Abstract
Transposable elements (TEs) and DNA repeats are commonly targeted by DNA and histone methylation to achieve epigenetic gene silencing. We isolated mutations in two Arabidopsis genes, AtMORC1 and AtMORC6, which cause de-repression of DNA-methylated genes and TEs, but no losses of DNA or histone methylation. AtMORC1 and AtMORC6 are members of the conserved Microrchidia (MORC) adenosine triphosphatase (ATPase) family, predicted to catalyze alterations in chromosome superstructure. The atmorc1 and atmorc6 mutants show decondensation of pericentromeric heterochromatin, increased interaction of pericentromeric regions with the rest of the genome, and transcriptional defects that are largely restricted to loci residing in pericentromeric regions. Knockdown of the single MORC homolog in Caenorhabditis elegans also impairs transgene silencing. We propose that the MORC ATPases are conserved regulators of gene silencing in eukaryotes.
Gene silencing in the Arabidopsis genome is highly correlated with DNA methylation, which is found in three different cytosine contexts. Methylation of symmetric CG and CHG sites (in which H = A, T, or C) are mediated by DNA METHYLTRANSFERASE1 (MET1) and CHROMOMETHYLASE3 (CMT3), respectively, while CHH methylation is mainly catalyzed by DOMAINS REARRANGED METHYLTRANSFERASE2 (DRM2) (1). Silent loci are also enriched in the repressive histone H3 lysine 9 dimethylation mark (H3K9me2) (2, 3).
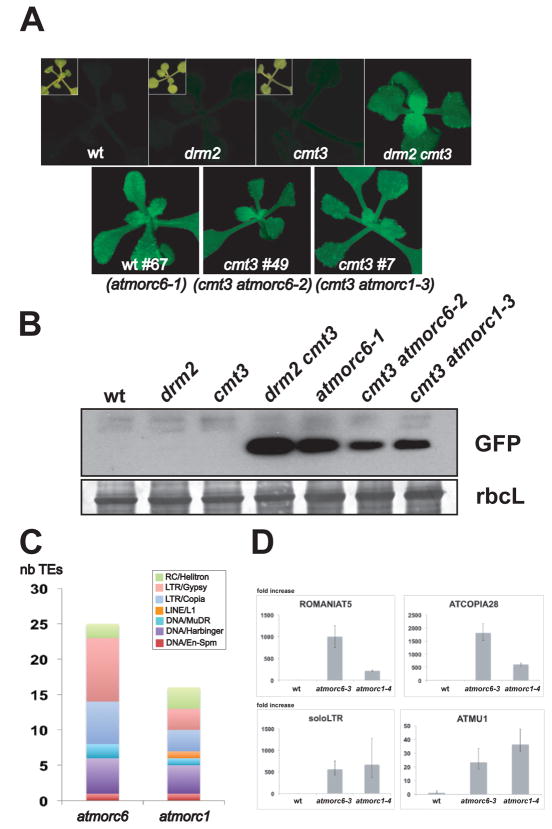
Suppressor of drm2 and cmt3 (SDC) is a gene whose repression in most tissues depends on the redundant activities of DRM2 and CMT3 (4, 5). Hence, a loss of SDC silencing is observed in the drm2 cmt3 double mutant but not in the drm2 or cmt3 single mutants. The SDC promoter carries seven tandem repeats, which recruit the DNA methylation machinery and cause transcriptional gene silencing. We engineered a GFP-based sensor construct controlled by the SDC promoter (fig. S1A). The SDC::GFP transgene behaves similarly to endogenous SDC, and GFP fluorescence is not detectable in wild-type, drm2 or cmt3 plants but is highly expressed in drm2 cmt3 double mutant (Fig. 1A).
We carried out ethyl methanesulfonate (EMS) mutagenesis screens in wild type or cmt3 backgrounds for mutants showing SDC::GFP overexpression, and identified the wt #67, cmt3 #7 and cmt3 #49 mutants (Fig. 1, A and B). Mapping experiments using bulk segregant analysis coupled to deep genome re-sequencing indicated that cmt3 #7 contained a mutation in At4g36290 (AtMORC1), previously also named COMPROMISED RECOGNITION OF TCV-1 (CRT1) (6, 7), while wt #67 and cmt3 #49 both contained mutations in At1g19100 (AtMORC6) (7) (figs. S1B, S2, and S3A). An atmorc1 allele was previously reported to show reduced resistance to the TCV virus (6, 7), suggesting that AtMORC1 is also involved in viral resistance, in addition to its role in gene silencing described in this study; whereas mutations in AtMORC6 have not been described. To ensure that atmorc1 and atmorc6 mutations were those responsible for the loss of SDC silencing, we isolated knock-out T-DNA insertion lines atmorc1-4 and atmorc6-3, and confirmed SDC overexpression in these two mutant alleles (fig. S3B, C and D). Genetic complementation crosses between the recessive EMS and T-DNA mutants confirmed AtMORC1 and AtMORC6 as the mutated genes responsible for SDC::GFP activation in the three EMS lines (fig. S3E). Therefore, #7, #67 and #49 were renamed atmorc1-3, atmorc6-1 and atmorc6-2, respectively.
Fig. 1.
Mutations of two MORC homologs induce SDC::GFP and TE overexpression. (A) Wild type (wt), drm2 mutant and cmt3 mutant plants carrying SDC::GFP showing no GFP fluorescence under UV light (insets show each plant under white light) and drm2 cmt3 double mutant and EMS-mutagenized lines wt #67, cmt3 #49, and cmt3 #7 plants showing strong GFP fluorescence. (B) Western blot using anti-GFP antibody confirms SDC::GFP overexpression in the EMS mutants. Coomassie staining of the large Rubisco subunit (rbcL) is used as loading control. (C) Number of TEs overexpressed in atmorc1 and atmorc6 and classified by superfamily. For each mutant, only TEs with a ≥ 4 fold increase in both the EMS and T-DNA alleles over wild type and with a p-value ≤ 0.05 are represented. (D) Relative fold increase of four TE transcripts in atmorc1-4 and atmorc6-3 over wild type assayed by Real-Time quantitative PCR (RT-qPCR) and normalized to ACTIN7. Errors bars indicate standard deviation based on three independent biological replicates.
Using RNA sequencing (RNA-seq) (8), we found that the majority of RNAs significantly affected in the atmorc1 and atmorc6 mutants showed up-regulation, and many of these TEs belong to various transposon super families including, among others, the LTR/Gypsy, LTR/Copia, DNA/MuDR and DNA/Harbinger families (Fig. 1C and D, fig. S4A, table S1). The expression defects in the atmorc1 and atmorc6 mutants were very similar, with all but two of the transposons upregulated in atmorc1 also upregulated in atmorc6 (fig. S4B). Protein-coding genes overexpressed in the atmorc1 and atmorc6 EMS and T-DNA mutants included endogenous SDC (table S2). There was a high degree of overlap between the genes upregulated in atmorc1 and atmorc6 (fig. S4C), most of them corresponding to DNA-methylated and silenced loci, derepressed in both atmorc1 and atmorc6 (fig. S4D and E). We also performed RNA-seq in the atmorc1 atmorc6 double mutant and found a very similar set of genes and transposons upregulated, with only a few genes upregulated in the double mutant that were not upregulated in each of the single mutants (table S3), suggesting that AtMORC1 and AtMORC6 may act together to enforce gene silencing.
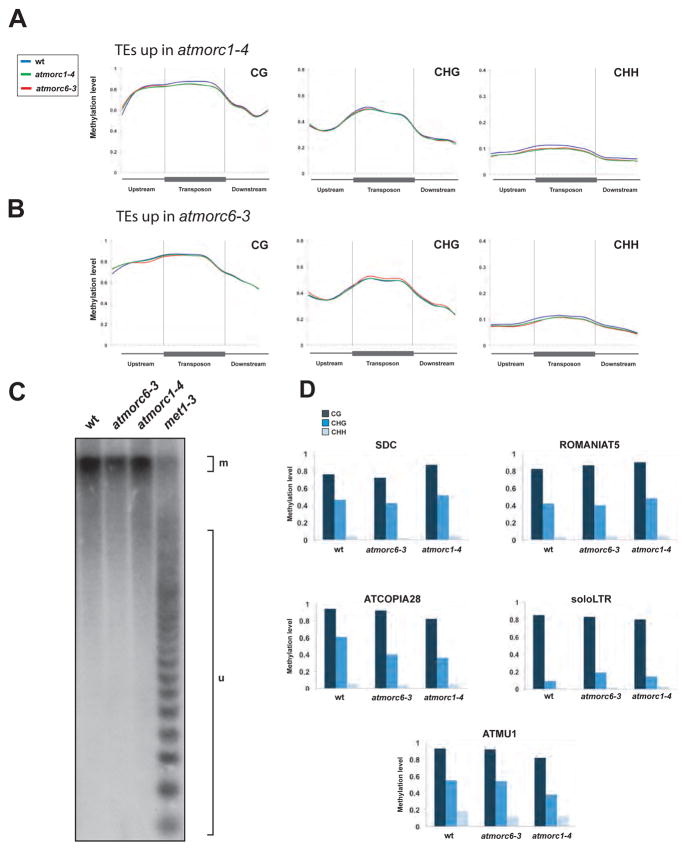
Whole genome bisulfite sequencing (BS-seq) (9) revealed that DNA methylation levels in all sequence contexts were unaltered in atmorc1 or atmorc6 relative to wild type at TEs upregulated in atmorc1 or atmorc6 (Fig. 2A and B) nor were there any bulk alterations in protein-coding genes or TEs in the genome (fig. S5A and B). In addition, analyses at the pericentromeric satellite CEN180 repeats and five loci upregulated in atmorc1 and atmorc6 showed that the DNA methylation patterns in atmorc1-4 and atmorc6-3 were similar to wild type (Fig. 2C and D). ChIP-seq analyses of H3K9me2 also did not reveal any changes in the atmorc1 or atmorc6 mutants at SDC or other upregulated locations (fig. S6A and B). Finally, small RNA sequencing analyses showed that elements up-regulated in atmorc1 and atmorc6 mutants were enriched in siRNAs, but these siRNA levels did not change in the mutants (fig. S7). Thus AtMORC1 and AtMORC6 are not required to maintain DNA methylation, H3K9me2, or siRNAs, suggesting that AtMORC1 and AtMORC6 are likely to either act downstream of DNA methylation or enforce silencing by a novel mechanism.
Fig. 2.
DNA methylation is not impaired in atmorc1 and atmorc6 mutants. (A and B) Metaplot analyses showing DNA methylation level in atmorc1-4, atmorc6-3 and wild type for the set of TEs upregulated in atmorc1-4 (A) and atmorc6-3 (B). The gray vertical lines mark the boundaries between 1 kilobase upstream and downstream regions of TEs. (C) Southern blot analyses assaying CG methylation level at CEN180 repeats using HpaII-treated genomic DNAs. m, methylated; u, unmethylated. met1-3 genomic DNA is used as positive control for loss of CG methylation (23). (D) Percent DNA methylation at SDC and 4 TEs overexpressed in atmorc1-4 and atmorc6-3 assayed by bisulfite sequencing. 24 clones were analyzed for each individual analysis.
AtMORC1 and AtMORC6 are homologs of mouse Microrchidia1 (MORC1) (10, 11), and contain Gyrase, Hsp90, Histidine Kinase and MutL (GHKL) and S5 domains, together comprising an ATPase module (6) in addition to a putative C terminal coiled-coil domain (fig. S1B). The EMS mutations found in atmorc1-3, atmorc6-1 and atmorc6-2 alleles all introduced premature stop codons within the GHKL domain (fig. S1B).
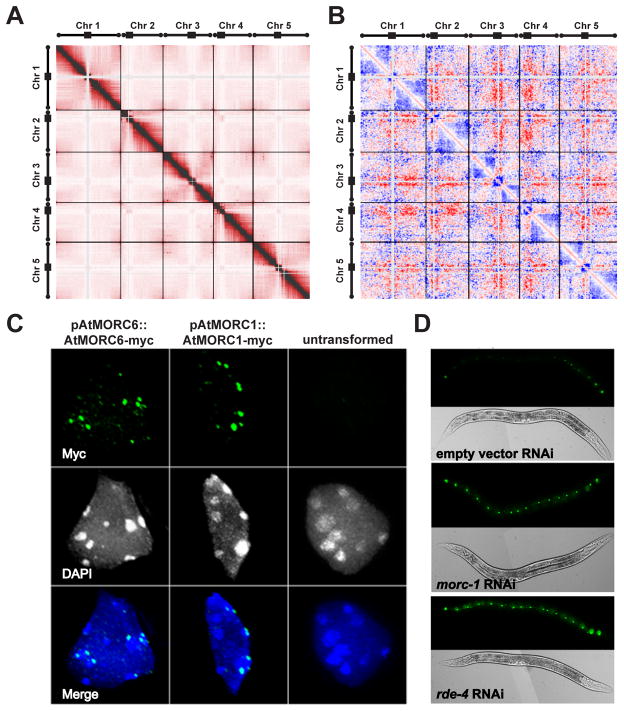
Because of the similarity of AtMORC1 and AtMORC6 to ATPases involved in manipulating chromatin superstructure (12), these proteins may affect gene silencing through higher order compaction of methylated and silent chromatin. In wild type nuclei, pericentromeric heterochromatin forms densely staining nuclear bodies called chromocenters that localize to the nuclear periphery (13). We observed decondensation of chromocenters in the atmorc1 and atmorc6 mutants (as well as in atmorc1 atmorc6 double mutant) (fig. S8-11), and also found that loci transcriptionally de-repressed in the mutants mostly localized to pericentromeric heterochromatin (fig. S12, tables S1 and S3). To directly examine whole genome chromatin interactions, we performed Hi-C analyses in wild type and atmorc6-1 (14). Consistent with previous cytological studies (13), the wild type genome showed interactions between telomeres as well as between euchromatic regions on the same chromosome arm (Fig. 3A). In contrast, pericentromeric heterochromatin regions interacted very weakly with the rest of the genome, consistent with their compaction in chromocenters (Fig. 3A). While atmorc6-1 showed a roughly similar chromatin architecture (fig. S13), plotting the differences between mutant and wild type showed that atmorc6-1 shows an increase in interactions between the pericentromeric regions of all chromosomes with the euchromatic arms of all chromosomes, and a corresponding depletion of interactions of euchromatic arms with themselves. Since the analysis reports relative changes, with the sum of differences set to zero, the most likely interpretation of these findings is that pericentromeric regions interact more strongly with the euchromatic arms in atmorc6-1, although we cannot exclude that the mutant also has effects on the euchromatic arms (Fig. 3B). This interpretation is consistent with the cytological observations showing that chromocenters expand out into a larger area of the nucleus in the mutants (fig. S8). We also found, using complementing myc-tagged transgenes, that AtMORC1 and AtMORC6 proteins formed small nuclear bodies that were usually adjacent to, but not within chromocenters (Fig. 3C and figs. S14-15). These results are all consistent with a model in which AtMORC1 and AtMORC6 enforce compaction and gene silencing of pericentromeric heterochromatin, although it is also possible that changes in chromatin and gene expression in the mutant secondarily lead to the observed changes in chromatin compaction. Mutation of the plant specific MOM1 gene has also been shown to affect gene silencing but not DNA methylation in Arabidopsis, however mom1 mutants do not show chromocenter decondensation and therefore are likely to act via a different mechanism (15, 16).
Fig. 3.
AtMORC1 and AtMORC6 are required for maintenance of chromatin architecture, form nuclear bodies near chromocenters, and morc-1 is involved in gene silencing in C. elegans. (A) Interaction matrix of the wild type Arabidopsis genome from Hi-C analysis. Positions along the 5 chromosomes are shown from left to right and top to bottom, and each pixel represents interactions from uniquely mapping paired end reads in 200 kilobase bins. Black bars and circles mark the positions of the pericentromeric and telomeric regions, respectively. Light grey regions represent areas masked out due to problematic mapping. Black bars show separation between chromosomes. (B) Difference plot showing enrichment of Hi-C interactions in atmorc6-1 in red and interactions depleted in atmorc6-1 in blue. (C) Anti-Myc immunostaining showing localization of pAtMORC6::AtMORC6-Myc and pAtMORC1::AtMORC1-Myc in nuclear bodies adjacent to chromocenters. AtMORC1 and AtMORC6 showed an average of 2.0 +/− 1.0 (average plus or minus standard deviation) and 2.5 +/− 1.2 bodies per chromocenter, respectively. DAPI staining shows chromocenter location. Bottom panels are merges. (D) A silenced seam cell-specific GFP transgene in the eri-1 (mg366) sensitized background is overexpressed in worms fed with bacteria expressing double-stranded RNA targeting morc-1 or rde-4 but not in worms fed with bacteria expressing a control empty vector. Results are representative of five independent replicates.
A single MORC homolog, morc-1, is present in the worm Caenorhabditis elegans, which is devoid of DNA methylation (17). To test if the C. elegans morc-1 (ZC155.3) is involved in gene silencing, we performed RNAi-mediated knock-down of morc-1 in the eri-1 sensitized background, in which a GFP transgene is silenced in most of the worm seam cells (Fig. 3D) (18). morc-1 depleted worms showed GFP reactivation similar to worms depleted of rde-4, an essential component of gene silencing in C. elegans (Fig. 3D) (19). These results suggest that MORCs may play an ancient and conserved role in gene silencing. In addition, the observation that morc-1 is required for gene silencing in C. elegans reinforces our view that MORCs in Arabidopsis are enforcing silencing by a mechanism that may not be directly linked with DNA methylation. It is interesting to note that the phenotype of the Morc1-knockout mouse resembles Miwi2- and Dnmt3L-knockout mouse phenotypes, showing male-specific meiotic defects during spermatogenesis (10, 20–22). Miwi2 and Dnmt3L are both required for TE silencing, and it is possible that Morc1 might be involved in transposon silencing in mammals as well. We propose that MORC family ATPases acts to regulate chromatin architecture and gene silencing in a wide variety of eukaryotes.
Supplementary Material
Acknowledgments
We thank M. Akhavan for sequencing, L. Goddard and L. Iruela-Arispe for assistance with confocal microscopy, and P. Fransz and I. Schubert for helpful discussions. S.F. is a Special Fellow of the Leukemia and Lymphoma Society. J.C. is supported by the Ruth L. Kirschstein National Research Service Award GM007185. R.P.M. is supported by the National Institute of General Medical Sciences (grant F32GM100617), and J.D. is supported by a W.M. Keck Foundation Distinguished Young Scholar in Medical Research Award. Research in the Jacobsen, Kim, Michaels, and Dekker laboratories was supported by NIH grants GM60398, GM088565, GM075060, and HG003143, respectively. Sequencing files have been deposited at GEO (accession code GSE24840). The authors declare no competing financial interests. S.E.J. is an investigator of the Howard Hughes Medical Institute.
Footnotes
This manuscript has been accepted for publication in Science. This version has not undergone final editing. Please refer to the complete version of record at http://www.sciencemag.org/. The manuscript may not be reproduced or used in any manner that does not fall within the fair use provisions of the Copyright Act without the prior, written permission of AAAS.
www.sciencemag.org/cgi/content/full/science.1221472/DC1
Materials and Methods
References and Notes
- 1.Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet. 2010;11:204. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jackson JP, Lindroth AM, Cao X, Jacobsen SE. Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature. 2002;416:556. doi: 10.1038/nature731. [DOI] [PubMed] [Google Scholar]
- 3.Malagnac F, Bartee L, Bender J. An Arabidopsis SET domain protein required for maintenance but not establishment of DNA methylation. EMBO J. 2002;21:6842. doi: 10.1093/emboj/cdf687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang X, et al. Genome-wide high-resolution mapping and functional analysis of DNA methylation in arabidopsis. Cell. 2006;126:1189. doi: 10.1016/j.cell.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 5.Henderson IR, Jacobsen SE. Tandem repeats upstream of the Arabidopsis endogene SDC recruit non-CG DNA methylation and initiate siRNA spreading. Genes Dev. 2008;22:1597. doi: 10.1101/gad.1667808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kang HG, Kuhl JC, Kachroo P, Klessig DF. CRT1, an Arabidopsis ATPase that interacts with diverse resistance proteins and modulates disease resistance to turnip crinkle virus. Cell Host Microbe. 2008;3:48. doi: 10.1016/j.chom.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 7.Kang HG, et al. Endosome-associated CRT1 functions early in resistance gene-mediated defense signaling in Arabidopsis and tobacco. Plant Cell. 2010;22:918. doi: 10.1105/tpc.109.071662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cokus SJ, et al. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature. 2008;452:215. doi: 10.1038/nature06745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Watson ML, et al. Identification of morc (microrchidia), a mutation that results in arrest of spermatogenesis at an early meiotic stage in the mouse. Proc Natl Acad Sci USA. 1998;95:14361. doi: 10.1073/pnas.95.24.14361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Inoue N, et al. New gene family defined by MORC, a nuclear protein required for mouse spermatogenesis. Hum Mol Genet. 1999;8:1201. doi: 10.1093/hmg/8.7.1201. [DOI] [PubMed] [Google Scholar]
- 12.Iyer LM, Abhiman S, Aravind L. MutL homologs in restriction-modification systems and the origin of eukaryotic MORC ATPases. Biol Direct. 2008;3:8. doi: 10.1186/1745-6150-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fransz P, De Jong JH, Lysak M, Castiglione MR, Schubert I. Interphase chromosomes in Arabidopsis are organized as well defined chromocenters from which euchromatin loops emanate. Proc Natl Acad Sci USA. 2002;99:14584. doi: 10.1073/pnas.212325299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lieberman-Aiden E, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Amedeo P, Habu Y, Afsar K, Mittelsten Scheid O, Paszkowski J. Disruption of the plant gene MOM releases transcriptional silencing of methylated genes. Nature. 2000;405:203. doi: 10.1038/35012108. [DOI] [PubMed] [Google Scholar]
- 16.Probst AV, Fransz PF, Paszkowski J, Mittelsten Scheid O. Two means of transcriptional reactivation within heterochromatin. Plant J. 2003;33:743. doi: 10.1046/j.1365-313X.2003.01667.x. [DOI] [PubMed] [Google Scholar]
- 17.Simpson VJ, Johnson TE, Hammen RF. Caenorhabditis elegans DNA does not contain 5-methylcytosine at any time during development or aging. Nucleic Acids Res. 1986;14:6711. doi: 10.1093/nar/14.16.6711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kennedy S, Wang D, Ruvkun G. A conserved siRNA-degrading RNase negatively regulates RNA interference in C. elegans. Nature. 2004;427:645. doi: 10.1038/nature02302. [DOI] [PubMed] [Google Scholar]
- 19.Tabara H, et al. The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell. 1999;99:123. doi: 10.1016/S0092-8674(00)81644-X. [DOI] [PubMed] [Google Scholar]
- 20.Carmell MA, et al. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev Cell. 2007;12:503. doi: 10.1016/j.devcel.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 21.Bourc’his D, Bestor TH. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature. 2004;431:96. doi: 10.1038/nature02886. [DOI] [PubMed] [Google Scholar]
- 22.Aravin AA, et al. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol Cell. 2008;31:785. doi: 10.1016/j.molcel.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kankel MW, et al. Arabidopsis MET1 cytosine methyltransferase mutants. Genetics. 2003;163:1109. doi: 10.1093/genetics/163.3.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kleinboelting N, Huep G, Kloetgen A, Viehoever P, Weisshaar B. GABI-Kat SimpleSearch: new features of the Arabidopsis thaliana T-DNA mutant database. Nucleic Acids Res. 2012;40(Database issue):D1211. doi: 10.1093/nar/gkr1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 26.Law JA, Vashisht AA, Wohlschlegel JA, Jacobsen SE. SHH1, a homeodomain protein required for DNA methylation, as well as RDR2, RDM4, and chromatin remodeling factors, associate with RNA polymerase IV. PLoS Genet. 2011;7:e1002195. doi: 10.1371/journal.pgen.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Greenberg MV, et al. Identification of genes required for de novo DNA methylation in Arabidopsis. Epigenetics. 2011;6:344. doi: 10.4161/epi.6.3.14242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 30.Benjamini Y, Hochberg Y. J R Stat Soc, B. 1995;57:12. [Google Scholar]
- 31.Chen PY, Cokus SJ, Pellegrini M. BS Seeker: precise mapping for bisulfite sequencing. BMC Bioinformatics. 2010;11:203. doi: 10.1186/1471-2105-11-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee J, et al. Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell. 2007;19:731. doi: 10.1105/tpc.106.047688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu C, Meyers BC, Green PJ. Construction of small RNA cDNA libraries for deep sequencing. Methods. 2007;43:110. doi: 10.1016/j.ymeth.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 34.Zhang Y, et al. Spatial organization of the mouse genome and its role in recurrent chromosomal translocations. Cell. 2012;148:908. doi: 10.1016/j.cell.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Soppe WJ, et al. DNA methylation controls histone H3 lysine 9 methylation and heterochromatin assembly in Arabidopsis. EMBO J. 2002;21:6549. doi: 10.1093/emboj/cdf657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim JK, et al. Functional genomic analysis of RNA interference in C. elegans. Science. 2005;308:1164. doi: 10.1126/science.1109267. [DOI] [PubMed] [Google Scholar]
- 37.Kamath RS, et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- 38.Polosina YY, Cupples CG. Wot the ’L-Does MutL do? Mutat Res. 2010;705:228. doi: 10.1016/j.mrrev.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 39.Mimura Y, Takahashi K, Kawata K, Akazawa T, Inoue N. Two-step colocalization of MORC3 with PML nuclear bodies. J Cell Sci. 2010;123:2014. doi: 10.1242/jcs.063586. [DOI] [PubMed] [Google Scholar]
- 40.Hirano T. The ABCs of SMC proteins: two-armed ATPases for chromosome condensation, cohesion, and repair. Genes Dev. 2002;16:399. doi: 10.1101/gad.955102. [DOI] [PubMed] [Google Scholar]
- 41.Blewitt ME, et al. SmcHD1, containing a structural-maintenance-of-chromosomes hinge domain, has a critical role in X inactivation. Nat Genet. 2008;40:663. doi: 10.1038/ng.142. [DOI] [PubMed] [Google Scholar]
- 42.Ausin I, Mockler TC, Chory J, Jacobsen SE. IDN1 and IDN2 are required for de novo DNA methylation in Arabidopsis thaliana. Nat Struct Mol Biol. 2009;16:1325. doi: 10.1038/nsmb.1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kanno T, et al. A structural-maintenance-of-chromosomes hinge domain-containing protein is required for RNA-directed DNA methylation. Nat Genet. 2008;40:670. doi: 10.1038/ng.119. [DOI] [PubMed] [Google Scholar]
- 44.Böhmdorfer G, et al. GMI1, a structural-maintenance-of-chromosomes-hinge domain-containing protein, is involved in somatic homologous recombination in Arabidopsis. Plant J. 2011;67:420. doi: 10.1111/j.1365-313X.2011.04604.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.