl hydroxylases targeting HIF1α may therefore act as the oxygen sensors that are inactive under hypoxia leading to stabilization of HIF1α and initiation of its transcription program. The HIF1α prolyl hydroxylases PHD1, PHD2, PHD3 are themselves under tight control. PHD1 and PHD3 are targets of HIF1 and establish a negative feedback loop. However, low oxygen induces degradation of PHD1 and PHD3 through the RING-type ubiquitin ligase Siah2 and reinforces the hypoxia response by further stabilization of HIF1α [71]. Prolyl hydroxylases are also targeted by several oncogenes, which may be important for tumor growth, and the VHL ubiquitin ligase is a well-known tumor suppressor protein [72].
The hypoxia response pathway exemplifies the central role of ubiquitin ligases in facilitating intimate interplay between stress sensing, signaling, and activation of response pathways.
4.2. Heat Shock
Already small changes in temperature pose a challenge to cells because many proteins are only stably folded within a narrow temperature window [73]. Cells are most likely unable to measure the temperature itself and react to the accumulation of unfolded proteins. The master regulator of this response is heat shock factor 1 (HSF1) [74]. Under normal conditions HSF1 is in an inactive complex with molecular chaperones. Chaperones assist proteins in folding by binding and thereby preventing unwanted intermolecular interaction. During heat stress the chaperones are titrated off HSF1 to refold unfolded proteins, which frees HSF1 and allows its trimerization. After phosphorylation and translocation to the nucleus, HSF1 activates transcription through binding to heat shock elements (HSE) in promoters of target genes [74]. This leads to an increase in chaperones, proteases, and other important proteins to meet the higher demand. Ubiquitylation is an important component of the heat stress response because the ubiquitin proteasome pathway efficiently clears terminally unfolded proteins through pathways involving the CHIP ubiquitin ligase as described above.
Recent results in yeast demonstrated an important role of the proteasome bound HECT-type ubiquitin ligase Hul5 in clearing of misfolded proteins in the cytoplasm [75]. A similar role for mammalian ortholog Hul5 remains to be demonstrated, but experiments in yeast indicate monoubiquitylated proteins as the primary substrates for Hul5 after heat stress [75]. This suggests that Hul5 functions as an E4 enzyme, a small group of ubiquitin ligases that recognize previously ubiquitylated proteins and extend ubiquitin chains [76]. The identity of the putative E3 partner for Hul5 is currently unknown.
The UPS plays a major role in clearing terminally unfolded proteins, but defined regulatory roles for ubiquitylation in the heat stress pathway are less clear. Interestingly, modification with the ubiquitin-like modifier SUMO2/3 seems to be particularly important for survival of heat stress [77, 78]. Accordingly, there is a marked increase in global SUMO2/3 conjugation during heat stress, and sumoylation of HSF1 is critical during recovery from the stress [79].
4.3. Stress Induced by Aneuploidy
Aneuploidy, i.e. an abnormal number of chromosomes, poses a substantial stress to cells and results in reduced rates of cell proliferation in normal cells, but paradoxically not in cancer cells [80]. Recent experiments suggest that proteotoxic stress is a major contributor to reduced organismal fitness caused by aneuploidy, and that the UPS plays a major role to cope with this stress. Mutations that overcome fitness defects of engineered disomic yeast cells affected several components of the UPS, including the proteasome associated deubiquitylating enzyme Ubp6 [81]. Ubp6 antagonizes proteasome activity [82], and it was thus suggested that increased proteasome activity in UBP6 deletion mutants allows cells to tolerate aneuploidy stress [81]. Consistent with the idea that proteasome activity is a limiting factor in aneuploidy cells, disomic yeast cells are hypersensitive to proteasome inhibitors and display synthetic lethality with hypomorphic proteasome mutants [81].
In addition to proteotoxic stress, aneuploidy also causes metabolic stress due to changes in metabolism and higher energy demand. However, the underlying mechanism is currently not understood.
Despite the generally negative effect of karyotypic imbalance on cell proliferation, aneuploidy is a hallmark of cancer cells, which are characterized by accelerated cell proliferation [83]. Aneuploid cancer cells remain under proteotoxic stress but have found ways to evade the associated antiproliferative effects. Whether changes in the UPS are involved remains to be shown.
5. Conclusion
Response to cellular stress is vital for cell survival and growth. Stress responses usually protect cellular and genetic integrity leading to recovery from stress situations. However, some stress response pathways can also be important contributors to disease progression. This is true, for example, for the cellular response to hypoxia. While the HIF pathway is critical to protect tissues from hypoxia caused by anemia or stroke, solid tumors frequently exploit this pathway to promote tumor growth trough vascularization [72]. Tumors often achieve that by inactivation of the VHL ubiquitin ligase.
The importance of the UPS in stress response for normal human physiology is evident from mutations of UPS components found in human disease. These include ubiquitin ligases involved in the DNA damage response such as the breast cancer susceptibility gene BRCA1, the p53 regulating E3 Mdm2, the Fanconi anemia ubiquitin ligase complex FANC, and the Cockayne syndrome ubiquitin ligase CRL4CSA [84–86]. In addition, protein quality control pathways that are almost universally required in response to stress are thought to play important roles in protection from neurodegenerative disease. The resulting protein aggregates that may occur as a consequence of inadequate protein quality control are characteristic for many neurodegenerative disorders and evoke a stress response pathway that stimulates clearance of these aggregates by the UPS and autophagy [87].
In conclusion, multicellular organisms exposed to stress need to find a fine balance between counter measures that aim to protect cellular integrity and induction of pathways that sacrifice cells in order to ensure survival of the organism. Ubiquitylation plays an important role in this fine-tuning and ubiquitin ligases are key factors coordinating stress response pathways.
Highlights.
Ubiquitylation regulates many aspects of stress response.
The Ubiquitin Proteasome System clears damaged proteins.
Ubiquitylation controls activation of stress response pathways.
Ubiquitin ligases are central for coordinated stress response.
Acknowledgments
Work in our laboratory is supported by NIH grants GM66164 and CA112560. We thank An Tyrrell for critical reading of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Buchberger A, Bukau B, Sommer T. Protein quality control in the cytosol and the endoplasmic reticulum: brothers in arms. Mol Cell. 2010;40:238–52. doi: 10.1016/j.molcel.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 2.Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kerscher O, Felberbaum R, Hochstrasser M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu Rev Cell Dev Biol. 2006;22:159–80. doi: 10.1146/annurev.cellbio.22.010605.093503. [DOI] [PubMed] [Google Scholar]
- 4.Pickart CM. Back to the future with ubiquitin. Cell. 2004;116:181–90. doi: 10.1016/s0092-8674(03)01074-2. [DOI] [PubMed] [Google Scholar]
- 5.Wilkinson KD. Ubiquitination and deubiquitination: targeting of proteins for degradation by the proteasome. Semin Cell Dev Biol. 2000;11:141–8. doi: 10.1006/scdb.2000.0164. [DOI] [PubMed] [Google Scholar]
- 6.Ciechanover A, Orian A, Schwartz AL. The ubiquitin-mediated proteolytic pathway: mode of action and clinical implications. J Cell Biochem Suppl. 2000;34:40–51. doi: 10.1002/(sici)1097-4644(2000)77:34+<40::aid-jcb9>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 7.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–79. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 8.Tokunaga F, Sakata S, Saeki Y, Satomi Y, Kirisako T, Kamei K, et al. Involvement of linear polyubiquitylation of NEMO in NF-kappaB activation. Nat Cell Biol. 2009;11:123–32. doi: 10.1038/ncb1821. [DOI] [PubMed] [Google Scholar]
- 9.Rahighi S, Ikeda F, Kawasaki M, Akutsu M, Suzuki N, Kato R, et al. Specific recognition of linear ubiquitin chains by NEMO is important for NF-kappaB activation. Cell. 2009;136:1098–109. doi: 10.1016/j.cell.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 10.Meierhofer D, Wang X, Huang L, Kaiser P. Quantitative analysis of global ubiquitination in HeLa cells by mass spectrometry. J Proteome Res. 2008;7:4566–76. doi: 10.1021/pr800468j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peng J, Schwartz D, Elias JE, Thoreen CC, Cheng D, Marsischky G, et al. A proteomics approach to understanding protein ubiquitination. Nat Biotechnol. 2003;21:921–6. doi: 10.1038/nbt849. [DOI] [PubMed] [Google Scholar]
- 12.Li W, Bengtson MH, Ulbrich A, Matsuda A, Reddy VA, Orth A, et al. Genome-wide and functional annotation of human E3 ubiquitin ligases identifies MULAN, a mitochondrial E3 that regulates the organelle’s dynamics and signaling. PLoS One. 2008;3:e1487. doi: 10.1371/journal.pone.0001487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ozkaynak E, Finley D, Solomon MJ, Varshavsky A. The yeast ubiquitin genes: a family of natural gene fusions. Embo J. 1987;6:1429–39. doi: 10.1002/j.1460-2075.1987.tb02384.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Finley D, Ozkaynak E, Varshavsky A. The yeast polyubiquitin gene is essential for resistance to high temperatures, starvation, and other stresses. Cell. 1987;48:1035–46. doi: 10.1016/0092-8674(87)90711-2. [DOI] [PubMed] [Google Scholar]
- 15.Fornace AJ, Jr, Alamo I, Jr, Hollander MC, Lamoreaux E. Ubiquitin mRNA is a major stress-induced transcript in mammalian cells. Nucleic Acids Res. 1989;17:1215–30. doi: 10.1093/nar/17.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haas AL, Bright PM. The dynamics of ubiquitin pools within cultured human lung fibroblasts. J Biol Chem. 1987;262:345–51. [PubMed] [Google Scholar]
- 17.Hanna J, Leggett DS, Finley D. Ubiquitin depletion as a key mediator of toxicity by translational inhibitors. Mol Cell Biol. 2003;23:9251–61. doi: 10.1128/MCB.23.24.9251-9261.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Crosas B, Hanna J, Kirkpatrick DS, Zhang DP, Tone Y, Hathaway NA, et al. Ubiquitin chains are remodeled at the proteasome by opposing ubiquitin ligase and deubiquitinating activities. Cell. 2006;127:1401–13. doi: 10.1016/j.cell.2006.09.051. [DOI] [PubMed] [Google Scholar]
- 19.Kimura Y, Yashiroda H, Kudo T, Koitabashi S, Murata S, Kakizuka A, et al. An inhibitor of a deubiquitinating enzyme regulates ubiquitin homeostasis. Cell. 2009;137:549–59. doi: 10.1016/j.cell.2009.02.028. [DOI] [PubMed] [Google Scholar]
- 20.Hanna J, Meides A, Zhang DP, Finley D. A ubiquitin stress response induces altered proteasome composition. Cell. 2007;129:747–59. doi: 10.1016/j.cell.2007.03.042. [DOI] [PubMed] [Google Scholar]
- 21.Zhang DD, Hannink M. Distinct cysteine residues in Keap1 are required for Keap1-dependent ubiquitination of Nrf2 and for stabilization of Nrf2 by chemopreventive agents and oxidative stress. Mol Cell Biol. 2003;23:8137–51. doi: 10.1128/MCB.23.22.8137-8151.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang DD, Lo SC, Cross JV, Templeton DJ, Hannink M. Keap1 is a redox-regulated substrate adaptor protein for a Cul3-dependent ubiquitin ligase complex. Mol Cell Biol. 2004;24:10941–53. doi: 10.1128/MCB.24.24.10941-10953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kobayashi A, Kang MI, Watai Y, Tong KI, Shibata T, Uchida K, et al. Oxidative and Electrophilic Stresses Activate Nrf2 through Inhibition of Ubiquitination Activity of Keap1. Mol Cell Biol. 2006;26:221–9. doi: 10.1128/MCB.26.1.221-229.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yamamoto T, Suzuki T, Kobayashi A, Wakabayashi J, Maher J, Motohashi H, et al. Physiological significance of reactive cysteine residues of Keap1 in determining Nrf2 activity. Mol Cell Biol. 2008;28:2758–70. doi: 10.1128/MCB.01704-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaiser P, Su NY, Yen JL, Ouni I, Flick K. The yeast ubiquitin ligase SCF-Met30: connecting environmental and intracellular conditions to cell division. Cell Div. 2006;1:16. doi: 10.1186/1747-1028-1-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barbey R, Baudouin-Cornu P, Lee TA, Rouillon A, Zarzov P, Tyers M, et al. Inducible dissociation of SCF(Met30) ubiquitin ligase mediates a rapid transcriptional response to cadmium. Embo J. 2005;24:521–32. doi: 10.1038/sj.emboj.7600556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yen JL, Su NY, Kaiser P. The Yeast Ubiquitin Ligase SCFMet30 Regulates Heavy Metal Response. Mol Biol Cell. 2005;16:1872–82. doi: 10.1091/mbc.E04-12-1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muckenthaler MU, Galy B, Hentze MW. Systemic iron homeostasis and the iron-responsive element/iron-regulatory protein (IRE/IRP) regulatory network. Annu Rev Nutr. 2008;28:197–213. doi: 10.1146/annurev.nutr.28.061807.155521. [DOI] [PubMed] [Google Scholar]
- 29.Vashisht AA, Zumbrennen KB, Huang X, Powers DN, Durazo A, Sun D, et al. Control of iron homeostasis by an iron-regulated ubiquitin ligase. Science. 2009;326:718–21. doi: 10.1126/science.1176333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stenkamp RE. Dioxygen and Hemerythrin. Chem Rev. 1994;94:715–26. [Google Scholar]
- 31.Marine JC, Lozano G. Mdm2-mediated ubiquitylation: p53 and beyond. Cell Death Differ. 2010;17:93–102. doi: 10.1038/cdd.2009.68. [DOI] [PubMed] [Google Scholar]
- 32.Perry ME. The regulation of the p53-mediated stress response by MDM2 and MDM4. Cold Spring Harb Perspect Biol. 2010;2:a000968. doi: 10.1101/cshperspect.a000968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Courtois-Cox S, Jones SL, Cichowski K. Many roads lead to oncogene-induced senescence. Oncogene. 2008;27:2801–9. doi: 10.1038/sj.onc.1210950. [DOI] [PubMed] [Google Scholar]
- 34.Sherr CJ. Divorcing ARF and p53: an unsettled case. Nat Rev Cancer. 2006;6:663–73. doi: 10.1038/nrc1954. [DOI] [PubMed] [Google Scholar]
- 35.Lipton JM, Ellis SR. Diamond-Blackfan anemia: diagnosis, treatment, and molecular pathogenesis. Hematol Oncol Clin North Am. 2009;23:261–82. doi: 10.1016/j.hoc.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gilkes DM, Chen L, Chen J. MDMX regulation of p53 response to ribosomal stress. Embo J. 2006;25:5614–25. doi: 10.1038/sj.emboj.7601424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shieh SY, Ikeda M, Taya Y, Prives C. DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2. Cell. 1997;91:325–34. doi: 10.1016/s0092-8674(00)80416-x. [DOI] [PubMed] [Google Scholar]
- 38.Meulmeester E, Maurice MM, Boutell C, Teunisse AF, Ovaa H, Abraham TE, et al. Loss of HAUSP-mediated deubiquitination contributes to DNA damage-induced destabilization of Hdmx and Hdm2. Mol Cell. 2005;18:565–76. doi: 10.1016/j.molcel.2005.04.024. [DOI] [PubMed] [Google Scholar]
- 39.Ulrich HD, Walden H. Ubiquitin signalling in DNA replication and repair. Nat Rev Mol Cell Biol. 2010;11:479–89. doi: 10.1038/nrm2921. [DOI] [PubMed] [Google Scholar]
- 40.Huen MS, Grant R, Manke I, Minn K, Yu X, Yaffe MB, et al. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell. 2007;131:901–14. doi: 10.1016/j.cell.2007.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kolas NK, Chapman JR, Nakada S, Ylanko J, Chahwan R, Sweeney FD, et al. Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science. 2007;318:1637–40. doi: 10.1126/science.1150034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mailand N, Bekker-Jensen S, Faustrup H, Melander F, Bartek J, Lukas C, et al. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell. 2007;131:887–900. doi: 10.1016/j.cell.2007.09.040. [DOI] [PubMed] [Google Scholar]
- 43.Stewart GS, Panier S, Townsend K, Al-Hakim AK, Kolas NK, Miller ES, et al. The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell. 2009;136:420–34. doi: 10.1016/j.cell.2008.12.042. [DOI] [PubMed] [Google Scholar]
- 44.Doil C, Mailand N, Bekker-Jensen S, Menard P, Larsen DH, Pepperkok R, et al. RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell. 2009;136:435–46. doi: 10.1016/j.cell.2008.12.041. [DOI] [PubMed] [Google Scholar]
- 45.Kim H, Chen J, Yu X. Ubiquitin-binding protein RAP80 mediates BRCA1-dependent DNA damage response. Science. 2007;316:1202–5. doi: 10.1126/science.1139621. [DOI] [PubMed] [Google Scholar]
- 46.Liu Z, Wu J, Yu X. CCDC98 targets BRCA1 to DNA damage sites. Nat Struct Mol Biol. 2007;14:716–20. doi: 10.1038/nsmb1279. [DOI] [PubMed] [Google Scholar]
- 47.Sobhian B, Shao G, Lilli DR, Culhane AC, Moreau LA, Xia B, et al. RAP80 targets BRCA1 to specific ubiquitin structures at DNA damage sites. Science. 2007;316:1198–202. doi: 10.1126/science.1139516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang B, Matsuoka S, Ballif BA, Zhang D, Smogorzewska A, Gygi SP, et al. Abraxas and RAP80 form a BRCA1 protein complex required for the DNA damage response. Science. 2007;316:1194–8. doi: 10.1126/science.1139476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gfeller A, Liechti R, Farmer EE. Arabidopsis jasmonate signaling pathway. Sci Signal. 2010;3:cm4. doi: 10.1126/scisignal.3109cm4. [DOI] [PubMed] [Google Scholar]
- 50.Devoto A, Turner JG. Regulation of jasmonate-mediated plant responses in arabidopsis. Ann Bot. 2003;92:329–37. doi: 10.1093/aob/mcg151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chini A, Fonseca S, Fernandez G, Adie B, Chico JM, Lorenzo O, et al. The JAZ family of repressors is the missing link in jasmonate signalling. Nature. 2007;448:666–71. doi: 10.1038/nature06006. [DOI] [PubMed] [Google Scholar]
- 52.Thines B, Katsir L, Melotto M, Niu Y, Mandaokar A, Liu G, et al. JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature. 2007;448:661–5. doi: 10.1038/nature05960. [DOI] [PubMed] [Google Scholar]
- 53.Yan Y, Stolz S, Chetelat A, Reymond P, Pagni M, Dubugnon L, et al. A downstream mediator in the growth repression limb of the jasmonate pathway. Plant Cell. 2007;19:2470–83. doi: 10.1105/tpc.107.050708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sheard LB, Tan X, Mao H, Withers J, Ben-Nissan G, Hinds TR, et al. Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature. 2010;468:400–5. doi: 10.1038/nature09430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Santner A, Estelle M. The ubiquitin-proteasome system regulates plant hormone signaling. Plant J. 2010;61:1029–40. doi: 10.1111/j.1365-313X.2010.04112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Itoh K, Mimura J, Yamamoto M. Discovery of the negative regulator of Nrf2, Keap1: a historical overview. Antioxid Redox Signal. 2010;13:1665–78. doi: 10.1089/ars.2010.3222. [DOI] [PubMed] [Google Scholar]
- 57.Kaelin WG. Proline hydroxylation and gene expression. Annu Rev Biochem. 2005;74:115–28. doi: 10.1146/annurev.biochem.74.082803.133142. [DOI] [PubMed] [Google Scholar]
- 58.Flick K, Raasi S, Zhang H, Yen JL, Kaiser P. A ubiquitin-interacting motif protects polyubiquitinated Met4 from degradation by the 26S proteasome. Nat Cell Biol. 2006;8:509–15. doi: 10.1038/ncb1402. [DOI] [PubMed] [Google Scholar]
- 59.Reed SI. Ratchets and clocks: the cell cycle, ubiquitylation and protein turnover. Nat Rev Mol Cell Biol. 2003;4:855–64. doi: 10.1038/nrm1246. [DOI] [PubMed] [Google Scholar]
- 60.Skaar JR, Pagano M. Control of cell growth by the SCF and APC/C ubiquitin ligases. Curr Opin Cell Biol. 2009;21:816–24. doi: 10.1016/j.ceb.2009.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gutierrez GJ, Ronai Z. Ubiquitin and SUMO systems in the regulation of mitotic checkpoints. Trends Biochem Sci. 2006;31:324–32. doi: 10.1016/j.tibs.2006.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bassermann F, Pagano M. Dissecting the role of ubiquitylation in the DNA damage response checkpoint in G2. Cell Death Differ. 2010;17:78–85. doi: 10.1038/cdd.2009.104. [DOI] [PubMed] [Google Scholar]
- 63.Arndt V, Rogon C, Hohfeld J. To be, or not to be--molecular chaperones in protein degradation. Cell Mol Life Sci. 2007;64:2525–41. doi: 10.1007/s00018-007-7188-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sermeus A, Michiels C. Reciprocal influence of the p53 and the hypoxic pathways. Cell Death Dis. 2011;2:e164. doi: 10.1038/cddis.2011.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wheeler GL, Trotter EW, Dawes IW, Grant CM. Coupling of the transcriptional regulation of glutathione biosynthesis to the availability of glutathione and methionine via the Met4 and Yap1 transcription factors. J Biol Chem. 2003;278:49920–8. doi: 10.1074/jbc.M310156200. [DOI] [PubMed] [Google Scholar]
- 66.Ouni I, Flick K, Kaiser P. A transcriptional activator is part of an SCF ubiquitin ligase to control degradation of its cofactors. Mol Cell. 2010;40:954–64. doi: 10.1016/j.molcel.2010.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ouni I, Flick K, Kaiser P. Ubiquitin and transcription: The SCF/Met4 pathway, a (protein-) complex issue. Transcription. 2011;2:135–9. doi: 10.4161/trns.2.3.15903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Majmundar AJ, Wong WJ, Simon MC. Hypoxia-inducible factors and the response to hypoxic stress. Mol Cell. 2010;40:294–309. doi: 10.1016/j.molcel.2010.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol. 2005;6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- 70.Alexandru G, Graumann J, Smith GT, Kolawa NJ, Fang R, Deshaies RJ. UBXD7 binds multiple ubiquitin ligases and implicates p97 in HIF1alpha turnover. Cell. 2008;134:804–16. doi: 10.1016/j.cell.2008.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nakayama K, Qi J, Ronai Z. The ubiquitin ligase Siah2 and the hypoxia response. Mol Cancer Res. 2009;7:443–51. doi: 10.1158/1541-7786.MCR-08-0458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kaelin WG., Jr The von Hippel-Lindau tumour suppressor protein: O2 sensing and cancer. Nat Rev Cancer. 2008;8:865–73. doi: 10.1038/nrc2502. [DOI] [PubMed] [Google Scholar]
- 73.Richter K, Haslbeck M, Buchner J. The heat shock response: life on the verge of death. Mol Cell. 2010;40:253–66. doi: 10.1016/j.molcel.2010.10.006. [DOI] [PubMed] [Google Scholar]
- 74.Anckar J, Sistonen L. Regulation of HSF1 function in the heat stress response: implications in aging and disease. Annu Rev Biochem. 2011;80:1089–115. doi: 10.1146/annurev-biochem-060809-095203. [DOI] [PubMed] [Google Scholar]
- 75.Fang NN, Ng AH, Measday V, Mayor T. Hul5 HECT ubiquitin ligase plays a major role in the ubiquitylation and turnover of cytosolic misfolded proteins. Nat Cell Biol. 2011;13:1344–52. doi: 10.1038/ncb2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Koegl M, Hoppe T, Schlenker S, Ulrich HD, Mayer TU, Jentsch S. A novel ubiquitination factor, E4, is involved in multiubiquitin chain assembly. Cell. 1999;96:635–44. doi: 10.1016/s0092-8674(00)80574-7. [DOI] [PubMed] [Google Scholar]
- 77.Flick K, Kaiser P. Proteomic revelation: SUMO changes partners when the heat is on. Sci Signal. 2009;2:pe45. doi: 10.1126/scisignal.281pe45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Golebiowski F, Matic I, Tatham MH, Cole C, Yin Y, Nakamura A, et al. System-wide changes to SUMO modifications in response to heat shock. Sci Signal. 2009;2:ra24. doi: 10.1126/scisignal.2000282. [DOI] [PubMed] [Google Scholar]
- 79.Brunet Simioni M, De Thonel A, Hammann A, Joly AL, Bossis G, Fourmaux E, et al. Heat shock protein 27 is involved in SUMO-2/3 modification of heat shock factor 1 and thereby modulates the transcription factor activity. Oncogene. 2009;28:3332–44. doi: 10.1038/onc.2009.188. [DOI] [PubMed] [Google Scholar]
- 80.Sheltzer JM, Amon A. The aneuploidy paradox: costs and benefits of an incorrect karyotype. Trends Genet. 2011;27:446–53. doi: 10.1016/j.tig.2011.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Torres EM, Dephoure N, Panneerselvam A, Tucker CM, Whittaker CA, Gygi SP, et al. Identification of aneuploidy-tolerating mutations. Cell. 2010;143:71–83. doi: 10.1016/j.cell.2010.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hanna J, Hathaway NA, Tone Y, Crosas B, Elsasser S, Kirkpatrick DS, et al. Deubiquitinating enzyme Ubp6 functions noncatalytically to delay proteasomal degradation. Cell. 2006;127:99–111. doi: 10.1016/j.cell.2006.07.038. [DOI] [PubMed] [Google Scholar]
- 83.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 84.Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–8. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hoeijmakers JH. DNA damage, aging, and cancer. N Engl J Med. 2009;361:1475–85. doi: 10.1056/NEJMra0804615. [DOI] [PubMed] [Google Scholar]
- 87.Rubinsztein DC. The roles of intracellular protein-degradation pathways in neurodegeneration. Nature. 2006;443:780–6. doi: 10.1038/nature05291. [DOI] [PubMed] [Google Scholar]